Significance
The α-subunit of heterotrimeric G proteins is a molecular switch that mediates a great number of physiological processes such as vision, smelling, and blood pressure regulation. A GTPase-activating protein (GAP) [e.g. regulator of G protein signaling 4 (RGS4) in the case of Gαi1] regulates the off-switch by catalyzing GTP hydrolysis. Here, we present the molecular reactions of GAP catalysis at atomic resolution using a combination of FTIR spectroscopy and biomolecular simulations. In contrast to X-ray structures, not GTP analogs but GTP itself is used. This approach is crucial to reveal now a previously undescribed GAP mechanism for Gα. A key player of the hydrolysis reaction, called the arginine finger, is pushed from a monodentate γ-GTP coordination toward a bidentate α-γ-GTP coordination by RGS4, and thereby catalyzes GTP-hydrolysis.
Keywords: GTPase, FTIR spectroscopy, QM/MM calculations, arginine finger, reaction mechanism
Abstract
Heterotrimeric G proteins are crucial molecular switches that maintain a large number of physiological processes in cells. The signal is encoded into surface alterations of the Gα subunit that carries GTP in its active state and GDP in its inactive state. The ability of the Gα subunit to hydrolyze GTP is essential for signal termination. Regulator of G protein signaling (RGS) proteins accelerates this process. A key player in this catalyzed reaction is an arginine residue, Arg178 in Gαi1, which is already an intrinsic part of the catalytic center in Gα in contrast to small GTPases, at which the corresponding GTPase-activating protein (GAP) provides the arginine “finger.” We applied time-resolved FTIR spectroscopy in combination with isotopic labeling and site-directed mutagenesis to reveal the molecular mechanism, especially of the role of Arg178 in the intrinsic Gαi1 mechanism and the RGS4-catalyzed mechanism. Complementary biomolecular simulations (molecular mechanics with molecular dynamics and coupled quantum mechanics/molecular mechanics) were performed. Our findings show that Arg178 is bound to γ-GTP for the intrinsic Gαi1 mechanism and pushed toward a bidentate α-γ-GTP coordination for the Gαi1·RGS4 mechanism. This movement induces a charge shift toward β-GTP, increases the planarity of γ-GTP, and thereby catalyzes the hydrolysis.
Heterotrimeric G proteins serve as a link between G protein-coupled receptors (GPCRs) and second messenger systems like adenylyl cyclases in the cell (1). The inactive trimeric form consisting of the GTPase Gα and the Gβγ complex gets activated by a GPCR that acts as a guanine nucleotide exchange factor (GEF). It promotes GDP release and enables GTP uptake at the active site of Gα, which results in structural changes in the switch I–III regions of the α-subunit (2), separation of the subunits, and signal transduction (3). Termination of the signal is initiated by GTP hydrolysis at the active center of Gα to GDP and Pi. This crucial mechanism is highly conserved among GTPases and requires numerous mechanistic features the protein has to provide [e.g., Mg2+ incorporation (4), substrate coordination (5), charge neutralization (6), positioning of the nucleophilic water (7)]. Some of these functions are maintained by two highly conserved residues: an arginine side chain (arginine “finger” in small GTPases) and a carboxyamide near the γ-phosphate (8). In contrast to small GTPases, where the arginine is provided by a GTPase-activating protein (GAP), heterotrimeric G proteins are equipped with an intrinsic arginine finger (Arg178 in Gαi1), which is located in switch I [residues 178–188 in Gαi1 (9)] and enables fast hydrolysis compared with small GTPases [factor of 50 (10, 11)]. A GAP protein [(e.g., regulator of G protein signaling 4 (RGS4) in the case of Gαi1] can further accelerate GTP hydrolysis (12). The importance of the arginine finger manifests in various diseases; for example, single point mutations in Gαs lead to McCune–Albright syndrome (13, 14) and ADP ribosylation of the arginine finger in Gαs by Vibrio cholerae leads to cholera disease (15).
We have demonstrated recently how FTIR spectroscopy on Gαi1 can monitor the GTPase reaction label-free with high spatiotemporal resolution (10). This approach was originally established to elucidate the proton-pump mechanism of bacteriorhodopsin via protein-bound water molecules (16). In this study, we will focus on the intrinsic arginine finger and elucidate its position and mechanism in intrinsic and RGS4-catalyzed Gαi1. Current models of the arginine finger mechanism rely on crystal structures that provide atomistic snapshots of the active GTP state using nonhydrolyzable GTP analogs (17, 18), the GDP-AlF4− intermediate state (17), and the inactive GDP state (19). Upon Gαi1 isoforms, the position of the arginine finger is variable; for example, in Gαt·Mg2+·GTPγS, it is hydrogen-bonded toward the β-γ–bridging oxygen (20), and in Gαi1·Mg2+·GppNHp and Gαi1·Mg2+·GTPγS, it is partially disordered, forming an ion pair with Glu43 (18). In all Gα isoforms resolved to date with GDP·AlF4−, the arginine finger is bound to the fluoride group, also facing the bridging β-γ-oxygen atom and the α-GTP group (2, 21–25). The arginine finger of the isoform Gαi1 seems to be flipped away from the nucleotide in both the GTP state and the GDP state, and it only participates in nucleotide binding during the intermediate AlF4− state (Fig. 1). However, active structures of Gαi1 were solved in presence of sulfur- or nitrogen-substituted GTP analogs only, which may influence the arginine finger position. Furthermore, the position of the arginine finger in the AlF4− intermediate state could also be influenced by the strong electronegativity of this intermediate state analog. To overcome this problem, we applied time-resolved FTIR spectroscopy with photocaged para-hydroxyphenacyl cgGTP (pHPcgGTP), which cleaves rapidly [107 s−1 (26)] and results in the natural GTP nucleotide that triggers the GTPase reaction label- and analog-free. The resulting photolysis and hydrolysis difference spectra reflect the reaction with subangstrom spatial and millisecond temporal resolution. We assigned individual phosphate and protein bands using isotopic labeling and site-directed mutagenesis, and were able to observe the arginine finger position and its mechanism with native GTP. Mutations were selected because of their catalytic relevance (e.g., Arg178, Gln204), effects in previous studies (e.g., Lys180), and spatial proximity to the active site. Positions of all point mutations are indicated in SI Appendix, Fig. 1. Molecular mechanics with molecular dynamics (MM-MD) and coupled quantum mechanics/molecular mechanics (QM/MM) simulations complemented the experiments and provided additional evidence for the arginine finger mechanism in heterotrimeric G proteins.
Fig. 1.
Position of the intrinsic arginine finger in crystal structures. In the active GTPγS-bound state (PDB ID code 1GIA), Arg178 is oriented away from the substrate toward a glutamate (Glu43). In the aluminium-tetrafluoride (AlF4−)-stabilized intermediate (PDB ID code 1GFI), Arg178 points toward the substrate. In the inactive GDP state (PDB ID code 1GP2), Arg178 is again flipped away from the substrate.
Results
Time-resolved FTIR spectroscopy monitors reactions label-free at atomic resolution and provides both rate constants and structural information that are coded into IR spectra. WT or mutant Gαi1 was loaded with pHPcgGTP, a photolabile GTP derivate that binds to Gαi1 but is not hydrolyzed. FTIR measurements were triggered by a laser flash that removes the pHP-caged group, resulting in the natural substrate GTP (result a0 of global fit as detailed in Materials and Methods, termed photolysis in the following) that is subsequently hydrolyzed by Gαi1 (result a1 of global fit, termed hydrolysis in the following). Both the photolysis and the hydrolysis reaction were monitored by time-resolved FTIR spectroscopy. The obtained data were evaluated using a global fit (Eq. 1) with one exponential function for Gαi1-WT [rate constant k = 0.02 s−1 at 15 °C (10)] or two exponential functions when intermediate formation occurred (Table 1). RGS4-catalyzed measurements were also evaluated using one exponential function. All resulting t1/2 values and their SEs are depicted in Table 1. Spectral information is coded into a photolysis spectrum (negative bands correspond to the caged GTP state, and positive bands correspond to the GTP state) and a hydrolysis spectrum (negative bands correspond to the GTP state, and positive bands correspond to the GDP + Pi state). Several Gαi1 mutants showed an intermediate during the hydrolysis reaction, and were therefore evaluated with an additional reaction rate. The importance of working label- and analog-free can be demonstrated when performing FTIR measurements using N-methylanthraniloyl (MANT)-GTP instead of natural GTP. Using MANT-GTP slows down hydrolysis kinetics by one order of magnitude and significantly alters, due to distortion of the catalytic center, the GTP and protein bands (SI Appendix, Fig. 2).
Table 1.
t1/2 values obtained from global fits in FTIR measurements
| Protein | Temperature | t1/2 values (global fit) |
| Gαi1-WT | 15 °C | 32.7 ± 2.5 s |
| Gαi1-WT | 5 °C | 68.2 ± 5.1 s |
| Gαi1-E43Q | 15 °C | t1/2 1 = 1.9 ± 0.8 s |
| t1/22 = 55.7 ± 6.2 s | ||
| Gαi1-T48A | 15 °C | t1/21 = 8.6 ± 1.2 s |
| t1/2 2 = 75.5 ± 9.4 s | ||
| Gαi1-D150N | 15 °C | t1/2 1 = 25.9 ± 6.2 s |
| t1/2 2 = 234.9 ± 44.0 s | ||
| Gαi1-R178S | 15 °C | t1/2 1 = 9.8 ± 0.9 s |
| t1/2 2 = 3437.7 ± 426.8 s | ||
| Gαi1-K180P | 15 °C | t1/2 1 = 68.9 s ± 26.0 s |
| t1/2 2 = 537.4 s ± 58.0 s | ||
| Gαi1-Q204A | 15 °C | t1/2 1 = 7.1 s ± 5.4 s |
| t1/2 2 = 3406 s ± 939.3 s | ||
| Gαi1-W211A | 15 °C | t1/2 1 = 4.4 s ± 0.4 s |
| t1/2 2 = 125.2 s ± 16.3 s | ||
| Gαi1-D229N | 15 °C | t1/2 1 = 14.7 s ± 1.0 s |
| t1/2 2 = 53.2 s ± 3.8 s | ||
| Gαi1-E236Q/D237Q | 15 °C | t1/2 1 = 6.2 s ± 0.9 s |
| t1/2 2 = 60.0 s ± 4.1 s | ||
| Gαi1-E245Q | 15 °C | t1/2 1 = 5.1 s ± 3.4 s |
| t1/2 2 = 53.9 s ± 6.1 s | ||
| Gαi1-WT·RGS4 | 5 °C | 1.4 ± 0.3 s |
| Gαi1-R178S·RGS4 | 5 °C | 12.6 ± 2.7 s |
| Gαi1-T48A·RGS4 | 5 °C | 9.6 ± 1.6 s |
Arg178 Is Bound to γ-GTP in the Active State of Gαi1.
