Significance
Ras-like small GTPases regulate a wide range of cellular processes involving signal transduction, cytoskeleton rearrangement, and membrane trafficking by switching between an active GTP-bound and an inactive GDP-bound form. In this study, we developed conformational sensors for GTPase activity (COSGAs) by combining protein engineering and chemical labeling. The COSGAs enable quantitative detection of the GDP/GTP binding state of Rab1 and K-Ras in living cells with spatiotemporal resolution to gain insights into Rab membrane cycling and Ras signaling at the plasma membrane. This approach paves a new avenue for imaging activity of signaling molecules in the cell, which could deliver valuable information regarding their function and dynamics.
Keywords: small GTPases, membrane trafficking, signal transduction, FLIM/FRET, protein chemical modification
Abstract
Ras-like small GTPases function as molecular switches and regulate diverse cellular events. To examine the dynamics of signaling requires spatiotemporal visualization of their activity in the cell. Current small GTPase sensors rely on specific effector domains that are available for only a small number of GTPases and compete for endogenous regulator/effector binding. Here, we describe versatile conformational sensors for GTPase activity (COSGAs) based on the conserved GTPase fold. Conformational changes upon GDP/GTP exchange were directly observed in solution, on beads, and in live cells by Förster resonance energy transfer (FRET). The COSGAs allow for monitoring of Rab1 and K-Ras activity in live cells using fluorescence lifetime imaging microscopy. We found that Rab1 is largely active in the cytoplasm and inactive at the Golgi, suggesting that the Golgi serves as the terminal of the Rab1 functional cycle. K-Ras displays polarized activity at the plasma membrane, with less activity at the edge of the cell and membrane ruffles.
GTP-binding proteins (also called GTPases or G proteins) are responsible for the regulation of a variety of biological processes, including signal transduction, cytoskeleton rearrangement, membrane trafficking, nuclear transport, and protein synthesis in eukaryotic cells. The Ras superfamily of small GTPases consists of >150 human members (1). They act as molecular switches by cycling between an active GTP-bound and an inactive GDP-bound form. This cycle is an earmark of their functional mode and is tightly regulated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) (2, 3). Only in the active GTP-bound form, small GTPases can bind to effector proteins, leading to the activation of downstream signaling.
Rab proteins are key regulators of intracellular vesicle transport in eukaryotic cells (4, 5). They comprise the largest subgroup of the Ras superfamily, with >60 members in mammals, and function at different cellular compartments to regulate the diverse transport stages. For example, Rab1 plays a key role in regulation of endoplasmic reticulum (ER)–Golgi trafficking, maintenance of Golgi structure, and autophagosome biogenesis during autophagy (6–8). Rab1 is hijacked, activated, and posttranslationally modified by Legionella effector proteins when the bacteria invade host cells (9–12). Regulation of spatial distribution of Rab proteins in the cell is a general question to be addressed (13).
Ras proteins function as master regulators of diverse signal transduction pathways that control processes such as cell proliferation, differentiation, adhesion, and migration. Ras is one of the most common oncogenes found in human tumors (14). Recent studies have shown that K-Ras is the most frequently mutated isoform (in 86% of Ras-driven cancers) (15). However, the exact timing and location of Rab1 and K-Ras activity have been largely undetermined because of the lack of appropriate sensors.
Spatiotemporal detection of the nucleotide-binding state of small GTPases in the cell provides valuable information regarding the function and dynamics of signal transduction. The existing approaches to monitor GTPase activation in cells rely mostly on specific binding of an effector domain to the active GTP-bound GTPase (16). Such interactions can be visualized by intermolecular (17) or intramolecular Förster resonance energy transfer (FRET) (e.g., Raichu sensors), as shown for H-Ras and Rap1 (18), Rho (19), and Rab5 (20). However, quantification of FRET requires calibration of the fluorophore concentrations and undesired signals from donor emission bleed-through, moreover, direct acceptor excitation have to be considered. In a FRET independent approach, the CRIB domain of the effector WASP labeled with an environment-sensitive dye displayed an increase in fluorescence intensity upon binding to the activated Cdc42 GTPase (21, 22).
Crucial for these strategies is the availability of a domain that binds specifically to the active GTPase. Consequently, these sensors remain limited to certain small GTPases, because for each target, a binding domain has to be identified and optimized (23, 24). In many cases, there are no suitable effectors available. Furthermore, such domains may have to compete with endogenous effectors. As a result, they either fail to bind to the activated GTPase or titrate out endogenous ligands (25, 26). An ideal probe would allow the direct observation of GTPase activation instead of indirect readout from the binding to effector domains. Here, we developed conformational sensors for GTPase activity (COSGAs) using a combination of protein engineering and chemical labeling. Based on the nucleotide-governed conformational change of the highly conserved GTPase fold, we generated versatile GTPase sensors. We used the COSGAs to visualize Rab1 and K-Ras activation, deactivation, and effector binding in solution, on beads and in live cells by fluorescence lifetime imaging microscopy (FLIM). This approach enabled us to quantitatively measure the GTP-/GDP-bound ratio of Rab1 in the intact cell with spatial and temporal resolution. Our studies provide insights into Rab membrane cycling and Ras signaling at the plasma membrane.
Results
FRET Sensor Design and Preparation.
