Abstract
Long non-coding RNA H19 is aberrantly expressed in multiple malignancies and its expression levels correlate with recurrence, metastasis, and patient survival. Despite numerous reports documenting the role of H19 in carcinogenesis, its contribution to cervical cancer development is still largely unknown. In this study, I observed that H19 expression was elevated in cervical cancer cell lines and could be detected in extracellular vesicles in the culture medium. In addition, I demonstrated, by overexpression and knockdown experiments, that H19 promoted cell proliferation and multicellular tumor spheroid formation without significantly affecting apoptosis and cell migration. Finally, treatment with transforming growth factor beta and hypoxia-mimetic CoCl2 could modulate H19 levels in a cell line-specific manner. These findings indicate that H19 promotes both anchorage-specific and -independent growth of cervical cancer cell lines and may serve as a potential target for cancer diagnosis and therapy.
Keywords: lncRNA, H19, cervical cancer, extracellular vesicles, transforming growth factor beta, hypoxia
Introduction
Cervical cancer is the second most frequently diagnosed cancer and the third leading cause of cancer death in women worldwide.1 The availability of human papillomavirus (HPV) vaccines has lowered the incidence of cervical cancer; yet, vaccines can only prevent infection by certain HPV types and have no therapeutic effect against existing HPV infections or disease.2,3 Therefore, early detection of treatable precancerous lesions is indispensable to reduce cancer incidence and increase life expectancy in patients.4 The existing Papanicolaou (Pap) test, albeit useful in reducing cancer incidence, can be inconclusive and exhibit a relatively high percentage of false-negatives.5 The more sensitive HPV-based test is often associated with a false-positive result, since the majority of HPV-positive women do not develop cervical cancer.6 A more precise cervical cancer test would be an invaluable tool in national screening programs.
Long non-coding RNAs (lncRNAs) are a new class of non-coding RNAs with more than 200 nucleotides in length and an estimated 15,000 lncRNAs in humans.7 Accumulating evidence indicates that lncRNAs are involved in a diverse range of biological functions, such as imprinting,8 epigenetic regulation,9 apoptosis and cell cycle control,10 transcriptional and translational regulation,11 RNA splicing,12 regulation of microRNA abundance,13 development and differentiation,14 aging,15 and disease pathogenesis.16 Dozens of lncRNAs have been shown to be aberrantly expressed in cancer and their expression levels are associated with recurrence, prognosis, metastasis, and predicting the response to therapy.16–19 Various lncRNAs have been reported to act as both oncogenes and tumor suppressors, playing an active role in modulating cancer progression.19,20
The imprinted, developmentally regulated lncRNA H19 is absent in most normal adult tissues; however, its re-expression has been detected in various cancers.21–23 Since the discovery of H19 in 1984,24 its function has remained enigmatic. Many studies using cell culture systems and mouse models have shown that H19 can function either as an oncogene25–27 or a tumor suppressor.28,29 Recently, H19 RNA transcripts have been detected in a majority of patient-derived premalignant lesions of cervical cancer.21 Whether H19 is actively involved in the development of cervical cancer and how it works remains to be determined.
In this study, I investigated whether increased H19 expression in both cancer cells and extracellular vesicles (EVs) was a characteristic molecular change in cervical cancer and elucidated the effect of altered H19 level on the phenotypes of cervical cancer cell lines. In addition, I examined the effect of epithelial–mesenchymal transition (EMT) inducers, transforming growth factor beta (TGF-β), and hypoxia, on the expression of H19 transcript. This study serves as a stepping-stone to unravel how lncRNA H19 regulates cervical carcinogenesis, which may help improve cervical cancer diagnosis and treatment.
Materials and methods
Reagents
Recombinant human TGF-β1 (#240-B-002) was purchased from R&D Systems, Minneapolis, MN, USA. Cobalt chloride hexahydrate (#C8661), puromycin (#P8833), and doxycycline hydrochloride (#D3447) were purchased from Sigma-Aldrich, St. Louis, MO, USA.
Plasmids and small interfering RNAs (siRNAs)
H19 expression vector, pCMV-Sport6-H19 (clone ID: 4993796); siRNAs targeting human H19 (SMARTpool: Lincode H19 siRNA); non-targeting siRNAs (Lincode Non-targeting Pool); and TRIPZ inducible lentiviral vectors expressing non-targeting small hairpin RNA (NT shRNA; #RHS4743), H19 shRNA1 (#RHS4696-200705182), or H19 shRNA2 (#RHS4696-200695573) were purchased from Dharmacon, Lafayette, CO, USA. pcDNA3.1 (gift from Janet Mertz) served as empty vector control. Plasmids encoding psPAX2 (#12260, Addgene; gift from Janet Mertz) and vesicular stomatitis virus G (VSVG) (gift from Bill Sugden) were used to produce lentiviruses. Plasmid DNA was purified using the GeneJET Plasmid Midiprep kit (Thermo Fisher Scientific, Waltham, MA, USA).
Cell culture
Human primary fibroblasts (ATCC® CRL-2708™) were obtained from the Nano-cosmeceuticals laboratory; HPV-positive cervical carcinoma HeLa (ATCC® CCL2™) and SiHa (ATCC® HTB-35™) cells were obtained from the Nano Delivery System laboratory; HPV-positive cervical carcinoma MS751 (ATCC® HTB-34™), HPV-negative cervical carcinoma C33A (ATCC® HTB-31™), and HPV-16 E6/E7 transformed ectocervical cell line Ect1/E6E730 (ATCC® CRL-2614™) were purchased from the American Type Culture Collection, Manassas, VA, USA. Primary fibroblasts, HeLa, SiHa, MS751, and C33A cell lines were grown in Dulbecco’s Modified Eagle Medium (DMEM; HyClone, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS; HyClone, Thermo Fisher Scientific) and 100 units/mL penicillin plus 100 µg/mL streptomycin, or 50 µg/mL Primocin™ (InvivoGen, San Diego, CA, USA). Ect1/E6E7 cells were grown in Keratinocyte serum-free medium (Gibco, Thermo Fisher Scientific) with 0.1 ng/mL human recombinant epidermal growth factor, 0.05 mg/mL bovine pituitary extract, 44.1 mg/L calcium chloride, and 50 µg/mL Primocin™. All cells were maintained at 37℃ in a 5% CO2 incubator.
