Significance
In Cambodia, where Plasmodium vivax and Plasmodium falciparum are coendemic and intense multimodal malaria-control interventions have reduced malaria incidence, P. vivax malaria has proven relatively resistant to such measures. We performed comparative genomic analyses of 150 P. vivax and P. falciparum isolates to determine whether different evolutionary strategies might underlie this species-specific resilience. Demographic modeling and tests of selection show that, in contrast to P. falciparum, P. vivax has experienced uninterrupted growth and positive selection at multiple loci encoding transcriptional regulators. In particular, a strong selective sweep involving an AP2 transcription factor suggests that P. vivax may use nuanced transcriptional approaches to population maintenance. Better understanding of P. vivax transcriptional regulation may lead to improved tools to achieve elimination.
Keywords: Plasmodium, malaria, vivax, transcription, genome
Abstract
Cambodia, in which both Plasmodium vivax and Plasmodium falciparum are endemic, has been the focus of numerous malaria-control interventions, resulting in a marked decline in overall malaria incidence. Despite this decline, the number of P. vivax cases has actually increased. To understand better the factors underlying this resilience, we compared the genetic responses of the two species to recent selective pressures. We sequenced and studied the genomes of 70 P. vivax and 80 P. falciparum isolates collected between 2009 and 2013. We found that although P. falciparum has undergone population fracturing, the coendemic P. vivax population has grown undisrupted, resulting in a larger effective population size, no discernable population structure, and frequent multiclonal infections. Signatures of selection suggest recent, species-specific evolutionary differences. Particularly, in contrast to P. falciparum, P. vivax transcription factors, chromatin modifiers, and histone deacetylases have undergone strong directional selection, including a particularly strong selective sweep at an AP2 transcription factor. Together, our findings point to different population-level adaptive mechanisms used by P. vivax and P. falciparum parasites. Although population substructuring in P. falciparum has resulted in clonal outgrowths of resistant parasites, P. vivax may use a nuanced transcriptional regulatory approach to population maintenance, enabling it to preserve a larger, more diverse population better suited to facing selective threats. We conclude that transcriptional control may underlie P. vivax’s resilience to malaria control measures. Novel strategies to target such processes are likely required to eradicate P. vivax and achieve malaria elimination.
During the last decade, western Cambodia has been the focus of numerous and multimodal interventions to control the spread of artemisinin-resistant Plasmodium falciparum (1, 2). Such interventions, including increased vector control, increased surveillance, and improved access to quality artemisinin-combination therapy (ACT), would be expected to curtail coendemic Plasmodium vivax as well. However, even as P. falciparum infections in Cambodia decreased by 81% between 2009 and 2013, P. vivax cases have increased, making it the predominant species in the Mekong region (3–6). This scenario, repeated in Brazil and other areas of coendemicity, has led to growing awareness that P. vivax, although infecting the same populations and transmitted by the same mosquito vectors, will likely be the more challenging species to eradicate (6–9). In this study, we use population genomics to gain insight into the evolutionary factors underlying P. vivax’s resilience to malaria control measures.
Population genetic studies have previously hinted at the resilience of P. vivax populations in comparison with P. falciparum. Studies of microsatellites and highly variable antigens of sympatric P. vivax and P. falciparum populations in Southeast Asia and the Southwest Pacific have consistently shown P. vivax populations to be more diverse, with a higher effective population size (Neff), more stable transmission, and increased gene flow between geographic islands, whereas P. falciparum populations tend to be clonal with episodic transmission and structure-by-geography (10–15). We hypothesized that the species have evolved disparate responses to selective pressures and that genomic studies of sympatric Plasmodium sp. populations would highlight key differences in their population structures, demographic histories, and genomic selective signatures, helping elucidate the basis for these observed differences.
To understand the genome-wide species-specific patterns of selection in sympatric P. vivax (n = 70) and P. falciparum (n = 80) populations in Cambodia, we conducted whole-genome sequencing of coendemic parasites sampled between 2009 and 2013 at a primary site and two satellite sites in western Cambodia (Fig. S1). These sites were designated as “zone 2” during the recent artemisinin-resistance containment campaign and as such were subject to intensified malaria control efforts (16). We initially assessed the relative diversity, within-host complexity, population structure, and demographic histories of the two parasite populations, finding that the P. vivax population remains less structured, more diverse, and more rapidly expanding than the sympatric P. falciparum population. We then evaluated genome-wide signatures of selection in both populations using haplotype-based tests of directional selection, allele frequency-based tests of selection, and copy-number analysis. Differences in genomic loci under directional selection in the two populations highlight different mechanistic responses to selective pressures, suggesting that more nuanced transcriptional control may underlie the resilience of the P. vivax population.
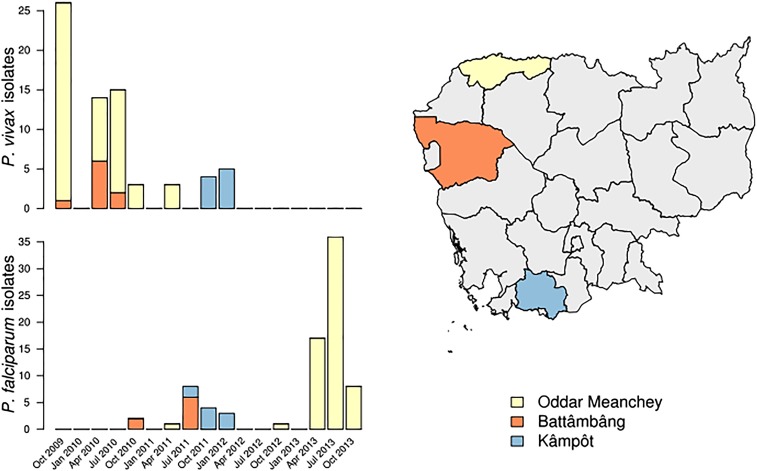
Fig. S1.
Collection dates and locations for samples included in this study. (Left) All P. vivax (Upper) and P. falciparum (Lower) clinical isolates were collected between October 2009 and October 2013, with P. vivax isolates collected, on average, earlier than P. falciparum isolates. (Right) P. vivax and P. falciparum isolates were collected from three unique provinces within Cambodia.
Results
Sequencing Sympatric P. vivax and P. falciparum Populations.
Whole-genome sequencing identified 61,448 high-quality P. vivax SNPs and 6,734 P. falciparum SNPs from 70 and 80 samples, respectively. All P. vivax isolates had fivefold or greater coverage in ≥99% of coding regions, whereas all P. falciparum isolates had fivefold or greater coverage in ≥94% of coding regions. Additional information about sequence quality and coverage is provided in SI Materials and Methods, SI Sequencing Sympatric P. vivax and P. falciparum Populations.
P. vivax Infections Have Higher Within-Host Diversity than Sympatric P. falciparum Infections.
Because Plasmodia infections are frequently multiclonal, we investigated the extent of multiclonality among our sequenced field isolates. When applied to whole-genome sequencing data, the FWS statistic accurately predicts polyclonality (17). P. vivax infections were more polyclonal (defined as FWS <0.95) than P. falciparum infections (P < 0.0001; Fisher’s exact test), a finding that remained unchanged after bootstrapping to account for the difference in the number of SNPs identified in the two species (Fig. 1A). Using the accepted standard of FWS <0.95 as the marker of a multiclonal infection, 60% of P. vivax isolates and 22.5% of P. falciparum isolates in our cohort were polyclonal (Fig. 1B). To confirm the reliability of this method for identifying polyclonal infections, we conducted amplicon deep sequencing of P. vivax merozoite surface protein 1 (pvmsp1) in 47 isolates, finding a high degree of agreement with FWS (SI Materials and Methods, SI P. vivax Infections Have Higher Within-Host Diversity than Sympatric P. falciparum Infections) (18). Subsequently, most analyses were performed both for the 28 P. vivax infections that were monoclonal by the FWS metric and for all P. vivax samples (i.e., multiclonal and monoclonal infections) to assess the effect of multiclonality on our results.
Fig. 1.
Higher MOI in P. vivax infections compared with P. falciparum infections. (A) FWS calculated for P. vivax and P. falciparum. Points represent the point estimate of FWS for each sample in the respective population. Vertical bars represent the maximum and minimum value in 1,000 bootstrap replicates, which downsampled the number of SNPs to be equal for P. vivax and P. falciparum, to correct for the increased number of P. vivax SNPs. (B) Summary bar graph representing the number of P. vivax (Pv) and P. falciparum (Pf) clinical isolates considered monoclonal or polyclonal, with a cutoff of FWS <0.95 being considered polyclonal.
P. vivax Has Less Population Substructuring than Sympatric P. falciparum.
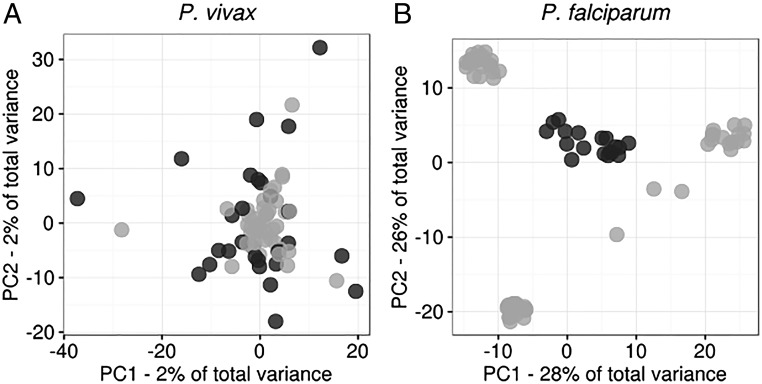
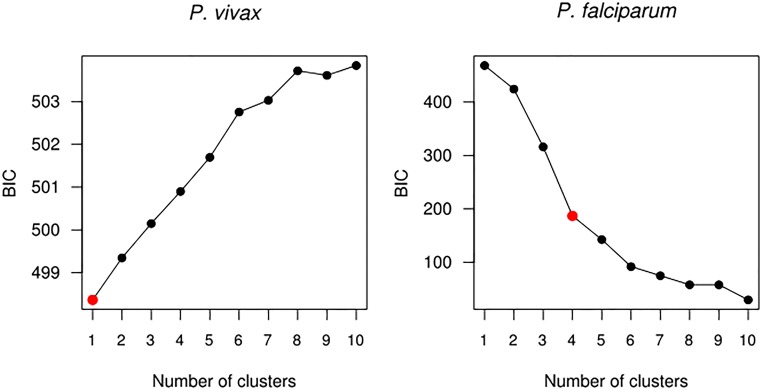
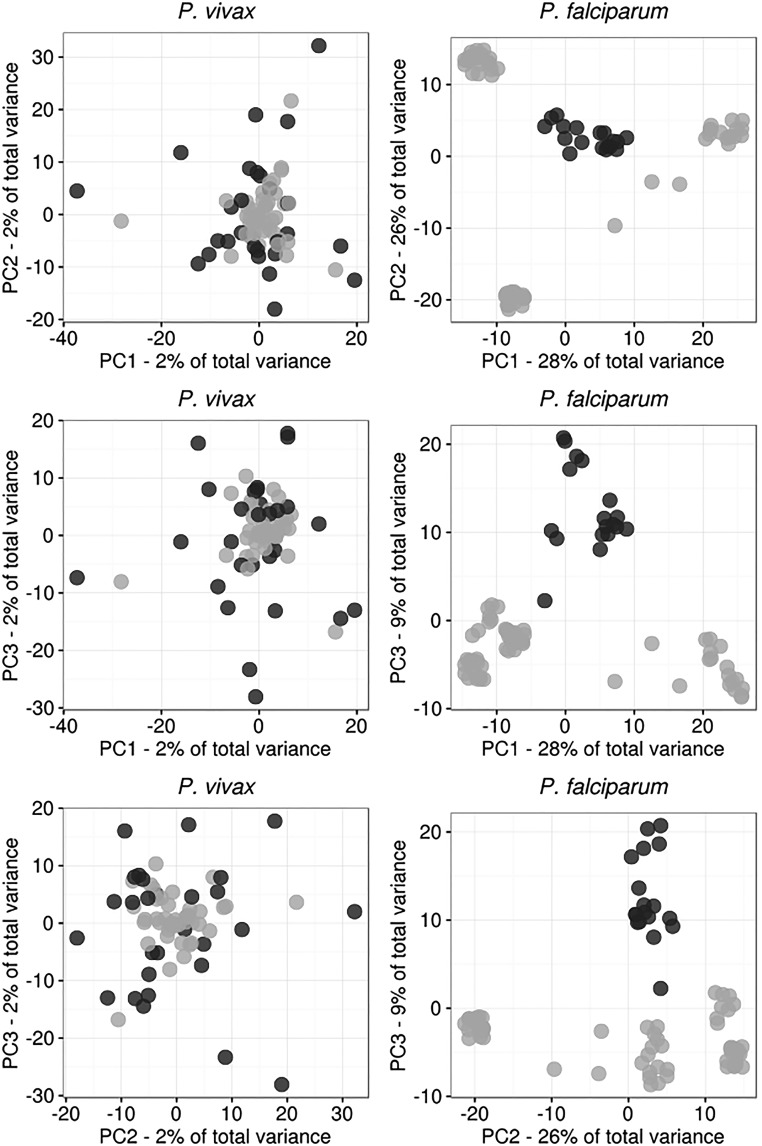
Principal component analysis (PCA) revealed no population substructuring among P. vivax isolates. In contrast, P. falciparum parasites were partitioned into subpopulations (Fig. 2). These partitions did not correspond to collection site, date of collection, or multiplicity of infection (MOI). We used k-means clustering to confirm that all P. vivax isolates were part of a single cluster and that the P. falciparum population was subdivided into four clusters (Fig. S2) (19). One cluster represents a central “ancestral-like” P. falciparum population from which subpopulations of drug-resistant parasites have undergone epidemic expansion. Analysis of additional projections supports these differences in population structure (Fig. S3). Therefore most analyses were performed using all P. falciparum samples and, separately, using the 18 parasites of the central ancestral-like population.
