Significance
Drug resistance is among the most critical problems in cancer treatment. This proof-of-principle study demonstrates that a combination of in vitro microecology and deep sequencing could provide an extremely efficient method to elucidate the in vivo resistance mechanisms of cancer drugs and predict if drug resistance is likely to occur. Remarkably, drug resistance was seen within 7 d in our microfluidic chip, enabling characterization of molecular mechanisms within a month. This would be of great value for clinicians in selecting drugs likely to be slow in drug resistance emergence, and choose therapies for resistant tumors more effectively. In principle, our microecology technology can be used for many combinations of cancer types and drugs.
Keywords: microhabitats, cancer, doxorubicin, evolution, resistance
Abstract
In vitro prediction of the probable rapid emergence of resistance to a drug in tumors could act to winnow out potential candidates for further costly development. We have developed a microfluidic device consisting of ∼500 hexagonal microcompartments that provides a complex ecology with wide ranges of drug and nutrient gradients and local populations. This ecology of a fragmented metapopulation induced the drug resistance in stage IV U87 glioblastoma cells to doxorubicin in 7 d. Exome and transcriptome sequencing of the resistant cells identified mutations and differentially expressed genes. Gene ontology and pathway analyses of the genes identified showed that they were functionally relevant to the established mechanisms of doxorubicin action. Specifically, we identified (i) a frame-shift insertion in the filamin-A gene, which regulates the influx and efflux of topoisomerase II poisons; (ii) the overexpression of aldo-keto reductase enzymes, which convert doxorubicin into doxorubicinol; and (iii) activation of NF-κB via alterations in the nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway from mutations in three genes (CARD6, NSD1, and NLRP13) and the overexpression of inflammatory cytokines. Functional experiments support the in silico analyses and, together, demonstrate the effects of these genetic changes. Our findings suggest that, given the rapid evolution of resistance and the focused response, this technology could act as a rapid screening modality for genetic aberrations leading to resistance to chemotherapy as well as counter selection of drugs unlikely to be successful ultimately.
A fundamental problem in chemotherapy is that cancer patients frequently develop drug resistance, limiting the survival benefit of therapeutic agents to the short term; this is true of traditional chemotherapeutic reagents as well as so-called second generation reagents that target specific oncogenes (1). Indeed, up to 80% of chronic myelogenous leukemia patients who received imatinib therapy, targeting the BCR-ABL1 (breakpoint cluster region-Abelson 1) fusion gene, eventually relapse even after apparent remission (2). Likewise, resistance to vemurafenib has been seen among melanoma patients with BRAF-V600 mutation, limiting its long-term efficacy (3, 4). Thus, prediction of the emergence of resistance to cancer drugs is a crucial clinical issue even in the era of targeted therapy.
A major bottleneck in studying the origins of cancer drug resistance is the inability to follow the progression of resistance at the molecular and cellular levels. Patient cells from drug-resistant tumors are the most relevant tissues in a clinical situation. However, they are difficult to obtain and limited in amount, making it difficult to use them for repetitive testing of different drugs. Alternatively, resistant cell lines can be established by serial passaging of cell cultures with drug concentration increasing in a stepwise manner, but this process typically requires several months of optimization. Therefore, faster ways of producing resistant cells where the progression could be tracked would be of great value for mechanistic studies of cancer drug resistance and rapid screening of new drugs for resistant tumors. Such a system could be used to counterselect given drugs for clinical use a priori based on rapid emergence of drug resistance in vitro.
A previous study showed that induction of bacterial resistance to antibiotics can be accelerated by an ecology consisting of thousands of interconnected microscale chambers with a concentration gradient of the antibiotic (5). More recently, a similar microfluidic perfusion cell culture device containing a single chamber of flow channels with stable gradients was used to induce resistance to an anticancer drug doxorubicin (DOX) within 72 h in metastatic breast cancer cells (6) and in multiple myeloma cancer cells (7). However, this linear ecology had a fixed drug gradient that fell short of fully reproducing the complexity of tumor environments. Furthermore, etching onto a silicon wafer does not permit a large-scale production for high-throughput screening.
Here, we describe the use of a modified ecology based on the bacterial work of Zhang et al. (5), which we call a Cancer Drug Resistance Accelerator (CDRA) chip, to rapidly induce DOX resistance in glioblastoma (U87) cells. Subsequent application of high-throughput sequencing revealed multiple mechanisms of DOX resistance development, including activation of NF-κB signaling via the nucleotide-binding oligomerization domain (NOD)-like receptor (NLR, a subset of pattern recognition receptors) pathway. Our results indicate the potential utility of CDRA chips in selecting and counterselecting therapeutic agents in a variety of cancer types.
Results
Microfluidic Chip Design and Cell Culture for Resistance Induction.
