The eukaryotic cell surface is composed of many distinct membrane domains that are formed by the cooperative interactions of different proteins and lipids. These domains are important for membrane trafficking and cell signaling and are modulated in turn by changes in the cell environment. Caveolae (“little caves”) are ∼60-nm membrane invaginations (Fig. 1) that are a dominant surface feature of many mammalian cells, including muscle fibers, endothelia, and adipocytes, where they play a role in membrane homeostasis, signaling, and cellular mechanoprotection. Formation of caveolae in vertebrate cells requires two distinct protein families: the membrane-embedded caveolins (CAV1–3) and the peripheral membrane cavins (Cavin1–4). Although the general morphology of caveolae has been known for decades, the atypical structures of the protein subunits has meant that progress has been slow with regards to the high-resolution studies of caveola architecture. By high-resolution scanning electron microscopy (EM) and frozen deep-etch transmission EM, caveolae have been shown to be coated with striations (1, 2) or to possess spike-like structures (3), very different from other well-characterized vesicle coats, such as clathrin (4, 5). In PNAS, Stoeber et al. use a combination of biochemical dissection and EM to provide important insights into the underlying architecture of the caveola protein coat (6).
Fig. 1.
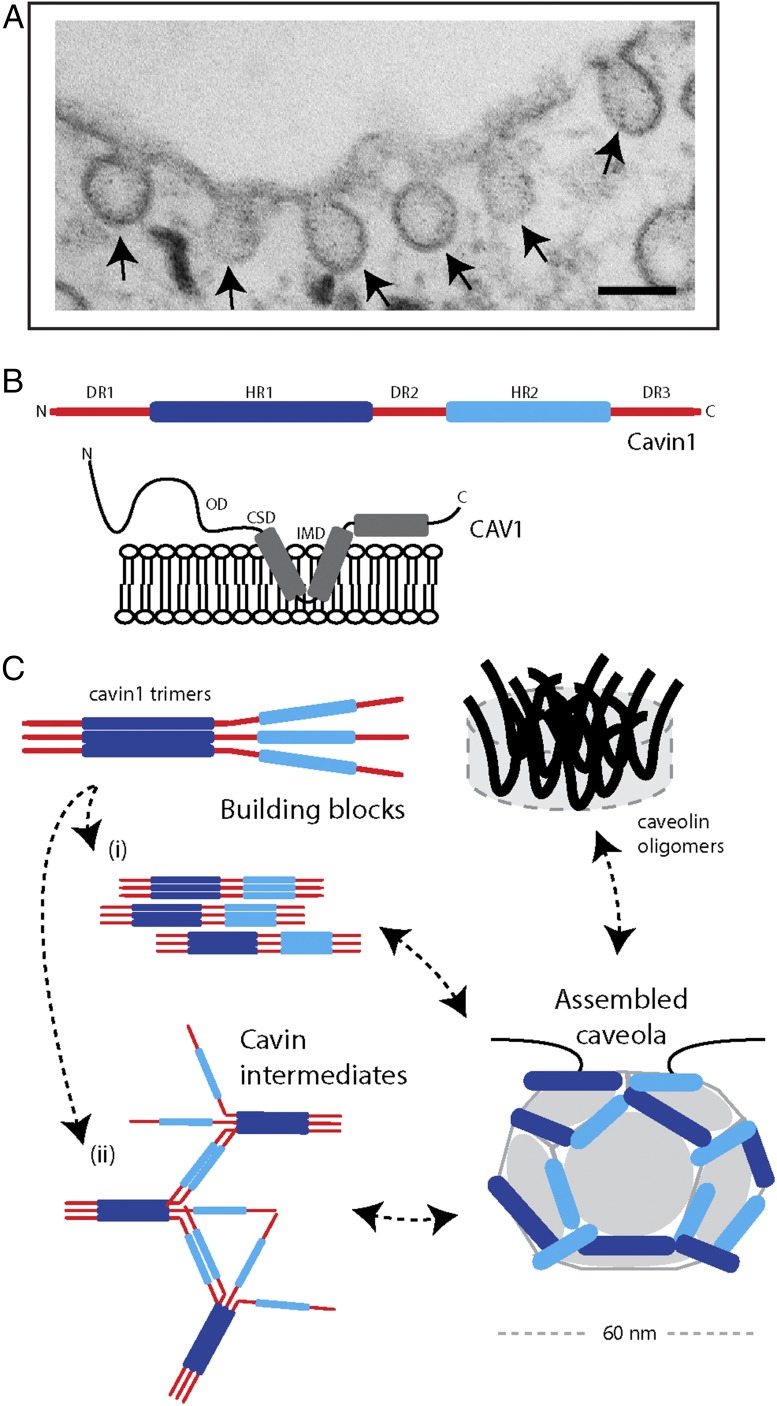
Proposed model for the assembly of caveolae. (A) Electron micrograph of caveolae in an adipocyte cell line. (Scale bar, 100 nm.) (B) Schematic diagrams depicting the main features of the CAV1 and Cavin1 proteins required for caveola formation. Cavins possess two regions of α-helical structure, termed HR1 and HR2, which are rich in basic residues. These are linked by disordered acidic sequences DR1–DR3. Caveolins are membrane-integral proteins that are embedded by a central helix–turn–helix. They also possess a conserved sequence suggested to be involved in oligomerization (oligomerization domain, OD) and a sequence with potential roles in protein–protein interactions (caveolin scaffolding domain, CSD), although this is controversial (20). (C) Proposed model for caveolar assembly by Stoeber et al. (6). CAV1 can be isolated as disk-shaped oligomers ∼15 nm in diameter that are proposed to be underlying building blocks of the caveolar coat. Cavin1 trimerizes through its N-terminal HR1 domain (10), and then can form oligomers arranged in a net-like structure. These oligomers organize into striated patterns on the surface of caveolae in cooperation with the CAV1 protein. (i) In the model proposed by Stoeber et al. (6) the cavin trimers form intermolecular networks via HR2 domain coiled-coil interactions. (ii) Alternate mechanisms for intramolecular cavin interactions, such as electrostatic interactions, have also been proposed (8).
