Significance
The surface of mammalian cells contains abundant plasma membrane invaginations termed “caveolae.” Caveolae are important for various cellular functions, e.g. signaling, membrane regulation, and vesicular trafficking. Assembly and stability of caveolae depends on a protein coat formed by complexes of cavin and caveolin proteins whose structure has remained elusive. To understand the architecture of caveolae, we structurally analyzed cavin and caveolin complexes and visualized caveolae in cells by electron cryomicroscopy. A regular polyhedron emerged as the most likely architectural principle of caveolae. Polyhedra provide the underlying principle in many biological structures in which spherical curvature is imposed by proteins. We suggest a model for the caveolar coat architecture based on regularly arranged, net-like cavin assemblies and disc-shaped caveolin oligomers.
Keywords: caveolae, coat proteins, coat assembly, membrane organization, electron cryomicroscopy
Abstract
Caveolae are invaginated plasma membrane domains involved in mechanosensing, signaling, endocytosis, and membrane homeostasis. Oligomers of membrane-embedded caveolins and peripherally attached cavins form the caveolar coat whose structure has remained elusive. Here, purified Cavin1 60S complexes were analyzed structurally in solution and after liposome reconstitution by electron cryotomography. Cavin1 adopted a flexible, net-like protein mesh able to form polyhedral lattices on phosphatidylserine-containing vesicles. Mutating the two coiled-coil domains in Cavin1 revealed that they mediate distinct assembly steps during 60S complex formation. The organization of the cavin coat corresponded to a polyhedral nano-net held together by coiled-coil segments. Positive residues around the C-terminal coiled-coil domain were required for membrane binding. Purified caveolin 8S oligomers assumed disc-shaped arrangements of sizes that are consistent with the discs occupying the faces in the caveolar polyhedra. Polygonal caveolar membrane profiles were revealed in tomograms of native caveolae inside cells. We propose a model with a regular dodecahedron as structural basis for the caveolae architecture.
Caveolae are protein-coated invaginations 50–80 nm in diameter that are present in the plasma membrane of many cell types (1, 2). In contrast to the polygonal and symmetrical clathrin, coat protein 1 (COPI), and coat protein 2 (COPII) coats, which have been studied in detail and solved to nearly atomic resolution (3–5), the architecture of the caveolar coat has remained unknown. The caveolar coat differs remarkably from clathrin and COP coats in being stable and long-lived without undergoing rounds of assembly and disassembly, even during vesicular transport (6–8). The membrane-embedded caveolins and the peripherally attached cavins are essential coat components. Each caveolar coat contains a defined number of both of these proteins that do not exchange with a free protein pool (9–11). Caveolins and cavins each form multimeric, independent assemblies with sedimentation coefficients of 70S and 60S, respectively (6, 12, 13).
Caveolins assemble into an integral membrane scaffold that contains an average of 144 caveolin molecules and defines the caveolar membrane domain (6, 8). The higher-order complex is assembled via lateral association of 8S caveolin oligomers in the course of their biosynthetic transit through the secretory pathway and requires the presence of cholesterol (6, 8, 14, 15). When the nascent caveolin scaffold has been delivered to the plasma membrane, cavins associate (6, 9, 13). Their role is to stabilize the caveolar domain and to promote the formation of the invaginated shape (16).
Cavins occur in homogenous 60S oligomers containing ∼50–60 cavin molecules (6, 17). It is likely that each caveolar coat consists of one of these complexes. Although the assembly of the 60S complex can occur independently of caveolin or caveolae, total internal reflection fluorescence (TIRF) microscopy has shown gradual attachment of cavins to nascent caveolae in the plasma membrane (6). In mammalian cells there are four cavin isoforms (Cavin1–4). Cavin1 is ubiquitously expressed and is essential for the formation and stability of caveolae. Cavin2 and 3 have roles in regulating caveolar morphology and trafficking, respectively (9, 18–20). Cavin4 is a muscle-specific isoform (13). Cavin1 is the most abundant protein in the cavin coat and is sufficient for its assembly. Cavin2 and Cavin3 require Cavin1 as a bridging factor for interaction and for recruitment to caveolae (10, 17, 20, 21). Thus, Cavin1 is the key structural component of the cavin coat.
Cavin1 is predicted to have two coiled-coil domains (10, 21). The crystal structure of the domain close to the N terminus (residues 45–155) exhibits a trimeric coiled-coil composed of parallel alpha helices. This coiled-coil mediates Cavin1 self-assembly in solution and association with phosphoinositides (21). A trimer formed by Cavin1 has been suggested to be the building block of the cavin coat (10). The second coiled-coil domain is close to the C terminus. It is α-helical and promotes higher-order assembly (21). Sequences outside the predicted coiled-coil regions are predicted to be disordered.
Cavin1 shows little or no direct interaction with caveolin (9, 20, 22). Although a protein complex containing caveolin and cavins can be isolated after chemical cross-linking before cellular lysis (10), it is likely that cavins associate with caveolae mainly via binding to membrane lipid head groups. Several lipid species (e.g., cholesterol, glycosphingolipids, and phosphatidylserine) are specifically enriched in caveolae and likely are immobilized through their interaction with caveolar proteins (23–25). Phosphatidylserine has been shown to interact with all cavin isoforms in vitro (9, 26, 27).
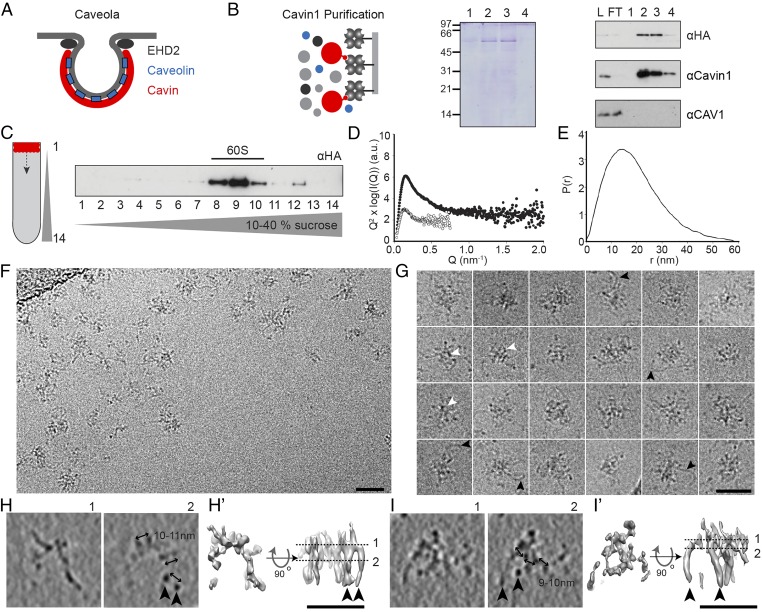
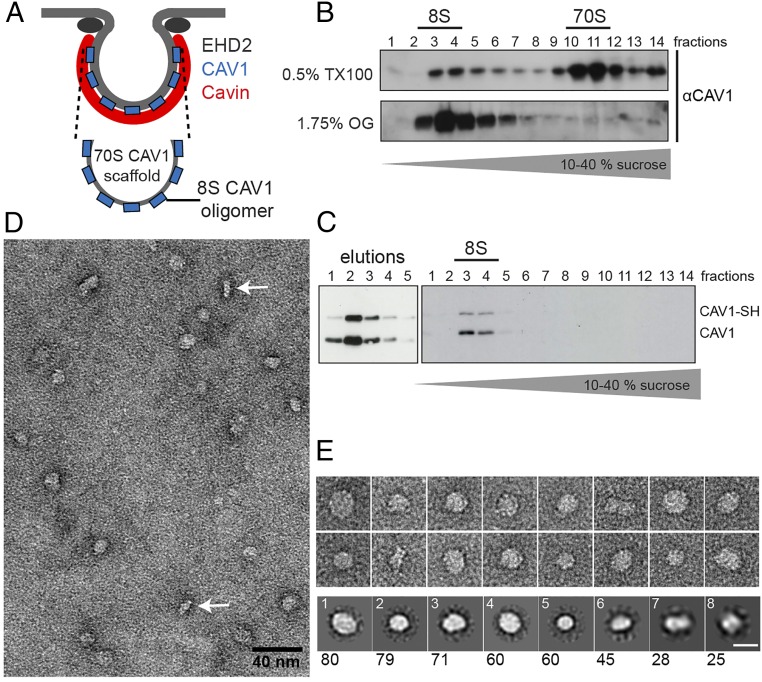
Even though the essential caveolar coat determinants, their stoichiometry, and oligomeric state in caveolae are known, a consensus regarding the structure of the caveolar coat is lacking. In EM images of heavy metal-shadowed, rapid-freeze, deep-etch membrane preparations, caveolae appear to bear a striated coat on their cytoplasmic surface. This coat consists of ridges that form meridian or spiral patterns (28, 29). Cellular electron tomography of fixed and stained plastic sections shows a sparse coat of cytoplasmic spikes and intramembrane densities (30). Immuno-EM and a tag-based staining method indicate that caveolins and cavins are distributed uniformly around the caveolar bulb, whereas EHD2, an ATPase that confines caveolae to the plasma membrane, localizes to the neck (Fig. 1A) (10).
Fig. 1.
Cavin1 60S complexes possess a filamentous net-like structure. (A) Schematic depicting the localization of major structural proteins at caveolae. (B) Purification of Strep-HA-Cavin1 expressed in HEK293 cells by Strep-Tactin affinity chromatography. Cavin1 appears as major band in Coomassie-stained SDS/PAGE gels; lanes 1–4 show eluted fractions. Western blots show the presence of Cavin1 but not CAV1 in purified samples (antibodies against HA, Cavin1, and CAV1). FT, flow through; L, lysate; 1–4, eluted fractions. (C) Fractions of 10–40% sucrose velocity gradient of purified Cavin1, analyzed by SDS/PAGE/Western blot. Cavin1 is present in an oligomer sedimenting at 60S. (D and E) SAXS analysis of purified Cavin1. (D) Kratky plots show the presence of both structured and intrinsically disordered regions. Shown are scattering profiles of Cavin1 complexes measured at concentrations of 0.1 mg/mL (open circles) and 0.5 mg/mL (filled circles). (E) Pair-distance distribution function P(r) giving the distribution of intramolecular distances within Cavin1 complexes. (F) CryoEM 2D projection image of Cavin1 60S complexes in vitreous ice. (G) Montage of cryoEM projection images of individual Cavin1 complexes showing a mesh core and extending protein filaments (black arrowheads). Central ring-shaped and polygonal arrangements can be observed (white arrowheads). (H and I) CryoET 3D reconstructions of two examples of Cavin1 60S complexes (∼50 nm diameter), showing two selected 12-Å-thick slices through the 3D volume. (H′ and I′) Surface renderings of the tomogram densities with top and side views of the complexes, contoured at two SD above the mean density value. Protein domains extend in the z axis as filaments (black arrowheads) that are connected in the top slices of the tomograms (section 1). Micrographs were recorded at 200 keV with a calibrated pixel size of 4 Å (F and G) and 5.9 Å (H and I) at defoci of −4 to −6 µm. (Scale bars in F–I: 50 nm.)