Transient transfections
For plasmid transfections, cells in six-well plates were transfected with 400 ng expression plasmids using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) or TurboFect (Thermo Fisher Scientific). For siRNA transfections, cells in six-well plates were transfected with 100 nM siRNA using DharmaFECT1 (Dharmacon). After incubation for 44–48 h, cells were ready for downstream characterization.
Infection of cell lines with lentivirus
Stable knockdown cell lines were generated as previously described, with some modifications.31 Tet-On-inducible TRIPZ lentiviral vectors expressing NT shRNA or shRNAs targeting H19 (H19 shRNA1 and H19 shRNA2) were used to produce lentiviruses. In brief, 293 T cells from 10 cm dishes were co-transfected with 4 µg lentiviral vector(s), 1.4 µg psPAX2, and 0.6 µg VSVG. Medium containing the viruses was harvested 72 h later, filtered through a 0.45 µm pore-size filter, and added to HeLa cells. After 72 h, infected cells were selected and shRNA expression was induced by incubation with medium containing 1 µg/mL puromycin and 1 µg/mL doxycycline for 7–10 days before harvesting.
RNA isolation from cells and EVs
Total RNA was extracted from cell lines using the RNeasy or miRNeasy mini kits (QIAGEN, Hilden, Germany) following the manufacturer’s protocols. For RNA isolation from EVs, cell lines were grown for two days in complete medium containing exosome-depleted FBS (prepared by ultracentrifugation at 120,000 × g for 2 h). The medium was harvested, spun down at 5251 × g for 20 min, and filtered through a 0.2 µm pore-size filter to remove cell debris. EVs were then collected by centrifugation at 200,000 × g for 2 h in a Type 100 Ti rotor using an Optima L-100 XP ultracentrifuge (Beckman-Coulter, Brea, CA, USA). After aspirating the supernatant, total RNA was extracted from the pellet using the miRNeasy mini kit (QIAGEN). RNA samples were quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific) and stored at −80℃ until use.
Quantitative reverse transcription PCR (RT-qPCR)
Quantification of gene expression was performed by RT-qPCR using a Luminaris Color HiGreen qPCR Master Mix (Thermo Fisher Scientific) or SsoAdvanced universal SYBR green supermix (Bio-Rad, Hercules, CA, USA) and a CFX96 Touch real-time PCR detection system (Bio-Rad). Briefly, 1.5–2.0 µg total RNA was incubated with RNase-free DNaseI and converted to cDNA with the Maxima cDNA synthesis kit (Thermo Fisher Scientific). The synthesized cDNA (10–20 ng/reaction) was used as template for qPCR. Amplification and detection were performed as described by the manufacturer, followed by melting curve analysis starting at 65℃ with increments of 0.5℃ per cycle (N = 60 cycles). Primers and annealing temperatures (Ta) were as follows: H19 (62.5℃): FWD (5′-ACTCAGGAATCGGCTCTGGAAG-3′), and REV (5′-GCTGCTGTTCCGATGGTGTC-3′); BAX (62.9℃): FWD (5'-CAGGATGCGTCCACCAAGAAG-3'), and REV (5'-AAAACATGTCAGCTGCCACTCG-3'); BCL2 (62.9℃): FWD (5'-CGACTTCGCCGAGATGTCC-3'), and REV (5'-CACACATGACCCCACCGAAC-3'); and BCL2L1 (62.9℃): FWD (5'-CACTGTGCGTGGAAAGCGT-3'), and REV (5'-CTCTAGGTGGTCATTCAGGTAAGTG-3'). Primers of the following target genes (Ta = 60℃) were purchased from Bio-Rad (assay ID): VIM (qHsaCED0042034), CDH1 (qHsaCED0042076), MYC (qHsaCID0012921), SNAI1 (qHsaCED0057267), SNAI2 (qHsaCID0011342), ZEB1 (qHsaCED0045418), ZEB2 (qHsaCED0038149), HIF1A (qHsaCED0042813), and VEGFA (qHsaCED0043454). Expression was normalized internally to that of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (62.5℃): FWD (5′-GGGAAACTGTGGCGTGATGG-3′), and REV (5'-TGGAGGAGTGGGTGTCGCTG-3') or U6 (62.5℃): FWD (5′-CTCGCTTCGGCAGCACATATAC-3′), and REV (5′-GGAACGCTTCACGAATTTGC-3′) using the ΔΔCt method. Assays were performed in triplicate and data were analyzed by the Bio-Rad CFX manager software.
Cell growth and proliferation assay using real-time cell analysis (RTCA)
Cell growth and proliferation kinetics were determined using RTCA (ACEA Biosciences, San Diego, CA, USA) as previously described, with some modifications.32 Briefly, 100 µL DMEM with 10% FBS and 50 µg/mL Primocin™ was added to each well of a gold microelectrode array integrated 16-well plate (E-Plate 16). The plate was connected to the system to measure background impedance. Cells were seeded at 10,000 cells/well and incubated at room temperature for 30 min. The E-Plate 16 was placed on the xCELLigence RTCA DP instrument (ACEA Biosciences) located in an incubator at 37℃ with 5% CO2. The impedance of each well was recorded every 30 min for 2–4 days and expressed as a cell index. The cell index value increased gradually as cells attached to the gold electrodes. The rate of cell proliferation was derived from the slope of the line between two given time points during log phase. All assays were performed in triplicate and data were analyzed by the RTCA data analysis software (version 1.0).
Multicellular tumor spheroid assay
Cell lines (3000–4000 cells/well) were seeded in 200 µL complete medium in ultra-low attachment 96-well plates (Corning, Thermo Fisher Scientific). Cells were grown for seven days with 50% medium changes every three days. Phase contrast and fluorescent images of tumor spheroids were recorded using an Olympus Is71 inverted fluorescence microscope (Olympus, Tokyo, Japan). The number of viable cells within spheroids was determined by measuring the amount of intracellular ATP using the CellTiter-Glo® 2.0 assay (Promega, Madison, WI, USA) according to the manufacture’s protocol. Assays were performed in triplicate.