The first protein component of caveolae to be identified and shown to be essential for caveola formation was CAV1 (7–9). Subsequently, it was found that both CAV2 and muscle-specific CAV3 homologs are also localized to caveolae. Caveolin proteins are ∼20 kDa and possess two membrane-inserted α-helices with cytoplasmic N- and C-terminal sequences. These proteins form oligomeric complexes at the plasma membrane that line the cytoplasmic leaflet of the invaginated caveola structures. Cavins, although equally important for caveola formation, were only identified in the past decade through proteomic and cellular studies from several groups. Cavins also possess a conserved and unique structural organization, in this case consisting of conserved N- and C-terminal α-helical domains (HR1 and HR2) interspersed by disordered linker sequences (DR1, DR2, DR3) (10). Although recent studies have begun to tease out the functions of these domains in membrane recruitment, remodeling, and protein–protein interactions, our understanding is still limited. For example, how do caveolin oligomers assemble within caveolae, and what drives cavin oligomerization and recruitment to caveolar domains at the cell surface?
In their report, Stoeber et al. (6) propose a model for the arrangement of the caveolin and cavin proteins that begins to address some of the ongoing questions in the field. Affinity purification of Cavin1 expressed in HEK293 cells revealed the presence of heterogeneous particles with a mesh-like appearance by cryoEM and 3D electron tomography. These particles bound robustly to liposomes containing phosphatidylserine (PS) or phosphatidic acid, as seen previously using recombinant cavin proteins (10, 11), and showed a propensity to form relatively well-ordered lattices of protein on the membrane surface. Interestingly, in this in vitro system the full-length Cavin1 protein could be observed on the interior surface of the vesicles, which Stoeber et al. (6) suggest may be because of the occurrence of frequent inward invagination of the PS-containing liposomes. Most remarkably, Cavin1 appeared to form membrane-dependent polygonal arrays, which begins to suggest a potential for protein–protein and protein–membrane interactions to generate a structural scaffold from a loose net-like assembly of soluble cavin oligomers. Next, Stoeber et al. (6) probed the importance of the HR1 and HR2 domains of Cavin1 for oligomerization, membrane association, and membrane remodeling activity. Both HR1 and HR2 were found to be equally important for membrane localization in cells; however, although removal of HR1 prevented oligomer formation altogether, removing HR2 still yielded partial oligomers, albeit smaller than the 60S particles of the wild-type Cavin1 protein. When the HR2-deleted Cavin1 protein was analyzed by negative-stain EM, elongated fiber-like particles were observed, which are also consistent with the crystal structure of cavin1 HR1 and EM studies of full-length Cavin1 purified from bacteria (10). Notably this protein could bind to PS-containing liposomes and generate membrane tubules that showed evidence of membrane-apposed filamentous arrays by cryoEM tomography.