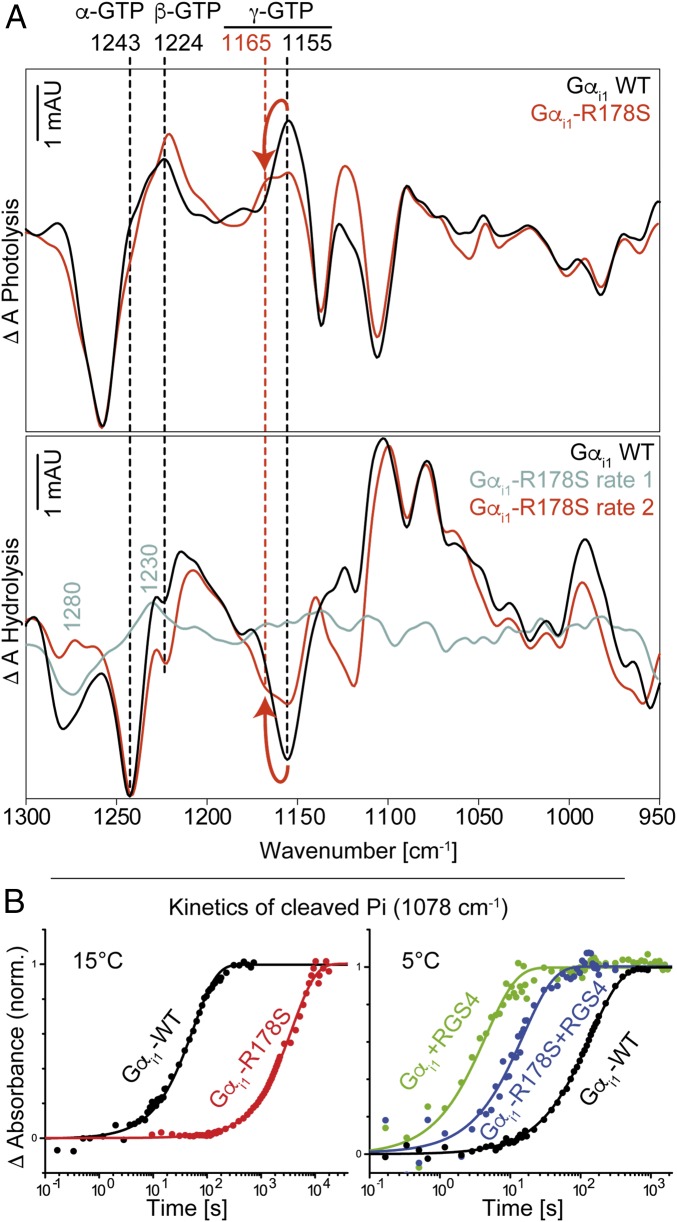
FTIR measurements of Gαi1-WT and Gαi1-R178S are depicted in Fig. 2. We recently assigned the bands for α-GTP, β-GTP, and γ-GTP (1,243 cm−1, 1,224 cm−1, and 1,156 cm−1, respectively); α-GDP and β-GDP (1,214 cm−1 and 1,134/1,103 cm−1, respectively); and free phosphate (1,078/991 cm−1) for Gαi1-WT (10). Surprisingly, in this study, the γ-GTP vibration of the mutant Gαi1-R178S appeared blue-shifted from 1,156 cm−1 to 1,165 cm−1 (Fig. 2), indicating changes in the direct environment of the γ-phosphate. Because the exchange of Arg178 to Ser178 was the only difference between the measurements, we hereby conclude that Arg178 forms a hydrogen bond to γ-GTP in the active state of Gαi1. The magnitude of the band shift is in agreement with the literature. A similar blue shift caused by removing a single hydrogen bond was recently described for the small GTPase Ran (4). The mutation Ran-Y32A caused a γ-GTP shift of 12 cm−1. For Gαi1-R178S, the band shift was observable in both the photolysis spectrum and the hydrolysis spectrum for measurements in H2O and measurements in D2O. The vibrations of α-GTP and β-GTP remained unaltered. To verify that the Gαi1-R178S mutation had no effect on α-GTP, we applied isotopic labeling using α18O2-pHPcgGTP in Gαi1-R178S and assigned the band at 1,243 cm−1 clearly to α-GTP, similar to Gαi1-WT (SI Appendix, Fig. 3). The vibration of α-GDP appeared slightly red-shifted from 1,214 cm−1 (Gαi1-WT) to 1,208 cm−1 (Gαi1-R178S). Mutation of the intrinsic arginine finger slowed down the reaction by almost two orders of magnitude as we previously reported (10), and is comparable to the Gαi1-Q204A mutation (Table 1). Kinetics of the cleaved phosphate product bands are depicted in Fig. 2B.
Fig. 2.
(A) Photolysis and hydrolysis FTIR difference spectra (global fit) of Gαi1-WT and the mutant Gαi1-R178S. (B) Kinetics of cleaved phosphate (1,078 cm−1) for Gαi1-WT, Gαi1-R178S, and Gαi1-R178S + RGS4 at 15 °C and 5 °C. In A, positive bands in the photolysis spectra and negative bands in the hydrolysis spectra correspond to the GTP state. Positive bands in the hydrolysis spectra correspond to the GDP state. Arrows indicate the γ-GTP shift caused by the mutation. In B, mutation of Arg178 caused a slowdown of the GTPase reaction by two orders of magnitude. RGS4 addition reversed this effect. Kinetic constants obtained from global fits are depicted in Table 1. mAU, milli absorbance units; norm., normalized.
Several Gαi1 Mutants Exhibit Intermediate Formation During Hydrolysis.
Mutation of the intrinsic arginine finger caused a rate separation during hydrolysis, showing an intermediate that preceded hydrolysis with spectral features at 1,280 cm−1 and 1,230 cm−1 (Fig. 2A, cyan). This intermediate was observable for various point mutations in Gαi1 (Gαi1-E43Q, Gαi1-K180P, Gαi1-D150N, Gαi1-E236Q/D237Q, Gαi1-D229N, Gαi1-W211A, and Gαi1-E245Q) and is not an exclusive feature of the arginine mutation. The t1/2 values of both rates are depicted in Table 1. We applied isotopic labeling of the Gαi1-E43Q mutant using α-18O2-pHPcgGTP and β-18O3-pHPcgGTP and found the band at 1,230 cm−1 to be caused by both α-GTP and β-GTP (SI Appendix, Fig. 4).
RGS4 Addition Accelerates GTP Hydrolysis in Gαi1 by Two Orders of Magnitude.
In addition to intrinsic FTIR measurements of Gαi1, we performed measurements with its GAP RGS4 in a 1:1 complex. Like in intrinsic Gαi1-WT measurements, no intermediate formation was observed. RGS4 addition accelerated the t1/2 value of the GTPase reaction in Gαi1 by almost two orders of magnitude from 68.2 ± 5.1 s (5 °C) to 1.4 ± 0.3 s (5 °C). Kinetics of the cleaved phosphate product bands are depicted in Fig. 2B.
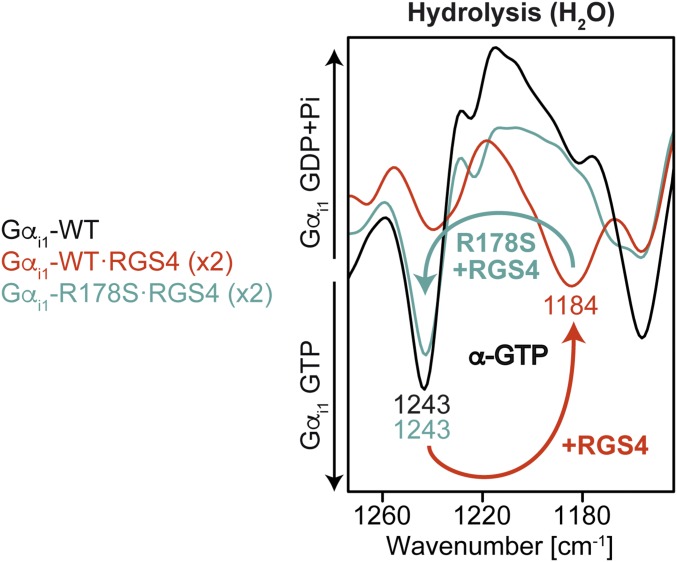
RGS4 Pushes Arg178 Toward a Bidentate α-γ-GTP Coordination.
The addition of RGS4 not only accelerated hydrolysis kinetics by two orders of magnitude but also changed the GTP and GDP vibrations. The most prominent change was a band at 1,184 cm−1 that appeared in the photolysis and disappeared in the hydrolysis (Fig. 3, red) (Complete photolysis and hydrolysis spectra from 1,800 cm−1 to 950 cm−1 are depicted in SI Appendix, Fig. 5). We applied isotopic labeling using α-18O2-pHPcgGTP (SI Appendix, Fig. 6) and β-18O3-pHPcgGTP (SI Appendix, Fig. 7), and could thereby determine that the α-GTP vibration caused the band at 1,182/1,184 cm−1 for RGS4-catalyzed Gαi1. Moreover, this effect was completely reversible when the intrinsic arginine finger was missing due to the R178S mutation (Fig. 3, cyan), so we can conclude that RGS4 induces a conformational change of Arg178 in such a way that it binds to α-GTP. In addition, Arg178 is simultaneously bound to γ-GTP, because for Gαi1-R178S·RGS4, it was seen that in both the photolysis and hydrolysis spectra, the γ-GTP vibration was shifted from 1,156 cm−1 to 1,165 cm−1 (Fig. 3, cyan), like in intrinsic Gαi1 (Fig. 2A). The magnitude of the RGS4-induced α-GTP shift indicates that RGS4 binding changes more than the α-GTP environment (e.g., induces structural changes like torsions or bond lengths and a different charge distribution). The α-GDP vibration was also shifted from 1,214 cm−1 to 1,219 cm−1 in RGS4-catalyzed measurements, which was again reversible when Arg178 was missing. For Gαi1-R178S·RGS4, no intermediate formation was observed. To exclude a temperature artifact, we also repeated these measurements at −5 °C and observed no spectral changes. Our measurements demonstrated that the intrinsic arginine finger is bound to γ-GTP for intrinsic Gαi1 and bound bidentately to α-GTP and γ-GTP for Gαi1·RGS4.
