Significance
Our work demonstrates that the growth hormone-releasing hormone receptor (GHRH-R) is highly expressed in human retinoblastoma (RB) cells, but not in other retinal cells. GHRH-R antagonists induced apoptosis in RB cells, but not other retinal cells, suggesting the specificity of their action of this class of analogs. GHRH-R antagonists also increased the expression of apoptotic genes and suppressed expression of cell proliferation genes. These studies indicate that GHRH-R is involved in the survival and proliferation of RB. GHRH-R antagonists could possibly be developed for RB therapy.
Keywords: GHRH pathway, GHRH-R antagonist, retinoblastoma, growth hormone-releasing hormone, apoptosis
Abstract
Retinoblastoma (RB) is the most common intraocular cancer in children worldwide. Current treatments mainly involve combinations of chemotherapies, cryotherapies, and laser-based therapies. Severe or late-stage disease may require enucleation or lead to fatality. Recently, RB has been shown to arise from cone precursor cells, which have high MDM2 levels to suppress p53-mediated apoptosis. This finding leads to the hypothesis that restoring apoptosis mechanisms in RBs could specifically kill the cancer cells without affecting other retinal cells. We have previously reported involvement of an extrapituitary signaling pathway of the growth hormone-releasing hormone (GHRH) in the retina. Here we show that the GHRH receptor (GHRH-R) is highly expressed in RB cells but not in other retinal cells. We induced specific apoptosis with two different GHRH-R antagonists, MIA-602 and MIA-690. Importantly, these GHRH-R antagonists do not trigger apoptosis in other retinal cells such as retinal pigmented epithelial cells. We delineated the gene expression profiles regulated by GHRH-R antagonists and found that cell proliferation genes and apoptotic genes are down- and up-regulated, respectively. Our results reveal the involvement of GHRH-R in survival and proliferation of RB and demonstrate that GHRH-R antagonists can specifically kill the RB cells.
Retinoblastoma (RB) is the most common childhood intraocular malignancy and accounts for approximately 3% of all childhood cancers (1). It has been reported to arise from the cone precursor cells of the retina that transduce light into electrical signals (2). RB is thought to result largely from the loss of function of both copies of RB1 gene located on human chromosome 13q14 (3, 4). Functional disruption of RB1 can be generated by the somatic inactivation of both RB1 alleles, or with a germline RB1 mutation in one allele and a somatic inactivation of the second RB1 allele (5). The RB1 protein acts as a signal transducer connecting cell cycle progression with the transcription machinery (6). There are four steps in the mitotic cycle of a cell: G1, S, G2, and cell division. In the G1 phase, cyclin D is highly expressed, which leads to activation of cyclin-dependent kinases (CDKs) 4 and 6. CDK4 and CDK6 then phosphorylate RB1, inhibiting RB1 binding to the transcription factor E2F (7, 8). As a result, the RB1-free E2F binds to promotors of several genes and turns on their expressions to induce cell cycle progression into S phase, the DNA synthesis phase. Similarly, cells carrying RB1 mutations would also progress into S phase. Normally, this premature progression into S phase would trigger apoptosis to prevent uncontrolled cell proliferation (9). However, it has been reported that the cone precursor cells express high levels of MDM2, a protein that suppresses apoptosis mediated by p53 (2). Therefore, cone precursor cells in patients carrying RB1 mutations pass through the cell cycle faster and without triggering apoptotic cell death. As a result, cone cells proliferate uncontrollably, leading to the development of RB. Based on this understanding of the molecular biology of RB, one effective treatment would be to identify a drug that can induce apoptosis despite the high MDM2 levels in cone precursor cells.
Current treatments of RB mainly involve combinations of chemotherapy, cryotherapy, and laser-based therapy (1). Early diagnosis is crucial. Severe or late-stage disease may require enucleation or lead to fatality. Despite treatment advances, delays in treatment may allow the RB to extend beyond the intraocular level. Also, treatments based on the concept of inducing apoptosis in a specific cell type should provide a high degree of effectiveness in treatment outcome. Consequently, we decided to investigate alternative treatments. Growth hormone (GH)-releasing hormone (GHRH) is a hypothalamic hormone, which binds to the GHRH receptor (GHRH-R) and triggers the synthesis and secretion of GH from the pituitary (10). Outside the pituitary, the GHRH–GH pathway also functions in normal and neoplastic peripheral tissues, and is mediated by, among others, insulin-like growth factor-1 (11). We have previously shown that GHRH-R antagonists play protective roles in the rat eye, suggesting that GHRH-R antagonists are potential therapeutic agents for ocular inflammation (12). Notably, we also found detectable levels of GHRH, GHRH-R, and GH expressions in the retina, indicating a role of GHRH-R antagonists in modulating functions in the retina at normal and pathological states (12). Notably, GHRH-R antagonists have been shown to trigger apoptosis and reduce the invasive and metastatic potential in late stage tumors, including glioblastoma, prostate, breast, and ovarian cancer (13, 14). We therefore hypothesized that GHRH-R antagonists can induce cell death specifically in RB cells.
Results
Specific Expression of GHRH-R in Y79 Cells.