Caspase-3/7 assay
Cells (8000–10,000 cells/well) were seeded in a 96-well black clear-bottom plate (Corning, Thermo Fisher Scientific) and incubated for 48 h. Caspase-3/7 activity was determined using the Caspase-Glo® 3/7 Assay (Promega) and a SpectraMax® L microplate luminometer (Molecular Devices, Sunnyvale, CA, USA) according to the manufacturer’s protocol. All assays were performed in triplicate.
Annexin V staining
Cells (8000–10,000 cells/well) were seeded in a 96-well black clear-bottom plate (Corning, Thermo Fisher Scientific) and incubated for 48 h. Phospholipid flipping during apoptosis was detected using the TACS Annexin V-FITC kit (R&D Systems) according to the manufacturer’s protocol. Fluorescent images were recorded using an Olympus Is71 inverted fluorescence microscope (Olympus).
Real-time cell migration assay by RTCA
Analysis of cell migration in real time was performed using the RTCA system and CIM-16 plates (ACEA Biosciences) as previously described, with some modifications.33 Briefly, CIM-16 plates comprise an upper and a lower chamber separated by a microporous membrane (8 µm pores) with microelectrodes at the underside for impedance-based detection of migrated cells. Initially, 175 µL complete medium and 50 µL serum-free medium were added to the lower and upper chambers of the CIM-16 plate, respectively. The plate was placed in the xCELLigence RTCA DP instrument at 37℃ and 5% CO2 for 1 h for equilibration and a background signal was recorded. Next, 50,000 cells in 100 µL serum-free medium were seeded in each well of the upper chamber. The electrical impedance, displayed as cell index, increased proportionally as cells migrated through the pore to the electrodes on the lower side of the membrane. Assays were performed in quadruplicate and the cell index was recorded every 15 min for 80 h.
Statistical analysis
Statistical analysis was performed using GraphPad InStat version 3 software (GraphPad Software, La Jolla, CA, USA). Unpaired Student’s t-test and one-way ANOVA with Tukey’s HSD test were used for comparing two or more groups, respectively. A P value < 0.05 was considered statistically significant.
Results
LncRNA H19 is overexpressed in cervical cancer cell lines and secreted into EVs
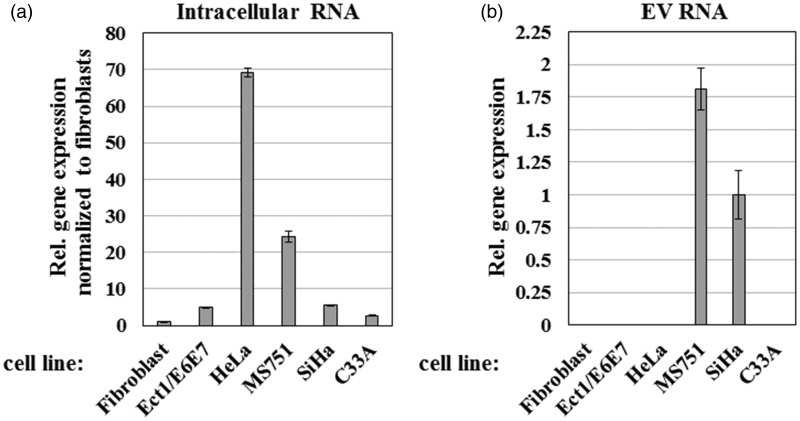
Numerous reports have shown that H19 RNA is essential for tumor growth and its expression is increased in a majority of human cancers.27 To test the role of H19 RNA in cervical carcinogenesis, I first used RT-qPCR to determine the relative levels of H19 in multiple cervical cancer cell lines, including an immortalized ectocervical epithelial cell line (Ect1/E6E7) and primary fibroblasts. As shown in Figure 1(a), H19 RNA was upregulated in all cell lines relative to the primary fibroblasts (3–6-fold in Ect1/E6E7, SiHa, and C33A; 24-fold in MS751; and 69-fold in HeLa), confirming a potential role for H19 in cervical cancer progression.
Figure 1.
Expression levels of lncRNA H19 in cervical cancer cell lines and secreted extracellular vesicles (EVs). Relative (Rel.) H19 transcript levels from: (a) human primary fibroblasts, immortalized ectocervical cell line (Ect1/E6E7), and four cervical cancer cell lines; and (b) EVs isolated from the culture medium of the same cell lines. RT-qPCR values were normalized internally to U6 levels in each cell line and externally to H19 expression in primary fibroblasts. Bars indicate means ± 1 SEM; assays were performed in triplicate. Results are representative of data obtained on two separate occasions
I also examined whether H19 RNA was secreted by cervical cancer cells into EVs. Thus, all cell lines were grown in complete medium with exosome-depleted FBS to minimize contamination with bovine exosomes. EVs were isolated from cell culture medium by ultracentrifugation and total EV RNA was purified. RT-qPCR indicated that H19 RNA was only detectable in EVs secreted by MS751 and SiHa cell lines (Figure 1(b)), suggesting a selective enrichment of lncRNA in EVs. Therefore, I conclude that H19 is significantly more expressed in cervical cancer cell lines and can be secreted in a cell line-specific manner.
H19 promotes proliferation of cervical cancer cells in vitro
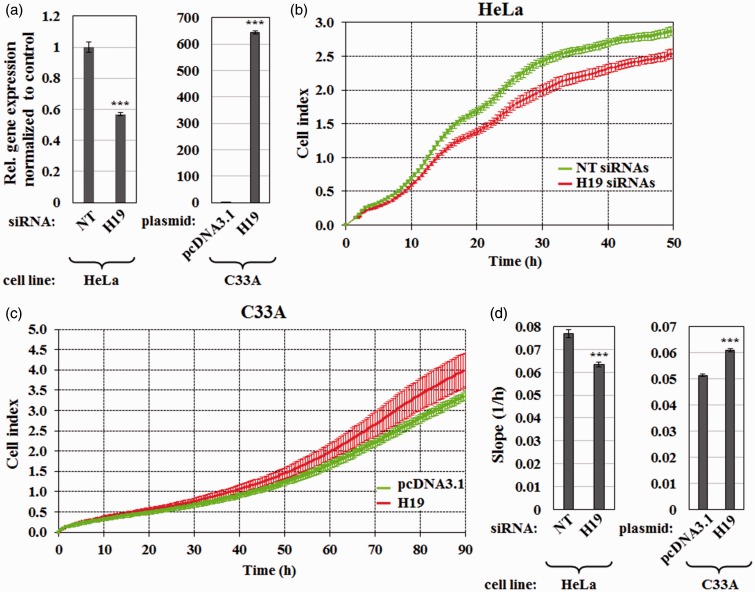
To assess the biological function of H19 in cervical cancer, I investigated whether H19 affected proliferation of cervical cancer cell lines. As HeLa cells express H19 at high level (Figure 1(a)), I first examined whether H19 knockdown affected cell proliferation. To do so, HeLa cells were transfected with siRNA targeting H19 or non-targeting siRNA control. I achieved H19 knockdown of 43% as determined by RT-qPCR (Figure 2(a)). Cell proliferation kinetics was then determined in real-time using the RTCA system. Figure 2(b) and (d) showed that downregulation of H19 significantly delayed growth of HeLa cells.
