Significance
Th17 cells accumulate in the gut, where they mediate barrier defenses and repair but can also provoke inflammatory disease. In mice, segmented filamentous bacteria (SFB) is sufficient to induce Th17 cells in the gut, but functionally analogous microbes in humans have not been defined. Here, we identified Bifidobacterium adolescentis as one of several human symbiont bacterial species that could, alone, induce Th17 cells in the small intestine of mice. B. adolescentis and SFB exhibited overlapping but also distinct activities, suggesting multiple routes to intestinal Th17 induction. Like SFB, B. adolescentis exacerbated autoimmune arthritis, arguing for its pathological relevance. Our results help to inform the search for therapeutic targets in diseases associated with Th17 responses and mucosal dysfunction.
Keywords: microbiota, Th17 cells, probiotic, mucosal immunology, intestine
Abstract
Th17 cells accrue in the intestine in response to particular microbes. In rodents, segmented filamentous bacteria (SFB) induce intestinal Th17 cells, but analogously functioning microbes in humans remain undefined. Here, we identified human symbiont bacterial species, in particular Bifidobacterium adolescentis, that could, alone, induce Th17 cells in the murine intestine. Similar to SFB, B. adolescentis was closely associated with the gut epithelium and engendered cognate Th17 cells without attendant inflammation. However, B. adolescentis elicited a transcriptional program clearly distinct from that of SFB, suggesting an alternative mechanism of promoting Th17 cell accumulation. Inoculation of mice with B. adolescentis exacerbated autoimmune arthritis in the K/BxN mouse model. Several off-the-shelf probiotic preparations that include Bifidobacterium strains also drove intestinal Th17 cell accumulation.
The mammalian gut harbors hundreds of species of symbiont bacteria that play a crucial function in various facets of host physiology, including metabolism, tissue development, and maturation of the immune system (1, 2). Germfree (GF) and antibiotic-treated mice have several defects in T-cell compartments of both their gut-associated and -distal organs, including a paucity of intestinal Th17 and Treg cells and a systemic skewing toward Th2 responses (3, 4). Importantly, specific members or subsets of the microbiota can rescue a dearth of Treg or Th17 cells. Although early reports argued that a consortium of Clostridium species from either the murine or human gut is needed to induce Treg cells in the murine colon (5, 6), more recent studies showed that an assortment of individual bacterial species, including Clostridium and Bacteroides family members, also possess this property (7, 8). Similarly, a single bacterial strain, segmented filamentous bacteria (SFB), is sufficient to drive the accumulation of Th17 cells in the small-intestinal lamina propria (SI-LP) of mice (9, 10); however, Th17-inducing microbes derived from the human gut have not yet been identified. A recent report did document an increase in colonic Th17 cells in GF mice inoculated with fecal material from healthy people and patients with ulcerative colitis, thus showing the existence of Th17-inducing species in the human microbiota (11). However, the microbiota composition differs substantially across both healthy individuals and colitis patients, and the symbionts responsible for Th17 cell induction at steady state remain uncharacterized (11).
In both mice (10, 12) and humans (13, 14), Th17 cells are normally at their highest levels in the SI-LP. They secrete the cytokines IL-17A, IL-17F, and IL-22, which induce the production of antimicrobial peptides and tight junction proteins from intestinal epithelial cells, thereby buttressing gut barrier integrity (15–17). Moreover, IL-17A and IL-17F promote the recruitment of neutrophils via the release of granulocyte colony-stimulating factor, thereby helping to defend the host against infections by fungi and extracellular bacteria (18). Consequently, humans genetically deficient in IL-17 signaling because of mutations in genes such as STAT3 and IL17RA suffer from an increased susceptibility to mucosal infections by Candida albicans and Staphylococcus aureus (18, 19). Overexuberant Th17 responses, however, have been implicated in various inflammatory and autoimmune disorders, including multiple sclerosis, rheumatoid arthritis (RA), and inflammatory bowel disease (IBD) (19, 20). Many of these disorders in both mice and humans are also associated with intestinal dysbiosis (21, 22). An example of the dichotomous effects of symbiont-dependent Th17 cells is provided by SFB, which confers resistance to the enteropathogen Citrobacter rodentium in mice but exacerbates disease severity in murine models of multiple sclerosis and RA (10, 23, 24). Hence, fluctuations in the human microbiome are likely to exert important effects on host mucosal defenses and the development of inflammatory conditions, in part via modulation of Th17 responses.
Therefore, we set out to identify bacterial species from the human gut microbiota capable of inducing Th17 cells in the mouse intestine. Focusing on the most robust inducer, Bifidobacterium adolescentis, we compared its activities and mechanisms with those of SFB, uncovering distinct modi operandi but similar promotion of autoinflammatory and inflammatory diseases. Several off-the-shelf probiotic preparations—touted to improve human gastrointestinal and metabolic health—promoted SI-LP Th17 cell accumulation in mice, highlighting the potential therapeutic application of Th17-inducing bacteria.
Results
B. adolescentis Is a Human Gut Symbiont That Strongly Induces Intestinal Th17 Cells in Mice.
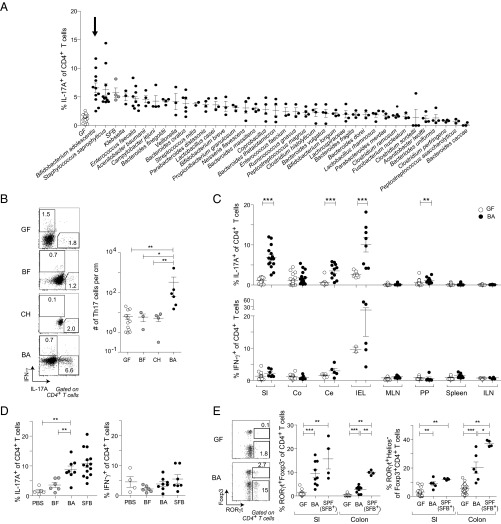
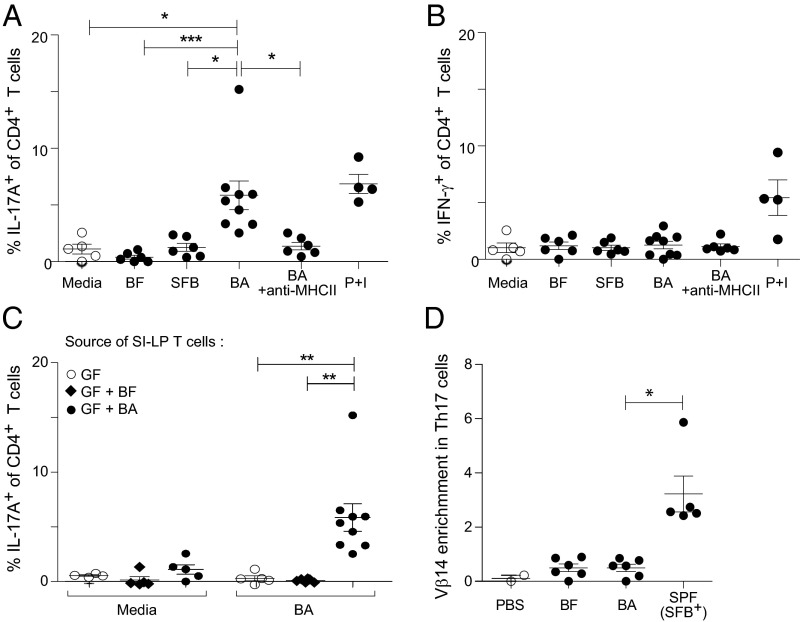
To identify human gut symbionts capable of influencing host immunity, we screened a large set of microbes by monocolonizing GF mice and evaluating a variety of immunologic parameters 2 wk later. The screen revealed a few phylogenetically diverse species that elicited SI-LP Th17 populations as large as those induced by SFB (Fig. 1A). Of the species examined, B. adolescentis (strain L2-32) promoted the greatest increase in Th17 cell frequencies and numbers in the SI-LP (Fig. 1 A and B). In addition to the SI-LP, colonization with B. adolescentis significantly increased Th17 cell levels in several other gut-associated organs, including the cecum, intraepithelial layer, and Peyer’s patches, although these effects were often much milder (Fig. 1C, Upper). In contrast, neither the percentages of Th17 cells in extraintestinal tissues (Fig. 1C, Upper) nor the fractions of Th1 cells in any organ (Fig. 1C, Lower) were substantially altered by B. adolescentis. Inoculation of specific-pathogen–free (SPF) mice with B. adolescentis recapitulated the immunologic phenotypes observed in monocolonized mice, augmenting Th17 frequencies in the SI-LP while leaving Th1 responses intact, in contrast to the lack of a significant response to a control microbe, Bacteroides fragilis (Fig. 1D). [Hereafter, we used either B. fragilis or Clostridium histolyticum as a control microbe because neither elicited significant SI-LP Th17 cell accumulation relative to GF mice (Fig. 1 A and B).]
Fig. 1.
B. adolescentis (BA) induces a robust intestinal Th17 population. (A) Frequency of SI-LP Th17 cells in GF mice monocolonized with individual symbiont bacteria as described in Materials and Methods. White and gray symbols represent GF and SFB-monocolonized mice, respectively. Arrow indicates BA-monocolonized mice. (B) Inflammatory cytokine production by SI-LP CD4+ T cells in mice colonized as indicated. (Left) Representative flow cytometric dot plot. (Right) Summary data. (C) Frequencies of (Upper) Th17 and (Lower) Th1 cells in various tissues of mice colonized as indicated. (D) Frequencies of (Left) Th17 and (Right) Th1 cells in the SI-LP of SPF mice gavaged as described in Materials and Methods with the indicated microbes. SFB+ SPF mice were bred at Harvard Medical School and naturally colonized with SFB. (E) Induction of intestinal RORγt expressers. (Left) Representative flow cytometric dot plot of SI-LP CD4+ T cells; summary data for frequencies of (Center) RORγt+ Foxp3− cells and (Right) RORγt+ Helios− Treg cells. Numbers in B and E refer to the fractions of cells in the identical gates. (B–E) Mean ± SEM pooled from two to four independent experiments. BF, B. fragilis; Ce, cecum; CH, C. histolyticum; Co, colon; IEL, intraepithelial lymphocyte layer; ILN, inguinal lymph node; MLN, mesenteric lymph nodes; PP, Peyer’s patches; SI, small intestine. *P < 0.05 (Mann–Whitney u test); **P < 0.01 (Mann–Whitney u test); ***P < 0.001 (Mann–Whitney u test).
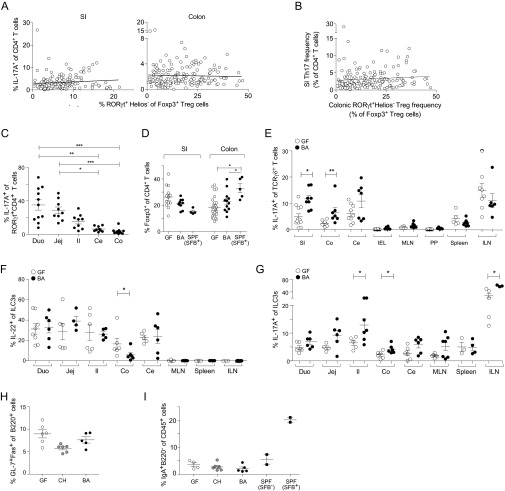
Because along with Th17 cells some intestinal Tregs express the transcription factor RORγt (8, 25), we looked more broadly at RORγt-expressing CD4+ T-cell populations in the gut. RORγt+ Foxp3− CD4+ T cells and RORγt+ Foxp3+ Tregs were both significantly enriched in the SI-LP and colonic lamina propria (LP) compartments of B. adolescentis-monocolonized mice (Fig. 1E). However, RORγt+ Treg induction was relatively modest, seldom reaching the levels found in SFB+ SPF mice or mice monocolonized with human-gut–derived Clostridium or Bacteroides species (Fig. 1E) (8). In addition, there was no correlation between RORγt+ Th17 and Treg cell induction across the panel of strains originally screened (Fig. S1 A and B). Interestingly, Th17 cell frequencies in the colonic LP were not elevated in B. adolescentis-monocolonized mice (Fig. 1C), despite a significant induction of RORγt in Foxp3− CD4+ T cells (Fig. 1E), implying a disjunction between RORγt expression and cytokine production (26). Indeed, the proportion of RORγt+ T cells producing IL-17A was diminished in the colon relative to the small intestine (Fig. S1C). Unlike the RORγt+ Treg subset, overall Foxp3+ Treg and IL-10+ T-cell frequencies were not altered by B. adolescentis (Fig. S1D). Hence, B. adolescentis preferentially induced Th17 cells in the intestine, with modest concomitant expansion of RORγt+ Treg cells.
Fig. S1.
Effects of BA monocolonization on other intestinal immunocyte populations. (A) No correlation between induction of intestinal Th17 and RORγt+ Treg cells after monocolonization with the panel of bacteria detailed in Fig. 1A. Lines represent linear regressions performed on the two immunocyte populations in the respective tissues. Pearson correlations are found to be not significant. (B) Data in A plotted as SI-LP Th17 vs. colonic RORγt+ Treg frequencies. Linear regression lines and correlations were calculated as in A (not significant). (C) Frequencies of IL-17A+ cells within the RORγt+ CD4+ T-cell population in various intestinal tissues in BA-monocolonized mice. (D) Frequencies of intestinal Foxp3+ CD4+ T cells in mice colonized as indicated. (E–G) Frequencies of (E) IL-17A–producing γδ T cells, (F) IL-22–producing Thy1+ RORγt+ type 3 innate lymphoid cells (ILC3s), and (G) IL-17A–producing ILC3s in various tissues of mice colonized as indicated. (H) Frequencies of GC B cells in the Peyer’s patches (PPs) of mice colonized as indicated. (I) Frequencies of IgA+ B220− plasma cells in the SI-LP of mice colonized as indicated. SPF mice from Jax were SFB− or colonized with SFB. Each symbol represents one mouse, and data were pooled from at least two independent experiments. Mean ± SEM. Ce, cecum; CH, Clostridium histolyticum; Co, colon; Duo, duodenum; Il, ileum; ILN, inguinal lymph node; Jej, jejunum; MLN, mesenteric lymph nodes; SI, small intestine. *P < 0.05 (Mann–Whitney u test); **P < 0.01 (Mann–Whitney u test); ***P < 0.001 (Mann–Whitney u test).
Apart from CD4+ T cells, an array of leukocyte subsets also secretes IL-17A and IL-22, often in response to similar cytokine cues, including IL-1 and IL-23 (27). We, thus, investigated the effects of B. adolescentis on cytokine production from other immunocyte populations. This microbe mildly increased cytokine production from intestinal γδ T cells (Fig. S1E) and RORγt+ type 3 innate lymphoid cells (Fig. S1 F and G), although this effect was also detected for other human symbiont bacteria that did not induce Th17 cells in our screen, in line with a previous report (11).