Fig. 3.
Hydrolysis FTIR spectra of intrinsic and RGS4-catalyzed Gαi1-WT and its mutant R178S. RGS4 shifted the α-GTP vibration from 1,243 cm−1 to 1,184 cm−1. The effect was reversible when Arg178 was mutated. Full spectra of the photolysis and the hydrolysis reaction are depicted in SI Appendix, Fig. 5.
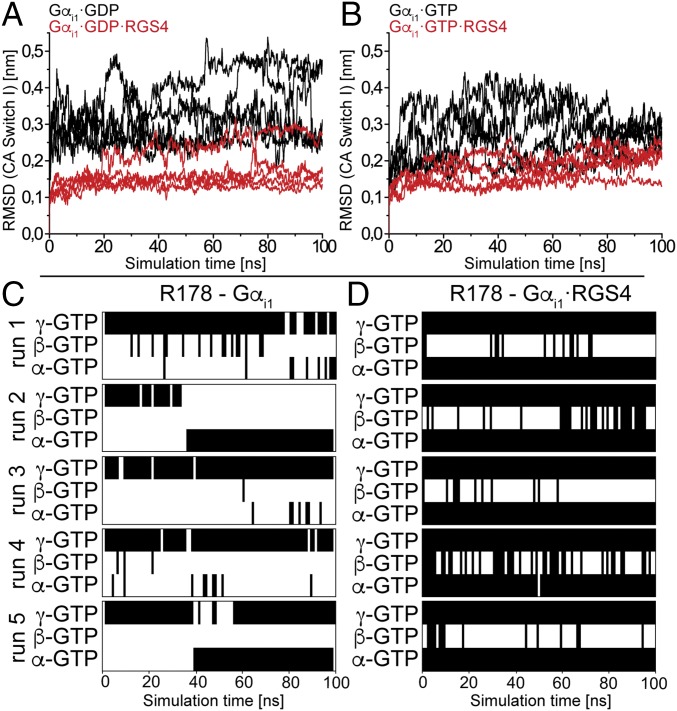
RGS4 Stabilizes Switch I in MD Simulations.
We performed MD simulations of intrinsic and RGS4-bound Gαi1 in the GDP- and GTP-bound states to gain a structural interpretation of the IR measurements. RGS4 does not contribute amino acids to the binding pocket of Gαi1 like, for example, RasGAP; rather, it stabilizes the switch regions of Gαi1 in a conformation that is probably favorable for GTP hydrolysis (27). We could confirm the proposed mechanism in our simulations. We focused on switch I, because it contains the intrinsic arginine finger, and found this region to be stabilized by RGS4 in both the GDP and GTP states within a simulation time of 100 ns (Fig. 4 A and B), probably in a conformation that is favorable for hydrolysis. In addition, intrinsic GTP-bound Gαi1 showed lower rmsd values than GDP bound Gαi1, which is consistent with the observation of increased thermostability of GTP-bound Gα subunits in previous studies (2, 20).
Fig. 4.
(A and B) The rmsd values of switch I Cα atoms (CA) during intrinsic (black) and RGS4-bound (red) MD simulations in comparison to the starting structures. Contact matrix analysis of the Arg178–GTP interaction for intrinsic (C) and RGS4-bound (D) Gαi1 in the MD simulations. In A and B, depicted data are 100-ns simulations averaged over 100-ps time windows. The stabilization of switch I by RGS4 is evident. The rmsd between switch I of intrinsic and RGS4-bound Gαi1 starting structures is 0.18 nm. In C and D, black bars indicate H-bonds and white spaces indicate no H-bond formation. Contacts were sampled in time windows of 1 ns. In the intrinsic case, a flexible R178 that is preferably bound to the γ-phosphate is found, whereas RGS4-bound Gαi1 shows a stable bidentate coordination to α-phosphate and γ-phosphate.
MD Simulations Confirm Monodentate (Intrinsic) and Bidentate (RGS4-Catalyzed) Coordination of Arg178.
In addition to rmsd calculations, we performed contact matrix analysis of the production runs with a focus on the interaction between Arg178 and the individual phosphate groups. For intrinsic Gαi1, the arginine finger was bound monodentately to γ-GTP in three of five simulation runs (Fig. 4C), but the coordination to α-GTP (Fig. 4C, run 2) and even a bidentate coordination to α-GTP and γ-GTP were also sampled (Fig. 4C, run 5) in replica runs. Simulations showed large conformational dynamics of Arg178, with the monodentate coordination at γ-GTP being the preferred interaction. It is notable that in all starting structures, the arginine pointed in the opposite direction, forming a stacked π-interaction to Glu43. This coordination was always disrupted, and Arg178 flipped down to the substrate in all simulations. We also repeated the simulations starting from intrinsic Gαi1·AlF4−·GDP [Protein Data Bank (PDB) ID code 1GFI, with GDP·AlF4− replaced by GDP or GTP], where the arginine is already oriented toward the substrate to exclude starting structure artifacts, and found similar behavior. With RGS4 bound to Gαi1 (PDB ID code 1AGR), the intrinsic arginine finger was always bound bidentately to α-GTP and γ-GTP. The terminal η-NH2 groups of the side chain were tightly bound to the oxygen atoms of the α- and γ-phosphate groups. This interaction was never interrupted in all five production runs (Fig. 4D).
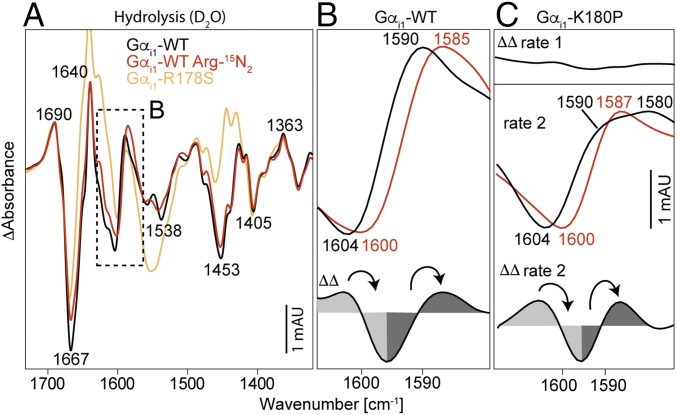
IR Band Assignment of Gαi1-Arg178.
Band assignments were performed using η-15N2 isotopically labeled arginine. Protein expression was performed in M9 minimal medium (28) with unlabeled arginine replaced by η-15N2 arginine. Purity and labeling efficiency of recombinant proteins were checked via SDS/PAGE and liquid chromatography (LC)-MS (minimum 92% labeling efficiency). Isotopic labeling results in a red shift of arginine side-chain vibrations due to the increased reduced masses. Because isotopic labeling of arginine amino acids affects all arginine side chains of Gαi1, we also performed measurements of the mutant Gαi1-R178S to ensure a site-specific assignment. In H2O, no isotopic shift could be determined because the ν(CN3H5+) vibration at 1,630–1,680 cm−1 (29) is superimposed by absorptions of water. We exchanged the solvent to D2O and observed isotopic shifts exclusively in the area between 1,580 cm−1 and 1,610 cm−1 that is described in the literature for the symmetrical and asymmetrical arginine side-chain vibrations (29) in the hydrolysis spectrum (Fig. 5A). The band at 1,604 cm−1 that belongs to the GDP state is shifted to 1,600 cm−1, and the band at 1,590 cm−1 that belongs to the GDP state is shifted to 1,585 cm−1 (Fig. 5B). These band shifts could not be observed in the photolysis spectrum, probably indicating that Arg178 is in the Gαi1 caged GTP state in a similar position as in the Gαi1 GTP state. To ensure a site-specific assignment, we also measured the mutant Gαi1-R178S (Fig. 5A, orange) and observed that the corresponding bands at 1,604 cm−1 and 1,590 cm−1 were missing. Additional changes at 1,553 cm−1 (negative, Amide II) and around 1,450 cm−1 (positive, Amide II*) are caused by deuterium exchange that follows the hydrolysis (increased flexibility of switch regions in the GDP state) and is more pronounced for Gαi1-R178S because of the slowed-down hydrolysis (two orders of magnitude). Therefore, the IR bands for Arg178 were assigned to 1,604 cm−1 (Gαi1·GTP) and 1,590 cm−1 (Gαi1·GDP). Complete hydrolysis spectra of unlabeled and labeled Gαi1-WT and the Gαi1-R178S mutants are depicted in SI Appendix, Fig. 8.
Fig. 5.
IR band assignment of Gαi1-Arg178. (A) Deuterated hydrolysis spectra of unlabeled (black), η15N2 arginine-labeled Gαi1 (red), and the site-specific mutant Gαi1-R178S (orange). (B) Detailed view and ΔΔ spectrum of the assignment for Gαi1-WT. The bands at 1,604 cm−1 (GTP state) and 1,590 cm−1 (GDP state) were assigned to Arg178. These bands are missing in Gαi1-R178S. (C) Same bands were assigned for the rate-separated mutant Gαi1-K180P. Rate 1 shows a zero-line, and rate 2 shows similar isotopic shifts as Gαi1-WT. Positive bands correspond to the GDP state, and negative bands correspond to the GTP state. Arrows indicate band shifts caused by the heavy isotopes. Full hydrolysis spectra from 1,800 to 950 cm−1 are depicted in SI Appendix, Fig. 8 (Gαi1-WT and Gαi1-R178S) and SI Appendix, Fig. 9 (Gαi1-K180P).
IR Band Assignment of Arg178 in Gαi1-K180P.