Here we used electron cryomicroscopy (cryoEM)/cryotomography (cryoET) to investigate the structure and assembly of the two major coat components, caveolin and cavin. First, we examined the peripheral coat formed by cavins and purified the intact 60S Cavin1 complex. When analyzing its structure in solution and after reconstitution with liposomes, we detected a flexible, net-like protein mesh capable of forming a polyhedral lattice on phosphatidylserine-containing vesicles of caveolar size. Mutant Cavin1 forms with deletions in the two coiled-coil domains yielded insights into assembly intermediates. Analysis of purified Caveolin-1 (CAV1) 8S complexes showed that they assume round, disc-shaped oligomers that fit and may occupy the faces of a polyhedral protein net formed by Cavin1. Finally, we analyzed native caveolae in situ in plunge-frozen cells by cryoET and found a closely appressed protein coat with striking polygonal profiles in slices through the caveolar bulb. Combining our data and previously published studies, we put forward a reconciling model for the overall architecture of caveolae that has a regular dodecahedron as its structural basis.
Results
The Cavin1 60S Complex Has a Filamentous Net-Like Structure.
To characterize the 60S complexes formed by cavins in cells, we developed a mammalian expression system using a Tet-inducible HEK293-FlpIn cell line expressing full-length mouse Cavin1 with an N-terminal Strep- and HA-tag. The complexes were purified using Strep-Tactin affinity chromatography after detergent solubilization (Materials and Methods). The mild, single-step purification protocol yielded an essentially pure preparation of Cavin1 as judged by Coomassie blue staining (Fig. 1B). When subjected to sucrose velocity gradient centrifugation in the absence of detergent, the purified Cavin1 sedimented as a sharp 60S peak (Fig. 1C). Mass spectrometry and Western blot analysis did not detect the presence of CAV1, Cavin2, or Cavin3 (Materials and Methods and Fig. 1B). It is noteworthy that, although Cavin1 and Cavin2 could be expressed in Escherichia coli, no 60S complexes were formed.
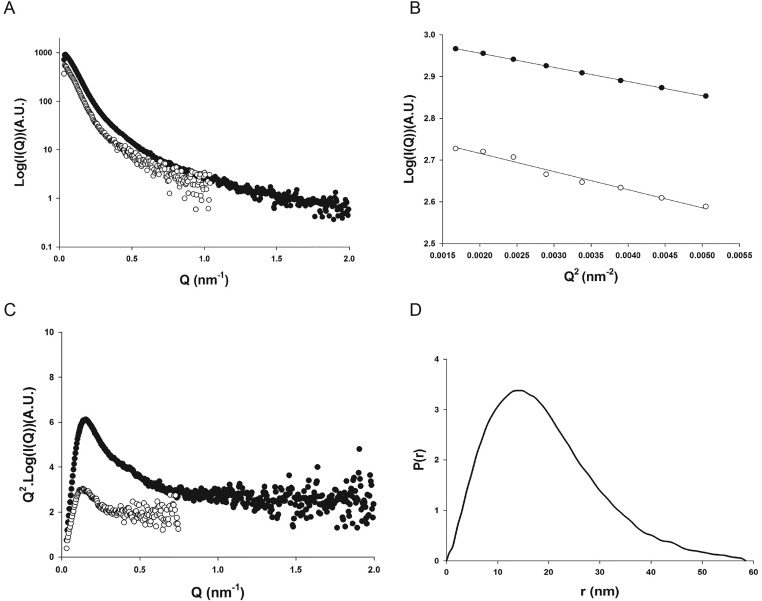
Small-angle X-ray scattering (SAXS) showed that the purified Cavin1 preparation was free of aggregates (Fig. 1 D and E and Fig. S1). Fitting of the linear Guinier plot yielded a radius of gyration of 16.0 (± 0.6) nm. Estimates of the mass indicated a 2.3-MDa complex, suggesting an average of ∼52 Cavin1 monomers per complex. Kratky plots assessing the level of intrinsic flexibility revealed significant deviation from the classic bell-shaped curve displayed by globular proteins (Fig. 1D). This deviation suggested the presence of both structured and intrinsically disordered regions. Similarly, analysis of the pair-distance distribution function suggested a rather extended molecule with a maximal intramolecular distance of about 60 nm (Fig. 1E).
Fig. S1.
SAXS analysis of the 60S Cavin1 complexes in solution. (A) SAXS scattering profiles of Cavin1 complexes measured at concentrations of 0.1 and 0.5 mg/mL are shown as white and black circles, respectively. (B) Guinier plots showing linear behavior in the low Q range. (C) Kratky plots showing the presence of both structured and intrinsically disordered regions. (D) Pair-distance distribution function P(r) reflecting the distribution of intramolecular distances within the Cavin1 complexes.
CryoEM projection images of 60S complexes showed filamentous assemblies with a mean diameter of 50 ± 10 nm (n = 30), in agreement with the values obtained by SAXS (Fig. 1 F and G). Although heterogeneous, the complexes shared a net-like appearance. They displayed a denser center and flexible filaments, ∼2–3 nm in diameter and varying in length, that extended outwards (Fig. 1G, black arrowheads). In the central domains we often observed several ring-shaped and polygonal profiles that were 6–8 nm in diameter (Fig. 1G, white arrowheads).
CryoET of the complexes revealed that, rather than being flat, they were three-dimensional with thicknesses >30 nm (Fig. 1 H and I). Analysis of tomograms showed that the complexes’ building blocks were interconnected filaments 2–3 nm thick and with interfilament spacing ranging from 5–11 nm (Fig. 1 H and I). Because of the thin nature of the filaments and the consequently limited signal-to-noise ratio in cryoET, we could not resolve the polygonal structures comprehensively in 3D. Taken together, soluble Cavin1 60S complexes resembled flexible, filamentous, net-like structures with polygonal or ring-shaped meshes.
Membrane-Bound Cavin1 Complexes Exhibit Locally Ordered Architecture.
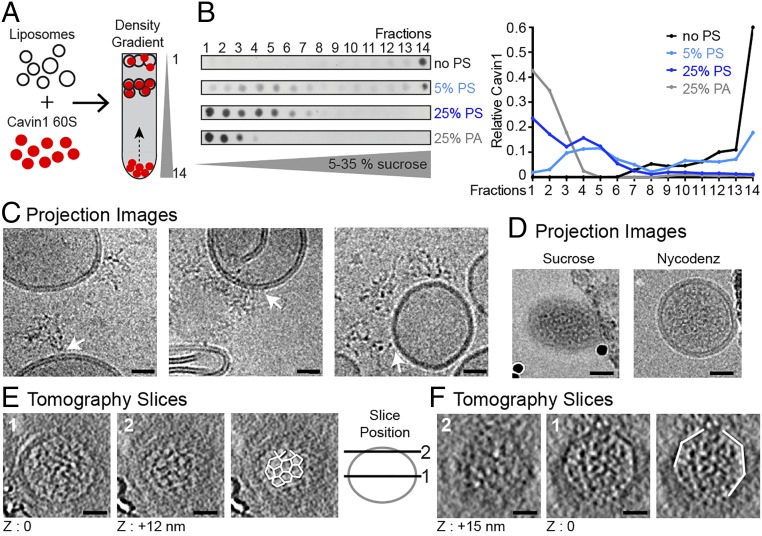
How do Cavin1 complexes arrange themselves to form a protein coat on membranes? To answer this question, we incubated purified 60S complexes with 80-nm liposomes. Using flotation in sucrose density gradients, buoyant liposome-bound complexes were separated from the unbound complexes (Fig. 2A).
Fig. 2.
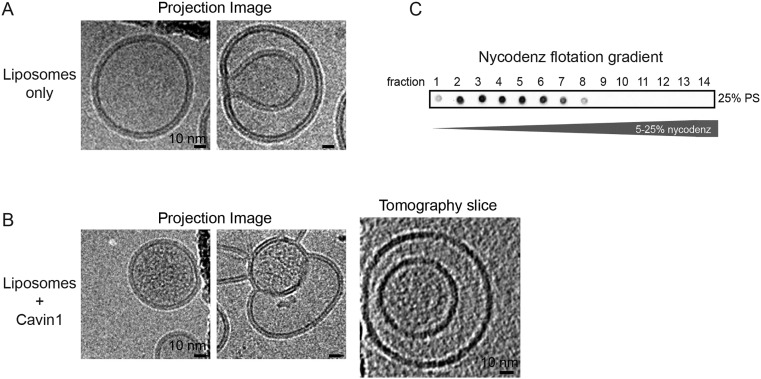
The Cavin1 complex associates with liposomes and exhibits locally ordered polygonal architecture. (A) Schematic of Cavin1–liposome incubation and subsequent proteoliposome flotation by density gradient centrifugation. (B) Cavin1 60S complexes were incubated with liposomes containing 5 or 25% PS or 25% PA (in the lipid mix: 25% DOPS, 15% DOPC, 15% DOPE, 45% cholesterol) and floated in 5–35% sucrose density gradients. Gradient fractions were analyzed by dot blot using an anti-HA antibody. The graph shows the quantification of Cavin1 distribution across fractions. (C–F) Cavin1 60S complexes incubated with 25% PS liposomes. (C) CryoEM projection images of liposomes incubated with Cavin1, before flotation. 60S complexes show connections to the liposome bilayer (white arrows). Images were taken at different defocus values. (D–F) CryoEM/ET analysis of proteoliposomes from fractions 4 and 5 of the gradients. (D) 2D projection images with sucrose or Nycodenz as the density medium. Liposomes appear naked on the outside of the membrane. The micrographs of sucrose gradients were recorded on a CCD camera at a higher defocus (−5 to −6 µm) than the micrographs of Nycodenz gradient samples acquired on a K2 summit direct detector (−2 to −4 µm). (E and F) CryoET of two exemplary proteoliposomes. For each, a central (1) and tangential (2) slice through the tomogram is shown. Slice position is depicted in the adjacent scheme. Cavin1 appears filamentous inside the liposome and adopts a polyhedral net-like structure in contact with the membrane (outlined in white in E). Angular shapes of proteoliposomes were visible (white outlines in F). Images were recorded at 200 or 300 keV with a calibrated pixel size of 5.9 Å or 4.9 Å at defoci of −3.5 to −5 µm. (Scale bars: 20 nm.) The thickness of the tomographic slice in E and F is 12 Å. Full tomographic volumes corresponding to E and F are shown in Movies S1 and S2.