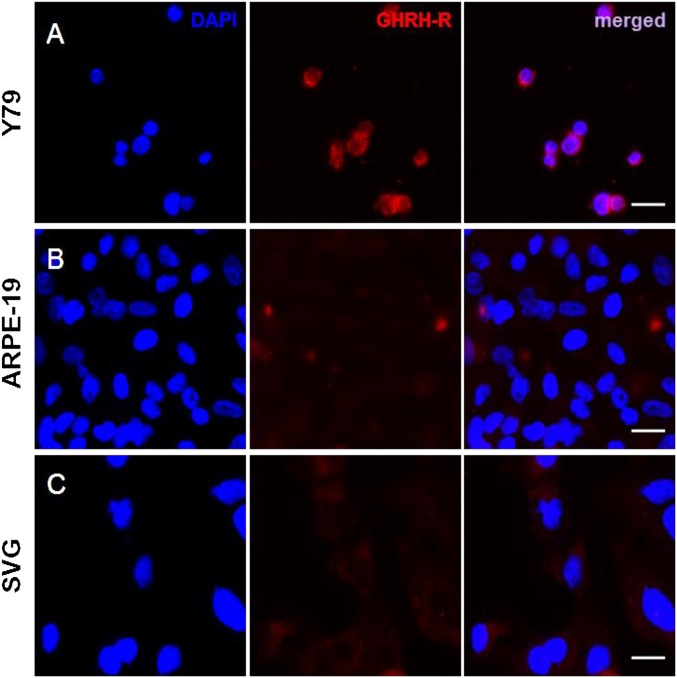
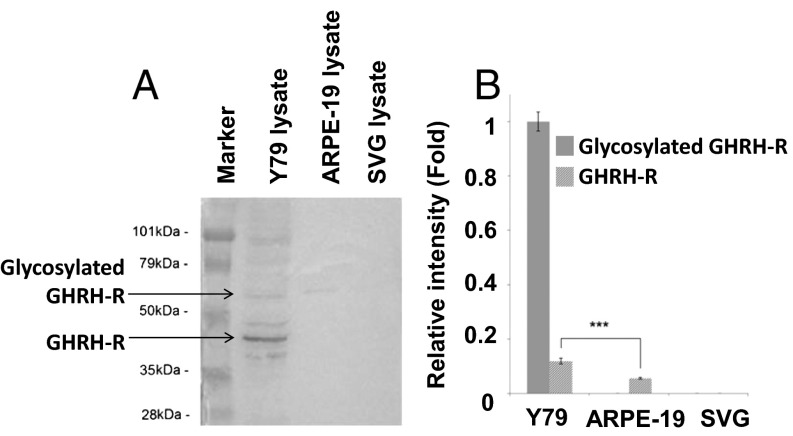
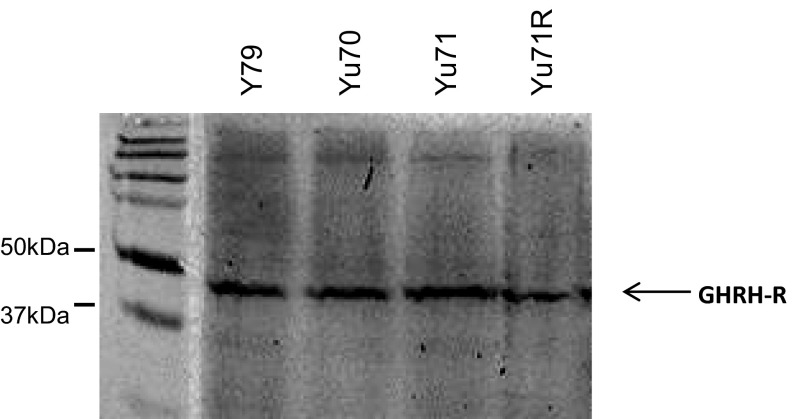
We used immunocytochemistry to investigate GHRH-R expression and cellular localization in RB cells of Y79, ARPE-19, or SVG. We found copious expression of GHRH-R in Y79 (Fig. 1A). The fluorescence signal indicated that GHRH-R was expressed in the nuclear and cytoplasmic regions of the Y79 cells, with a slightly stronger signal in the cytoplasm. The two noncancerous cell lines, ARPE-19 (from retinal pigment epithelium) and SVG (an astroglia cell line), however, exhibited barely detectable levels of GHRH-R expression, compared with Y79 (Fig. 1 B and C). Western blotting to quantify GHRH-R levels in these cell lines confirmed quantitatively that GHRH-R was selectively expressed in Y79, which showed a strong band with molecular weight of approximately 45 kDa, consistent with the predicted size of GHRH-R (Fig. 2A). GHRH-R protein in the lysates of ARPE-19 and SVG was undetectable, with absence of the appropriate bands on Western blot. A minor band, at approximately 70 kDa, was detected in Y79 and ARPE-19 lysates, indicating the presence of glycosylated GHRH-R in these two cell lines but not in the SVG cells (Fig. 2A). Quantification by densitometry showed that expression of unglycosylated GHRH-R was 10 times higher than that of the glycosylated form in Y79 cells. The glycosylated GHRH-R was also present in the ARPE-19 cells but at a significantly (P < 0.001) lower level, at approximately 50% of that in Y79 (Fig. 2B). We also found comparable GHRH-R expressions in the Y79 cells with three other primary cells isolated from human RB tissues (Yu70, Yu71, and Yu71R; Fig. S1).
Fig. 1.
(A) GHRH-R is expressed copiously in Y79 RB cells as detected by immunofluorescence. Fluorescence signals were barely detected in the two noncancerous cell lines, (B) ARPE-19 and (C) SVG cells. GHRH-R was stained with specific antibody (red). Nuclei were stained with DAPI (blue). (Scale bar: 20 µm.)
Fig. 2.
Cellular proteins from Y79, ARPE-19, and SVG cell lines were extracted and resolved on 10% SDS gel. GHRH-R was detected with anti–GHRH-R antibody. (A) A representative image of Western blot membrane probed with primary anti–GHRH-R antibody (1:1,000; Abcam) and then secondary anti–rabbit-HRP antibody. (B) Relative expression levels of unglycosylated GHRH-R (predicted size, 47 kDa) and glycosylated GHRH-R (predicted size, 70 kDa) were determined by densitometry. Gel images were analyzed with ImageJ software. The means of three independent experiments are shown. P values were evaluated statistically by using an unpaired t test. Error bars represent SDs. Asterisks indicate statistical significance (P < 0.001).
Fig. S1.
Cellular proteins from Y79, Yu70, Yu71, and Yu71R were extracted and resolved on 10% SDS gel. GHRH-R was detected with anti–GHRH-R antibody.