Figure 2.
H19 induces cell proliferation of cervical cancer cell lines. (a) RT-qPCR results showing relative expression of H19 in HeLa cells transfected with siRNAs targeting H19 or non-targeting (NT) siRNAs and in C33A cells transfected with a plasmid expressing H19 (pCMV-SPORT6-H19) or empty vector pcDNA3.1. Data were normalized internally to GAPDH and externally to the control. Bars indicate means ± 1 SEM; assays were performed in triplicate. (b and c) Real-time cell index values showing the proliferation rates of (b) HeLa cells transfected with H19 or NT siRNAs and (c) C33A cells transfected with a plasmid expressing H19 or pcDNA3.1. (d) Proliferation rates obtained by analyzing the slope during log phase between time points 10 and 34 h for (b) and 50–74 h for (c). Data are shown as mean ± 1 SD; assays were performed in triplicate. ***, P < 0.001. (A color version of this figure is available in the online journal.)
On the other hand, I also tested whether the overexpression of H19 could give the opposite phenotype. As C33A cells express H19 at relatively low level, comparable to the level in a noncancerous ectocervical epithelial cell line (Ect1/E6E7) (Figure 1(a)), C33A cells were used to determine effects of H19 gain of function. The reason for choosing cells with low H19 for overexpression was to minimize possible artifact generated by overexpression.34 RT-qPCR confirmed a 644% overexpression of H19 in the transfected cells (Figure 2(a)). As expected, ectopic expression of H19 enhanced the proliferation rate of C33A cells (Figure 2(c) and (d)). It should be noted, however, that forced overexpression of H19 did not affect the proliferation rates of HeLa cells, probably because they already expressed a high level of endogenous H19 (data not shown). Hence, I conclude that H19 promotes the proliferation of cervical cancer cells.
H19 enhances tumor spheroid forming ability of cervical cancer cells
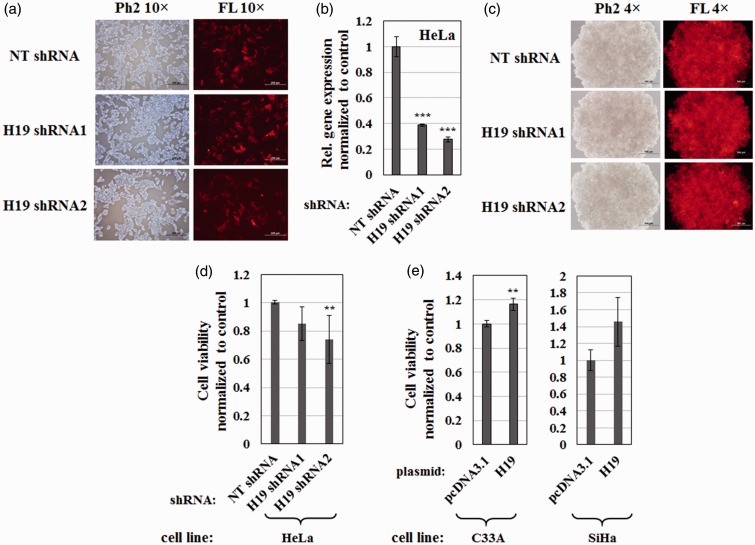
Next, I explored whether H19 affected the ability of cervical cancer cells to form tumor spheroids and took advantage of high H19 expression levels in HeLa cells (Figure 1(a)). To establish stable H19-knockdown cell lines, HeLa cells were infected with lentiviruses expressing two individual shRNAs (H19 shRNA1 or H19 shRNA2) or NT shRNA. Transduced cells were selected by growth in medium containing 1 µg/mL puromycin and expression of shRNA was induced by adding 1 µg/mL doxycycline. Given that a tetracycline-inducible promoter on the TRIPZ lentiviral vector drove the expression of turboRFP along with a target shRNA, turboRFP levels served as a marker for shRNA expression. As shown in Figure 3(a), HeLa cells infected with lentivirus bearing all three shRNAs strongly expressed turboRFP, confirming the successful generation of stable cell lines and shRNA expression. In addition, RT-qPCR indicated 60 and 70% knockdown of H19 in stable cell lines expressing H19 shRNA1 and H19 shRNA2, respectively (Figure 3(b)).
Figure 3.
H19 affects the formation of multicellular tumor spheroids from cervical cancer cells. (a) HeLa stable cell lines, containing lentivirus expressing non-targeting shRNA (NT shRNA) or shRNA targeting H19 (H19 shRNA1 or H19 shRNA2), were incubated for seven days in the presence of 1 µg/mL puromycin and 1 µg/mL doxycycline. TurboRFP served as a marker for shRNA expression. Paired phase contrast (Ph2) and fluorescence (FL) images obtained with a 10 × objective are shown. Scale bars = 200 µm. (b) RT-qPCR results showing relative levels of H19 following shRNA knockdown in HeLa stable cell lines. Values were normalized internally to GAPDH and externally to expression of H19 in the NT shRNA control. Bars indicate means ± 1 SEM; assays were performed in triplicate. ***, P < 0.001. (c) Multicellular tumor spheroids of HeLa stable cells expressing NT shRNA, H19 shRNA1, or H19 shRNA2 grown in ultra-low attachment plates for seven days in the presence of 1 µg/mL puromycin and 1 µg/mL doxycycline. TurboRFP indicated shRNA expression as shown in paired Ph2 and FL images obtained with a 4 × objective. Scale bars = 500 µm. Cell viability within (d) tumor spheroids of HeLa stable cell lines with H19 knockdown or control, and (e) tumor spheroids of C33A or SiHa cells overexpressing H19 or empty vector control, as determined by the CellTiter-Glo 2.0 assay. Values were normalized to the cell number of the control cell line. Bars indicate means ± 1 SD; assays were performed in triplicate. **, P < 0.01. (A color version of this figure is available in the online journal.)