Fig. 2.
PCA reveals striking differences in population structure between Cambodian P. vivax and P. falciparum. PCA was performed from single-nucleotide genetic variant data for each of 70 P. vivax clinical isolates (A) and 80 P. falciparum clinical isolates (B). Monoclonal P. vivax isolates are shaded dark gray. Among P. falciparum isolates, members of the ancestral-like population are shaded dark gray. Noise was added to the P. falciparum panel to mitigate overplotting.
Fig. S2.
Nonparametric k-means clustering of total genetic diversity in P. vivax and P. falciparum isolates identifies distinct subpopulations among P. falciparum but not among P. vivax. For P. vivax, clustering suggests that all parasites are drawn from the same population. For P. falciparum, increasing the number of clusters from n = 3 to n = 4 yields a substantial improvement in Bayesian information criterion (BIC). Although subsequent increases do yield BIC improvements, the improvement in goodness of fit is not commensurate with the cost associated with increased degrees of freedom.
Fig. S3.
Additional projections of P. vivax and P. falciparum genetic differentiation. Additional projections for the top three principal components are shown for the P. vivax (Left) and P. falciparum (Right) populations. Noise was added to the P. falciparum dataset to mitigate overplotting.
P. vivax Has Undergone More Rapid Expansion than Sympatric P. falciparum.
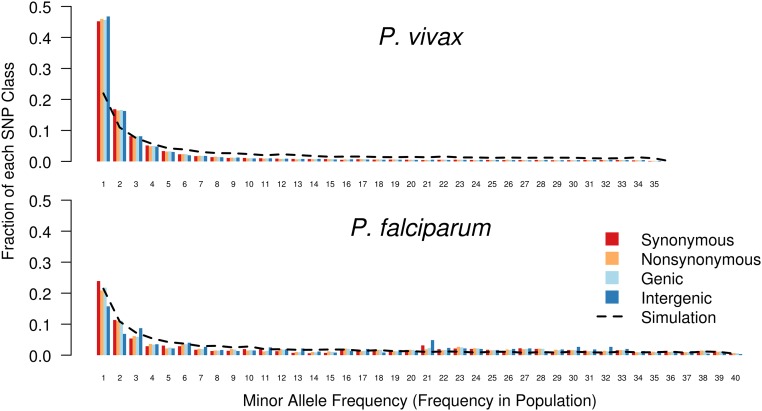
To explore the differences in demographic histories, we examined the allele-frequency spectra (AFS) of the P. vivax and P. falciparum populations. Spectra were calculated by variant type and compared with the spectrum expected in a simulated coalescent population with no natural selection, constant population size, and complete random mating (Fig. 3). We observed an excess of low-frequency derived alleles in the Cambodian P. vivax AFS [both for the entire sample and for the monoclonal infections only (Fig. 3 and Fig. S4)], suggesting population expansion. In contrast, the overall P. falciparum population had no excess of low-frequency alleles, suggesting limited or absent population expansion. However the overall P. falciparum population did exhibit an excess of intermediate-frequency derived alleles, which likely reflected the presence of multiple P. falciparum subpopulations and which disappeared upon analysis of only the central, ancestral-like population (Fig. 3 and Fig. S4).
Fig. 3.
Allele-frequency spectra suggest greater population expansion among P. vivax than among P. falciparum. Each column indicates the fraction of the total SNPs that fall into a particular frequency class. Columns are color coded by type of SNP. The x axis represents the number of samples within which each SNP occurs. (Upper) We observed a preponderance of low-frequency alleles in the P. vivax population, compared with coalescent simulations of a no-growth population (black line). Under a Wright–Fisher model of genetic evolution, these data suggest that the P. vivax population has undergone a recent expansion. (Lower) In contrast, for P. falciparum, we observed an excess in intermediate-frequency minor alleles, reflecting the subdivided population structure.
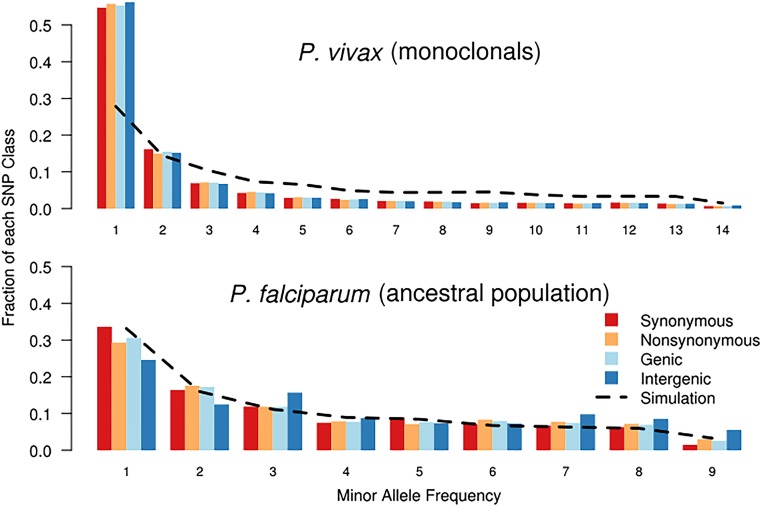
Fig. S4.
AFS for P. vivax monoclonal isolates and the P. falciparum ancestral population. Similar to the entire P. vivax population, the P. vivax monoclonal isolates have a strong overrepresentation of low-frequency alleles, suggesting an expanding population size. When considered in isolation, the P. falciparum ancestral population shows no increase in intermediate-frequency alleles.
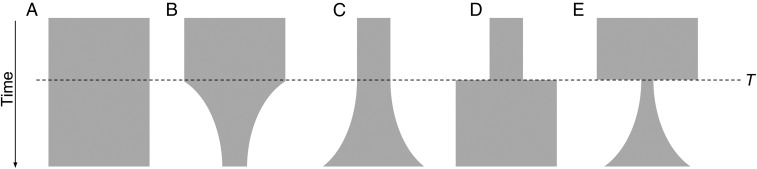
Next, we fit various demographic scenarios to the observed allele-frequency spectra to identify a best-fit model and specific population parameters. Using a diffusion approximation paradigm, we tested scenarios of constant population size, population decline, exponential increase, two-epoch increase, and bottleneck with subsequent exponential growth (Fig. 4) (20). We used the Akaike information criterion (AIC) to inform model selection. For P. vivax (all samples and monoclonals only), models of parasite expansion strongly outperformed the other models, with the model of positive exponential growth having the best fit (Table S1). For the ancestral-like P. falciparum population, exponential growth models marginally outperformed the other models, suggesting only modest expansion of this parasite population (Table S2). Comparison of best-fit models suggests that P. vivax has expanded more dramatically [factor of population contraction (ηG) = 20.00] and over a short time span (T = 1.03) than the ancestral-like P. falciparum population (ηG = 1.94, T = 4.99), resulting in a larger Neff for P. vivax than for P. falciparum [ancestral mutation rate (θ) = 850 and 240, respectively] (Tables S1 and S2).
Fig. 4.
Five one-population models were used for demographic inference. The following models were fitted to observed P. vivax and P. falciparum allele-frequency spectra using a diffusion approximation of population evolution: (A) a model of no change in Neff through time; (B) a model of population decline beginning at time T; (C) a model of exponential population expansion, beginning at time T; (D) a two-epoch model of sudden population expansion at time T; (E) a model of rapid population decline followed by exponential growth, beginning at time T. In all cases, the exponential-growth model (C) was at least marginally the best fit scenario. For the P. vivax population, growth models (C–E) far outperformed the static (A) or decline (B) models. See Tables S1 and S2.
Table S1.
Demographic models strongly support positive growth in the P. vivax population
| Model | θ | ηD* (0.001–1.0) | ηG* (1.0–100) | T* (0.01–5.0) | Log likelihood | AIC |
| P. vivax monoclonal infections | ||||||
| Static | 1,868.499 | — | — | −911.145 | 1,392.57 | |
| NegG | 1,617.421 | 0.00115 | — | 3.351 | −913.524 | 1,831.05 |
| PosG-Epoch | 898.556 | — | 9.819 | 0.672 | −30.838 | 65.68 |
| PosG-Exp | 850.687 | — | 20.000 | 1.032 | −28.077 | 60.15 |
| BG | 5,865.229 | 0.0184 | 2.808 | 0.261 | −28.020 | 62.04 |
| P. vivax all infections | ||||||
| Static | 1,568.496 | — | — | — | −601.569 | 1,203.14 |
| NegG | 1,406.454 | 0.00111 | — | 4.755 | −602.773 | 1,209.55 |
| PosG-Epoch | 848.495 | — | 7.798 | 0.547 | −31.500 | 67.00 |
| PosG-Exp | 816.378 | — | 14.342 | 0.830 | −27.594 | 59.19 |
| BG | 816.373 | 0.972 | 14.332 | 0.839 | −27.593 | 61.19 |
Log-likelihoods and AIC were determined for the following five models as seen in Fig. 4: constant population size (Static), population decline (NegG), two-epoch increase (PosG-Epoch), exponential increase (PosG-Exp), and bottleneck with subsequent exponential growth (BG). Models were fit using only the monoclonal P. vivax isolates or the entire sample. Model parameters included (i) θ, the ancestral mutation rate which is a proxy for the Neff; (ii) ηD, the factor of population contraction; (iii) ηG, the factor of population expansion; and (iv) T, the time in the past of change in population. Best-fit (i.e., lowest AIC) parameter sets were selected from 100 model-fit replicates. AIC aids in scenario selection by penalizing highly parameterized models while seeking to minimize information loss. The extent of information loss between the best-fit and other models is described by , where AICmin is AIC for the best-fit model (i.e., lowest AIC), and AICi represents AIC for the ith model.
Ranges explored for optimization are provided for optimizable parameters.
Table S2.