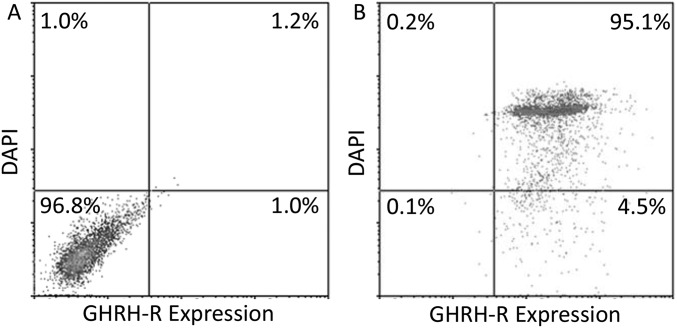
On flow cytometry, the density plot indicated a detectable and drastic shift of cells stained with GHRH-R antibody in Y79 cells, compared with the negative control stained without primary antibody or DAPI (Fig. S2A). Moreover, approximately 95.1% of DAPI-positive cells were GHRH-R–positive, showing that nearly all of the Y79 cells expressed detectable levels of GHRH-R (Fig. S2B).
Fig. S2.
Y79 cells were (A) stained without primary antibody or DAPI (negative control), or (B) stained with anti–GHRH-R antibody (1:1,000; Abcam) and DAPI. Both samples were then stained with anti-rabbit Alexa 488 antibody. Cells were subjected to flow cytometry analysis. Representative density plots from three independent flow cytometric runs are shown.
Apoptosis Inducing Effects of GHRH-R Antagonists in Y79 Cells.
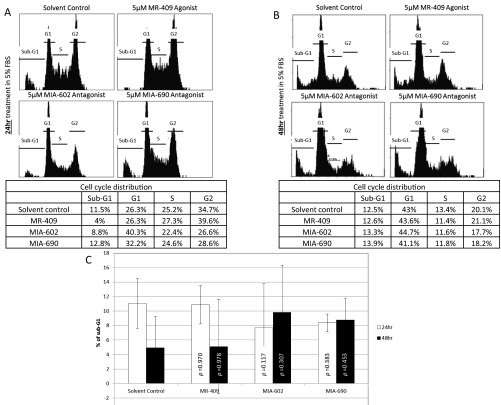
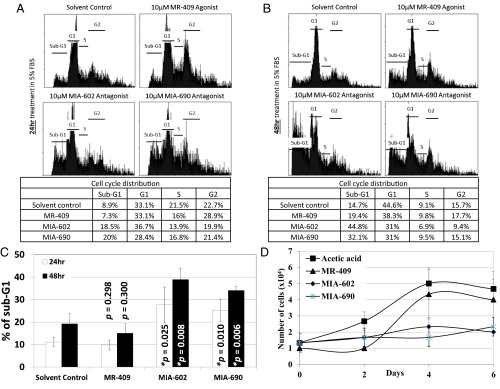
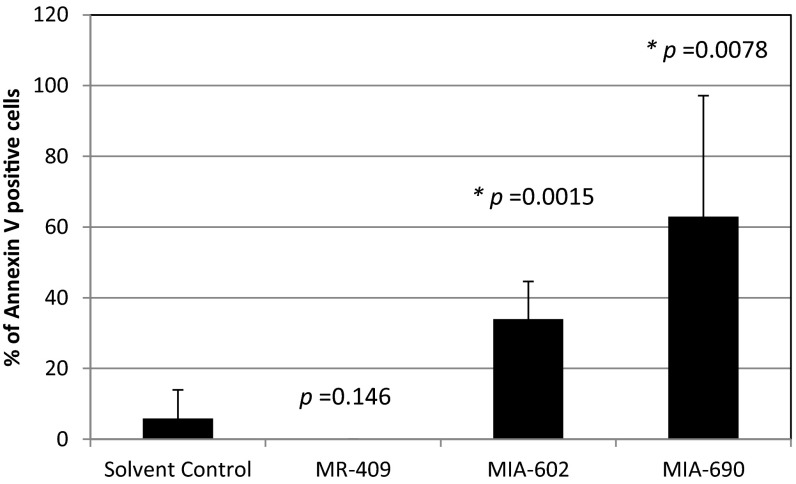
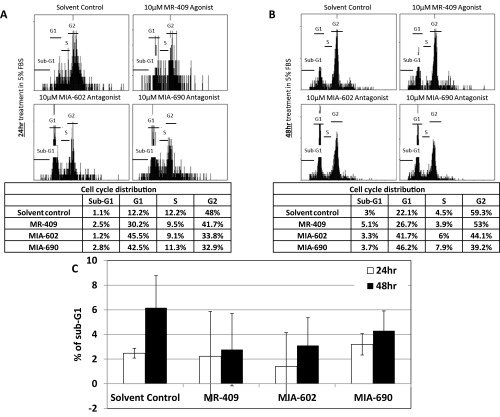
As GHRH-R is highly expressed in Y79 cells, we hypothesized that interfering with the GHRH signaling pathway would affect the cell cycle of the RB cells. We therefore treated these cells with 5 µM of the GHRH-R agonist MR-409, which reportedly enhanced the GHRH signaling pathway (13), for 24 or 48 h. These cells were also treated separately with similar concentration of GHRH-R antagonists MIA-602 or MIA-690, which suppressed the GHRH signaling pathway (14). There was no significant difference in the cell cycle stages at G1, S, and G2 between the solvent control and MR-409–, MIA-602–, or MIA-690–treated cells (Fig. S3). In addition, there was a slight but insignificant variation in the sub-G1 population between these treatments. We then increased the concentration of MR-409, MIA-602, and MIA-690 to 10 µM and treated cells for 24 or 48 h. Under this treatment condition, MR-409 had no significant effect on the cell cycle distribution. Notably, both GHRH-R antagonists, MIA-602 and MIA-690, significantly enhanced the sub-G1 population, which is a hallmark of apoptosis (Fig. 3 A–C). After the 24-h treatment, both GHRH-R antagonists, MIA-602 and MIA-690, increased the sub-G1 population by approximately twofold. After 48-h treatment, the sub-G1 population was further increased by more than threefold. Apart from using the sub-G1 method, we also quantified the apoptotic cells by the Annexin V assay to detect the translocation of phosphatidylserine from the inner to the outer leaflet of the plasma membrane of apoptotic cells. After the 48 h treatment, both GHRH-R antagonists, MIA-602 and MIA-690, significantly enhanced the Annexin V-positive population by more than sixfold (Fig. S4).