To generate tumor spheroids, HeLa stable cell lines were seeded in 96-well ultra-low attachment plates and cultured for seven days. Figure 3(c) showed that all HeLa stable cell lines could form tumor spheroids and sustain shRNA expression, as evidenced by TurboRFP signal. I compared the number of viable cells within spheroids and observed that H19 knockdown decreased tumor spheroid formation in HeLa cells (Figure 3(d)). Additionally, effects of H19 gain of function on tumor spheroid formation were investigated in C33A and SiHa cells which expressed low levels of endogenous H19 (Figure 1(a)). As expected, H19 RNA overexpression increased tumor spheroid growth in both cell lines (Figure 3(e)). Thus, I conclude that H19 promotes tumor spheroid formation of cervical cancer cells in vitro.
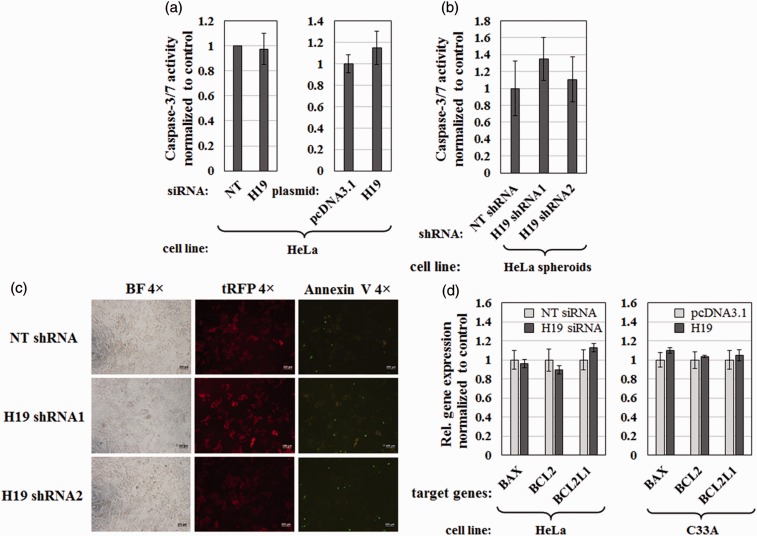
H19 does not affect apoptosis in cervical cancer cells
Given that H19 has been reported to regulate apoptosis in cancer cells,35 I tested whether changes in H19 expression affected apoptosis by measuring the combined activity of the major effector caspases, caspase-3 and -7. Neither siRNA-mediated knockdown of H19 nor ectopic expression of H19 for two days yielded any change in caspase-3/7 activity in HeLa cells (Figure 4(a)). The same result was obtained with spheroids of HeLa cells infected with lentiviruses expressing H19 shRNA1, H19 shRNA 2, or NT shRNA (Figure 4(b)). In addition, the Annexin V staining assay was performed to determine phosphatidylserine (PS) externalization during apoptosis. As shown in Figure 4(c), shRNA-mediated knockdown of H19 did not affected the proportion of Annexin V-staining positive cells in HeLa cells.
Figure 4.
Effect of lncRNA H19 on caspase-3/7 activation, PS externalization, and expression of selected genes involved in apoptosis. (a) Caspase-3/7 activities of HeLa cells transfected with H19 siRNAs or non-targeting (NT) siRNAs (left panel), or transfected with H19 expression plasmid or pcDNA3.1 control (right panel). (b) Caspase-3/7 activities of tumor spheroids grown from HeLa stable cells expressing NT shRNA, H19 shRNA1, or H19 shRNA2 cultured in ultra-low attachment plates for seven days in the presence of 1 µg/mL puromycin and 1 µg/mL doxycycline. (a and b) Data were normalized to caspase-3/7 activity of the control cell line in each set of experiments and shown as mean ± 1 SD; assays were performed in triplicate. (c) Annexin V staining of HeLa stable cells expressing NT shRNA, H19 shRNA1, or H19 shRNA2. Panels on the left show cells in bright field (BF) images, middle panels show TurboRFP (tRFP) as a marker for shRNA expression, and right panels show Annexin V labeled with fluorescein isothiocyanate (green fluorescence). Images were obtained with a 4 × objective. Scale bars = 200 µm. (d) Effect of H19 on expression of apoptosis-related genes BAX, BCL2, and BCL2L1 as measured by RT-qPCR. HeLa cells were transfected with H19 siRNAs or NT siRNAs while C33A cells were transfected with H19 expression plasmid or the pcDNA3.1 control. RNA levels were normalized to GAPDH. Bars represent mean ± 1 SEM; assays were performed in triplicate. (A color version of this figure is available in the online journal.)
I also examined whether H19 affected the expression of three apoptotic regulators, BAX, BCL2, and BCL2L1, previously reported to be controlled by H19.35,36 As shown in Figure 4(d), neither H19 knockdown in HeLa cells nor H19 overexpression in C33A cells altered mRNA levels of the aforementioned genes. Therefore, I conclude that H19 does not significantly affect apoptosis in cervical cancer cell lines.
