Abstract
Gastric cancer is a common malignancy, and is one of the most frequent causes of cancer deaths worldwide. Recently, members of the transglutaminases (TGM) family, especially TGM2, have been implicated in the progression and drug resistance of cancers, but the function of TGM1 in cancer development has been largely overlooked. In this study, we demonstrate the roles of TGM1 in development of gastric cancer. We found that expression levels of TGM1 were upregulated in both gastric cancer tissues and cultured gastric cancer cells, and that TGM1 expression levels were correlated with patient survival. In cultured gastric cancer cells, loss of TGM1 expression inhibited cell proliferation and promoted apoptosis, as well increased gastric cancer cell sensitivity to chemotherapeutic drugs and reducing stemness. These results strongly supported the participation of TGM1 in the regulation of gastric cancer development. In addition, we found evidence that the mechanism of action of TGM1 in regulating gastric cancer cell might involve the Wnt signaling pathway, as loss of TGM1 expression in gastric cancer cells led to a significant suppression of Wnt signaling activities.
Keywords: Gastric cancer, transglutaminase-1, Wnt signaling, stemness, chemoresistance
Introduction
Gastric cancer is a common malignancy and is one of the most frequent causes of cancer deaths worldwide.1,2 The high mortality rates of gastric cancer are partially due to it usually being diagnosed in later stages, resulting in poor prognosis and therapeutic outcomes.3 Like many other cancer types, initiation and progression of gastric cancer involves multistep processes of various genetic and molecular factors, including activation of multiple oncogenes and suppression of tumor suppressor genes.4,5 Molecular pathways that affect cell proliferation and migration, including the integrin/focal adhesion kinase,6 mammalian target of rapamycin (mTOR) signaling,7 mitogen-activated protein kinases (MAPK),8 and nuclear factor-κB (NF-κB) pathways,9 have been demonstrated to play roles in development of gastric cancer.
Recently, members of the transglutaminases (TGM) family, especially tissue TGM, have been implicated in progression and drug resistance of cancers. TGM catalyze Ca2+-dependent post-translational protein modifications by inserting irreversible ε-(γ-glutamyl)-lysine cross-links between polypeptide chains. Tissue TGM are primarily localized within the compartments of cell structures, but they can also be secreted into extracellular spaces.10 Recent studies have shown that tissue TGM have been associated with several biological functions including signal transduction, apoptosis, cell adhesion, cell migration, and extracellular matrix formation.11,12 In the context of cancer development, overexpression of tissue TGM has been observed in breast,13,14 pancreatic,15 ovarian,16 colon,17 non-small cell lung cancers,18 as well as melanoma19 and glioblastoma.20 Elevated levels of TGM expression have been postulated to be a potential negative prognostic marker and are associated with advanced stages of cancer, metastatic migration and chemoresistance.19–22 There are several members of the TGM family, including TGM1 and TGM2. The majority of previous studies regarding the roles of TGM in cancer development were focused on TGM2, whereas the functions of TGM1 were mostly investigated in an epidermal condition known as ichthyosis.23 Because expression of TGM1 is not restricted to skin tissue and TGM1 possesses enzymatic functions similar to TGM2, the possibility that TGM1 is also involved in regulating progression and drug resistance of cancer can not be excluded.
For the first time we demonstrated that the expression levels of TGM1 are regulated in both gastric cancer tissues and cells, and that TGM1 levels are associated with patient survival. Furthermore, loss of TGM1 expression in gastric cancer cell lines inhibited cell proliferation and promoted cell apoptosis. Loss of TGM1 expression also increased chemosensitivity and reduced stemness of gastric cancer cells. In addition, we found evidence that the mechanism of action of TGM in regulating gastric cancer might involve the Wnt signaling pathway, as loss of TGM1 expression in gastric cancer cells leads to significant suppression of Wnt signaling.
Materials and methods
Tissue samples and cell lines
Ninety-six pairs of primary gastric cancer samples and their matched adjacent normal gastric tissues were obtained from patients who underwent surgical resections at the Department of General Surgery, Daqing Oilfield General Hospital. The fresh samples were snap-frozen in liquid nitrogen and stored at −80 ℃ for future use. Prior patient consent and approval from the Institutional Research Ethics Committee were obtained. Histologic diagnosis of gastric normal and cancer tissues was confirmed by three pathologists.
Cell culture and treatments
Four gastric cancer cell lines (BGC823, SGC7901, MGC803 and AGS) and one human normal gastric epithelial cell line (GES-1) were grown in Dulbecco's Modified Eagle Medium (DMEM; Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 50 units/mL penicillin and streptomycin. All cell lines were maintained in a humidified incubator with 5% CO2 at 37 ℃. Small interfering RNA (siRNA) were transfected into gastric cancer cells using the Lipofectamine® RNAiMAX Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Gastric cancer cells were treated with 5-Fu (0, 4, 8, 12 or 16 µg/mL) (Sigma, St Louis, MO), cisplatin (0, 3, 6, 9 or 12 µg/mL) (Sigma) or vincristine (0, 1.5, 3.0, 4.5 or 6.0 µg/mL) (Sigma) for 48 h. All cell lines used were routinely tested for mycoplasma contamination by the Mycoplasma Detection Kit (Thermo-Fisher Scientific, Pittsburgh, PA). An authentication of all cell lines used was confirmed by short tandem repeat DNA profiling.