Cavin1 complexes did not associate with liposomes composed of phosphatidylcholine (DOPC):phosphatidylethanolamine (DOPE):cholesterol; they remained in the bottom fraction of the gradient (Fig. 2B). Because Cavin1 is a phosphatidylserine (PS)-binding protein (9), we included 5 or 25 mol% PS in the DOPC:DOPE:cholesterol liposomes. With 5 mol% PS, half of the Cavin1 floated and formed a peak in fractions 3–5 corresponding to a sucrose density of 1.03–1.05 g/cm3. With 25 mol%, all the Cavin1 floated, and a second peak of lower density was observed in fractions 1 and 2 (Fig. 2B).
PS is an abundant, negatively charged phospholipid found enriched in caveolae (24, 25). To test whether the Cavin1 60S complex has a specific affinity for this phospholipid, as opposed to a general affinity for negatively charged membrane surfaces, we replaced PS with phosphatidic acid (PA) at 25 mol%. Strikingly, all Cavin1 proteins bound to PA liposomes and floated to the top of the gradient, indicating that Cavin1 interaction with lipids was mediated by electrostatic interactions with charged lipid head groups (Fig. 2B). However, the flotation pattern was different: Cavin1 was detected only in the top fractions 1–3. The denser vesicles in fractions 4–5 were not present.
First, we analyzed the mixture of 25 mol% PS liposomes and Cavin1 before flotation by cryoEM. Projection images showed Cavin1 60S complexes loosely attached to the liposome surfaces (Fig. 2C). The multifilamentous complexes appeared as the 60S complexes seen in solution (Fig. 1 F and G).
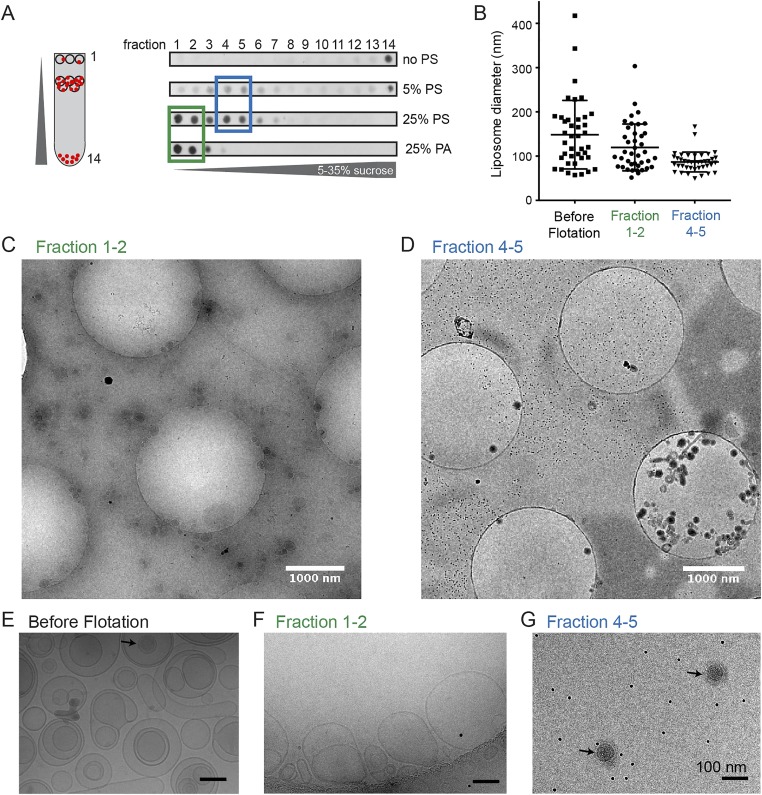
Next, we analyzed the population of liposomes found in fractions 1 and 2 and in fractions 4 and 5 after sucrose density gradient centrifugation. Vesicles in fractions 4 and 5 were more homogenous and smaller than liposomes detected in fractions 1 and 2 or preflotation (Fig. S2 B–G). Notably, proteoliposomes in fractions 4 and 5 appeared electron dense, i.e., protein-rich (Fig. 2D and Fig. S2 D and G), whereas the majority of liposomes in fractions 1 and 2 and preflotation were naked (Fig. S2 C, E, and F).
Fig. S2.
Characterization of liposome sizes before and after flotation following incubation with 60S Cavin1 complexes. (A) Gradient fractions analyzed by dot blot using an anti-HA antibody. (B) Diameter of the liposomes measured from cryoEM 2D projection images show a significant reduction of liposome size in fractions 4 and 5 with a mean diameter of 87 nm (n = 40) compared with fraction 1 and 2 or before flotation. (C and D) Representative fields of view of fractions 1 and 2 (green box in A) (C) and fractions 4 and 5 (blue box in A) (D) by cryoEM; defocus −70 µm. Tiny black dots in background are 10-nm gold fiducial markers. (E and F) Higher-magnification projection images. (E) Before flotation. (F) Pooled fractions 1 and 2. (Scale bars: 100 nm.) (G) Pooled fractions 4 and 5 after flotation. Liposomes are more heterogeneous in size and less electron dense in E and F; denser proteoliposomes are found in G (black arrow).
CryoET analysis revealed that the Cavin1 proteoliposomes were not empty but seemed to contain densities on the inside of liposomes (Fig. 2 E and F). A dense meshwork of filamentous protein was detected in all slices through the vesicles (Movies S1 and S2). To confirm that this observation was not caused by the remaining sucrose affecting the detection of cavin on the outside of the membrane, we performed the flotation assay with Nycodenz density medium instead of sucrose. This test also resulted in naked membranes on the outside of the liposomes (Fig. 2D and Fig. S3). Although the occurrence of cavin densities on the inside was puzzling at first, frequent observation of inward-folding intermediates of PS-containing liposomes may explain the mode of access to the liposome inside (Fig. S2E and Fig. S3).
Fig. S3.
(A) CryoEM 2D projection images of 25 mol% PS-containing liposomes that were floated in a Nycodenz density gradient and collected from the top fraction. Round liposomes with a single membrane and liposomes with more complex shapes and inward-folding intermediates can be observed. (B) CryoEM projection images and cryoET tomogram slices of Cavin1 proteoliposomes that were floated in a Nycodenz density gradient and collected from fractions 3–5. Cavin1 densities are localized to the inside of the vesicle. Inward-folding events of PS-containing liposomes may explain the mode by which Cavin1 accesses the liposomes inside. (C) Cavin1 proteoliposome flotation in 5–35% Nycodenz density gradients. Gradient fractions were analyzed by dot blot using an anti-HA antibody. Images were recorded at 200 or 300 keV with a calibrated pixel size of 6 Å or 5 Å at defoci of −3.5 to −5 µm. The thickness of the tomographic slices in B is 20 Å.
Strikingly, in top and bottom tomogram slices through proteoliposomes we occasionally observed patches of planar polygonal protein lattices on the inner face of the membrane (Fig. 2E and Movie S1). Individual pentagonal and hexagonal polygons were ∼10–12 nm in diameter with edge lengths of ∼6–7 nm. Such regular polygonal arrangements were not found independent of membranes. Additionally, in several cases we detected that the 60S cavin complex induced membrane deformation leading to liposomes with an angular shape consistent with the properties of a polyhedron (Fig. 2F and Movie S2).
Taken together, these observations show that Cavin1 60S complexes could associate with PS-containing liposomes to generate proteoliposomes. Although it was not possible to reveal the molecular details of the full coat architecture of the proteins on the membrane, it was evident that the organization was consistent with the formation of polyhedral structures. The lattice observed in selected tomogram slices resembled a regular polyhedron, e.g., a dodecahedron or a truncated icosahedron.
Cavin1 60S Complexes Assemble via Predicted Coiled-Coil Domains.
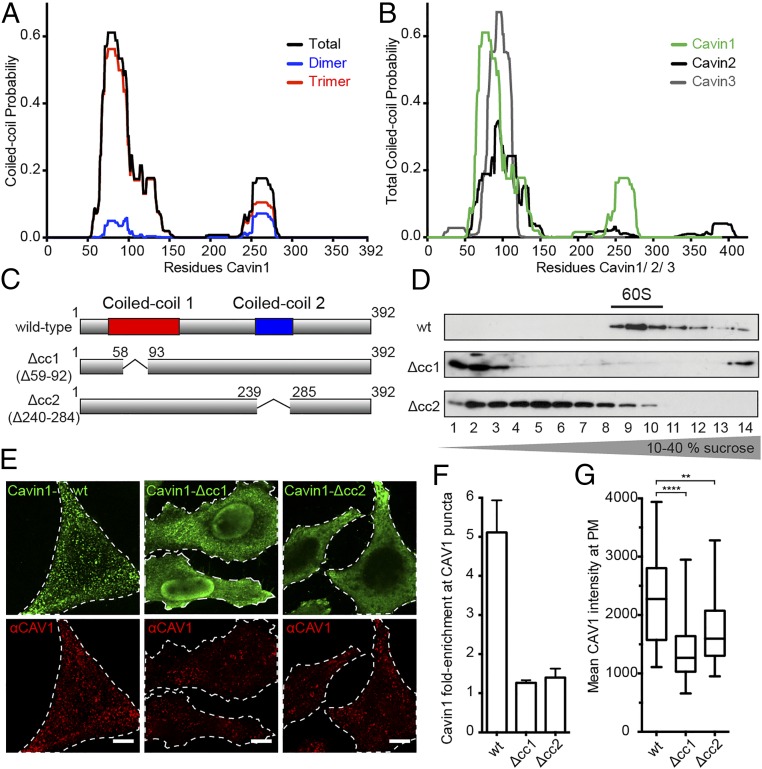
To reveal how homo-oligomerization of Cavin1 could lead to a polyhedral net-like complex, we constructed deletion mutants in the two Cavin1 coiled-coil regions (cc1 and cc2), i.e., Δcc1 (with amino acids 59–92 deleted) and Δcc2 (with amino acids 240–284 deleted) that lacked coiled-coil region 1 and coiled-coil region 2, respectively (Fig. 3 A and C). The design was based on secondary structure prediction for the mouse/human Cavin1 sequence (Fig. 3A). Residues 50–95 of cc1 exhibited a high likelihood for a trimeric arrangement, consistent with recent data indicating that peptides enclosing this domain can form parallel trimers when expressed in bacteria (21). Domain cc2 showed a lower probability for coiled-coil conformation with no predicted preference for trimers vs. dimers (Fig. 3A). Interestingly, the sequence predictions indicated that both Cavin2 and Cavin3 contain cc1 but lack the predicted cc2 domain (Fig. 3B).