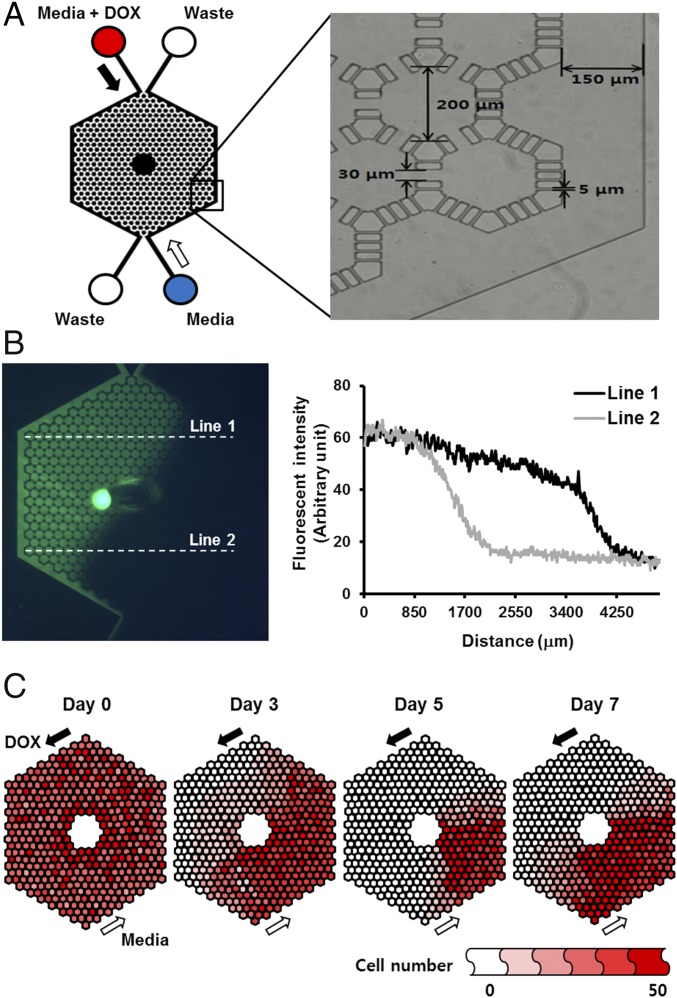
The CDRA chip contained 488 hexagonal microchambers (diameter 200 μm, height 40 μm) surrounded by two microchannels (width 150 μm) for long-term cell culture under antiparallel overlapping gradients of DOX and nutrients (Fig. 1A). The concentration gradients on the chip were successfully shown by flowing fluorescent material from one of the inlets and measuring the fluorescence intensity (Fig. 1B). The chip operates without the aid of additional instrumentation for the generation of concentration gradient in typical cell culture incubators. Culture requires minimal manipulation with daily resupply of DOX and nutrients whose flow is maintained by gravity only. The handy operation is schematically depicted along with the actual image of the CDRA chip (SI Appendix, Fig. S1). The chip, made from polydimethylsiloxane (PDMS), can be produced in large scale at low cost via routine soft-microfabrication replica processes.
Fig. 1.
Design and properties of the CDRA chip. (A) Schematic diagram of the chip and a microscopic image showing chip dimensions. Filled arrow indicates the flow of 5 μM DOX and nutrients, whereas empty arrow represents the flow of nutrients only. (B) A fluorescence image of the concentration gradient. The plot shows the fluorescence intensity profile along two horizontal lines. (C) Distribution of viable cells in the chip over a period of 7 d. Cell numbers in each hexagonal are indicated in red color coded in six steps.
U87 cancer cells were distributed homogeneously throughout the microchambers before flowing DOX and media, and cell numbers were counted over 7 d from microscopic images of the CDRA chip (SI Appendix, Fig. S2). We observed a dramatic decrease in cell numbers in the chambers near the DOX channel at day 3, and, apparently, three-quarters of the chambers became empty at day 5 owing to the cytotoxic effect of DOX (Fig. 1C). Cell repopulation was evident by day 7 (Fig. 1C), indicating the development of resistance and migration of resistant cells to areas with higher DOX concentration.
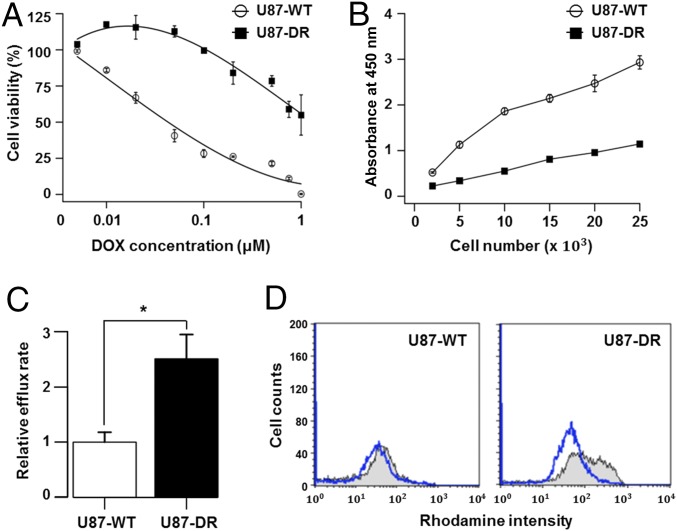
We performed several experiments to validate the DOX resistance. Cells harvested from the chip at day 7 (U87-DR) showed ∼30 times greater resistance to DOX than wild-type U87 cells (U87-WT) as indicated by IC50 (Fig. 2A), although the proliferation activity was reduced by half in the resistant cells compared with the WT cells (Fig. 2B). One of the main causes of drug resistance is the change in drug pump activity. In a Rhodamine 123 efflux assay, reduction in fluorescence intensity reflecting dye exclusion was ∼2.5-fold higher in the resistant cells (Fig. 2C). Also, a drug efflux assay using flow cytometry showed a substantial increase in efflux activity after acquisition of the resistance (Fig. 2D). Together, these data indicate that the cells acquired distinct attributes as DOX-resistant cells within 7-d-long drug exposure.
Fig. 2.
Characterization of DOX-resistant cells from the CDRA chip. (A) Cell viability of resistant (U87-DR) and WT (U87-WT) cells at various concentrations (0 μM to 1 μM) of DOX. IC50 values were estimated to be 0.05 μM and 1.47 μM for the WT and resistant cells, respectively. Note that the x-axis scale is logarithmic. (B) Cell proliferation activity of the resistant and WT cells in a drug-free condition. (C) Relative efflux rate measured by the reduced fluorescence intensity of Rhodamine 123 due to dye efflux (P value = 0.008 with one-tailed t test, from triplicate experiments). (D) Estimation of drug efflux activities of the WT and resistant cells. Cell counts were measured before (gray line) and after (blue line) the 1 h-incubation in flow cytometry.
Identification of Gene Alterations for DOX Resistance.