H19 does not significantly affect migration ability of cervical cancer cell lines
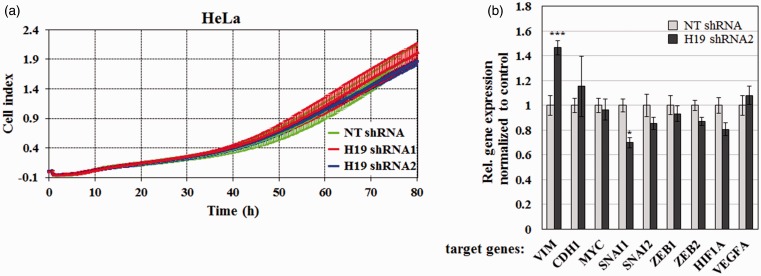
Numerous reports have documented a crucial role for lncRNA H19 in tumor metastasis through regulation of EMT.37 To assess whether H19 altered migration of cervical cancer cells, I used the RTCA system to measure the migratory activity of HeLa knockdown stable cell lines. I observed no significant change in migration rate (Figure 5(a)), even when H19 expression was significantly decreased (Figure 3(b)).
Figure 5.
Effect of H19 on cervical cancer cell migration and expression of selected genes involved in epithelial–mesenchymal transition (EMT) and metastasis. (a) Real-time cell migration of HeLa stable cells expressing NT shRNA, H19 shRNA1, or H19 shRNA2 over 80 h. Bars represent mean ± 1 SD; assays were performed in quadruplicate. (b) Effect of H19 on mRNA levels of key genes involved in EMT and metastasis. HeLa stable cells expressing NT shRNA or H19 shRNA2 were analyzed for relative levels of VIM, CDH1, MYC, SNAI1, SNAI2, ZEB1, ZEB2, HIF1A, and VEGFA transcripts by RT-qPCR. RNA levels were normalized internally to GAPDH and externally to expression of the target gene in the NT shRNA cell line. Bars represent mean ± 1 SEM; assays were performed in triplicate. *, P < 0.05, ***, P < 0.001. (A color version of this figure is available in the online journal.)
Moreover, I profiled the expression of nine key genes involved in EMT and metastasis using RT-qPCR. As shown in Figure 5(b), H19 silencing decreased SNAI1 expression without significantly affecting mRNA levels of CDH1, MYC, SNAI2, ZEB1, ZEB2, HIF1A, and VEGFA. These findings indicate that H19 does not significantly alter the motility of cervical cancer cells, even though it positively regulates the expression of SNAI1.
TGF-β1 and hypoxia regulate H19 expression in cervical cancer cells
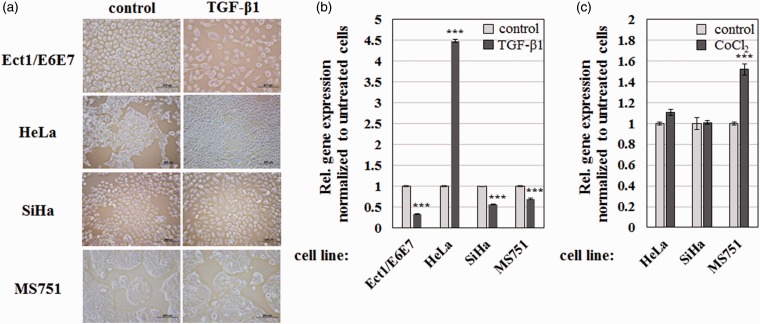
Several reports have demonstrated that EMT modulators such as TGF-β and hypoxia can induce H19 expression in various carcinoma models.37,38 To test this possibility in cervical cancer, I determined the effect of TGF-β1 on the expression of H19 in various cervical cancer cell lines. As expected from the tumor suppressive role of TGF-β during early tumorigenesis,39 incubating the non-cancerous Ect1/E6E7 cell line with TGF-β1 led to apoptotic cell death (Figure 6(a)). In contrast, TGF-β1 treatment induced spindle cell-like morphology in HeLa cells, but not in SiHa and MS751 cells (Figure 6(a)). These findings were confirmed by RT-qPCR, which revealed that incubation with TGF-β1 for 48 h greatly elevated H19 expression in HeLa cells while significantly downregulating it in Ect1/E6E7, SiHa, and MS751 cells (Figure 6(b)). Treatment with 100 µM CoCl2, a known hypoxia inducer,40 stimulated H19 expression in MS751 but not in HeLa and SiHa cell lines (Figure 6(c)). I did not observe any morphological change when these cells were treated with CoCl2 (data not shown). Therefore, I conclude that TGF-β1 and hypoxia regulate H19 expression in a cell line-specific manner.
Figure 6.
Changes in H19 expression in response to TGF-β1 and hypoxia. (a) Phase contrast micrographs of the indicated cell lines in the absence or presence of 10 ng/mL TGF-β1 obtained using a 10 × objective. Scale bars = 200 µm. (b and c) RT-qPCR results showing relative expression of H19 following incubation with (b) 10 ng/mL TGF-β1 and (c) 100 µM CoCl2 for two days. Data were normalized internally to GAPDH and externally to H19 expression in non-treated cells. Bars indicate means ± 1 SEM; assays were performed in triplicate. ***, P < 0.001. (A color version of this figure is available in the online journal.)
Discussion
Here, I demonstrate that lncRNA H19 is upregulated in cervical cancer cell lines, especially HeLa and MS751 (Figure 1(a)) and can be found in EVs outside the cells (Figure 1(b)). Interestingly, the intracellular level of H19 did not necessarily correlate with that in secreted EVs, suggesting selective packaging of H19 during vesicle formation. Given that EVs have been detected in many different body fluids, circulating H19 could serve as a biomarker for non-invasive cervical cancer screening, akin to the use of plasma H19 in gastric cancer diagnosis.41
A growing number of reports have explored the effect of H19 loss- and gain-of-function in various malignancies.27,42–44 However, data on the role of H19 in cervical cancer remain limited. Here, knockdown and overexpression of H19 confirmed a role for H19 in promoting cervical cancer cell proliferation (Figure 2). I further investigated the effect of H19 using the multicellular tumor spheroid model. Spheroids exhibit a gradient of proliferating cells in the outer cell layers and quiescent cells in the core, where they usually die from oxygen and glucose deprivation. This heterogeneity recapitulates more accurately the micro-regions of tumors than monolayer cell cultures.45,46 In agreement with 2D cultures, I demonstrated that H19 moderately induced tumor spheroid growth in cervical cancer cell lines (Figure 3).