In rodents, SFB colonization leads to robust germinal center (GC) B-cell responses in the Peyer’s patches and the subsequent accumulation of T-cell–dependent IgA-producing plasma cells in the SI-LP (28, 29). Additionally, Th17 cells promote IgA class switching in the Peyer’s patches (30). We, therefore, assessed the impact of B. adolescentis on intestinal B-cell responses but found enhancement of neither the Peyer’s patch GC B-cell levels (Fig. S1H) nor SI-LP IgA-producing plasma cell frequencies (Fig. S1I). Taken together, our findings indicate that B. adolescentis exerted a potent and specific effect only on CD4+ T cells, primarily in the small intestine.
B. adolescentis Does Not Provoke Either Intestinal or Systemic Inflammation.
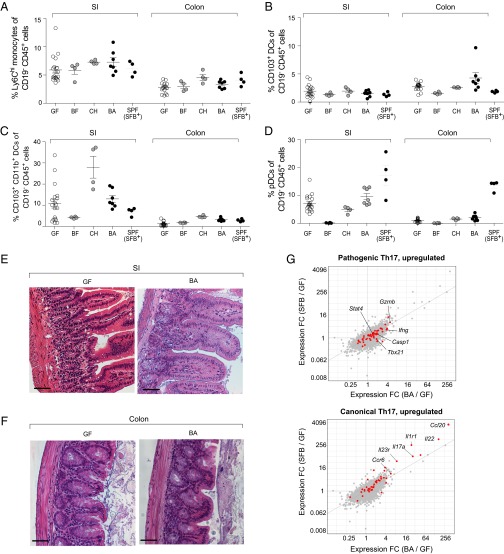
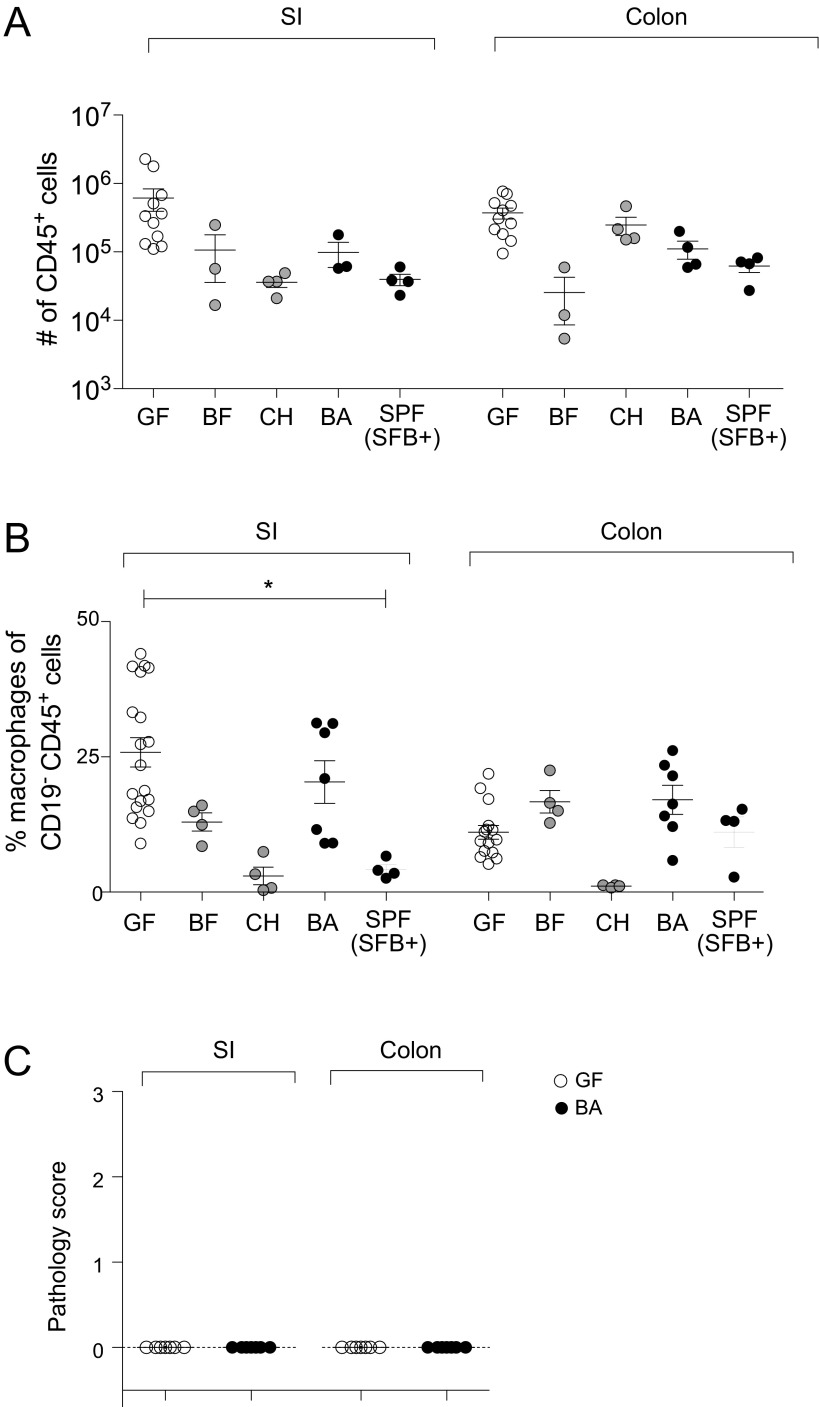
Because Th17 cells manifest potent proinflammatory effector functions and have been associated with both intestinal and systemic inflammatory diseases, we sought to determine whether expansion of the intestinal Th17 compartment in B. adolescentis-monocolonized mice was accompanied by inflammation in the gut or extragut organs. Multiple findings argue against B. adolescentis triggering generalized inflammation. First, Th1 cell numbers are often elevated in cases of inflammation and immunopathology, but we saw no increase in gut or systemic Th1 responses (Fig. 1C). Second, the number of CD45+ leukocytes and frequencies of various intestinal myeloid subsets that typically expand during colitis or gut infections (31) remained similar in GF and B. adolescentis-monocolonized mice (Fig. 2 A–D and Fig. S2 A and B). Although Ly6Chi monocytes were slightly expanded by B. adolescentis in the SI-LP, this increase was not significant, and several other intestinal symbionts produced the same effect without attendant histological signs of inflammation (Fig. 2A). Thus, the modest increase in Ly6Chi monocytes driven by B. adolescentis likely reflects a physiological response to colonization by broad classes of microbes. Third, histological examination revealed the absence of gross signs of inflammation in the small intestine and colon (Fig. 2 E and F and Fig. S2C). Fourth, transcriptional profiling of small-intestinal CD4+ T cells showed only a modest up-regulation of genes associated with pathogenic Th17 cells (32) in GF mice on colonization with B. adolescentis; this increase was comparable with that elicited by SFB and weaker than the up-regulation of canonical Th17 transcripts observed for both microbes (e.g., Rorc and Ccr6) (33) (Fig. 2G). Therefore, B. adolescentis seems to be a bona fide intestinal symbiont akin to SFB in mice, capable of peaceful coexistence in the gut of a healthy host, despite its profound impact on the Th17 compartment.
Fig. 2.
B. adolescentis (BA) does not provoke intestinal inflammation. (A–D) Myeloid cells. Frequencies of the indicated myeloid cell populations in the intestines of mice colonized as indicated. Each symbol represents one mouse. Mean ± SEM. Data for GF and BA are pooled from at least two independent experiments. P value was not significant for all comparisons (Kruskal–Wallis test and Dunn’s multiple comparisons test). (E and F) Histopathology. H&E staining of representative sections of (E) the small intestine (SI) and (F) the colon. (Scale bar: 50 μm.) (G) Th17 cell phenotype. Fold change (FC)/FC plots comparing transcripts induced by BA vs. SFB in SI-LP CD4+ T cells. Red indicates transcripts up-regulated in (Upper) pathogenic Th17 cells or (Lower) canonical Th17 cells. Example genes induced for each Th17 phenotype are indicated (n = 3–4 per group). BF, B. fragilis; CH, C. histolyticum.
Fig. S2.
BA does not provoke intestinal inflammation. (A) Numbers of CD45+ cells and (B) frequencies of CD11b+F4/80+CD103− macrophages in the intestines of mice colonized as indicated. Each symbol represents one mouse. Data for GF and BA pooled from at least two independent experiments. *P < 0.05 (Kruskal–Wallis test and Dunn’s multiple comparisons test). (C) Pathology scores of ileum and colon sections from GF and BA-monocolonized mice. Scoring method is detailed in SI Materials and Methods. Each symbol represents one mouse, and data were pooled from two independent experiments. BF, Bacteroides fragilis; CH, Clostridium histolyticum; SI, small intestine.
Gut Th17 Cells Expanded by B. adolescentis Are Symbiont-Specific.
Recent studies have shown that gut Th17 cells in SFB-bearing hosts are specific for SFB-derived antigens (34, 35). To determine if B. adolescentis-induced intestinal Th17 cells are analogously specific for B. adolescentis, we isolated CD4+ T cells from the small intestine of monocolonized mice and measured their cytokine responses to stimulation by lysates from various bacterial species. IL-17A production was markedly enhanced on restimulation by B. adolescentis lysate to levels comparable with those provoked by activation with phorbol 12-myristate 13-acetate (PMA) plus ionomycin but was not augmented by restimulation by B. fragilis or SFB lysates (Fig. 3A). The increased Th17 response to B. adolescentis was also dependent on MHC class II (MHC-II) molecules (Fig. 3A). Consistent with the lack of Th1 cell induction by B. adolescentis, IFN-γ production was uniformly low in response to all bacteria tested and remained unaltered by antibody blockade of MHC-II molecules (Fig. 3B). B. adolescentis-specific Th17 responses were detected only in mice colonized with B. adolescentis but not in GF or B. fragilis-colonized mice (Fig. 3C). Moreover, Th17 cells in SPF mice gavaged with B. adolescentis did not display preferential Vβ14 T-cell receptor (TCR) chain use as exhibited by SFB-specific Th17 cells (Fig. 3D) (35), suggesting that the gut Th17 cells elicited by B. adolescentis and SFB were not recognizing a common immunodominant microbial antigen. Collectively, the data indicate that intestinal Th17 cells induced by B. adolescentis were symbiont-specific.
Fig. 3.
B. adolescentis (BA)-driven intestinal Th17 responses are cognate. (A and B) BA-specific Th17 cells. Frequencies of SI-LP CD4+ T cells producing (A) IL-17A or (B) IFN-γ on overnight stimulation by splenic dendritic cells incubated with media alone or lysates prepared from the indicated bacterial species. +Anti–MHC-II (the blocking antibody M5/114.15.2) was added. (C) Frequency of SI-LP CD4+ T cells producing IL-17A on stimulation with media alone or BA lysate when T cells were isolated from mice colonized as indicated. (D) The degree of TCRVβ14 enrichment in SI-LP CD4+ T cells from SPF mice colonized as indicated, calculated as (percent of Vβ14+ cells in Th17 fraction)/(percent of TCRVβ14+ cells in non-Th17 fraction). Each symbol represents one mouse. Data are pooled from two to four independent experiments. Mean ± SEM. BF, B. fragilis; P + I, PMA and ionomycin. *P < 0.05 (Kruskal–Wallis test and Dunn’s multiple comparisons test); **P < 0.01 (Kruskal–Wallis test and Dunn’s multiple comparisons test); ***P < 0.001 (Kruskal–Wallis test and Dunn’s multiple comparisons test).
B. adolescentis Colonizes the Gastrointestinal Tract Widely and Closely Associates with the Epithelium.
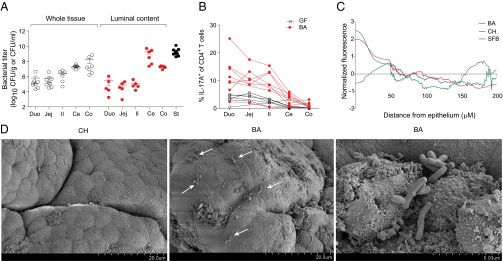
The enrichment of Th17 cells in the ileum of SFB-harboring mice corresponds to an overrepresentation of SFB in that intestinal segment (36, 37) and the ability of SFB to form intimate associations with the intestinal epithelium (11). To determine whether B. adolescentis occupied an intestinal niche similar to that of SFB, we measured bacterial titers in various intestinal compartments of mice monocolonized with the former. B. adolescentis was found in both the gut mucosa and lumen, with the overall bacterial load in the lumen progressively increasing from the duodenum to the colon, reflecting the distribution of overall bacterial burden in SPF mice (38) (Fig. 4A). Bacterial loads did not, however, correlate with Th17 levels, because the colon harbored relatively few Th17 cells while hosting very high quantities of B. adolescentis (Figs. 1C and 4 A and B), although the uniformly high B. adolescentis titers throughout the small intestine might explain the increase in Th17 cells in the duodenum and jejunum as well as the ileum (Fig. 4B).
Fig. 4.
B. adolescentis (BA) colonizes the entire length of the intestines, closely associating with the ileal epithelium. (A) Titers of bacteria associated with various segments of the intestinal mucosa or shed into the corresponding lumen or the stool (St). Normalized to tissue or St weight or the volume of luminal wash. Each symbol represents one mouse. Data are pooled from three to four independent experiments. Mean ± SEM. (B) Frequencies of Th17 cells along the length of the intestinal LP in mice colonized as indicated. Each line represents frequencies from one mouse. Data are pooled from three independent experiments. (C) FISH quantification. Normalized bacterial fluorescence vs. distance from the epithelial surface of the terminal ileum (Il) from mice monocolonized as indicated. Data are plotted as the average fluorescence from six to eight total images from two to three fields of view per section from two to four mice per microbe. (D) Representative SEM photograph of the ileal surface in mice colonized as indicated. Arrows indicate sites of bacterial association with the intestinal epithelium. Ce, cecum; CH, C. histolyticum; Co, colon; Duo, duodenum; Jej, jejunum.
To visualize the intestinal niche of B. adolescentis, we performed FISH on ileum and colon sections from B. adolescentis-monocolonized mice and quantified bacterial densities in relation to their distance from the intestinal epithelium (Fig. 4C). Like SFB, B. adolescentis associated closely with the ileal but not the colonic epithelium. In contrast, C. histolyticum, a human symbiont that did not promote Th17 cell expansion, was found primarily in the ileal lumen. SEM revealed B. adolescentis but not C. histolyticum to be localized close to the ileal surface in mice, corroborating the FISH findings (Fig. 4D). The capacity for tight association with the epithelium may thus represent a conserved feature of Th17 cell-inducing microbes.