qRT-PCR
Total RNA was isolated from gastric cancer tissues or cell lines using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. One µg of total RNA was reverse transcribed to cDNA using a reverse transcription kit (Takara, Dalian, China). Real-time polymerase chain reaction (PCR) was performed with the SYBR Green Premix EX Taq kit (Takara) and the ABI 7500 real time PCR System (Applied Biosystems, Carlsbad, CA). β-actin was used as an internal control. Primer sequences in our study are as follows: TGM1 forward 5′-CCCCCGCAATGAGATCTACA-3′ and reverse 5′-ATCCTCATGGTCCACGTACACA-3′; Axin2 forward 5′-GGTGTTTGAGGAGATCTGGG-3′, and reverse 5′-TGCTCACAGCCAAGACAGTT-3′; c-myc forward 5′-AAGAGGACTTGTTGCGGAAA-3′ and reverse 5′-CTCAGCCAAGGTTGTGAGGT-3′; CCND1 forward 5′-AAGGCCTGAACCTGAGGAG-3′ and reverse 5′-CTTGACTCCAGCAGGGCTT-3′; TCF4 forward 5′-TCTTCACAGTAGTGCCATGGAG-3′ and reverse 5′-CTTGCTGATGGAGCATAGACTG-3′; β-actin forward 5′-GA AAATCTGGCACCACACCTT-3′ and reverse 5′-GTTGAAGGTAGTTTCGTGGAT-3′.
Western blot assays
Gastric cancer tissues or cultured gastric cancer cells were homogenized in radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime Biotechnology, Beijing, China) on ice, and centrifuged at 12,000 × g for 30 min. The cytoplasmic and nuclear protein fractions were extracted using a nuclear and cytoplasmic protein extraction kit (Beyotime Biotechnology) according to the manufacturer's instructions. Protein concentrations were determined by the bicinchoninic acid (BCA) method (Beyotime Biotechnology), and 20–40 mg of protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) onto nitrocellulose membrane (Bio-Rad, Hercules, CA). Western blots were performed with primary antibodies: anti-TGM1 (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA), anti-Bcl-2 (1:2000; Santa Cruz Biotechnology), anti-Bax (1:1000; Santa Cruz Biotechnology), anti-β-catenin (1:500; Abcam, Cambridge, MA), anti-LaminA (1:1000; Santa Cruz Biotechnology), and anti-β-actin (1:1000; Santa Cruz Biotechnology) along with their corresponding secondary antibodies. The signals were visualized with enhanced chemiluminescence (ECL) reagent (Pierce Biotechnology, Rockford, IL).
MTT assays
We used the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay according to the manufacturer's instructions to determine cell growth and viability. Briefly, the cells were seeded into 96-well plates at 4000 cells/well after 48 h incubation. Twenty microliters of MTT solution (5 mg/mL, Sigma, Grand Island, NY) were added to each well at different time points, and the cells were incubated at 37 ℃ for 4 h. The supernatant was removed and 150 mL of dimethyl sulfoxide (DMSO) was added to each well. The plate was gently shaken on a shaker for 10 min, and the absorbance was measured at 570 nm using a spectrophotometer.
CCK-8 assays
The CCK-8 kit (Beyotime Biotechnology) was used to analyze cell proliferation. Gastric cancer cells were seeded at a density of 4000 cells/well into 96-well plates and incubated at 37 ℃, 5% CO2 for different periods of time. CCK-8 solution (100 µL) was added into each well and incubated at 37 ℃ for 4 h, then shaken for 1 min. Absorbance at 450 nm was measured using a microplate reader.
Sphere formation assays
Gastric cancer cells were plated into wells of ultra-low attachment surface six-well culture plates (Corning, Corning, NY) at a density of 3000 cells/mL in DMEM/F12 medium (Gibco) supplemented with 20 ng/mL epidermal growth factor (EGF), 20 ng/mL basic fibroblast growth factor (bFGF), and 2% B27. Diameters of spheres were measured seven days post seeding.
Flow cytometry
Flow cytometric analysis was used to determine cell surface expression of CD44 + and CD133 + markers from gastric cancer sphere cells. Briefly, gastric cancer sphere cells were harvested, washed twice and resuspended in Hanks' balanced salt solution (HBSS). Cells were then incubated with antibodies in the dark for 30 min at room temperature. After three washes, the cell surface markers were detected using a flow cytometer (BD Biosciences, San Jose, CA), and analyzed with Flow Jo software (BD Biosciences). CD133-PE and CD44-APC antibodies were obtained from Miltenyibiotec (Auburn, CA).
Cell cycling was examined by flow cytometry. Gastric cells (1 × 106) were washed twice with ice-cold phosphate-buffered saline (PBS), treated with trypsin, fixed with 70% cold ethanol at 4 ℃ for 1 h, centrifuged at 1500 r/min for 5 min to remove ethanol, again washed with PBS, and incubated in 25 µg/mL of RNase for 1 h at 37 ℃. Before analysis, cells were stained with 50 µg/mL of propidium iodide (PI) at room temperature for 30 min in the dark, then subjected to flow cytometric analysis.
