Significance
Chromosomal DNA replication is initiated by unwinding duplex DNA via formation of dynamic higher-order protein–DNA complexes. Whereas biochemical analyzes have been performed for a long time, structural dynamics in those processes still remain elusive due to their complex nature. Here, using hybrid approaches of computational modeling and biochemical assays, we explore dynamic structures of the entire machinery and significance of spatial arrangement of every component in the replication initiation complex of Escherichia coli. We show the structural model of the crucial core part of the complex at near-atomic resolution. The complex is composed of three subcomplexes, and spatial arrangements of those as well as their components are important for efficient replication initiation.
Keywords: DnaA, molecular simulation, coarse-grained model, oriC
Abstract
Upon DNA replication initiation in Escherichia coli, the initiator protein DnaA forms higher-order complexes with the chromosomal origin oriC and a DNA-bending protein IHF. Although tertiary structures of DnaA and IHF have previously been elucidated, dynamic structures of oriC–DnaA–IHF complexes remain unknown. Here, combining computer simulations with biochemical assays, we obtained models at almost-atomic resolution for the central part of the oriC–DnaA–IHF complex. This complex can be divided into three subcomplexes; the left and right subcomplexes include pentameric DnaA bound in a head-to-tail manner and the middle subcomplex contains only a single DnaA. In the left and right subcomplexes, DnaA ATPases associated with various cellular activities (AAA+) domain III formed helices with specific structural differences in interdomain orientations, provoking a bend in the bound DNA. In the left subcomplex a continuous DnaA chain exists, including insertion of IHF into the DNA looping, consistent with the DNA unwinding function of the complex. The intervening spaces in those subcomplexes are crucial for DNA unwinding and loading of DnaB helicases. Taken together, this model provides a reasonable near-atomic level structural solution of the initiation complex, including the dynamic conformations and spatial arrangements of DnaA subcomplexes.
Chromosomal DNA replication is initiated by unwinding the dsDNA of the replication origin, which requires formation of higher-order protein–DNA complexes, typically referred to as the initiation complexes (1). DnaA is a major replication initiation protein conserved in the initiation complex of most eubacterial species. In a model organism, Escherichia coli, DnaA forms homooligomers on the replication origin oriC, which promotes dsDNA unwinding. The resulting ssDNA is captured by DnaB helicase, followed by formation of the replisomes (2–5). Molecular mechanisms of how DnaA facilitates dsDNA unwinding are still unclear, although some models have been proposed (1, 6–10). A high-resolution structure model of the initiation complex, discovered using computational modeling based on experimental data, would provide significant insight into the molecular mechanism.
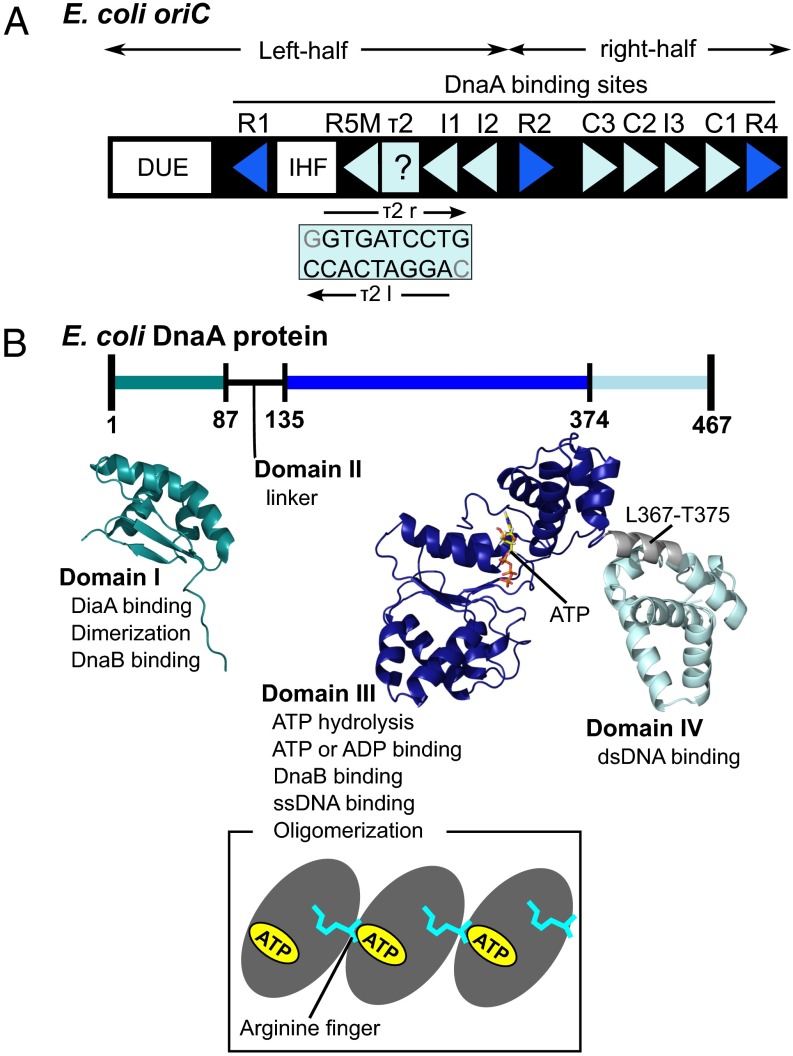
The E. coli minimal oriC region contains the AT-rich DNA unwinding element (DUE), at least 11 DnaA-binding motifs (termed DnaA boxes) and a single binding site for the integration host factor (IHF) (Fig. 1A) (1–5, 11–15). The DnaA boxes contain 9-mer nucleotides (consensus sequence TTATNCACA, where N can be any base) (16). The 11 DnaA boxes (R1–2, R4, R5M, I1–3, C1–3, and τ2) have differing affinities to DnaA and motif orientations (indicated by triangles in Fig. 1A): The two terminal boxes R1 and R4 have especially high affinities (dissociation constants 1–6 nM for R1 and ∼1 nM for R4), whereas others have modest (R2) to low affinities (I1–3, C1–3, and R5M and τ2; dissociation constants for R5M are >200 nM) (12–19). The 11 DnaA boxes have been divided into two groups on the left- and right-half oriC. The six right boxes (R2, C3, C2, I3, C1, and R4) share the same orientation of the motifs, whereas the four motifs in the left half, i.e., R1, R5M, and I1–2, have a motif orientation opposite to that of the right half. The orientation of the remaining low-affinity site τ2 in the left half has yet to be examined because of sequence degeneracy (14, 15). DnaA binding to the τ1 box, which is located at the left side of R5M, is observed with a low affinity only for a linear form of oriC but not for a replication-active supercoiled form (14, 15); we thus excluded this box in this study. IHF is one of the bacterial architectural proteins and sharply bends dsDNA (20).
Fig. 1.
oriC and DnaA in E. coli. (A) The oriC region contains one DUE, an IHF-binding site (IHF), and 11 DnaA boxes (binding sites). At each DnaA box, the orientation of the binding motif is indicated by the triangle, with the exception of τ2. The blue and light-blue triangles mean high- and low-affinity boxes, respectively. For the τ2 site, two possibilities of binding, τ2l (AGGATCACC) and τ2r (GTGATCCTG), were considered as previously described in the literature. (B) Domain architecture of DnaA protein. (Top) DnaA contains four domains, for which functions and structures are summarized. Domain I is NMR structure from Protein Data Bank 2E0G (26), and domain III–IV is a homology model (37). (Bottom) DnaA domain III homooligomerizes in a head-to-tail manner. The ATP-bound side of one molecule contacts with the arginine-finger side of another molecule.
DnaA is composed of four domains, in which domain IV is the dsDNA-binding domain that recognizes DnaA boxes (Fig. 1B) (1–5, 21, 22). A crystal structure model is available for the complex of domain IV with dsDNA for E. coli DnaA (21). Domain III is the multifunctional ATPases associated with various cellular activities (AAA+) domain, which tightly binds ATP and ADP (1–5, 23–25). The ATP-bound AAA+ domain can form stable helical homooligomers by assembling in a head-to-tail manner (Fig. 1B, Bottom cartoon) (14, 24). The arginine-finger in an interface (Arg-finger interface) of one AAA+ domain coordinates to the ATP molecule bound on another interface (ATP-bound interface) of an adjacent AAA+ domain, which explains the nucleotide-dependent head-to-tail oligomer formation on oriC (Fig. 1B) (14, 24). The helical oligomer of a AAA+ domain can bind ssDNA by specific residues exposed on its central path (6, 8). Crystal structures have been solved for helically arranged homooligomer of domains III and IV of Aquifex aeolicus DnaA with and without bound ssDNA (8, 24). Domain IV in these complexes, however, does not accommodate dsDNA. A rigid body rotation at the linker between domains III and IV has been suggested to enable this homooligomer to bind to dsDNA at domain IV (7, 23, 24). The N-terminal domain I contains a DnaB-binding site as well as a site for low-affinity domain I–domain I interaction (Fig. 1B) (2, 26). Domain II is a flexible linker (Fig. 1B) (26).