B. adolescentis and SFB Induce Largely Distinct Transcriptional Programs in the Intestine.
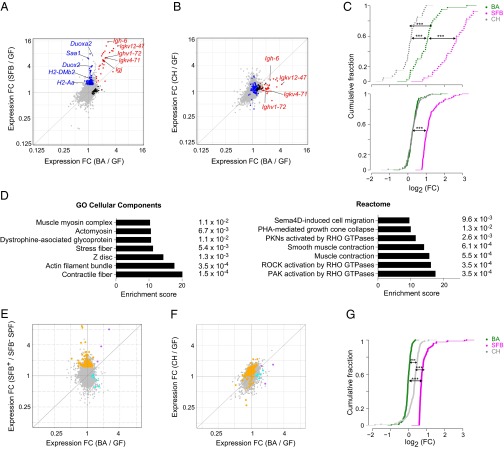
Because the effects of SFB and B. adolescentis on the host immune system seemed similar, and because both microbes interacted closely with the small-intestinal epithelium, we next profiled gene expression in whole ileal tissue from GF mice and mice monocolonized with B. adolescentis, control C. histolyticum, or SFB to determine whether B. adolescentis and SFB triggered overlapping intestinal gene programs that might account for their ability to elicit robust Th17 populations (Fig. 5 A and B). A substantial number of genes was up- or down-regulated in common by the Th17-cell–inducing bacteria, but we also detected a microbe-specific transcriptional imprint for each of them (Fig. 5A and Tables S1, S2, and S3). RNAs up-regulated by both SFB and B. adolescentis were dominated by Ig gene transcripts (Fig. 5A, red symbols). This induction likely reflected a robust IgA response in the case of SFB (9, 39), but we did not observe any signs of an enhanced IgA response (Fig. S1G) or a change in B-cell numbers in mice monocolonized with B. adolescentis. In this case, the induction of Ig transcripts was likely to be a consequence of increased intestinal IgM+ plasma cells, because Igj and Igh-6, encoding the J chain of secretory IgA/IgM and the constant region of the IgM heavy chain, respectively, were among the Ig transcripts most strongly induced. Quantitative comparison revealed that the Ig transcripts up-regulated by SFB were significantly enriched in B. adolescentis- and SFB-colonized mice compared with mice colonized with C. histolyticum (Fig. 5C, Upper).
Fig. 5.
B. adolescentis (BA) and SFB both induce B-cell transcripts, but otherwise, they trigger distinct transcriptional programs. (A and B) Whole-tissue transcriptomes. Fold change (FC)/FC plots comparing ileum tissue transcripts induced by (A) BA vs. SFB (Th17 inducing) or (B) BA vs. C. histolyticum (CH; Th17 noninducing). In A, blue indicates genes up-regulated primarily by SFB, black indicates genes up-regulated primarily by BA, and red indicates genes up-regulated by both bacteria. Certain transcripts previously associated with SFB-mediated induction of Th17 cells or encoding Igs are indicated. In B, the genes highlighted in A are again highlighted in the same colors. (C) Statistics for A and B. FC distribution of (Upper) SFB-induced Ig or (Lower) non-Ig RNAs in the ileal transcriptomes of mice colonized as indicated. Only transcripts increased by SFB by at least ≥1.6-fold with a P value of <0.05 were plotted. ***P < 0.001 (Kolmogorov–Smirnov test). (D) Pathways (from the two databases as indicated) enriched in transcripts specifically up-regulated in the ileum by BA relative to GF mice were determined using Enrichr. P values are indicated to the right. (E and F) S-IEC transcriptomes. As per A and B, except that transcripts from isolated S-IECs were examined. In E, orange indicates genes up-regulated primarily by SFB, cyan indicates genes up-regulated primarily by BA, and purple indicates genes up-regulated by both bacteria. In F, the genes highlighted in E are again highlighted in the same colors. (G) Statistics for E and F. Calculated as per C. n = 3 (A–D) or 2–4 (E and F) per group. ***P < 0.001.
Table S1.
Transcripts up-regulated by BA only in the ileum
| Gene symbol | Mean expression, BA | FC (BA/GF) | FC (SFB/GF) | FC (CH/GF) | P value (BA/GF) |
| Chrm2 | 2,039.042584 | 0.297165938 | 0.053122127 | 0.185615884 | 0.026213408 |
| Kcnmb1 | 834.4396439 | 0.228469362 | 0.039914337 | 0.079625566 | 0.018217538 |
| Fam129a | 764.6772109 | 0.214364536 | 0.131626283 | 0.065367649 | 0.006844059 |
| Pgm5 | 2,134.984612 | 0.192459299 | 0.017546134 | 0.051828909 | 0.034880569 |
| Apod | 362.3731772 | 0.191477129 | 0.049334306 | 0.041225626 | 0.037431459 |
| Synpo2 | 1,474.051482 | 0.182828291 | 0.005696728 | 0.090915475 | 0.022833095 |
| Sparcl1 | 5,159.684931 | 0.170390627 | 0.044770166 | 0.120997438 | 0.049877649 |
| Ppp1r12b | 993.5207571 | 0.167169822 | 0.040088844 | 0.068432048 | 0.032989101 |
| Grem1 | 2,168.058013 | 0.161476564 | 0.038689727 | 0.063985708 | 0.022960986 |
| Cald1 | 1,324.791217 | 0.161178373 | 0.065615106 | 0.095031404 | 0.049492381 |
| Rgs5 | 1,122.44556 | 0.160219601 | 0.011599174 | 0.085212925 | 0.029286996 |
| Dub2a | 193.3454068 | 0.160188985 | 0.078216464 | 0.094889908 | 0.018184781 |
| Akap6 | 208.3059232 | 0.150834887 | 0.069175681 | 0.115824707 | 0.046885373 |
| Myl9 | 7,540.219454 | 0.148399949 | 0.030194409 | 0.045405107 | 0.02711294 |
| Plagl1 | 390.7836096 | 0.148319413 | 0.040171994 | 0.084845769 | 0.000811109 |
| Foxp2 | 245.1713851 | 0.146732002 | 0.018165906 | 0.065862053 | 0.012876574 |
| Tmod1 | 332.9137681 | 0.145300461 | 0.044415606 | 0.130662223 | 0.043049236 |
| Sst | 2,023.937403 | 0.144544791 | 0.011278214 | 0.074130018 | 0.025429215 |
| Cryab | 980.9726133 | 0.135183373 | 0.052100227 | 0.045000354 | 0.045110064 |
| Htr2b | 120.5591971 | 0.130358626 | 0.006860536 | 0.152167738 | 0.048073693 |
| Ces1c | 273.6391882 | 0.12619939 | −0.030248251 | −0.05605925 | 0.022343221 |
| Cpe | 780.6046894 | 0.125643916 | 0.017200609 | 0.138914779 | 0.031929232 |
| Dkk2 | 236.2097351 | 0.120376423 | 0.014055521 | 0.044619085 | 0.008056348 |
| Kcna2 | 408.4921803 | 0.118636839 | 0.03191973 | 0.074397744 | 0.026272408 |
| Rtn1 | 482.167881 | 0.115100537 | 0.031102746 | 0.093735511 | 0.048734668 |
| Emp2 | 685.3317379 | 0.110883823 | −0.046298901 | 0.016606248 | 0.025409771 |
| Sgcd | 310.1193808 | 0.110858929 | −0.008436168 | 0.037070979 | 0.014906528 |
| Myh11 | 4,334.468243 | 0.108964868 | −0.005791837 | 0.039228198 | 0.045319729 |
| Sncg | 573.2725556 | 0.101656186 | 0.019457406 | 0.055514698 | 0.04463545 |
| Tnnt2 | 471.0495548 | 0.087179771 | −0.039245536 | 0.005405056 | 0.028160534 |
| Mbnl1 | 3,516.801186 | 0.085151777 | −0.000793497 | 0.020569128 | 0.037218281 |
| Sostdc1 | 130.6652929 | 0.083012532 | −0.001587572 | 0.045023847 | 0.031984525 |
| Calcb | 144.7821575 | 0.082393173 | −0.029647663 | 0.052611952 | 0.001675199 |
Log2 (FC) values are shown; P values are calculated by Student's t test. CH, Clostridium histolyticum.
Table S2.
Transcripts up-regulated by SFB only in the ileum
| Gene symbol | Mean expression, SFB | FC (BA/GF) | FC (SFB/GF) | FC (CH/GF) | P value (SFB/GF) |
| Igk-V19-14 | 853.4267302 | 0.110261244 | 2.988557144 | −1.376516717 | 0.006500653 |
| Duoxa2 | 665.8108381 | 0.306584696 | 2.838439663 | 0.460138644 | 0.000283233 |
| Gm16848 | 261.5989275 | −0.055555362 | 2.663868886 | −0.638987242 | 0.01286801 |
| Saa1 | 26.00806464 | 0.000723811 | 2.598610916 | 0.003127799 | 0.011514492 |
| Nos2 | 666.5484364 | 0.27184227 | 2.053561217 | −0.18707337 | 0.003574072 |
| Duox2 | 921.5562128 | 0.229189845 | 2.036918302 | 0.047404289 | 0.000825664 |
| Fut2 | 193.3725294 | 0.189200961 | 1.960981174 | 0.322889829 | 0.04051458 |
| Tat | 105.3294106 | 0.068066196 | 1.864156053 | 0.137433476 | 0.000766139 |
| Smpdl3b | 379.0667855 | 0.103363249 | 1.703118975 | 1.171393754 | 0.022155074 |
| Upp1 | 1,100.373966 | 0.17593797 | 1.692262354 | 0.332276036 | 0.007958242 |
| 2010001M09Rik | 233.3053017 | 0.141494283 | 1.63084607 | −0.015283966 | 0.000888497 |
| Pla2g5 | 220.9362025 | 0.135648046 | 1.602086072 | 0.079130172 | 3.21368E-05 |
| Zbp1 | 440.1568157 | 0.11858657 | 1.542272461 | 0.299552881 | 0.020338646 |
| Stom | 1,280.475012 | 0.115229948 | 1.411132497 | 0.389142369 | 0.00430015 |
| Il18 | 290.0474797 | 0.214639793 | 1.294489296 | 0.490521638 | 0.03713033 |
| Ptk6 | 622.6759816 | 0.170309651 | 1.274728699 | 0.121509504 | 0.031550056 |
| Prdm1 | 199.5191639 | 0.195016726 | 1.25581363 | 0.626627876 | 0.004514204 |
| Derl3 | 243.9642226 | 0.082054973 | 1.166445329 | −0.056050993 | 0.001277782 |
| Cd7 | 363.7810359 | 0.224355143 | 1.132213065 | 0.54358203 | 0.004272466 |
| Dmbt1 | 5,071.396852 | 0.166475941 | 1.131090173 | 0.289636614 | 0.002590963 |
| Psmb8 | 310.7899556 | 0.046296448 | 1.101849779 | 0.452490375 | 0.004822295 |
| Trp53i11 | 1,474.848073 | 0.124066019 | 1.024766917 | 0.234757761 | 0.003944383 |
| Pou2af1 | 285.3608501 | 0.222996292 | 0.997894796 | 0.029802605 | 0.00825597 |
| Cd274 | 250.3089872 | 0.243929123 | 0.976944452 | 0.290112001 | 0.002209936 |
| H2-DMb2 | 383.5890869 | 0.237152621 | 0.957862477 | 0.174433294 | 0.010548807 |
| Nlrc5 | 217.2882591 | 0.233973524 | 0.94749075 | 0.152698667 | 0.001627023 |
| Capg | 265.8165695 | 0.128175453 | 0.94705339 | −0.102103848 | 0.000276135 |
| Hk2 | 393.4849339 | 0.146949907 | 0.93364982 | 0.133185761 | 0.007799811 |
| Ifi47 | 178.0393997 | 0.145281576 | 0.927939241 | 0.271351842 | 0.001837908 |
| Mfsd2a | 296.8908829 | 0.128438275 | 0.926244391 | 1.098395273 | 0.021505061 |
| AA467197 | 1,453.467133 | −0.052688895 | 0.92183718 | 0.153596259 | 0.049380592 |
| Psmb9 | 649.696286 | 0.135138631 | 0.913310425 | 0.344513587 | 0.000930324 |
| H2-gs10 | 509.7286242 | 0.065015432 | 0.89771305 | 0.552932722 | 0.01838137 |
| Prss27 | 347.7431618 | 0.08194731 | 0.897219636 | 0.019211733 | 0.004778358 |
| Gm12250 | 214.7544773 | 0.326061195 | 0.891062269 | 0.388392121 | 0.001361823 |
| 2210407C18Rik | 1,658.390203 | −0.041923819 | 0.880681682 | 1.01260504 | 0.03454189 |
| Mafb | 405.7671552 | −0.150637129 | 0.876432509 | 0.541579007 | 0.047145682 |
| Slc40a1 | 485.7287587 | −0.2832559 | 0.870688425 | −0.29689716 | 0.020210258 |
| Herc6 | 215.6600076 | 0.243997375 | 0.865545115 | 0.462827494 | 0.021314191 |
| Cd28 | 93.06588762 | 0.174280638 | 0.861591875 | −0.054186363 | 0.026116773 |
| Rabl5 | 444.4059466 | 0.0274047 | 0.854330423 | 0.207181681 | 0.010381511 |
| Ociad2 | 1,025.05628 | −0.005388622 | 0.852159465 | 0.38443276 | 0.012371807 |
| Casp4 | 751.1385615 | −0.045686437 | 0.829900103 | 0.179037389 | 0.003306849 |
| Ceacam10 | 223.3571229 | −0.038221539 | 0.822062891 | 0.13183611 | 0.013837654 |
| Gm10384 | 118.0494671 | 0.136910477 | 0.819371836 | 0.097155785 | 0.028751693 |
| Ikzf3 | 130.2163585 | 0.14639016 | 0.809794649 | 0.359101281 | 0.004060268 |
| Cd38 | 3,538.761494 | 0.137103726 | 0.807769453 | −0.085486731 | 0.011668035 |
| 1810065E05Rik | 1,332.481968 | −0.153522166 | 0.803854949 | −1.087182267 | 0.01789105 |
| AI504432 | 105.5326384 | 0.216285755 | 0.803686059 | 0.110420276 | 0.003062097 |
| Nlrc5 | 175.6758691 | 0.190266595 | 0.801752572 | 0.230269194 | 0.000813309 |
| Tgm2 | 1,254.452033 | 0.074964528 | 0.79962031 | 0.028863601 | 0.005017818 |