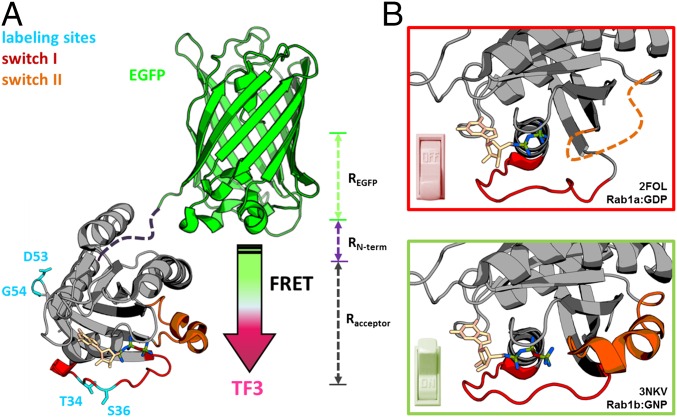
An N-terminal enhanced green fluorescent protein (EGFP) or yellow fluorescent protein (mCitrine) served as the FRET donor. The FRET acceptor Tide Fluor 3 (TF3) or Tide Fluor 4 (TF4) was introduced by site-specific cysteine labeling at a particular position within the GTPase fold (Fig. 1A) (27). These positions should undergo significant conformational changes upon nucleotide exchange and effector binding, but should not be directly involved in the GTPase function and interactions with GEFs, GAPs, and effectors, so that the GTPase sensor can report molecular events without impeding its functionality. To this end, we carried out a rational design of the labeling site on Rab1. We identified residues involved in large conformational changes by molecular dynamics (MD) simulations based on the root mean square fluctuation (RMSF) in GTP- and GDP-bound Rab1 structures (Protein Data Bank ID code 3NKV; Fig. 1B and SI Appendix, Fig. S1). The binding between small GTPases and their regulators and effectors largely involve interactions with the switch I and II regions (28). Based on the crystal structures of Rab complexes, we excluded positions that are crucial for the protein function and chose T34 and S36 in the switch I region and D53 and G54 in the interswitch region for chemical labeling (Fig. 1A and SI Appendix, Fig. S2).
Fig. 1.
COSGA design. (A) The intramolecular FRET sensor combines an N-terminal fluorescent protein with a small organic acceptor dye that is introduced in the protein by site-specific cysteine labeling. (B) Significant conformational changes in the switch regions of Rab1 in GTP- and GDP-bound states.
Quantitative and homogenous labeling was confirmed by liquid chromatography (LC)-electrospray ionization (ESI) mass spectrometry (MS) and SDS/PAGE (SI Appendix, Figs. S3 and S4). C-terminal prenylatable cysteines were truncated and reconstituted after labeling through native chemical ligation (29)/expressed protein ligation (30) with a dicysteine peptide to allow membrane targeting in cells (SI Appendix, Fig. S5).
In Vitro Validation of Rab1 GTPase Sensors.
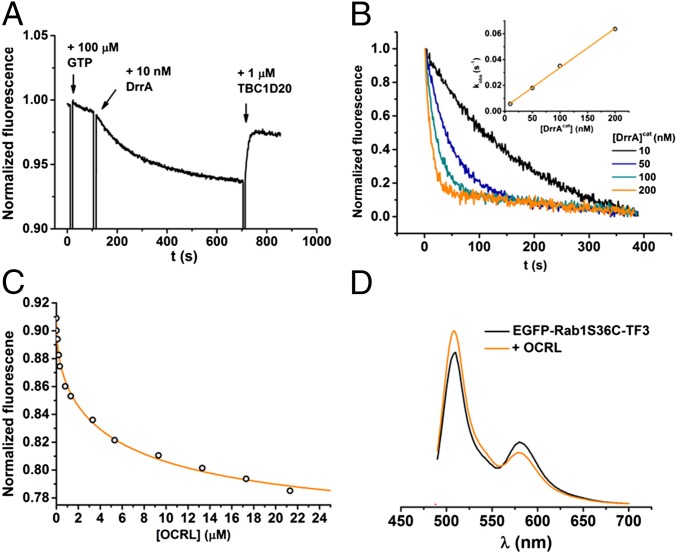
We first tested the activity of the sensors toward GEF-mediated nucleotide exchange and GAP-induced GTP hydrolysis. The nucleotide exchange was initiated upon addition of the GEF domain of the Legionella protein DrrA (DrrA340–533; referred to as DrrAcat) (Fig. 2 A and B). To confirm the specificity of the FRET signal toward the nucleotide-binding state, more DrrAcat or excess GDP was added after GDP-to-GTP exchange. In the former case, no further change of FRET signal was observed (SI Appendix, Fig. S6). In the latter case, the inverse change of FRET signal was observed, suggesting rebinding of GDP (SI Appendix, Fig. S7). Interestingly, the FRET signal decreased in the switch I constructs (T34C and S36C), but showed an increase in the interswitch constructs (D53C and G54C) (SI Appendix, Fig. S8). The FRET signal change was reversed through addition of the GAP domain of TBC1D20 (TBC1D201–362) (Fig. 2A and SI Appendix, Fig. S10). To further confirm that the observed FRET responses were indeed caused by GTP hydrolysis, the experiments were repeated using the nonhydrolyzable GTP analog GppNHp (SI Appendix, Fig. S10). In this case, TBC1D201–362 failed to reverse the FRET signal change in all Rab1 constructs. We determined the catalytic efficiencies (kcat/Km) of DrrAcat-mediated nucleotide exchange and TBC1D201–362-mediated GTP hydrolysis for the Rab1 constructs, which were in good agreement with previous reports for wild-type Rab1 (Table 1 and SI Appendix, Figs. S9 and S10) (11, 12). These results demonstrate that the EGFP–Rab1–TF3 constructs serve as FRET sensors for Rab1 activation and deactivation and that the introduction of organic dyes to specific positions in the protein fold does not disrupt the interactions with GEFs and GAPs.
Fig. 2.