As mentioned above, DnaA forms at least two subcomplexes corresponding to the left and right halves of oriC (9, 10, 15). The left-half oriC subcomplex sustains the entire activity in DUE unwinding; i.e., deletion of the region spanning R2 to R4 does not impair the unwinding activity (9). Each subcomplex binds a DnaB helicase for loading onto the single-stranded region (9, 10). DnaA oligomers are formed on the regions spanning R1–I2 and R4–C3 on oriC in a manner depending on the Arg finger–ATP interaction and other specific interactions (9, 10, 14, 15, 27). DnaA binding to the two terminal, high-affinity DnaA boxes has been proposed to elicit sequential binding of ATP–DnaA molecules in low-affinity binding sites (15, 18, 27). The Arg-finger interface, but not the ATP-bound interface, of the terminal DnaA protomers orients inward within oriC and interacts with the ATP-bound interface of the adjacent DnaA protomer (27). However, the entire oriC–DnaA complex structure model is still unknown. There thus remain important questions about how DnaA box-bound DnaA molecules assemble to form helical oligomers using the AAA+ domain in oriC, and how oriC DNA is curved and/or supercoiled in such complexes. Also, there remains a question concerning the importance of specific DnaA–DnaA interactions to depositing DnaA with specific directionality on low-affinity binding sites. In the right-half oriC, the R2 box is 9 and 20 bp apart from the left and right adjacent DnaA boxes, whereas most of the boxes are only 2 bp apart from each other, which is the most favored distance between two DnaA boxes in DnaA–DnaA interactions (15). These provoke a question of whether the R2-bound DnaA is involved in helical oligomer formation. These questions are essential to understand the efficacy of DNA unwinding. Establishing the entire structure may further provide insights into the extent of evolutionary conservation in molecular mechanisms of replication initiation.
We used a cooperative approach between computational modeling and biochemical assays to obtain a model for the E. coli initiation complex with atomic resolution. Given that high-resolution structures are known for many parts of the systems and that numerous biochemical functional assays have already been accumulated (8, 16, 20, 21, 27), we set up molecular simulations to integrate these data for generating structures of the initiation complex. Because the entire system is too large to be modeled by straightforward atomistic molecular dynamics (MD) simulations, we took a multiscale approach. We started by using coarse-grained molecular models to broadly sample structures. The coarse-grained molecular simulation is particularly useful to efficiently obtain a broad range of potentially stable structures (28–32). We have previously developed coarse-grained simulation models for protein–DNA complexes (32, 33); we used these models in this work. The coarse-grained models were back transferred into fully atomistic models to examine the stability of the structures. The resulting models were then further tested by biochemical assays using reconstituted reactions with purified proteins and various mutant oriCs.
Based on these results, we propose a model structure composed of three subcomplexes; the left oriC region bearing pentameric DnaA and IHF, the middle oriC region bearing monomeric DnaA, and the right oriC region bearing pentameric DnaA. We also explored the importance of the spaces in DnaA boxes and regulated DnaA–DnaA interactions in construction of functional oriC–DnaA complexes.
Results
Construction of Model Structures and Interactions.
Our simulations in the initial part used coarse-grained models where each amino acid in proteins was modeled by one particle located at Cα position (34) and each nucleotide in DNA was represented by three particles, each representing sugar, phosphate, and base (35, 36). The protein energy function is based on native structure information at atomic resolution (SI Appendix, SI Methods provides details). For DnaA, we constructed a model structure of the E. coli ATP–DnaA domain III–IV using a domain III–IV homology model by I-TASSER (37). Interactions between DnaA box and DnaA domain IV were defined by cocrystal structure of E. coli domain IV and a DnaA box R1 (21). This homology model has similar structure to the A. aeolicus ADP–DnaA domain III–IV monomer (23). The IHF structure was based on its complex with DNA (20). Even though we use MD approaches, our main purpose here is not to clarify the dynamics of complex formation, but to obtain the complex structure. The interactions between molecules consist of specific and generic interactions. The former are structure-based energy: DNA–protein interactions were set between the DnaA domain IV and each DnaA box as well as between IHF and its binding site. Assuming head-to-tail docking of DnaA domain III (14, 24), we set specific protein–protein interactions between the Arg-finger side and the ATP side of the adjacent protomer (Fig. 1B, Bottom cartoon). The generic interactions included electrostatic interactions between charged particles of DNA and protein as well as excluded volume interactions that avoid steric overlaps (SI Appendix, SI Methods provides details).
Modeling the E. coli Left-Half oriC–DnaA–IHF Subcomplex.
We began with simulations of the left-half subcomplex, which contains the left-half oriC DNA (excluding the DUE region and including the five DnaA boxes, R1, R5M, τ2, I1, and I2), one IHF as a dimer, and five copies of the ATP–DnaA domain III–IV. The simulations start in a configuration where five DnaA domains IV are bound to the five DnaA boxes, R1, R5M, τ2, I1, and I2 in the left-half oriC via specific interaction. For the five boxes, we set the DnaA interaction at the R1 box to be strong and those at the other boxes to be weak (dissociation constants ∼200 nM), based on experimental observation; the interaction strengths were tuned to reproduce measured dissociation constants (16). Using enhanced attractive interactions between the Arg-finger side and ATP-bound side of neighboring DnaA domain IIIs, we performed coarse-grained MD simulations to generate the complex structures with several different setups described below.
During MD simulations, complex formation was monitored by the following two integer-valued measures. (i) Npp, the number of DnaA–DnaA molecular interfaces through their domain III (0 ≤ Npp ≤ 4; p, protein). (ii) Npd, the number of native-like DnaA–DNA molecular interactions at DnaA domain IV (0 ≤ Npd ≤ 5; d, DNA). More precise definitions are given in SI Appendix, SI Methods. We judge that the replication initiation complex forms when both of the two measures have their maximum values.
In the first simulation, we used a model with rigid L367–T375 of DnaA domains III and IV for simplicity, although the short linker (residues L367–T375, Fig. 1B) between the two domains is suggested to be flexible (see below). During simulations at physiological temperature, the rigid model fluctuated around the reference structure, but few large-amplitude motions were observed due to many native contacts at the linker region. Next, we analyzed complex formation. In the τ2 box, due to degenerated similarity to the consensus sequence, two definitions of the alignments are possible. τ2l has the same direction as the other left-half DnaA boxes, whereas τ2r has the same direction as the right-half DnaA boxes (Fig. 1A and SI Appendix, Table S1). These alignments have opposite directions from and certain similarities to the consensus sequence (14, 15). We note that, once we choose one alignment, the structure-based interaction in our simulations, only the specific DnaA–DNA interactions in that alignment are taken into account. Thus, for each of two τ2 alignments, we performed 108-step MD simulations 10 times with different stochastic forces. Across the 20 total trajectories, however, we did not find any single structure that had maximal values of both Npp (= 4) and Npd (= 5) simultaneously. In these simulations, some DnaA molecules often dissociated from DNA, suggesting that DnaA domain III disturbs interactions between domain IV and DNA, which is consistent with the necessity of rigid body rotation of domain III upon binding to DNA (7, 23, 24).
These results motivated us to make the domain III–IV linker region flexible (38). The flexible linker is further justified by the following arguments: (i) When we constructed the E. coli DnaA domain III–IV homology model, the secondary structure prediction of the I-TASSER modeling server suggests that the linker tends to form loops rather than an α-helix (37). The α-helix structure was derived from a homology model because the crystal structure of this region was unavailable. (ii) The linker region has a specific trypsin-sensitive site, a biochemical indication of a flexible region (39).
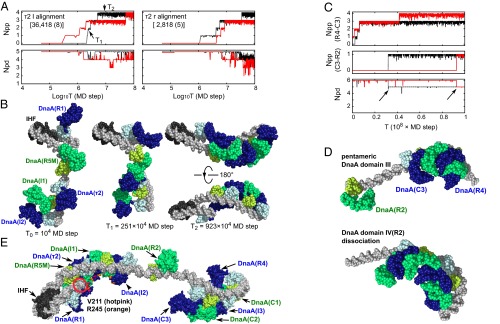
For each of two alignments in the τ2 box, we illustrate the evolution of Npp and Npd for 2 of the 10 trajectories in Fig. 2A, red and black curves). In the case of τ2l alignment (Fig. 2A, Left), both trajectories reached the Npp = 4 and Npd = 5 state. From the trajectory with a black curve, we show three snapshots at t0 = 1, t1 =251, and t2 = 923 (× 104-MD step) in Fig. 2B. The Npp grew from 0 at t0, to 2 at t1, and to 4 at t2. Interestingly, when the Npp became 4, the DNA was bent along the helical curve of the DnaA domain III pentamer, as in Fig. 2B, Right. In the right snapshots of Fig. 2B, AAA+ domains of DnaA are apart from dsDNA. Whereas DnaA domain III pentamer approached dsDNA occasionally during simulations, it did not bind dsDNA. Statistics for all 10 trajectories are summarized in SI Appendix, Table S2. Of the 10 total trajectories, 8 formed the complete complex. Once the complete complex formed, it was mostly stable; the complex could be transiently perturbed, but it returned to the complete complex soon after. Thus, the accumulative time that the system resided in the complex was rather long (36% of the entire time). In contrast, for the τ2r alignment, the complete complex formed in 5 trajectories, and accumulative time was less than 1/10 of the τ2l case (2.8% of the entire time). We found that maintaining Npd = 5 was more difficult than the τ2l case. This result suggests that, for the left-half subcomplex, the complex is more stable with the same orientations of all five bindings, which is in agreement with the report by Rozgaja et al. (15).