After the successful band assignment of Arg178 in Gαi1-WT, we were also interested in which step the arginine finger takes action in rate-separated mutants. For this purpose, we performed measurements with η-15N2 arginine-labeled Gαi1-K180P (Fig. 5C). The bands at 1,230 cm−1 and 1,280 cm−1 that appeared in the first rate in H2O were shifted to 1,238 cm−1 and 1,275 cm−1 in D2O. Isotopic labeling of the first rate showed no band shifts [Fig. 5C, double-difference (ΔΔ) rate 1], whereas the second rate showed that the same bands at 1,604 cm−1 and 1,590 cm−1 were shifted to 1,600 cm−1 and 1,587 cm−1 like in WT Gαi1 (Fig. 5C, ΔΔ rate 2). Hence the arginine finger remains bound to GTP in the first rate and changes its conformation exclusively in the second rate of rate-separated Gαi1 mutants. The t1/2 values of the rates were t1/2 1 = 69 ± 26 s and t1/2 2 = 537 ± 58 s at 15 °C. Complete hydrolysis spectra of unlabeled and labeled Gαi1-K180P are depicted in SI Appendix, Fig. 9.
IR Bands of Gαi1-Arg178 Differ from the Bands of the Arginine Finger in Ras·RasGAP.
The arginine finger mechanism is conserved among many small GTPases. For the RasGAP neurofibromin 1 (NF1), the band assignment of Arg1276 was already performed (30). A band at 1,589 cm−1 was assigned to arginine in a deuterated water environment, and a band at 1,571 cm−1 was assigned to arginine within the binding pocket of Ras·GAP. It was not possible to observe the transient state, where Arg1276 is coupled to GTP, because bond breakage is faster than the movement of Arg1276 into the binding pocket. Gαi1 is equipped with an intrinsic arginine finger, and we could observe here the arginine finger coupled to GTP (SI Appendix, Fig. 10).
IR Bands of the Arginine Finger in Gαi1·RGS4 Differ from Intrinsic Gαi1.
After the observation that Arg178 is coupled bidentately to α-GTP and γ-GTP upon RGS binding, we examined whether the vibration of Arg178 in Gαi1·RGS4 also differs from intrinsic Gαi1. Therefore, we performed FTIR measurements with η-15N2 arginine-labeled Gαi1 in a 1:1 complex with RGS4. Indeed, the bands of Arg178 were no longer found at 1,604 cm−1 and 1,590 cm−1 as assigned for intrinsic Gαi1 (SI Appendix, Fig. 11). Isotopic labeling revealed that for both the GTP and GDP states, two bands could be assigned. In the GTP state, bands at 1,601 cm−1 and 1,583 cm−1 were shifted to 1,596 cm−1 and 1,580 cm−1, respectively. In the GDP state, bands at 1,613 cm−1 and 1,593 cm−1 were shifted to 1,610 cm−1 and 1,590 cm−1, respectively. The difference in arginine vibrations supports the monodentate vs. bidentate binding mode we proposed for intrinsic and RGS4-catalyzed Gαi1. We also performed measurements of the complex Gαi1-R178S·RGS4 in D2O to assign the site-directed arginine bands; however, in this case, the mutant is too invasive. The resulting photolysis and hydrolysis difference spectra differed significantly from Gαi1·RGS4, disallowing specific assignments. The hydrolysis spectrum of Gαi1-R178S·RGS4 matched the hydrolysis spectrum of intrinsic Gαi1 with missing bands at 1,604 cm−1 and 1,590 cm−1.
Spectra from QM/MM Simulations Reproduce FTIR Experiments.
To decode the spectral data further, we performed coupled QM/MM simulations and calculated theoretical IR spectra of the individual phosphate groups. We calculated an ensemble of 15 snapshots of a 100-ns MD trajectory (details are provided in Materials and Methods) with the functionals B3LYP, M06, and PBE with the basis set 6-31G*. The geometry of the binding pocket, including the intrinsic arginine finger, is depicted in Fig. 6A. As shown via MD simulations and experiments, Arg178 is coupled to γ-GTP. Mean values and SDs of each of 15 QM/MM simulations for the functional M06 are depicted in Fig. 6E and compared with peak maxima and full width at half-maximum values of the experiment (FTIR). The functional M06/6-31G* exactly reproduces experimental peaks and band widths for νAS Pα-O2, νAS Pβ-O2, and νAS Pγ-O3. Calculated IR vibrations of B3LYP/6-31G* and PBE/6-31G* showed slightly decreased absolute wavenumbers, but with comparable spacing and SDs, indicating the calculated geometry matches the experimental situation (SI Appendix, Fig. 12).
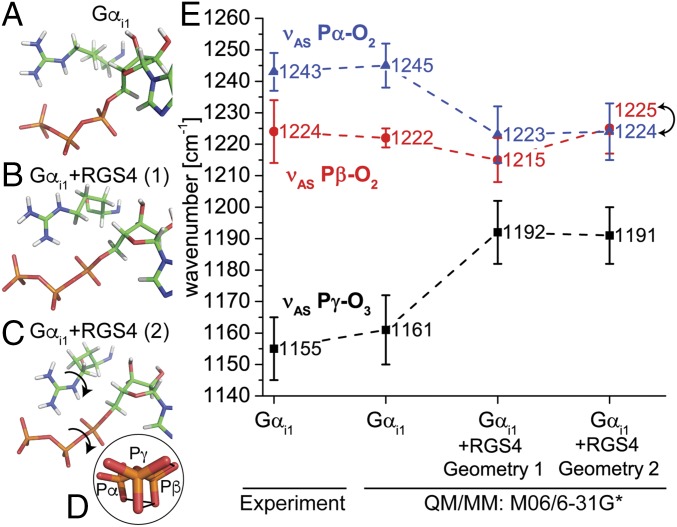
Fig. 6.
QM/MM calculations of Gαi1 and Gαi1 + RGS4. Binding pocket of intrinsic (A) and RGS4-catalyzed Gαi1 with two different positions of the arginine finger (B and C) and the corresponding calculated IR spectra from QM/MM calculations in comparison to the experiment (E). Shown are mean values and SDs for 15 snapshots of a 100-ns MD simulation. Calculation of geometry 2 (C) resulted in eclipsed (α-β-γ)-GTP (D). Although calculation of geometry 1 (B) with RGS4 resulted in higher α-GTP vibrations in comparison to the β-GTP vibrations, this trend is reversed for the eclipsed geometry 2 (C), which is in line with the experiment (E). Spectra were scaled according to the Computational Chemistry Comparison and Benchmark Database (M06/6-31G*: 0.95). Results with the functionals B3LYP and PBE were similar and are depicted in SI Appendix, Fig. 12.
Bidentate Coupling of Arg178 Causes Eclipsed (α-β-γ)-GTP Conformation.
Because the experiments predicted only a bidentate coupling of Arg178 to α-GTP and γ-GTP, we calculated different starting positions of Arg178, with Nε oriented away from α-GTP (PDB ID code 1AGR) (Fig. 6B) and Nε oriented toward α-GTP (Fig. 6C), as suggested by 19F NMR of Rho-GAP (31). Both conformations caused significant shifts of the individual GTP vibrations (Fig. 6E), but only the latter conformation formed α-β-γ eclipsed GTP (Fig. 6D). The eclipsed α-β-oxygens in this geometry were proposed to stretch the bridging P-O-P bonds, and thereby catalyze GTP hydrolysis (11). Experimental band assignments for the Gαi1–RGS4 complex were already performed, showing a slight RGS4-induced blue shift (+6 cm−1) for β-GTP (SI Appendix, Fig. 7) and a large red shift of α-GTP of −59 cm−1, resulting in a shift of the α-GTP vibration between the β-GTP and γ-GTP vibrations (Fig. 3 and SI Appendix, Fig. 6). This behavior was not reproduced in calculations with the Nε group of Arg178 turned away from α-GTP (Fig. 6E). In this case, the β-GTP band was shifted to the opposite direction than in the experiments and the α-GTP band showed higher wavenumbers than the β-GTP band (Fig. 6E). Only with the Nε group of Arg178 pointing toward α-GTP (Fig. 6C) could the experimental behavior be reproduced with a small blue shift of β-GTP and a large red shift of α-GTP below the β-GTP frequency (Fig. 6E). However, the experimental α-GTP shift of −59 cm−1 could not be reproduced quantitatively. The calculated average α-GTP shift was only −20 cm−1, which still reproduced the experimental behavior qualitatively.
Origin of the −59 cm−1 α-GTP Shift.
The QM/MM calculations suggested a tight coordination of α-GTP between Arg178 on one side and Thr48 on the other side. Therefore, we performed FTIR measurements of intrinsic and RGS4-catalyzed Gαi1-T48A and assigned the α-GTP band via isotopic labeling (SI Appendix, Fig. 14), resulting in an α-GTP band of 1,270 cm−1 for intrinsic Gαi1-T48A and a red shift to 1,222 cm−1 for Gαi1-T48A-RGS4. This point mutation decreased the RGS4-induced α-GTP shift from −59 cm−1 (Gαi1-WT) to −48 cm−1 (Gαi1-T48A), which is representative of a strong hydrogen bond. Hydrolysis kinetics of the Gαi1-T48A mutant with and without RGS4 appeared slightly slowed down to t1/2 = 9.6 ± 1.6 s (5 °C), which is one order of magnitude slower than Gαi1-RGS4 (t1/2 = 1.4 ± 0.3 s) and in the same range as Gαi1-R178S-RGS4 (t1/2 = 12.6 ± 2.7 s) (Table 1), demonstrating that Thr48 is relevant for RGS4-catalyzed hydrolysis of Gαi1.
QM/MM Simulations Show Charge Shifts and Structural Rearrangements That Assist GTP Hydrolysis.