Luciferase assays
Wnt signaling activity was examined using the TOPflash/FOPflash luciferase assay. Briefly, cells were transfected with TOPFlash or FOPflash firefly luciferase reporter vector (Upstate, Chicago, IL) and Renilla luciferase vectors (Promega, Madison, WI) according to the recommended protocol for the Lipofectamine 2000 transfection system. Post incubation (48 h), luciferase activity was detected using the dual-luciferase reporter assay system (Promega).
Statistical analysis
Data are presented as mean ± SD. Statistical significance was determined using two-tailed Student's t tests between the means of the control and experimental groups. The overall survival curves were drawn using the Kaplan–Meier method. All statistical calculations were analyzed using GraphPad Prism 5.0 software (GraphPad Software Inc., San Diego, CA). P < 0.05 was considered statistically significant.
Results
TGM1 is upregulated in gastric cancer
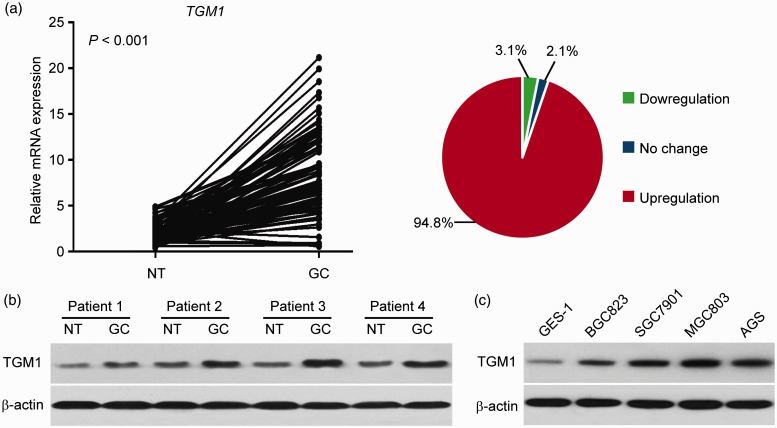
To determine the expression pattern of TGM1 in gastric cancer, we performed qRT-PCR to examine the expression levels of TGM1 in clinical gastric cancer specimens. In the 96 samples of gastric cancer tissues and matched adjacent normal gastric tissues, we found that TGM1 was significantly upregulated in gastric cancer tissues (P < 0.001). Overexpression of TGM1 was confirmed in 94.8% of the patient samples (Figure 1(a)). We further performed Western blot analysis to examine the protein levels of TGM1 in randomly selected gastric cancer samples, and found that TGM1 levels were higher than in all normal gastric tissues tested (Figure 1(b)). We next compared the expression levels of TGM1 in four gastric cancer cell lines (BGC823, SGC7901, MGC803 and AGS), to those in human normal GES-1 and found that TGM1 was consistently upregulated in all four gastric cancer cell lines (Figure 1(c)). BGC823 and MGC803 cells are poorly differentiated, SGC7901 cells are moderately differentiated and AGS cells were poorly to moderately differentiated,24,25 but there were no significant differences between poorly and moderately differentiated cells. Because SGC7901 and MGC803 cells expressed relatively high levels of TGM1, they were selected for further functional analyses. We divided the patient samples equally to ‘high’ and ‘low’ groups based on the average values of TGM1 mRNA expression and found that the expression levels of TGM1 were significantly associated with tumor lymph node metastasis, TNM stages, and cancer-related deaths (Table 1 and Figure 2). Taken together, these results indicated that TGM1 was upregulated in both gastric cancer tissues and cells, and that TGM1 levels were associated with cancer progression.
Figure 1.
TGM1 is upregulated in gastric cancer. (a) Expression levels of TGM1 as determined by qRT-PCR in gastric cancer tissues (GC) and compared with matched adjacent normal gastric tissues (NT) from the same patient (P < 0.001). (b) Protein levels of TGM1 as determined by Western blot analysis in GC and matched NT from four randomly selected patient samples. (c) Protein levels of TGM1 as determined by Western blot analysis in one human normal gastric epithelial cell line (GES-1) and four gastric cancer cell lines (BGC823, SGC7901, MGC803 and AGS). (A color version of this figure is available in the online journal.)
Table 1.
Comparison of clinicopathological features of patients with TGM1 low and high gastric cancer
| Factors | Total | TGM1 expression |
P value | |
|---|---|---|---|---|
| Low (%) | High (%) | |||
| Gender | 0.423 | |||
| Male | 79 | 38 (48.1) | 41 (51.9) | |
| Female | 17 | 10 (58.8) | 7 (41.2) | |
| Age (years) | 0.679 | |||
| <60 | 40 | 19 (47.5) | 21 (52.5) | |
| ≥60 | 56 | 29 (51.8) | 27 (48.2) | |
| Location | 0.214 | |||
| Upper and middle stomach | 40 | 23 (57.5) | 17 (42.5) | |
| Lower stomach | 56 | 25 (44.6) | 31 (55.4) | |
| Tumor size (cm) | 0.138 | |||
| <6 | 61 | 34 (55.7) | 27 (44.3) | |
| ≥6 | 35 | 14 (40.0) | 21 (60.0) | |
| Differentiation | 0.088 | |||
| High/moderate | 34 | 21 (61.8) | 13 (38.2) | |
| Low/undifferentiated | 62 | 27 (43.5) | 35 (56.5) | |
| Gastric wall invasion | 0.083 | |||
| T1–T2 | 32 | 12 (37.5) | 20 (62.5) | |
| T3–T4 | 64 | 36 (56.3) | 28 (43.7) | |
| Lymph node metastasis | 0.021* | |||
| Yes | 59 | 24 (40.7) | 35 (59.3) | |
| No | 37 | 24 (64.9) | 13 (35.1) | |
| TNM stage | 0.028* | |||
| I–II | 30 | 20 (66.7) | 10 (33.3) | |
| III–IV | 66 | 28 (42.4) | 38 (57.6) | |
P value < 0.05 was considered statistically significance.