Stoeber et al. (6) next extended their analyses to the other major structural component of caveolae, the integral membrane protein, CAV1. Although this necessitated the use of detergents and negative staining, rather than cryoEM, the detergent solubilized 8S CAV1 oligomers were found to form relatively homogeneous disk-shaped particles ∼15 nm in diameter, which also resemble particles of recombinant CAV3 seen previously when expressed in insect cells (12). Finally, cryoEM tomography of caveolae in situ suggested that rather than being purely spherical, caveola bulbs possess inherent polygonal symmetries. Interestingly, identical polygonal symmetry is observed by cryoEM in a model system in which CAV1 alone drives formation of caveola-like vesicles in bacterial membranes (13). This finding strongly suggests that CAV1 is responsible for this polygonal architecture, rather than the peripheral cavin coat, although in mammalian cells both components are required to form a stable caveola.
Putting these results together, Stoeber et al. (6) propose a working model for the assembly of the cavin and caveolin proteins required to form functional caveolae (Fig. 1). In this model, the Cavin1 HR1 forms a trimeric coiled-coil structure as an intermediate to forming cavin oligomers. The assembly of the cavin oligomer and presentation of basic patches within HR1 and HR2 promote membrane recruitment in conjunction with the membrane-embedded CAV1. Based on the dimensions of the purified CAV1 structures observed by EM, it is proposed that the ∼12 faces of the caveolar polygon may be occupied by individual CAV1 disk-shaped oligomers. This is an intriguing and highly novel suggestion, and certainly merits further study to determine its validity. Such an arrangement then allows formation of a meshwork of associated Cavin1 trimers at the membrane surface, where cooperative cavin–caveolin–lipid interactions then promote membrane invagination. Based on the importance of the HR2 domain in promoting the oligomerization of Cavin1, Stoeber et al. (6) evoke a model where this α-helical region, which is predicted to form coiled-coil structures, forms linkages between Cavin1 trimeric HR1 domains. Whether cavins self-associate via these proposed intratrimer linkages or via alternative mechanisms, such as lateral electrostatic interactions (8), however, will require further study. Why Cavin1, rather than other cavins, is essential for caveola formation was suggested to be because of a region in the Cavin1 HR2 domain with a higher propensity to form coiled-coils than other cavins, an interesting proposal that remains to be tested.
In comparison with clathrin-coated endocytic pits and other protein-coated vesicles, such as COPI and COPII, studies of caveolar architecture are still in their infancy. Studies by many laboratories in recent years are now beginning to bridge this gulf using approaches that include biochemical isolation, NMR spectroscopy, EM, and X-ray crystallography (6, 10, 12–17). A picture is now emerging of a vesicular structure that possesses much greater complexity than originally thought. In combination with an enormous literature on the cellular function and regulation of caveolae, however, many questions still remain to be answered. How does this organization fit with the observed striations associated with the cytoplasmic face of caveolae and shown to comprise cavins (15, 18)? The critical role of lipids, such as PS, phosphoinositides, and cholesterol in caveolae is an outstanding issue, and the structural models that are emerging may begin to suggest testable predictions about the importance of lipid–lipid and lipid–protein interactions in caveola formation, or indeed whether caveolae represent structures that function to organize lipid microdomains that can respond to cellular stimuli (19). As Stoeber et al. (6) suggest, the architecture of caveolae must also allow for extensive posttranslational modification by phosphorylation and ubiquitination that is known to occur in specific cavin and caveolin sequences. Finally, the structures of caveolae must allow them to rapidly and reversibly respond to changes in membrane signaling and mechanical tension. These new findings suggest that the principles involved in caveola formation may be more similar to those involved in clathrin-coated vesicle formation than previously imagined, despite their fundamental molecular differences.
Acknowledgments
This work was supported by National Health and Medical Research Council of Australia (NHMRC) Grants APP569542 and APP1037320 (to R.G.P.); and Australian Research Council Grants DP120101298 (to B.M.C. and R.G.P.) and CE141000036 (to R.G.P.). R.G.P. is supported by NHMRC Senior Principal Research Fellowship APP1058565, and B.M.C. is supported by NHMRC Career Development Fellowship APP1061574.