GEF/GAP-mediated GTPase switching and effector binding monitored by Rab1 sensors. (A) Addition of 10 nM DrrAcat to 200 nM EGFP–Rab1T34C–TF3 induces rapid nucleotide exchange in the presence of excess GTP. The addition of TBC1D201–362 restores the FRET signal through GAP-induced GTP hydrolysis. (B) Rate constants of nucleotide exchange depend on DrrAcat concentration. (B, Inset) Plot of observed rate constant against DrrAcat concentration, yielding the second-order rate constant for nucleotide exchange (kcat/Km). (C) Titration of OCRL1539–901 to 200 nM EGFP–Rab1S36C–TF3. Binding was monitored by FRET. (D) Fluorescence spectra of EGFP–Rab1S36C–TF3 before and after binding to OCRL1539–901.
Table 1.
Summary of catalytic efficiencies (kcat/Km) of DrrAcat and TBC1D201-362 for Rab1 sensor constructs
| Rab1 | kcat /Km, M·s-1 |
| GEF assay | |
| WT | (1.38 ± 0.04) × 105 |
| T34C | (3.06 ± 0.08) × 105 |
| S36C | (1.73 ± 0.02) × 105 |
| D53C | (5.44 ± 0.62) × 105 |
| G54C | (4.23 ± 0.27) × 105 |
| GAP assay | |
| WT | (2.85 ± 0.4) × 105 |
| T34C | (1.24 ± 0.07) × 105 |
Errors represent fitting errors.
To confirm that labeling does not interfere with native effector binding, we determined dissociation constants (KD) for the interaction of GTP-bound Rab1 sensors with the Rab-binding domains of OCRL1 (OCRL1539–901) (31) and the bacterial Legionella effector LidA (LidA201–583) (32), using both FRET and fluorescence polarization (Fig. 2C and SI Appendix, Figs. S11 and S12). Because of its exceptionally high affinity toward Rab proteins, even in their inactive GDP form, LidA has been described as a Rab “supereffector” (32). The observed KD values are summarized in Table 2. Fluorescence spectra collected before and after effector binding showed an increase in donor emission with concomitant decrease in acceptor emission, which is a characteristic feature for FRET (Fig. 2D and SI Appendix, Figs. S12 and S13). The obtained dissociation constants are in good agreement with reported values for the interaction of Rab1 with OCRL1 and LidA (31, 32). Accurate determination of the KD (far below 10 nM) for high-affinity LidA binding was not possible by titration.
Table 2.
Summary of dissociation constants (KD) for interaction of Rab1 constructs with effectors
| Rab1 | Effector binding | |
| OCRL, KD, μM | LidA, KD, nM | |
| T34C | — | <10 |
| S36C | 1.5 ± 0.3 | <10 |
| D53C | 2.2 ± 0.4 | <10 |
| G54C | 2.9 ± 0.7 | <10 |
FRET was monitored with excitation at 480 nm and emission at 580 nm. Errors represent fitting errors.
Interestingly, we observed significant changes in the FRET signal upon OCRL1 binding to the S36C, D53C, and G54C constructs, but not for the T34C construct (SI Appendix, Fig. S11). Interaction measurements by fluorescence polarization excluded the impeded effector binding as a cause for the lack of FRET response (SI Appendix, Fig. S11B). In keeping with previous studies, OCRL1 binding was nucleotide-dependent, whereas Legionella effector LidA bound both GDP– and GTP–Rab1 with high affinity (SI Appendix, Figs. S11–S13) (32). Therefore, the EGFP–Rab1–TF3 constructs are effective reporters for the Rab1 nucleotide-binding state and effector binding, while maintaining their native interactions with GEFs, GAPs, and effector proteins.
FLIM-FRET of Rab1 Activity on Beads.
FLIM is a robust tool for detecting FRET and is now routinely used for dynamic measurements in live cells (33, 34). Energy transfer from a donor fluorophore to an acceptor fluorophore decreases the average fluorescence lifetime of the donor. The FRET efficiency can be extracted from the fluorescence lifetime of the donor fluorophore. A significant advantage of FLIM in comparison to conventional FRET imaging is that it does not rely on changes in fluorescence intensity and thus does not need to be corrected for artifacts resulting from changes in local fluorophore concentration and emission intensity (33). To validate whether our Rab1 sensors are suited for FLIM measurements, we tested the sensors immobilized via an N-terminal His-tag on Ni–nitrilotriacetic acid (Ni-NTA) beads (SI Appendix, Fig. S14).
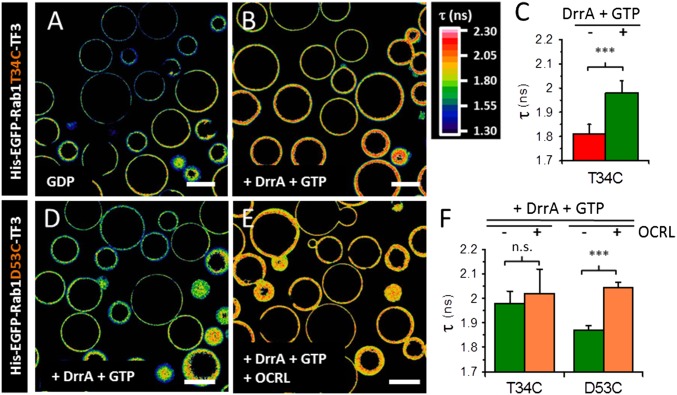
Upon GEF-mediated GDP/GTP exchange, the T34C construct exhibited a significant increase in EGFP fluorescence lifetime, which is correlated with the decrease in FRET signal observed in the sensitized emission measurements (Fig. 3 A–C). Accordingly, binding of OCRL1 led to a substantial increase of the fluorescence lifetime in the D53C construct, but not in the T34C construct (Fig. 3 D–F). The donor-only probe did not exhibit any lifetime changes upon nucleotide exchange and effector binding (SI Appendix, Fig. S15). Therefore, T34C and D53C constructs can be used for the measurements of nucleotide exchange and effector binding on beads, respectively.