Demographic models show equivocal evidence for growth among the P. falciparum ancestral-like subpopulation
| Model | θ | ηD* (0.001–1.0) | ηG* (1–100) | T* (0.01–5.0) | Log likelihood | AIC |
| Static | 396.183 | — | — | — | −27.336 | 54.67 |
| NegG | 396.188 | 0.999 | — | 0.308 | −27.337 | 58.67 |
| PosG-Epoch | 297.831 | — | 1.515 | 1.262 | −24.747 | 53.49 |
| PosG-Exp | 240.479 | — | 1.943 | 4.998 | −24.736 | 53.47 |
| BG | 272.881 | 0.999 | 1.714 | 3.457 | −24.737 | 55.47 |
Log-likelihoods and AIC were determined for the following five models as seen in Fig. 4: constant population size (Static), population decline (NegG), two-epoch increase (PosG-Epoch), exponential increase (PosG-Exp), and bottleneck with subsequent exponential growth (BG). Models were fit to the P. falciparum central (i.e., ancestral) population, because it comprises a single population for comparison with P. vivax scenarios. Output variables determined included (i) θ, the ancestral mutation rate which is a proxy for the Neff; (ii) ηD, the factor of population contraction; (iii) ηG, the factor of population expansion; and (iv) T, the time in the past of change in population. Best-fit (i.e., lowest AIC) parameter sets were selected from 100 model-fit replicates. AIC aids in scenario selection by penalizing highly parameterized models while seeking to minimize information loss. The extent of information loss between the best-fit and other models is described by , where AICmin is AIC for the best-fit model (i.e., lowest AIC), and AICi represents AIC for the ith model.
Ranges explored for optimization are provided for optimizable parameters.
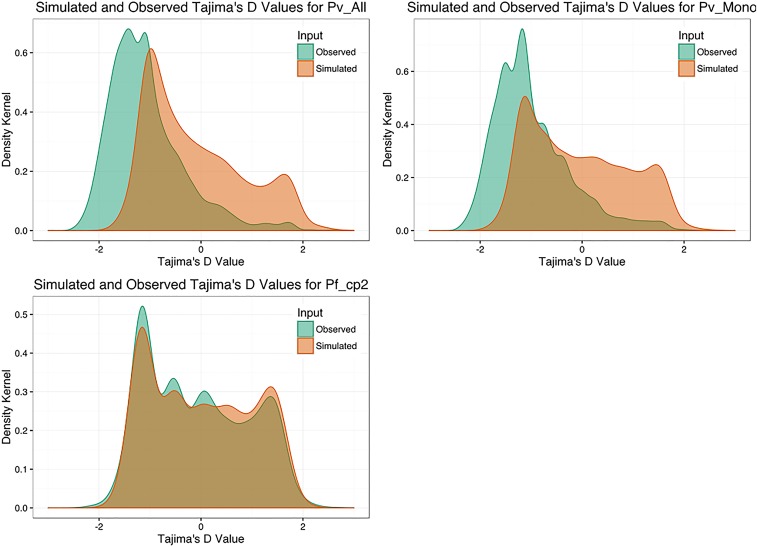
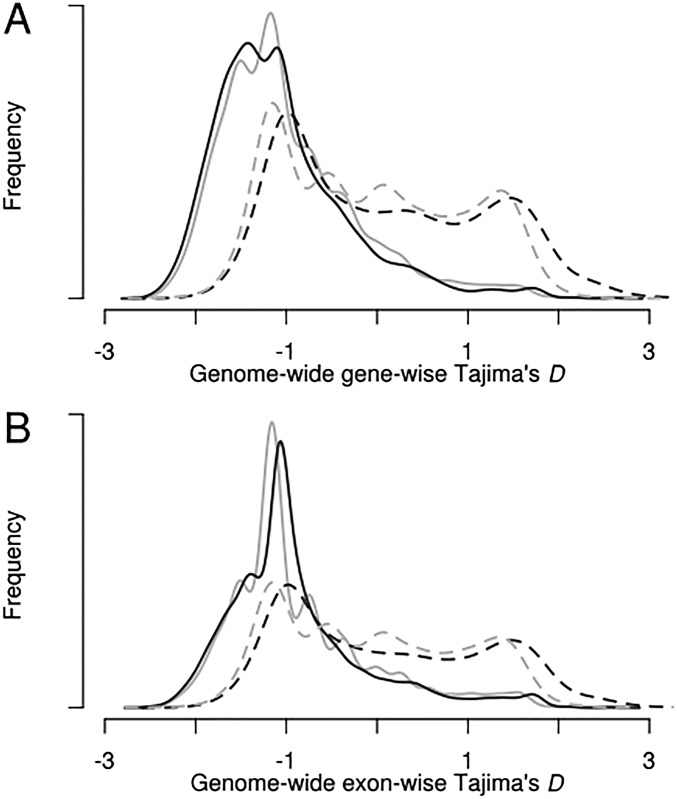
To assess the goodness-of-fit of these inferred parameters, we selected the best-fit models to parameterize coalescent simulations. Tajima’s D was calculated for each simulated gene and compared with observed values of Tajima’s D (Fig. S5). Simulated and observed values of Tajima’s D for P. vivax and P. falciparum were concordant, with a negative mean value and a strong right skew, supporting the inferred population histories for both P. vivax and P. falciparum (Fig. S6). Cell-surface proteins in P. falciparum have been described previously as being under strong balancing selection (high Tajima’s D) because of selection by human immunity. Our analysis confirmed enrichment for cell-surface protein exons among targets of strong balancing selection after Bonferroni correction (P = 0.00124) (Table 1). Genome-wide assessment of balancing selection with Tajima’s D for P. vivax has not been reported previously, and we found several instances of modest Gene Ontology (GO)-term enrichment among targets of strong balancing selection, including chromatin modifiers (Table 1). Additional details about the assessment of Tajima’s D are provided in SI Materials and Methods, SI Assessment of Tajima’s D.
Fig. S5.
Observed and simulated distributions of population summary statistics. Observed (orange) and simulated (green) Tajima’s D values for the ancestral P. falciparum population (Pf_cp2), the monoclonal P. vivax clinical isolates (Pv_Mono), and all P. vivax isolates (Pv_All). Modeling parameters were inferred from one-model best-fit demographic scenarios.
Fig. S6.
Observed genewise and exonwise Tajima’s D distributions for P. vivax and P. falciparum. (A) Whole-genome genewise Tajima’s D was calculated for the entire P. vivax isolate set (n = 70; solid gray line), the monoclonal P. vivax isolates (n = 28; solid black line), the entire P. falciparum isolate set (n = 80; dashed black line), and the ancestral P. falciparum subpopulation (n = 18; dashed gray line). Distributions of whole and subset datasets for both species show close correlation, providing evidence that subsetting these genetic data does not bias allele frequencies. (B) Distributions of exonwise Tajima’s D demonstrate characteristics similar to the genewise analysis.
Table 1.
GO analysis of observed Tajima’s D values uncovers evidence for enrichment of GO terms in genes under strong directional and balancing selection
| Dataset* | Feature† | D percentile‡ | GO term | GO ID | P value§ |
| P. vivax (M/A) | Exon | First | Histone deacetylase complex | GO:0000118 | 0.0423 |
| P. vivax (M) | Gene | First | ATP binding | GO:0005524 | 0.0493 |
| P. vivax (A) | Gene | First | Cellular component | GO:0005575 | 0.00941 |
| P. vivax (M) | Exon | 99th | Metallo-sulfur cluster assembly | GO:0031163 | 0.0440 |
| P. vivax (M) | Exon | 99th | Iron-sulfur cluster assembly | GO:0016226 | 0.04402 |
| P. vivax (M/A) | Gene/exon | 99th | Plastid large ribosomal subunit | GO:0000311 | 0.0166 |
| P. vivax (M/A) | Gene/exon | 99th | Plastid ribosome | GO:0009547 | 0.0306 |
| P. vivax (M/A) | Gene/exon | 99th | Plastid part | GO:0044435 | 0.0306 |
| P. vivax (M/A) | Gene/exon | 99th | Plastid stroma | GO:0009532 | 0.0306 |
| P. falciparum | Gene | First | Retrograde transport, endosome to Golgi | GO:0042147 | 0.0431 |
| P. falciparum | Gene | First | Endosomal transport | GO:0016197 | 0.0431 |
| P. falciparum | Gene/exon | 99th | Cell surface | GO:0009986 | 0.00124 |
Tajima’s D was calculated on a per-gene and per-exon basis for the entire P. vivax population sample, the P. vivax monoclonal subset, and the central, ancestral-like P. falciparum subpopulation. For genewise and exonwise Tajima’s D values, the first percentile (largest negative values) and 99th percentile (largest positive values) were included in GO term enrichment analysis.
(M) indicates the result was found among P. vivax monoclonals; (A) indicates result found among the entire P. vivax population sample.
GO-term enrichment was found in exonwise analysis, genewise analysis, or both exon- and genewise analyses.
First percentile indicates largest negative Tajima’s D values, consistent with directional selection; 99th percentile indicates largest positive Tajima’s D values, consistent with balancing selection.
Bonferroni-corrected P values; if a term was significant (to a Bonferroni-corrected P ≤ 0.05) in both monoclonal (M) and all (A) analyses, the more conservative (higher) of the two P values is reported.
P. vivax Shows Stronger Evidence of Recent Directional Selection on Transcriptional Regulators than P. falciparum.
The population structure of P. falciparum in Cambodia has been shaped by the intensive use of artemisinins and their partner drugs, bed nets, and improved diagnostics (21, 22). However, little is known about how coendemic P. vivax populations have responded to these same selective forces. We used linkage disequilibrium-based tests to identify genomic regions that have undergone recent directional selection consistent with selective sweeps. Because these tests are haplotype based, we focused on the monoclonal P. vivax isolates (n = 28) and the ancestral-like P. falciparum group (n = 18), which were predominantly monoclonal (15/18).