Fig. S3.
Y79 cells were treated with 5 µM of the GHRH-R agonist MR-409 or GHRH-R antagonists MIA-602 or MIA-690 for (A) 24 h or (B) 48 h. (Top) Representative cell cycle distribution profiles of these treatments. (Bottom) Tables showing quantifications of the cell population in the sub-G1, G1, S, and G2 phases of the corresponding representative cell cycle distribution profiles. (C) Quantifications of the sub-G1 population of Y79 cells treated with 5 µM MR-409, MIA-602, or MIA-690 for 24 or 48 h. The means for three independent experiments are shown. P values were evaluated statistically by using an unpaired t test. Error bars represent SD.
Fig. 3.
Y79 cells were treated with 10 µM of the GHRH-R agonist MR-409 or the GHRH-R antagonists MIA-602 or MIA-690 for (A) 24 h or (B) 48 h. (Top) Representative distribution profiles in cell cycles of these treatments. (Bottom) Tables show quantifications of cell population percentages in the sub-G1, G1, S, and G2 phases of the corresponding representative cell cycle distribution profiles. (C) Quantifications of the sub-G1 population of Y79 cells treated with 10 µM MR-409, MIA-602, or MIA-690 for 24 or 48 h. The means for three independent experiments are shown. P values were evaluated by using an unpaired t test. Asterisks indicate statistical significance (P < 0.05), and error bars indicate SD. (D) Cells were treated with 10 µM MR-409, MIA-602, or MIA-690 for 48 h, and plated into fresh culture medium to study the cell proliferation at 0, 2, 4, or 6 d after the treatment. Points represent the mean of three determinations, and error bars represent SD.
Fig. S4.
Quantifications of the Annexin V-positive cells of Y79 treated with 10 µM MR-409, MIA-602, or MIA-690 for 48 h. At least 20 cells were quantified in each group. P values were evaluated statistically by using an unpaired t test. Error bars represent SDs.
Subsequently, we treated the primary cells Yu71R, which were isolated from a human RB tissue, with 10 µM MR-409, MIA-602, or MIA-690 for 48 h. Similar to Y79 cells, both GHRH-R antagonists, MIA-602 and MIA-690, increased the sub-G1 population by approximately twofold after 48-h treatment (Fig. S5). To evaluate the impact of these GHRH-R antagonists on cell proliferation, we treated Y79 cells with 10 µM MR-409, MIA-602, or MIA-690 for 48 h. The cells were then plated into fresh culture medium to study their proliferation. Cells treated with the solvent control or the GHRH-R agonist MR-409 resumed cell proliferation with a doubling time of approximately 2 d (Fig. 3D). In contrast, the cells treated with the GHRH-R antagonists MIA-602 or MIA-690 exhibited limited cell proliferation and the cell number remained the same throughout the 6-d incubation period (Fig. 3D).
Fig. S5.
Yu71R cells were treated with 10 µM of the GHRH-R agonist MR-409 or GHRH-R antagonists MIA-602 or MIA-690 for 48 h. Representative cell cycle distribution profiles of these treatments are shown on the left. (Left) Table shows quantifications of cell population percentages in the sub-G1, G1, S, and G2 phases of the corresponding representative cell cycle distribution profiles (Right).
GHRH-R Antagonists Did Not Induce Apoptosis in RPE Cell Line.
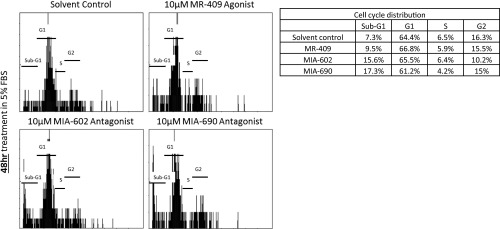
To test whether the apoptosis-inducing properties of GHRH-R antagonists are specific to RB but not to other cell types in the eye, we treated ARPE-19 cells with 10 µM MR-409, MIA-602, or MIA-690 for 24 or 48 h. In contrast to the results obtained in Y79 cells, the sub-G1 populations of ARPE-19 cells were not significantly altered by the treatment with MR-409, MIA-602, or MIA-690 for the 24- or 48-h test periods (Fig. 4). Instead, the G1 population in ARPE-19 was enriched by the GHRH-R antagonists, MIA-602 or MIA-690, for 24 or 48 h (Fig. 4). These data showed that the GHRH-R antagonists induced apoptotic cell death specifically in RB cells, but not in the noncancerous retinal cells.
Fig. 4.
ARPE-19 cells were treated with 10 µM of the GHRH-R agonist MR-409 or GHRH-R antagonists MIA-602 or MIA-690 for (A) 24 h or (B) 48 h. (Top) Representative cell cycle distribution profiles of these treatments. (Bottom) Tables show quantifications of cell population percentages in the sub-G1, G1, S, and G2 phases of the corresponding representative cell cycle distribution profiles. (C) Quantifications of the sub-G1 population of ARPE-19 cells treated with 10 µM MR-409, MIA-602, or MIA-690 for 24 or 48 h. The means for three independent experiments are shown. P values were evaluated statistically by unpaired t test. Error bars represent SD.
Effects of GHRH-R Antagonists on Cell Proliferation and Apoptosis Related Genes.