Fig. 3.
FLIM-FRET on Ni-NTA beads. (A) FLIM image of GDP-bound His–EGFP–Rab1T34C–TF3 immobilized on Ni-NTA beads. (B) FLIM image of GTP-bound His–EGFP–Rab1T34C–TF3 immobilized on Ni-NTA beads after DrrAcat-mediated nucleotide exchange. (C) Quantification of EGFP lifetime for GDP- and GTP-bound His–EGFP–Rab1T34C–TF3 (n = 6 images with >15 beads, mean ± SD, two-tailed t test). ***P < 0.001. (D) FLIM image of GTP-bound His–EGFP–Rab1D53C–F3 immobilized on Ni-NTA beads. (E) FLIM image of His–EGFP–Rab1D53C–TF3 immobilized on Ni-NTA beads after binding of the OCRL1539–901 effector domain. (F) Quantification of EGFP lifetime for T34C and D53C constructs before and after OCRL binding (n = 5 images with >15 beads, mean ± SD, two-tailed t test). ***P < 0.001. n.s., not significant (P > 0.2). (Scale bars: 100 μm.)
Rab1 Molecules Are Inactivated at the Golgi.
The positive results obtained in vitro prompted us to further examine whether the Rab1 sensors are suitable for visualizing Rab1 activity in live cells. As described above, a EGFP–Rab1T34C–TF3–CC construct containing two C-terminal prenylatable cysteines was used for cellular studies. The Rab1 construct was microinjected into HeLa cells. The sensor located correctly to the Golgi apparatus as expected for Rab1 protein (SI Appendix, Fig. S16). Membrane cycling is a key function of Rab GTPases (5), which is regulated by the GDP dissociation inhibitor (GDI), GEFs, GAPs, and effectors (35, 36). To further demonstrate the functionality of the Rab1 construct in cells, we investigated the dynamics of the Rab1 construct at the Golgi by fluorescence recovery after photobleaching measurements. We found that the Rab1 construct undergoes fast cycling between the Golgi apparatus and cytoplasm (SI Appendix, Fig. S17). These results suggest that the Rab1 construct is prenylated and functions properly in the cell.
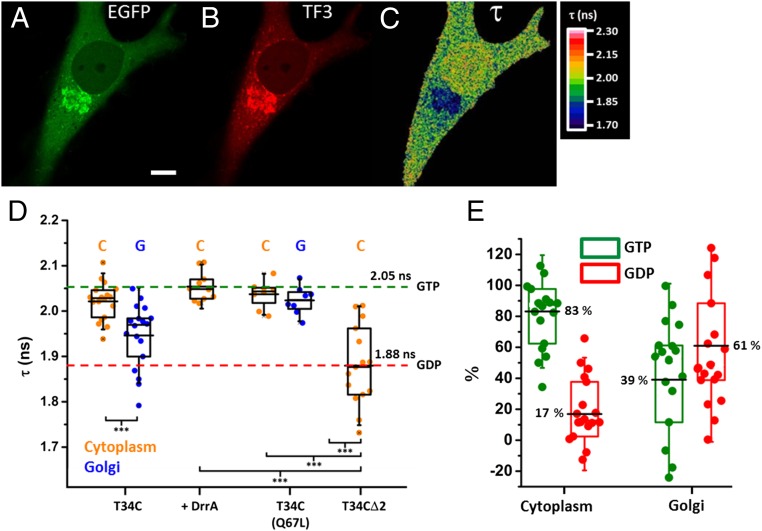
To correlate the FLIM results with the nucleotide-binding states, we measured the lifetime of the constitutively active GTP-bound Rab1T34C(Q67L) mutant and the wild-type Rab1T34C construct in the non-Golgi cytoplasm (hereafter referred to as cytoplasm) and the Golgi region (Fig. 4). The wild-type T34C construct showed a significantly lower (P < 0.001; two-tailed t test) fluorescence lifetime at the Golgi than in the cytoplasmic fraction. However, the T34C(Q67L) construct displayed similar fluorescence lifetime in the Golgi and the cytoplasm, which is comparable with that of the cytoplasmic fraction of the T34C construct. In cells expressing BFP–DrrA340–533, Rab1 was universally activated and displayed the same fluorescence lifetime as the Q67L construct. The GDP-bound Rab1T34CΔ2 construct lacking the C-terminal prenylatable cysteines serves as the reference for the inactive state (Fig. 4D and SI Appendix, Fig. S18). Without the C-terminal cysteines, the protein is not functional and therefore not subject to regulation by GEFs and GAPs. The fluorescence lifetime observed for the GDP-bound state was significantly lower than that of the GTP-bound state (P < 0.001; two-tailed t test). Using these measurements, we determined that ∼83% of the Rab1 molecules in the cytoplasm are GTP-bound and that ∼61% are GDP-bound at the Golgi region (Fig. 4E and SI Appendix, Fig. S19).
Fig. 4.
Imaging of Rab1 activity in cells. (A–C) Confocal EGFP (A), TF3 (B), and FLIM (C) images of EGFP–Rab1T34C–TF3–CC microinjected into HeLa cells. (Scale bar: 10 μm.) (D) Quantification of EGFP lifetime for T34C sensor constructs in cells. C and G denote cytoplasmic and Golgi-localized proteins, respectively (n = 7–25 cells; individual data points are plotted; box plots: -, median; --, mean; upper hinge, 75th percentile; lower hinge, 25th percentile; whiskers, SD; two-tailed t test). ***P < 0.001. (E) Quantification of GTP- and GDP-bound fraction of cytoplasmic and Golgi-localized Rab1 as described in SI Appendix, Fig. S19.