Figure 2.
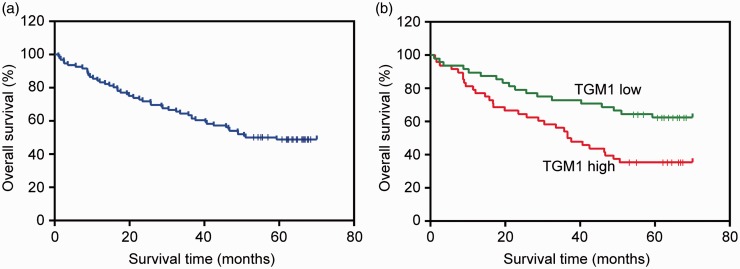
Expression levels of TGM1 are associated with survival of gastric cancer patients. (a) The Kaplan–Meier survival curves of 96 gastric cancer patients. (b) The Kaplan–Meier survival curves of patients in (a) are categorized by low and high TGM1 expression levels (n = 96; P < 0.001). (A color version of this figure is available in the online journal.)
Loss of TGM1 inhibits gastric cancer cell proliferation and promotes apoptosis
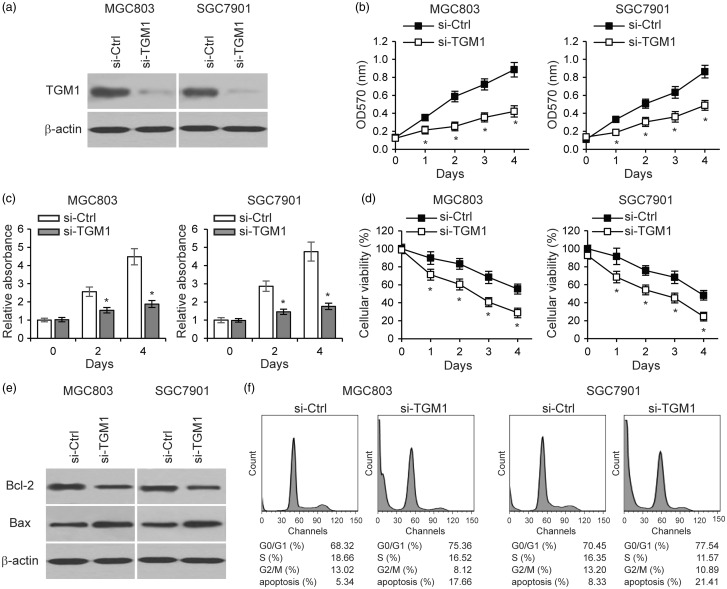
To explore the function of TGM1 in gastric cancer development, we knocked down TGM1 expression in SGC7901 and MGC803 gastric cancer cells by transfecting siRNA targeting TGM1 (Figure 3(a)). Using the MTT cellular proliferation assay, we found that TGM1 knockdown significantly reduced cell proliferation of both SGC7901 and MGC803, as compared to gastric cancer cells transfected with control scrambled siRNA (Figure 3(b)). Using the cell counting kit-8 (CKK-8) assay, we consistently confirmed decreases in proliferation of TGM1 knockdown cells (Figure 3(c)). We performed MTT cellular viability assays by culturing cells in serum free medium with glucose deprivation, and found that knockdown of TGM1 expression led to significantly decreased cell viability (Figure 3(d)). We postulated that the reduced cell viability was due to increased apoptosis. Therefore we examined the expression levels of Bcl-2 and Bax in TGM1 knockdown cells, and found that TGM1 knockdown resulted in increased Bcl-2 and decreased Bax levels, consistent with elevated cell apoptosis (Figure 3(e)). Furthermore, cell cycle analysis showed that TGM1 downregulation arrested more gastric cancer cells in the G0/G1 phase and led to further cells apoptosis (Figure 3(f)). These results collectively indicate that loss of TGM1 inhibits gastric cancer cell proliferation and promotes apoptosis.
Figure 3.