| B3gnt7 | 453.8156872 | 0.010985062 | 0.791711888 | 0.006540703 | 0.020735661 |
| Selplg | 328.4064854 | 0.259187786 | 0.787528997 | 0.236169353 | 0.010638523 |
| Stk10 | 317.0814944 | −0.030477066 | 0.774196696 | 0.564182614 | 0.031996061 |
| Lgmn | 1,566.386308 | 0.21281686 | 0.769491493 | 0.365697531 | 0.000115332 |
| Lrg1 | 121.2524078 | 0.222870817 | 0.749145127 | 0.1430782 | 0.019811432 |
| Reg3b | 11,101.33873 | 0.284081772 | 0.747342427 | −0.156209311 | 0.001661347 |
| Asns | 1,434.581547 | 0.080252656 | 0.741259277 | −0.105818096 | 0.044625471 |
| St3gal1 | 414.3189977 | 0.209970335 | 0.737906365 | 0.510967996 | 0.00326104 |
| Stat1 | 955.3798545 | 0.310438807 | 0.733851126 | 0.298675517 | 0.027991341 |
| Nlrc5 | 246.7167169 | 0.243963257 | 0.733514055 | 0.120431888 | 0.012482394 |
| Pla2g2a | 1,267.478799 | 0.322326643 | 0.731566764 | −0.0425949 | 0.022015169 |
| Myl7 | 199.7102766 | −0.12486677 | 0.73017749 | 0.057705155 | 0.00874179 |
| Gimap6 | 344.4959519 | 0.308114583 | 0.726718786 | 0.270842634 | 0.019472668 |
| Bhmt | 134.8292305 | 0.096917242 | 0.722770695 | 0.010943844 | 0.000683747 |
| Il12rb2 | 85.40593392 | 0.163310668 | 0.718114362 | 0.214559594 | 0.004065423 |
| Bhmt | 98.84113938 | 0.108827688 | 0.716185492 | 0.077385429 | 0.002031448 |
| Endod1 | 827.8649249 | 0.007403989 | 0.71300915 | 0.118108166 | 0.042632407 |
| St6gal1 | 288.1671864 | 0.14116588 | 0.712785754 | −0.040176173 | 0.02554139 |
| Barx2 | 255.3722715 | 0.21750414 | 0.704020052 | 0.493159202 | 0.005805398 |
| Cfi | 75.30842049 | 0.098843456 | 0.703514107 | 0.189498165 | 0.021058133 |
| Pfkfb3 | 202.4205171 | 0.218276151 | 0.703204147 | 0.635880759 | 0.000285107 |
| H2-Aa | 3,698.470247 | 0.301759713 | 0.69934207 | −0.067194315 | 0.040362026 |
| Mfsd4 | 541.0736322 | 0.077640246 | 0.696209578 | 0.173072728 | 0.043306204 |
| Itgam | 334.915123 | 0.185467634 | 0.691039761 | −0.169259504 | 0.009938058 |
| Gimap4 | 244.2536863 | 0.264307854 | 0.690126653 | 0.267964615 | 0.017901949 |
| Unc5cl | 500.2622859 | −0.03710276 | 0.686866838 | 0.364319145 | 0.011640721 |
| Serpina3f | 113.8261155 | 0.232269265 | 0.686538056 | 0.201577533 | 0.012034229 |
| Cd79b | 255.304071 | 0.065934588 | 0.684774566 | 0.029415705 | 0.029836091 |
| Gch1 | 96.54637783 | −0.111868491 | 0.684119427 | 0.165463347 | 0.034776573 |
| Alpk1 | 308.9951264 | 0.214644533 | 0.681968079 | −0.180876815 | 0.009157362 |
| Nxf7 | 167.6610135 | 0.08937288 | 0.67746157 | 0.135004618 | 0.027199713 |
| Muc4 | 455.7438252 | 0.073473018 | 0.675703922 | 0.254889469 | 0.032263311 |
| Itgal | 181.6486518 | 0.21087756 | 0.669096914 | 0.212685791 | 0.002301608 |
| Nlrc5 | 82.91887014 | 0.342300649 | 0.669006382 | 0.267637632 | 0.000144179 |
| 9930111J21Rik2 | 355.7561139 | 0.287354354 | 0.662332235 | 0.336324909 | 0.024774891 |
| Tmem173 | 332.9206654 | 0.266926837 | 0.658114938 | −0.001165033 | 0.015214991 |
| Gm1965 | 630.4514691 | 0.154048636 | 0.658047981 | −0.023995136 | 0.004048673 |
| Tnfsf13b | 461.5308732 | 0.138433901 | 0.65586992 | 0.655461998 | 0.027428361 |
| H2-D1 | 3,369.916118 | 0.031611759 | 0.655666978 | 0.355051344 | 0.016218375 |
| Fah | 215.4706342 | 0.214319266 | 0.65384814 | 0.215162646 | 0.001198049 |
| Ccnd1 | 1,748.865661 | 0.209333742 | 0.647239044 | −0.098793355 | 0.007926484 |
| Zc3h12a | 312.3430679 | −0.046646432 | 0.634041124 | 0.061635625 | 0.016670529 |
| Trp53inp1 | 998.20059 | −0.194029153 | 0.632220939 | −0.233697893 | 0.032612192 |
| Tmem50b | 1,927.976387 | 0.006210008 | 0.625928581 | 0.298653291 | 0.000762451 |
| Irf1 | 1,262.626635 | 0.110061307 | 0.613485967 | 0.323835995 | 0.001393747 |
| Ehd4 | 730.7094354 | 0.290509571 | 0.603753454 | 0.420399619 | 0.014812246 |
| Cxcl13 | 764.5884394 | 0.327007914 | 0.598709686 | −0.292033319 | 0.023829593 |
| Tigit | 196.0336814 | 0.108702112 | 0.598202564 | 0.124200721 | 0.002423849 |
| Nlrc5 | 144.1034463 | 0.187870451 | 0.597781203 | 0.147835161 | 0.009141048 |
| Cyr61 | 308.6350772 | 0.057401631 | 0.596513639 | 0.085894925 | 0.005199748 |
| Sbno2 | 461.0022599 | −0.053220029 | 0.59125327 | 0.187734982 | 0.03080481 |
| Cd96 | 123.7371438 | 0.182993928 | 0.586774016 | 0.10762491 | 0.016158982 |
| Irf8 | 1,497.074584 | 0.098273904 | 0.58647347 | 0.073806163 | 0.004742564 |
| Irgm1 | 657.8123267 | 0.270432787 | 0.585964499 | −0.159198396 | 0.009259095 |
Log2 (FC) values are shown; P values are calculated by Student's t test. CH, Clostridium histolyticum.
Table S3.
Transcripts up-regulated by both SFB and BA only in the ileum
| Gene symbol | Mean expression, BA | FC (BA/GF) | FC (SFB/GF) | FC (CH/GF) | P value (BA/GF) |
| Gm5574 | 497.935906 | 2.181026802 | 3.606735017 | 0.530237169 | 0.028679178 |
| LOC672291 | 1,186.935899 | 1.862651377 | 3.406671763 | 0.800997014 | 0.182004174 |
| C3 | 1,209.754191 | 1.81768711 | 1.595027061 | 1.024557787 | 0.0065743 |
| LOC672291 | 1,265.056443 | 1.815213996 | 3.209373596 | 0.332041866 | 0.194663618 |
| LOC382693 | 562.9076023 | 1.735634641 | 3.241078263 | 0.897740884 | 0.004169052 |
| LOC382693 | 306.5810376 | 1.567718195 | 3.096028826 | 0.960518713 | 0.009995508 |
| Gm189 | 780.8833733 | 1.450384534 | 3.921440889 | −0.721220137 | 0.154572337 |
| Igkv4-71 | 2,814.225382 | 1.368658339 | 2.66899264 | −0.009794598 | 0.119051201 |
| Igh-6 | 170.6047943 | 1.320938525 | 3.607537877 | 0.877992857 | 0.064266343 |
| Ighv1-72 | 1,133.354834 | 1.26255382 | 2.681077317 | 0.487228036 | 0.009312949 |
| Gm1419 | 2,426.512632 | 1.241339959 | 2.655497177 | −0.002618509 | 0.104512792 |
| Serpina3n | 921.6573503 | 1.231287844 | 0.857347674 | 0.438402658 | 0.057343383 |
| IghmAC38.205.12 | 2,430.658219 | 1.218847523 | 3.200774456 | 0.221519086 | 0.241490411 |
| Ighv1-72 | 2,389.246958 | 1.168766397 | 2.385874484 | 0.311739353 | 0.062030116 |
| Ighg | 65.42361335 | 1.152077675 | 2.46944295 | 0.176777336 | 0.000756468 |
| Gm1524 | 200.997573 | 1.144439173 | 2.137813281 | −0.049552347 | 0.161116037 |
| Rprl1 | 397.8824815 | 1.120176455 | 2.491489342 | −0.049488908 | 0.072668128 |
| LOC435333 | 611.3550219 | 1.118939124 | 3.091746432 | 0.237322504 | 0.065912473 |
| Igl-V2 | 366.6977629 | 1.091295884 | 1.887907342 | 0.490791199 | 0.119915948 |
| Gm10880 | 2,030.493833 | 1.087120949 | 2.429797762 | −0.1863013 | 0.251200331 |
| Igj | 1,969.788621 | 1.062435211 | 2.520184559 | 0.133343753 | 0.218206742 |
| AI324046 | 1,817.164917 | 1.034117057 | 2.979410803 | −0.00843333 | 0.26975465 |
| Ighv1-72 | 1,847.434115 | 1.026107116 | 2.20707766 | 0.199097931 | 0.112943721 |
| LOC435333 | 1,376.962395 | 0.976627632 | 2.831748286 | 0.229350659 | 0.066097013 |
| Cxcl9 | 550.7686812 | 0.916645903 | 1.430366788 | 0.507487904 | 0.162776747 |
| Gm1077 | 487.9218827 | 0.862581419 | 2.098298235 | 0.056218229 | 0.121944706 |
| Igk-V28 | 467.7104548 | 0.792520742 | 2.016268915 | 0.08342139 | 0.109984118 |
| Igk-V19-20 | 269.3210399 | 0.771466499 | 1.806137593 | 0.038992127 | 0.238274696 |
| Cxcl10 | 512.6406491 | 0.720678061 | 0.887339027 | 0.399431159 | 0.223939927 |
| Ly6a | 2,946.003557 | 0.599307721 | 0.786074961 | 0.267616912 | 0.135665822 |
| Tcrg-V3 | 686.3986057 | 0.590679546 | 1.205704976 | 0.610480274 | 0.163537077 |
| Ccr9 | 177.4158173 | 0.585883681 | 1.073338917 | 0.218089151 | 0.039873999 |
Log2 (FC) values are shown; P values are calculated by Student t test. CH, Clostridium histolyticum.
RNAs specifically enriched in the intestines of SFB+ mice included several previously imputed to SFB, such as MHC-II transcripts (40) and mRNAs encoding molecules that augment Th17 responses (e.g., the serum amyloid A family of proteins), Duox2, and Duoxa2 (11, 26) (Fig. 5A, blue symbols). Expression of these transcripts was increased by B. adolescentis as well, but the level of induction was much lower than that for SFB and comparable with that provoked by non–Th17-inducing C. histolyticum (Fig. 5 B and C, Lower). Parsing of the genes specifically induced by B. adolescentis (Fig. 5A, black symbols) revealed an enrichment in nonimmunologic, particularly muscle-related, pathways that suggested a role for nonhematopoietic intestinal cells in relaying bacterial signals to the host immune system (Fig. 5D).
A clear divergence in gene expression profiles was also observed in small-intestinal epithelial cells (S-IECs) purified from mice colonized with B. adolescentis vs. SFB (Fig. 5E and Tables S4–S6). Indeed, the S-IEC transcriptome responses elicited by B. adolescentis and C. histolyticum were more concordant with each other than with those provoked by SFB (Fig. 5 E and F), and RNAs up-regulated by SFB were significantly more enriched in SFB- and C. histolyticum-colonized mice compared with B. adolescentis-colonized mice (Fig. 5G). Many of the SFB-specific transcripts in S-IECs (e.g., Duoxa2 and MHC-II transcripts) were unique to SFB in the ileum as well, suggesting a primary role for S-IECs in coordinating the host response to SFB but not to the other two microbes. Accordingly, the number of transcripts in S-IECs with expression that was differentially regulated by bacterial colonization was far higher for SFB than for B. adolescentis, and the identities of B. adolescentis-specific RNAs in the ileum and S-IECs were distinct (Tables S1–S6).
Table S4.
Transcripts up-regulated by BA only in S-IECs
| Gene symbol | Mean expression, BA | FC (BA/GF) | FC (SFB/GF) | FC (CH/GF) | P value (BA/GF) |
| Hist1h1d | 299.9153877 | 0.722765445 | −0.671737773 | 0.378796338 | 0.030997824 |
| Gstk1 | 1,764.3438 | 0.56052946 | −0.265096505 | 0.135195298 | 0.011314565 |
| Akr1c19 | 1,027.760121 | 0.499015696 | −0.724274897 | −0.126180154 | 0.037393782 |
| Lactb2 | 490.7820923 | 0.406886708 | −0.111160571 | 0.131812813 | 0.023069152 |
| Chac2 | 421.9886096 | 0.370999613 | 0.066632265 | 0.042416863 | 0.028647008 |
| Txndc9 | 831.592786 | 0.296193174 | −0.023473122 | 0.028513794 | 0.042011026 |
Log2 (FC) values are shown; P values are calculated by Student's t test. CH, Clostridium histolyticum.
Table S6.
Transcripts up-regulated by both SFB and BA only in S-IECs
| Gene symbol | Mean expression, BA | FC (BA/GF) | FC (SFB/GF) | FC (CH/GF) | P value (BA/GF) |
| Reg3b | 343.0416203 | 1.336901521 | 2.985827449 | 0.748474908 | 0.128857563 |
| Ly6a | 134.9492536 | 0.856763519 | 1.893017353 | −0.47483637 | 0.127183484 |
| Mrpl18 | 1,841.110202 | 0.63579989 | 0.921343524 | 0.661464462 | 0.180724598 |
Log2 (FC) values are shown; P values are calculated by Student's t test. CH, Clostridium histolyticum.
Table S5.