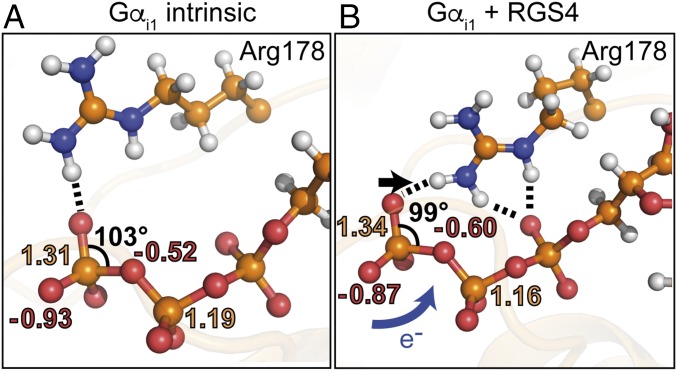
We evaluated the QM/MM Gαi1-GTP geometries and calculated Merz–Kollman partial charges [electrostatic potential fitting (ESP)] for each GTP atom. Charge distribution showed that upon RGS4 binding and rearrangement of Arg178, charges are transferred from γ-GTP to the bridging β-γ-oxygen (Fig. 7 and SI Appendix, Fig. 13). The bridging oxygen becomes more negative from −0.5 e− (intrinsic) to −0.6 e− (RGS4-catalyzed). Upon product formation, one net charge is transferred to the β-phosphate (qβ-GTP = −1 e−; qβ-GDP = −2 e−). RGS4 binding already transfers 10% of this charge in QM/MM calculations of the educt state (Fig. 7). In addition, the terminal Pγ-oxygen atom that binds the nucleophilic attacking water becomes more positive, which might decrease the negative γ-GTP barrier the attacking water has to cross upon hydrolysis. Analysis of the γ-GTP angles showed that upon RGS4 binding and Arg178 movement, the γ-GTP group becomes more planar (Fig. 7). The corresponding angle is shifted below 100°, facilitating Walden inversion of the γ-GTP group that is needed for a second-order kinetics nucleophilic substitution (SN2) hydrolysis mechanism.
Fig. 7.
RGS4-catalyzed GTP hydrolysis model in Gαi1. (A) Binding pocket, electrostatic potential fitting (ESP) partial charges, and bond angle of GTP in intrinsic Gαi1 obtained from QM/MM calculations on the level M06/6-31G*. Shown are mean values of each of 15 snapshots. (B) Binding pocket, ESP partial charges, and bond angle of GTP in Gαi1-RGS4. Shown are mean values of each of 15 snapshots. RGS4 pushes Arg178 from a monodentate coordination of GTP (A) to a bidentate coordination of GTP (B). Thereby, charges are shifted to the bridging β-γ-oxygen (B, blue arrow), which stabilizes the product state, and the Pγ-group becomes more planar (B, black arrow), which stabilizes the intermediate state and thereby catalyzes GTP hydrolysis. Detailed charge distributions on the levels B3LYP/6-31G* and PBE/6-31G* show the same trend and are depicted in SI Appendix, Fig. 13.
RGS4 Pushes Gαi1-Arg178 from a Monodentate to a Bidentate GTP Coordination.
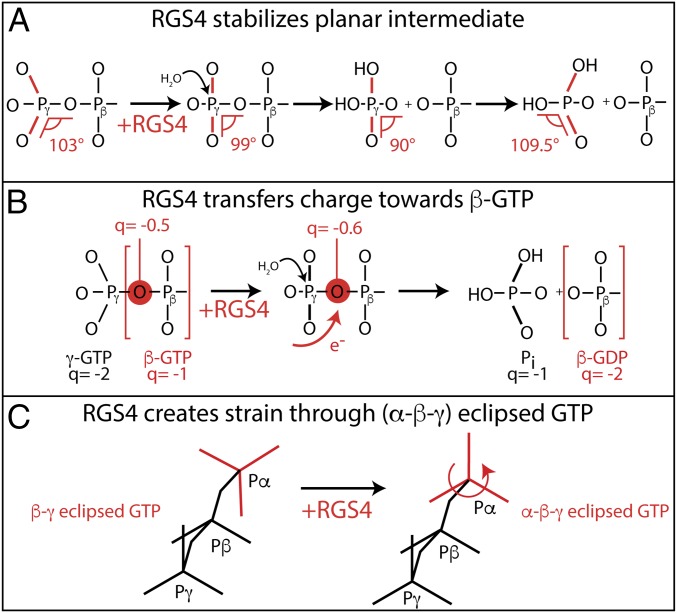
Our findings are summarized in Figs. 7 and 8. In contrast to crystal structures, Arg178 is bound to γ-GTP in the active state of Gαi1 in solution. RGS4 catalyzes GTP hydrolysis in Gαi1 by pushing Arg178 from a monodentate γ-GTP coordination to a bidentate α-γ-GTP coordination. This change is accompanied by increased planarity of the γ-GTP group (Fig. 8A), charge transfer toward β-GTP (Fig. 8B), and formation of (α-β-γ) eclipsed GTP (Fig. 8C).
Fig. 8.
Schematic representation of the mechanisms in Gαi1-RGS catalysis. (A) RGS4 stabilizes the planar intermediate that corresponds to the GDP-AlF4− crystal structure. (B) RGS4 transfers charges toward β-GTP. Upon hydrolysis, the β-phosphate gets more negative from −1 to −2 e0. RGS4 binding already transfers 0.1 e0 in the prehydrolysis state. (C) RGS4 puts GTP into an (α-β-γ) eclipsed conformation. This eclipsed conformation creates a strain in the substrate, and thereby facilitates hydrolysis.
Discussion
Here, we have elucidated the intrinsic and GAP-catalyzed molecular reaction of the intrinsic arginine finger of Gαi1 label-free with high spatiotemporal resolution by a combination of FTIR spectroscopy and molecular simulations. GTP itself, and not GTP analogs as in X-ray structure analysis, was used. The arginine finger of heterotrimeric G proteins is hydrogen-bonded to γ-GTP in the active state of intrinsic Gαi1. RGS4 binding causes stabilization of the switch areas, and thereby pushes the arginine finger to a bidentate α-γ-GTP coordination. GTP hydrolysis in GTPases (e.g., EF-G, EF-Tu, Ras, Ran, Gαi1) is proposed to take place in an SN2 mechanism via in-line attack of a water molecule at γ-GTP, with inversion of coordination around the phosphate (32–37). Many of the proposed aspects were directly observed by combination of FTIR spectroscopy and QM/MM calculations in the present study. First, the inversion of γ-GTP (38) is prepared by the Gαi1–RGS4 complex (Fig. 8A). The O-Pγ-O angle is twisted from a tetrahedral geometry toward a planar geometry that is proposed to be the transition state before inversion. A corresponding pentavalent planar transition state analog is resolved in a number of crystal structures with aluminum fluoride and GDP (2, 6, 39, 40). Displacement of Arg178 in Gαi1-RGS4 toward α-GTP pulls the hydrogen-bonded γ-oxygen atom toward a more planar geometry (Fig. 8A). Second, displacement of the arginine finger resulted in reorganization of the charge distribution: β-GTP becomes more negative (Fig. 8B). While in the GTP state, the β-phosphate moiety has a formal charge of −1, but its charge is increased to −2 in the GDP state. Thus, the RGS4-induced movement of the intrinsic arginine finger from a monodentate γ-GTP toward a bidentate α-γ-GTP coordination, which transfers a negative charge to β-GTP, induces a more product-like charge distribution. A shift toward the charge distribution of the transition state lowers the activation barrier, and thereby catalyzes the reaction. The more dissociative the character of the transition state is, the more its charge distribution resembles the charge distribution of the product (41). This mechanism is in excellent agreement with previous studies (2, 42, 43) that suggest charge transfer via the arginine finger as an important mechanism of GTP hydrolysis in small GTPases. Third, RGS4 coupling to Gαi1 twists GTP from a β-γ eclipsed conformation to a α-β-γ eclipsed conformation (Fig. 8C). It was shown previously for the small GTPase Ras-GAP system that this mechanism creates a strain in GTP, and thereby catalyzes the hydrolysis of GTP (11). Bidentate substrate binding caused a red shift of the α-GTP band (Fig. 3). The extent of the IR shift (−59 cm−1) that exceeds the expected shift of an additional hydrogen bond indicated that further structural alterations and charge shifts are caused by Arg178. To exclude the possibility that the α-GTP band is influenced by the second critical amino acid of Gαi1, Gln204, we also performed FTIR measurements of the mutant Gαi1-Q204A and found almost no spectral changes (SI Appendix, Fig. 15). Biomolecular simulations revealed that the extent of the α-GTP shift is caused by eclipsed (α-β-γ)-GTP and tight coordination of α-GTP between Arg178 and Thr48. Accordingly, Thr48 is also relevant for RGS4-catalyzed GTP hydrolysis in Gαi1 because it slowed down hydrolysis by one order of magnitude (Table 1). This finding is contrary to the small GTPase Ras, where the corresponding inverse mutation, Ras-A18T, was described to cause mild forms of cancer (44), demonstrating the GAP mechanism of small GTPases and heterotrimeric GTPases is different. The presented model could not be deduced from a number of Gαi1 crystal structures that were solved with sulfur- or nitrogen-substituted, nonhydrolyzable GTP analogs. Interestingly the position that is occupied by Arg178 in our simulations of intrinsic Gαi1 is identical to the position of the O→S substitution at the γ-phosphate in various structures (2, 45–48). Sulfur substitution causes altered biochemical and biophysical properties of the γ-GTP group (e.g., an increased van der Waals radius), probably hindering the Arg178 binding. However, some crystal structures of Gα isoforms [e.g., Gαi1-T329A (49)] feature a different sulfur position. By rotation around the Pβ-O-Pγ-O torsion angle, the terminal γ-GTPγS group is rotated and faces the side chain of Lys46 and the backbone of Gly203. The rotation of the γ-GTPγS group is accompanied by the binding of Arg178 to α-GTP and γ-GTP exactly as demonstrated here for Gαi1·RGS4 (Fig. 6A). Similar behavior of the intrinsic arginine finger is also observable in the Gα isoform Gαt (PDB ID code 1TND), where Arg174 is pointing toward β-γ-GTP, interestingly also with Nε oriented toward α-GTP as suggested by our QM/MM simulations (Fig. 6C). Several crystal structures that were stabilized with GDP-AlF4− also show that Arg178 is oriented toward the nucleotide (e.g., PDB ID code 1GFI). The combination of FTIR experiments and QM/MM simulations presented here was able to distinguish between this broad ensemble of crystal structure conformations and to choose the conformation that Arg178 adopts in solution. We also performed FTIR measurements of the conservative mutant Gαi1-R178K (SI Appendix, Fig. 16), which showed an almost unperturbed vibration of γ-GTP at 1,155 cm−1, probably indicating that Lys178 is also bound to γ-GTP. However, the kinetics of Gαi1-R178K are slowed down to the level of Gαi1-R178S, demonstrating that not only the charge of the arginine but also the precise geometry that enables bidentate binding is important for hydrolysis. Hydrolysis kinetics of WT or mutant Gαi1 in FTIR measurements agree with previous studies using other methods on WT Gαi1 (50–52), the R178S mutant (51), and the K180P mutant (53). The acceleration by RGS4 by approximately two orders of magnitude is reproduced, as well as the ability of RGS4 to accelerate Gαi1-R178S to nearly WT kinetics (51). For the related isoform Gαs, no RGS protein is known that can accelerate GTP hydrolysis, preventing the corresponding arginine finger mutants of Gαs from regaining activity due to GAP binding; it might also explain now why mutation of the arginine finger in Gαs-affected patients much more compared to Gαi1 (13, 14) and might open new roads for reversing the effect of these mutations by small molecules. For Gαi1-K180P, we observed a kinetic uncoupling between the movement of switch areas and hydrolysis. In agreement, Sprang and coworkers (53) observed conformational changes that preceded the hydrolysis reaction for this mutant. This rate is accompanied by structural changes in switch I, and can therefore be characterized by a change in fluorescence properties (53). We identified label-free spectral marker bands, indicating that the conformational change is accompanied by changes of α-GTP (SI Appendix, Figs. 3 and 4) and β-GTP (SI Appendix, Fig. 4). One might assume that this conformational change is also caused by rotation of the α-β-GTP torsions toward α-β-γ eclipsed GTP. Upon RGS4 addition, the monodentate vs. bidentate binding model is further supported by the assignment of individual IR bands of Arg178. The assigned band at 1,604 cm−1 for intrinsic Gαi1·GTP disappears when RGS4 binds Gαi1 and shows an altered band pattern at 1,601 cm−1 and 1,583 cm−1 when Arg178 is bound to α-GTP and γ-GTP (SI Appendix, Fig. 11). The arginine vibration coupled to GTP could previously not be determined because this state is not sufficiently populated in Ras·GAP hydrolysis (SI Appendix, Fig. 10). In conclusion we demonstrated that the intrinsic arginine finger is bound to γ-GTP in the intrinsic active state of Gαi1. RGS4 binding pushes the arginine finger toward a bidentate α-γ-GTP coordination, and thereby facilitates GTP hydrolysis by shifting charges toward β-GTP and increased planarity of the γ-GTP group. We expect this mechanism to be conserved among heterotrimeric GTPases.