Fig. 3.
Two coil-coil domains determine Cavin1 complex assembly. (A) Coiled-coil prediction for Cavin1 displaying total, dimer, and trimer probability. Structure prediction was according to the MultiCoil program (Materials and Methods) for the human/mouse Cavin1 protein sequence. Coiled-coil domains at the N and C termini are predicted. (B) The comparison of total coiled-coil probabilities for human Cavin1, Cavin2, and Cavin3 sequences shows that Cavin2 and Cavin3 lack the predicted C-terminal coiled-coil domain. (C) Details of Cavin1-Δcc1 and -Δcc2 coiled-coil–deletion mutants that were designed according to the highest computed coiled-coil probability scores, i.e., deletion of residues 59–92 for cc1 and of residues 240–284 for cc2. (D) Cell lysates prepared from HeLa cells expressing Flag-Cavin1-WT, -Δcc1, or -Δcc2 were run through 10–40% sucrose velocity gradients, and fractions were analyzed by SDS–PAGE/Western blot with an anti-Flag antibody. Cavin1 mutants are deficient in 60S assembly and show distinct sedimentation peaks. (E) Confocal fluorescence microscopy images of HeLa cells transiently expressing Flag-Cavin1-WT, -Δcc1, or -Δcc2 for 48 h. Cells were fixed and stained for endogenous CAV1 (red) and Flag (green). (Scale bars: 10 μm.) (F) WT and mutant Cavin1 enrichment at caveolae (CAV1 puncta). Data are shown as mean value ± SD; n = 9. (G) CAV1 intensity at the plasma membrane in cells expressing Cavin1-WT, -Δcc1, or -Δcc2. Whiskers indicate minimum and maximum values. ****P < 0.0001, WT vs. Δcc1; **P < 0.0012, WT vs. Δcc2; Mann–Whitney test. n = 40 cells for each condition.
We expressed the Cavin1 deletion mutants in HeLa cells and tested their assembly status by sucrose velocity gradient centrifugation (Fig. 3D). Whereas full-length Cavin1 was present as 60S complexes, Δcc1 remained in the top fractions 1–3 with a sedimentation coefficient less than 6S corresponding to a mass below 150 kDa (Fig. 3D). However, Δcc2 assembled into larger complexes peaking in fractions 4 and 5 (corresponding to about 25S). Whereas both of the coiled-coil regions were thus needed for the formation of the native 60S complex, the mutant lacking cc2 was capable of partial assembly.
Instead of colocalizing with endogenous CAV1 in plasma membrane puncta like the full-length Cavin1, both Δcc1 and Δcc2 failed to associate efficiently with caveolae in HeLa cells (Fig. 3 E and F). In addition to being diffusely distributed in the cytosol, Δcc1 was enriched in the nucleus. Δcc2 was distributed throughout the cytosol with weak staining in a few CAV1-containing spots. We noticed that the number of CAV1-positive puncta decreased in cells expressing Δcc1 or Δcc2, and the total plasma membrane signal of CAV1 was reduced by 40% and 25%, respectively (Fig. 3G), consistent with the established role of Cavin1 as a stabilizer of caveolae.
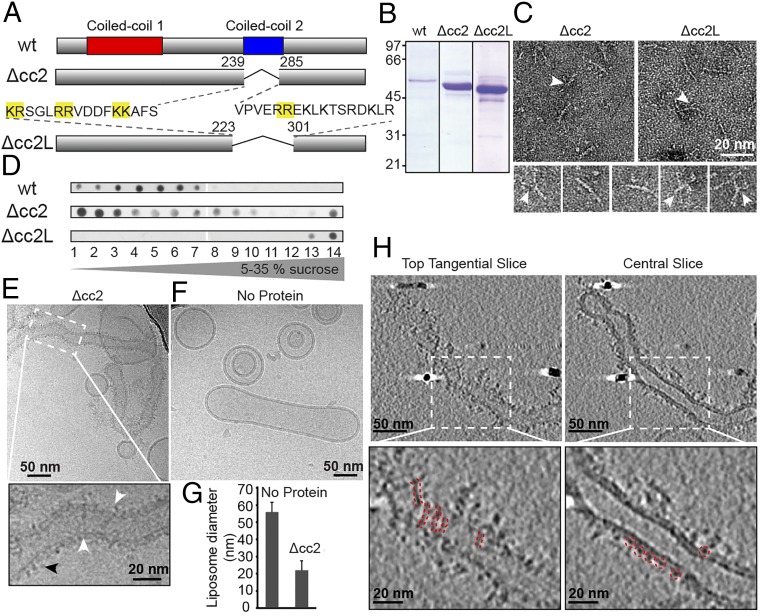
Cavin1 Δcc2 purified from HEK293 cells (Fig. 4 A and B) was analyzed by EM after negative staining (Fig. 4C). The Δcc2 complexes were detected as filamentous particles of similar overall size. The common features included the presence of rather uniform filaments about 3 nm wide and 17 ± 2 nm long (n = 30). The filaments often had slight curvature, and many showed one or more thinner branches (Fig. 4C, arrowheads).
Fig. 4.
Cavin1–liposome interaction requires a stretch of basic residues and induces membrane tubulation. (A) Schematic of Cavin1-Δcc2 and -Δcc2L. Pairs of basic residues are highlighted in yellow. (B) Coomassie-stained SDS/PAGE gel of purified WT and mutant proteins. (C, Upper) Negative-stained transmission electron microscopy (TEM) images of mutants show filaments with an average length of 17 nm. (Lower) Individual Cavin1-Δcc2 particles. White arrowheads indicate the presence of thinner branches extending from the main filament. (D) Cavin1-WT, -Δcc2, or -Δcc2L were incubated with 25% PS-liposomes and were floated in 5–35% sucrose density gradients. Gradient fractions were analyzed by dot blot using an anti-HA antibody. (E) CryoEM 2D projection image of Cavin1-Δcc2 proteoliposomes. The enlargement of the area indicated by the dotted white box shows filaments across the liposome surface (white arrowheads) and protein domains extending outside the liposome membrane (black arrowhead). Cavin1-Δcc2 proteoliposomes appear elongated and tubulated. (F) Control liposomes without protein. (G) Diameter of Cavin1-Δcc2–bound liposomes and liposomes in the absence of protein. Data are shown as the mean liposome diameter ± SD, n = 10. (H) CryoET performed on Cavin1-Δcc2 proteoliposomes. (Upper) Tangential and central slices (thickness 20 Å); (Lower) Enlarged views of the boxed areas. Cavin1-Δcc2 is tightly associated with liposomes, forming a 3- to 4-nm coat (central slice). The presence of filaments (red dashed lines) is visible in tangential slices. Images were recorded at 120 keV (C) and 300 keV (E, F, and H) with a final calibrated pixel size of 9 Å (C) or 5 Å (E, F, and H) at defoci of −0.75 to −2 µm (C) and −3.5 to −6 µm (E, F, and H).
We could not determine the mass of the complexes because concentrating Δcc2 caused aggregation. However, the dimensions of the filamentous part of the complexes were consistent with those of the crystal structure of the trimeric coiled-coil region of cc1 (21), reported to be 16 nm long and 2.5 nm wide. Therefore the flexible branches likely represent the N-terminal sequences (amino acids 1–50) and the C-terminal domains devoid of the cc2, both predicted to be unfolded. The results demonstrated that both coiled-coil domains were required for 60S complex formation. However, their respective roles were different. The results were consistent with the observations of Kovtun et al. (21), who found that cc1 is responsible for the trimerization of Cavin1 fragments in vitro and cc2 for the formation of higher-order oligomers.
Δcc2 Binds to and Tubulates Liposomes.
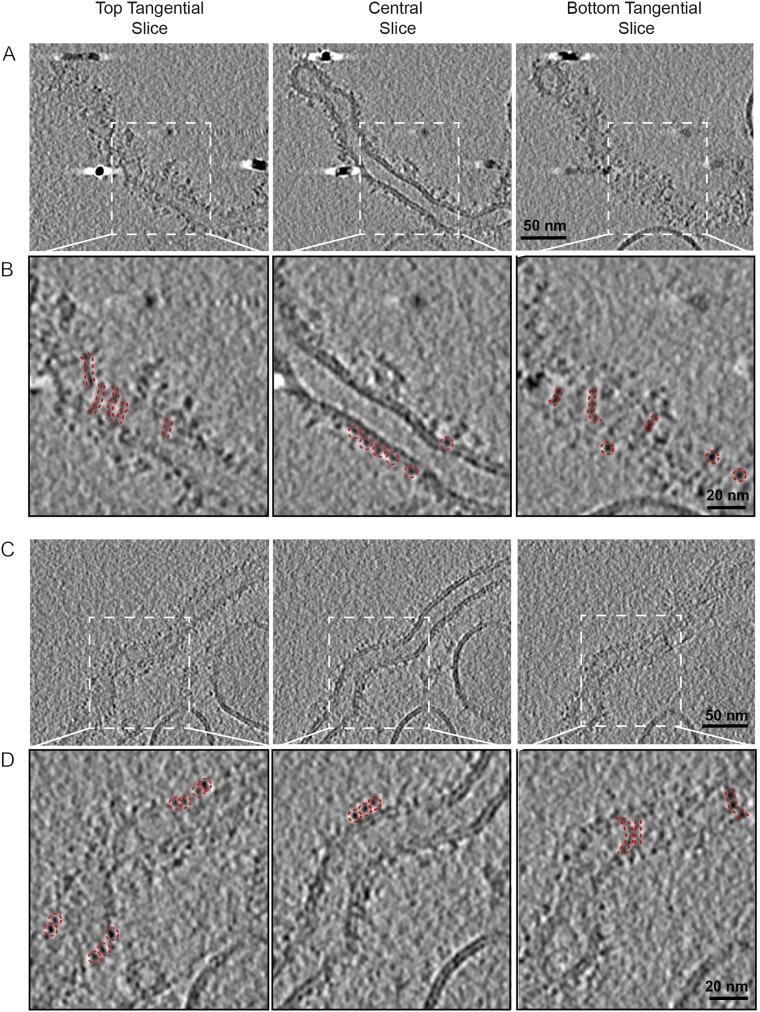
The purified Δcc2 mutant protein bound efficiently to PS-containing liposomes. Upon density ultracentrifugation, most of the cc2 mutant protein floated with the liposomes to the top of the gradient (Fig. 4D). We imaged the Δcc2–lipid complexes by cryoEM and cryoET and observed protein-coated liposomes of variable shapes and sizes (Fig. 4 E–H and Fig. S4). Liposome sizes were much larger than observed for wild-type Cavin1. They typically were elongated, tubular, and narrower than control liposomes that only occasionally had a tubular shape (Fig. 4F), suggesting that Δcc2 had membrane-remodeling activity (Fig. 4 E–H).