Spatiotemporal Imaging of K-Ras Activity in Live Cells.
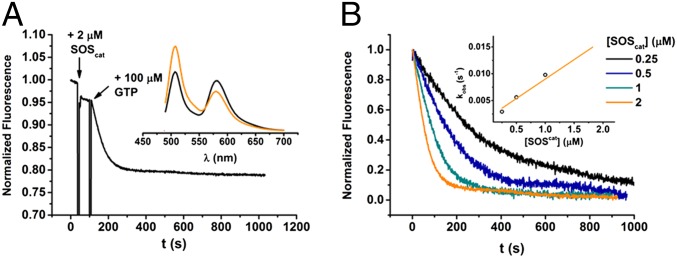
Because of the versatility of this strategy, we were able to generate a K-Ras sensor. Based on a structural alignment with the Rab1 sensor (SI Appendix, Fig. S20) and MD simulations (SI Appendix, Fig. S1), six K-Ras sensor constructs were generated. The constructs were tested in a nucleotide-exchange assay and a GAP assay using the GEF domain of SOS1 (SOS1564–1049; referred to as SOScat) and the GAP domain of p120GAP (RasGAP714–1047), respectively (Fig. 5 and SI Appendix, Figs. S21 and S23). The D30C and E31C constructs showed a substantial decrease in FRET signal upon SOS-mediated nucleotide exchange, which was reversed by RasGAP-mediated GTP hydrolysis (Fig. 5A and SI Appendix, Figs. S22 and S23). In comparison with K-Ras wild type, the observed nucleotide-exchange rate and GTP hydrolysis rate were reduced by a factor of 7 and 2 for the D30C construct and a factor of 20 and 31 for the E31C construct, respectively (Fig. 5 and Table 3). The impairment occurred in both activation and deactivation, which might explain the similar kinetics observed for the Ras sensor activity compared with the endogenous Ras activity in cells (shown below).
Fig. 5.
GEF-mediated GTPase switching monitored by K-Ras sensor. (A) Addition of SOScat to 200 nM EGFP–K-RasD30C–TF3 induces rapid nucleotide exchange in the presence of excess GTP (100 μM). (A, Inset) Fluorescence spectra of EGFP–K-RasD30C–TF3 before (black line) and after (orange line) nucleotide exchange. (B) Rate constants of nucleotide exchange depend on SOScat concentration. (B, Inset) Plot of observed rate constant against SOScat concentration, yielding the second-order rate constant for nucleotide exchange (kcat/Km). FRET was monitored by excitation set at 480 nm and emission at 580 nm.
Table 3.
Summary of catalytic efficiencies (kcat/KM) of SOScat and RasGAP714–1047 for K-Ras sensor constructs
| K-Ras | Signal | kcat/KM, M·s−1 |
| GEF assay | ||
| WT | Mant-GDP | (5.2 ± 0.2) × 104 |
| D30C | EGFP-TF3 | (0.73 ± 0.06) × 104 |
| E31C | mCitrine-TF4 | (0.26 ± 0.02) × 104 |
| GAP assay | ||
| WT | Mant-GTP | (1.37 ± 0.3) × 105 |
| D30C | EGFP-TF3 | (0.79 ± 0.11) × 105 |
| E31C | mCitrine-TF4 | (0.43 ± 0.04) × 104 |
Errors represent fitting errors.
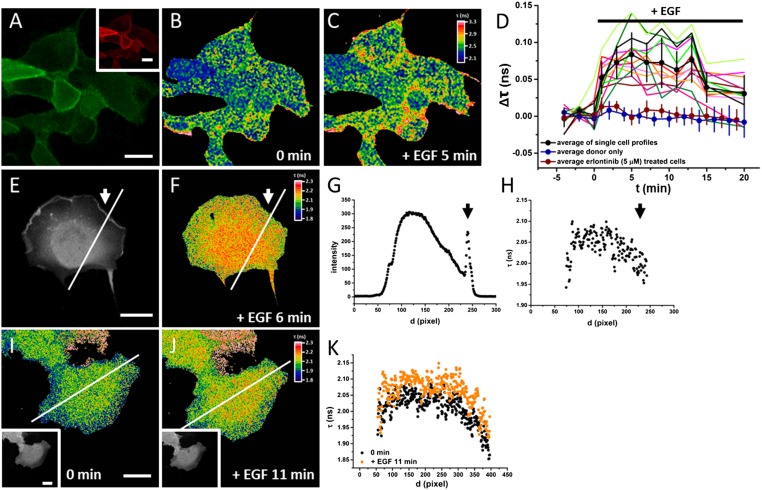
For cellular experiments, the K-Ras C terminus was reconstituted by native chemical ligation with the CVIM peptide (SI Appendix, Figs. S4 and S5). After microinjection into MDCK and COS-7 cells, K-Ras constructs enriched at the plasma membrane as expected for the native protein, suggesting that they were prenylated and function properly in cells (Fig. 6A) (37, 38). EGF stimulation led to rapid activation of K-Ras at the periphery of the MDCK cells (Fig. 6 A and C). K-Ras activation peaked within 5 min and then decreased, suggesting a negative feedback in EGFR–Ras signaling (Fig. 6D) (39). The kinetics of K-Ras activation were comparable with endogenous Ras, where the peak activation was observed at 4 min (40). In control experiments, no signal responses were observed in cells injected with donor-only constructs (SI Appendix, Fig. S24), constructs lacking the CVIM prenylation motif (SI Appendix, Fig. S25), or cells treated with the EGFR kinase inhibitor Erlotinib (SI Appendix, Fig. S26). The dynamic range of the K-Ras sensor can be obtained by evaluation of the FRET efficiency (EFRET) before and after activation. Analysis of the fluorescence lifetime in cells and sensitized emission in vitro showed a decrease of EFRET by 25% after EGF stimulation and by 51% after SOS-mediated GTP/GDP exchange, respectively. The in vitro measurement reveals the maximal dynamic range of the sensor, because GDP-bound Ras is quantitatively converted to GTP-bound Ras in this case (SI Appendix, Fig. S27).