Materials and Methods
Chemicals.
The pHPcgGTP and the isotopologs α-18O2-pHPcgGTP and β-18O3-pHPcgGTP were synthesized as described previously (17, 26, 54, 55). The η-15N2–labeled arginine was purchased from Cambridge Isotope Laboratories. Deuterium oxide was purchased from Deutero GmbH.
Cloning.
Human Gαi1 (UniProtKB P63096-1) and human RGS4 (UniProtKB P49798) were amplified as described previously (10). Briefly, genes were cloned into the vector pET27bmod with N-terminal 10× His-tag and tobacco etch virus site, and transformed into Escherichia coli DH5α for amplification. Point mutants of Gαi1 were created using overlap extension PCR. Each construct was verified by sequencing.
Protein Expression.
The plasmid encoding Gαi1 was transformed into E. coli Rosetta2(DE3) (Novagen, Merck) and incubated at 37 °C overnight on LB agar plates supplemented with 0.2% (wt/vol) glucose, 50 μg/mL kanamycin, and 20 μg/mL chloramphenicol. Precultures were incubated overnight at 37 °C and shaking at 160 rpm in LB supplemented with the same components. The plasmid encoding RGS4 was transformed into E. coli BL21(DE3) (Novagen, Merck) under identical conditions using only kanamycin for plasmid selection. For Gαi1 main cultures, 1.5 L of M9 medium (50 mM glucose, 2 mM MgSO4, 0.2 mM CaCl2, 49 mM Na2HPO4, 22 mM KH2PO4, 11.5 mM NaCl, 23 mM NH4Cl, 0.2 mM thiamine-HCl, 0.5 mM thymine, and 0.62 mM each standard amino acid) supplemented with 50 μg/mL kanamycin was inoculated with the preculture and grown at 37 °C and shaking at 100 rpm to an A600 of 0.5 absorbance units (AU). For isotopic labeling, unlabeled arginine was exchanged for η-15N2 arginine in the main culture. Protein expression was induced at 18 °C by the addition of isopropyl 1-thio-β-d-galactopyranoside (IPTG) overnight. For RGS4, main cultures contained 18 L of LB supplemented with 50 μg/mL kanamycin and 0.2% glucose. Cultures were incubated at 37 °C, shaking at 100 rpm, and 20 L/min−1 airflow in a Biostat C20-3 Fermenter (Sartorius), induced with IPTG, and grown at 18 °C for 18 h. Cells were harvested by centrifugation at 5,000 × g and 4 °C. Gαi1 was suspended in buffer A containing 20 mM Tris (pH 8), 300 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, and 5 mM d-norleucine; RGS4 was suspended in buffer B containing 50 mM Tris (pH 8), 150 mM NaCl, 0.5 mM EDTA, and 5 mM d-norleucine. Cells were flash-frozen and stored at −80 °C until protein purification.
Protein Purification.
Purification was performed as described (10). Briefly, cells were thawed, disrupted with a microfluidizer M-110L (Microfluidics Corp.), and centrifuged for 45 min at 45,000 × g and 4 °C to remove cell fragments. In contrast, RGS4-containing cells were centrifuged with an additional low-speed step for 15 min at 18,000 × g and 4 °C, followed by a high-speed step for 45 min at 75,000 × g and 4 °C. Supernatants were applied to a 25-mL nickel-nitrilotriacetic acid superflow columm (Qiagen) and eluted with buffers containing 200 mM imidazole. Fractions containing Gαi1 or RGS4 were screened via SDS/PAGE, pooled, concentrated to 5 mL using a 10,000 molecular weight cutoff (MWCO) concentrator (Amicon Ultra-15; Merck Millipore), and applied to an Illustra HiLoad 26/600 Superdex 200-pg column (GE Healthcare Life Sciences). Peak fractions were collected and concentrated to ca. 20 mg/mL for Gαi1 or ca. 10 mg/mL for RGS4. Protein concentrations were determined using Bradford reagent in triplicate. Proteins were aliquoted, flash-frozen in liquid nitrogen, and stored at −80 °C until utilization.
Control of Labeling Efficiency with LC-MS.
Efficiency of arginine η-15N2 labeling was checked via LC-MS in cooperation with Claudia Lindemann (Medical Proteome Center, Ruhr University, Bochum, Germany). Labeling efficiency was determined for four digested peptide fragments [AVVYSNTIQSIIAIIR (92.6% labeled), EYQLNDSAAYYLNDLDR (93.0% labeled), QLFVLAGAAEEGFMTAELAGVIKR (92.6% labeled), and IAQPNYIPTQQDVLR (93.2% labeled)] and resulted in an overall label efficiency of 92.8% ± 0.3%. Gαi1-K180P was arginine η-15N2–labeled with an efficiency of 96.7 ± 0.5%. Both Gαi1-WT and Gαi1-K180P showed no scattering of heavy isotopes to amino acids other than arginine.
Nucleotide Exchange to pHPcgGTP.
Nucleotide exchange reactions of WT and mutant Gαi1 were performed in the presence of alkaline phosphatase coupled to agarose beads (Sigma–Aldrich Chemie GmbH,). Phosphatase beads were washed five times with 50 mM Tris (pH 7.5) and 100 μM ZnSO4, with each washing step followed by centrifugation at 10,000 × g until the supernatant was free of phosphatase. Five milligrams of WT or mutant Gαi1 was supplemented with 50 mM Tris (pH 7.5), 10 μM ZnSO4, and a twofold molar excess of unlabeled or oxygen-labeled pHPcgGTP. Samples were incubated for 3 h and analyzed via reversed-phase HPLC (LC-2010; Shimadzu) [mobile phase: 50 mM Pi (pH 6.5), 5 mM tetrabutylammoniumbromide, 7.5% (vol/vol) acetonitrile; stationary phase: ODS-Hypersil C18 column] for remaining GDP. After >95% of GDP was hydrolyzed to guanosine, samples were centrifuged at 10,000 × g for 2 min and the supernatant was rebuffered using a Nap5 column (GE Healthcare Life Sciences) to 10 mM Hepes (pH 7.5), 7.5 mM NaCl, 0.25 mM MgCl2, and 1 mM DTT at 7 °C. Fractions containing the highest protein concentrations were pooled and concentrated using a 10,000 MWCO concentrator (Amicon Ultra-0.5; Merck Millipore). Protein concentrations were determined using Bradford reagent in triplicate, and samples were aliquoted into 107.5-μg portions (5 mM final concentration in FTIR samples). Nucleotide exchange rates to pHPcgGTP were again analyzed via HPLC (always >95% cgGTP). Samples were flash-frozen in liquid nitrogen, lyophilized light-protected for 3 h at −55 °C and 0.05 mbar in a Christ Alpha-1-2 LDPlus lyophilizer (Martin Christ GmbH), and stored packed in parafilm and aluminum foil at −20 °C until utilization. For RGS4-catalyzed measurements, RGS4 was rebuffered using a Micro Bio-Spin P-6 column (Biorad) to 4 mM Tris (pH 7.5), 3 mM NaCl, 0.1 mM MgCl2, and 0.4 mM DTT. The buffer was prepared at 5 °C to guarantee pH stability in FTIR measurements. Concentrations were determined using Bradford reagent in triplicate, and for each FTIR sample, 65 μg of RGS4 was joined with 107.5 μg of Gαi1 (1:1 complex; lyophilization of isolated RGS4 leads to loss of protein function). Samples were flash-frozen in liquid nitrogen, lyophilized, and stored light-protected at −20 °C.
FTIR Sample Preparation.
FTIR sample preparations of intrinsic Gαi1 were made as described (10). Briefly, lyophilized samples were resuspended in individual buffers to match a final concentration of 200 mM Hepes (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 200 mM DTT, and 0.1% (vol/vol) ethylene glycol (5 mM protein concentration). Samples were packed between two CaF2 windows, one with a 10-μm groove, that were sealed with silicon grease; fixed in a metal sample holder; and mounted in a spectrometer (Bruker IFS 66v/S or Vertex 80v; Bruker). Gαi1·RGS4 was measured in 100 mM Hepes (pH 7.5), 100 mM Tris (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 20 mM DTT, and 0.1% (vol/vol) ethylene glycol. For deuterated measurements, all stock solutions were prepared in D2O instead of H2O. Sample preparation was performed under nitrogen airflow under a transparent plastic cap in order to maximize hydrogen–deuterium (HD) exchange. Samples were constrained under nitrogen airflow and resuspended in D2O five times before the sample was sealed. Efficiency of deuteration was checked by integration of the H2O and D2O stretching vibrations in the spectrometer (>90% D2O).