Although H19 has been shown to inhibit apoptotic cell death,35,36 changes in H19 levels failed to affect the activities of effector caspases-3 and -7 (Figure 4(a) and (b)). At present, I cannot exclude the possibility that H19 regulates apoptosis in cervical cancer cells via a caspase-independent mechanism. However, Annexin V-FITC staining did not reveal any difference in the early stages of apoptosis in HeLa cells knocked down with H19 shRNA1 or H19 shRNA2 compared with the NT shRNA control (Figure 4(c)). Nor did I observe any change in mRNA expression of the pro-apoptotic gene BAX and antiapoptotic genes BCL2 and BCL2L1 upon H19 knockdown or overexpression (Figure 4(d)). Hence, my findings suggest that inhibition of apoptosis is not the primary mechanism by which H19 promotes cervical cancer development.
Accumulating evidence indicates that H19 and miR-675, which is processed from it, play key roles in metastasis, by regulating EMT and mesenchymal–epithelial transitions (MET).37,47 Using the state-of-the-art RTCA system, I failed to observe any real-time change in cell motility, even when H19 expression was downregulated by 60–70% and the mRNA level of the EMT transcription factor Snail (SNAI1) was significantly decreased (Figure 5). However, I cannot exclude the possibility that the observed phenotype is cell line specific. To this end, H19 appears to have a smaller effect on cervical cancer cells than on other types of cancer.42,48,49 Further studies will determine whether H19 plays a predominant role during early cervical cancer events or it only marginally regulates cervical carcinogenesis.
Several EMT modulators, including TGF-β, hepatocyte growth factor, hypoxia, and Wnt/β-catenin signaling pathway, have been shown to induce H19/miR-675 expression, indicating that H19/miR-675 upregulation is a common downstream event of EMT inducers.37,50 Here, I report that TGF-β1 treatment strongly upregulated H19 expression in HeLa cells while CoCl2, a hypoxia-mimetic agent, increased H19 expression in MS751 cells (Figure 6(b) and (c)). Interestingly, downregulation of H19 expression upon TGF-β1 treatment was evident in both SiHa and MS751 cell lines (Figure 6(b)), suggesting that H19 might function primarily as a MET inducer in these cell lines. The fact that H19 silencing induced expression of vimentin, a known marker of EMT, also supports the dual role of H19 in promoting either EMT or MET processes (Figure 5(b)).37 The molecular event that dictates the role of H19 as EMT/MET inducer will require further exploration.
In summary, lncRNA H19 is upregulated in cervical cancer cell lines and can be secreted into EVs. Its expression can be modulated by both TGF-β1 and hypoxia in a cell line-specific manner. H19 functions in cervical cancer cells primarily by promoting cell proliferation and anchorage-independent multicellular spheroid growth, without significantly affecting apoptosis and cell migration. Further studies are required to analyze molecular mechanisms of H19 in cervical tumorigenesis in vivo as well as determining the possibility of using cellular or circulating H19 RNA as a cervical cancer biomarker from clinical samples.
Acknowledgements
I thank Bill Sugden and Janet Mertz (University of Wisconsin-Madison, USA) for supplying plasmids. I am also indebted to the members of the Nano-cosmeceuticals and Nano Delivery System laboratories for cell lines, and members of the Nanomolecular Target Discovery group and Prof. Dr. Tararaj Dharakul for helpful suggestions and discussions. This work was funded by the National Nanotechnology Center (NANOTEC), Thailand.
Author contribution
TI designed, conducted all experiments, analyzed the data, and wrote the manuscript. No other person is entitled to authorship.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease C, Prevention. FDA licensure of bivalent human papillomavirus vaccine (HPV2, Cervarix) for use in females and updated HPV vaccination recommendations from the Advisory Committee on Immunization Practices (ACIP). MMWR Morb Mortal Wkly Rep 2010; 59: 626–9. [PubMed] [Google Scholar]
- 3.McCormack PL, Joura EA. Quadrivalent human papillomavirus (types 6, 11, 16, 18) recombinant vaccine (Gardasil(R)): a review of its use in the prevention of premalignant genital lesions, genital cancer and genital warts in women. Drugs 2010; 70: 2449–74. [DOI] [PubMed] [Google Scholar]
- 4.Jin XW, Lipold L, McKenzie M, Sikon A. Cervical cancer screening: what’s new and what’s coming? Cleve Clin J Med 2013; 80: 153–60. [DOI] [PubMed] [Google Scholar]
- 5.Zielinski GD, Snijders PJ, Rozendaal L, Voorhorst FJ, van der Linden HC, Runsink AP, de Schipper FA, Meijer CJ. HPV presence precedes abnormal cytology in women developing cervical cancer and signals false negative smears. Br J Cancer 2001; 85: 398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin CJ, Lai HC, Wang KH, Hsiung CA, Liu HW, Ding DC, Hsieh CY, Chu TY. Testing for methylated PCDH10 or WT1 is superior to the HPV test in detecting severe neoplasms (CIN3 or greater) in the triage of ASC-US smear results. Am J Obstet Gynecol 2011; 204: 21 e1–7. [DOI] [PubMed] [Google Scholar]
- 7.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012; 22: 1775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeon Y, Sarma K, Lee JT. New and Xisting regulatory mechanisms of X chromosome inactivation. Curr Opin Genet Dev 2012; 22: 62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays 2009; 31: 51–9. [DOI] [PubMed] [Google Scholar]
- 10.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011; 21: 354–61. [DOI] [PubMed] [Google Scholar]
- 11.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010; 143: 46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zong X, Tripathi V, Prasanth KV. RNA splicing control: yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol 2011; 8: 968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorn GW, 2nd, Matkovich SJ. Menage a Trois: intimate relationship among a microRNA, long noncoding RNA, and mRNA. Circ Res 2014; 114: 1362–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol 2011; 22: 366–76. [DOI] [PubMed] [Google Scholar]