Fig. 6.
Imaging of K-Ras activity in cells. (A–C) Confocal mCitrine (A) TF4 (A, Inset) and FLIM images of serum-starved MDCK cells microinjected with mCitrine–K-RasE31C–TF4–CVIM before (B) and 5 min after (C) EGF stimulation. (D) Quantification of fluorescence lifetime change in cells. Colored lines represent individual cell measurements, and the black line with error bars represents mean ± SD (n = 13 cells). Blue and red lines with error bars represent control measurements of donor only (mean ± SD; n = 14 cells) and in the presence of Erlotinib (mean ± SD; n = 14 cells), respectively (two-tailed t test). *P < 0.05; **P < 0.01. (E and F) Intensity (E) and FLIM image (F) of EGFP–K-RasD30C–TF3–CVIM after EGF stimulation in serum-starved COS-7 cells. Arrows indicate membrane ruffling. (G and H) Profiles of fluorescence intensity (G) and fluorescence lifetime (H) along the indicated line in E and F. (I and J) FLIM image of EGFP–K-RasD30C–TF3–CVIM in serum-starved COS-7 cells before (I) and 11 min after (J) EGF stimulation. (K) Quantification of the fluorescence lifetime along the line shown in I and J. Black and orange profiles indicate before and after EGF stimulation, respectively.
In COS-7 cells, K-Ras displayed a gradient of activity, with less activity at the edge of the cell (Fig. 6 F and I). The gradient was not altered after EGF stimulation, although the overall K-Ras activity increased (Fig. 6 J and K). In the EGF-induced membrane ruffles at the edge of the cell, K-Ras was recruited, but showed less activity than at other regions of the plasma membrane (Fig. 6 E–H and SI Appendix, Fig. S28).
Discussion
We rationally designed conformational sensors for nucleotide exchange and effector binding based on differences in backbone flexibility between GDP- and GTP-bound small GTPases examined by computational simulations. Because the GTPase fold is highly conserved, these conformational sensors should be applicable to the whole small GTPase family. This approach is superior to classical GTPase activation sensors in terms of versatility. In those sensors, specific effector domains have to be identified and optimized for each target. By combining protein engineering, site-specific chemical labeling, and in vitro protein ligation, we were able to generate a set of dual-labeled GTPase conjugates. The small organic dye was introduced at specific position within the protein by cysteine labeling using commercial probes. A single labeling step is sufficient to generate the sensor for a wide range of in vitro applications. Through immobilization on Ni-NTA beads, a high local protein concentration can be achieved, leading to a robust signal while using only small amounts of protein. GEFs have been considered promising therapeutic targets, because of their function in regulating disease-relevant GTPase signaling (15, 41). By using FLIM-FRET, the GTPase sensor-loaded beads can be readily used to screen for inhibitors of GTPase activation and effector binding. FRET is exceptionally sensitive to changes in distance and dipole orientation and therefore allows detection of conformational changes upon nucleotide exchange as well as effector binding. In this regard, the COSGA approach is advantageous to the classical GTPase activation sensors. The latter rely on indirect readout from effector binding, leading to competition with endogenous effector interactions.
We showed that site-specific labeling of Rab1 at positions T34, S36, D53, and G54 yields EGFP–Rab1–TF3 conjugates that maintain their native functionality. Although rational design and careful validation in vitro were carried out, we cannot exclude the possibility that some unknown regulators or effectors may bind in a significantly different manner. Complete validation in vitro is difficult and out of the range of this study. However, this is a common problem for all biosensors. Therefore, validation of the sensor in living cells is necessary. Here, we have examined the dynamics of Rab1 localization and K-Ras activity to confirm their proper function in cells. Moreover, to quantify the nucleotide-binding state in the cell, we included positive (GTP-bound, constitutively active) and negative (GDP-bound) controls as references (Fig. 4 and SI Appendix, Fig. S18). In these measurements, the effects from all native factors were included, because they were measured in the same cellular environment (i.e., associated with their endogenous regulators and effectors). Therefore, the sensors, albeit exogenous, reported on the activity of endogenous GEFs and GAPs.
By using the COGSAs, it was possible to determine the ratio of GTP/GDP-bound Rab molecules in intact cells with spatial resolution. In this study, we observed that cytoplasmic and Golgi-localized Rab1 molecules are largely in the GTP- and GDP-bound state, respectively. The cytoplasmic GTP–Rab1 molecules presumably localize on small vesicles and mediate vesicular transport between the ER and Golgi (42). The rest (17%) of cytoplasmic Rab1 molecules are in the GDP-bound state, presumably bound to GDI (43, 44). Consistently, ∼10% of prenylated Rab1 molecules in the whole cell were soluble in the cytosol, likely associated with GDI in GDP-bound form (45). This result suggests that only a small fraction of Rab1 molecules undergo GDI-mediated recycling. It is conceivable that there is high GAP activity at the Golgi membrane, leading to Rab1 deactivation. Therefore, the Rab1 functional cycle ends at the Golgi, and the GDP-bound Rab1 is then recycled via GDI (SI Appendix, Fig. S29) (46). Further studies are required to elucidate how exactly GEF and GAP activity coordinate the spatial cycle of Rab1.