FTIR Measurements.
Measurements were carried out as described (10). Briefly, after background spectra were taken (400 scans), photolysis of the caged compounds was initiated with a laser flash at 308 nm with an LPX 240 XeCl excimer laser (Lambda Physics; 80 flashes within 160 ms). The time point zero is defined after 40 laser flashes. The reaction was followed in the rapid scan mode of the spectrometer at 5 °C or 15 °C for intrinsic Gαi1 or at 5 °C for Gαi1·RGS4. Data were analyzed via global fit (56). The time-resolved absorbance change ΔA(ν,t) is described by the absorbance change induced by photolysis a0(ν) followed by a number n of exponential functions fitting the amplitudes a for each wavenumber ν. In the case of n = 1, a1 corresponds to the hydrolysis spectrum:
| [1] |
In the figures, disappearing bands face downward and appearing bands face upward. Data were averaged over at least three measurements. Evaluation was performed in MATLAB R2012a (The MathWorks, Inc.) and OPUS (Bruker Corp).
MD Simulations and Evaluation.
Structures of active Gαi1 (PDB ID code 1GIA), inactive Gαi1 (PDB ID code 1GIA, chain A), GDP·AlF4−-bound Gαi1 (PDB ID code 1GFI), and Gαi1·RGS4 (PDB ID code 1AGR) were prepared in the Moby program suite (57). Structure preparation included dihedral, angle, and bond corrections according to the UA amber84 force field (58), protonation according to pKa calculations using the PKA,Max,UH,JAB3 algorithm in Moby, and replacement of nucleotide analogs with natural GTP or GDP. Systems were initially solvated according to the Vedani algorithm (59), and thoroughly solvated in a cubic simulation cell with transferable intermolecular potential with 4 points (TIP4P) water and 154 mM NaCl in GROMACS 4.0.7 (60–63). Systems were energy-minimized using steepest descent and heated to 310 K using the Berendsen thermostat and barostat with a time step of 1 fs for 25 ps with, restrained protein backbone positions in the optimized potentials for liquid simulations all atom (OPLS/AA) force field (64). Electrostatics were calculated using PME (0.9 nm) and a van der Waals cutoff of 1.4 nm. Production runs were carried out without restraints for 100 ns in five replicas per system (total simulation time of 2 μs). Evaluation was performed using the GROMACS package (g_rms) and the contact matrix algorithm implemented in Moby. Pictures were created using PyMOL (Schrödinger LLC) and Gnuplot 4.4 (65).
QM/MM Calculations.
Snapshots from equilibrated MM simulations (25–100 ns in 5-ns steps for each system) were truncated substructure-based around 1.5 nm of the QM core (GTP + Mg2+ + Mg2+-coordinating water molecules). Nearest Na/Cl ions were taken into account to ensure the total charge of the system was zero. The QM core region (51 atoms) was embedded in a mobile MM region (0.5 nm), which was embedded in a fixed MM region containing protein and solvent centers (1.5 nm) that was again embedded by a fixed MM region containing protein centers beyond 1.5 nm of the QM core (MM layer ca. 6,000 atoms). Initially, a single point calculation was performed using our own N-layered integrated molecular orbital and molecular mechanics (ONIOM) QM/MM embedded method (66–68) implemented in Gaussian09 (69). Calculated Merz–Kollman (ESP) charges were transferred to an external quasi-newton Broyden–Fletcher–Goldfarb–Shanno minimizer (70, 71) implemented in the Moby program suite. The mobile MM layer was optimized, and the updated coordinates were retransferred to the Gaussian program system, where a full optimization of the QM part with QM/MM embedding according to the ONIOM scheme was performed. This procedure was repeated two times, followed by spectra calculation in the Gaussian program using normal mode analysis. No imaginary frequencies were observed for each calculation, indicating a minimum structure was always reached. Even a normal mode analysis of the MM part showed no imaginary frequencies, indicating that a minimum structure for both parts was reached successfully. QM calculations were performed with the density functionals B3LYP, M06, and PBE and the basis set 6-31G*. Calculations using the B3LYP functional are well-characterized in the literature. Additional functionals were chosen because of their strengths in dispersion (M06) and small IR scaling factors (PBE). IR frequencies were scaled according to the Computational Chemistry Comparison and Benchmark Database of the National Institute of Standards and Technology. IR frequencies for each vibration were averaged over 15 snapshots for each simulation system, and the SE was calculated for comparison with the experimental band widths. We depicted only the asymmetrical vibrations of the individual phosphate groups, because their high transition dipole moment causes IR intensities that dominate the experimental spectrum. QM/MM calculations showed two distinct νAS(Pγ-O3) bands for γ-GTP, whereas the experiments showed only γ-GTP vibration, which was previously described for small GTPases in the literature (2, 72). Therefore, mean values for νAS(Pγ-O3) were depicted to enable comparison with the experiment. The whole QM/MM calculation procedure for one snapshot was performed within a single day on eight parallel central processing unit cores (1.9 GHz; AMD Opteron).
Supplementary Material
Acknowledgments
We thank Prof. Dr. Katrin Marcus and Dr. Claudia Lindemann for LC-MS measurements at the Medical Proteome Center (Ruhr University). We thank PD Dr. Udo Höweler [chemistry-oriented program system (CHEOPS)] for helpful discussions. We further thank Dr. Jonas Schartner and Dr. Yan Suveyzdis for synthesis of the caged compounds, Iris Bourdos for excellent technical support, and the Deutsche Forschungsgemeinschaft Grant SFB 642, TP A1 for financial support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.F.P. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612394113/-/DCSupplemental.
References
- 1.Northup JK, et al. Purification of the regulatory component of adenylate cyclase. Proc Natl Acad Sci USA. 1980;77(11):6516–6520. doi: 10.1073/pnas.77.11.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB. GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin α-GDP-AIF-4. Nature. 1994;372(6503):276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- 3.Bokoch GM, Katada T, Northup JK, Ui M, Gilman AG. Purification and properties of the inhibitory guanine nucleotide-binding regulatory component of adenylate cyclase. J Biol Chem. 1984;259(6):3560–3567. [PubMed] [Google Scholar]
- 4.Rudack T, et al. Catalysis of GTP hydrolysis by small GTPases at atomic detail by integration of X-ray crystallography, experimental, and theoretical IR spectroscopy. J Biol Chem. 2015;290(40):24079–24090. doi: 10.1074/jbc.M115.648071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia F, Rudack T, Cui Q, Kötting C, Gerwert K. Detailed structure of the H2PO4(-)-guanosine diphosphate intermediate in Ras-GAP decoded from FTIR experiments by biomolecular simulations. J Am Chem Soc. 2012;134(49):20041–20044. doi: 10.1021/ja310496e. [DOI] [PubMed] [Google Scholar]
- 6.Scheffzek K, et al. The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277(5324):333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 7.Pai EF, et al. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: Implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9(8):2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kötting C, Gerwert K. What vibrations tell us about GTPases. Biol Chem. 2015;396(2):131–144. doi: 10.1515/hsz-2014-0219. [DOI] [PubMed] [Google Scholar]
- 9.Baltoumas FA, Theodoropoulou MC, Hamodrakas SJ. Interactions of the α-subunits of heterotrimeric G-proteins with GPCRs, effectors and RGS proteins: A critical review and analysis of interacting surfaces, conformational shifts, structural diversity and electrostatic potentials. J Struct Biol. 2013;182(3):209–218. doi: 10.1016/j.jsb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Schröter G, Mann D, Kötting C, Gerwert K. Integration of Fourier transform infrared spectroscopy, fluorescence spectroscopy, steady-state kinetics and molecular dynamics simulations of Gαi1 distinguishes between the GTP hydrolysis and GDP release mechanism. J Biol Chem. 2015;290(28):17085–17095. doi: 10.1074/jbc.M115.651190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rudack T, Xia F, Schlitter J, Kötting C, Gerwert K. Ras and GTPase-activating protein (GAP) drive GTP into a precatalytic state as revealed by combining FTIR and biomolecular simulations. Proc Natl Acad Sci USA. 2012;109(38):15295–15300. doi: 10.1073/pnas.1204333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandt DR, Ross EM. Catecholamine-stimulated GTPase cycle. Multiple sites of regulation by beta-adrenergic receptor and Mg2+ studied in reconstituted receptor-Gs vesicles. J Biol Chem. 1986;261(4):1656–1664. [PubMed] [Google Scholar]
- 13.McCune DJ, Bruch H. Osteodystrophia fibrosa: Report of a case in which the condition was combined with precocious puberty, pathologic pigmentation of the skin and hyperthyroidism, with a review of the literature. Am J Dis Child. 1937;54(4):806–848. [Google Scholar]
- 14.Albright F, Butler AM, Hampton AO, Smith P. Syndrome characterized by osteitis fibrosa disseminata, areas of pigmentation and endocrine dysfunction, with precocious puberty in females. N Engl J Med. 1937;216(17):727–746. [Google Scholar]
- 15.O’Neal CJ, Jobling MG, Holmes RK, Hol WGJ. Structural basis for the activation of cholera toxin by human ARF6-GTP. Science. 2005;309(5737):1093–1096. doi: 10.1126/science.1113398. [DOI] [PubMed] [Google Scholar]
- 16.Gerwert K. Molecular reaction mechanism of photosynthetic proteins as determined by FTIR-spectroscopy. Biochimica et Biophysica Acta-Bioenergetics. 1992;1101(2):147–153. [Google Scholar]
- 17.Gavriljuk K, et al. Catalytic mechanism of a mammalian Rab·RabGAP complex in atomic detail. 109(52):21348–21353. doi: 10.1073/pnas.1214431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman DE, Sprang SR. Structure of Gialpha1.GppNHp, autoinhibition in a galpha protein-substrate complex. J Biol Chem. 1999;274(24):16669–16672. doi: 10.1074/jbc.274.24.16669. [DOI] [PubMed] [Google Scholar]