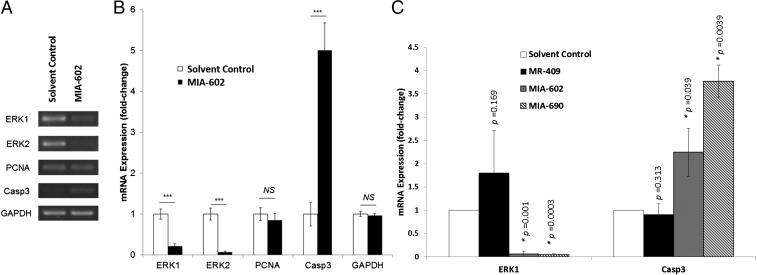
We analyzed the changes in mRNA expression after treating Y79 cells with 10 µM MIA-602 for 6 h (Fig. 5 A and B). Genes related to cell cycle progression and proliferation (extracellular signal-regulated kinases ERK1 and ERK2), apoptotic cell death [cysteine-aspartic protease 3 (caspase 3)], and cell proliferation [proliferating cell nuclear antigen (PCNA)] were studied. We found significant down-regulation of the expressions of proliferation mediators by MIA-602. ERK1 was significantly suppressed by 4.76-fold (P < 0.001), and ERK2 showed an even more drastic suppression (15.13-fold; P < 0.001). The proliferative marker PCNA demonstrated a slight reduction of approximately 15% in mRNA expression, although the change was not statistically significant (P > 0.05). On the contrary, the expression of the apoptotic marker, Caspase 3, was significantly (P < 0.001) up-regulated by MIA-602, with an increase of 5.01-fold.
Fig. 5.
Y79 cells were treated with 10 µM of the GHRH-R antagonist MIA-602 for 6 h. Total mRNA was extracted and reverse-transcribed. Gene expressions were determined by RT-PCR. (A) A representative image of RT-PCR of ERK1, ERK2, PCNA, and Casp3. GAPDH was the housekeeping gene. (B) Gel images were analyzed by ImageJ software. Relative mRNA expression levels were determined by densitometry. The means for three independent experiments are shown. P values were evaluated statistically by unpaired t test. Error bars represent SD. Asterisks indicate statistical significance (P < 0.001). (C) Y79 cells were treated with 10 µM of the GHRH-R agonist MR-409 or the GHRH-R antagonists MIA-602 or MIA-690 for 48 h. Relative mRNA expression levels of ERK1 and Casp3 were quantified by real-time PCR. The means for two independent experiments are shown. P values were evaluated statistically by unpaired t test. The asterisks indicate statistical significance (P < 0.05). Error bars represent SD.
To understand the prolonged effects of GHRH-R antagonists in ERK1 and Caspase 3 expression, Y79 cells were treated with 10 µM MR-409, MIA-602, or MIA-690 for 48 h (Fig. 5C). ERK1 was significantly suppressed by 17.8-fold or 25.0-fold by treatment with MIA-602 or MIA-690, respectively. Caspase 3 was significantly up-regulated by 2.2-fold or 3.8-fold by MIA-602 or MIA-690, respectively. These results suggest that GHRH-R antagonists can down-regulate cell proliferation pathways and enhance the cell death pathway, confirming our findings in the cell proliferation and apoptosis assays.
Discussion
Our understanding of the pathogenesis of RB has been greatly improved in the past few years. RB has been shown to arise from the cone precursor cells, which have high MDM2 levels that suppress p53-mediated apoptosis (2). In these cone precursor cells, mutation in the RB1 gene would lead to E2F activation (7, 8). Activating the E2F pathway causes a depletion of cellular nucleotide levels, which in turn generates DNA replication stress and DNA damage (15). Fortunately, excessive DNA replication stress and DNA damage are known to trigger apoptosis and restriction of cellular growth (16, 17). However, in the cone precursor cells, p53-mediated apoptosis is suppressed by the highly expressed MDM2 (2). Therefore, these DNA replication stress and DNA damages are accumulated and undermine genomic stability, which leads to tumorigenesis.
Although p53 is considered as a key regulator of apoptosis, it has been reported that apoptosis can also be induced in p53-negative cells (18). Despite the fact that cone precursor cells have high level of MDM2, these findings suggest that apoptosis may still possibly be induced in RB. Our previous work indeed demonstrated that GHRH-R antagonists are able to inhibit cell proliferation in the small-cell lung carcinoma expressing mutated p53 (19). Therefore, we chose to study the effects of GHRH-R antagonists on RB cells.
Our findings indicate that GHRH-R is differentially expressed in the RB cell line Y79, which was the first human RB cell line established more than 40 y ago (20). It has been widely used for the study of RB, including by direct injection into nude mice (21). In this reported study, 67.5% of the Y79-injected eyes developed RB after 12 wk, suggesting that Y79 is a good cellular model of RB (21). Y79 cells express GHRH-R throughout the population, with subcellular localizations in the nucleus and cytoplasm. GHRH-R was originally identified as a membrane receptor protein. However, GHRH-R has been reported to internalize upon GHRH binding (22). Our immunocytochemistry data suggest presence of some active GHRH signaling pathways in RB cells. Immunoblotting detected the existence of multiple variants of GHRH-R, which is consistent with previous reports in which the glycosylated form of GHRH-R was identified at approximately 70 kDa, in addition to unglycosylated GHRH-R (23). In contrast, APRE-19 cells expressed only a low level of glycosylated GHRH-R and SVG cells showed no detectable GHRH-R. GHRH-R has been reported to be N-linked glycosylated on amino acid asparagine at position 50 (24). This glycosylated GHRH-R was not detected in Western blot after treatment with endoglycosidase H (24). The functional consequence of this GHRH-R glycosylation is currently unknown. However, this glycosylation site is in close proximity to a key amino acid, aspartate at position 60, which is essential for the downstream GHRH signaling pathway (24). In conclusion, these results on GHRH-R expression suggest that the GHRH-R antagonists could induce apoptosis specifically in RB but not in other (i.e., normal) retinal cells. GHRH-R expression in other human ocular tissues has not been reported to our knowledge. In our previous study in rats, GHRH-R could be detected in cornea, iris, and ciliary body (12). In addition to the GHRH-R expression, whether Y79 cells express its ligand GHRH is currently unknown. Notably, Y79 cells have been reported to secret a growth factor to support the cell growth in a serum-free environment for more than 5 mo (25). The identity of this growth factor was not determined, but it has a molecular weight of 38 kDa, which is similar to the size of GHRH.