Fig. S4.
Structural characterization by cryoET of Cavin1-Δcc2 liposome binding. Liposomes were incubated with Cavin1-Δcc2 for 60 min and were imaged by cryoET. Panels A and B and panels C and D show two representative events of Cavin1-Δcc2 liposome tubulation. The top, central, and bottom sections of each tomogram are shown. Zoomed views of the boxed areas in the upper rows are displayed in the lower rows. The presence of filaments is seen on the top and bottom tangential views (red dashed lines). The central sections show the cavin coat tightly associated with the membrane (red dashed circle), with individual average densities of 3–4 nm. Liposome composition was 25% DOPS, 15% DOPC, 15% DOPE, and 45% cholesterol. Images were recorded at 300 keV with final calibrated pixel size of 5 Å at defoci of −3.5 to −5 µm.
In 2D projection images (Fig. 4E) and tomogram slices (Fig. 4H), we observed a tight association of Δcc2 with the membrane forming a 3- to 4-nm thick, uneven protein coat. In some cases, protein extended up to 6 nm from the membrane. Because the cc1 trimeric coiled-coil is 16 nm long, it was likely that Δcc2 filaments were closely associated and arranged parallel to the membrane (Fig. S4).
Δcc2 mutant protein binding to PS liposomes was unexpected because the deleted cc2 domain contained a sequence with highly basic and positively charged amino acids thought to mediate electrostatic interaction with PS (21, 27). However, we noticed an abundance of double arginine-lysine clusters on either side of the cc2 sequence. We therefore expanded the deletion by an additional 16 and 17 amino acids toward the N and C termini, respectively (Fig. 4A). We expressed and purified this mutant protein (in which amino acids 224–300 were deleted) and termed it “Δcc2L.” Although negative staining showed Δcc2L to be similar in shape to the Δcc2 mutant (Fig. 4C), Δcc2L failed to associate with liposomes and stayed at the bottom of the flotation gradient (Fig. 4D), indicating that the positively charged sequences surrounding the borders of the predicted cc2 are required for the binding of Cavin1 complexes to PS-containing liposomes.
CAV1 8S Oligomers Are Round Protein Discs.
How would oligomers of the integral membrane protein caveolin form a single caveola together with the filamentous, net-like cavin coat? To answer this question, we isolated and analyzed CAV1 oligomers. Two major oligomeric species of CAV1 can be detected after cell solubilization: (i) an intermediate-size 8S complex consisting of 7–14 CAV1 monomers and (ii) a large 70S complex composed of 8S CAV1 subunits, which corresponds to the mature caveolar scaffold (Fig. 5A) (6). In addition to CAV1 8S building blocks, it is likely that the 70S complex contains lipids and/or detergent molecules because cholesterol and the choice of detergent are critical for preserving the large complex (6). We could obtain total cellular CAV1 as 8S complexes when lysis was performed with 1.75% octyl glucoside instead of 0.5% Triton X-100 (Fig. 5B). To purify the 8S CAV1 building block, we created a Tet-inducible HEK293-FlpIn cell line expressing full-length CAV1 with an N-terminal Strep-Tag. Epitope-tagged CAV1 copurified together with endogenous CAV1, indicating its incorporation into CAV1 oligomers (Fig. 5C). We subjected purified CAV1 to gradient centrifugation and found that all CAV1 sedimented as a sharp 8S peak, as expected (Fig. 5C). Aliquots of fractions 3 and 4 were negatively stained and analyzed by EM. CAV1 oligomers were observed as homogenous, well-separated particles (Fig. 5 D and E). Most particles displayed a circular shape; some particles showed a more elongated appearance (Fig. 5E), potentially presenting tilted or side views of the complex. The diameter of the circular CAV1 particles agreed with the maximum length of the more elongated particles, the latter having a minimal thickness of ∼4–5 nm. Hence, the two distinct views were consistent with being top and side views of flat discs. We next performed unbiased reference-free 2D class averaging of the CAV1 particles. Calculations were performed using 4, 8, or 12 as the parameter for the output number of classes; best results were obtained for eight classes as judged by small number of class outliers, i.e., <3% of particles per class. Averaged classes (Fig. 5E, Bottom Row) were similar in shape overall and showed circular particles with a diameter between 11.7 nm (class 5) and 17 nm (class 1). There were fewer side views than disc-shaped top views of the complexes, and the side views did not appear as a separate and well-defined 2D class (classes 6 and 7 are representative), likely because of the variation in the disc angle to the carbon support.
Fig. 5.
CAV1 8S oligomers are round protein discs. (A) Schematic of the CAV1 70S and 8S complex. (B) Cellular extracts were prepared using 0.5% TX100 or 1.75% octyl-glucoside (OG) and were run through 10–40% sucrose velocity gradients. CAV1 is present in 8S and 70S complexes in TX100 extracts. All CAV1 sediments as 8S complexes in OG extracts. (C) Purification of Strep-HA-CAV1 expressed in HEK293 cells. (Right) SH-CAV1 is coeluted and forms complexes with endogenous CAV1 (eluted fractions 1–5). (Left) Fractions of 10–40% sucrose velocity gradient of purified CAV1, analyzed by SDS/PAGE/Western blot. All CAV1 is present in the 8S oligomer. (D and E) Negative-stain TEM of purified CAV1. (D) Micrograph of CAV1 particles in negative stain. Side views are indicated by white arrows. (E, Upper) Gallery of particles from TEM images. (Lower) 2D class averages from 450 particles, numbered according to the abundance of particles in each class (given below). The majority of CAV1 8S complexes display a circular shape ∼15 nm in diameter. (Scale bar in E: 15 nm.)
Considering the 2D averages and the projection images together, we could conclude that CAV1 8S oligomers resembled protein discs with a width of 15–17 nm and a thickness of 5 nm. Our results were strikingly consistent with recent findings regarding the structure of oligomers formed by Caveolin-3, the muscle-specific caveolin family member (31).
CryoET of Native Caveolae Reveals Polygonal Membrane Profiles.
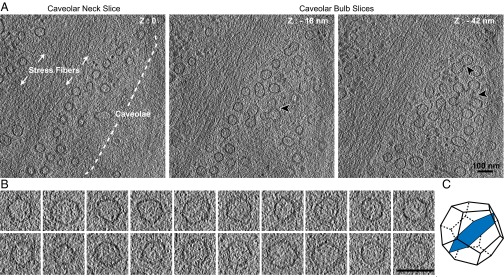
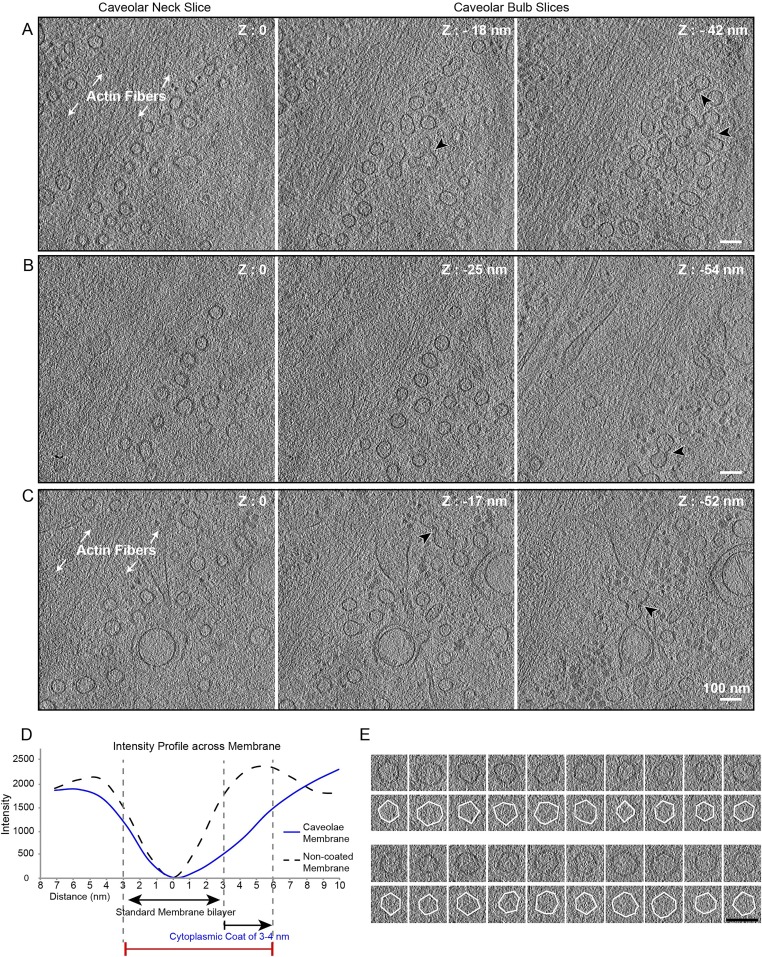
To characterize caveolae in a native, hydrated state, we performed cryoET on vitrified mammalian human fibroblasts. Tomograms were recorded in areas densely packed with caveolae often found in proximity to stress fibers (Fig. 6A, Fig. S5 A–C and Movie S3). Caveolae at the plasma membrane were identified according to their size, morphology, and structural similarity to proximal characteristic clusters of caveolae protruding into the cytoplasm (30, 32). The diameter of individual caveolae (61 nm ± 5.6 nm, n = 41) was consistent with published measurements (30). A thin, closely appressed membrane coat, rather than a prominent extending one, was found on the cytosolic face of the caveolar bulb (Fig. S5D). In tomogram slices oriented through central parts of the caveolar bulb we detected membranes with striking polygonal profiles (Fig. 6 B and C). The angular membranes had five or six planar surfaces with an edge length of 30–40 nm (Fig. S5E) and an internal angle of 118.1 ± 9.9° (n = 108). Clear polygonal membrane profiles were detected in tomogram slices through all caveolar bulbs. Thus, the cellular caveolar coat had the capacity to deform the underlying membrane into a polyhedral structure.
Fig. 6.
In situ cryoET of caveolae in human foreskin fibroblasts reveals polygonal membrane profiles. (A) Caveolae in a nonfixed, nonstained human foreskin fibroblast cells imaged by cryoET. Tomogram slices of 14-Å thickness at different heights in the same cellular area are shown. Z-height is defined relative to zero being the slice in which caveolar necks appear at the plasma membrane. Actin stress fibers and clusters of caveolae (black arrowheads) are highlighted. (Scale bar: 100 nm.) For the full tomogram see Movie S3. (B) Gallery of central tomogram slices (bulb region) of individual caveolae that show polygonal, often hexagonal profiles. (Scale bar: 100 nm.) Tilt series were recorded at 300 keV at a calibrated pixel size of 7.1 Å and a defocus of −4 µm. (C) Schematic of a polyhedron, here a dodecahedron, displaying a 2D slice (blue) through the 3D volume that resembles the angularity observed in caveolae.