Transcripts up-regulated by SFB only in S-IECs
| Gene symbol | Mean expression, SFB | FC (BA/GF) | FC (SFB/GF) | FC (CH/GF) | P value (SFB/GF) |
| H2-Ab1 | 864.3036064 | −0.324408008 | 3.202256552 | −1.521163198 | 0.029287522 |
| Cd74 | 1,060.953036 | −0.290844415 | 3.092857404 | −1.862282404 | 0.049742042 |
| H2-Eb1 | 322.4872275 | −0.112400906 | 2.164104766 | −0.693531819 | 0.003597187 |
| Duox2 | 394.0993067 | −0.374474092 | 2.124737032 | −0.063176123 | 0.036657598 |
| Ptk6 | 161.0831133 | −0.046995572 | 1.538463048 | 0.089640331 | 0.017248875 |
| Eif5a | 1,789.643526 | 0.120287287 | 1.529926134 | 1.095302679 | 0.02900951 |
| H2-DMa | 264.8838672 | −0.106568444 | 1.528387337 | −0.472898048 | 0.042888668 |
| Tifa | 346.9209079 | −0.044207107 | 1.378854466 | 0.474167511 | 0.001057428 |
| Tgtp1 | 148.0728486 | −0.3028355 | 1.366861107 | −0.763370742 | 0.042711938 |
| Fut2 | 197.5802056 | −0.058696529 | 1.35730567 | −0.563572643 | 0.035898416 |
| Psmb8 | 188.9961694 | −0.006376721 | 1.352040665 | −0.223229797 | 0.00267256 |
| Ifi44 | 125.8182951 | 0.035393366 | 1.340878176 | −0.097493679 | 0.003726934 |
| Ceacam10 | 428.9394856 | −0.092720287 | 1.307346599 | 0.237103626 | 0.00661915 |
| Tnf | 147.9902206 | −0.179116865 | 1.263680788 | 0.204376572 | 0.008512408 |
| Aldh2 | 445.8218127 | −0.033655259 | 1.224499514 | 0.631294649 | 0.03253427 |
| Tfam | 194.231684 | 0.372769881 | 1.191469727 | 1.099633904 | 0.040669346 |
| Hnrnpa0 | 121.1726667 | −0.02709674 | 1.133682113 | 0.490963364 | 0.048613253 |
| Egr2 | 190.7897547 | −0.949001585 | 1.104808708 | −0.564805376 | 0.024198664 |
| Tap1 | 294.5812203 | −0.084711809 | 1.066470549 | −0.331071012 | 0.009052195 |
| H2-gs10 | 227.4762465 | −0.248390999 | 1.063330737 | 0.041060278 | 0.011698569 |
| Ctss | 316.265238 | −0.103108139 | 1.059735387 | 0.040686248 | 0.043712925 |
| Aldh1b1 | 1,181.764867 | −0.037675832 | 1.058305525 | 0.852257884 | 0.032582083 |
| Psmb9 | 548.6074749 | 0.165314259 | 1.051021468 | 0.001376265 | 0.02211841 |
| Tapbp | 926.2655353 | −0.060938018 | 1.049338905 | 0.174487227 | 0.041701087 |
| Irgm2 | 171.433705 | 0.032383042 | 1.016344167 | −0.477902879 | 0.037031526 |
| Alpk1 | 69.58134627 | −0.092606928 | 0.998955566 | 0.288675235 | 0.046187952 |
| Akr1b8 | 100.4186767 | 0.152094934 | 0.997275275 | 0.156184575 | 0.032250424 |
| Muc3 | 2,184.638659 | −0.53508588 | 0.980763441 | 0.086414667 | 0.021203783 |
| Cd14 | 91.75652206 | 0.041077225 | 0.974792703 | 0.415377042 | 0.01424996 |
| Lmna | 311.6235193 | −0.073409921 | 0.969193306 | 0.488004825 | 0.04298178 |
| Alpk1 | 98.2839131 | 0.006417528 | 0.963044435 | 0.208108539 | 0.002476056 |
| Alpk1 | 146.0583271 | 0.048409094 | 0.942259672 | 0.437523607 | 0.011033926 |
| Slc44a4 | 927.7208509 | −0.059362631 | 0.926918264 | 0.181395166 | 0.048147332 |
| Pim1 | 254.4340622 | −0.162779001 | 0.917345308 | 0.085768861 | 0.049978741 |
| H2-D1 | 1,937.54729 | −0.011714872 | 0.911668241 | 0.107462751 | 0.020469557 |
| Vwa5a | 223.007149 | −0.048470117 | 0.888440394 | 0.131417184 | 0.03464325 |
| Slc35f2 | 508.3488969 | 0.106158917 | 0.871959093 | 0.613080369 | 0.007770322 |
| 2510006D16Rik | 649.7924109 | −0.12253481 | 0.870618264 | 0.436073434 | 0.033801007 |
| Auh | 276.4461284 | −0.039341574 | 0.859762059 | 0.367844796 | 0.001951987 |
| Gsr | 1,179.717001 | 0.151263834 | 0.853983138 | 0.388838783 | 0.031737108 |
| Gm12250 | 159.6970376 | 0.144950057 | 0.841071934 | −0.421698502 | 0.006214875 |
| Gstt3 | 492.295427 | −0.050177331 | 0.835219462 | 0.591549427 | 0.039551792 |
| Thoc6 | 258.5126681 | 0.073053376 | 0.827155367 | 0.621163556 | 0.040358674 |
| Acadl | 427.7414258 | 0.097334204 | 0.824597087 | −0.025807889 | 0.000242 |
| Nox1 | 420.7207819 | −0.544232253 | 0.818516656 | −0.23672549 | 0.008838847 |
| Trabd | 428.7785606 | 0.206983161 | 0.81770812 | 0.693625145 | 0.026975323 |
| Cdc42ep1 | 125.1954183 | −0.078511793 | 0.812228257 | 0.372161391 | 0.007554632 |
| Mknk2 | 427.8455383 | −0.048961505 | 0.808081014 | 0.517224158 | 0.040824685 |
| Gabarapl1 | 159.4988978 | −0.386049463 | 0.806750452 | 0.053744159 | 0.030099425 |
| Cd177 | 190.3525363 | −0.138960558 | 0.804304905 | 0.04765301 | 0.014124953 |
| Rnf213 | 428.6160414 | 0.149077594 | 0.803138423 | 0.427420259 | 0.03015489 |
| Tmem54 | 1,556.331592 | −0.080430896 | 0.80138319 | 0.144172177 | 0.047677122 |
| Gpd1 | 958.6161025 | 0.078939306 | 0.799998057 | 0.763680793 | 0.01572532 |
| Fos | 1,012.46335 | −0.502899921 | 0.799891127 | 0.033073799 | 0.037216162 |
| Nans | 520.9687845 | −0.032188415 | 0.797706676 | 0.510260701 | 0.022532429 |
| Cox15 | 354.4671488 | −0.078967981 | 0.78723932 | 0.294362139 | 0.04652294 |
| Eif2s1 | 870.3708021 | 0.270712235 | 0.780815701 | 0.613135834 | 0.002927944 |
| 1110004F10Rik | 276.258603 | 0.136545142 | 0.779370563 | 0.234205413 | 0.026115963 |
| Irgm1 | 436.9936141 | 0.022953476 | 0.767844473 | 0.032999897 | 0.043537383 |
| Ctdsp2 | 354.6803291 | −0.134851284 | 0.762115889 | 0.349000016 | 0.040534857 |
| Nrm | 264.3525319 | −0.107569376 | 0.758593624 | 0.364641645 | 0.027205895 |
| Rnf213 | 475.7625362 | 0.153283375 | 0.755046347 | 0.288497253 | 0.029001776 |
| Lonp1 | 225.368496 | −0.082415528 | 0.753098706 | 0.390154269 | 0.039925285 |
| Dut | 293.3013282 | 0.1478784 | 0.749990752 | 0.521535105 | 0.038543958 |
| Prmt1 | 184.8512311 | −0.068883333 | 0.748625298 | 0.436335165 | 0.049424595 |
| Ubl4 | 377.5721773 | −0.08631798 | 0.74533314 | 0.494452358 | 0.039431507 |
| Ppif | 1,484.442256 | −0.181518144 | 0.736359998 | 0.397556836 | 0.019067053 |
| Rab22a | 346.7216227 | −0.058613386 | 0.73181034 | 0.512294298 | 0.040701397 |
| Samm50 | 981.3431587 | 0.004678751 | 0.729188612 | 0.541250807 | 0.005356581 |
| Rnf213 | 500.3337491 | 0.204938717 | 0.729139015 | 0.284488796 | 0.018276385 |
| Ddah1 | 394.6985054 | −0.201062995 | 0.728563348 | −0.139752555 | 0.006829513 |
| Psmd2 | 1,629.116733 | 0.182903541 | 0.725788525 | 0.639655525 | 0.038941761 |
| Lasp1 | 1,438.808897 | −0.046897057 | 0.724701446 | 0.315574911 | 0.035365309 |
| Leng8 | 398.088658 | −0.147218858 | 0.724057166 | 0.304223185 | 0.012437372 |
| Kcnn4 | 497.9095778 | −0.305017138 | 0.722907244 | 0.230757235 | 0.034070832 |
| Socs3 | 213.3785358 | 0.040385098 | 0.716144074 | −0.019219586 | 0.027892414 |
| Kcnk5 | 229.8222278 | −0.19951165 | 0.715284854 | 0.323019117 | 0.003772795 |
| Scpep1 | 701.0298425 | −0.091097416 | 0.71479258 | 0.33176566 | 0.033927504 |
| Hnrnpa1 | 1,691.089506 | 0.076112414 | 0.714337572 | 0.411684837 | 0.044690053 |
| Fen1 | 207.8829216 | 0.257123807 | 0.713508095 | 0.645303865 | 0.034303371 |
| R3hcc1 | 213.7382317 | −0.409700376 | 0.712688034 | −0.049408817 | 0.021286932 |
| Copg | 891.3751921 | 0.047894604 | 0.710999521 | 0.352746329 | 0.037563249 |
| Slc25a39 | 1,294.016431 | 0.011854634 | 0.710766031 | 0.53880392 | 0.033789153 |
| Aamp | 519.6517943 | 0.073177813 | 0.709429088 | 0.385657771 | 0.036701433 |
| Fbln1 | 527.1451112 | −0.293228548 | 0.706770494 | 0.094155104 | 0.018277207 |
| Aldh7a1 | 199.6616417 | −0.256083301 | 0.706430978 | 0.294917092 | 0.02794333 |
| Tbcd | 453.2641058 | −0.057241827 | 0.705437646 | 0.360166169 | 0.038873727 |
| Cops7a | 1,149.741735 | −0.111092425 | 0.703262746 | 0.440175499 | 0.024442374 |
| Impa2 | 249.4182071 | 0.077742565 | 0.700950722 | 0.474141683 | 0.040908808 |
| Racgap1 | 528.3065635 | −0.030730307 | 0.700852217 | 0.466463015 | 0.021909785 |
| Etv3 | 485.2732922 | −0.106407706 | 0.699935981 | 0.299574569 | 0.006419214 |
| Sema4a | 760.1822621 | −0.150575351 | 0.699101852 | 0.05214455 | 0.030507252 |
| Pmpcb | 518.7810892 | 0.187429534 | 0.698939592 | 0.287910919 | 0.027711762 |
| H2-Q7 | 1,971.686228 | −0.033540965 | 0.690903317 | 0.025694482 | 0.004849169 |
| Hectd3 | 430.9718012 | −0.109229763 | 0.690883598 | 0.146149911 | 0.024814191 |
| Srp9 | 499.0198435 | 0.229177668 | 0.688200498 | 0.334171806 | 0.044594811 |
| Rnf5 | 886.4360861 | −0.01573157 | 0.682411901 | 0.385386178 | 0.039489694 |
| Tmem159 | 224.6584435 | −0.054349086 | 0.680913795 | 0.487061289 | 0.007390561 |
| Mmab | 188.3914654 | −0.32132729 | 0.679072169 | 0.182068085 | 0.003570257 |
| B4galt1 | 484.955027 | 0.111191495 | 0.675134042 | 0.303147065 | 0.020525324 |
| Mcm7 | 809.3391936 | 0.002188805 | 0.673962008 | 0.434255828 | 0.013275378 |
| 2810004N23Rik | 186.6069081 | 0.018845814 | 0.670973219 | 0.440425515 | 0.034863951 |
| Gm9853 | 350.343605 | −0.076880902 | 0.670785426 | 0.554783111 | 0.025135628 |
| Rnf213 | 637.7170264 | 0.135818983 | 0.668351521 | 0.286844412 | 0.023099091 |
| Mlkl | 151.8417788 | 0.039600514 | 0.667837795 | −0.187098566 | 0.014360993 |
| Dohh | 79.93536678 | −0.059310754 | 0.66723907 | 0.342580879 | 0.02297723 |
| Stk40 | 394.1437763 | 0.003021076 | 0.666463647 | 0.292515622 | 0.043656577 |
| Mll2 | 87.77335576 | 0.109687448 | 0.664809663 | 0.575329282 | 0.03633027 |
| Got1 | 1,210.059853 | 0.029434154 | 0.664012926 | 0.367844574 | 0.016071957 |
| Golm1 | 1,026.300637 | 0.061833541 | 0.663179322 | 0.497176818 | 0.009197693 |
| Chmp1a | 1,101.3044 | −0.165412399 | 0.661201119 | 0.112992356 | 0.037081577 |
| Casp7 | 1,343.017393 | −0.101534238 | 0.660842228 | 0.153197299 | 0.036818891 |
| Snhg1 | 107.8751458 | 0.342038891 | 0.659426609 | 0.728286178 | 0.015673423 |
| Cnbp | 1,197.039079 | 0.090710185 | 0.659226362 | 0.442899127 | 0.033526808 |
| Rnf213 | 254.0058019 | 0.087598008 | 0.656994567 | 0.304157037 | 0.0423432 |
| Rnps1 | 765.7892573 | −0.206239926 | 0.652304911 | 0.313675873 | 0.011386319 |
| Ttc4 | 296.9581144 | −0.021245529 | 0.649211001 | 0.354772919 | 0.045727215 |
| Dbnl | 366.8144839 | −0.044944534 | 0.646592237 | 0.273874227 | 0.043625112 |
| Mcm2 | 594.1872909 | −0.017589881 | 0.646189203 | 0.592592361 | 0.007915774 |
| Rnps1 | 684.9402054 | −0.208401579 | 0.645659064 | 0.294948937 | 0.013693934 |