Treatments of the RB cells with two different GHRH-R antagonists at 10 µM for 48 h induced three times higher apoptosis values compared with control. Notably, the same treatment protocol did not induce significant apoptosis in RPE cells. These results indicate that an extrapituitary signaling pathway, in which the GHRH pathway plays an important role, regulates cell death within an ocular environment. Furthermore, our results show a RB-specific killing effect of the GHRH-R antagonists, which can be partially explained by the interference with relevant biological signaling pathways. The RAF/MEK/ERK pathway has been known for its vital role in mediating diverse cellular signaling pathways including cell survival, proliferation, metabolism, migration, and cell cycle progression (26). In particular, ERK1 and ERK2 expressions were reduced in response to the treatment by GHRH-R antagonist, suggesting a corresponding reduction in biological signaling involving proliferation and survival. Caspase 3 expression has been established as a hallmark of apoptosis (27). Thus, the generation of a high proportion of apoptotic cells in the population, as was observed in our cell cycle analysis, is well explained by the significant up-regulation of Caspase 3 expression. Many pathways are involved in regulating Caspase 3 expression. RB has been reported to show a high MDM2 expression level. GHRH-R antagonists might suppress MDM2 to restore p53 activity, which subsequently induces Caspase 3 expression (2). However, several global gene expression studies did not identify Caspase 3 as a direct transcriptional target of p53 (28, 29), although the Caspase 3 protein was up-regulated upon p53 activation (30). These studies suggest that p53 induces Caspase 3 expression indirectly. On the contrary, an inflammatory cytokine, IFN-γ, has been reported to induce Caspase 3 expression (31). It is noted that we have previously shown alleviation of ocular inflammation by GHRH-R antagonist in the rat (12). The possibility of GHRH-R antagonists enhancing Caspase 3 expression through the modulation of ocular inflammation remains to be determined.
In conclusion, the GHRH-R antagonists show specific apoptosis-inducing effects on RB cells with minimal effects on the surrounding noncancerous retinal cells. Our study reveals previously unidentified information on RBs and suggests a promising approach consisting of therapy targeted to GHRH receptors to accomplish a significant decrease in tumor cells through the mechanism of apoptosis.
Materials and Methods
Cell Lines and Culture.
Y79 [American Type Culture Collection (ATCC)] was maintained in RPMI 1640 medium with 20% (vol/vol) FBS. Primary cells Yu70, Yu71, and Yu71R were maintained in RPMI 1640 medium with 20% (vol/vol) FBS. ARPE-19 (ATCC) was grown in DMEM/F12 with 10% (vol/vol) FBS. SVG (ATCC) was grown in MEM with 10% (vol/vol) FBS (SI Materials and Methods).
Quantification of GHRH-R Expression by Immunocytochemistry.
Cells were stained with primary antibody against GHRH-R (1:1,000 dilution; Abcam) and then were incubated with DAPI stain and secondary antibody conjugated to Rhodamine Red-X (1:1,000 dilution; Jackson ImmunoResearch). Fluorescence signals were visualized by using a fluorescence microscope (SI Materials and Methods).
Quantification of GHRH-R Expression by Western Blot.
A 30-µg portion of cellular protein was resolved in 10% (wt/vol) SDS polyacrylamide gel. Expression of GHRH-R was determined by using primary antibody against GHRH-R (1:1,000 dilution; Abcam). Expression levels were analyzed by ImageJ software (SI Materials and Methods).
Quantification of the Y79 Subpopulation Expressing GHRH-R by Flow Cytometry.
Y79 cells were stained with or without primary antibodies against GHRH-R (1:1,000 dilution; Abcam) and then were incubated with secondary antibody conjugated to Alexa 488 (1:1,000 dilution; Jackson ImmunoResearch). Cells were counterstained with DAPI and analyzed with a BD FACS Fortessa Flow Cytometer (BD Bioscience; SI Materials and Methods).
GHRH-R Agonist and GHRH-R Antagonists.
The GHRH-R agonist MR-409 and GHRH-R antagonists MIA-602 and MIA-690 were synthesized and purified as described previously (13, 14) in the laboratory of A.V.S. The lyophilized synthetic neuropeptides were dissolved in 50% (vol/vol) acetic acid and then diluted 1,000-fold in corresponding culture medium before use.
Cell Cycle Distribution Analysis.
After treatment, cells were stained with propidium iodide and analyzed with an FC500 flow cytometer (Beckman Coulter). Sub-G1 data were measured for the evaluation of the apoptotic population (SI Materials and Methods).
Annexin V Assay.
After treatment, cells were stained freshly with Annexin V Apoptosis Detection Kit (Invitrogen) according to the manufacturer’s instructions. Cells were analyzed by a fluorescence microscope (Nikon; SI Materials and Methods).
Cell Proliferation Assay.
After the treatment, equal numbers of cells were seeded into 24-well plates in RPMI 1640 medium supplemented with 20% (vol/vol) FBS. After 0, 2, 4, and 6 d of incubation, the numbers of cells were counted by using a hemocytometer (SI Materials and Methods).
Real-Time PCR.
Relative expressions of mRNA were detected and semiquantitatively determined by using specific primers amplifying ERK1, ERK2, PCNA, Caspase 3, and GAPDH. PCR products were resolved in 1.2% (wt/vol) agarose gel electrophoresis, visualized by UV, and analyzed with ImageJ software. Relative expressions of ERK1 and Caspase 3 were further quantified by LightCycler 480 II real-time PCR (Roche Applied Science; SI Materials and Methods).
SI Materials and Methods
Cell Lines and Culture.
Y79 (ATCC), a human RB cell line, was maintained as a nonadherent culture in RPMI 1640 medium with 20% (vol/vol) FBS. Primary Yu70, Yu71, and Yu71R cells were isolated from human RB tissues treated with 0.075% type IV collagenase. These primary cells were maintained in RPMI 1640 medium with 20% (vol/vol) FBS. ARPE-19 (ATCC), a retinal pigmented epithelial cell line, was grown in DMEM/F12 with 10% (vol/vol) FBS. SVG (ATCC), an astroglia cell line, was grown in MEM with 10% (vol/vol) FBS. The cultures were supplemented with 100 U/mL penicillin and 100 µg/mL streptomycin, and were incubated at 37 °C in a humidifying incubator with 5% (vol/vol) CO2 and 95% (vol/vol) O2.