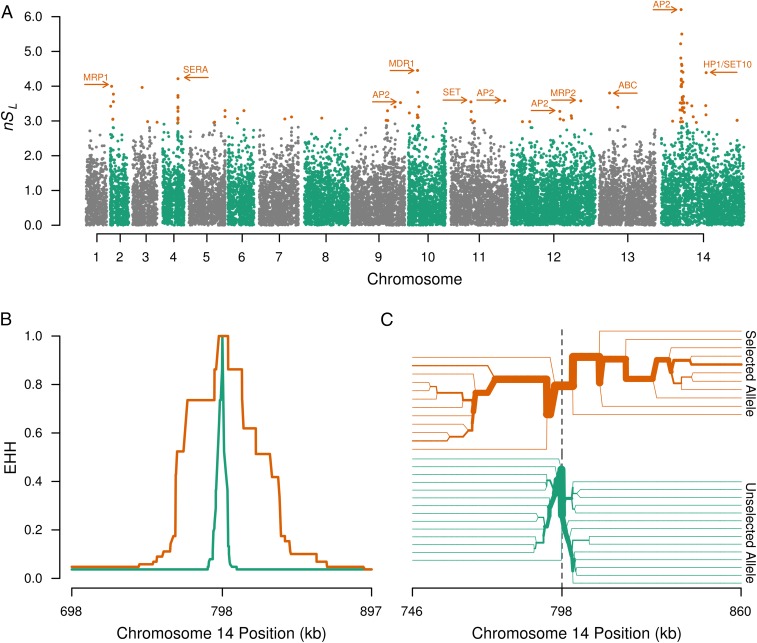
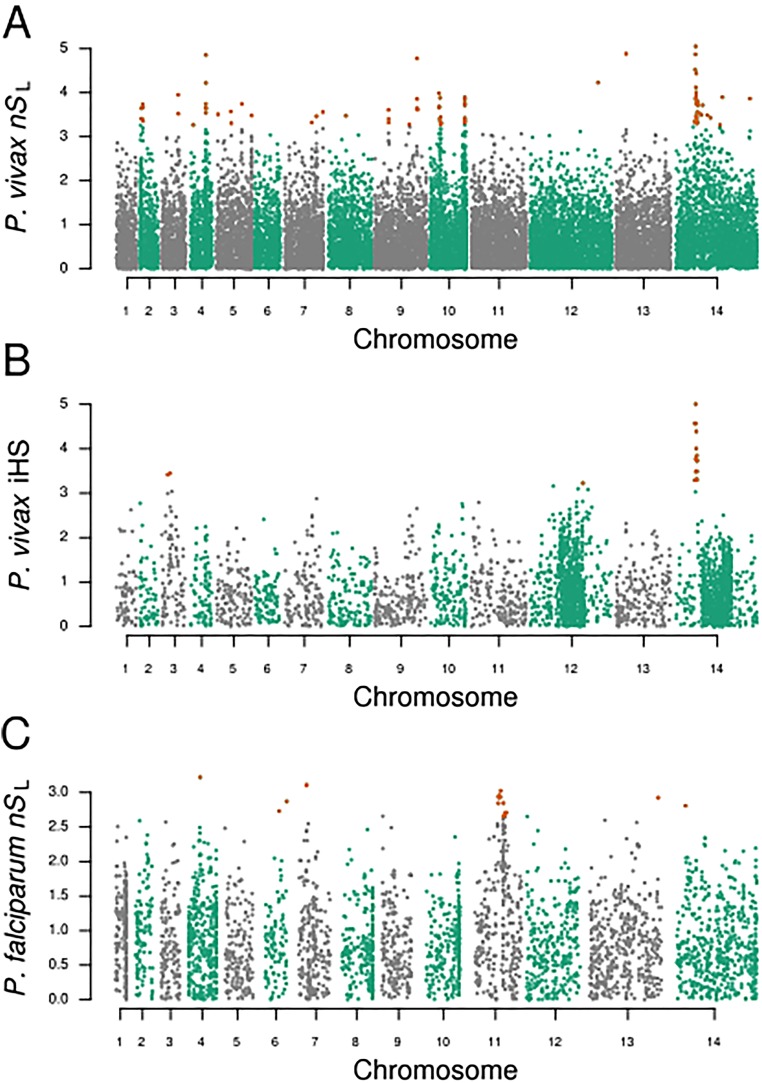
Using 45,701 high-quality SNPs occurring in the monoclonal P. vivax infections, we performed the nSL test for directional selection (23). This haplotype-based testing offers the advantage of not requiring a genome recombination map and has proven sensitive for detecting incomplete selective sweeps. Strikingly, among the 15 strongest signals of directional selection, six were in close proximity to regulators of gene expression (four AP2 domain-containing transcription factors and two proteins containing the SET domain, which is a histone modulator) (Fig. 5A and Table 2). The strongest signal was on chromosome 14, in close proximity to an AP2 domain-containing transcription factor (PVX_122680). Analysis of the entire P. vivax population using nSL yielded qualitatively similar, although blunted, results because of the artifactual breakdown of linkage disequilibrium caused by multiple infections (Fig. S7). Similar results were found when the integrated haplotype score (iHS), which has been used extensively in malaria studies to assess directional selection, was calculated for these same loci (Fig. S7). In contrast, when nSL testing was performed for the 5,158 SNPs in the P. falciparum ancestral-like population, only a single transcription factor was identified as being under moderate directional selection (Table 3 and Fig. S7). This difference does not appear to be attributable to the smaller number of SNPs used in the P. falciparum analysis (likely resulting from a more clonal population as well as from a decreased ability to call SNPs in intergenic regions of the AT-rich P. falciparum genome). As evidence that results are not significantly impacted by detection bias between genic and intergenic SNPs, when we repeated nSL testing in the P. vivax population using only the 28,746 genic SNPs, no appreciable differences in nSL signals were identified. The high level of coverage achieved in the coding regions of both populations suggests an equal opportunity to detect signatures of selection among transcription factors in both species.
Fig. 5.
Evidence for strong selective sweeps in Cambodian P. vivax. (A) A Manhattan plot of normalized nSL values. Each point corresponds to an SNP, and the top 0.5% values (under strong directional selection) are rendered in orange. Polymorphisms without evidence of strong directional selection are rendered in either gray or green, according to chromosome. This view suggests that several genomic regions are under positive selection, including areas near transcription factors (AP2 domain-containing), chromatin regulators (SET10 and HP1), antigens under known positive selection (SERA4 and 5), and drug-resistance genes (MDR1 and MRP1). (B and C) The extent of EHH in the strongest sweep, within chromosome 14 in A. (B) EHH decay for the haplotypes around selected (orange) and unselected (green) alleles. (C) A haplotype bifurcation diagram is shown centered on the focal variant. Line thickness represents the number of identical haplotypes flanking the selected (orange) or unselected (green) alleles. Linkage breaks down with increasing distance from the focal variant. B and C provide evidence that the strong sweep on chromosome 14 extends ∼50 kb in each direction.
Table 2.
Top hits in nSL analysis of P. vivax monoclonal infections
| Chr | Focal SNP statistics | Sweep region statistics | |||
| Location | nSL | Closest plausible genetic driver | Gene ID | Distance* | |
| 14 | 797,870 | 6.203 | Transcription factor with AP2 domain(s), putative (apiap2) | PVX_122680 | −4,462 |
| 10 | 351,691 | 4.453 | Multidrug resistance protein 1, putative (MDR1) | PVX_080100 | 10,010 |
| 14 | 1,650,897 | 4.389 | Heterochromatin protein 1 (HP1); H3 lysine methyltransferase (SET10) | PVX_123682; PVX_123685 | 999; 9,918 |
| 04 | 577,543 | 4.215 | Serine-repeat antigen 4 (SERA) | PVX_003825; PVX_003830 | 789; −1,691 |
| Serine-repeat antigen 5 (SERA) | |||||
| 02 | 85,446 | 3.997 | Multidrug resistance-associated protein 1, putative (MRP1) | PVX_097025 | −68,196 |
| 03 | 368,737 | 3.964 | Hypothetical proteins | — | — |
| 13 | 396,427 | 3.805 | ABC transporter B family member 7, putative (ABCB7) | PVX_084521 | −7,326 |
| 11 | 1,900,445 | 3.581 | Transcription factor with AP2 domain(s), putative (apiap2) | PVX_113370 | −31,184 |
| 12 | 2,421,187 | 3.578 | Multidrug resistance protein 2 (MDR2) | PVX_118100 | −4,361 |
| 11 | 749,030 | 3.549 | SET domain containing protein | PVX_114585 | 46,877 |
| 09 | 1,693,066 | 3.527 | Transcription factor with AP2 domain(s), putative (AP2-O) | PVX_092760 | 7,201 |
| 14 | 1,002,439 | 3.515 | Hypothetical proteins | — | — |
| 09 | 1,506,397 | 3.403 | Transcription factor with AP2 domain(s), putative (apiap2) | PVX_092570 | 9,327 |
| 13 | 676,297 | 3.393 | Hypothetical protein | — | — |
| 05 | 1,303,321 | 3.299 | Vir and fam antigens | — | — |
Haplotype-based tests of selection were performed in monoclonal samples to avoid false breakdown of linkage caused by mixed genotypes from sequencing. Results are presented by ranking on absolute normalized nSL score.
Distance in bases from focal SNP to the putative driver gene; a negative sign indicates putative driver gene occurs upstream of the focal SNP.
Fig. S7.
Additional haplotype-based tests of selection confirm the identity of different types of loci under selection in P. vivax and P. falciparum. In each panel, all points correspond to normalized nSL or iHS values, with the 0.5% highlighted. (A) The haplotype-based selection statistic nSL was determined for all P. vivax cases (n = 70) (i.e., including multiclonal infections). Because of haplotype-based tests rely on single-clone haplotypes for selection, this analysis is secondary to the monoinfection-based analysis presented in the main text. Nonetheless, the results of this analysis are largely similar to the monoinfection-based analysis. (B) The iHS was calculated for P. vivax monoinfections (n = 28). This analysis identified the same chromosome 14 linkage group as being under strong directional selection. Because iHS depends on a genetic map, and because a curated genetic map of the P. vivax Sal1 genome is not available at present, this analysis was secondary to the nSL test, which is a map-free statistic. (C) The haplotype-based selection statistic nSL was also determined for the ancestral P. falciparum cases (n = 18), revealing several loci under strong directional selection, including crt, ama1, and kelch K13 (Table 3).
Table 3.
Top hits in nSL analysis of P. falciparum ancestral-like population
| Chr | Focal SNP statistics | Sweep region statistics | |||
| Location | nSL | Closest plausible genetic driver | Gene ID | Distance* | |
| 04 | 531,138 | 3.220 | Hypothetical proteins | — | — |
| 07 | 384,302 | 3.106 | Chloroquine resistance transporter (CRT) | PF3D7_0709000 | 18,920 |
| 11 | 1,089,187–1,347,798 | 3.024 | Phosphatidylinositol-4-phosphate 5-kinase; apical membrane antigen 1 (AMA1) | PF3D7_1129600; PF3D7_1133400 | 51,935; −93,151 |
| 13 | 2,550,178 | 2.924 | Hypothetical proteins | — | — |
| 06 | 1,093,476 | 2.871 | Hypothetical proteins | — | — |
| 14 | 610,371 | 2.805 | Hypothetical proteins | — | — |
| 06 | 825,473 | 2.730 | Merozoite surface protein 10 (MSP10) | PF3D7_0620400 | −25,905 |
| 09 | 203,308 | 2.656 | Hypothetical proteins | — | — |
| 12 | 124,649 | 2.652 | Dynein heavy chain, putative | PF3D7_1202300 | 0 |
| 13 | 639,213 | 2.597 | Transcription factor MYB1 (MYB1) | PF3D7_1315800 | −20,805 |
| 02 | 374,197 | 2.589 | Serine repeat antigen 1 (SERA1) | PF3D7_0208000 | 48,474 |
| 03 | 361,403 | 2.571 | Hypothetical proteins | — | — |
| 13 | 1,775,996 | 2.563 | Kelch protein K13 (K13) | PF3D7_1343700 | 48,999 |
| 01 | 226,924 | 2.505 | Hypothetical proteins | — | — |
| 09 | 513,729 | 2.489 | Cysteine repeat modular protein 1 (CRMP1) | PF3D7_0912000 | 0 |
Results are presented by ranking on absolute normalized nSL score.
Distance in bases from focal SNP to the putative driver gene; a negative sign indicates putative driver gene occurs upstream of the focal SNP; a zero indicates focal SNP occurs within the putative driver gene.
To determine the extent of haplotype homozygosity around the predominant selective sweep, we performed extended haplotype homozygosity (EHH) testing, centering our analysis on the SNP with the highest nSL score. We identified a 100-kb region of strong linkage disequilibrium around the principle chromosome 14 locus (Fig. 5 B and C) in isolates with the selected allele. In our Cambodian P. vivax population sample, this ApiAP2 transcription factor contains 25 polymorphisms (Table S3), including two high-frequency nonsynonymous changes, one of which occurs within the AP2 DNA-binding domain, based on comparisons with its P. falciparum ortholog (PF3D7_1317200) (24). In addition to the AP2-domain transcription factor, this region contains 25 genes (Table S4).
Table S3.