Loss of TGM1 inhibits gastric cancer cell proliferation and promotes apoptosis. (a) Western blot of TGM1 expression in MGC803 and SGC7901 gastric cancer cells transfected with siRNA targeting TGM1 (si-TGM1), (b) MTT cell proliferation assay, (c) CKK-8 assay, and (d) MTT cell viability assays in MGC803 and SGC7901 cells transfected with si-TGM1, and compared to cells transfected with a control scramble siRNA (si-Ctrl). (e) Protein levels of Bcl-2 and Bax as determined by Western blot analysis in MGC803 and SGC7901 cells transfected with si-TGM1 and compared to cells transfected with a control scrambled siRNA. (f) Cell cycle analysis of MGC803 and SGC7901 cells transfected with si-TGM1 as compared to si-Ctrl. Each bar represents the mean ± SEM of three independent experiments. *P < 0.001
Loss of TGM1 increases chemosensitivity and reduces stemness of gastric cancer cells
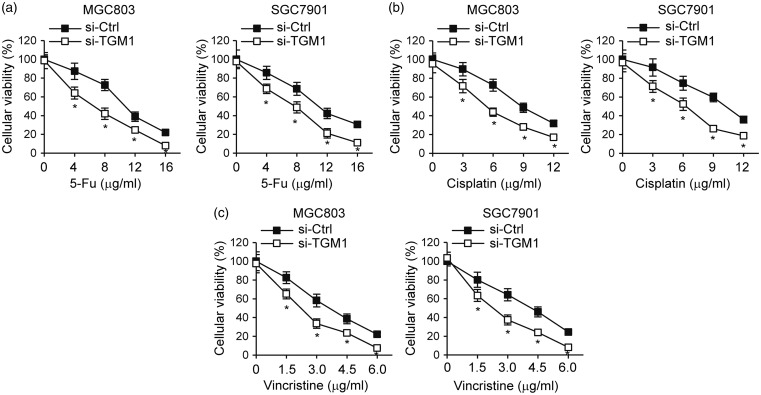
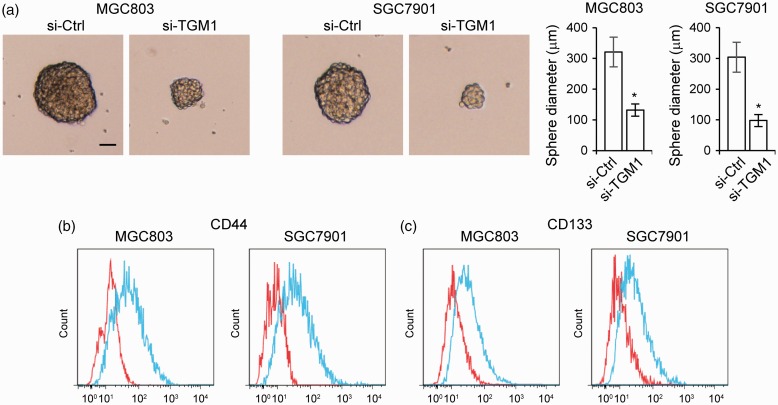
We next investigated the role of TGM1 in regulating gastric cancer drug resistance. We treated the TGM1 knockdown cell lines, SGC7901 and MGC803, with different concentrations of 5-Fu (Figure 4(a)), cisplatin (Figure 4(b)), or vincristine (Figure 4(c)). Knockdown of TGM1 significantly increased chemosensitivity to all three of these anticancer drugs in both SGC7901 and MGC803 cell lines, compared to cells transfected with control scrambled siRNA (Figure 4). Since cancer cell stemness is one of the determinants of drug resistance and TGM is known to participate in regulating cell stemness,26,27 we determined whether TGM1 regulates the stemness of gastric cancer cells. We performed cell sphere formation assays with TGM1 knockdown cell lines, SGC7901 and MGC803, and found that knockdown of TGM1 led to markedly decreased sphere diameters (Figure 5(a)). TGM1 knockdown also significantly reduced CD44 + (Figure 5(b)) and CD133 +(Figure 5(c)) cell numbers in SGC7901 and MGC803 cell spheres. Taken together, these results suggest that loss of TGM1 increases chemosensitivity and reduces stemness of gastric cancer cells.
Figure 4.
Loss of TGM1 increases chemosensitivity of gastric cancer cells. MTT cell proliferation assays in MGC803 and SGC7901 gastric cancer cells transfected with siRNA targeting TGM1 (si-TGM1) as compared to cells transfected with a control scrambled siRNA (si-Ctrl). Cells were treated with different concentrations of (a) 5-Fu, (b) cisplatin or (c) vincristine. Each bar represents the mean ± SEM of three independent experiments. *P < 0.001
Figure 5.
Loss of TGM1 reduces stemness of gastric cancer cells. (a) Representative images of mammospheres formed by MGC803 and SGC7901 cells transfected with siRNA targeting TGM1 (si-TGM1) as compared to those formed by cells transfected with a control scrambled siRNA (si-Ctrl), left panel; quantification of sphere diameters, right panel. Each bar represents the mean ± SEM of three independent experiments. *P < 0.001. (b) CD44 + and (c) CD133 + cells in spheres derived from MGC803 and SGC7901 cells transfected with siRNA targeting TGM1 (si-TGM1) as compared to cells transfected with a control scrambled siRNA (si-Ctrl). Red, si-TGM1; blue, si-Ctrl. (A color version of this figure is available in the online journal.)