The conformational sensor was also successfully applied to K-Ras. Our measurements showed that K-Ras activation constitutes a pattern of a “string of beads” at the plasma membrane (Fig. 6C). This finding is in keeping with the proposed nanoclusters of Ras signaling (47). Moreover, we found that K-Ras displays a distinct pattern of polarized activity at the plasma membrane in COS-7 cells in comparison to H-Ras reported before (18). H-Ras exhibits higher activity at the free edge of the cell after EGF stimulation, whereas K-Ras activity is minimal at the cell edge, where membrane ruffling is induced by EGF. This activity gradient indicates that there may be a negative feedback to K-Ras from the signaling of actin rearrangement. The opposing gradients of H- and K-Ras activity imply different mechanisms for regulating each Ras isoform in the cell.
The principle of COSGAs should be readily applicable to other processes involving conformational changes of a signaling protein, and in particular to other small GTPases. Further development of the conformational sensors will include application of unnatural amino acids (UAAs) mutagenesis and bioorthogonal chemistry, which allow cotranslational residue-specific incorporation of UAAs and chemoselective labeling of small GTPases in the cell (48).
Materials and Methods
Preparation of COSGAs.
All proteins were expressed in Escherichia coli BL21(DE3) and purified Ni-NTA affinity chromatography and by gel filtration chromatography. For labeling, Rab1 or K-Ras constructs were incubated with 1.5–2.5 eq TF3/TF4 maleimide (AAT Bioquest) at 25 °C for 30–120 min. For reconstitution of the C-terminal prenylation motif, Rab1 or K-Ras thioester proteins (5–10 mg/mL) were incubated with 2 mM CC or CVIM peptide in the presence of 20 mM Tris(2-carboxyethyl)phosphine and 50 mM 4-mercaptophenylacetic acid at 4 °C overnight. Detailed methods can be found in the SI Appendix, SI Methods.
Cell Culture and Microinjection.
The sensor constructs were concentrated to 6–10 mg/mL and microinjected into 40–70 cells by using an Eppendorf Transjector 5246 and Eppendorf Micromanipulator 5171. Before imaging, the microinjected cells were incubated at 37 °C for 1 h. Details of cell culture can be found in SI Appendix, SI Methods.
Fluorescence and Fluorescence Polarization Measurements.
Spectra acquisition, time-dependent sensitized emission, and fluorescence polarization measurements were performed with a FluoroMax-3 spectrofluorometer (Horiba Jobin Yvon). Details can be found in SI Appendix, SI Methods.
Confocal Microscopy and FLIM.
Confocal microscopy and FLIM were carried out by using the laser scanning confocal microscope FlouView FV1000 (Olympus Deutschland GmbH) equipped with a time-correlated single-photon counting LSM Upgrade Kit (PicoQuant). Cell images were collected through a 60×/1.35 UPlanSApo oil immersion objective (Olympus Deutschland GmbH). Ni-NTA bead images were collected through a 40×/1.35 UPlanSApo air objective. For FLIM measurements, the samples were excited with a 470-nm pulsed diode laser (LDH 470; PicoQuant) at a repetition rate of 40 MHz. The photons were collected in a single-photon counting avalanche photodiode (PDM Series, MPD; PicoQuant) and timed by using a time-correlated single-photon counting module (PicoHarp 300; PicoQuant) after being spectrally filtered using a narrow-band emission filter (HQ 525/15; Chroma). All measurements were carried out in a live-cell imaging chamber with control of humidity, temperature of 37 °C, and 5% CO2.
MD Simulations.
MD simulation details are described in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank Lei Zhao for providing the CVIM peptide; Emerich Mihai Gazdag for providing SOScat and K-Ras wild-type protein; and Philippe Bastiaens and Sven Müller for the support in the FLIM microscope. This work was supported by Deutsche Forschungsgemeinschaft, Grants SPP 1623 and SFB 642; Behrens-Weise-Stiftung; and European Research Council (ChemBioAP) (Y.-W.W.). O.K. is supported by the German Federal Ministry for Education and Research (Medizinische Chemie, TU Dortmund University) Grant BMBF 1316053.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613999113/-/DCSupplemental.