We used a combination of exome and transcriptome sequencing to elucidate resistance mechanisms and shed light on the development of DOX resistance in cancer cells. We note that, like many cancers, the mutational spectrum of the U87 cells before incubation in the chip is extraordinarily complex, with hundreds of single-nucleotide variants (SNVs), small indels, large microdeletions, and interchromosomal translocations (8). Despite this complexity, the U87 cells come into our chip highly sensitive to DOX, but come out highly resistant.
To identify genomic aberrations responsible for the development of DOX resistance, we performed exome sequencing of WT and DOX-resistant U87 cells obtained after 7 d of growth on a CDRA chip and compared the results. We combined the results from four software programs [MuTect (9), Strelka (10), VarScan2 (11), and Virmid (12)] for detecting somatic mutations and identified 107 variants with high confidence (i.e., called by multiple programs), including 2 indels, 5 stopgain mutations, and 54 missense mutations (SI Appendix, Table S1; see Materials and Methods for details). A statistical functional analysis of these nonsynonymous variants using Web-based Gene Set Analysis Toolkit (WebGestalt) (13) showed that three representative terms were enriched in the Gene Ontology (GO) molecular function category, namely “carbohydrate derivative binding,” “nucleoside binding,” and “ATP-dependent helicase activity” (SI Appendix, Table S2). These findings further support the findings of previous studies that DOX inhibits the enzyme topoisomerase II by intercalating between two base pairs of the DNA double helix (14).
The helicase activity of topoisomerase II was shown to be potently inhibited by anthracycline antibiotics such as DOX (15). Moreover, we observed a mutation in the CHD1 (chromodomain helicase DNA binding protein 1) gene, which is a known target of epirubicin, the 4′-epi-isomer of DOX (16). Another notable mutation was a frame-shift insertion in the filamin-A (FLNA) gene, whose loss is known to promote DOX resistance by regulating the influx and efflux of topoisomerase II poisons (17).
Transcriptome Sequencing and Pathway Analysis for Resistance Development.
To further identify the altered pathways and molecular processes responsible for emergence of drug resistance, we generated RNA-seq data for the U87 WT and resistant cell lines obtained after 7 d of growth on a CDRA chip. Our computational pipeline identified 83 differentially expressed genes (DEGs), of which 77 were up-regulated and 6 were down-regulated (SI Appendix, Table S3; see Materials and Methods for details). A gene set analysis of GO terms for these 83 DEGs yielded many terms related to immune responses, including “response to wounding,” “inflammatory response,” “leukocyte chemotaxis,” “response to hypoxia,” “regulation of cell proliferation,” and “cytokine activity’ (SI Appendix, Table S4). The most relevant term was “DOX metabolic process” [false discovery rate (FDR) = 3.89E-6], which was represented by four aldo-keto reductases (AKRs; AKR1B1, AKR1C1, AKR1C2, and AKR1C3) that convert DOX to doxorubicinol. Compared with DOX, doxorubicinol shows dramatically reduced cytotoxicity, reduced DNA-binding activity, and strong localization to extranuclear lysosomes (18).
The Kyoto Encyclopedia of Genes and Genomes pathway analysis of 83 DEGs with WebGestalt (13) yielded the NOD-like receptor signaling pathway (FDR = 1.33E-6) and cytokine−cytokine receptor interaction pathway (FDR = 2.46E-8). The DEGs involved in these pathways were mostly chemokines and proinflammatory cytokines (e.g., CCL2, CXCL1, CXCL2, CXCL3, IL1B, IL6, IL8, and IL11) that were regulated by the nuclear factor kappa B (NF-κB) pathway.
Notably, NF-κB activation is known to be essential for the cytotoxic effects of DOX and its analogs (19). From the exome sequencing data, we further identified three mutated genes (CARD6, NSD1, and NLRP13) that are associated with NF-κB activation. CARD6 (caspase activation and recruitment domain family, member 6) negatively regulates NF-κB activation by NOD1 (nucleotide-binding oligomerization domain containing 1) receptor in human embryonic kidney-derived HEK293T cells (20, 21) whereas the effect is reversed in Huh7 cells of hepatocarcinoma origin (22), thus indicating that the role of CARD6 in NF-κB activation may be cell-type specific.
Experimental Validation for Key Regulators of DOX Resistance.