Fig. 2.
Simulation-based models of the replication initiation complex. (A) For the left-half subcomplex, formation trajectories of oriC–DnaA–IHF complexes in two representative coarse-grained simulation trajectories (red and black) with two setups about τ2 site, τ2l (Left), and τ2r (Right) alignments. The number of formed DnaA–DnaA molecular interfaces at domain III (Npp, Top) and the number of formed DNA–DnaA interfaces (Npd, Bottom) are plotted. Npp and Npd are moving averaged over nine frames. (B) Snapshots in a trajectory (black curve in A, Left) with the τ2l alignment setup. DNA, gray; IHF, black. ATP–DnaA is drawn in alternative choices of two color schemes: one in dark blue (domain III) and light blue (domain IV) and the other in green (domain III) and yellowish green (domain IV). The MD step of each snapshot is also depicted. The Right structure is drawn from two orientations. The IHF is spatially aligned for Left, Middle, and Right-Top images. (C) For the right-half subcomplex, formation trajectories of oriC–DnaA complexes in two representative coarse-grained simulation trajectories (red and black). In this figure, the number Npp is divided into two parts: Npp (R4–C3), the number of formed DnaA–DnaA molecular interfaces of five DnaA domain III (DnaA binding to the R4, C1, I3, C2, or C3 box) and Npp (C3–R2). Npp (R4–C3), Npp (C3–R2), and Npd are moving averaged over nine frames. (D) Snapshots in a trajectory (red curve in C). (Top) Snapshot where the DnaA domain III formed a pentamer. (Bottom) Snapshot at the end of simulation. The R2 box-side terminal of DNA is spatially aligned. (E) Atomic model that connects the left- and middle-right-half subcomplexes after short full-atomistic MD simulations. The same color code applies as in B. V211 (hotpink) and R245 (orange) are ssDNA-binding residues (6) and indicated for the five left-half DnaA molecules. The Left end of DNA is encircled in red. Symbols in the parentheses represent the DnaA boxes to which DnaA is bound.
The above results clarified that flexibility in the domain III–IV linker is crucial to form the complete complex. Upon the complex formation, the linkers, at least in some molecules, need to change their conformations. The details of the conformational change will be analyzed later.
With τ2l alignment, the rmsds between a representative DnaA pentamer structure and other pentamer structures were distributed around 6 Ǻ, which is rather small for this size of protein complexes (SI Appendix, Fig. S1). This finding indicates each subcomplex is similar to the representative structure. Some snapshots are shown in SI Appendix, Fig. S2. In the formed complex, we analyzed the interfaces between DnaA domain IV and DNA using the interface Q score, which is the fraction of formed intermolecular contacts to those in the reference structure. The Q score for the left-half subcomplex was distributed mainly around 0.5–0.9, which is similar to the system of the DnaA domain III–IV binding 13-bp DNA containing the R1 box (SI Appendix, Fig. S3). In contrast, the Q score of τ2r DnaA box–DnaA domain IV was exceptionally low, mainly between 0.1 and 0.2. This finding suggests that the τ2r box–DnaA interaction is not very stable while maintaining other box–DnaA interactions. This tendency is also consistent with Npd transition in Fig. 2A, Right. With τ2r alignment, the structures of DnaA pentamer were more diverse (SI Appendix, Figs. S1 and S4).
Modeling the Middle and Right oriC–DnaA Subcomplexes.
Next, we modeled the formation of oriC subcomplexes next to the left-half subcomplex. Here, the modeling system consisted of the oriC region containing the six DnaA boxes R2, C3, C2, I3, C1, and R4, along with six copies of ATP–DnaA domain III–IV. We set C1–3 and I3 boxes as low-affinity sites and R4 and R2 boxes as high-affinity sites; DnaA binds to the latter two boxes independently of other boxes (13, 14). We started the coarse-grained MD simulations with the configuration binding the six DnaA molecules to the six DnaA boxes on oriC. The simulations were used to induce head-to-tail oligomerization of DnaA domain III.
The goal of the simulations was to test whether R2-bound DnaA participates in the oligomerization or not. In the setup, domain III (the ATP-bound side) of R2-bound DnaA has attractive interface interactions to domain III (the Arg-finger side) of the C3-bound DnaA. With this setup, we expected to see hexameric helical arrangement of domain III. In this case, the maximal value of Npp is 5. To focus on the involvement of R2-bound DnaA, we divided Npp into two parts, Npp (R4–C3) (≤4) and Npp (C3–R2) (0 or 1). The former represents the Npp of the pentamer part of R4-bound DnaA to C3-bound DnaA. The latter represents the interface between R2-bound DnaA and C3-bound DnaA. We illustrate 2 of the total of 10 trajectories in Fig. 2C. Npp (R4–C3) can reach the maximal value of 4 and maintain it (the red curve). However, when Npp (C3–R2) reached 1, Npd immediately dropped to 5 (Fig. 2C, Bottom). The drop was independent of Npp (R4–C3). The dropping Npd was caused by dissociations of DnaA from the R2 box (Fig. 2D, Bottom). The result suggests that DnaA domain III can form a pentamer, but not a hexamer in the right-half oriC and DnaA domain III–IV system. Representative structures of pentameric DnaA domain III on the right-half oriC are shown in the Upper image of Fig. 2D. In a similar manner to the left-half case, DNA curved along with the helical shape of DnaA domain III pentamer. Statistics for all 10 trajectories are summarized in SI Appendix, Table S3. Of the 10 trajectories, 7 formed the domain III pentameric complex. The domain III hexameric complex rarely appeared.
To confirm that the domain III pentamer between DnaAs was bound to the R4 and C2 boxes, we also conducted a complex formation simulation without an attractive potential between R2-bound DnaA and C3-bound DnaA. In simulations, the system resided in the pentameric complex 62% of the entire time, which is rather high. Like the left-half subcomplex, the rmsds between the representative right-half DnaA pentamer and other right-half DnaA pentamers were small (around 6 Ǻ) (SI Appendix, Fig. S5). All right-half DnaA pentamers had similar structure (SI Appendix, Fig. S2). The Q score for the interfaces between the DnaA-binding box in the pentameric domain III complex was approximately similar to that of the DnaA monomer 13-bp system (SI Appendix, Fig. S6).
Considering the above results together with the fact that DnaA–R2 box binding occurs with no cooperative mechanism (13, 14) and the biochemical data described below, we propose redefining the subcomplexes; i.e., the R2-bound DnaA complex as the middle subcomplex and the C3–R4 region bound pentameric DnaA complex as the right-half subcomplex (also see below). Comparing the left- and right-half oriC DNA bound to the pentameric DnaA, we see that the DNA is bent more strongly in the right-half subcomplex.
Modeling oriC–DnaA–IHF Complex Structure at Atomic Resolution.
To obtain models at atomic resolution for the left-half subcomplex and the middle-right-half subcomplexes, we performed reverse mapping from a coarse-grained model to a fully atomistic model, followed by short-time atomistic MD simulation. During atomistic simulations, the overall structures did not change much (SI Appendix, Fig. S7). Then, we connected the left- and the middle-right-half subcomplexes to make the whole oriC–DnaA–IHF complex (Fig. 2E, the coordinates file in Dataset S1). Notice that the R2-bound DnaA is closer to the left-half DnaA pentamer than the right-half pentamer. The DNA terminal of the left-half subcomplex (Fig. 2E, red circle) orients inward of the DnaA domain III pentamer on which the ssDUE-binding residues V211 and R245 are exposed (6). This figure might be relevant to the DUE unwinding mechanism. In contrast, DnaA domain III hexameric complex on the middle-right-half oriC collapsed when we performed a similar simulation (SI Appendix, Fig. S7). This result is consistent with coarse-grained simulations.
The τ2 Box Alignment: Computational Analysis.