Fig. S5.
Caveolae in nonfixed, nonstained human foreskin fibroblasts imaged by cryoET. (A–C) Slices through three different tomograms that show caveolae located near actin stress fibers. Clusters of caveolae (black arrowhead) are present in the lower slice of the tomograms. Z height is set as zero for the first slice in which caveolae appear at the plasma membrane of the cell. (Scale bars: 100 nm.) (D) A thin, closely appressed coat is detected on the cytosolic face of caveolae. Intensity profiles across caveolar membrane and noncaveolar membrane (vesicles present in the same tomogram). Line scans were measured on filtered tomograms with ImageJ using a 10-nm line width. Distances were set to zero at minimal intensity (darker) to allow comparison of the caveolae membrane and noncaveolar membrane intensity profiles. (E, Upper Rows) Gallery of central tomogram slices of individual caveolae (as shown in Fig. 6B). (Lower Rows) White outlines highlight polyhedral angularity. (Scale bar: 100 nm.)
Discussion
In this study we isolated and analyzed cavin and caveolin complexes that together form the caveolar coat and visualized native caveolae in vitrified cells by EM. A regular polyhedron emerged as the most likely architectural principle of the caveolar coat structure.
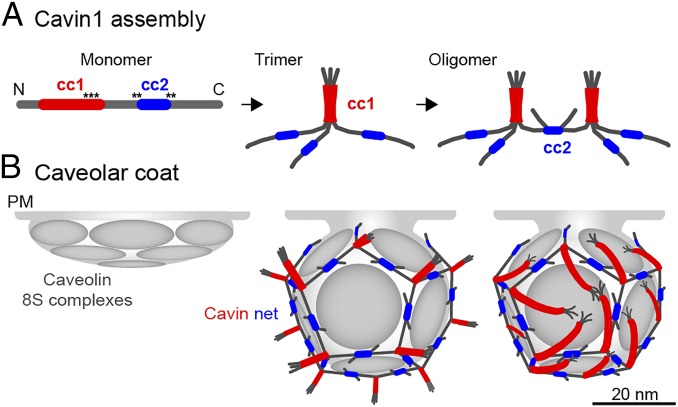
Our EM and SAXS analyses of Cavin1 60S complexes isolated from mammalian cells indicated that the cavin coats are homogenous in size, flexible, and net-like. Characterization of Cavin1 mutants showed that the 60S assembly is based on nonstructured polypeptide filaments joined by coiled-coil segments. The basic structural unit is a cavin trimer in which three subunits are joined by a parallel triple coiled-coil between the cc1 sequences (10, 21). The cc2 domains, on the other hand, are responsible for connecting trimers with each other, perhaps by forming dimeric coiled-coils (Fig. 7A). With the estimate of about 50–60 cavins per cavin complex (6, 17), equivalent to ∼18–20 trimers, the overall organization of the 60S cavin coat corresponds to a polyhedral “nano-net” peripherally attached to the cytoplasmic surface of the caveolar invagination. The function of the net is not only to stabilize the microdomain by trapping caveolin complexes and other proteins in the meshes but also to help define and maintain the size, curvature, and hence the shape of the caveolar indentation or caveolar vesicle (9, 19). Of note, regularity in the cavin coat also has been observed by Ludwig and colleagues who determined the periodic spacings between Cavin molecules (10) that match the polygon sizes of the 60S complexes determined in this study.
Fig. 7.
A model for Cavin1 complex assembly and caveolar coat architecture. In our speculative model, Cavin1 monomers form trimers and then assemble into larger, interconnected oligomers (A) to form a dodecahedron around the caveolar membrane (B). Stoichiometry and estimated particle size of the Cavin1 60S complex suggest that caveolae contain one such complex each. Trimers formed by cc1 interactions are localized to the 20 vertices and are connected through dimeric cc2 interactions along the edges. The cc1 domains (red) form a parallel trimeric coiled-coil generating a 17-nm-long, relatively stiff rod-like structure with a PIP2 binding region about 2–3 nm from the C-terminal end of the coil (black asterisks). There is limited information for cc2 (blue), and we present dimeric, antiparallel interactions in our model. Contacts with the membrane are mediated by PS-binding through residues that flank and possibly include cc2 and by PIP2 binding to cc1 (black asterisks). In addition, there possibly are weak interactions with CAV1. The trimers are likely positioned parallel to the surface of the caveolar membrane and could generate the striations seen on the surface of caveolae in previous EM studies. The model leaves the sequences N-terminal to cc1 (amino acids 1–57) and C-terminal to cc2 (amino acids 285–392) free; they are external to the coat. Because they contain many potential phosphorylation sites, they likely are involved in the interaction with additional proteins. Cavin2 and Cavin3 (not depicted) may induce breaks in the polyhedron because they do not contain cc2 and thereby could allow the formation of the neck of caveolae. The 12 faces of the dodecahedron are likely to be occupied by disc-shaped caveolin oligomers that float in the underlying membrane and thus can interact with each other, lipids, and the appressed cavin coat. It is likely that the caveolin domain requires the cavin coat to induce and maintain the curvature and hence the invaginated shape of caveolae.
Our reconstitution experiments with isolated, preformed Cavin1 60S complexes and liposomes indicated that in an extended, membrane-associated conformation, Cavin1 can define the diameter of a vesicle to a range characteristic of caveolae. In the reconstituted vesicles, we observed a polygonal lattice structure where Cavin1 was closely associated with the membrane. In our experiments attachment occurred through interactions with PS, although in situ interactions with phosphatidylinositides and contacts with caveolin domains are also likely to contribute to the specific association with caveolae (9, 13, 21). Given the gradual association of Cavin1 with nascent caveolin spots in the plasma membrane of cells over a period of 25 min (6), it is likely that Cavin1 initially binds as smaller building blocks that subsequently establish higher-order connections. In the reconstituted vesicles, however, the cavin complex was observed on the inside of the liposomes. Such a net-like coat can be formed inside liposomes as well as on the outside, and it should not change the imposed structure. In fact, a collapsed coat resulting from the detergent extraction-based purification protocol might be disentangled more easily if it is surrounded by lipid interactions than if it has only a single interaction with a liposome. Such a scenario of surrounding lipid interactions was favored by the observed tendency of PS-rich liposomes for spontaneous invaginations.
Analysis of Cavin1 mutants in which coiled-coil domains were individually removed showed that both domains cc1 and cc2 are essential for 60S complex formation and for Cavin1 association with caveolae. In the absence of cc1, no defined Cavin1 complexes were observed. Removal of cc2 resulted in heterogeneously sedimenting complexes most likely corresponding to trimers (21). Cavin1 mutants devoid of cc2 still associated with PS-containing liposomes. Forming a protein layer on the liposome surface (3–4 nm thick), they caused membrane tubulation. Removal of additional positively charged sequences abolished PS–liposome binding, indicating that sequences flanking cc2 are required for Cavin1 binding to PS-containing vesicles.
A single, uniform structure for the cavin complexes is unlikely, given the flexibility of the cavin molecules, the presence of different cavin species, and caveolar deformability and plasticity (2, 33). However, an underlying polygonal architecture is suggested by the profiles of caveolae in cells and reconstituted Cavin1 cages here revealed. Polyhedra provide the underlying principle in many biological structures in which spherical curvature is imposed by proteins. Examples range from viruses to clathrin, COPI, and COPII vesicles. It is not clear from our images exactly what type of polyhedra the Cavin1 complexes represent. However, the earlier reported stoichiometry of the involved proteins would fit best with a dodecahedron. With its 20 vertices, a regular dodecahedron would contain 20 cavin trimers and thus a total of 60 cavin molecules, precisely matching the estimated number of cavin molecules per 60S complex (6, 17).
Our analysis of full-length CAV1 8S complexes showed that the caveolin oligomer exhibits a homogenous, disc-like shape with a diameter of 15–17 nm and a thickness of 4–5 nm. This shape and size is strikingly similar to the structure of Caveolin-3 complexes presented by Whiteley and colleagues (31). Caveolin 8S complexes contain an estimated 7–14 caveolin molecules and assemble into higher-order 70S caveolar scaffolds by lateral interaction with each other and cholesterol. The dimensions of the 8S caveolin complexes suggest that they may occupy the empty faces of a dodecahedron formed by cavins. The fivefold flat faces in a dodecahedron could provide space for a total of up to 12 8S caveolin complexes and thus about 120–144 CAV molecules, again agreeing well with the previously estimated number of caveolin molecules per caveola (8). The interaction between the Cavin and caveolin complexes would not necessarily be direct but might be mediated by lipids.
When native caveolae were imaged in plunge-frozen cells (i.e., omitting any artifacts potentially caused by staining or sectioning), the bulbous part of the caveolae exhibited a coat that appeared tightly appressed to the membrane. In the bulb region most of the in situ caveolae exhibited polygonal membrane profiles with five or six edges ∼30–40 nm in length and with a dihedral angle of 118° suggesting a 3D polygonal shape. Analysis of the reconstituted Cavin1 vesicles indicated that the Cavin1 coat alone can cause such polygonal shapes. In this context it is interesting that, when expressed in bacteria in the absence of cavins, caveolin generates vesicles of similar size and shape (34, 35). Thus, the polygonal shape of caveolae is probably favored, generated, and maintained by both caveolins and cavins.
Our hypothetical, idealized model of the Cavin1 complex (Fig. 7) shows a regular dodecahedron. We place the cc1 coiled-coils of cavin trimers (Fig. 7A) at the threefold vertices (Fig. 7B). The unstructured middle sequences, the cc2 coiled-coils, and the unstructured C-terminal segments generate thin, flexible filaments that connect the vertices in a net-like web (Fig. 7A). According to our sequence predictions, Cavin2 and Cavin3, which make up about one-fourth of the cavins in caveolae (10), lack the cc2 domain. Thus, although Cavin2 and Cavin3 are part of cavin trimers (10, 17), they may resemble the Cavin1 Δcc2 mutant in being unable to connect neighboring cavin trimers with each other. Inclusion of Cavin2 and Cavin3 therefore will create breaks and openings in the polygonal net. Hence they could be involved in the formation of the neck in caveolae residing at the plasma membrane. This notion is consistent with earlier reports linking the function of Cavin2 to the formation of the caveolar neck suggested by the observation that caveolae devoid of Cavin2 are shallow and have a half-dome rather than a flask shape (12, 19).