| Unc5cl | 252.9362913 | 0.082447253 | 0.644983456 | 0.451801592 | 0.012956353 |
| H2-K1 | 1,368.187133 | −0.018428994 | 0.643732116 | 0.024828837 | 0.012258779 |
| Kras | 629.1931375 | 0.088494776 | 0.643056201 | 0.303088339 | 0.02051213 |
| Isyna1 | 603.2600294 | −0.177302398 | 0.638367971 | 0.577992681 | 0.036398881 |
| Noxa1 | 256.9485434 | −0.206569569 | 0.637308355 | 0.504169302 | 0.016249313 |
| Shmt2 | 438.1671668 | −0.091060342 | 0.636971684 | 0.407756037 | 0.026514947 |
| Nop58 | 303.554552 | 0.100885414 | 0.635744513 | 0.587497909 | 0.000488 |
| Atl3 | 355.8091855 | 0.189316164 | 0.633175638 | 0.638234261 | 0.039815571 |
| D17H6S56E-5 | 1,106.335903 | −0.069207838 | 0.632635585 | 0.508384111 | 0.022594284 |
| Usp19 | 373.2959307 | 0.057641983 | 0.63261347 | 0.57363622 | 0.038364891 |
| Vars | 639.8954975 | −0.06370674 | 0.631092949 | 0.364381912 | 0.006650937 |
| Acad8 | 278.8564257 | −0.057436283 | 0.630919079 | 0.41928084 | 0.048064907 |
| Lmnb1 | 411.4337005 | 0.05995137 | 0.630150495 | 0.562437006 | 0.021640015 |
| Acsf2 | 195.8420184 | −0.264737513 | 0.628705114 | −0.180684276 | 0.033104218 |
| Snx17 | 468.1081772 | −0.053003167 | 0.62809884 | 0.462831237 | 0.008606001 |
| Rad54l | 259.2303557 | 0.173002395 | 0.627591526 | 0.595451281 | 0.049746761 |
| Slc35a4 | 333.2359761 | 0.077048315 | 0.623074016 | 0.489892598 | 0.000612 |
| Acsf2 | 197.7962242 | −0.225709506 | 0.622492477 | −0.121141893 | 0.043280743 |
| Psap | 2,745.853589 | −0.090384654 | 0.620567115 | 0.274588744 | 0.046600158 |
| Ctdsp2 | 916.699924 | −0.196246003 | 0.620492826 | 0.119113471 | 0.003716511 |
| Nup62 | 619.3168435 | −0.043234174 | 0.620223638 | 0.461350352 | 0.019059225 |
| Abcb6 | 203.458556 | 0.008991337 | 0.619759588 | 0.484789377 | 0.014386861 |
| Ppp2r4 | 365.9775948 | 0.068514234 | 0.619527862 | 0.492320706 | 0.016769666 |
| Htra2 | 277.2271786 | −0.012135448 | 0.619131166 | 0.334845608 | 0.021588638 |
| Aldh9a1 | 2,201.734416 | −0.124961377 | 0.617004325 | 0.312163687 | 0.015026998 |
| Rnf213 | 322.0485884 | 0.043709194 | 0.616805024 | 0.183848045 | 0.030731796 |
| Erbb3 | 1,475.562755 | −0.136046921 | 0.613193891 | 0.175663551 | 0.043574987 |
| Nucb2 | 106.0905564 | 0.083630076 | 0.611634017 | −0.03008157 | 0.023765684 |
| Eno1 | 2,254.463021 | 0.182980226 | 0.607740308 | 0.509971208 | 0.029883759 |
| Med18 | 88.44012961 | 0.030032512 | 0.607380112 | 0.570561665 | 0.006240166 |
| Manbal | 572.2236175 | −0.137865806 | 0.607070424 | 0.077584808 | 0.045687528 |
| Ehd4 | 260.0985443 | 0.03585688 | 0.60636848 | 0.349965453 | 0.02878 |
| Stat1 | 443.4290062 | 0.113087296 | 0.604897523 | 0.251959655 | 0.002445993 |
| Stat3 | 688.5619236 | 0.076477235 | 0.604874347 | −0.071808598 | 0.007663359 |
| Hyou1 | 882.3158196 | 0.324694142 | 0.604147114 | 0.698535354 | 0.005343729 |
| Cdt1 | 375.4455878 | −0.114929499 | 0.603348393 | 0.357730751 | 0.030283926 |
| Dusp5 | 388.669155 | −0.5278057 | 0.602567275 | −0.275468382 | 0.027346573 |
| Grhpr | 411.0792627 | −0.051414792 | 0.602434861 | 0.278468251 | 0.015321714 |
| Srp68 | 446.876007 | −0.01995338 | 0.600220985 | 0.5080635 | 0.011879957 |
| Dhdds | 357.7781692 | 0.292109256 | 0.600124859 | 0.871087434 | 0.029011737 |
| Abhd14b | 356.2234469 | −0.06764228 | 0.599412258 | 0.387927264 | 0.040367048 |
| Tmem50a | 854.0278889 | −0.008021259 | 0.599229446 | 0.27154395 | 0.039130855 |
| Hnrnpa1 | 1,302.407765 | 0.002947635 | 0.595822632 | 0.25907306 | 0.044027028 |
| Hk2 | 243.0198342 | −0.046563933 | 0.595064847 | 0.377234187 | 0.031246932 |
| Lysmd2 | 89.20163748 | 0.150543621 | 0.594789486 | 0.363204035 | 0.031877078 |
| Afg3l1 | 260.7971973 | 0.042963439 | 0.593257537 | 0.313427813 | 0.010897723 |
| Plekhb2 | 1,513.949932 | 0.000346045 | 0.592187847 | 0.373997045 | 0.02391808 |
| Pttg1ip | 1,025.152765 | −0.19922309 | 0.591688742 | 0.348753223 | 0.028336477 |
| Suv420h2 | 553.5969436 | 0.021210265 | 0.590706624 | 0.243571094 | 0.004588532 |
| Slc39a7 | 481.7370152 | 0.063900487 | 0.589637545 | 0.327927016 | 0.029739369 |
| Exph5 | 128.6397063 | −0.032969069 | 0.589389164 | 0.268808852 | 0.027005604 |
| BC031781 | 227.5235025 | −0.137178336 | 0.587145375 | 0.390982787 | 0.02438911 |
| Smu1 | 515.2426612 | 0.080531536 | 0.586560034 | 0.467572388 | 0.01434829 |
| Tcof1 | 329.2185576 | −0.012537114 | 0.585094494 | 0.46523729 | 0.023529582 |
Log2 (FC) values are shown; P values are calculated by Student's t test. CH, Clostridium histolyticum.
B. adolescentis Exacerbates Autoimmune Arthritis in a Mouse Model.
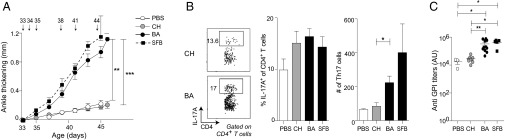
Elevated Th17 cell responses have been associated with autoimmune/inflammatory disease in both mice and humans (19, 20). For example, SFB colonization promotes disease in the K/BxN mouse model of RA, in part by inducing SI-LP Th17 cells that emigrate from the gut to the spleen, where they promote production of autoantibodies against glucose-6-phosphate isomerase (24, 41). Such autoantibodies in and of themselves can induce arthritis and, after they reach high levels, require no further input from Th17 cells. To assess the role of symbiont-induced Th17 populations in autoimmune arthritis, we gavaged SPF K/BxN mice with B. adolescentis, C. histolyticum, SFB, or PBS. Similar to SFB, B. adolescentis, but not PBS or the control microbe C. histolyticum, drove more severe arthritis, as evidenced by increased joint thickening (Fig. 6A). Heightened disease in B. adolescentis-treated mice was associated with increased numbers (although not frequencies) of SI-LP Th17 cells (Fig. 6B) and elevated titers of antiglucose-6-phosphate isomerase autoantibodies (Fig. 6C). Thus, B. adolescentis drives gut-distal Th17-cell–associated disease progression.
Fig. 6.
B. adolescentis (BA) exacerbates K/BxN arthritis. K/BxN mice were pretreated with antibiotics from 22 to 32 d of age and subsequently gavaged with PBS, C. histolyticum (CH), or BA over 12–13 d. Arrows indicate gavage time points. (A) Ankle-thickening values over the course of bacterial gavage. (B, Left) Representative flow cytometric plot of IL-17A production from SI-LP CD4+ T cells 11–13 d after the initial bacterial gavage. Summary data of (B, Center) frequencies and (B, Right) numbers of SI-LP Th17 cells at the same time point. (C) Antiglucose-6-phosphate isomerase titers 11–13 d after initial bacterial gavage. Each symbol represents one mouse [n = 3 (PBS), 8 (CH), 11 (BA), and 2–5 (SFB)]. Data for CH and BA are pooled from two to three independent experiments. Mean ± SEM. GPI, glucose-6-phosphate isomerase. *P < 0.05 (Kruskal–Wallis test with Dunn’s multiple comparisons test); **P < 0.01 (Kruskal–Wallis test with Dunn’s multiple comparisons test); ***P < 0.001; (Kruskal–Wallis test with Dunn’s multiple comparisons test).
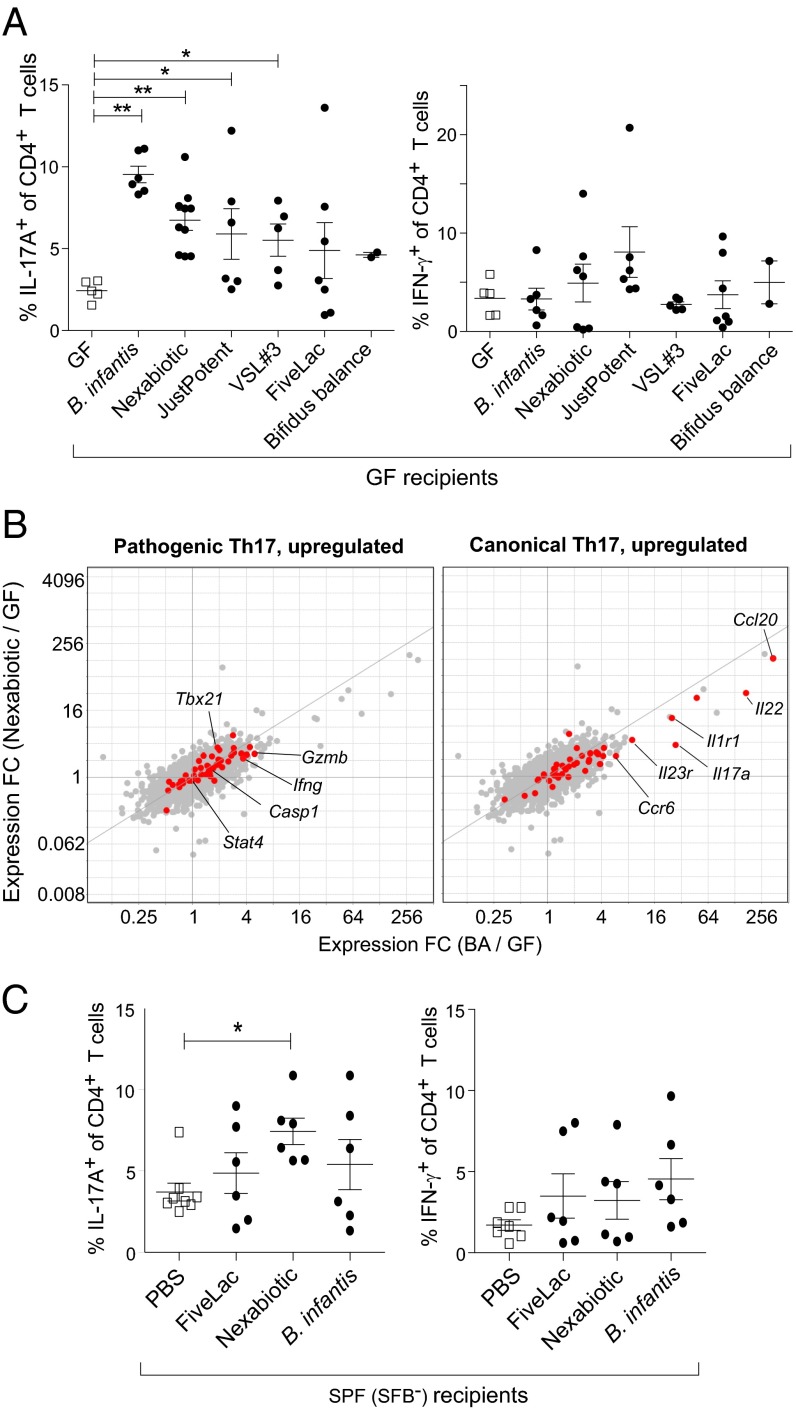
Probiotics Containing Bifidobacterium Species Can Augment Intestinal Th17 Cell Compartments.
The genus Bifidobacterium, of which B. adolescentis is a member, is a common component of healthy infant and adult microbiotas and is often included in probiotic formulations because of its purported benefits in promoting gastrointestinal health. The question arose whether such probiotic preparations share B. adolescentis’ ability to induce intestinal Th17 cells. We evaluated six probiotic formulations available online, all but two of which (FiveLac and Bifidobacterium infantis, which both contain one Bifidobacterium strain) contained two or more Bifidobacterium species (Table S7). When introduced into GF mice, four of the preparations significantly induced Th17 but not Th1 cell accumulation in the SI-LP (Fig. 7A) comparing values from GF mice. Similar to what was seen with B. adolescentis, gavage of Nexabiotic highly induced canonical but not pathogenic signature genes in whole SI-LP tissue. We also tested three of the probiotic preparations in SPF-housed mice, given their complex, more natural microbiotas. One of two Th17-inducing probiotic mixes reproducibly induced the accumulation of SI-LP Th17 cells in SPF mice without significantly altering Th1 cell frequencies (Fig. 7B). Thus, it seems that Th17 cell induction in the gut may be a feature widely shared by probiotics.
Table S7.
Bacterial composition of probiotic formulations used in this study
| Nexabiotic | JustPotent | B. infantis |
| Bacillus coagulans | Bifidobacterium bifidium | B. infantis 35624 |
| Bacillus subtilis | Bifidobacterium lactis | |
| Bifidobacterium animalis lactis | Bifidobacterium longum | |
| Bifidobacterium bifidum | Lactobacillus acidophilus | |
| Bifidobacterium breve | Lactobacillus bulgaricus | |
| Bifidobacterium lactis | Lactobacillus plantarum | |
| Bifidobacterium longum | Lactobacillus rhamnosus | |
| Enterococcus faecium | Lactobacillus salivarus | |
| Lactobacillus acidophilus | ||
| Lactobacillus brevis | ||
| Lactobacillus casei | ||
| Lactobacillus delbrueckii LE | ||
| Lactobacillus gasseri | ||
| Lactobacillus helveticus | ||
| Lactobacillus lacris | ||
| Lactobacillus paracasei | ||
| Lactobacillus plantarum | ||
| Lactobacillus plantarum LM | ||
| Lactobacillus rhamnosus | ||
| Lactobacillus rhamnosus LB3 | ||
| Lactobacillus salivarius | ||
| Saccharomyces boulardii | ||
| Streptococcus thermophilus | ||
| VSL#3 | Bifidus Balance | FiveLac |
| Bifidobacterium breve | Bifidobacterium breve BR03 | Bacillus coagulans |
| B. infantis | Bifidobacterium bifidum BB01 | Bacillus subtilis |
| Bifidobacterium longum | Bifidobacterium lactis BI-04 | Bifidobacterium longum |
| Lactobacillus acidophilus | Bifidobacterium longum BB536 | Enterococcus faecalis |
| Lactobacillus bulgaricus | Lactobacillus acidophilus | |
| Lactobacillus paracasei | ||
| Lactobacillus plantarum | ||
| Streptococcus thermophilus |
Full names of probiotics can be found in SI Materials and Methods.
Fig. 7.