Mutations occurring within the ApiAP2 gene (PVX_122680) under strong directional selection
| Chromosome | Position | Ref | Alt | Gene ID | Type | Detail | Frequency |
| Pv_Sal1_chr14 | 785,752 | C | T | PVX_122680 | Syn | NA | 0.014 |
| Pv_Sal1_chr14 | 785,785 | C | A | PVX_122680 | Syn | NA | 0.029 |
| Pv_Sal1_chr14 | 786,028 | C | G | PVX_122680 | Nonsyn | N107K | 0.114 |
| Pv_Sal1_chr14 | 786,152 | C | T | PVX_122680 | Nonsyn | P149S | 0.014 |
| Pv_Sal1_chr14 | 786,192 | C | G | PVX_122680 | Nonsyn | P162R | 0.029 |
| Pv_Sal1_chr14 | 786,199 | C | A | PVX_122680 | Syn | NA | 0.029 |
| Pv_Sal1_chr14 | 786,647 | T | C | PVX_122680 | Nonsyn | Y314H | 0.029 |
| Pv_Sal1_chr14 | 787,039 | C | A | PVX_122680 | Nonsyn | N444K | 0.014 |
| Pv_Sal1_chr14 | 788,105 | A | G | PVX_122680 | Nonsyn | S800G | 1.000 |
| Pv_Sal1_chr14 | 788,143 | C | A | PVX_122680 | Syn | NA | 0.014 |
| Pv_Sal1_chr14 | 788,229 | G | T | PVX_122680 | Nonsyn | G841V | 0.100 |
| Pv_Sal1_chr14 | 788,674 | C | T | PVX_122680 | Syn | NA | 0.043 |
| Pv_Sal1_chr14 | 788,965 | C | T | PVX_122680 | Syn | NA | 0.471 |
| Pv_Sal1_chr14 | 789,092 | C | A | PVX_122680 | Nonsyn | H1129N | 0.014 |
| Pv_Sal1_chr14 | 789,621 | T | A | PVX_122680 | Nonsyn | F1305Y | 0.943 |
| Pv_Sal1_chr14 | 789,675 | A | G | PVX_122680 | Nonsyn | K1323R | 0.014 |
| Pv_Sal1_chr14 | 790,131 | G | A | PVX_122680 | Nonsyn | G1475D | 0.071 |
| Pv_Sal1_chr14 | 791,180 | G | A | PVX_122680 | Nonsyn | A1825T | 0.014 |
| Pv_Sal1_chr14 | 791,472 | G | A | PVX_122680 | Nonsyn | R1922Q | 0.243 |
| Pv_Sal1_chr14 | 791,486 | C | T | PVX_122680 | Syn | NA | 0.057 |
| Pv_Sal1_chr14 | 791,949 | C | T | PVX_122680 | Nonsyn | A2081V | 0.043 |
| Pv_Sal1_chr14 | 792,052 | C | T | PVX_122680 | Syn | NA | 0.043 |
| Pv_Sal1_chr14 | 792,078 | T | G | PVX_122680 | Nonsyn | I2124S | 0.057 |
| Pv_Sal1_chr14 | 792,562 | T | G | PVX_122680 | Syn | NA | 0.500 |
| Pv_Sal1_chr14 | 792,677 | C | T | PVX_122680 | Nonsyn | L2324F | 0.014 |
In the Cambodian P. vivax population (n = 70), this AP2-domain transcription factor contained numerous nonsynonymous mutations, two of which occurred at high frequency (>50%), and one of which occurred within the gene’s sole DNA-binding AP2 domain. Alt, alternate allele; Nonsyn, nonsynonymous; Ref, reference alelle; Syn, synonymous.
Table S4.
Genes falling within the principal P. vivax selective sweep occurring on chromosome 14
 |
Identifier, description, and location of genes that occur within the sweep. Although the sweep haplotype extends ∼50 kb in each direction, genes occurring within 5 kb of the focal SNP are highlighted.
+/− refer to the positive and negative strand of DNA, respectively.
We found complementary evidence that P. vivax transcriptional regulators are under strong directional selection using allele frequency-based testing for selection. We calculated genewise Tajima’s D (an allele frequency-based test, in contrast to nSL and iHS, which are haplotype-based tests of selection) for P. vivax and P. falciparum. We selected the first and 99th percentile of observed genewise Tajima’s D values and investigated these genes for functional enrichment. We observed enrichment of histone deacetylase complex members among the first percentile of genes, but this observation did not reach significance after Bonferroni correction. Because strong local values of Tajima’s D can be obscured when considering an entire gene, we also performed this analysis in an exonwise manner for both P. vivax and P. falciparum (18). In this way we found statistically significant enrichment for histone deacetylase complex members among the first-percentile exons after strict Bonferroni correction (P = 0.0423) (Table 1).
Both P. vivax and P. falciparum Show Evidence of Recent Directional Selection on Known and Putative Drug-Resistance Genes.
Four of the strongest 15 strongest nSL signals in P. vivax were in close proximity to transporters (pvmdr1, pvmdr2, pvmrp1, and an ABC transporter), all of which are potential drug-resistance loci (Table 2). A prominent sweep did encompass the pvmdr1 locus. We compared key drug-resistance SNP frequencies in pvmdr1 to frequencies in Cambodian isolates collected several years earlier, during 2006 and 2007, from Kâmpôt province (25). Two key mutations (Y976F and F1076L) existed at roughly the same frequency (89% in previously collected samples vs. 77% in more recently collected samples and 87% in previously collected samples vs. 90% in more recently collected samples, respectively). This high similarity in allele frequency in samples collected before and during the artemisinin-resistance containment efforts suggests that the sweep encompassing the pvmdr1 locus was not driven by ACT drug pressure or resistance-containment efforts and may result from more longstanding chloroquine pressure (similar to pfmdr1 mutations in P. falciparum) that preceded containment efforts. Allele frequencies for all polymorphisms in known or putative drug-resistance genes are summarized in Table S5.
Table S5.
Previously described drug-resistance mutations among P. vivax parasites
| Gene | Resistance variant | Prevalence among P. vivax monoclonal isolates, % |
| pvdhps | M205I | 96.4 |
| A383G | 96.4 | |
| pvdhfr | S58R | 100 |
| S117N | 100 | |
| pvmdr1 | S513R | 14.3 |
| G698S | 96.4 | |
| M908L | 100 | |
| T958M | 100 | |
| Y976F | 75 | |
| F1076L | 85.7 | |
| pvmdr2 | L43V* | 85.7 |
| V76A* | 10.7 | |
| S224C* | 7.14 | |
| Y514F* | 7.14 | |
| I852L* | 39.3 | |
| V1467A* | 57.1 | |
| L1471P* | 28.6 | |
| T1473A* | 32.1 | |
| T1473I* | 14.3 | |
| L1477P* | 53.6 | |
| T1479A* | 50.0 | |
| L1507P* | 17.9 | |
| T1509A* | 32.1 | |
| L1513P* | 14.3 | |
| T1515A* | 17.9 | |
| T1515I* | 10.7 | |
| L1519P* | 46.4 | |
| T1521A* | 46.4 | |
| T1521I* | 35.7 | |
| pvmrp1 | T234M | 85.7 |
| T259I* | 3.57 | |
| T259R | 96.4 | |
| Q906E | 92.9 | |
| L1207I* | 96.4 | |
| Y1393D | 100 | |
| V1478I | 100 |
Reported polymorphisms occurred in at least two samples. Although the strict coverage and quality filters which are otherwise applied throughout this work are appropriate and necessary for demographic modeling and for detecting selective sweeps, they are too restrictive for reliable locus genotyping. Thus, these predictions of variant effects on known drug-resistance genes were derived from variant calls that are unfiltered for coverage and quality.
Mutation not previously described.
Similar to previous studies of directional selection in P. falciparum, we identified drug-resistance genes (e.g., pfcrt and kelch K13) with evidence of strong directional selection (Table 3) (26–29). Allele frequencies for all polymorphisms in known drug-resistance genes are summarized in Table S6. Of note, both nSL and iHS statistics revealed a chromosome 11 locus with strong and extended directional selection (Table 3 and Fig. S7). This locus encompassed the pfama1 and phosphatidylinositol-4-phosphate 5-kinase genes (PF3D7_1129600). Interestingly, PF3D7_1129600 catalyzes the phosphorylation of phosphatidylinositol 4-phosphate to form phosphatidylinositol 4,5-bisphosphate (30). The drug target PI(4)K (PF3D7_0509800) alters the intracellular distribution of phosphatidylinositol-4-phosphate in the parasite, placing these genes on the same biologic pathway (31). Although the functional significance of this signal is unclear, genome scans in other populations also have identified strong directional selection at this locus (27, 32).
Table S6.
SNPs among known drug-resistance genes in P. falciparum ancestral-like parasites
| Gene | Resistance variant | Prevalence among ancestral* P. falciparum isolates, % |
| kelch K13 | R539T | 0 |
| C580Y | 61.1 | |
| pfcrt | K76T | 100 |
| dhps | S436A | 38.9 |
| K540E | 61.1 | |
| K540N | 38.8 | |
| A581G | 50 | |
| dhfr | N51I | 100 |
| C59R | 100 | |
| S108N | 100 | |
| I164L | 50 | |
| pfmdr1 | Y184F | 72.2 |
| F1068L† | 22.2 | |
| pfmdr2 | T484I | 72.2 |
| F423Y | 100 | |
| G299D† | 83.3 | |
| S208N | 100 | |
| pfmrp1 | H191Y | 94.4 |
| K202E† | 44.4 | |
| N325S | 11.1 | |
| S437A | 94.4 | |
| I876V | 83.3 | |
| T1007M | 16.7 | |
| F1390I | 72.2 |
Reported polymorphisms occurred in at least two samples. Although the strict coverage and quality filters which are otherwise applied throughout this work are appropriate and necessary for demographic modeling and for detecting selective sweeps, they are too restrictive for reliable locus genotyping. Thus, these predictions of variant effects on known drug-resistance genes were derived from variant calls that are unfiltered for coverage and quality.
The central P. falciparum population.
Mutation not previously described.
Copy-Number Variants Are Not Associated with Detected Selective Sweeps.
Copy-number variants (CNVs) are important in the evolution of parasite populations (33). Using two complementary in silico methods, we identified a handful of high-confidence CNVs in the P. vivax and P. falciparum populations (Table S7). Notably, we identified Duffy-binding protein (DBP) duplication events in 17 of 70 P. vivax isolates and multidrug-resistance protein 1 (MDR1) duplication events in 12 of 80 P. falciparum isolates. As in previous work in P. falciparum (33), no high-confidence CNV occurred in close proximity with the selective sweeps identified above.
Table S7.
Limited copy-number variation in P. vivax and P. falciparum clinical isolates
| Species | Gene(s) | GeneID | Breakpoints* (Chr:start-end) | Copy number | Frequency |
| P. vivax | Duffy-binding protein (dbp) | PVX_110810 | Chr06 | 2× | 16/70 |
| 974,753 (± 3) | |||||
| 982,117 (± 1) | |||||
| 4× | 1/70 | ||||
| P. vivax | Cytoadherence-linked asexual protein (clag); exported protein, unknown function | PVX_094265; PVX_094270 | Chr08 | 0׆ | 6/70 |
| 73,105 (± 2) | |||||
| 82,397 (± 6) | |||||
| P. falciparum | Multidrug-resistance protein 1 (pfmdr1) | PF3D7_0523000 | Chr05 | 2× | 5/80 |
| 946,536 (± 3124) | |||||
| 967,973 (± 3065) | |||||
| 3× | 5/80 | ||||
| 4× | 2/80 | ||||
| P. falciparum | Plasmepsin II | PF3D7_1408000 | Chr14 | 2× | 16/80 |
| 289,558 (± 34) | |||||
| 298,819 (± 16) | |||||
| 3× | 16/80 |
Copy-number duplication events are reported where detected by two methodologically distinct copy-number–calling software programs.
Breakpoints were predicted probabilistically for each sample and in some cases differ among isolates. It is currently unclear whether these differences represent unique duplication events or imperfect accuracy in predicting breakpoints. Mean breakpoints with SDs are reported.
A copy number of zero indicates a deletion.
Discussion
In this study we compared whole-genome sequence data of 70 P. vivax and 80 coendemic P. falciparum infections to show that the P. vivax population in western Cambodia has experienced more rapid and uninterrupted growth than the sympatric P. falciparum population, resulting in less substructuring and a larger Neff. Clues to the causes underlying this population-level resilience of P. vivax in the face of malaria-control measures can be found in the different genomic signatures of selection between the two species. Evidence for the importance of transcriptional regulation to the success of P. vivax was found both by haplotype-based and allele frequency-based tests of selection. Although known and putative drug-resistance genes were found at the center of selective sweeps in both P. falciparum and P. vivax in this and other studies (Tables 2 and 3), the strongest selective sweep in the P. vivax population occurred in close proximity to an AP2 transcription factor (PVX_122680), suggesting that P. vivax is responding to selective pressures by altering its transcriptional profile (29, 34, 35).