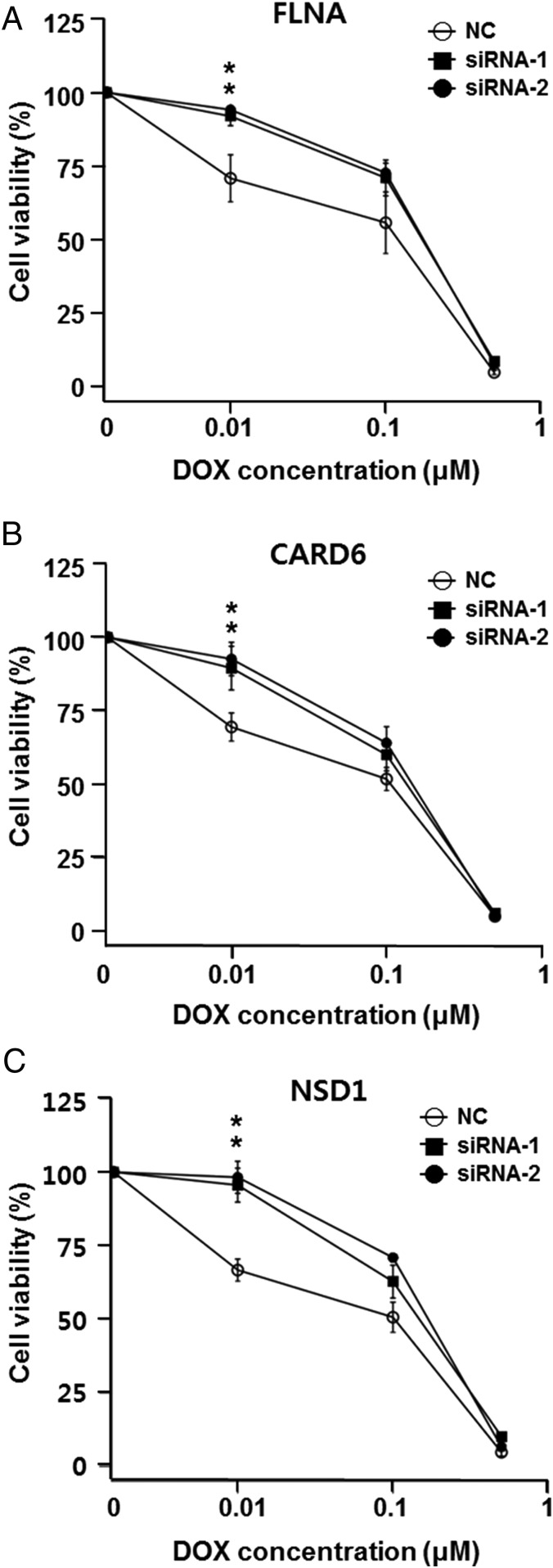
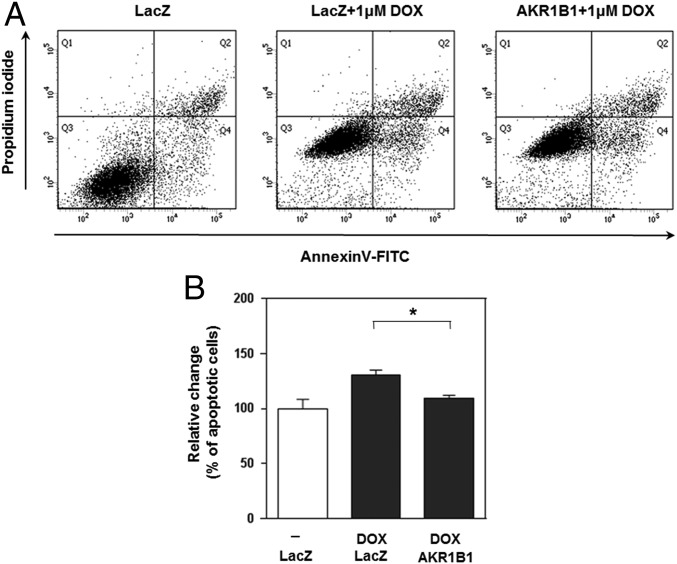
We next sought to carry out preliminary validation of our in silico analyses. First, we attempted to mimic the effect of the frame-shift mutation in FLNA by knocking down with specific siRNAs. Consistent with predicted loss of function from a frame-shift mutation, it was verified that the down-regulation of FLNA by siRNA caused a 1.5-fold increase in DOX resistance at the 10 nM concentration in the U87 cancer cell line (Fig. 3A). We also observed a stopgain mutation in CARD6 in resistant cells (c.G2014T of NM_032587 encoding p.Gly672*) and performed a similar knockdown experiment for CARD6. The treatment of U87 cells with siRNA specific for CARD6 elevated DOX resistance consistent with the down-regulation of NF-κB pathway signaling in these cells (Fig. 3B). In addition, we identified a missense mutation (c.T3380G of NM_02245 encoding p.Leu1127Arg) in NSD1 (nuclear receptor binding SET domain protein 1), reduced expression of which is known to decrease NF-κB activation (23). Again, the knockdown of NSD1 increased DOX resistance consistent with reduced NF-κB activation, in turn, leading to reduced apoptosis (Fig. 3C). Finally, we identified a missense mutation (c.T1067A of NM_176810 encoding p.Leu365Gln) in NLRP13 (NOD-like receptor family, pyrin domain containing 13). Notably however, RNA-seq data indicated that NLRP13 is not expressed in U87 cells at a detectable level, thereby rendering the functional assignment of the mutation difficult. Overall, both the transcriptome and exome sequencing data indicate that NF-κB activation via NOD-like receptor signaling and subsequent cytokine/chemokine production are important factors in the development of DOX resistance. Our work establishes the relevance of NOD-like receptor signaling in DOX resistance. Finally, we verified that AKR family 1, member B1 (AKR1B1), was up-regulated in DOX-resistant U87 cells and that the ectopic expression of AKR1B1 reduced apoptosis upon DOX treatment (Fig. 4).
Fig. 3.
Knockdown of FLNA, CARD6, and NSD1 by siRNAs in U87 cells leads to increased resistance to DOX. Cell survival curves of 72-h DOX-treated cells transfected with control siRNAs (NC) or gene-specific siRNAs are shown. Graphs show the representative results of three independent cell viability assays after knockdown of each of three genes, (A) FLNA, (B) CARD6, and (C) NSD1. Knockdown of FLNA, CARD6, and NSD1 led to increased resistance to DOX, where the average *P = 0.023, 0.004, and 0.004, respectively, at 0.01 μM DOX concentration. The knockdown efficiency of each siRNA is shown in SI Appendix, Fig. S3.
Fig. 4.
AKR1B1 overexpression inhibits DOX-induced apoptosis in U87 cells. (A) AKR1B1-overexpressing U87 cells were treated with DOX (1.0 μM) for 24 h. Subsequent apoptosis was measured by dual staining with Annexin V-FITC and propidium iodide. Representative dot plots show the distinct populations of viable cells (FITC−PI−, lower left Q3), early apoptotic (FITC+PI−, lower right quadrant Q4) and late apoptotic or necrotic cells (FITC+PI+, upper right quadrant Q2). (B) Graphs show the relative change in percentage of cells in early apoptosis and late apoptosis/necrosis compared with lacZ-transfected cells as a negative control, set to 100. Bars indicate the means of the relative changes in percentages of total apoptosis from the triplicate experiments. *P = 0.0022 with one-tailed t test.