Some probiotic formulations containing bifidobacterial species also elicit Th17 populations in the SI-LP. (A) Frequencies of SI-LP (Left) Th17 or (Right) Th1 cells in GF mice colonized with the indicated probiotic mixes. (B) Th17 cell phenotype. Fold change (FC)/FC plots comparing transcripts induced by B. adolescentis (BA) vs. the probiotic mix, Nexabiotic, in SI-LP CD4+ T cells. Symbols in red and labels are as per Fig. 2G (n = 2–3 per group). (C) Frequencies of SI-LP (Left) Th17 and (Right) Th1 cells in SPF (SFB−) mice gavaged with PBS or one of three other probiotic preparations in A. Full names and corresponding abbreviations of probiotics can be found in SI Materials and Methods. Each symbol represents one mouse. Data are pooled from two to three independent experiments. Mean ± SEM. *P < 0.05 (Mann–Whitney u test); **P < 0.01 (Mann–Whitney u test).
Discussion
We have identified individual symbiont microbes from the human gut that can induce robust Th17 populations in the murine intestine. Interestingly, the mechanisms used by the most potent inducer, B. adolescentis, differed from those of the well-known Th17-promoting mouse symbiont SFB. B. adolescentis exacerbated autoimmune arthritis, arguing for its pathological relevance. These findings raise several interesting issues meriting additional elaboration.
First, bifidobacteria seem to be common inducers of intestinal Th17 cells. A complex microbial community was insufficient for generating a robust population of Th17 cells in the gut of SPF mice lacking SFB (9, 10) and gnotobiotic mice colonized long term with human fecal contents (42). Although SFB has been detected in multiple vertebrate species (43), there exists only sparse evidence of a related microbe colonizing humans (43–46). A recent study showed that a consortium of 20 symbionts from the feces of an IBD patient could induce Th17 cells in mice but failed to identify the active microbes in healthy people (11).
Although B. adolescentis was 1 of 3 microbes (of a total of 39) in our screen that was able to robustly induce intestinal Th17 cells, we think it highly plausible that many other symbiont species, including other bifidobacteria, can act singly or in concert with other microbes to promote Th17 cell accumulation in the human gut. Given the tremendous diversity of the human microbiota (47), our screen was perforce limited to testing a fraction of it. Even with this restricted scope, we succeeded in identifying three bacterial species spanning distinct phyla that could induce intestinal Th17 cells to a degree comparable with that of SFB. Wider sampling would almost certainly unveil more microbes with this property. Indeed, most of the bifidobacteria-containing probiotics that we tested potently expanded Th17 cells, arguing that this property might be a relatively common bifidobacterial trait. Relatedly, as exemplified by SFB in rodents, some intestinal symbionts show host specificity, consequent to millennia of coevolution (11, 42). By design, our screen elides symbiont strains capable of inducing Th17 cells in humans but unable to do so in mice. Moreover, some symbionts might exert their effects on the host immune system only in the presence of other microbes, a restriction that might apply to certain bifidobacterial species, which could account for the prevalence of Th17 induction by probiotic mixes. Hence the results from our screen likely underestimate the true number and diversity of Th17-promoting human symbionts.
Bifidobacteria are ubiquitous symbionts, well-represented in the gut microbiota of healthy humans across age and geography (47). In infants, they are among the first colonizers of the intestine, and their abundance serves as a biomarker of a healthy microbiota (47, 48). With age, the frequency of bifidobacteria in the gut wanes, and the dominant species change, although members of the genus remain a substantial component in the adult (48, 49). A metagenomic sequencing study of gut microbes from 124 adults identified several Bifidobacterium strains as dominant symbionts, with B. adolescentis exceeding 10% in relative abundance in two-thirds of the individuals (50). Thus, B. adolescentis, along with other bifidobacteria, is well-poised to be a universal Th17-inducing symbiont in humans throughout ontogeny into adulthood.
Second, B. adolescentis induced Th17 cells by a mechanism that clearly diverged from that of SFB. Overall, SFB triggered more pronounced transcriptional changes than those elicited by B. adolescentis, C. histolyticum, or the vast majority of human symbionts that we have tested. A prosaic explanation for this divergence is that SFB is a murine symbiont and thus better adapted to interact with the mouse host, thereby inducing more profound gene expression changes. Related or not to the host species, B. adolescentis was located in considerable quantities in the intestinal lumen in close association with the epithelium, whereas SFB was found almost exclusively attached to the ileal surface. In addition, the association of SFB seemed tighter, actually penetrating the epithelium in places. SFB and B. adolescentis seemed to mobilize distinct cell types and transcriptional programs to induce Th17 responses. The transcriptional changes effected by B. adolescentis on S-IECs were relatively subtle and distinct from those on whole ileal tissue, where the up-regulated RNAs were enriched in non–S-IEC–related pathways, such as muscle contraction and interactions with the ECM. These pathways could potentially regulate the activity of mechanosensitive integrins and cytokines (e.g., TGF-β) relevant to Th17 cell differentiation and trafficking. Interestingly, DNA from SFB-like microbes was recently enriched in the gut of human IBD patients, associated with cavernous fistulous tracts running between muscle bundles (46). The enrichment for muscle-related pathways in whole-tissue SI-LP preparations from mice colonized with B. adolescentis hints at a more general relationship between Th17-inducing microbes and intestinal muscle tissue. In contrast, S-IECs seemed to be critical drivers of the ileal transcriptional response to SFB, in line with previous studies (10, 11). Gut microbes also produce metabolites that can access the stroma and immunocytes in the LP without directly interacting with the gut epithelium (51), and intestinal antigen presenting cells can extend their dendrites into the lumen to sample bacteria directly (52). These mechanisms, in particular those accomplished by intestinal immunocytes, might explain the relatively modest impact of B. adolescentis on the transcriptomes of S-IECs and the ileum (where leukocytes are vastly outnumbered by nonhematopoietic cells).
Third, Th17 cells seem to have a yin–yang role in human health. Mice devoid of IL-17 signaling manifest alterations in their microbiotas and suffer from increased intestinal permeability and bacterial translocation to systemic sites after infectious insults of the gut (15, 16, 53). Additionally, loss of Th17 populations during infections by either simian virus or HIV has been associated with intestinal dysbiosis, systemic microbial translocation, and disease progression (53–55). Moreover, SFB confers heterologous protection from the murine enteropathogen C. rodentium (10). Hence, symbiont-driven intestinal Th17 cells seem to bolster host mucosal defenses via various mechanisms, including the augmentation of barrier integrity, the provision of cross-protective defenses against pathogens during early stages of infection, and sculpting of the gut microbiota.
However, microbiota-dependent Th17 responses have been implicated in IBD and other extraintestinal autoimmune disorders, including psoriasis, multiple sclerosis, and RA. Elevated Th17 frequencies have been observed in the intestinal mucosa of IBD patients (20), and increased IL-17A titers can be detected in the synovial fluid of people afflicted with RA (56, 57). Variants in genes important for Th17 cell differentiation and function (e.g., IL23R and CCR6) have also been associated with the severity of these diseases (58–60). Furthermore, dysbiosis is concomitant with new-onset, treatment-naïve IBD and RA, implying a potential etiological role for the intestinal microbiota (21, 22). Of note, the relative abundances of B. adolescentis and several Bifidobacterium species were profoundly altered in the microbiotas of pediatric (22) and adult IBD subjects (Fig. S3) (50), albeit in opposite directions, with an enrichment of bifidobacteria in the microbiotas of the latter cohort of patients. In support of a pathogenic role for symbiont-driven Th17 cells in inflammatory diseases, we observed an exacerbation of spontaneous autoimmune arthritis in mice gavaged with B. adolescentis but not with the non–Th17-inducing microbe, C. histolyticum.
Fig. S3.
BA and related species are enriched in the microbiotas of IBD patients. Comparison of the microbiotas of 85 healthy vs. 39 IBD subjects in the MetaHIT database using LEfSe as outlined in SI Materials and Methods. Microbial clades with the largest effect sizes (>3.5) with a false discovery rate q < 0.05 (Kruskal–Wallis test with Benjamini–Hochberg correction) are shown. (Left) Negative and (Right) positive values correspond to healthy controls (HCs) and IBD patients, respectively. BA and other bifidobacterial species are represented as white bars.
In this context, how should one interpret the induction of Th17 cells by several probiotic formulations in widespread use as ad hoc nutritional supplements? Our in vivo data are in concert with findings on cultured human immunocytes (61). One interpretation is that this induction is part of their favorable action in the setting of gastrointestinal infection and dysbiosis (62), where Th17 cells elicited by probiotics might evince antiinfectious benefits. However, one might also consider that these Th17 cells contribute in an unrecognized manner to the frequency or exacerbation of chronic inflammatory diseases linked to Th17 responses, such as RA or multiple sclerosis.
Materials and Methods
Mice.
Unless otherwise stated, SPF C57BL/6J (B6) mice were obtained from the Jackson Laboratory and housed under SPF conditions at Harvard Medical School. GF mice were bred and maintained in sterile isolators at Harvard Medical School. Manipulations of mice are detailed in SI Materials and Methods. Experiments were conducted according to the guidelines of the Harvard Medical School Institutional Animal Care and Use Committee.
Bacteria and Probiotics.
Bacteria were cultured as previously described (8). Bacteria and probiotics used are detailed in SI Materials and Methods.
Isolation of S-IECs, Intraepithelial Lymphocytes, and Intestinal LP Leukocytes.
S-IECs, intraepithelial lymphocytes, and leukocytes were processed as previously described (8) and are further detailed in SI Materials and Methods.
Antibodies and Flow Cytometry.
Single-cell suspensions from intestinal tissues and lymphoid organs were stained with antibodies for flow cytometry and analyzed as detailed in SI Materials and Methods.
Antigen Presentation Assays.
Antigen presentation assays are detailed in SI Materials and Methods.
Measurement of Bacterial Titers.
Bacterial titers from monocolonized mice were measured as detailed in SI Materials and Methods.
Histopathology.
Histopathology of intestinal sections was scored as detailed in SI Materials and Methods.
FISH and SEM.
FISH and SEM were performed on intestinal sections as previously described (63, 64) and are detailed in SI Materials and Methods.
K/BxN Murine Arthritis Model and ELISA.
Three-week-old K/BxN mice of both sexes were pretreated with antibiotics [1 g/L ampicillin (Sigma), 1 g/L neomycin (Fisher Scientific), 1 g/L metronidazole (Sigma), 0.5 g/L vancomycin (Amresco)] for 10 d, rested for 1 d, and subsequently gavaged with PBS or bacteria (108 cfu of C. histolyticum or B. adolescentis) for 3 consecutive days and every 3 d thereafter until the time of euthanasia. Ankle thickness was measured with a caliper (J15 Blet Micrometer) as previously described (24). All mice were housed at the SPF animal facility at the University of Arizona. Antiglucose-6-phosphate isomerase antibody titers were measured as previously described and further detailed in SI Materials and Methods. (41).
Gene Expression Profiling and Analysis.
Microarray or RNA sequencing analysis was performed on whole ileal tissue, S-IECs, or SI-LP CD4+ T cells of monocolonized mice as detailed in SI Materials and Methods.
Comparison of the Microbiotas of Healthy Vs. IBD Subjects.
Publicly available metagenomic profiling reads from 124 adults from the MetaHIT database (50) were analyzed as detailed in SI Materials and Methods.
Statistics.
Unless otherwise stated, significance was assessed using the Mann–Whitney u test or the Kruskal–Wallis test with Dunn’s multiple comparisons test (Prism 6; Graph-Pad). P values were deemed significant if less than 0.05. To compare ankle thickening, the area under the curve was calculated for each mouse followed by the Mann–Whitney u test between groups. Mean ± SEM was routinely used.
SI Materials and Methods
Mice.
Gnotobiotic mice were generated by gavaging 4-wk-old GF mice once with 108–109 cfu designated bacteria and housing them in sterile isolators for 2–3 wk before euthanasia. SPF B6 females were gavaged starting at 4 wk of age with 108–109 cfu bacteria every other day for 2 wk. For probiotic experiments, the contents of one probiotic capsule or sachet were resuspended in 3–5 mL sterile PBS, and each mouse was gavaged with 200 μL bacterial suspension, which generally corresponded to 109 cfu bacteria based on plating of the inocula, either once (for gnotobiotic mice) or every other day for 2 wk (for SPF mice). Both male and female mice were used for gnotobiotic experiments, whereas only females were used for SPF experiments.
Bacteria and Probiotics.
Bacteria were obtained from the American Type Culture Collection, Biological and Emerging Infections Resources, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, or laboratory collections (D. L. Kasper and A. Onderdonk, Brigham and Women’s Hospital Clinical Laboratories, Boston). The following probiotics and their corresponding abbreviations were used in this study: Nexabiotic 23 Probiotics (Nexabiotic; ProVita Labs, Speedway, IN), Align Probiotic Supplement Capsules (Bifidobacterium infantis; Align, Cincinnati, OH), JustPotent Probiotic Supplement (JustPotent, Kent, WA), Bifidus Balance + Fos (Bifidus Balance; Jarrow Formulas, Los Angeles), FiveLac (Global Health Trax, Vista, CA), and VSL#3 (VSL Pharmaceuticals Gaithersburg, MD). Bacterial composition of probiotics is listed in Table S7.
Isolation of S-IECs, Intraepithelial Lymphocytes, and Intestinal LP Leukocytes.
Intestines were cut lengthwise into short segments and shaken in RPMI-1640 (Corning Cellgro) containing 1 mM DTT, 2 mM EDTA, and 2% (vol/vol) FCS for 15 min at 37 °C to remove the epithelial layer. S-IECs were enriched by collecting the flow through and washing the cells one to two times with RPMI-1640 containing 1% FCS. To harvest intraepithelial lymphocytes (IELs) from the epithelial layer, cells were further spun through a 40:70 Percoll gradient, and IELs were isolated from the interphase. To obtain LP leukocytes, the tissue leftover from epithelial stripping was minced and digested in RPMI-1640 containing 1.5 mg/mL collagenase II (Gibco), 50 μg/mL DNase I (Sigma), and 1% FCS for 40–45 min at 37 °C. Digested tissue was washed and filtered at least twice to obtain a single-cell suspension.
Antibodies and Flow Cytometry.
Cells were stained with antibodies against CD4 (GK1.5), CD8α (53-6.7), TCRβ (H57-597), TCRγδ (UC7-13D5), CD90.2 (30-H12), CD45 (30-F11), CD11b (M1/70), CD11c (N418), CD19 (6D5), CD103 (2E7), B220 (RA3-6B2), Ly6C (HK1.4), F4/80 (BM8), PDCA-1 (927), GL7 (GL7), and EpCAM (G8.8) from Biolegend and Fas (Jo2) and TCRVβ14 (14-2) from BD Biosciences. To detect intracellular transcription factors or IgA, we fixed and permeabilized cells using the Intracellular Fixation & Permeabilization Buffer Set from eBioscience and stained them with anti-Foxp3 (FJK-16s), anti-RORγt (AFKJS-9), anti-Helios (22F6), and anti-IgA (mA-6E1) antibodies (all from eBioscience). To assess cytokine production, we restimulated cells with 10 ng/mL phorbol 12-myristate 13-acetate (Sigma) and 1 μM ionomycin (Sigma) in the presence of GolgiPlug (BD Biosciences) for 3.5 h at 37 °C. Cells were then stained for surface markers as described above, fixed and permeabilized using the BD Cytofix/Cytoperm Kit per the manufacturer’s instructions, and stained with antibodies against IFN-γ (XMG1.2), IL-10 (JES5-16E3), IL-17A (TC11-18H10.1), and IL-22 (poly5164; all from Biolegend). Cell viability was determined using the LIVE/DEAD Fixable Dead Cell Stain Kit (Invitrogen). Samples were acquired on an LSR II (BD Biosciences) and analyzed using FlowJo (Tree Star).