- 19.Wall MA, et al. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83(6):1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 20.Noel JP, Hamm HE, Sigler PB. The 2.2 A crystal structure of transducin-alpha complexed with GTP gamma S. Nature. 1993;366(6456):654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 21.Slep KC, et al. Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 A. Nature. 2001;409(6823):1071–1077. doi: 10.1038/35059138. [DOI] [PubMed] [Google Scholar]
- 22.Waldo GL, et al. Kinetic scaffolding mediated by a phospholipase C-beta and Gq signaling complex. Science. 2010;330(6006):974–980. doi: 10.1126/science.1193438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreutz B, et al. A new approach to producing functional G alpha subunits yields the activated and deactivated structures of G alpha(12/13) proteins. Biochemistry. 2006;45(1):167–174. doi: 10.1021/bi051729t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z, Singer WD, Danesh SM, Sternweis PC, Sprang SR. Recognition of the activated states of Galpha13 by the rgRGS domain of PDZRhoGEF. Structure. 2008;16(10):1532–1543. doi: 10.1016/j.str.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sprang SR. Invited review: Activation of G proteins by GTP and the mechanism of Gα-catalyzed GTP hydrolysis. Biopolymers. 2016;105(8):449–462. doi: 10.1002/bip.22836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park C-H, Givens RS. New photoactivated protecting groups. 6. p-Hydroxyphenacyl: A phototrigger for chemical and biochemical probes. J Am Chem Soc. 1997;119(10):2453–2463. [Google Scholar]
- 27.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4--activated G(i alpha1): Stabilization of the transition state for GTP hydrolysis. Cell. 1997;89(2):251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 28.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 29.Barth A. The infrared absorption of amino acid side chains. Prog Biophys Mol Biol. 2000;74(3-5):141–173. doi: 10.1016/s0079-6107(00)00021-3. [DOI] [PubMed] [Google Scholar]
- 30.Kötting C, Kallenbach A, Suveyzdis Y, Wittinghofer A, Gerwert K. The GAP arginine finger movement into the catalytic site of Ras increases the activation entropy. Proc Natl Acad Sci USA. 2008;105(17):6260–6265. doi: 10.1073/pnas.0712095105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Y, Molt RW, Jr, Waltho JP, Richards NGJ, Blackburn GM. (19)F NMR and DFT analysis reveal structural and electronic transition state features for RhoA-catalyzed GTP hydrolysis. Angew Chem Int Ed Engl. 2016;55(10):3318–3322. doi: 10.1002/anie.201509477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb MR, Eccleston JF. The stereochemical course of the ribosome-dependent GTPase reaction of elongation factor G from Escherichia coli. J Biol Chem. 1981;256(15):7734–7737. [PubMed] [Google Scholar]
- 33.Eccleston JF, Webb MR. Characterization of the GTPase reaction of elongation factor Tu. Determination of the stereochemical course in the presence of antibiotic X5108. J Biol Chem. 1982;257(9):5046–5049. [PubMed] [Google Scholar]
- 34.Feuerstein J, Goody RS, Webb MR. The mechanism of guanosine nucleotide hydrolysis by p21 c-Ha-ras. The stereochemical course of the GTPase reaction. J Biol Chem. 1989;264(11):6188–6190. [PubMed] [Google Scholar]
- 35.Sprang SR. G protein mechanisms: Insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 36.Wittinghofer A. Phosphoryl transfer in Ras proteins, conclusive or elusive? Trends Biochem Sci. 2006;31(1):20–23. doi: 10.1016/j.tibs.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Li G, Zhang XC. GTP hydrolysis mechanism of Ras-like GTPases. J Mol Biol. 2004;340(5):921–932. doi: 10.1016/j.jmb.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 38.Walden P. Ueber die gegenseitige Umwandlung optischer Antipoden. Chem Ber. 1896;29(1):133–138. German. [Google Scholar]
- 39.Coleman DE, et al. Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science. 1994;265(5177):1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 40.Rittinger K, Walker PA, Eccleston JF, Smerdon SJ, Gamblin SJ. Structure at 1.65 A of RhoA and its GTPase-activating protein in complex with a transition-state analogue. Nature. 1997;389(6652):758–762. doi: 10.1038/39651. [DOI] [PubMed] [Google Scholar]
- 41.Carvalho ATP, Szeler K, Vavitsas K, Åqvist J, Kamerlin SCL. Modeling the mechanisms of biological GTP hydrolysis. Arch Biochem Biophys. 2015;582:80–90. doi: 10.1016/j.abb.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 42.Klähn M, Schlitter J, Gerwert K. Theoretical IR spectroscopy based on QM/MM calculations provides changes in charge distribution, bond lengths, and bond angles of the GTP ligand induced by the Ras-protein. Biophys J. 2005;88(6):3829–3844. doi: 10.1529/biophysj.104.058644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grigorenko BL, Nemukhin AV, Shadrina MS, Topol IA, Burt SK. Mechanisms of guanosine triphosphate hydrolysis by Ras and Ras-GAP proteins as rationalized by ab initio QM/MM simulations. Proteins. 2007;66(2):456–466. doi: 10.1002/prot.21228. [DOI] [PubMed] [Google Scholar]
- 44.Demunter A, et al. A novel N-ras mutation in malignant melanoma is associated with excellent prognosis. Cancer Res. 2001;61(12):4916–4922. [PubMed] [Google Scholar]
- 45.Raw AS, Coleman DE, Gilman AG, Sprang SR. Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42--> Val mutant of Gialpha1. Biochemistry. 1997;36(50):15660–15669. doi: 10.1021/bi971912p. [DOI] [PubMed] [Google Scholar]
- 46.Posner BA, Mixon MB, Wall MA, Sprang SR, Gilman AG. The A326S mutant of Gialpha1 as an approximation of the receptor-bound state. J Biol Chem. 1998;273(34):21752–21758. doi: 10.1074/jbc.273.34.21752. [DOI] [PubMed] [Google Scholar]
- 47.Thaker TM, Sarwar M, Preininger AM, Hamm HE, Iverson TM. A transient interaction between the phosphate binding loop and switch I contributes to the allosteric network between receptor and nucleotide in Gαi1. J Biol Chem. 2014;289(16):11331–11341. doi: 10.1074/jbc.M113.539064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaya AI, et al. A conserved phenylalanine as a relay between the α5 helix and the GDP binding region of heterotrimeric Gi protein α subunit. J Biol Chem. 2014;289(35):24475–24487. doi: 10.1074/jbc.M114.572875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kapoor N, Menon ST, Chauhan R, Sachdev P, Sakmar TP. Structural evidence for a sequential release mechanism for activation of heterotrimeric G proteins. J Mol Biol. 2009;393(4):882–897. doi: 10.1016/j.jmb.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 50.Zielinski T, et al. Two Galpha(i1) rate-modifying mutations act in concert to allow receptor-independent, steady-state measurements of RGS protein activity. J Biomol Screen. 2009;14(10):1195–1206. doi: 10.1177/1087057109347473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell. 1996;86(3):445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 52.Linder ME, Ewald DA, Miller RJ, Gilman AG. Purification and characterization of Go alpha and three types of Gi alpha after expression in Escherichia coli. J Biol Chem. 1990;265(14):8243–8251. [PubMed] [Google Scholar]
- 53.Thomas CJ, et al. Uncoupling conformational change from GTP hydrolysis in a heterotrimeric G protein alpha-subunit. Proc Natl Acad Sci USA. 2004;101(20):7560–7565. doi: 10.1073/pnas.0304091101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goody RS. A simple and rapid method for the synthesis of nucleoside 5′-monophosphates enriched with 17O or 18O on the phosphate group. Anal Biochem. 1982;119(2):322–324. doi: 10.1016/0003-2697(82)90592-9. [DOI] [PubMed] [Google Scholar]
- 55.Hecht SM, Kozarich JW. A chemical synthesis of adenosine 5′-(gamma-32P)triphosphate. Biochim Biophys Acta. 1973;331(3):307–309. doi: 10.1016/0005-2787(73)90015-4. [DOI] [PubMed] [Google Scholar]
- 56.Gerwert K. Molecular reaction mechanisms of proteins monitored by time-resolved FTIR-spectroscopy. Biol Chem. 1999;380(7-8):931–935. doi: 10.1515/BC.1999.115. [DOI] [PubMed] [Google Scholar]
- 57.Höweler U. 2007. MAXIMOBY 11.1 (CHEOPS, Altenberge, Germany)
- 58.Case DA, et al. 2014. AMBER 14 (University of California, San Francisco)
- 59.Vedani A, Huhta DW. Algorithm for the systematic solvation of proteins based on the directionality of hydrogen bonds. J Am Chem Soc. 1991;113(15):5860–5862. [Google Scholar]
- 60.Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91(1-3):43–56. [Google Scholar]
- 61.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. Mol Model Annu. 2001;7(8):306–317. [Google Scholar]
- 62.Van Der Spoel D, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 63.Pronk S, et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110(6):1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 65.Williams T, Kelley C. 2015 Gnuplot 4.5: An interactive plotting program. Available at www.gnuplot.info. Accessed September 12, 2016.
- 66.Dapprich S, Komáromi I, Byun KS, Morokuma K, Frisch MJ. A new ONIOM implementation in Gaussian98. Part I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. Theochem. 1999;461-462:1–21. [Google Scholar]
- 67.Vreven T, Morokuma K, Farkas O, Schlegel HB, Frisch MJ. Geometry optimization with QM/MM, ONIOM, and other combined methods. I. Microiterations and constraints. J Comput Chem. 2003;24(6):760–769. doi: 10.1002/jcc.10156. [DOI] [PubMed] [Google Scholar]
- 68.Vreven T, Morokuma K. Hybrid Methods: ONIOM(QM:MM) and QM/MM. Annual Reports in Computational Chemistry. Elsevier; Amsterdam: 2006. pp. 35–51. [Google Scholar]
- 69.Frisch MJ, et al. 2009. Gaussian 09, Rev. A.02 (Gaussian, Inc., Wallingford, CT)
- 70.Broyden CG. The convergence of a class of double-rank minimization algorithms 1. General considerations. IMA J Appl Math. 1970;6(1):76–90. [Google Scholar]
- 71.Fletcher R. A new approach to variable metric algorithms. Comput J. 1970;13(3):317–322. [Google Scholar]
- 72.Xia F, Rudack T, Kötting C, Schlitter J, Gerwert K. The specific vibrational modes of GTP in solution and bound to Ras: A detailed theoretical analysis by QM/MM simulations. Phys Chem Chem Phys. 2011;13(48):21451–21460. doi: 10.1039/c1cp22741f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.