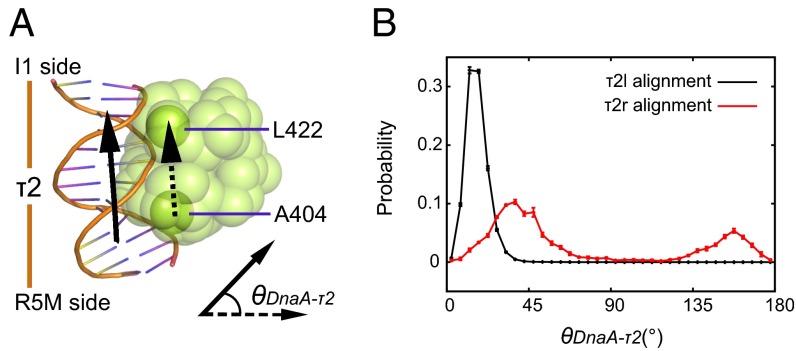
We further analyzed the molecular mechanisms that determine alignment of τ2 box, i.e., τ2l or τ2r, using coarse-grained simulations. As mentioned above, DnaA–τ2 box interactions were insufficient in the left-half complex with τ2r, but not τ2l. The representative complex structures obtained were similar between the τ2l case and the τ2r case (SI Appendix, Fig. S1). Visually inspecting the structure, we noticed that the DnaA molecule bound to the τ2 box flipped from the orientation of the τ2r alignment to that of the τ2l alignment during the MD simulations (SI Appendix, Fig. S4). Thus, the interface Q score to the reference τ2r alignment is low, but the structure apparently resembles that obtained in the τ2l-based simulations. To quantify this, we defined an orientation of DnaA domain IV relative to the bound DNA (Fig. 3 A and B). The angle is set so that it is zero near the τ2l alignment, whereas the τ2r alignment corresponds to about 180°. We found that, in the τ2r-based simulations, DnaA domain IV bound to the τ2 box flipped with significant probability (∼70%).
Fig. 3.
The orientation of DnaA domain IV binding to τ2 box. (A) The angle θDnaA-τ2 is defined by the two vectors that monitor the local DNA long axis (arrow with solid line) and the DnaA domain IV (arrow with dotted line). (B) Distribution of the angle θDnaA-τ2 for complete complexes with two sets of the τ2 alignments. Error bars express sample SD.
Altogether, our computational analysis suggests that DnaA bound to τ2l is more stable than to τ2r and that the homooligomerization of DnaA domain III disrupts the DnaA–τ2r interfaces. Because this disruption caused the DnaA on the τ2 box to flip to the τ2l-like orientation in our τ2r-based simulation, we suggested that oligomerization of DnaA domain III assists unification of the DnaA domain IV binding orientations within the left-half subcomplex.
The τ2 Box Alignment: Biochemical Assays.
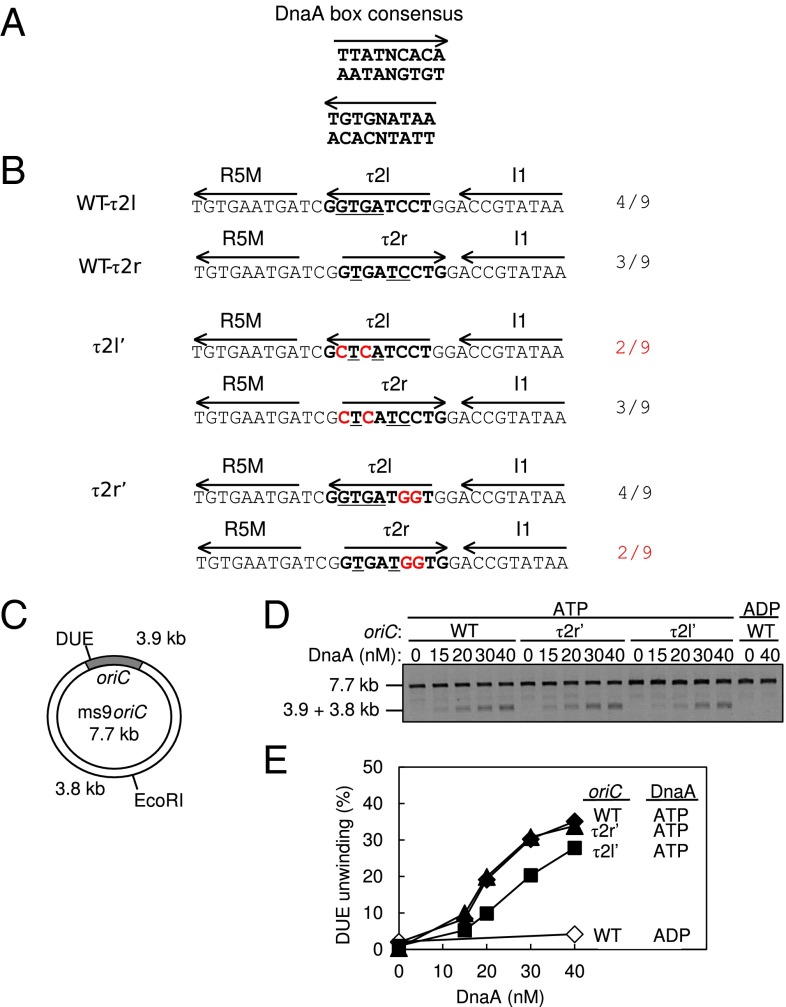
Next, we performed biochemical experiments to analyze the DUE unwinding activity of oriC mutants bearing altered τ2 box sequences. This box resides in the left-half oriC, which sustains the entire activity in DUE unwinding (9, 19). To identify which definition, i.e., τ2l or τ2r, is most appropriate, we designed two τ2 mutant sequences τ2l′ and τ2r′ that reduce the identities to the consensus 9-mer in the definitions of τ2l and τ2r, respectively, retaining the identities in the other definition (Fig. 4 A and B). These substitutions should abolish DnaA binding to the sequences, as those substitutions should inhibit interaction between the DNA and DnaA His434/Thr435 (21), which is crucial to DnaA box–DnaA binding (17, 40).
Fig. 4.
DUE activity of oriC bearing mutant τ2 sequences. (A) DnaA box consensus. The 9-mer consensus sequence with both orientations (arrow) are shown. (B) WT and mutant sequences including the R5M and I1 boxes. The Upper two lines show the WT sequence and each definition of the τ2 box (boldface) with the orientation (arrow). Identical bases to the 9-mer consensus are indicated by underlining; the number is also shown. The Middle two lines show the τ2l′ mutant sequence. The Bottom two lines show the τ2r′ mutant sequence. Each definition of the τ2 box is also shown. The mutations are highlighted by red letters. (C) Structure of an oriC plasmid M13oriCMS9 (ms9oriC). The positions of oriC, DUE, and the EcoRI site are indicated. (D and E) DUE assay. The given amounts of ATP–DnaA or ADP–DnaA were incubated with M13oriCMS9 or its derivative bearing τ2l′ or τ2r′ mutation, followed by analysis using ssDNA-specific nuclease P1, EcoRI, and agarose gel electrophoresis (D). Band intensities of the gel image were quantified and the relative amounts of the 3.8- and 3.9-kb bands were plotted as DUE (%) (E).
DUE unwinding activity of these oriC sequences was analyzed using ssDNA-specific nuclease P1. In this assay, the unwound DUE is cut by P1 nuclease, which produces 3.8- and 3.9-kb fragments following EcoRI digestion (Fig. 4C). We found that the unwinding activity of oriC bearing τ2r′ was similar to WT oriC, whereas that of oriC bearing τ2l′ was moderately reduced (Fig. 4 D and E). These results suggest that τ2l is the proper τ2-box definition (Fig. 4B). Also, the considerable activity exhibited even with the oriC τ2l′ is consistent with the idea that affinity for the τ2 box is not a crucial determinant of DnaA binding to this site. Homooligomeric interactions of DnaA domain III bound to the τ2 site with those bound to the flanking sites would effectively assist proper DnaA binding to τ2. Thus, these results are consistent with those of the computational analysis described above.
Conformational Change Between Domains III and IV upon Complex Formation.
Based on crystal structures, researchers suggested that the relative orientation of DnaA domain III to domain IV has change upon binding to dsDNA and again upon the replication initiation complex formation (23, 24). Consistently, we needed to use a flexible linker between the domains III and IV to form the complete complex in the coarse-grained MD simulations. Here, we use coarse-grained simulations to analyze how the interdomain orientation changed upon the complex formation.