Antigen Presentation Assays.
Small-intestinal tissue was processed as described, and SI-LP CD4+ T cells were sorted using a MoFlo Sorter (Beckman Coulter). Dendritic cells (DCs) were enriched from splenocytes using anti-CD11c magnetic beads and two rounds of selection on the autoMACS Pro Separator (Miltenyi Biotec). T cells and DCs were seeded in a 96-well round-bottom plate at a 1:10 ratio with 4,000–5,000 T cells per well in the presence of 20 U/mL IL-2 (Proleukin; Chiron) for 20–24 h at 37 °C. GolgiPlug was added during the last 4 h of culture. In some cases, bacterial lysates, prepared by resuspending bacterial monocultures in PBS and autoclaving at 121 °C for 20 min, were added to wells at a final concentration of 250 μg/mL. To obtain SFB lysates, we collected stool pellets from SFB-monocolonized mice, homogenized them in PBS, and filtered them through a 40-μM strainer before autoclaving. Where indicated, blocking antibody to MHC-II (M5/114.15.2; Biolegend) was added at a final concentration of 2.5 μg/mL. At the end of the incubation, cells were stained for surface markers and intracellular cytokines as described above.
Measurement of Bacterial Titers.
Bacterial titers from monocolonized mice were assessed by plating stool or tissue homogenates and luminal washes in serial dilutions on Brucella blood agar plates (BD Biosciences). Briefly, intestinal segments were flushed with 1 mL sterile PBS, and the flow through was collected as luminal washes for quantification. A 3- to 5-mm segment of the washed tissue was weighed, transferred into 1 mL sterile PBS, and homogenized using 3.2-mm (diameter) metal beads (Biospec). Bacterial colonies were counted after 48 h of incubation in an anaerobic chamber.
Histopathology.
Sections of 3–5 mm of the ileum or colon were fixed for at least 48 h in Bouin’s solution (Sigma) before embedding in paraffin and sectioning for H&E staining. The degree of inflammation in the tissues was scored blind by two investigators according to the following criteria: zero, normal intact structure; one, mild inflammation with intact structure, two, infiltration of leukocytes with some damage to structure; three, severe inflammation accompanied by complete loss of structure; four, necrosis of the tissue.
FISH.
Intestinal tissues were fixed in Carnoy’s solution (Electron’s Microscopy Sciences), embedded in paraffin, sectioned, and stained for bacteria. Briefly, sections were deparaffinized using EZ-DeWax Solution (BioGenex) and washed successively with 100% ethanol, PBS, and hybridization buffer (20 mM Tris⋅HCl, 0.9 M NaCl, 0.01% SDS, pH 7.4). Slides were then incubated overnight at 50 °C with 1 μM Cy5-conjugated probe (5′-GCTGCCTCCCGTAGGAGT-3′) directed toward the 16S rDNA of all bacteria, washed three times with hybridization buffer and once with PBS, and mounted using Vectashield Antifade Mounting Medium with DAPI (Vector Laboratories). Images were acquired on an Olympus FluoView Confocal Microscope. Quantification of bacterial fluorescence vs. distance from epithelium was performed using the BacSpace package (65).
SEM.
Intestinal tissues were fixed in 2.5% (vol/vol) glutaraldehyde in buffer containing 0.1 M sodium cacodylate (pH 7.2) and subsequently processed by the Electron Microscopy Core at Northeastern University. Images were acquired on a Hitachi S-4800 Field Emission Scanning Electron Microscope.
ELISA.
ELISA plates were coated with recombinant mouse glucose-6-phosphate isomerase at 5 μg/mL, and diluted mouse sera were added. Subsequently, plates were washed, and alkaline-phosphatase (AP)-conjugated anti-mouse IgG antibodies (Jackson ImmunoResearch) were added. After the final wash, AP substrate was added, and titers were quantified as OD values via an ELISA reader. The antibody titers were expressed as arbitrary units, which were calculated from serial dilutions of sample serum and defined as the reciprocal of the highest dilution that gave a background OD value set as 0.1.
Comparison of the Microbiotas of Healthy vs. IBD Subjects.
Publicly available metagenomic profiling reads from 124 adults [85 healthy controls and 39 IBD (ulcerative colitis and Crohn’s disease) patients] were obtained from the MetaHIT database (50) and processed using MetaPhlAn v2.0 (66) to derive the microbiota composition for each individual. Microbiotas between healthy and IBD subjects were then compared using LEfSe (67) to identify the bacterial taxa that were statistically different (Kruskal–Wallis test with Benjamini–Hochberg multiple testing correction) between the two groups of people. Results were expressed as effect sizes for each group of subjects.
Gene Expression Profiling and Analysis.
Microarray or RNA sequencing analysis was performed on whole ileal tissue, S-IECs, or SI-LP CD4+ T cells of monocolonized mice. In brief, whole ileal tissue was cleaned of luminal debris and homogenized in TRIzol (Invitrogen), whereas 20,000–30,000 S-IECs (EpCAM+ CD45−) or 1,000 CD4+ T cells were double-sorted into TRIzol and TCL buffer (Qiagen) containing 1% (vol/vol) 2-mercaptoethanol, respectively, using a MoFlo Sorter (Beckman Coulter) before RNA extraction. Sample processing, hybridization onto Affymetrix Mouse Genome M1.0 ST arrays, and data normalization were performed as previously described (68). Transcripts up-regulated uniquely or in tandem by SFB and/or Bifidobacterium adolescentis (BA) (Fig. 5 and Tables S1–S6) were determined as follows: SFB-specific transcripts − fold change (FC; SFB/GF) > 1.5, FC (SFB/BA) > 1.2, P < 0.05 followed by manual curation on the FC/FC plot to remove transcripts lying on the SFB vs. BA diagonal; BA specific − FC (BA/GF) > 1.2, FC (BA/SFB) and FC (BA/Clostridium histolyticum) > 1.2, P < 0.05; both SFB and BA − FC (SFB/GF) > 1.5 and FC (BA/GF) > 1.5. Analysis of normalized microarray data was conducted using MultiplotStudio v1.5.29 and GENE-E (www.broadinstitute.org/cancer/software/GENE-E/). Pathway enrichment analysis was performed using Enrichr (69), which yields the adjusted P value, z score, and combined enrichment score (integrating both P value and z-score information) for each pathway. RNA sequencing was performed, and data were normalized as previously described (8). Transcript signatures up-regulated in pathogenic (32) or canonical (33) Th17 cells were previously described.
Acknowledgments
We thank Drs. A. Onderdonk and L. Bry for microbial strains; S. Edwards, A. T. Sherpa, J. Ramos, and K. Hattori for help with mice; A. Rhoads, L. Yang, and G. Gopalan for help with gene expression profiling and microbiota analysis; R. Bronson for help with scoring histopathology; and G. Buruzula and C. Araneo for help with cell sorting. This work was funded by a Sponsored Research Agreement from UCB (Union Chimique Belge) and NIH Grant R01AI107117 (to H.-J.J.W.). T.G.T. was supported by an Agency for Science, Technology, and Research Graduate Scholarship Fellowship. E.S. was supported by a fellowship from the Boehringer Ingelheim Fonds. N.G.-Z. was supported by Human Frontier Science Program Fellowship LT00079/2012, European Molecular Biology Organization Fellowship ALTF 251-2011, a Fulbright Award, a United Nations Educational, Scientific, and Cultural Organization L’Oreal National and International Women in Science Award, and the Weizmann Institute of Science National Postdoctoral Award Program for Advancing Women in Science. L.K. was supported by the National Science Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo/ (accession nos. GSE87676, GSE87678, and GSE89261).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617460113/-/DCSupplemental.
References
- 1.Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14(7):685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535(7610):75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 4.Smith PM, Garrett WS. The gut microbiota and mucosal T cells. Front Microbiol. 2011;2:111. doi: 10.3389/fmicb.2011.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 7.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6(220):220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sefik E, et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ⁺ regulatory T cells. Science. 2015;349(6251):993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 10.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atarashi K, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163(2):367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinschek MA, et al. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J Exp Med. 2009;206(3):525–534. doi: 10.1084/jem.20081712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pène J, et al. Chronically inflamed human tissues are infiltrated by highly differentiated Th17 lymphocytes. J Immunol. 2008;180(11):7423–7430. doi: 10.4049/jimmunol.180.11.7423. [DOI] [PubMed] [Google Scholar]
- 15.Kumar P, et al. Intestinal interleukin-17 receptor signaling mediates reciprocal control of the gut microbiota and autoimmune inflammation. Immunity. 2016;44(3):659–671. doi: 10.1016/j.immuni.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxwell JR, et al. Differential roles for interleukin-23 and interleukin-17 in intestinal immunoregulation. Immunity. 2015;43(4):739–750. doi: 10.1016/j.immuni.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 17.Lee JS, et al. Interleukin-23-independent IL-17 production regulates intestinal epithelial permeability. Immunity. 2015;43(4):727–738. doi: 10.1016/j.immuni.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirota K, Ahlfors H, Duarte JH, Stockinger B. Regulation and function of innate and adaptive interleukin-17-producing cells. EMBO Rep. 2012;13(2):113–120. doi: 10.1038/embor.2011.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: From mechanisms to therapeutic testing. Nat Rev Immunol. 2014;14(9):585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanai T, Mikami Y, Sujino T, Hisamatsu T, Hibi T. RORγt-dependent IL-17A-producing cells in the pathogenesis of intestinal inflammation. Mucosal Immunol. 2012;5(3):240–247. doi: 10.1038/mi.2012.6. [DOI] [PubMed] [Google Scholar]
- 21.Scher JU, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu HJ, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohnmacht C, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORγt⁺ T cells. Science. 2015;349(6251):989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- 26.Sano T, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell. 2015;163(2):381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cua DJ, Tato CM. Innate IL-17-producing cells: The sentinels of the immune system. Nat Rev Immunol. 2010;10(7):479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- 28.Bunker JJ, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity. 2015;43(3):541–553. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lécuyer E, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40(4):608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Hirota K, et al. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. 2013;14(4):372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zigmond E, et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 2012;37(6):1076–1090. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 32.Lee Y, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13(10):991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nurieva RI, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29(1):138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goto Y, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40(4):594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510(7503):152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klaasen HL, Koopman JP, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria. FEMS Microbiol Rev. 1992;8(3-4):165–180. doi: 10.1111/j.1574-6968.1992.tb04986.x. [DOI] [PubMed] [Google Scholar]
- 37.Farkas AM, et al. Induction of Th17 cells by segmented filamentous bacteria in the murine intestine. J Immunol Methods. 2015;421:104–111. doi: 10.1016/j.jim.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14(10):667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 39.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun. 1999;67(4):1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol. 1995;39(8):555–562. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- 41.Teng F, et al. Gut microbiota drive autoimmune arthritis by promoting differentiation and migration of Peyer’s Patch T follicular helper cells. Immunity. 2016;44(4):875–888. doi: 10.1016/j.immuni.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149(7):1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klaasen HL, et al. Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab Anim. 1993;27(2):141–150. doi: 10.1258/002367793780810441. [DOI] [PubMed] [Google Scholar]
- 44.Yin Y, et al. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 2013;7(3):615–621. doi: 10.1038/ismej.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caselli M, Tosini D, Gafà R, Gasbarrini A, Lanza G. Segmented filamentous bacteria-like organisms in histological slides of ileo-cecal valves in patients with ulcerative colitis. Am J Gastroenterol. 2013;108(5):860–861. doi: 10.1038/ajg.2013.61. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Palacios A, et al. Stereomicroscopic 3D-pattern profiling of murine and human intestinal inflammation reveals unique structural phenotypes. Nat Commun. 2015;6:7577. doi: 10.1038/ncomms8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bäckhed F, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17(5):690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 49.Bottacini F, Ventura M, van Sinderen D, O’Connell Motherway M. Diversity, ecology and intestinal function of bifidobacteria. Microb Cell Fact. 2014;13(Suppl 1):S4. doi: 10.1186/1475-2859-13-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin J, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shapiro H, Thaiss CA, Levy M, Elinav E. The cross talk between microbiota and the immune system: Metabolites take center stage. Curr Opin Immunol. 2014;30:54–62. doi: 10.1016/j.coi.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 52.Cerovic V, Bain CC, Mowat AM, Milling SW. Intestinal macrophages and dendritic cells: What’s the difference? Trends Immunol. 2014;35(6):270–277. doi: 10.1016/j.it.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 53.Raffatellu M, et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14(4):421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ryan ES, et al. Loss of function of intestinal IL-17 and IL-22 producing cells contributes to inflammation and viral persistence in SIV-infected rhesus macaques. PLoS Pathog. 2016;12(2):e1005412. doi: 10.1371/journal.ppat.1005412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stieh DJ, et al. Th17 cells are preferentially infected very early after vaginal transmission of SIV in Mmcaques. Cell Host Microbe. 2016;19(4):529–540. doi: 10.1016/j.chom.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chabaud M, et al. Human interleukin-17: A T cell-derived proinflammatory cytokine produced by the rheumatoid synovium. Arthritis Rheum. 1999;42(5):963–970. doi: 10.1002/1529-0131(199905)42:5<963::AID-ANR15>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 57.Kotake S, et al. IL-17 in synovial fluids from patients with rheumatoid arthritis is a potent stimulator of osteoclastogenesis. J Clin Invest. 1999;103(9):1345–1352. doi: 10.1172/JCI5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314(5804):1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stahl EA, et al. BIRAC Consortium; YEAR Consortium Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4(3):e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanabe S. The effect of probiotics and gut microbiota on Th17 cells. Int Rev Immunol. 2013;32(5-6):511–525. doi: 10.3109/08830185.2013.839665. [DOI] [PubMed] [Google Scholar]
- 62.Ritchie ML, Romanuk TN. A meta-analysis of probiotic efficacy for gastrointestinal diseases. PLoS One. 2012;7(4):e34938. doi: 10.1371/journal.pone.0034938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334(6053):255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howitt MR, et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 2016;351(6279):1329–1333. doi: 10.1126/science.aaf1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Earle KA, et al. Quantitative Imaging of Gut Microbiota Spatial Organization. Cell Host Microbe. 2015;18(4):478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9(8):811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cipolletta D, et al. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486(7404):549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuleshov MV, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]