Discussion
This proof-of-principle study shows that the CDRA chip consisting of a complex microecology with concentration gradients of mutagenic drugs such as DOX can induce DOX resistance rapidly in glioblastoma cells in vitro and that subsequent analysis of mutation and expression data can identify the molecular mechanisms involved in drug resistance development.
With an optimization in chip design and fabrication processes to allow for mass production at low cost, the throughput of analyzing drugs that rapidly lead to resistant clones under conditions of heterogeneous stress and fragmented cell populations can be increased to analyze dozens or even hundreds of cancer cell lines and drugs in a month. Combined with deep sequencing, we should be able to obtain a list of causal mutations reflecting diverse tumor contexts of many cell lines efficiently. Compilation of mutations with respect to cancer types and primary chemotherapeutic reagents that had been applied could lead to a large-scale, genetically based profiling of resistance mechanisms; this should represent a database for high-throughput mechanistic studies.
Our system can also provide guidance for avoiding certain combinations of drugs and cancer types. Specifically, grade IV glioma, also called glioblastoma multiforme (GBM), is the most common primary malignant brain tumor and also the most deadly, with a 1-y survival rate of only 30% (24). Temozolomide is the first-line treatment for chemotherapy of GBM patients used concomitantly with radiotherapy. The CDRA chip typically runs with the drug concentration in the inlet reservoirs at 50 to 100 times the IC50 value. Because the IC50 value of temozolomide is unusually high, the DMSO solvent itself is toxic to cells at such concentration. Thus, we chose DOX instead. DOX is used, at present, to treat many cancers with some success, but it has not been used in treating GBM, because of the poor penetration of the blood−brain barrier. There are efforts underway to develop innovative means for delivery of DOX to GBMs (25), in the hopes that DOX can be effective in this grim cancer. We suggest here that, even if it is possible for DOX to be delivered across the blood–brain barrier, it will not be an effective GBM therapy, at least not by itself, because of induced resistance.
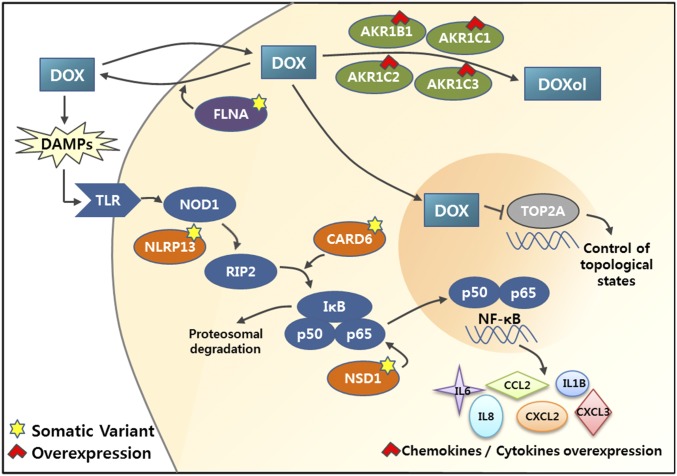
In this work on U-87 cells under DOX treatment, three molecular mechanisms were identified (see schematic diagram in Fig. 5), namely (i) an FLNA mutation affecting DOX influx and efflux, (ii) overexpression of AKR, which converts DOX into the less toxic doxorubicinol, and (iii) NF-κB activation via NOD-like receptor signaling and production of inflammatory cytokines and chemokines. Of note, although the drug efflux activity increased by 2.5-fold (Fig. 2C), transcriptome sequencing data did not show up-regulation of transporters associated with DOX pathway, such as ABCC1 (MRP1), ABCC2, ABCG2, RALBP1, and ABCB1 (MDR1) (26). One possible explanation may have to do with the mutation of FLNA, the deficiency of which alters the transport of DOX via yet uncharacterized mechanisms (17). Interestingly, Campos et al. (27) reported recently that FLNA negatively regulated NF-κB activation in melanoma cell by inhibition of Akt signaling, which may further link the loss-of-function mutation of FLNA to NF-κB activation.
Fig. 5.
Molecular mechanisms of DOX resistance. Functional network involving genes with mutations and altered expression is shown. Genes with loss-of-function mutations are marked with yellow stars, and genes with elevated gene-expression are marked with red arrowheads. AKR1 family members are involved in metabolic conversion of DOX to doxorubicinol, and FLNA regulates its efflux out of the cells. CARD6, NSD1, and NLRP13 are involved in regulation of NF-κB activation (see Results and Discussion for details).
Although we provide preliminary functional validation, several outstanding issues remain for further investigation. First, it is unclear whether these mechanisms are independent or cooperative, because we sequenced a mixture of cells from the entire chip in this study. Sequencing cells in each compartment separately, which requires modification of the chip design to allow access to cells in individual compartments, would be useful to determine the independence or cooperativeness of resistance mechanisms. Another important issue in the resistance development is the origin of the resistant clones. Two contrasting scenarios are clonal expansion of resistant clones that were present in the initial population in extreme minor population and acquisition of de novo mutations after drug treatment and subsequent clonal expansion. Single-cell sequencing of resistant products from the CDRA chip would be a viable option to investigate the evolutionary process of tumor cells for developing resistance.
The mechanisms involved in the development of drug resistance vary greatly, depending on the cancer types and cancer drugs (28). This, in turn, implies that there may be some level of specificity due to personal variations, including the genotype and clinical and treatment history as well as the cancer type in question (29). In this regard, as well, our chip system could be of value, as our system could be also applied to patient-derived primary tumor cells. Culturing patient-derived primary cells on the CDRA chip should make it possible to investigate the drug resistance mechanisms that are specific to the patient within a few weeks. In addition, the requirement for a small number of cells per chip implies that multiple parallel prospective studies can be carried out in clinical settings to identify the likely response/resistance course over time and the mechanisms of tumor cell adaption. Driver mutations thus identified may serve as predictive markers for monitoring of resistance development and/or as targets for therapeutic agents that can bypass the relevant drug resistance mechanisms. This represents the potential to greatly advance studies of the molecular mechanisms of resistance development and thereby to allow the development of more targeted, personalized, and effective cancer treatments.