We first looked into the interdomain orientation change upon binding to dsDNA, before domain III oligomerization, finding that the change is not addressable for the following reasons. In the case of A. aeolicus DnaA domains III and IV, which we used as the template for homology modeling, a rigid docking to a 9-bp dsDNA produced steric clashes (23). The steric clashes are removed by a small conformational change (23). Similarly, in our model, a small orientation change of domain III against domain IV was necessary to stabilize domain IV–dsDNA interactions (SI Appendix, Fig. S8A). However, because the sidechain orientation is not well represented by our coarse-grained modeling, the influence of the clashes is underestimated. In addition, the linker sequence between domains III and IV is distinct between E. coli and A. aeolicus. Thus, the homology model mostly obtained from A. aeolicus DnaA cannot be very reliable in the linker region. With atomic resolution modeling of interdomain interactions, some, albeit small, changes could be attributed.
In contrast, the interdomain orientation change was clearly seen upon oligomerization at the DnaA domain III of oriC. For each of the DnaA molecules, we calculated the rmsd of domain III from the reference (initial) structure when domain IV was superimposed (Fig. 5A). As a control, we plotted the rmsd distributions for monomeric DnaA (black) and for DnaA bound to the R2 box without forming an oligomer state to adjacent monomers (red). Clearly, the interdomain orientation change is markedly larger for DnaA molecules in the pentameric oligomer than the isolated molecules. Interestingly, the behaviors are somewhat similar between the left and right halves; looking from outmost ends, i.e., R1 and R4, the rmsd distributions in two halves resemble each other for the corresponding positions, i.e., R1 and R4, τ2 and I3, I1 and C2, and I2 and C3. The rmsd is larger for the central three monomers in the pentameric form. We further calculated the mean distance between the centers of mass of domain III at two positions relative to domain IV for each monomer (SI Appendix, Fig. S8B). The results suggest that the interdomain orientations are relatively uniform among the central three monomers in each pentameric subcomplex, whereas the terminal monomers R1, I2, and C3 take diverse conformations.
Fig. 5.
Analysis of the simulated complex structures. (A) An analysis of the conformational change in each DnaA molecule in the complete complex. The distributions of the rmsd of individual DnaA domain III from the reference DnaA structure are plotted. The rmsd is calculated after domain IV is superimposed. Error bars express sample SD of trajectories. (B) Representative conformations of DnaA before and after formation of the pentameric complex (I1 box as an example). Domain IV (gray) is spatially aligned for all images. Domain III is rainbow colored from blue (K135) to red (L373).
Representative snapshots are depicted in Fig. 5B and SI Appendix, Fig. S8C. The structural changes in the central three protomers of each subcomplex are essentially the same as those modeled in A. aeolicus DnaA domain III–IV oligomers formed with dsDNA (7). However, the left- and the right-half subcomplexes take different forms, primarily at the terminal monomers. This finding is consistent with our previous work, suggesting structural differences in the two subcomplexes (10) (Discussion).
Role of Spacing Between the IHF-Binding Site and the R1 Box in the Left-Half Subcomplex.
As the R1 box is distant from the R5M box, and the IHF-binding site is located between the R1 and R5M boxes, the position of the R1 box could be important for stability of the left-half subcomplex. Thus, we examined the significance of the space between the IHF-binding site and the R1 box by biochemical assays and computational modeling.
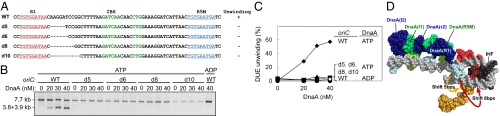
We first analyzed DUE unwinding activity of a mutant oriC bearing a 5-, 6-, 8-, or 10-bp deletion between the IHF-binding site and the R1 box (Fig. 6A, d5, d6, d8, and d10). Results of the P1 nuclease assay showed that all mutant plasmids were inactive in unwinding DUE (Fig. 6 B and C). The residual activities were only faint as was the case where ADP–DnaA and WT oriC was used. These results indicate that the length of spacing between the IHF-binding site and the R1 box is of critical importance. Consistent with this finding, a 10-bp insertion between the the R1 site and the IHF-biding site was previously suggested to inhibit replication of oriC plasmid in vivo (41).
Fig. 6.
DUE activity of oriC with deletions between R1 and the IHF-binding site. (A) DNA sequence covering from R1 to R5M. WT sequence and deleted sequences (d5–d10) are shown. Sequences of the R1, R5M, and IHF-binding site (IBS) are highlighted by red, cyan, and green, respectively. Deleted sequences are indicated by hyphens. DUE activities of each oriC are also shown based on the data shown in B and C. +, WT level; −, inactive. (B and C) DUE assay. The given amounts of ATP/ADP–DnaA were incubated with plasmid M13oriCMS9 or its derivatives bearing deletions as shown in A, followed by analysis using ssDNA-specific nuclease P1, EcoRI, and 1% agarose gel electrophoresis (B). Percentages of P1 nuclease-digested oriC DNA molecules per that of input DNA are shown as “DUE unwinding (%)” (C). (D) Positional examination of the R1-bound DnaA in the R1–IBS region-deleted oriC. In a representative structure of left-half subcomplex, the R1-bound DnaA domain III–IV was relocated toward the IHF-binding site by 5 base pair (bp) (orange) or 8 bp (red).
Next, we examined whether these mutant oriCs can form a DnaA pentamer via computational modeling. First, based on the WT left-half subcomplex model, we shifted the R1 site by a number of deleted base pairs (5, 6, 8, or 10 bp) and relocated the R1-bound DnaA (Fig. 6D, d5 and d8). For d8 and d10 mutants, the relocated R1-bound DnaA sterically overlapped with IHF, suggesting difficulty in stable binding of both the R1-bound DnaA and IHF. For d5 and d6 mutants, DnaA was placed on the relocated R1 site without any steric clashes, suggesting a possibility to form the complete left-half subcomplex. To test this possibility, we conducted coarse-grained MD simulations for modeling the DnaA pentamer on the d5 mutant oriC. However, the complex structure satisfying both the Npp, Npd criteria was not observed during the course of the simulations, primarily because DnaA on the R1 box did not come close enough to DnaA on the R5M box.
Taken together, the space between the IHF-binding site and the R1 box is stringently designed for the DUE unwinding activity of the left-half oriC. This is consistent with the proposed mechanism for DUE unwinding (Discussion).
Significance of the Space Between R2 and C3 Boxes: Biochemical Assay.
The oriC left-half subcomplex has DUE unwinding activity in addition to a basal activity of DnaB helicase loading (9). The activity of the full-length oriC in DnaB loading is about twofold higher than that of the left-half subcomplex, indicating that the oriC middle- to right-half subcomplexes stimulate DnaB helicase-loading activity. For bidirectional replication from oriC, a pair of DnaB helicases is loaded on the unwound DNA of the oriC region in opposite directions (42). Each subcomplex might exert specific dynamics to coordinately load a pair of DnaB helicases (9, 27).
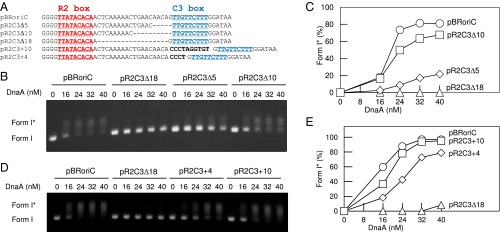
The vicinity of the R2 box has exceptionally long intervening spaces, which do not have DnaA binding sequences; the space to the I2 box is 9 bp, whereas that to the C3 box is 20 bp (Fig. 7A). These spaces might be important for coordinated construction of the DnaA complexes. To analyze the importance of the 20-bp space from R2 to C3, we constructed oriC plasmid mutants bearing a 5-, 10-, or 18-bp deletions (denoted as pR2C3Δ5, pR2C3Δ10, and pR2C3Δ18), or a 4- or 10-bp insertion (denoted as pR2C3+4, and pR2C3+10) into this space. The left-half oriC subregion was intact in these mutants, and thus DUE unwinding activity should be preserved (9, 10). Indeed, in a biochemical assay using P1 nuclease, DUE unwinding activities of all of the oriC mutants were not substantially inhibited (SI Appendix, Fig. S9). We analyzed the DnaB helicase-loading activity of these mutant oriC using the form I* assay (9, 27, 43). In this assay, if DnaB helicase is loaded onto the unwound oriC region of supercoiled (i.e., form I) oriC plasmid, then it expands the unwound region, positive supercoiling is increased, and DNA gyrase counteracts to introduce negative supercoiling, resulting in a hyper supercoiled form, form I*. Form I and form I* have different migration rates in agarose gel electrophoresis (44).
Fig. 7.
DnaB loading activity of oriC with altered R2–C3 spacing. (A) DNA sequence covering R2–C3. The WT sequence in pBRoriC and mutant sequences in pBRoriC-derivative plasmids are shown. Sequences of the R2 and C3 boxes are highlighted by red and cyan letters, respectively. Deleted sequences (hyphen) and inserted sequences (black boldface) are indicated. (B–E) Form I* assay. The given amounts of DnaA were incubated with pBRoriC or its derivatives in the presence of DnaB, DnaC, SSB, IHF, and gyrase, followed by analysis using agarose gel electrophoresis (B and D). Band intensities of the gel image were quantified and the relative amounts of form I* were plotted as form I* (%) (C and E).