Loss of TGM1 suppresses Wnt/β-catenin signaling
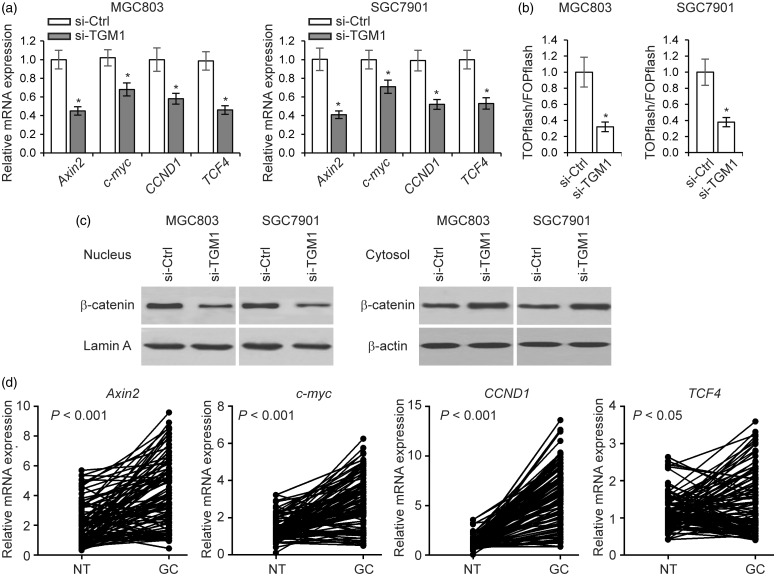
The Wnt/β-catenin signaling pathway is known to play an important role in maintaining gastric cancer cell stemness and drug resistance.28 Since increased levels of TGM1 have been reported to associate with higher levels of nuclear β-catenin,29 we investigated whether TGM1 regulates Wnt/β-catenin signaling in gastric cancer cells. We first examined the expression levels of a series of genes related to Wnt/β-catenin signaling (Axin2, c-myc, CCND1 and TCF4), and found that the levels of these genes were significantly lower in TGM1 knockdown cells than in cells transfected with control scrambled siRNA (Figure 6(a)), suggesting that TGM1 could be involved in regulating Wnt/β-catenin signaling in gastric cancer cells. In addition, knockdown of TGM1 significantly reduced Wnt/β-catenin luciferase reporter activity (Figure 6(b)). Furthermore, we found that nuclear β-catenin levels were decreased in TGM1 knockdown gastric cancer cells, while cytosolic β-catenin levels were elevated (Figure 6(c)). Upregulation of Wnt/β-catenin signaling downstream genes in clinical samples confirmed activation of Wnt/β-catenin signaling in gastric cancer (Figure 6(d)). Collectively, these results support the hypothesis that loss of TGM1 expression led to reduced Wnt/β-catenin signaling activity in gastric cancer cells.
Figure 6.
Loss of TGM1 suppresses Wnt/β-catenin signaling. (a) Expression levels of Axin2, c-myc, CCND1, and TCF4, as determined by qRT-PCR in MGC803 and SGC7901 cells transfected with siRNA targeting TGM1 (si-TGM1) as compared to cells transfected with a control scrambled siRNA (si-Ctrl). (b) TOPflash/FOPflash luciferase assay activities in MGC803 and SGC7901 cells transfected with siRNA targeting TGM1 (si-TGM1) as compared to those in cells transfected with a control scrambled siRNA (si-Ctrl). Each bar represents the mean ± SEM of three independent experiments. *P < 0.001. (c) β-catenin levels in the nucleus and cytosol in MGC803 and SGC7901 cells transfected with siRNA targeting TGM1 (si-TGM1) compared to cells transfected with a control scrambled siRNA (si-Ctrl). (d) mRNA expression levels of Axin2, c-myc, CCND1, and TCF4 as determined by qRT-PCR in 96 clinical gastric cancer tissue samples
Discussion
We demonstrated the roles of TGM1 in development of gastric cancer cell in vitro, with expression levels of TGM1 being upregulated in both gastric cancer tissues and cultured gastric cancer cells. TGM1 expression levels were correlated with patient survival. In detailed cell culture studies, loss of TGM1 expression in gastric cancer cell lines inhibited proliferation and promoted apoptosis, as well as increasing chemosensitivity and reducing stemness of gastric cancer cells. These results strongly indicate participation of TGM1 in regulating gastric cancer cell malignancy. Most previous research has focused on involvement of TGM2 in carcinogenesis, whereas the function of TGM1 in cancer development has been largely overlooked. Given that TGM1 is expressed in tissues other than the skin, and that TGM1 and TGM2 are functionally redundant, it is important to also explore the roles of TGM1 in progression and drug resistance of gastric cancer.
We provide evidence that the mechanism of action of TGM1 in regulating gastric cancer might involve the Wnt signaling pathway, because loss of TGM1 expression in gastric cancer cells led to significant suppression of Wnt signaling. Although a previous study suggested that TGM1 could promote Wnt/β-catenin signaling,29 it is not clear whether this is also true in gastric cancer cells. Loss of TGM1 inhibited both nuclear accumulation of β-catenin and transcriptional activities of the Wnt/β-catenin signaling pathway in gastric cancer cells, consistent with the function of Wnt signaling in cancer development. It has been shown that activation of Wnt signaling is associated with increased cell proliferation and decreased cell apoptosis.30,31 To further confirm our hypothesis that TGM1 promotes gastric cancer development by activating the Wnt signaling pathway, we are investigating need to determine whether TGM1 can regulate proliferation, apoptosis, and stemness in cells lacking functional Wnt/β-catenin signaling. In addition to Wnt/β-catenin signaling, numerous other factors are involved in gastric cancer stemness and chemoresistance.32,33 Further investigations of other potential mechanisms of TGM1 function are necessary.