Materials and Methods
Fabrication and Operation of a CDRA Chip.
The chip was designed by AutoCAD software (student version 2012) and fabricated in a chrome mask. Soft lithography was used to make the chip in PDMS (Sylgard 184 silicone elastomer; Dow Corning Co.) (30). In detail, the negative-tone epoxy photoresist, SU-8 2025 (MicroChem), was spin-coated on a 4-inch silicon wafer for 30 s at 2,000 rpm to obtain a 40-µm-thick layer. The SU-8-coated wafer was exposed to UV through the mask by a contact aligner (MDA-400M; MIDAS). The exposed wafer was developed by immersing it into a glass container filled with SU-8 developer (Microchem). The patterned mold was coated with trichloro(1H,1H,2H,2H-perfluorooctyl)silane (Sigma-Aldrich) under vacuum conditions at room temperature for 2 h. To make a PDMS replica of the mold, a mixture of the PDMS prepolymer and curing agent in a 10:1 (w:w) ratio was cast onto the mold and cured at 80 °C for 2 h. Then, the polymerized PDMS layer was peeled off of the mold. A single cell loading hole (0.9 mm diameter) and four reservoirs (6 mm diameter) were punched through the cured PDMS layer (31). Afterward, the layer was treated with O2 plasma for 30 s at 90 watts and then bound to a freshly cleaned glass slide (76 mm × 26 mm × 1 mm) (Marienfeld).
Dimensions of the completed chip are 32 mm (length) × 20 mm (width) × 9 mm (height). The CDRA chip contained an array of 488 hexagonal microchambers (diameter 200 μm, height 40 μm). The peripheral microchambers had sides of microslits with a gap of 5 μm to allow drugs and nutrients to flow into the interior microchambers, and the other interior microchambers had sides with three posts with the largest gap of 30 μm to allow cells to freely migrate into their neighboring microchambers.
Cell Culture in CDRA Chip.
The human glioma cell line U87 was obtained from the commercial provider ATCC (American Type Culture Collection) and did not require institutional review board (IRB) approval. It was cultured in MEM (minimum essential media) (Life Technologies) supplemented with 10% (vol/vol) FBS (HyClone Laboratories), 100 units per milliliter of penicillin (Life Technologies) and 100 µg/mL of streptomycin (Life Technologies).
For sanitization, 70% ethanol was carefully injected into the chip via the cell loading hole in the center of the chip using a needle-free syringe; 70% ethanol was then rinsed with PBS and MEM in a sequential manner. The interior surface of the chip was filled with PBS containing 10 μg/mL of fibronectin (Sigma-Aldrich) and incubated at 37 °C for 1 h, and unbound fibronectin was washed away three times with PBS.
At first, 100 μL of MEM were individually loaded into the four reservoirs (DOX+ and DOX− inlets, and waste 1 and waste 2 outlets) of the chip. About 50,000 cells in MEM were then loaded into the chip through the cell loading hole in the center of the chip using a pipette. Once the array of hexagonal microchambers was filled with cells, the hole was sealed with a metal pin. DOX+ and DOX− inlet reservoirs were filled with 150 μL of MEM, and the waste outlet reservoirs were filled with 50 μL of MEM. A pressure difference between each pair of the inlet and outlet induces gravity-driven perfusion to feed cells in the microchambers with nutrients and oxygen (32). The chip was incubated 24 h. The next day, the media in all of the reservoirs was removed using a pipette. DOX+ and DOX− inlet reservoirs were filled with 150 μL of MEM containing 5 μM DOX and MEM, respectively, and waste 1 and waste 2 outlet reservoirs were filled with 50 μL of MEM containing 5 μM DOX and MEM, respectively. This generated overlapping gradients of DOX and nutrients over the hexagonal chambers. The reservoir contents were replaced daily with freshly prepared MEM containing 5 μM DOX or MEM in the same manner and incubated for 7 d.
Once the culture was finished, cells were trypsinized, collected, and transferred to a new culture dish to keep culturing them for 1 wk under MEM to obtain sufficient numbers of cells for further characterization.
Demonstration of Concentration Gradients in the Chip.
The cell loading hole in the chip was sealed with the metal pin. DOX+ and DOX− inlet reservoirs in a diagonal position were filled with 150 μL of MEM containing 1 μM fluorescein isothiocyanate (FITC) (Sigma-Aldrich) and MEM, and waste 1 and waste 2 outlet reservoirs were filled with 50 μL of MEM containing 1 μM FITC and MEM, respectively. Fluorescence images were taken under an SMZ-1500 stereoscope with fluorescence filters (Olympus Co.) 6 h after loading MEM containing 1 μM FITC into the reservoirs. Fluorescence intensity profiles over the chip were analyzed by Image J program.
Cell Viability Analysis for U87 Cells.
To assess viability of U87-WT and U87-DR cells, 3,000 cells were seeded in 96 wells without or with DOX. After 72-h incubation, viable cells were counted in each well. The percentage of cell viability was calculated by dividing the viable cell number at each drug concentration by the number of cells cultured without drugs.
Drug Efflux Assay.
Multidrug resistance was determined by assaying the ability of the cell to extrude fluorescent dye, Rhodamine 123 (Sigma-Aldrich), which is a substrate of MDR1 (multidrug resistance protein 1) and MRP1 (multidrug resistance-associated protein 1) genes. Cells were preloaded with Rhodamine 123 for 30 min on ice, and then incubated in a 37 °C for 1 h to allow MDR protein-mediated efflux of the dye. The cells were stained with propidium iodide (PI; Sigma-Aldrich) and maintained on ice until analysis. The dye efflux was analyzed by flow cytometry (BD Biosciences) excluding PI-positive dead cells from the analysis. We also evaluated the efflux rate of Rhodamine 123 by a fluorometric plate reader (Molecular Devices). Cell suspensions were dispensed in the wells of a 96-well plate before and after dye efflux. Fluorescence intensities were measured at 485 nm (excitation) and 530 nm (emission).