Quantification of the GHRH-R Expression by Immunocytochemistry.
The cells were seeded onto glass slides, followed by fixation with 2% (wt/vol) paraformaldehyde for 2 h. For the Y79 cells, 5 µg/cm2 of poly-d-lysine was used for coating of the culture plate to enhance their adhesion onto the planar surface. Cells were blocked in assay diluent containing 2% (wt/vol) BSA, 1% dimethyl sulfoxide, and 0.5% Triton X-100 in PBS solution. Cells on glass slides were then incubated with primary antibody against GHRH-R (1:1,000 dilution; Abcam) for 1 h at room temperature. After repeated washes with PBS solution, cells were incubated with DAPI stain and anti-rabbit secondary antibody conjugated to Rhodamine Red-X (1:1,000 dilution; Jackson ImmunoResearch) for 30 min at room temperature in darkness. Fluorescence signals were visualized by using a fluorescence microscope.
Quantification of the Expression of GHRH-R by Western Blot.
Briefly, the Y79, primary human RB cells (Yu70, Yu71, and Yu71R), ARPE-19, and SVG cells were washed in ice-cold PBS solution and harvested in ice-cold fresh RIPA buffer containing 150 mM NaCl, 50 mM Tris·HCl, 0.1% SDS, 0.1% Triton X-100, and 0.5% sodium deoxycholate (pH 8.0). Cell lysate was incubated at 4 °C for 1 h and was then centrifuged at 10,000 × g to obtain protein extract from the supernatant fraction. A total of 30 µg of cellular protein was resolved in 10% (wt/vol) SDS polyacrylamide gel for 2.5 h at 80 V. Expression of GHRH-R was determined by using primary antibody against GHRH-R (1:1,000 dilution; Abcam). Expression levels were visualized under enhanced chemiluminescence, analyzed by the ImageJ software. Relative expression levels of GHRH-R were determined by densitometry.
Quantification of the Y79 Subpopulation Expressing GHRH-R by Flow Cytometry.
Briefly, the Y79 cells were harvested, washed, and stained with or without primary antibodies against GHRH-R (1:1,000 dilution; Abcam) for 1 h at room temperature. After repeated PBS solution washes, cells were incubated with anti-rabbit secondary antibody conjugated to Alexa 488 (1:1,000 dilution; Jackson Immunoresearch) for 30 min at room temperature in darkness. Cells were counterstained with DAPI and analyzed by using an FACS Fortessa Flow Cytometer (BD Bioscience). The percentages of cells with peak shift in fluorescence signal were analyzed to determine the subpopulation of GHRH-R–expressing cells.
Cell Cycle Distribution Analysis.
Y79 cells, Yu71R cells, and ARPE-19 cells were treated with GHRH analogs MR-409, MIA-602, and MIA-690 in the corresponding medium supplemented with 5% (vol/vol) FBS. In each treatment, the solvent control consisted of 0.05% acetic acid. At the end point, cells were fixed and permeabilized with ice-cold ethanol for at least 24 h, followed by staining with propidium iodide at 37 °C. Cells were analyzed by the Flow Cytometer FC500 (Beckman Coulter). The percentage of cells at G1, S, and G2 phases was measured to determine cell cycle changes. Sub-G1 data were measured for the evaluation of the apoptotic population.
Annexin V Assay.
Y79 cells were treated with GHRH analogs MR-409, MIA-602, and MIA-690 in RPMI 1640 medium supplemented with 5% (vol/vol) FBS. In each treatment, the solvent control consisted of 0.05% acetic acid. At the end point, cells were stained freshly with Annexin V Apoptosis Detection Kit (Invitrogen) according to the manufacturer’s instructions. Cells were analyzed by a fluorescence microscope (Nikon).
Cell Proliferation Assay.
Y79 cells were treated with 10 µM MR-409, MIA-602, or MIA-690, respectively, in RPMI 1640 medium supplemented with 5% (vol/vol) FBS for 48 h. Cells were then collected and washed with PBS solution. Equal numbers of cells were seeded into 24-well plates in RPMI 1640 medium supplemented with 20% (vol/vol) FBS. After 0, 2, 4, and 6 d of incubation, the numbers of cells were counted by using a hemocytometer.
Real-Time PCR.
Y79 cells were treated with MR-409, MIA-602, or MIA-690, all at 10 µM concentration, in RPMI 1640 medium supplemented with 5% (vol/vol) FBS for 6 h. Cells were then harvested and lysed completely in TRIzol reagent. RNA was extracted with an RNA Extraction Kit (Qiagen), followed by cDNA conversion by a standard reverse-transcription protocol. Relative expressions of mRNA were detected and semiquantitatively determined by using the following primers: ERK1, forward, 5′-AACCACATTCTGGGCATCCTG-3′, reverse, 5′-AGCCGCTCCTTAGG TAGGTCA-3′; ERK2, forward, 5′- TTCCAAGGGCTACACCAAGTC-3′, reverse, 5′-TGTCT GAGCACGTCCAGTCC-3′; PCNA, forward, 5′-ACGTCTCTTTGGTGCAGCTCA-3′, reverse, 5′-CATTGCCGGCGCATTTTA-3′; Caspase 3, forward, 5′-TATTCTTGGGGAAAT TCAAAGGAT-3′, reverse, 5′-AAAGTAGCGTCAAAGGAAAAGGAC-3′; and GAPDH, forward, 5′-ACCACAGTCCATGCCATCAC-3′, reverse, 5′-TCCACCACCCTGTTGCTGTA-3′. PCR products were resolved in 1.2% (wt/vol) agarose gel electrophoresis, visualized by UV, and quantified by densitometry by using ImageJ software. Relative expressions of ERK1 and Caspase 3 in Y79 cells treated with MR-409, MIA-602, or MIA-690, all at 10 µM concentration for 48 h, were further quantified by LightCycler 480 II real-time PCR (Roche Applied Science).