Activities of pR2C3Δ10 and pR2C3+10 in form I* production were only slightly inhibited or even similar compared with that of WT oriC plasmid, pBRoriC (Fig. 7 B–E). In contrast, the activities of pR2C3Δ5 and pR2C3Δ18 were severely inhibited (Fig. 7 B and C). The activity of pR2C3+4 was moderately inhibited (Fig. 7 D and E). These results indicated that the length of the intervening space between R2 and C3 is important in DnaB helicase loading. In addition, the relative orientation of the right-half complex to the left-half complex also might be relevant because a single pitch of DNA strand contains 10 bp, and the deletion and insertion of 10 bp had minimum inhibitions.
Next, we analyzed DnaB helicase binding to oriC–DnaA complexes using a pull-down assay (SI Appendix, Fig. S10). As the first step of DnaB helicase loading onto unwound DUE, complexes of DnaB helicase–DnaC loader bind to DnaA complexes bound to oriC (2). The DnaA N terminus has the specific site for DnaB binding (26). In this experiment, ATP–DnaA, DnaB helicase, and DnaC loader were incubated with the biotin-tagged oriC DNA with and without the Δ18 mutation. Our previous study demonstrates that recovery of DnaB depends on DnaA in this experiment (10, 43). Recoveries of DnaA and DnaB were not significantly changed by the Δ18 (SI Appendix, Fig. S10). These results suggest that inhibition of DnaB helicase loading exhibited in the mutant oriCs is not due to a decrease in DnaA or DnaB binding to the oriC.
Taken together, a plausible idea is that in the pR2C3 bearing the Δ5, Δ18, or +4 mutation, the right-half DnaA subcomplex has a conformation that prevents the bound DnaB from approaching ssDUE. In our previous paper, we showed that activity of the left-half oriC in form I* production was about 50% of that of the full-length oriC (9). Based on this fact, the DnaA subcomplexes formed on the oriC bearing the Δ5 or Δ18 mutation might be inactive in DnaB loading on ssDUE because of not only improper position of right-half DnaB–DnaA complex but also steric clash/interference between the left-half and right-half DnaB–DnaA complexes.
Biochemical Analysis of R2-Bound DnaA.
We first analyzed using EMSA DnaA binding to the WT R2–C3 region and its Δ5, Δ10, and Δ18 mutants (SI Appendix, Fig. S11). DnaA binding was not inhibited in these mutants, consistent with the idea described for Fig. 7. In addition, DnaA-binding levels to both sites in the WT fragment (i.e., complex II formation) were similar between ATP–DnaA and ADP–DnaA, consistent with the idea that domain III–domain III interaction does not occur in this case, as suggested from previous footprint analyses (13, 14). This feature in DnaA binding was basically similar even in the Δ5 and Δ10 mutants. However, in the Δ18 mutant, complex II formation was stimulated in an ATP–DnaA-dependent manner (SI Appendix, Fig. S11), which is consistent with the idea that specific domain III–domain III interaction was allowed in this mutant. These results are also consistent with the previous report indicating that efficient binding between DnaA molecules requires 2- to 5-bp spacing between the DnaA boxes (15). The space between the R2 and C3 sites of the Δ10 mutant was 10 bp, whereas that of the Δ18 mutant is 2 bp.
In addition, when the WT fragment was used, complexes with three DnaA molecules (i.e., complex III) were only slightly detected (SI Appendix, Fig. S11). This result might indicate a possibility of the presence of a DnaA molecule that bridges DnaAs bound to the specific DNA sites and does not directly bind to the DNA (see below).
Next, we analyzed the R2-bound DnaA, using chimeric DnaA and oriC (SI Appendix, Fig. S12). The chimeric DnaA (chiDnaA) consisted of E. coli DnaA domain I–III and Thermotoga maritima DnaA domain IV (27). The chimeric R2–R4 oriC region (R2R4–R2tma) carried the R2 site substituted with T. maritima-specific DnaA binding sequence (TmaDnaA box), which differs in sequence from the E. coli DnaA box. Our previous study demonstrates that the chiDnaA specifically binds to the TmaDnaA box, but not the E. coli DnaA box, and sustains basic activities required for replication initiation (27). Also, the orientation of DnaA bound to the DnaA box has been determined and the bound ATP/ADP side (but not the Arg-finger side) of the R2-bound DnaA is oriented toward the C3 site (27). When chiDnaA was coincubated with E. coli DnaA (EcoDnaA) and the R2R4–R2tma fragment, formation of the DnaA oligomers were stimulated depending on the dose of chiDnaA, and this stimulation occurred similarly with the ATP and ADP forms of chiDnaA (SI Appendix, Fig. S12). These results consistent with the idea that domain III of the R2-bound DnaA does not interact with domain III of another protomer, at least in a ATP-specific manner. The stimulation might depend on domain I–domain I interaction involving the R2-bound DnaA (Fig. 1B and Discussion). These results are consistent with the data of EMSA using the WT R2–C3 region (SI Appendix, Fig. S11).
Significance of the Space Between R2 and C3 Boxes: Computational Modeling.
To understand the above biochemical results from a structural perspective, we modeled these mutant middle-right-half subcomplexes with the same coarse-grained MD simulations of the corresponding mutants. Considering the results of the EMSA assay, we first examined the possibility of DnaA domain III hexamers. Indeed, the Δ10 and Δ18 mutants formed domain III hexamer complexes more than 50% of the time during simulations. For the Δ5 mutant, the hexameric complex was formed but only for less than 1% of the simulation time. For the +4 mutant, oriC–DnaA domain III–IVs satisfied the formation criteria of a hexamer in only 1 of 10 trajectories. The domain III hexameric complex did not form for the +10 mutant (SI Appendix, Table S3 for statistics). For the Δ18 and Δ5 mutants, simulation results were well consistent with the EMSA analysis. However, for the Δ10 mutant, hexameric complexes formed in simulations, but this was not supported in EMSA (SI Appendix, Fig. S11). Some simulation settings, e.g., flexibility of the L367–T375 linker, may make hexameric complex in simulations more stable than in reality. To examine whether pentameric domain III complexes form or not, we also conducted complex formation simulations without an attractive potential between C3-bound DnaA and R2-bound DnaA. As a result, domain III pentameric complex formed for all mutants (SI Appendix, Table S3).
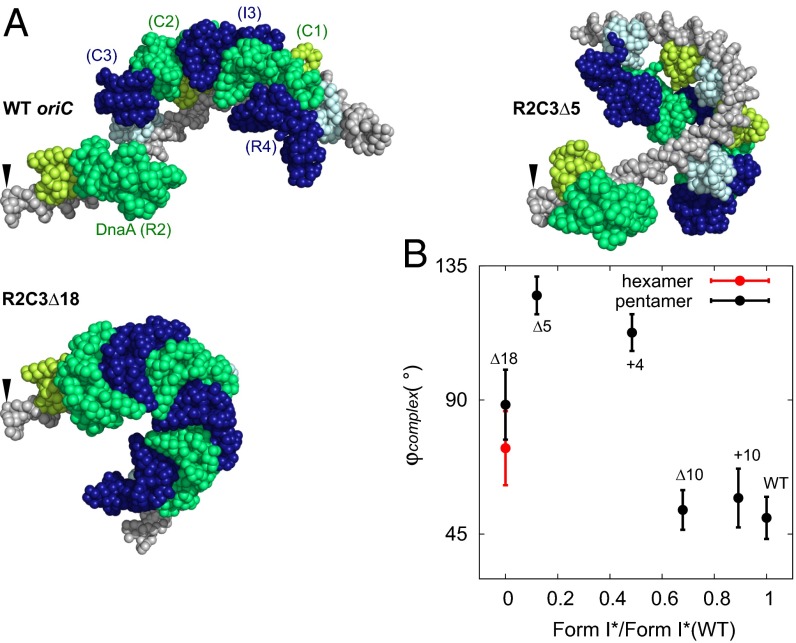
To investigate these complexes, we further simulated complexes formed on the mutant right-half oriCs. Representative complex structures obtained for the mutants, together with that of the WT, are depicted in Fig. 8A and SI Appendix, Fig. S13. Here, we aligned the structures at the R2-side terminus of DNA (indicated by arrow). Comparing the right-end orientation, we notice that the Δ10 and +10 mutants resemble the WT. Form I* activities of these two were similar to WT (Fig. 7). The other mutants show larger deviation in the orientation of the right end, which is correlated with the low form I* activities. In addition, the hexameric DnaA formed on the Δ18 mutant resulted in much higher bending of the DNA, which made this complex rather different from the WT, which probably severely affected the form I* activity. Thus, the overall structures of the mutant complexes explain the form I* assay data nicely.
Fig. 8.