It has been proposed that the striations seen on the surface of caveolae by EM after deep etching and platinum coating (28) are composed of cavin trimers (10, 17, 21). The stripes could contain the trimeric cc1 coiled-coils that form rigid rods 16 nm long and 2.5 nm wide. We observed filaments with these dimensions both after negative staining and cryoET of the Δcc2 mutant Cavin1. If positioned parallel to the surface of the caveolar membrane, the filaments could indeed generate striation patterns (Fig. 7B, Right). The location of residues in the trimeric cc1 coiled-coil responsible for binding to phosphatidylinositide head groups (21) suggests that, to reach the phosphatidylinositol bisphosphate (PIP2) head groups, the rigid cc1 coiled-coils need to be tilted parallel to the membrane. Interestingly, Richter and colleagues detected a spiked coat instead of membrane-parallel striations extending from the caveolar surface into the cytosol (30). Based on our model, Cavin1 trimers can form such cytoplasmic spikes when pointing away from the membrane (Fig. 7B, Middle). The occurrence of spikes vs. striations may be regulated by the phospholipid content in the caveolar domain.
In summary, our results support a model for caveolar architecture that is based on a constraining web composed of thin, flexible, unstructured cavin filaments. These filaments are connected by coiled-coils into a polygonal, net-like complex that provides stability to this functionally specialized membrane domain. Caveolin protein discs may occupy the faces of the dodecahedron, thus allowing lateral interaction, predominantly mediated by the lipid environment, between caveolin oligomers embedded in the membrane bilayer and the peripherally attached cavin coat.
Materials and Methods
Expression Constructs and Mammalian Cell Lines.
Full-length human Cavin1 and CAV1 sequences were obtained from the ORFeome collection (V3.1 Open Biosystems) and were cloned into the Gateway-compatible destination vector pcDNA5/FRT/TO/SH/GW (36). Cavin1 or CAV1 vectors for tetracycline-controlled expression of proteins N-terminally tagged with a streptavidin-binding peptide and hemagglutinin epitope tag (SH) were cotransfected with pOG44 Flp-Recombinase Expression vector into Flp-In T-REx HEK293 cells (Invitrogen) to integrate SH-Cavin1 or SH-CAV1 stably into the genome via FRT sites. Transfected cells were selected with 100 μg/mL hygromycin, and expression of the target protein was assessed by immunoblotting and immunofluorescent imaging. SH-Cavin1/CAV1 HEK293 cells were grown in DMEM supplemented with 10% (vol/vol) FBS and 1% GlutaMAX (Invitrogen) containing 15 mg/mL blasticidin and 100 μg/mL hygromycin. SH-Cavin1/-CAV1 expression was induced by adding 1 μg/mL tetracycline 48 h before lysis. The coiled-coil deletion mutants Cavin1-Δcc1 (amino acids 59–92 deleted), Cavin1-Δcc2 (amino acids 240–284 deleted), and Cavin1-Δcc2L (amino acids 224–300 deleted) were generated by QuikChange deletion mutagenesis based on the SH-Cavin1 expression vector. For transient expression, HEK293 cells (ATCC CRL-1573) were transfected with SH-Cavin1 constructs using Lipofectamine 2000 (Invitrogen), and cells were lysed 48 h after transfection. The N-terminally Flag-tagged Cavin1 construct in pRc/CMV (from Ingrid Grummt, Division of Molecular Biology of the Cell II, German Cancer Research Center, Heidelberg, Germany) contained full-length mouse Cavin1; mutants were generated by QuikChange deletion mutagenesis. For transient expression, HeLa cells (ATCC) were transfected by electroporation (AMAXA), and cells were analyzed after 24 h. HeLa and HEK293 cells were grown in DMEM supplemented with 10% (vol/vol) FBS and 1% GlutaMAX (Invitrogen).
Protein Purification and Velocity Gradient Centrifugation.
Cavin1-expressing HEK293 cells were lysed in 1% TX100 in TNE buffer [20 mM Tris (pH 7.4), 250 mM NaCl, and 5 mM EDTA supplemented with Complete protease inhibitors (Roche)] for 30 min on ice and were passed five times through a 25-gauge needle. Insoluble material was removed by a 5-min spin at 1,100 × g to obtain postnuclear supernatants followed by a 15-min spin at 14,000 × g. Cleared lysate was loaded consecutively on 5-mL gravity-flow columns (Bio-Rad) containing 100 μL Strep-Tactin Sepharose beads (IBA) per 5 mL lysate. Beads were washed five times with 10 bed volumes of TNE-TX100 buffer followed by five washes in TNE without detergent. Proteins were eluted from Strep-Tactin beads with five bed volumes of 2.5 mM desthiobiotin in TNE. Samples were analyzed immediately or were stored at 4 °C for not longer than 1 wk. CAV1 purification was performed as above but with all buffers containing 1.78% (wt/vol) octyl glucoside instead of TX100. Sucrose velocity gradient centrifugation was performed as described previously (6). Eluate or lysate samples were loaded onto 10–40% (wt/vol) linear sucrose gradients prepared in TNE buffer and were spun in a rotor (SW41Ti or SW55Ti; Beckman Coulter) at 151,263 × g for 737 min (SW41) or at 237,020 × g for 255 min (SW55) at 4 °C. Gradients were fractionated top to bottom, and fractions were analyzed by SDS/PAGE/Western blotting, Coomassie staining, or EM.
Liposome Preparation, Binding of Cavin1 to Liposomes, and Flotation.
DOPC, DOPE, phosphatidylserine (DOPS), and cholesterol (Avanti Polar Lipids) at concentrations of 10–20 mg/mL in chloroform were dried under a stream of argon. Lipids were solubilized in TNE buffer at 1.25 mM and vortexed to emulsify the lipid mixture. Repetitive freeze-thaw cycles were performed (liquid nitrogen followed by 3 min at 37 °C, repeated 10 times), and liposomes subsequently were passed through 80-nm cutoff membranes using an Avanti miniextruder. Liposome concentrations with the defined mol% of the respective lipid species were prepared as indicated for each experiment.
For binding assays, 25–75 μg purified Cavin1 was incubated with 200 μg liposomes (protein: lipid ratio between 1:10–1:2) in a final volume of 250 μL TNE buffer for 30 min at room temperature. Samples either were used directly for EM analysis or proteoliposomes were floated in sucrose density gradients. For the latter, the incubated mixture was adjusted to 40% (wt/vol) sucrose with 70% (wt/vol) sucrose in TNE, and a 500-µL sample was layered under a 5–35% (wt/vol) linear sucrose gradient in TNE. Centrifugation was performed in a SW55Ti rotor for 16 h at 151,693 × g at 4 °C. Gradients were fractionated top to bottom. When indicated, Nycodenz was used as the density medium instead of sucrose, and 30–60% (vol/vol) linear Nycodenz gradients were prepared. Gradients were fractionated top to bottom, and fractions were analyzed by SDS/PAGE/Western blotting, EM, or dot blot. For the preparation of samples of floated liposomes for cryoEM, fractions 4 and 5 were mixed together, and samples on Quantifoil R2/1 holey carbon-coated EM grids were plunge-frozen after washing of excess sucrose or Nycodenz twice on 500-µL droplets of buffer.
Antibodies.
Rabbit polyclonal antibody (pAb) anti-Cavin1 was from Abcam (ab48824); mAb anti-HA was from Covance (MMS-101P); mAb anti-Flag (M2) was from Sigma; and rabbit pAb anti-CAV1 was from Santa Cruz (N20, sc-894). Alexa Fluor-conjugated secondary antibodies for immunofluorescence were from Molecular Probes, Invitrogen.
Fluorescence Imaging and Colocalization Analysis.
Cells grown on coverslips were fixed using 4% (vol/vol) formaldehyde in PBS. Cells were permeabilized with 0.05% saponin in PBS with 1% BSA and were incubated with primary (1:1,000) and secondary (1:1,000) antibodies. Coverslips were mounted on slides using Immu-Mount (Thermo Scientific). Imaging was performed on an inverted scanning confocal microscope system (LSM 510 Meta; Carl Zeiss, Inc.). Colocalization of CAV1 spots with full-length or mutant Cavin1 or mutants was quantified using ImageJ. A threshold was applied to the CAV1 channel. Pixel coordinates of caveolar spots were extracted with the implemented Analyze particles function. The Cavin1 intensity at these pixels was divided by the total cellular Cavin1 intensity to present Cavin1 enrichment at caveolae.
Mass Spectrometry Analysis.
Mass spectrometry analysis was performed on floated fractions 4 and 5 after incubation of Cavin1 60S complexes with liposomes and ultracentrifugation. MS/MS spectra were searched against the UniProt database in Mascot (37), with 1% false discovery rates (ions score cutoff superior to 20). As expected, the main constituent was Cavin1, with peptide coverage of 38%. Most importantly Cavin2, Cavin3, and caveolin peptides were not identified. Other protein hits were ribosomal proteins, heat-shock protein, and keratin contaminations.
Negative Staining and 2D Averaging.
Δcc2 and Δcc2L particles and CAV1 8S complexes were negatively stained with 0.75% Uranyl formate, pH 5. Images were recorded on an FEI F12 microscope at 120 kV using an FEI Eagle Camera at a calibrated pixel size of 0.2 nm and on a JEOL1400-Plus transmission electron microscope using a Gatan OneView camera at a calibrated pixel size of 0.22 nm and defoci ranging from −0.75 to −1.5 µm. Images from the JEOL1400-Plus microscope were used for 2D averaging of CAV1 complexes. Four hundred fifty articles were picked using Boxer [part of EMAN (38)] and were classified in Relion (39) within 25 iterations into eight classes.
CryoEM Grid Preparation of Cavin Samples.
Protein complexes were applied to copper EM grids with holey carbon support films (Protochips C2/1) after a glow-discharge of 30 s. Vitrification was performed using a liquid nitrogen-cooled ethane/propane mixture and single-side manual blotting (40).
CryoEM and CryoET Data Collection.
CryoEM imaging was performed on vitrified specimens at liquid nitrogen temperature under low-dose conditions on either a FEI F30 microscope at 200 keV using a Gatan UltraScan 4000 CCD camera or a FEI F30 Polara microscope at 300 keV equipped with a Gatan 964 Quantum energy filter operated with a 20-eV-wide energy-selecting slit centered on the zero-loss peak and a K2 summit direct detector at calibrated pixel sizes, ranging from 0.46 nm to 0.71 nm (see the legends of Figs. 1, 2, 4, and 6). For cryoEM projection images the total dose was kept below 15 electrons/Å2. Tilt series were recorded with 2° to 3° increments covering an angular range from −48/50° to +48/50° using SerialEM (41). The total electron doses for tilt series were kept between 60 and 80 electrons/Å2. K2 Summit images were acquired in counting mode at approximately 10 electrons per pixel per second at 0.4 frames per second in dose-fractionation mode. On-the-fly frame alignment was performed using the SerialEM combined filter.