Cell Proliferation Assay in Drug-Free Condition.
Cell proliferation of U87-WT and U87-DR cells was measured using EZ-Cytox cell viability assay kit (Daeillab Service). Briefly, the cells (1 × 103 to 3 × 104) were seeded in 96 wells containing 180 μL of media and cultured for 6 h. Then, 20 μL of EZ-Cytox reagents were added to each well and incubated for 1 h. Absorbance of water-soluble tetrazolium salt (WST)-formazan produced by mitochondrial dehydrogenase was measured at 450 nm using a microplate reader (Molecular Devices).
Gene-Specific siRNAs and siRNA Transfection.
For the knockdown experiments of FLNA, predesigned siRNAs and the scrambled negative control siRNAs were obtained from Genolution Pharmaceuticals. For the knockdown experiments of CARD6 and NSD1, predesigned siRNAs (FlexiTube GeneSolution GS84674 for CARD6 with IDs of Hs_CARD6_5 and Hs_CARD6_6; FlexiTube GeneSolution GS64324 for NSD1 with IDs of Hs_NSD1_3 and Hs_NSD1_8) and the scrambled negative control siRNAs were purchased from Qiagen. The siRNA duplexes were transfected to U87 cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions.
Cell Proliferation Assay for the Gene Knockdown.
The cell proliferation was determined by using the EZ-Cytox cell viability assay kit (Daeillab Service). Briefly, 3,000 cells of U87 were seeded to each well of a 96-well plate. The next day, 20 nM of target siRNAs were transfected. After 6 h, cells were treated with DOX in culture medium and incubated for 3 d. EZ-Cytox solution was added to each well and incubated at 37 °C for 3 h. Absorbance was measured at 450 nm using a microplate reader (Molecular Devices). The percentage viability was determined by normalizing the value at the condition without DOX.
Annexin V Apoptosis Analysis.
For apoptosis analysis, pcDNA3.1-AKR1B1 or pcDNA3.1-lacZ (a negative control) was transfected into U87 cells in a 35-mm dish using Lipofectamine 3000 (Life Technologies). At 24 h posttransfection, the cells were treated with 1.0 μM DOX for 24 h. Apoptosis was determined by dual staining with Annexin V-FITC and propidium iodide using a kit purchased from BD Biosciences. Flow cytometric analysis was carried out on a BD LSRFortessa cell analyzer (BD Biosciences). The fluorescence distribution of a total of 10,000 nuclei was analyzed using BD FACSDiva software. The relative proportion of Annexin V-positive cells was determined and counted as apoptotic cells.
Exome Sequencing Data Processing and Quality Control.
Total DNA was extracted from U87-WT and U87-DR cells with a genomic DNA extraction kit (Qiagen) following manufacturer’s instructions. Exome enrichment was achieved using Agilent SureSelect Human All Exon V4 with postcapture kit (Agilent). Exome sequencing was carried out with Illumina HiSeq-2000 sequencing platform (Illumina) using 101 base pair paired-end runs. Fastx-Toolkit (version 0.0.13.2) (hannonlab.cshl.edu/fastx_toolkit/) was used for preprocessing of the raw sequencing data such as trimming the adapter sequences, filtering artifacts, and discarding low-quality reads with a quality score less than 20. The reads were aligned to the human reference genome (hg19) using bowtie2 (version 2.1.0) with the default options (33). Genome Analysis Toolkit (version 2.5.2) (34) and Picard (version 1.92) (broadinstitute.github.io/picard/) were used for removal of duplicate reads, local realignment, and recalibration of base quality scores. The average number of aligned reads was 81 million, and the mean read depth of the target region was 187×.
Mutation Calling and Annotation.
To identify variations in DOX-resistant cells, we used four programs for identifying somatic mutations using the U87-WT cells as a control sample: (i) MuTect (version 1.1.4) (9) with final judgment of site, KEEP, (ii) Strelka (version 1.0.7) (10) with default parameters, (iii) VarScan2 (version 2.3.5) (11) with somatic P value < 0.05, and (iv) Virmid (version 1.1.0) (12) with passed variants. Somatic variants identified by multiple (≥2) programs were cataloged as reliable substitutions, and we have obtained 107 highly confident variants that included 2 indels, 5 stopgain mutations, and 54 missense mutations (SI Appendix, Table S1). Insertions and deletions were identified by Strelka and VarScan2. Functional variants were annotated using the ANNOVAR program (35), and Integrative Genomics Viewer was used for visual examination of read alignment (36).
RNA-Seq Data Processing.
Total RNA was extracted from U87-WT and U87-DR cells using a Direct-zol RNA MiniPrep kit (Zymo Research) following manufacturer’s instructions. RNA sequencing was performed with Illumina HiSeq-2500 sequencing platform (Illumina) using 101-bp paired-end runs. Fastx-Toolkit (version 0.0.13.2) was used for preprocessing of the raw sequencing data. The reads were aligned to the human reference genome (hg19) using MapSplice 2 (version 2.1.7) (37). RNA Sequencing by Expectation Maximization (RSEM) (version 1.2.12) (38) was used to quantify the mRNA abundance. Differential expressions between U87 and U87-DR cells were obtained by edgeR program (version 3.8.6) (39). DEGs were selected under the threshold of P value < 0.01 (SI Appendix, Table S3).
GO and Pathway Analysis.
Gene set analysis for GO terms and pathways was performed using WebGestalt web server (13). Somatic mutations and DEGs were included in the analysis (SI Appendix, Tables S2 and S4, respectively).