Since we do not have data to indicate whether loss of TGM1 inhibits cell proliferation, promotes chemosensitivity, or inhibits Wnt/β-catenin signaling in other cancer types, this study is not yet applicable to other cancer types. It is also important to note that the function of Wnt signaling is not limited to regulating cell apoptosis and stemness. Recent studies have elucidated important roles of the Wnt/β-catenin signaling pathway in regulating mesenchymal changes.34–36 One of the primary mechanisms by which the Wnt/β-catenin signaling pathway regulates mesenchymal changes is modulation of levels of key cell adhesion molecules, such as E-cadherin. E-cadherin is negatively controlled at the transcriptional level by transcription factors induced by Wnt/β-catenin signaling.37 Transcription factors such as Twist38 and Slug39 play fundamental roles in regulating E-cadherin. As epithelial to mesenchymal transition is a hallmark of the malignancy processes of many cancer types, we are actively exploring whether TGM1 can also function in the cellular processes related to the mesenchymal transition in gastric cancer development.
Author Contributions
XN designed the experiments and reviewed manuscript; HH and ZC conducted the experiments and collected the data; XN and HH analyzed the data; HH wrote the manuscript.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1.Bertuccio P, Chatenoud L, Levi F, Praud D, Ferlay J, Negri E, Malvezzi M, La Vecchia C. Recent patterns in gastric cancer: a global overview. Int J Cancer 2009; 125: 666–73. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- 3.Hohenberger P, Gretschel S. Gastric cancer. Lancet 2003; 362: 305–15. [DOI] [PubMed] [Google Scholar]
- 4.Kim K, Chun KH, Suh PG, Kim IH. Alterations in cell proliferation related gene expressions in gastric cancer. Crit Rev Eukaryot Gene Expr 2011; 21: 237–54. [DOI] [PubMed] [Google Scholar]
- 5.Milne AN, Carneiro F, O'Morain C, Offerhaus GJ. Nature meets nurture: molecular genetics of gastric cancer. Hum Genet 2009; 126: 615–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng YJ, Zhu ZX, Zhou JS, Hu ZQ, Zhang JP, Cai QP, Wang LH. Silencing profilin-1 inhibits gastric cancer progression via integrin beta1/focal adhesion kinase pathway modulation. World J Gastroenterol 2015; 21: 2323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Syu LJ, Zhao X, Zhang Y, Grachtchouk M, Demitrack E, Ermilov A, Wilbert DM, Zheng X, Kaatz A, Greenson JK, Gumucio DL, Merchant JL, di Magliano MP, Samuelson LC, Dlugosz AA. Invasive mouse gastric adenocarcinomas arising from Lgr5 + stem cells are dependent on crosstalk between the Hedgehog/GLI2 and mTOR pathways. Oncotarget 2016;7:10255–70. [DOI] [PMC free article] [PubMed]
- 8.Wei L, Li Y, Suo Z. TSPAN8 promotes gastric cancer growth and metastasis via ERK MAPK pathway. Int J Clin Exp Med 2015; 8: 8599–607. [PMC free article] [PubMed] [Google Scholar]
- 9.Hao NB, Tang B, Wang GZ, Xie R, Hu CJ, Wang SM, Wu YY, Liu E, Xie X, Yang SM. Hepatocyte growth factor (HGF) upregulates heparanase expression via the PI3K/Akt/NF-kappaB signaling pathway for gastric cancer metastasis. Cancer Lett 2015; 361: 57–66. [DOI] [PubMed] [Google Scholar]
- 10.Folk JE, Finlayson JS. The epsilon-(gamma-glutamyl)lysine crosslink and the catalytic role of transglutaminases. Adv Protein Chem 1977; 31: 1–133. [DOI] [PubMed] [Google Scholar]
- 11.Mehta K, Kumar A, Kim HI. Transglutaminase 2: a multi-tasking protein in the complex circuitry of inflammation and cancer. Biochem Pharmacol 2010; 80: 1921–9. [DOI] [PubMed] [Google Scholar]
- 12.Park D, Choi SS, Ha KS. Transglutaminase 2: a multi-functional protein in multiple subcellular compartments. Amino Acids 2010; 39: 619–31. [DOI] [PubMed] [Google Scholar]
- 13.Grigoriev MY, Suspitsin EN, Togo AV, Pozharisski KM, Ivanova OA, Nardacci R, Falasca L, Piacentini M, Imyanitov EN, Hanson KP. Tissue transglutaminase expression in breast carcinomas. J Exp Clin Cancer Res 2001; 20: 265–8. [PubMed] [Google Scholar]
- 14.Mehta K, Fok J, Miller FR, Koul D, Sahin AA. Prognostic significance of tissue transglutaminase in drug resistant and metastatic breast cancer. Clin Cancer Res 2004; 10: 8068–76. [DOI] [PubMed] [Google Scholar]
- 15.Iacobuzio-Donahue CA, Ashfaq R, Maitra A, Adsay NV, Shen-Ong GL, Berg K, Hollingsworth MA, Cameron JL, Yeo CJ, Kern SE, Goggins M, Hruban RH. Highly expressed genes in pancreatic ductal adenocarcinomas: a comprehensive characterization and comparison of the transcription profiles obtained from three major technologies. Cancer Res 2003; 63: 8614–22. [PubMed] [Google Scholar]