Acknowledgments
We thank the Li Ka Shing Foundation for generous support, and Ms. Pancy Tam and Dr. Benjamin Li for excellent technical support. This work was supported by the Li Ka Shing Foundation and a block grant from the University Grants Committee Hong Kong (to C.P.P.). The work in Miami was supported by the Medical Research Service of Veterans Affairs Department, South Florida Veterans Affairs Foundation for Research and Education, and the University of Miami Miller School of Medicine (A.V.S. and N.L.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617427113/-/DCSupplemental.
References
- 1.Rodriguez-Galindo C, et al. Treatment of metastatic retinoblastoma. Ophthalmology. 2003;110(6):1237–1240. doi: 10.1016/S0161-6420(03)00258-6. [DOI] [PubMed] [Google Scholar]
- 2.Xu XL, et al. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell. 2009;137(6):1018–1031. doi: 10.1016/j.cell.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friend SH, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323(6089):643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 4.Fan BJ, et al. Molecular diagnostics of genetic eye diseases. Clin Biochem. 2006;39(3):231–239. doi: 10.1016/j.clinbiochem.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Choy KW, et al. Loss of heterozygosity and mutations are the major mechanisms of RB1 gene inactivation in Chinese with sporadic retinoblastoma. Hum Mutat. 2002;20(5):408. doi: 10.1002/humu.9077. [DOI] [PubMed] [Google Scholar]
- 6.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81(3):323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 7.Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7(3):331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 8.Ewen ME, et al. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73(3):487–497. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- 9.Qin XQ, Livingston DM, Kaelin WG, Jr, Adams PD. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91(23):10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frohman LA, Kineman RD. Growth hormone-releasing hormone and pituitary somatotrope proliferation. Minerva Endocrinol. 2002;27(4):277–285. [PubMed] [Google Scholar]
- 11.Kiaris H, Schally AV, Kalofoutis A. Extrapituitary effects of the growth hormone-releasing hormone. Vitam Horm. 2005;70:1–24. doi: 10.1016/S0083-6729(05)70001-7. [DOI] [PubMed] [Google Scholar]
- 12.Qin YJ, et al. Antagonist of GH-releasing hormone receptors alleviates experimental ocular inflammation. Proc Natl Acad Sci USA. 2014;111(51):18303–18308. doi: 10.1073/pnas.1421815112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai R, et al. Synthesis of new potent agonistic analogs of growth hormone-releasing hormone (GHRH) and evaluation of their endocrine and cardiac activities. Peptides. 2014;52:104–112. doi: 10.1016/j.peptides.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szalontay L, et al. Novel GHRH antagonists suppress the growth of human malignant melanoma by restoring nuclear p27 function. Cell Cycle. 2014;13(17):2790–2797. doi: 10.4161/15384101.2015.945879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bester AC, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145(3):435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 17.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 18.Strasser A, Harris AW, Jacks T, Cory S. DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell. 1994;79(2):329–339. doi: 10.1016/0092-8674(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 19.Kanashiro CA, et al. Inhibition of mutant p53 expression and growth of DMS-153 small cell lung carcinoma by antagonists of growth hormone-releasing hormone and bombesin. Proc Natl Acad Sci USA. 2003;100(26):15836–15841. doi: 10.1073/pnas.2536558100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid TW, et al. Characteristics of an established cell line of retinoblastoma. J Natl Cancer Inst. 1974;53(2):347–360. doi: 10.1093/jnci/53.2.347. [DOI] [PubMed] [Google Scholar]
- 21.Tschulakow AV, Schraermeyer U, Rodemann HP, Julien-Schraermeyer S. Establishment of a novel retinoblastoma (Rb) nude mouse model by intravitreal injection of human Rb Y79 cells - Comparison of in vivo analysis versus histological follow up. Biol Open. 2016 doi: 10.1242/bio.019976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veyrat-Durebex C, Pomerleau L, Langlois D, Gaudreau P. Internalization and trafficking of the human and rat growth hormone-releasing hormone receptor. J Cell Physiol. 2005;203(2):335–344. doi: 10.1002/jcp.20233. [DOI] [PubMed] [Google Scholar]
- 23.Lee EJ, Duan WR, Kotlar T, Jameson JL. Restoration of growth hormone-releasing hormone (GHRH) responsiveness in pituitary GH3 cells by adenovirus-directed expression of the human GHRH receptor. Endocrinology. 2001;142(1):414–420. doi: 10.1210/endo.142.1.7896. [DOI] [PubMed] [Google Scholar]
- 24.Gaylinn BD, et al. The mutant growth hormone-releasing hormone (GHRH) receptor of the little mouse does not bind GHRH. Endocrinology. 1999;140(11):5066–5074. doi: 10.1210/endo.140.11.7092. [DOI] [PubMed] [Google Scholar]
- 25.Rubin NA, Tarsio JF, Borthwick AC, Gregerson DS, Reid TW. Identification and characterization of a growth factor secreted by an established cell line of human retinoblastoma maintained in serum-free medium. Vision Res. 1981;21(1):105–112. doi: 10.1016/0042-6989(81)90142-5. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Ilasaca M. Signaling from G-protein-coupled receptors to mitogen-activated protein (MAP)-kinase cascades. Biochem Pharmacol. 1998;56(3):269–277. doi: 10.1016/s0006-2952(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 27.Porter AG, Jänicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6(2):99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- 28.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124(1):207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 29.Allen MA, et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife. 2014;3:e02200. doi: 10.7554/eLife.02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman-Roblick R, et al. p53 targets identified by protein expression profiling. Proc Natl Acad Sci USA. 2007;104(13):5401–5406. doi: 10.1073/pnas.0700794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai C, Krantz SB. Interferon gamma induces upregulation and activation of caspases 1, 3, and 8 to produce apoptosis in human erythroid progenitor cells. Blood. 1999;93(10):3309–3316. [PubMed] [Google Scholar]