Supplementary Material
Acknowledgments
This work was supported by the Technology Innovation Program of the Ministry of Trade, Industry and Energy, Republic of Korea (Grant 10050154 to S.L. and S.P.), the National Research Foundation of Korea (Grant NRF-2014M3C9A3065221 to S.L. and Grant NRF-2015K1A4A3047851 to J.K. and S.L.) funded by the Ministry of Science, ICT, and Future Planning.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the NCBI Sequence Read Archive database (accession no. SRP066879).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1614898113/-/DCSupplemental.
References
- 1.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: An evolving paradigm. Nat Rev Cancer. 2013;13(10):714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 2.Hochhaus A, La Rosée P. Imatinib therapy in chronic myelogenous leukemia: Strategies to avoid and overcome resistance. Leukemia. 2004;18(8):1321–1331. doi: 10.1038/sj.leu.2403426. [DOI] [PubMed] [Google Scholar]
- 3.Chapman PB, et al. BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagle N, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29(22):3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, et al. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science. 2011;333(6050):1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 6.Wu A, et al. Cell motility and drug gradients in the emergence of resistance to chemotherapy. Proc Natl Acad Sci USA. 2013;110(40):16103–16108. doi: 10.1073/pnas.1314385110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu A, et al. Ancient hot and cold genes and chemotherapy resistance emergence. Proc Natl Acad Sci USA. 2015;112(33):10467–10472. doi: 10.1073/pnas.1512396112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark MJ, et al. U87MG decoded: The genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6(1):e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saunders CT, et al. Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28(14):1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 11.Koboldt DC, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S, et al. Virmid: Accurate detection of somatic mutations with sample impurity inference. Genome Biol. 2013;14(8):R90. doi: 10.1186/gb-2013-14-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013;41(Web Server issue):W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang F, Teves SS, Kemp CJ, Henikoff S. Doxorubicin, DNA torsion, and chromatin dynamics. Biochim Biophys Acta. 2014;1845(1):84–89. doi: 10.1016/j.bbcan.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachur NR, et al. Helicase inhibition by anthracycline anticancer agents. Mol Pharmacol. 1992;41(6):993–998. [PubMed] [Google Scholar]
- 16.Rask-Andersen M, Almén MS, Schiöth HB. Trends in the exploitation of novel drug targets. Nat Rev Drug Discov. 2011;10(8):579–590. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- 17.Yue J, Lan S, Yuan C, Shen Z. Prognostic values of filamin-A status for topoisomerase II poison chemotherapy. Int J Biol Sci. 2012;8(4):442–450. doi: 10.7150/ijbs.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heibein AD, Guo B, Sprowl JA, Maclean DA, Parissenti AM. Role of aldo-keto reductases and other doxorubicin pharmacokinetic genes in doxorubicin resistance, DNA binding, and subcellular localization. BMC Cancer. 2012;12:381. doi: 10.1186/1471-2407-12-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashikawa K, Shishodia S, Fokt I, Priebe W, Aggarwal BB. Evidence that activation of nuclear factor-kappaB is essential for the cytotoxic effects of doxorubicin and its analogues. Biochem Pharmacol. 2004;67(2):353–364. doi: 10.1016/j.bcp.2003.08.039. [DOI] [PubMed] [Google Scholar]
- 20.Strober W, Murray PJ, Kitani A, Watanabe T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat Rev Immunol. 2006;6(1):9–20. doi: 10.1038/nri1747. [DOI] [PubMed] [Google Scholar]
- 21.Stehlik C, Hayashi H, Pio F, Godzik A, Reed JC. CARD6 is a modulator of NF-kappa B activation by Nod1- and Cardiak-mediated pathways. J Biol Chem. 2003;278(34):31941–31949. doi: 10.1074/jbc.M300009200. [DOI] [PubMed] [Google Scholar]
- 22.Dufner A, Pownall S, Mak TW. Caspase recruitment domain protein 6 is a microtubule-interacting protein that positively modulates NF-κB activation. Proc Natl Acad Sci USA. 2006;103(4):988–993. doi: 10.1073/pnas.0510380103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu T, et al. Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci USA. 2010;107(1):46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Central Brain Tumor Registry of the United States . Statistical Report: Primary Brain Tumors in the United States, 2000–2004. Central Brain Tumor Reg United States; Hinsdale, IL: 2008. [Google Scholar]
- 25.Whittle JR, et al. First in human nanotechnology doxorubicin delivery system to target epidermal growth factor receptors in recurrent glioblastoma. J Clin Neurosci. 2015;22(12):1889–1894. doi: 10.1016/j.jocn.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 26.Thorn CF, et al. Doxorubicin pathways: Pharmacodynamics and adverse effects. Pharmacogenet Genomics. 2011;21(7):440–446. doi: 10.1097/FPC.0b013e32833ffb56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campos LS, et al. Filamin A expression negatively regulates sphingosine-1-phosphate-induced NF-κB activation in melanoma cells by inhibition of Akt signaling. Mol Cell Biol. 2015;36(2):320–329. doi: 10.1128/MCB.00554-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garraway LA, Jänne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2012;2(3):214–226. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- 29.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia Y, Whitesides GM. Soft lithography. Angew Chem Int Ed Engl. 1998;37(5):550–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 31.Choi J, et al. Wnt5a-mediating neurogenesis of human adipose tissue-derived stem cells in a 3D microfluidic cell culture system. Biomaterials. 2011;32(29):7013–7022. doi: 10.1016/j.biomaterials.2011.05.090. [DOI] [PubMed] [Google Scholar]
- 32.Chen SY, Hung PJ, Lee PJ. Microfluidic array for three-dimensional perfusion culture of human mammary epithelial cells. Biomed Microdevices. 2011;13(4):753–758. doi: 10.1007/s10544-011-9545-3. [DOI] [PubMed] [Google Scholar]
- 33.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K, et al. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38(18):e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.