Computational modeling of deletion/insertion mutants of oriC. (A) Representative complex models obtained for WT oriC and mutant sequences. For R2C3Δ5, we chose a pentameric oligomer model. For R2C3Δ18, we chose a hexameric model. The models are spatially aligned by the Left end (shown by the arrow). The color scheme is identical to Fig. 2. (B) The correlation plot between the experimental form I* values from 24-nM DnaA condition normalized by that of WT (horizontal axis) and the mean angle φcomplex of DnaA oligomer in simulated models (vertical axis). Here, φcomplex represents the mean tilt of each complex relative to WT (SI Appendix, SI Methods). Error bars express sample SD of nine individual experiments.
To quantify these effects, we defined the rotation angle φcomplex of the DnaA complex relative to the WT complex (SI Appendix, SI Methods). We found that small φcomplex values are associated with high form I* activity (Fig. 8B) except for Δ18. This correlation suggests that, if the mutant right-half DnaA subcomplex is rotated further away from the WT, DnaB helicase loading to ssDNA may become harder.
Together, these deletion/insertion mutant experiments suggest that spatial arrangement between the left-half and right-half subcomplexes has functional relevance. It also suggests that DnaA oligomers with proper spatial arrangement function as a molecular machine that loads DnaB helicase onto ssDNA efficiently.
Discussion
In the initiation complex of bacterial genome replication, multiple molecules of DnaA bind to oriC via its domain IV and form a helical shape through homooligomerization of DnaA’s domain III (1–5). Despite the fact that high-resolution structures have previously been solved for DnaA, the entire complex structure remains to be explored. Here, combining MD simulations with biochemical assays, we propose a model for fundamental structures and their roles in the E. coli initiation complex. In this model, the left-half, middle, and right-half subcomplexes contain helical DnaA pentamer and IHF, DnaA monomer, and helical DnaA pentamer (Fig. 2), respectively. This model explains important dynamics for construction of specific DnaA oligomers on oriC at a fairly high resolution and will be a common basis for future studies of structural and functional analyses of the eubacterial replication initiation complex. In addition, we do not exclude the possibility that DnaA molecules, which bridge the DNA-bound DnaAs but do not directly bind to the DNA, are included in the initiation complex. However, even in this case, the fundamental frame of the initiation complex structure would be determined by the DNA-bound DnaAs as shown in this study.
Form I* and pull-down assays using insertion/deletion mutants of oriC together with computational modeling suggested that the relative positions of the left-half, middle, and right-half subcomplexes are important for efficient DnaB loading to ssDNA. As the initial single-stranded region in DUE is limited in length, the arrangement of three subcomplexes may have to be well designed for precisely delivering DnaB to the ssDNA region in addition to loading of a pair of DnaB helicases in opposing directions. We propose that each of the left-half and right-half DnaA subcomplexes binds a DnaB helicase, resulting in highly organized, two DnaA–DnaB–DnaC complexes that are competent to DnaB loading onto ssDNA (SI Appendix, Fig. S14). DnaA domain I has a primary DnaB-binding site and binding of multiple (more than a single) DnaA molecules to a DnaB helicase is required for formation of stable DnaA–DnaB complexes (26, 43). DnaB helicase forms a stable complex with DnaC-loader protein before loading onto ssDNA (2). Distance between and relative orientations of the two DnaA–DnaB–DnaC complexes would be crucial in precise delivering of DnaB helicases to the ssDNA region and in dynamic DnaA–DnaB interactions during DnaB loading.
The middle subcomplex is composed of DnaA bound to the R2 box, which is separated from the adjacent C3 box by 20 bp. Our simulations suggested that the R2-bound DnaA is not involved in the AAA+ domain III oligomers. This suggestion is in agreement with the form I* data where the 10-bp insertion between the R2 box and C3 box did not greatly change the form I* activity. Results of EMSA analyses are also consistent (SI Appendix, Figs. S11 and S12). Notably, when the R1 box is deleted, the R2-bound DnaA stimulates DnaA assembly in the left-half oriC (9, 45). DnaA–DnaA interaction depending on domain I homooligomerization rather than domain III might be important for this stimulation (46, 47). Based on the opposite orientation and significant distance between DnaA boxes I2 and R2, head-to-tail domain III–domain III interaction between DnaAs bound to these sites seems unlikely.
In a crystal structure of the DNA-free oligomers of A. aeolicus DnaA domains III and IV, there exist interactions between domain III of one molecule and the domain III/IV interface of an adjacent molecule (which we label the “secondary” interaction), in addition to the domain III–domain III interaction mediated by the arginine finger and ATP (the “primary” interaction) (14). Importantly, in our model of the oriC-bound DnaA complex, the relative orientation of domains III and IV was changed markedly from that found in the crystal structure, eliminating the secondary interactions. The existence of the primary interactions and absence of the secondary interactions further highlights the importance of the domain III–domain III interaction in formation of functional initiation complexes. The secondary interaction might be important for possible DnaA binding, which bridges the DNA-bound DnaAs without direct DNA binding.
Ala-substitution mutations at R227 and L290, which are located at the domain III–domain III interfaces and termed ATP–DnaA-specific interactive locus for DUE unwinding (AID) motifs, moderately inhibit formation of the left-half subcomplex, but not the right-half subcomplex (10). Our computational models showed that DnaA pentamer structures are distinct at the terminal monomers between the left- and right- half subcomplexes. These differences may explain the different responses to the mutations that perhaps weakened the interface interactions.
In addition, we infer a possibility that DnaA R285A, in which the arginine finger Arg285 is substituted by Ala, and WT ADP–DnaA may be respectively homooligomerized on oriC depending on the secondary type of interactions. ATP–DnaA R285A and WT ADP–DnaA can respectively form a homooligomer on oriC, but binding to the low-affinity sites is inhibited, resulting in a similar footprint pattern as an oriC complex inactive in DUE unwinding (14). Thus, it is proposed that DnaA Arg285 is crucial to support proper head-to-tail domain III–domain III interactions, which are required for the active initiation complex. It is thus conceivable that DnaA R285A forms an oligomer, depending on the secondary interactions rather than the primary interaction. This view explains a probable mechanism that DnaA R285A is impaired in binding to the low-affinity sites on oriC, resulting in inactive complexes. Basically the same mechanism can be applied to ADP–DnaA. However, it should be noted that DnaA R285A oligomers on oriC could also be supported by other interactions, such as those depending on DnaA Arg281, which stabilizes DnaA–DnaA interaction and indirect stabilization through DnaA domain I homodimerization (46, 47).
We demonstrated that the low-affinity site τ2 shares its binding orientation with other sites in the left-half subcomplex, which is in agreement with a recent DMS footprint assay (15). It is suggested in ref. 15 that DnaA loading to τ2 is assisted by cooperative interaction of DnaA molecules. Interactions through domain III, which forms homooligomers, would restrain binding sites and the orientation of domain IV of τ2-binding DnaA. This indirect support of the domain IV binding to DNA via the domain III oligomer may have functional roles in the formation of the initiation complex.
Regarding the mechanisms of how DnaA facilitates unwinding of dsDNA, two models have been proposed. In one model, DnaA molecules, which do not take part in the replication initiation complex, bind to ssDUE and retain the unwound state (7, 8). The other model is that DnaA molecules composing left-half subcomplex possess dual functions of dsDNA binding and ssDUE binding (9, 10). Our current model can be used to test the latter model. In our simulation model of the left-half subcomplex, the modeled DNA terminus proximal to DUE orients toward the pentameric DnaA AAA+ helix, despite the absence of an explicit DUE region. Additionally there seems to be enough space for ssDUE entering around DnaA V211 and R245, which are known to be important residues for ssDUE binding (6). Thus, the current model suggests that the second model is possible in E. coli. Of note, this modeled structure does not rule out the first model.
Finally, we note that the current coarse-grained modeling has potential limitations. First, we used a flexible linker model (38) for the linker between domains III and IV. The flexibility could be somewhat overestimated. We note, however, that the current modeling provided well-converged models, suggesting that interactions between DnaA domain IV and dsDNA as well as those in AAA+ domain III oligomerization provided sufficient restraints to uniquely determine the whole complex structure. Second, we used the complex crystal structure of the R1 box and DnaA domain IV as the reference structures for all of the interactions in the DnaA boxes. The difference in affinity was only represented by the interaction strength. The docked structure may be distinct between the strong and weak binding cases. Currently, the binding modes at low-affinity boxes have not been structurally characterized.
Methods
All detailed methods are described in SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
We thank Dr. Masayuki Su’etsugu for construction of M13oriCMS9; Dr. Matthew J. McGrath for revising the English; and the Research Support Center, Graduate School of Medical Sciences (Kyushu University) for DNA sequencing. This work was funded by Japan Society for the Promotion of Science (JSPS) KAKENHI Grants 15H01351, 26104517, 25251019, 16J02075, 26291004, and 16H00775; and the Strategic Programs for Innovative Research “Supercomputational Life Science” of the Ministry of Education, Culture, Sports, Science and Technology, Japan. Y.S. was supported by a JSPS predoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.H. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609649113/-/DCSupplemental.