Cell Preparation and Cellular Tomography.
Human foreskin fibroblasts were grown directly on the holey carbon film of gold grids in DMEM and 10% (vol/vol) FCS for 24 h and then were vitrified using a liquid nitrogen-cooled ethane/propane mixture and single-side manual blotting as previously described (40). CryoET of whole vitrified cells was performed as described above, at 300 keV in low-dose mode using a F30 Polara electron microscope (FEI). Images were recorded at nominal magnifications of 61,000× and calibrated pixel sizes of 0.71 at the specimen level. Tomographic tilt series were collected at defoci of −3.5 µm to −5 µm. The total electron doses for the tilt series were kept between 80–90 electrons/Å2.
Image Processing and Visualization.
Tomograms were reconstructed and visualized as reported previously (42). To improve visualization, tomograms and 2D projection images were binned 2× or 4× for direct detection device (DDD) images acquired at high magnification and were low-pass filtered to remove high-frequency noise. Dimensions measurements were performed in Amira (FEI) and ImageJ (43).
SAXS Experiments.
SAXS measurements of purified Cavin1 homo-oligomeric complexes were performed at the BM29 beamline in the European Synchrotron Radiation Facility (ESRF), Grenoble, France. Data were collected at 20 °C, a wavelength of 0.0995 nm, and a sample-to-detector distance of 1 m. 1D scattering profiles were generated, and blank subtraction was performed by the data-processing pipeline available at BM29 at the ESRF. Guinier analysis and pair-distance distribution functions were calculated using the programs Primus and GNOM (44). Molecular weights were estimated based on ref. 45.
Supplementary Material
Acknowledgments
We thank Daven Vasishtan for help with image processing, helpful insights, and critical discussion; Arnold Hayer for experimental ideas and helpful advice; Cathy Whittle for assistance with tissue culture; and B. Kessler and R. Konietzny for expert advice on mass spectrometry analysis. This work was supported by Wellcome Trust Senior Research Fellowships 090895/Z/09/Z and 107806/Z/15/Z (to K.G.); Wellcome Trust Equipment Grant 093305/Z/10/Z to the Oxford Particle Imaging Centre; and Wellcome Trust Core Award 090532/Z/09/Z to the Wellcome Trust Centre for Human Genetics. M.S. is the recipient of European Molecular Biology Organisation (EMBO) Short-Term Fellowship 36-2013, and P.S. is the recipient of EMBO Long-Term Fellowship ALTF 642-2011.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 14170.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616838113/-/DCSupplemental.
References
- 1.Parton RG, del Pozo MA. Caveolae as plasma membrane sensors, protectors and organizers. Nat Rev Mol Cell Biol. 2013;14(2):98–112. doi: 10.1038/nrm3512. [DOI] [PubMed] [Google Scholar]
- 2.Nassoy P, Lamaze C. Stressing caveolae new role in cell mechanics. Trends Cell Biol. 2012;22(7):381–389. doi: 10.1016/j.tcb.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Fotin A, et al. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432(7017):573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- 4.Stagg SM, et al. Structural basis for cargo regulation of COPII coat assembly. Cell. 2008;134(3):474–484. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodonova SO, et al. VESICULAR TRANSPORT. A structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science. 2015;349(6244):195–198. doi: 10.1126/science.aab1121. [DOI] [PubMed] [Google Scholar]
- 6.Hayer A, Stoeber M, Bissig C, Helenius A. Biogenesis of caveolae: Stepwise assembly of large caveolin and cavin complexes. Traffic. 2010;11(3):361–382. doi: 10.1111/j.1600-0854.2009.01023.x. [DOI] [PubMed] [Google Scholar]
- 7.Tagawa A, et al. Assembly and trafficking of caveolar domains in the cell: Caveolae as stable, cargo-triggered, vesicular transporters. J Cell Biol. 2005;170(5):769–779. doi: 10.1083/jcb.200506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelkmans L, Zerial M. Kinase-regulated quantal assemblies and kiss-and-run recycling of caveolae. Nature. 2005;436(7047):128–133. doi: 10.1038/nature03866. [DOI] [PubMed] [Google Scholar]
- 9.Hill MM, et al. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 2008;132(1):113–124. doi: 10.1016/j.cell.2007.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig A, et al. Molecular composition and ultrastructure of the caveolar coat complex. PLoS Biol. 2013;11(8):e1001640. doi: 10.1371/journal.pbio.1001640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoeber M, et al. Oligomers of the ATPase EHD2 confine caveolae to the plasma membrane through association with actin. EMBO J. 2012;31(10):2350–2364. doi: 10.1038/emboj.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen CG, Shvets E, Howard G, Riento K, Nichols BJ. Deletion of cavin genes reveals tissue-specific mechanisms for morphogenesis of endothelial caveolae. Nat Commun. 2013;4:1831–13. doi: 10.1038/ncomms2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bastiani M, et al. MURC/Cavin-4 and cavin family members form tissue-specific caveolar complexes. J Cell Biol. 2009;185(7):1259–1273. doi: 10.1083/jcb.200903053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sargiacomo M, et al. Oligomeric structure of caveolin: Implications for caveolae membrane organization. Proc Natl Acad Sci USA. 1995;92(20):9407–9411. doi: 10.1073/pnas.92.20.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monier S, et al. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol Biol Cell. 1995;6(7):911–927. doi: 10.1091/mbc.6.7.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen CG, Nichols BJ. Exploring the caves: Cavins, caveolins and caveolae. Trends Cell Biol. 2010;20(4):177–186. doi: 10.1016/j.tcb.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Gambin Y, et al. Single-molecule analysis reveals self assembly and nanoscale segregation of two distinct cavin subcomplexes on caveolae. eLife. 2013;3(0):e01434. doi: 10.7554/eLife.01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMahon K-A, et al. SRBC/cavin-3 is a caveolin adapter protein that regulates caveolae function. EMBO J. 2009;28(8):1001–1015. doi: 10.1038/emboj.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen CG, Bright NA, Howard G, Nichols BJ. SDPR induces membrane curvature and functions in the formation of caveolae. Nat Cell Biol. 2009;11(7):807–814. doi: 10.1038/ncb1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohan J, Morén B, Larsson E, Holst MR, Lundmark R. Cavin3 interacts with cavin1 and caveolin1 to increase surface dynamics of caveolae. J Cell Sci. 2015;128(5):979–991. doi: 10.1242/jcs.161463. [DOI] [PubMed] [Google Scholar]
- 21.Kovtun O, et al. Structural insights into the organization of the cavin membrane coat complex. Dev Cell. 2014;31(4):405–419. doi: 10.1016/j.devcel.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Pilch PF. A critical role of cavin (polymerase I and transcript release factor) in caveolae formation and organization. J Biol Chem. 2008;283(7):4314–4322. doi: 10.1074/jbc.M707890200. [DOI] [PubMed] [Google Scholar]
- 23.Ortegren U, et al. Lipids and glycosphingolipids in caveolae and surrounding plasma membrane of primary rat adipocytes. Eur J Biochem. 2004;271(10):2028–2036. doi: 10.1111/j.1432-1033.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 24.Wanaski SP, Ng BK, Glaser M. Caveolin scaffolding region and the membrane binding region of SRC form lateral membrane domains. Biochemistry. 2003;42(1):42–56. doi: 10.1021/bi012097n. [DOI] [PubMed] [Google Scholar]
- 25.Fairn GD, et al. High-resolution mapping reveals topologically distinct cellular pools of phosphatidylserine. J Cell Biol. 2011;194(2):257–275. doi: 10.1083/jcb.201012028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gustincich S, Schneider C. Serum deprivation response gene is induced by serum starvation but not by contact inhibition. Cell Growth Differ. 1993;4(9):753–760. [PubMed] [Google Scholar]
- 27.Burgener R, Wolf M, Ganz T, Baggiolini M. Purification and characterization of a major phosphatidylserine-binding phosphoprotein from human platelets. Biochem J. 1990;269(3):729–734. doi: 10.1042/bj2690729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothberg KG, et al. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68(4):673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- 29.Anderson RG. The caveolae membrane system. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 30.Richter T, et al. High-resolution 3D quantitative analysis of caveolar ultrastructure and caveola-cytoskeleton interactions. Traffic. 2008;9(6):893–909. doi: 10.1111/j.1600-0854.2008.00733.x. [DOI] [PubMed] [Google Scholar]
- 31.Whiteley G, Collins RF, Kitmitto A. Characterization of the molecular architecture of human caveolin-3 and interaction with the skeletal muscle ryanodine receptor. J Biol Chem. 2012;287(48):40302–40316. doi: 10.1074/jbc.M112.377085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lebbink MN, et al. Spiral coating of the endothelial caveolar membranes as revealed by electron tomography and template matching. Traffic. 2010;11(1):138–150. doi: 10.1111/j.1600-0854.2009.01008.x. [DOI] [PubMed] [Google Scholar]
- 33.Echarri A, Del Pozo MA. Caveolae - mechanosensitive membrane invaginations linked to actin filaments. J Cell Sci. 2015;128(15):2747–2758. doi: 10.1242/jcs.153940. [DOI] [PubMed] [Google Scholar]
- 34.Walser PJ, et al. Constitutive formation of caveolae in a bacterium. Cell. 2012;150(4):752–763. doi: 10.1016/j.cell.2012.06.042. [DOI] [PubMed] [Google Scholar]
- 35.Ariotti N, et al. Molecular Characterization of Caveolin-induced Membrane Curvature. J Biol Chem. 2015;290(41):24875–24890. doi: 10.1074/jbc.M115.644336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glatter T, Wepf A, Aebersold R, Gstaiger M. An integrated workflow for charting the human interaction proteome: Insights into the PP2A system. Mol Syst Biol. 2009;5:237. doi: 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackeen MM, et al. Small-molecule-based inhibition of histone demethylation in cells assessed by quantitative mass spectrometry. J Proteome Res. 2010;9(8):4082–4092. doi: 10.1021/pr100269b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schellenberger P, et al. High-precision correlative fluorescence and electron cryo microscopy using two independent alignment markers. Ultramicroscopy. 2014;143(c):41–51. doi: 10.1016/j.ultramic.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Martinez R, et al. The amphipathic helix of adenovirus capsid protein VI contributes to penton release and postentry sorting. J Virol. 2015;89(4):2121–2135. doi: 10.1128/JVI.02257-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25(4):495–503. [Google Scholar]
- 45.Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496(7446):477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.