Footnotes
The authors declare no conflict of interest.
See companion article on page E8069.
References
- 1.Peters KR, Carley WW, Palade GE. Endothelial plasmalemmal vesicles have a characteristic striped bipolar surface structure. J Cell Biol. 1985;101(6):2233–2238. doi: 10.1083/jcb.101.6.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothberg KG, et al. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68(4):673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- 3.Richter T, et al. High-resolution 3D quantitative analysis of caveolar ultrastructure and caveola-cytoskeleton interactions. Traffic. 2008;9(6):893–909. doi: 10.1111/j.1600-0854.2008.00733.x. [DOI] [PubMed] [Google Scholar]
- 4.Faini M, Beck R, Wieland FT, Briggs JAG. Vesicle coats: Structure, function, and general principles of assembly. Trends Cell Biol. 2013;23(6):279–288. doi: 10.1016/j.tcb.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Kirchhausen T, Owen D, Harrison SC. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb Perspect Biol. 2014;6(5):a016725. doi: 10.1101/cshperspect.a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoeber M, et al. Model for the architecture of caveolae based on a flexible, net-like assembly of Cavin1 and Caveolin discs. Proc Natl Acad Sci USA. 2016;113:E8069–E8078. doi: 10.1073/pnas.1616838113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echarri A, Del Pozo MA. Caveolae—Mechanosensitive membrane invaginations linked to actin filaments. J Cell Sci. 2015;128(15):2747–2758. doi: 10.1242/jcs.153940. [DOI] [PubMed] [Google Scholar]
- 8.Kovtun O, Tillu VA, Ariotti N, Parton RG, Collins BM. Cavin family proteins and the assembly of caveolae. J Cell Sci. 2015;128(7):1269–1278. doi: 10.1242/jcs.167866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parton RG, del Pozo MA. Caveolae as plasma membrane sensors, protectors and organizers. Nat Rev Mol Cell Biol. 2013;14(2):98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- 10.Kovtun O, et al. Structural insights into the organization of the cavin membrane coat complex. Dev Cell. 2014;31(4):405–419. doi: 10.1016/j.devcel.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Hill MM, et al. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 2008;132(1):113–124. doi: 10.1016/j.cell.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whiteley G, Collins RF, Kitmitto A. Characterization of the molecular architecture of human caveolin-3 and interaction with the skeletal muscle ryanodine receptor. J Biol Chem. 2012;287(48):40302–40316. doi: 10.1074/jbc.M112.377085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ariotti N, et al. Molecular characterization of caveolin-induced membrane curvature. J Biol Chem. 2015;290(41):24875–24890. doi: 10.1074/jbc.M115.644336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludwig A, et al. Molecular composition and ultrastructure of the caveolar coat complex. PLoS Biol. 2013;11(8):e1001640. doi: 10.1371/journal.pbio.1001640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludwig A, Nichols BJ, Sandin S. Architecture of the caveolar coat complex. J Cell Sci. 2016;129(16):3077–3083. doi: 10.1242/jcs.191262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plucinsky SM, Glover KJ. Secondary structure analysis of a functional construct of Caveolin-1 reveals a long C-terminal helix. Biophys J. 2015;109(8):1686–1688. doi: 10.1016/j.bpj.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rui H, Root KT, Lee J, Glover KJ, Im W. Probing the U-shaped conformation of caveolin-1 in a bilayer. Biophys J. 2014;106(6):1371–1380. doi: 10.1016/j.bpj.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gambin Y, et al. Single-molecule analysis reveals self assembly and nanoscale segregation of two distinct cavin subcomplexes on caveolae. eLife. 2013;3:e01434. doi: 10.7554/eLife.01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ariotti N, et al. Caveolae regulate the nanoscale organization of the plasma membrane to remotely control Ras signaling. J Cell Biol. 2014;204(5):777–792. doi: 10.1083/jcb.201307055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins BM, Davis MJ, Hancock JF, Parton RG. Structure-based reassessment of the caveolin signaling model: Do caveolae regulate signaling through caveolin-protein interactions? Dev Cell. 2012;23(1):11–20. doi: 10.1016/j.devcel.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]