References
- 1.Costa A, Hood IV, Berger JM. Mechanisms for initiating cellular DNA replication. Annu Rev Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaguni JM. Replication initiation at the Escherichia coli chromosomal origin. Curr Opin Chem Biol. 2011;15(5):606–613. doi: 10.1016/j.cbpa.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: Conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol. 2010;8(3):163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 4.Leonard AC, Grimwade JE. Regulation of DnaA assembly and activity: Taking directions from the genome. Annu Rev Microbiol. 2011;65:19–35. doi: 10.1146/annurev-micro-090110-102934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozaki S, Katayama T. DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid. 2009;62(2):71–82. doi: 10.1016/j.plasmid.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Ozaki S, et al. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J Biol Chem. 2008;283(13):8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 7.Duderstadt KE, et al. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J Biol Chem. 2010;285(36):28229–28239. doi: 10.1074/jbc.M110.147975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duderstadt KE, Chuang K, Berger JM. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478(7368):209–213. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozaki S, Katayama T. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2012;40(4):1648–1665. doi: 10.1093/nar/gkr832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozaki S, Noguchi Y, Hayashi Y, Miyazaki E, Katayama T. Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J Biol Chem. 2012;287(44):37458–37471. doi: 10.1074/jbc.M112.372052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bramhill D, Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 12.Grimwade JE, Ryan VT, Leonard AC. IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol Microbiol. 2000;35(4):835–844. doi: 10.1046/j.1365-2958.2000.01755.x. [DOI] [PubMed] [Google Scholar]
- 13.McGarry KC, Ryan VT, Grimwade JE, Leonard AC. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc Natl Acad Sci USA. 2004;101(9):2811–2816. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawakami H, Keyamura K, Katayama T. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J Biol Chem. 2005;280(29):27420–27430. doi: 10.1074/jbc.M502764200. [DOI] [PubMed] [Google Scholar]
- 15.Rozgaja TA, et al. Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol Microbiol. 2011;82(2):475–488. doi: 10.1111/j.1365-2958.2011.07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaper S, Messer W. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J Biol Chem. 1995;270(29):17622–17626. doi: 10.1074/jbc.270.29.17622. [DOI] [PubMed] [Google Scholar]
- 17.Blaesing F, Weigel C, Welzeck M, Messer W. Analysis of the DNA-binding domain of Escherichia coli DnaA protein. Mol Microbiol. 2000;36(3):557–569. doi: 10.1046/j.1365-2958.2000.01881.x. [DOI] [PubMed] [Google Scholar]
- 18.Margulies C, Kaguni JM. Ordered and sequential binding of DnaA protein to oriC, the chromosomal origin of Escherichia coli. J Biol Chem. 1996;271(29):17035–17040. doi: 10.1074/jbc.271.29.17035. [DOI] [PubMed] [Google Scholar]
- 19.Weigel C, Schmidt A, Rückert B, Lurz R, Messer W. DnaA protein binding to individual DnaA boxes in the Escherichia coli replication origin, oriC. EMBO J. 1997;16(21):6574–6583. doi: 10.1093/emboj/16.21.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: A protein-induced DNA U-turn. Cell. 1996;87(7):1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 21.Fujikawa N, et al. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003;31(8):2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roth A, Messer W. The DNA binding domain of the initiator protein DnaA. EMBO J. 1995;14(9):2106–2111. doi: 10.1002/j.1460-2075.1995.tb07202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erzberger JP, Pirruccello MM, Berger JM. The structure of bacterial DnaA: Implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002;21(18):4763–4773. doi: 10.1093/emboj/cdf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol. 2006;13(8):676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- 25.Sekimizu K, Bramhill D, Kornberg A. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50(2):259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- 26.Abe Y, et al. Structure and function of DnaA N-terminal domains: Specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J Biol Chem. 2007;282(24):17816–17827. doi: 10.1074/jbc.M701841200. [DOI] [PubMed] [Google Scholar]
- 27.Noguchi Y, Sakiyama Y, Kawakami H, Katayama T. The Arg fingers of key DnaA protomers are oriented inward within the replication origin oriC and stimulate DnaA subcomplexes in the initiation complex. J Biol Chem. 2015;290(33):20295–20312. doi: 10.1074/jbc.M115.662601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potoyan DA, Zheng W, Komives EA, Wolynes PG. Molecular stripping in the NF-κB/IκB/DNA genetic regulatory network. Proc Natl Acad Sci USA. 2016;113(1):110–115. doi: 10.1073/pnas.1520483112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra G, Levy Y. Molecular determinants of the interactions between proteins and ssDNA. Proc Natl Acad Sci USA. 2015;112(16):5033–5038. doi: 10.1073/pnas.1416355112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potoyan DA, Savelyev A, Papoian GA. Recent successes in coarse-grained modeling of DNA. WIREs Comput Mol Sci. 2013;3(1):69–83. [Google Scholar]
- 31.Takada S, et al. Modeling structural dynamics of biomolecular complexes by coarse-grained molecular simulations. Acc Chem Res. 2015;48(12):3026–3035. doi: 10.1021/acs.accounts.5b00338. [DOI] [PubMed] [Google Scholar]
- 32.Kenzaki H, et al. CafeMol: A coarse-grained biomolecular simulator for simulating proteins at work. J Chem Theory Comput. 2011;7(6):1979–1989. doi: 10.1021/ct2001045. [DOI] [PubMed] [Google Scholar]
- 33.Terakawa T, Kenzaki H, Takada S. p53 searches on DNA by rotation-uncoupled sliding at C-terminal tails and restricted hopping of core domains. J Am Chem Soc. 2012;134(35):14555–14562. doi: 10.1021/ja305369u. [DOI] [PubMed] [Google Scholar]
- 34.Li W, Terakawa T, Wang W, Takada S. Energy landscape and multiroute folding of topologically complex proteins adenylate kinase and 2ouf-knot. Proc Natl Acad Sci USA. 2012;109(44):17789–17794. doi: 10.1073/pnas.1201807109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knotts TA, 4th, Rathore N, Schwartz DC, de Pablo JJ. A coarse grain model for DNA. J Chem Phys. 2007;126(8):084901. doi: 10.1063/1.2431804. [DOI] [PubMed] [Google Scholar]
- 36.Sambriski EJ, Schwartz DC, de Pablo JJ. A mesoscale model of DNA and its renaturation. Biophys J. 2009;96(5):1675–1690. doi: 10.1016/j.bpj.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang J, et al. The I-TASSER Suite: Protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terakawa T, Takada S. Multiscale ensemble modeling of intrinsically disordered proteins: p53 N-terminal domain. Biophys J. 2011;101(6):1450–1458. doi: 10.1016/j.bpj.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garner J, Crooke E. Membrane regulation of the chromosomal replication activity of E. coli DnaA requires a discrete site on the protein. EMBO J. 1996;15(13):3477–3485. [PMC free article] [PubMed] [Google Scholar]
- 40.Sutton MD, Kaguni JM. Threonine 435 of Escherichia coli DnaA protein confers sequence-specific DNA binding activity. J Biol Chem. 1997;272(37):23017–23024. doi: 10.1074/jbc.272.37.23017. [DOI] [PubMed] [Google Scholar]
- 41.Messer W, et al. The complex for replication initiation of Escherichia coli. Chromosoma. 1992;102(1) Suppl:S1–S6. doi: 10.1007/BF02451779. [DOI] [PubMed] [Google Scholar]
- 42.Fang L, Davey MJ, O’Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol Cell. 1999;4(4):541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- 43.Keyamura K, Abe Y, Higashi M, Ueda T, Katayama T. DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J Biol Chem. 2009;284(37):25038–25050. doi: 10.1074/jbc.M109.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker TA, Sekimizu K, Funnell BE, Kornberg A. Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell. 1986;45(1):53–64. doi: 10.1016/0092-8674(86)90537-4. [DOI] [PubMed] [Google Scholar]
- 45.Kaur G, et al. Building the bacterial orisome: High-affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol Microbiol. 2014;91(6):1148–1163. doi: 10.1111/mmi.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simmons LA, Felczak M, Kaguni JM. DnaA Protein of Escherichia coli: Oligomerization at the E. coli chromosomal origin is required for initiation and involves specific N-terminal amino acids. Mol Microbiol. 2003;49(3):849–858. doi: 10.1046/j.1365-2958.2003.03603.x. [DOI] [PubMed] [Google Scholar]
- 47.Felczak MM, Simmons LA, Kaguni JM. An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J Biol Chem. 2005;280(26):24627–24633. doi: 10.1074/jbc.M503684200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.