References
- 1.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118(Pt 5):843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 2.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell. 2007;129(5):865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 3.Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93(1):269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 4.Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91(1):119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 6.Davis S, Ferro-Novick S. Ypt1 and COPII vesicles act in autophagosome biogenesis and the early secretory pathway. Biochem Soc Trans. 2015;43(1):92–96. doi: 10.1042/BST20140247. [DOI] [PubMed] [Google Scholar]
- 7.Plutner H, et al. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J Cell Biol. 1991;115(1):31–43. doi: 10.1083/jcb.115.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, et al. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc Natl Acad Sci USA. 2013;110(24):9800–9805. doi: 10.1073/pnas.1302337110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee S, et al. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature. 2011;477(7362):103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller MP, et al. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329(5994):946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 11.Schoebel S, Oesterlin LK, Blankenfeldt W, Goody RS, Itzen A. RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol Cell. 2009;36(6):1060–1072. doi: 10.1016/j.molcel.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 12.Goody PR, et al. Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J. 2012;31(7):1774–1784. doi: 10.1038/emboj.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly EE, Horgan CP, Goud B, McCaffrey MW. The Rab family of proteins: 25 years on. Biochem Soc Trans. 2012;40(6):1337–1347. doi: 10.1042/BST20120203. [DOI] [PubMed] [Google Scholar]
- 14.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7(4):295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 15.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13(11):828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Shaughnessy EC, Yi JJ, Hahn KM. Biosensors of small GTPase proteins for use in living cells and animals. In: Zhang J, Mehta S, Schultz C, editors. Optical Probes in Biology. CRC; Boca Raton, FL: 2015. pp. 137–166. [Google Scholar]
- 17.Kraynov VS, et al. Localized Rac activation dynamics visualized in living cells. Science. 2000;290(5490):333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 18.Mochizuki N, et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411(6841):1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- 19.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440(7087):1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 20.Kitano M, Nakaya M, Nakamura T, Nagata S, Matsuda M. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. 2008;453(7192):241–245. doi: 10.1038/nature06857. [DOI] [PubMed] [Google Scholar]
- 21.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305(5690):1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 22.MacNevin CJ, et al. Ratiometric imaging using a single dye enables simultaneous visualization of Rac1 and Cdc42 activation. J Am Chem Soc. 2016;138(8):2571–2575. doi: 10.1021/jacs.5b09764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura T, Kurokawa K, Kiyokawa E, Matsuda M. Analysis of the spatiotemporal activation of rho GTPases using Raichu probes. Methods Enzymol. 2006;406:315–332. doi: 10.1016/S0076-6879(06)06023-X. [DOI] [PubMed] [Google Scholar]
- 24.Yoshizaki H, et al. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J Cell Biol. 2003;162(2):223–232. doi: 10.1083/jcb.200212049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pertz O, Hahn KM. Designing biosensors for Rho family proteins—deciphering the dynamics of Rho family GTPase activation in living cells. J Cell Sci. 2004;117(Pt 8):1313–1318. doi: 10.1242/jcs.01117. [DOI] [PubMed] [Google Scholar]
- 26.Yasuda R, et al. Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat Neurosci. 2006;9(2):283–291. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- 27.Voss S, Zhao L, Chen X, Gerhard F, Wu YW. Generation of an intramolecular three-color fluorescence resonance energy transfer probe by site-specific protein labeling. J Pept Sci. 2014;20(2):115–120. doi: 10.1002/psc.2590. [DOI] [PubMed] [Google Scholar]
- 28.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294(5545):1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 29.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266(5186):776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 30.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 31.Hou X, et al. A structural basis for Lowe syndrome caused by mutations in the Rab-binding domain of OCRL1. EMBO J. 2011;30(8):1659–1670. doi: 10.1038/emboj.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoebel S, Cichy AL, Goody RS, Itzen A. Protein LidA from Legionella is a Rab GTPase supereffector. Proc Natl Acad Sci USA. 2011;108(44):17945–17950. doi: 10.1073/pnas.1113133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berezin MY, Achilefu S. Fluorescence lifetime measurements and biological imaging. Chem Rev. 2010;110(5):2641–2684. doi: 10.1021/cr900343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y, Day RN, Periasamy A. Investigating protein-protein interactions in living cells using fluorescence lifetime imaging microscopy. Nat Protoc. 2011;6(9):1324–1340. doi: 10.1038/nprot.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F, et al. The role of the hypervariable C-terminal domain in Rab GTPases membrane targeting. Proc Natl Acad Sci USA. 2014;111(7):2572–2577. doi: 10.1073/pnas.1313655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu YW, et al. Membrane targeting mechanism of Rab GTPases elucidated by semisynthetic protein probes. Nat Chem Biol. 2010;6(7):534–540. doi: 10.1038/nchembio.386. [DOI] [PubMed] [Google Scholar]
- 37.Schmick M, et al. KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell. 2014;157(2):459–471. doi: 10.1016/j.cell.2014.02.051. [DOI] [PubMed] [Google Scholar]
- 38.Bar-Sagi D, Feramisco JR. Microinjection of the ras oncogene protein into PC12 cells induces morphological differentiation. Cell. 1985;42(3):841–848. doi: 10.1016/0092-8674(85)90280-6. [DOI] [PubMed] [Google Scholar]
- 39.Sasagawa S, Ozaki Y, Fujita K, Kuroda S. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat Cell Biol. 2005;7(4):365–373. doi: 10.1038/ncb1233. [DOI] [PubMed] [Google Scholar]
- 40.Qiu MS, Green SH. NGF and EGF rapidly activate p21ras in PC12 cells by distinct, convergent pathways involving tyrosine phosphorylation. Neuron. 1991;7(6):937–946. doi: 10.1016/0896-6273(91)90339-2. [DOI] [PubMed] [Google Scholar]
- 41.Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582(14):2093–2101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 42.Alvarez C, Garcia-Mata R, Brandon E, Sztul E. COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol Biol Cell. 2003;14(5):2116–2127. doi: 10.1091/mbc.E02-09-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soldati T, Riederer MA, Pfeffer SR. Rab GDI: A solubilizing and recycling factor for rab9 protein. Mol Biol Cell. 1993;4(4):425–434. doi: 10.1091/mbc.4.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ullrich O, et al. Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J Biol Chem. 1993;268(24):18143–18150. [PubMed] [Google Scholar]
- 45.Wilson AL, Erdman RA, Maltese WA. Association of Rab1B with GDP-dissociation inhibitor (GDI) is required for recycling but not initial membrane targeting of the Rab protein. J Biol Chem. 1996;271(18):10932–10940. doi: 10.1074/jbc.271.18.10932. [DOI] [PubMed] [Google Scholar]
- 46.Wu YW, Tan KT, Waldmann H, Goody RS, Alexandrov K. Interaction analysis of prenylated Rab GTPase with Rab escort protein and GDP dissociation inhibitor explains the need for both regulators. Proc Natl Acad Sci USA. 2007;104(30):12294–12299. doi: 10.1073/pnas.0701817104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plowman SJ, Muncke C, Parton RG, Hancock JF. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc Natl Acad Sci USA. 2005;102(43):15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lang K, Chin JW. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem Rev. 2014;114(9):4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.