Although studies of transcriptional regulation in P. vivax are just beginning (36–38), some of the key biological processes that are thought to help P. vivax evade traditional control measures may be under tight transcriptional control. Unlike P. falciparum, P. vivax gametocytogenesis occurs early and concomitantly with the asexual cycle, so by the time a person is symptomatic or is diagnosed with P. vivax malaria, that person already harbors infectious gametocytes capable of infecting mosquitoes (39). It appears that a key step in Plasmodium gametocytogenesis is accomplished by transcriptional repression via an AP2 family transcription factor that blocks asexual replication and promotes conversion to the sexual stage (40). In fact, the P. falciparum ortholog of the principal AP2 gene identified in our study has been associated with transcriptional regulation of gametocytogenesis (41). Similarly, processes governing hypnozoite dormancy and activation that underlie P. vivax relapse may be under epigenetic regulation. Histone deacetylase inhibitors accelerated the rate of hypnozoite activation in long-term primary simian hepatocyte cultures, suggesting that histone methylation may maintain hypnozoite dormancy by suppressing transcription (42, 43). Finally, P. vivax chloroquine resistance has not been found to correlate with DNA changes in pvcrt but may correlate with the expression of this transporter protein, whose ortholog mediates chloroquine resistance in P. falciparum (44–46).
Thus, an ability to modulate transcriptional regulation may be more central to P. vivax’s adaptation to selective pressures than for P. falciparum. Notably, a previous comparison of a 1990s era Peruvian P. vivax isolate to the reference Sal1 strain collected 35 y earlier found an increased dN/dS signal at two AP2-containing transcription factors, suggesting that evolutionary changes in these genes are not confined to P. vivax in Cambodia (47). Because this was a within-species comparison of nonsynonymous to synonymous SNP ratio (dN/dS), it is difficult to determine the time-scale (e.g., whether ancient or recent) associated with this selection (48). The top hit in our selective sweep, a sign of recent and strong directional selection, was an AP2 factor that was specifically cited among genes with >1.0 dN/dS in this previous study.
The potential reliance on transcriptional changes is not an absolute difference between species, however. CNV of pfmdr1, which likely leads to increased gene transcription, is known to modulate drug efficacy. However, a closer look at previously published P. falciparum scans for selection in multiple locations in Africa also reveals a role for adaptation through the modification of transcriptional regulation. The P. falciparum ortholog of the SET-domain protein on chromosome 11 (PVX_114585) identified in our nSL analysis lies near the center of a selective sweep that also has occurred in Senegal, the Gambia, and Ghana (16, 49, 50). In addition, it is known that transcriptional timing can affect P. falciparum drug-resistance responses, in particular to artemisinins (51). These findings suggest an underappreciated role for the modification of transcriptional regulation in P. falciparum fitness.
Further investigation of the potential role of transcriptional modification in P. vivax drug resistance and other biological processes will require continued development of in vitro models. Such research strategies include modifications to Plasmodium knowlesi homologs, the use of humanized mice infected with transgenic P. vivax, allowing transcriptional analysis through the liver stage, and the use of monkeys infected with transgenic P. vivax, allowing transcriptional analysis through the blood stage (52–56). To date, descriptions of transcriptional modification in Plasmodium sp. appear to be limited to associations with genomic structural variants, for example, gene amplification as in pfmdr1 and deletions or repeat-length polymorphisms in promoter regions (57, 58). One could hypothesize that SNPs in an AP2 transcription factor DNA-binding domain may alter its motif binding and stage-specific expression of downstream genes. More likely, we have not identified causal variants; rather, our selective sweep is detecting variants that are in linkage disequilibrium with the true functional variants, which could be larger deletions or insertions not readily detected by short-read sequencing. If the selected variants found in our analyses are replicated in other cohorts, further in vitro experimentation is needed to determine whether they associate with transcriptional regulation. Such findings would provide key evidence of an advanced parasitic response to selective pressure in P. vivax. They also would suggest that tracking genetically fit parasites could be complicated, supporting the idea that P. vivax will be the more challenging species to eliminate.
Our findings of a diverse P. vivax population without a discernible population structure are similar to microsatellite- and gene-based studies in Papua New Guinea, Indonesia, Venezuela, and Cambodia that have observed more genetic structuring among P. falciparum populations than among coendemic P. vivax populations (Fig. 2) (10–15). We also have previously characterized the within-host diversity of coendemic P. vivax and P. falciparum in this region by amplicon deep sequencing, with results supporting the difference in polyclonality (59, 60). The high proportion of polyclonal P. vivax infections and the lack of parasite substructuring also were observed in a recent report from Cambodia that used deep sequencing of more than 100 SNPs across the P. vivax genome (18, 61).
Whole-genome sequencing allowed us to extend beyond descriptions of diversity and to make inferences about the demographic history of sympatric Plasmodium in the region. The best-fit demographic models indicated that P. vivax in western Cambodia has undergone steady exponential growth, expanding more rapidly than the ancestral-like P. falciparum population, for which exponential growth was only marginally the best-fit model (Tables S1 and S2). The demography-adjusted estimate of the ancestral mutation rate (θ), a proxy for Neff, found that the P. vivax Neff is substantially larger than the ancestral-like P. falciparum population. These demographic scenarios match epidemiological observations that P. vivax cases have increased even while P. falciparum incidence has rapidly declined over the same time period (6). Interestingly, although the total number of P. falciparum cases in Cambodia has decreased, our demographic models show a slowly increasing Neff, at least for the ancestral-like population. Although it is possible that our models are detecting ancient rather than recent trends, another study of isolates in this area reached similar conclusions, lending credence to our findings and suggesting that recent control efforts have not significantly decreased P. falciparum genetic diversity in the region (62).
We should note that we have largely assumed that selective pressures have been imposed by human control interventions, primarily antimalarial treatment. However, we cannot rule out the possibility that other selective pressures have also affected parasite population structuring, for example, widespread deforestation and climate change altering the diversity of Anopheline vectors or evolving host–parasite interactions within mosquito vectors. In P. vivax, a strong New World vs. Old World divide correlates with genetic variation in pvs47, the ortholog of pfs47, which has been associated with differential infectivity in different mosquito species (63, 64). However, both Anopheles dirus s.s. and Anopheles minimus s.s., the main malaria vectors in Cambodia, are adept at transmitting both P. falciparum and P. vivax (65, 66). Therefore we do not think that the species-specific differences observed in population structure and signatures of selection are attributable solely to unmeasured mosquito factors.
Finally, the population structuring we observed among contemporary Cambodian P. vivax and P. falciparum isolates should be interpreted in light of their ancient demographic histories (67). Evidence has shown that both species of malaria are significantly less diverse than primate malarias and likely have undergone genetic bottlenecks associated with host switching and emergence out of Africa, likely less than 10,000 y ago (63, 68–72). As these species have emerged from the bottleneck, they likely have undergone both sustained and recent selection. In the case of P. vivax, previous work has demonstrated that long-term diversifying and directional selection has shaped its genetic diversity (63, 68). Our data are consistent with this finding, showing minimal structuring in the P. vivax population but evidence of strong directional selection in multiple regions of the genome. Additional investigation is required to determine if these adaptive signatures are geographically restricted (68) or occurred as or before P. vivax expanded out of Africa. In the case of P. falciparum, reductions in diversity immediately around loci associated with drug resistance (73, 74) are commonly reported, and the current substructuring seen in Cambodian P. falciparum parasites has been linked to drug-resistant subtypes (22). This evidence suggests that more recent selective forces have shaped population structure in this species. Previous studies also suggest that P. falciparum infections in Southeast Asia were more multiclonal two decades ago, again suggesting that recent forces have constrained its genetic diversification (75–77). This evidence is consistent with our findings that P. falciparum in Cambodia has undergone large demographic shifts much more recently than P. vivax.
Applying population genomic tools to Plasmodium parasites comes with caveats. Malaria parasites have a complex lifecycle, including human and mosquito stages, with multiple clonal generations occurring within the human bloodstream and frequent bottlenecks during transmission (78). Such realities violate the assumptions of the Wright–Fisher model and complicate inference from genetic data. These peculiarities of the malaria lifecycle may skew the allele-frequency spectrum toward increased singletons, even at neutral sites, leading to quantitatively or even qualitatively inappropriate demographic conclusions (79). However, our demographic conclusions are supported by epidemiologic observation as well as by the results of other population genetic studies (6, 13). Because extensive recognized and unrecognized paralogous families in the P. vivax genome present significant mapping and variant-calling challenges, we curated our data carefully, performing extensive tests to determine the best alignment, filtering, and variant-calling approaches. In addition, none of our samples underwent hybrid selection, giving us greater confidence in the quantitative accuracy of calls in mixed infections and structural variants (80, 81).
In summary, we present evidence that sympatric P. vivax and P. falciparum populations in Cambodia have responded in substantively different ways to the intense selective pressure imposed by the recent artemisinin-resistance containment campaign and national antimalarial drug policies. Although P. falciparum has experienced population splitting, the P. vivax population remains admixed, with strong growth, high genetic diversity, and frequent polyclonal infections. These findings match epidemiologic observations of relative P. vivax resilience to current control measures. Our comparative genomic analysis hints at the mechanisms behind these different responses. Although we found that both P. vivax and P. falciparum have experienced selective sweeps around known or putative antimalarial-resistance genes, the strongest signatures of directional selection in P. vivax occur near genes involved in transcriptional regulation. These findings highlight important differences between P. vivax and P. falciparum biology that are relevant to the direction of future malaria elimination efforts. P. vivax elimination will require a deeper understanding of the ways in which this species exerts transcriptional control. A clearer picture of P. vivax adaptive mechanisms will guide drug and vaccine strategies that target early gametocytogenesis, hypnozoite biology, and the next generation of drug resistance.
Materials and Methods
Sample Collection.
Clinical isolates were collected between 2009 and 2013 by the Armed Forces Research Institute of Medical Sciences in three Cambodian provinces, Oddar Meanchey, Battâmbâng, and Kâmpôt (Fig. S1). Uncomplicated P. vivax or P. falciparum malaria patients presenting to study-site clinics gave written informed consent for their participation in this study. Study staff collected and leukodepleted venous blood and administered treatment in accordance with Cambodian National Malaria Control Program guidelines. Molecular studies were approved by the Institutional Review Board at the University of North Carolina, the Walter Reed Army Institute of Research Institutional Review Board, and the Cambodian National Ethical Committee for Health Research. Additional details of the participants are included in SI Materials and Methods, SI Participant Characteristics and Table S8.
Table S8.
Baseline characteristics of subjects who donated isolates for this study
| P. falciparum median (N)* | P. vivax median (N)* | P value | |
| Age | 26.5 (80) | 27 (70) | 0.45 |
| Female sex (%) | 6.3 (80) | 15.7 (70) | 0.07 |
| Parasite density (per microliter) | 31,937 (80) | 7,401 (70) | –† |
| Days of illness | 3 (78) | 3 (70) | 0.11 |
| Fever, ≥38 °C (%) | 70 (80) | 75 (69) | 0.46 |
| Hematocrit | 42.0 (73) | 41.0 (70) | 0.81 |
| Mixed infection (%) | 6.3 (80) | 0 (70) | – |
N is the number of isolates contributing data.
Comparison not performed, because P. vivax and P. falciparum are known to cause different levels of parasitemia.
P. vivax and P. falciparum Sample Sequencing.
For P. vivax, whole blood was leukodepleted using Plasmodipur filters (Euro-Diagnostica). The ratio of parasite DNA to host DNA was determined using a quantitative PCR (qPCR) assay, and isolates with ≥20% P. vivax DNA were sequenced (82). Clinical isolates with high plasmodium:human DNA content were sequenced on the HiSeq 2000 or HiSeq 2500 sequencing system (Illumina) using 100- or 125-bp paired-end chemistry. Data are available at the Sequence Read Archive, and accession numbers are listed in Table S9. For P. falciparum, data from previously reported isolates as well as from two previously unidentified isolates were used and reanalyzed in this study to allow comparable analysis methods between species.