- 15.Rando TA, Chang HY. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell 2012; 148: 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serviss JT, Johnsson P, Grander D. An emerging role for long non-coding RNAs in cancer metastasis. Front Genet 2014; 5: 234–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003; 22: 8031–41. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, Wiewrodt R, Muller-Tidow C. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol 2011; 6: 1984–92. [DOI] [PubMed] [Google Scholar]
- 19.Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol 2013; 34: 613–20. [DOI] [PubMed] [Google Scholar]
- 20.Sun M, Xia R, Jin F, Xu T, Liu Z, De W, Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol 2014; 35: 1065–73. [DOI] [PubMed] [Google Scholar]
- 21.Feigenberg T, Gofrit ON, Pizov G, Hochberg A, Benshushan A. Expression of the h19 oncofetal gene in premalignant lesions of cervical cancer: a potential targeting approach for development of nonsurgical treatment of high-risk lesions. ISRN Obstet Gynecol 2013; 2013: 137509–137509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanos V, Prus D, Ayesh S, Weinstein D, Tykocinski ML, De-Groot N, Hochberg A, Ariel I. Expression of the imprinted H19 oncofetal RNA in epithelial ovarian cancer. Eur J Obstet Gynecol Reprod Biol 1999; 85: 7–11. [DOI] [PubMed] [Google Scholar]
- 23.Matouk I, Raveh E, Ohana P, Lail RA, Gershtain E, Gilon M, De Groot N, Czerniak A, Hochberg A. The increasing complexity of the oncofetal h19 gene locus: functional dissection and therapeutic intervention. Int J Mol Sci 2013; 14: 4298–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pachnis V, Belayew A, Tilghman SM. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proc Natl Acad Sci USA 1984; 81: 5523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010; 31: 350–8. [DOI] [PubMed] [Google Scholar]
- 26.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett 2013; 333: 213–21. [DOI] [PubMed] [Google Scholar]
- 27.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One 2007; 2: e845–e845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, Yu J. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J 2014; 281: 3766–75. [DOI] [PubMed] [Google Scholar]
- 29.Steenman MJ, Rainier S, Dobry CJ, Grundy P, Horon IL, Feinberg AP. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumour. Nat Genet 1994; 7: 433–9. [DOI] [PubMed] [Google Scholar]
- 30.Fichorova RN, Rheinwald JG, Anderson DJ. Generation of papillomavirus-immortalized cell lines from normal human ectocervical, endocervical, and vaginal epithelium that maintain expression of tissue-specific differentiation proteins. Biol Reprod 1997; 57: 847–55. [DOI] [PubMed] [Google Scholar]
- 31.Iempridee T, Reusch JA, Riching A, Johannsen EC, Dovat S, Kenney SC, Mertz JE. Epstein-Barr virus utilizes Ikaros in regulating its latent-lytic switch in B cells. J Virol 2014; 88: 4811–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ke N, Wang X, Xu X, Abassi YA. The xCELLigence system for real-time and label-free monitoring of cell viability. Methods Mol Biol 2011; 740: 33–43. [DOI] [PubMed] [Google Scholar]
- 33.Nugitrangson P, Puthong S, Iempridee T, Pimtong W, Pornpakakul S, Chanchao C. In vitro and in vivo characterization of the anticancer activity of Thai stingless bee (Tetragonula laeviceps) cerumen. Exp Biol Med (Maywood) 2016; 241: 166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghazal S, McKinnon B, Zhou J, Mueller M, Men Y, Yang L, Mueller M, Flannery C, Huang Y, Taylor HS. H19 lncRNA alters stromal cell growth via IGF signaling in the endometrium of women with endometriosis. EMBO Mol Med 2015; 7: 996–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Z, Song L, He J, Sun Y, Liu X, Zou X. Ectopic expressed long non-coding RNA H19 contributes to malignant cell behavior of ovarian cancer. Int J Clin Exp Pathol 2015; 8: 10082–91. [PMC free article] [PubMed] [Google Scholar]
- 36.Guo G, Kang Q, Chen Q, Chen Z, Wang J, Tan L, Chen JL. High expression of long non-coding RNA H19 is required for efficient tumorigenesis induced by Bcr-Abl oncogene. FEBS Lett 2014; 588: 1780–6. [DOI] [PubMed] [Google Scholar]
- 37.Matouk IJ, Halle D, Raveh E, Gilon M, Sorin V, Hochberg A. The role of the oncofetal H19 lncRNA in tumor metastasis: orchestrating the EMT-MET decision. Oncotarget 2016; 7: 3748–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matouk IJ, Raveh E, Abu-lail R, Mezan S, Gilon M, Gershtain E, Birman T, Gallula J, Schneider T, Barkali M, Richler C, Fellig Y, Sorin V, Hubert A, Hochberg A, Czerniak A. Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys Acta 2014; 1843: 1414–26. [DOI] [PubMed] [Google Scholar]
- 39.Pardali K, Moustakas A. Actions of TGF-beta as tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta 2007; 1775: 21–62. [DOI] [PubMed] [Google Scholar]
- 40.Wu D, Yotnda P. Induction and testing of hypoxia in cell culture. J Vis Exp 2011; 54: e2899–e2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep 2015; 5: 11516–11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014; 5: 2318–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J 2013; 280: 1709–16. [DOI] [PubMed] [Google Scholar]
- 44.Berteaux N, Lottin S, Monte D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T, Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem 2005; 280: 29625–36. [DOI] [PubMed] [Google Scholar]
- 45.Dufau I, Frongia C, Sicard F, Dedieu L, Cordelier P, Ausseil F, Ducommun B, Valette A. Multicellular tumor spheroid model to evaluate spatio-temporal dynamics effect of chemotherapeutics: application to the gemcitabine/CHK1 inhibitor combination in pancreatic cancer. BMC Cancer 2012; 12: 15–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutherland RM. Cell and environment interactions in tumor microregions: the multicell spheroid model. Science 1988; 240: 177–84. [DOI] [PubMed] [Google Scholar]
- 47.Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis – a proposed unifying theory. Mol Cancer 2015; 14: 184–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan L, Zhou J, Gao Y, Ghazal S, Lu L, Bellone S, Yang Y, Liu N, Zhao X, Santin AD, Taylor H, Huang Y. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene 2015; 34: 3076–84. [DOI] [PubMed] [Google Scholar]
- 49.Shi Y, Wang Y, Luan W, Wang P, Tao T, Zhang J, Qian J, Liu N, You Y. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One 2014; 9: e86295–e86295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, Fellig Y, Degroot N, Galun E, Hochberg A. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta 2010; 1803: 443–51. [DOI] [PubMed] [Google Scholar]