- 16.Satpathy M, Cao L, Pincheira R, Emerson R, Bigsby R, Nakshatri H, Matei D. Enhanced peritoneal ovarian tumor dissemination by tissue transglutaminase. Cancer Res 2007; 67: 7194–202. [DOI] [PubMed] [Google Scholar]
- 17.Miyoshi N, Ishii H, Mimori K, Tanaka F, Hitora T, Tei M, Sekimoto M, Doki Y, Mori M. TGM2 is a novel marker for prognosis and therapeutic target in colorectal cancer. Ann Surg Oncol 2010; 17: 967–72. [DOI] [PubMed] [Google Scholar]
- 18.Martinet N, Bonnard L, Regnault V, Picard E, Burke L, Siat J, Grosdidier G, Martinet Y, Vignaud JM. In vivo transglutaminase type 1 expression in normal lung, preinvasive bronchial lesions, and lung cancer. Am J Respir Cell Mol Biol 2003; 28: 428–35. [DOI] [PubMed] [Google Scholar]
- 19.Fok JY, Ekmekcioglu S, Mehta K. Implications of tissue transglutaminase expression in malignant melanoma. Mol Cancer Ther 2006; 5: 1493–503. [DOI] [PubMed] [Google Scholar]
- 20.Zhang R, Tremblay TL, McDermid A, Thibault P, Stanimirovic D. Identification of differentially expressed proteins in human glioblastoma cell lines and tumors. Glia 2003; 42: 194–208. [DOI] [PubMed] [Google Scholar]
- 21.Herman JF, Mangala LS, Mehta K. Implications of increased tissue transglutaminase (TG2) expression in drug-resistant breast cancer (MCF-7) cells. Oncogene 2006; 25: 3049–58. [DOI] [PubMed] [Google Scholar]
- 22.Mehta K, Fok JY, Mangala LS. Tissue transglutaminase: from biological glue to cell survival cues. Front Biosci 2006; 11: 173–85. [DOI] [PubMed] [Google Scholar]
- 23.Russell LJ, DiGiovanna JJ, Rogers GR, Steinert PM, Hashem N, Compton JG, Bale SJ. Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat Genet 1995; 9: 279–83. [DOI] [PubMed] [Google Scholar]
- 24.Guo P, Zhu Z, Sun Z, Wang Z, Zheng X, Xu H. Expression of legumain correlates with prognosis and metastasis in gastric carcinoma. PloS One 2013; 8: e73090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Lu J, Hua Y, Zhang P, Liang Z, Ruan L, Lian C, Shi H, Chen K, Tu Z. Targeting heat-shock protein 90 with ganetespib for molecularly targeted therapy of gastric cancer. Cell Death Dis 2015; 6: e1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher ML, Keillor JW, Xu W, Eckert RL, Kerr C. Transglutaminase is required for epidermal squamous cell carcinoma stem cell survival. Mol Cancer Res 2015; 13: 1083–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee CH, Yu CC, Wang BY, Chang WW. Tumorsphere as an effective in vitro platform for screening anti-cancer stem cell drugs. Oncotarget 2016; 7: 1215–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song X, Xin N, Wang W, Zhao C. Wnt/beta-catenin, an oncogenic pathway targeted by H. pylori in gastric carcinogenesis. Oncotarget 2015; 6: 35579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hampton PJ, Ross OK, Reynolds NJ. Increased nuclear beta-catenin in suprabasal involved psoriatic epidermis. Br J Dermatol 2007; 157: 1168–77. [DOI] [PubMed] [Google Scholar]
- 30.Sherwood V. WNT signaling: an emerging mediator of cancer cell metabolism? Mol Cell Biol 2015; 35: 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pai P, Rachagani S, Dhawan P, Batra SK. Mucins and Wnt/beta-catenin signaling in gastrointestinal cancers: an unholy nexus. Carcinogenesis 2016; 37: 223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuo Q, Liu J, Zhang J, Wu M, Guo L, Liao W. Development of trastuzumab-resistant human gastric carcinoma cell lines and mechanisms of drug resistance. Sci Rep 2015; 5: 11634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Guo W, Wang X, Liu X, Huang M, Gan L, Cheng Y, Li J. Antitumor activity and inhibitory effects on cancer stem cell-like properties of Adeno-associated virus (AAV)-mediated Bmi-1 interference driven by Bmi-1 promoter for gastric cancer. Oncotarget. Epub ahead of print 2016. [DOI] [PMC free article] [PubMed]
- 34.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer 2001; 1: 55–67. [DOI] [PubMed] [Google Scholar]
- 35.Huelsken J, Vogel R, Brinkmann V, Erdmann B, Birchmeier C, Birchmeier W. Requirement for beta-catenin in anterior-posterior axis formation in mice. J Cell Biol 2000; 148: 567–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell 1996; 87: 159–70. [DOI] [PubMed] [Google Scholar]
- 37.Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature 2003; 422: 317–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howe LR, Watanabe O, Leonard J, Brown AM. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res 2003; 63: 1906–13. [PubMed] [Google Scholar]
- 39.Vallin J, Thuret R, Giacomello E, Faraldo MM, Thiery JP, Broders F. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/beta-catenin signaling. J Biol Chem 2001; 276: 30350–8. [DOI] [PubMed] [Google Scholar]