Table S9.
Sequence Read Archive accession numbers
| Species | Province | SRA accession nos. |
| P. vivax | Battâmbâng | SRR2315729, SRR2315849, SRR2316038, SRR2316105, SRR2316478, SRR2316872, SRR2316895, SRR2317109, SRR2317489 |
| P. vivax | Kâmpôt | SRR2317560, SRR2315958, SRR2315988, SRR2316010, SRR2316011, SRR2316013, SRR2316015, SRR2316017, SRR2316032 |
| P. vivax | Oddar Meanchey | SRR2316034, SRR2316036, SRR2316041, SRR2316044, SRR2316048, SRR2316051, SRR2316083, SRR2316087, SRR2316091, SRR2316095, SRR2316098, SRR2316101, SRR2316108, SRR2316111, SRR2316115, SRR2316117, SRR2316172, SRR2316252, SRR2316299, SRR2316344, SRR2316398, SRR2316422, SRR2316531, SRR2316564, SRR2316625, SRR2316658, SRR2316724, SRR2316755, SRR2316807, SRR2316839, SRR2316865, SRR2316867, SRR2316875, SRR2316879, SRR2316883, SRR2316887, SRR2316889, SRR2316890, SRR2316891, SRR2316892, SRR2316893, SRR2316894, SRR2316920, SRR2316921, SRR2316922, SRR2316923, SRR2316924, SRR2316925, SRR2316926, SRR2316970, SRR2317000, SRR2317001, SRR2317142, SRR2317171, SRR2317223, SRR2317268, SRR2317321, SRR2317322, SRR2317410, SRR2317444 |
| P. falciparum | Battâmbâng | SRR2317584, SRR2317585, SRR2317700, SRR2317711, SRR2317722, SRR2318021, SRR2318061, SRR2318484, SRR2318682, SRR2318694 |
| P. falciparum | Kâmpôt | SRR2317586, SRR2317587, SRR2317692, SRR2317693, SRR2317694, SRR2317695, SRR2317696, SRR2317697, SRR2317698 |
| P. falciparum | Oddar Meanchey | SRR2317699, SRR2317701, SRR2317702, SRR2317703, SRR2317704, SRR2317705, SRR2317706, SRR2317707, SRR2317708, SRR2317709, SRR2317710, SRR2317712, SRR2317713, SRR2317714, SRR2317715, SRR2317716, SRR2317717, SRR2317718, SRR2317719, SRR2317720, SRR2317721, SRR2317723, SRR2317724, SRR2317725, SRR2317726, SRR2317727, SRR2317728, SRR2317729, SRR2317730, SRR2318019, SRR2318020, SRR2318023, SRR2318024, SRR2318031, SRR2318033, SRR2318034, SRR2318035, SRR2318039, SRR2318040, SRR2318041, SRR2318056, SRR2318126, SRR2318130, SRR2318177, SRR2318219, SRR2318297, SRR2318319, SRR2318368, SRR2318415, SRR2318446, SRR2318479, SRR2318576, SRR2318622, SRR2318670, SRR2318675, SRR2318676, SRR2318677, SRR2318678, SRR2318679, SRR2318680, SRR2318681, SRR2318683, SRR2318684, SRR2318685, SRR2318686, SRR2318688, SRR2318689, SRR2318690, SRR2318691, SRR2318692, SRR2318693, SRR2318702, SRR2318703, SRR2318704 |
Accessions are classified according to species (P. vivax or P. falciparum) and province of collection (Battâmbâng, Kâmpôt, or Oddar Meanchey).
Sequence Analysis.
Sequence reads were aligned to the P. falciparum 3D7 (v3) and P. vivax Sal1 (v3) genomes using bwa mem, which allows a hybrid end-to-end and local alignment approach (83). To increase sensitivity through hypervariable regions, we raised the base-match bonus (A = 2) and the clip penalty for local alignment (L = 15). PCR and optical duplicates were removed from alignments using the Picard Tools MarkDuplicates utility, and local realignment of highly entropic regions was performed using the GATK IndelRealigner utility (broadinstitute.github.io/picard/). Isolates with fivefold coverage at ≥80% of the genome and ≥90% of genes in the case of P. vivax and with ≥60% of the genome and ≥90% of genes in the case of P. falciparum were considered for variant calling and further analyses.
Variant calling for both species is described in SI Materials and Methods, SI Sequencing Sympatric P. vivax and P. falciparum Populations. Variants were called for each species independently but jointly for all samples using the GATK UnifiedGenotyper (84, 85). Additional details of sequencing and bioinformatic analysis are provided in SI Materials and Methods, SI Sequencing Sympatric P. vivax and P. falciparum Populations.
Within-Host Diversity.
To test whether infections were monoclonal or polyclonal, we used the FWS statistic (86, 87). To enable direct FWS comparisons between P. falciparum and P. vivax isolates, which had different numbers of loci, we bootstrapped each calculation 1,000 times, randomly selecting 5,000 variable sites for each isolate and each replicate. The maximum and minimum FWS bootstrap values were identified to provide a generous upper and lower confidence interval for each FWS point estimate. Additionally, P. vivax isolates were screened for MOI using ultra-deep sequencing of the highly polymorphic pvmsp1 locus (60). This screening was performed using the Ion Torrent platform, and the reads for each individual were clustered using SeekDeep, an iterative clustering algorithm (baileylab.umassmed.edu/SeekDeep/). Experimental procedures were performed as previously described (60). For the purposes of the present study, a sample was deemed multiclonal if a minor clone existed at ≥10% read frequency.
Population Structure and Demographic Inferences.
We determined population substructuring using PCA, and cluster assignments were determined using a nonparametric k-means approach. PCA was calculated using adegenet (19). Five one-population demographic scenarios were fit to the observed site-frequency spectra at synonymous sites in both the P. vivax and P. falciparum populations (20). For each model, 100 independent runs were performed for each dataset. Interrun parameter values were compared to assess model convergence, and the iteration with the highest log-likelihood was selected. Details pertaining to the models and parameter space explored are available in SI Materials and Methods, SI Demographic Modeling Using a Diffusion Approximation Paradigm.
Assessment of Tajima’s D.
Gene loci with extreme high and low Tajima’s D were identified for both P. vivax and P. falciparum, excluding genes from highly paralogous families and in chromosomal telomeres (80). Nonexcluded genes were simulated for corresponding Tajima’s D values using the R package coala, a wrapper for ms (88, 89). Inferred parameters for the best-fit one-population demographic scenario for the P. falciparum ancestral-like population, the entire P. vivax population, and the P. vivax monoclonal population were used to parameterize coalescent simulations. Mutation rates per gene were determined from gene length and the calculated genome mutation rate, because the mutation rate per gene considered in the ms model is proportional to gene length (26). From these simulations, we established a null distribution of Tajima’s D values for both P. vivax populations and for the P. falciparum ancestral-like population to aid in identifying genes under unexpectedly strong balancing or directional selection. Genes identified in the first and 99th percentile of the observed Tajima’s D distribution for each of the three populations were investigated with the GO analysis tool from PlasmoDB. Genes with a Bonferroni-corrected P value <0.05 were considered significant by the GO analysis.
Haplotypic Scans of Positive Selection.
Because there is as yet no fine-scale map of recombination for the P. vivax genome, we initially sought a map-independent haplotype-based approach. nSL, a modification of iHS, obviates the need for a genetic map, reduces its dependence on recombination and demographic events, and may afford increased sensitivity to detect soft selective sweeps (23). This statistic has proved sensitive in identifying selection in other nonmodel organisms (90). As a secondary test, we constructed recombination maps using LDhat interval and performed the iHS haplotype-based test for directional selection (91, 92). iHS was calculated using iHH0/iHH1, in which the subscripts 0 and 1 denote alleles, agnostic to ancestral or derived status. For both nSL and iHS, because of the lack of outgroup sequences, we discarded the sign. We plotted iHS and nSL scores that were normalized according to allele frequency bins. For selected intervals, EHH was calculated for the selected and unselected allele. These three haplotype-based tests for selection were performed using the program selscan (93).
Copy Number Analysis.
We identified CNVs using a tailored two-step approach for identifying segmental duplications using a custom search approach tuned to the AT-rich genome of Plasmodium sp. followed by a probabilistic framework for detecting variants that incorporates multiple types of evidence (SI Materials and Methods, SI CNV in P. vivax and P. falciparum). A subset of P. falciparum md1 CNVs was confirmed using qPCR as described previously (1, 94).
SI Materials and Methods
SI Participant Characteristics
Study samples were collected from patients with uncomplicated malaria between 2009 and 2013. Plasmodium vivax infections tended to be collected earlier in this period than P. falciparum infections (Fig. S1). There were no significant differences between the two groups in terms of age, sex, hematocrit, days of illness, or presence of fever (Table S9). All three study sites, Oddar Meanchey, Battâmbâng, and Kâmpôt, were located in zone 2 artemisinin-resistance containment areas, likely resulting in increased bed net use, increased diagnostic availability, and improved access to ACT (16). During this period, national drug policy dictated that P. falciparum was to be treated with artesunate-mefloquine (AS-MQ) until 2012 and then with dihydroartemisinin-piperaquine (DHA-PPQ) after 2012. Although chloroquine was the primary antimalarial used for P. vivax monoinfection during this period, concerns regarding chloroquine resistance in Cambodia led to a change to DHA-PPQ as the treatment of choice for P. vivax malaria in 2012. Some infections contained both species, a common finding in this region, likely resulting in frequent treatment of P. vivax malaria with ACT rather than with chloroquine even before 2012 (95).
SI Sequencing Sympatric P. vivax and P. falciparum Populations
We generated whole-genome short-sequence reads for 78 Cambodian P. vivax field isolates and 93 P. falciparum field isolates and aligned these sequence data to the Sal1 and 3D7 reference genomes, respectively. Of the P. vivax isolates, 70 (90%) had fivefold or greater coverage across at least 80% of the genome, and 80 (86%) of the P. falciparum isolates had fivefold or greater coverage across at least 60% of the genome. All P. vivax isolates had fivefold or greater coverage in ≥99% of coding regions, and all P. falciparum isolates had fivefold or greater coverage in ≥94% of coding regions.
Variants were filtered stringently using cutoffs responsive to the underlying distribution of quality scores: quality-by-depth ≥25, mapping quality ≥55, Fisher score (mapping quality) ≤10, map-quality rank sum equal to or greater than −5.0, read position rank sum equal to or greater than −5.0, extreme filtered depth (≤10th and ≥90th percentile), and an overall quality below the log-scaled inflection point. In addition, only biallelic variant records with at least fivefold coverage in 100% of isolates were considered. Variants in low-complexity regions were identified using tandem repeat finder and were excluded. Members of highly paralogous gene families were excluded (80, 96). We also excluded 60,000 bases of sequence space from the beginning and end of each chromosome. These masking and exclusion steps totaled ∼4 Mb of P. vivax and P. falciparum sequence space, containing ∼300 and 400 genes, respectively. These regions were primarily subtelomeric regions containing members of highly paralogous gene families. In total we identified 61,448 high-quality P. vivax SNPs and 6,734 P. falciparum SNPs. This result is consistent with previous reports that P. vivax has greater genetic diversity than sympatric P. falciparum (80). Pipeline management was facilitated by snakemake (97).
SI P. vivax Infections Have Higher Within-Host Diversity than Sympatric P. falciparum Infections
To confirm the extreme polyclonality predicted for P. vivax by FWS, we deep sequenced the hypervariable region of the 42-kDa domain of pvmsp1 (identified in ref. 18 and used in ref. 60). Amplicon deep sequencing confirmed the highly multiclonal nature of P. vivax infections: Of the 47 (out of 52) P. vivax isolates from Oddar Meanchey province that were deep sequenced, 24 (51%) were considered multiclonal based on a minor allele frequency of ≥10%, similar to a previous study (61). These results had high concordance with FWS results: 17 of 21 isolates with an FWS ≥0.95 were monoclonal by pvmsp1 sequencing, whereas 19 of 26 isolates with an FWS <0.95 were multiclonal (Fisher’s exact test P = 0.0004).
The above approach provided reliable groupings for the purpose of whole-genome analyses. However, to define the extent of multiclonality in these samples more sensitively, we assessed MOI using more sensitive cutoffs, which we previously defined for P. vivax infections in Cambodia (60). Applying a 0.5% detection cutoff to all samples with ≥1,000-fold coverage in two replicates, our samples had a mean MOI of 3.6 (range 1–15), nearly identical to our previous findings in a contemporaneous Cambodian P. vivax cohort (60).
SI Demographic Modeling Using a Diffusion Approximation Paradigm
Five one-population demographic scenarios were fit to the observed site-frequency spectra at synonymous sites in both the P. vivax and P. falciparum populations (20). For P. vivax, we fit models against the allele-frequency spectra derived from our entire population sample (n = 70) and also against the spectra derived from the subset of monoclonal infections (n = 28) to test the consequences of including polyclonal samples in analysis. P. falciparum demographic analysis was performed on isolates of the central (i.e., ancestral) population to facilitate direct comparisons with one-population P. vivax demographic tests. First, a model of constant population size was fit, in which there is no increase or decline in the Neff over time. Second, a model of population decline was fit, in which the Neff began to decrease at some time, T, in the past. Third and fourth, exponential and two-epoch growth models were fit, in which the Neff either began exponential growth or experienced a sudden change in size at T. Finally, a model of sudden decline (“bottleneck”) followed by exponential growth was fit. For each model, as applicable, the following parameter space was explored: time (T, 0.01–5.0), the factor of population contraction (ηD, 0.001–1.0) and expansion (ηG, 1–100), and the ancestral mutation rate (θ). Our sample sizes allowed robust detection of ancient demographic events and reasonable sensitivity for recent demographic events (98).
SI Assessment of Tajima’s D
We investigated the genomic signatures of directional and balancing selection in coendemic P. vivax and P. falciparum by calculating Tajima’s D for every analyzable not-masked gene, totaling 4,909 P. vivax genes and 5,293 P. falciparum genes. Because strong local values of Tajima’s D can be missed when considering an entire gene, we also performed these analyses in an exonwise manner (18). In our data, there were 13,034 analyzable P. vivax exons and 13,951 analyzable P. falciparum exons. We selected the first and 99th percentile of observed genewise and exonwise Tajima’s D values and investigated these for functional enrichment.
Because Tajima’s D is sensitive to demographic history, we used the best-fit demographic models, determined as described above, to parameterize genewise coalescent simulations. Inferred values for the best-fit models were used to parameterize coalescent simulations for the 4,909 P. vivax genes and 5,293 P. falciparum genes that were not masked during alignment or variant calling. A coalescent simulation was performed for each gene, adjusting for gene length, as part of the P. vivax monoclonal population sample (n = 28), the entire P. vivax population sample (n = 70), and the ancestral P. falciparum population sample (n = 18). Finally, Tajima’s D was calculated for each gene in each simulation, producing simulated distributions of Tajima’s D. Simulated D values were compared with observed D values and demonstrated good concordance, providing evidence that the selected models and inferred parameters used indeed reflect the population history of P. vivax and P. falciparum (Fig. S5). Notably, we saw excellent concordance between simulated and observed D distributions for P. falciparum, with less perfect concordance for P. vivax. This finding may be attributable to lifecycle differences between P. vivax and P. falciparum. Specifically, P. vivax, with its latent hypnozoite stage, may violate assumptions of nonoverlapping generations. Work is ongoing to determine the unique effects of Plasmodia lifecycles on these species’ allele-frequency spectra (79, 99). Another explanation for this finding could be that our inference and modeling methods failed to capture the full extent of the dramatic and rapid growth experienced by the P. vivax population in recent history.
SI CNV in P. vivax and P. falciparum
To identify CNVs in these genome data, we used a two-step approach: (i) a custom search algorithm that accommodates the high AT content of Plasmodium species and (ii) a probabilistic framework for detecting variants that incorporates multiple types of evidence (94). Briefly, our custom search algorithm entailed (i) filtering out simple tandem repeats and transposable elements, (ii) realigning the repeat-free genome against itself to identify paralogous regions, (iii) unmasking high-copy repeats, and (iv) refining CNV alignment boundaries. To validate our CNV-calling approach, we performed qPCR on 54 of the whole-genome–sequenced P. falciparum isolates to determine copy number at pfmdr1, a known CNV locus in Cambodia. We identified seven isolates with a segmental duplication encompassing this locus. In comparison with experimental results, we found that both genome-wide CNV-detection methods identified five of these seven isolates and detected no false positives, demonstrating reasonable sensitivity and good specificity.
We identified limited CNV in our 70 P. vivax genomes (Table S7). Two loci were found to have multiple copies. Sixteen isolates had a 2× duplication encompassing the Duffy-binding protein (dbp; PVX_110810), which previously was described in P. vivax isolates worldwide (81, 100). However, the inferred breakpoints in our samples are several hundred base pairs removed from those described by Menard et al. (100), providing evidence for multiple pvdbp duplication events. We also discovered one P. vivax isolate that contained a 4× pvdbp duplication event. Additionally, we identified 13 isolates with a deletion that includes the two 5′-most exons of the cytoadherence-linked asexual protein (clag; PVX_094265) as well as an exported protein of unknown function (PVX_094270). Deletions of clag are common in cultured P. falciparum strains (101). Notably, the predicted breakpoints for both the dbp- and the clag-encompassing CNVs were nearly identical in all isolates, providing evidence that either (i) these specific genomic positions are prone to duplication events or (ii) a single event occurred and expanded through the population. There was no evidence of segmental duplications occurring near selective sweeps in the P. vivax population. Finally, in accordance with another study, we did not find any evidence of CNVs encompassing the pvmdr1 locus in Cambodian isolates (25).
Similarly, we identified limited copy-number variation among the 80 P. falciparum genomes (Table S7). These CNVs included segmental duplications spanning the pfmdr1 locus: 12 such events were detected, including five 2× duplications, five 3× duplications, and two 4× duplications. Beyond pfmdr1, we identified high-confidence duplication events encompassing the plasmepsin II gene (PF3D7_1408000, chromosome 14). For plasmepsin II, 32 samples had a duplication event, including 16 2× duplications and 16 3× duplications. In contrast to P. vivax duplication events, analysis of predicted event breakpoints for P. falciparum provided evidence of multiple-origin events. As in the most comprehensive P. falciparum CNV analysis to date (33), we found no evidence of selective sweeps around CNVs.
Segmental duplications in Plasmodia species can be associated with increased fitness in certain settings (e.g., the P. falciparum multidrug resistance transporter), and studies of copy-number changes have proved instrumental in uncovering unique P. vivax biology (81, 100). A striking finding was the highly duplicated nature of pvdbp in this Cambodian population. A conserved segmental duplication encompassing the pvdbp locus was previously described in a handful of globally sourced P. vivax isolates, suggesting a recent duplication event and rapid sweep, perhaps because it conferred an advantage in RBC invasion. That the pvdbp duplication is so common in our sample (17 of 70 isolates, including a single 4× duplication) suggests an important mechanism that may transcend invasion of Duffy-negative human hosts.
Acknowledgments
We thank the study participants and Kristina De Paris, Corbin Jones, and Praveen Sethupathy for review of the manuscript. This research was supported by NIH Grants R01AI089819 and R21AI111108 (to J.J.J.) and R01AI099473 (to J.A.B.). C.M.P. was supported by NIH Training Grants T32GM007092, T32GM008719, and F30AI109979. J.T.L. was supported by NIH Grant K08AI110651. The views expressed in this presentation are those of the authors and do not reflect official policy of the Department of the Army, Department of Defense, or the United States Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA) database. For a list of SRA accession numbers, see Table S9.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608828113/-/DCSupplemental.
References
- 1.Spring MD, et al. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: An observational cohort study. Lancet Infect Dis. 2015;15(6):683–691. doi: 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- 2.Dondorp AM, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361(5):455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou G, et al. Spatio-temporal distribution of Plasmodium falciparum and p. Vivax malaria in Thailand. Am J Trop Med Hyg. 2005;72(3):256–262. [PubMed] [Google Scholar]
- 4.Cui L, et al. Malaria in the Greater Mekong subregion: Heterogeneity and complexity. Acta Trop. 2012;121(3):227–239. doi: 10.1016/j.actatropica.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wangroongsarb P, Sudathip P, Satimai W. Characteristics and malaria prevalence of migrant populations in malaria-endemic areas along the Thai-Cambodian border. Southeast Asian J Trop Med Public Health. 2012;43(2):261–269. [PubMed] [Google Scholar]
- 6.Maude RJ, et al. Spatial and temporal epidemiology of clinical malaria in Cambodia 2004-2013. Malar J. 2014;13:385. doi: 10.1186/1475-2875-13-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization . Control and Elimination of Plasmodium vivax Malaria – A Technical Brief. WHO; Geneva: 2015. [Google Scholar]
- 8.Ould Ahmedou Salem MS, et al. Increasing prevalence of Plasmodium vivax among febrile patients in Nouakchott, Mauritania. Am J Trop Med Hyg. 2015;92(3):537–540. doi: 10.4269/ajtmh.14-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vitor-Silva S, et al. Declining malaria transmission in rural Amazon: Changing epidemiology and challenges to achieve elimination. Malar J. 2016;15(1):266. doi: 10.1186/s12936-016-1326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray K-A, et al. Population genetics of Plasmodium falciparum and Plasmodium vivax and asymptomatic malaria in Temotu Province, Solomon Islands. Malar J. 2013;12:429. doi: 10.1186/1475-2875-12-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jennison C, et al. Plasmodium vivax populations are more genetically diverse and less structured than sympatric Plasmodium falciparum populations. PLoS Negl Trop Dis. 2015;9(4):e0003634. doi: 10.1371/journal.pntd.0003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noviyanti R, et al. Contrasting transmission dynamics of co-endemic Plasmodium vivax and P. falciparum: Implications for malaria control and elimination. PLoS Negl Trop Dis. 2015;9(5):e0003739. doi: 10.1371/journal.pntd.0003739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orjuela-Sánchez P, et al. Higher microsatellite diversity in Plasmodium vivax than in sympatric Plasmodium falciparum populations in Pursat, Western Cambodia. Exp Parasitol. 2013;134(3):318–326. doi: 10.1016/j.exppara.2013.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ord RL, Tami A, Sutherland CJ. ama1 genes of sympatric Plasmodium vivax and P. falciparum from Venezuela differ significantly in genetic diversity and recombination frequency. PLoS One. 2008;3(10):e3366. doi: 10.1371/journal.pone.0003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnott A, et al. Distinct patterns of diversity, population structure and evolution in the AMA1 genes of sympatric Plasmodium falciparum and Plasmodium vivax populations of Papua New Guinea from an area of similarly high transmission. Malar J. 2014;13(1):233. doi: 10.1186/1475-2875-13-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization . Global Plan for Artemisinin Resistance Containment. WHO; Geneva: 2011. [Google Scholar]
- 17.Murray L, et al. Microsatellite genotyping and genome-wide single nucleotide polymorphism-based indices of Plasmodium falciparum diversity within clinical infections. Malar J. 2016;15(1):275. doi: 10.1186/s12936-016-1324-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parobek CM, et al. Differing patterns of selection and geospatial genetic diversity within two leading Plasmodium vivax candidate vaccine antigens. PLoS Negl Trop Dis. 2014;8(4):e2796. doi: 10.1371/journal.pntd.0002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jombart T, Ahmed I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27(21):3070–3071. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5(10):e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miotto O, et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet. 2013;45(6):648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miotto O, et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet. 2015;47(3):226–234. doi: 10.1038/ng.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrer-Admetlla A, Liang M, Korneliussen T, Nielsen R. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol Biol Evol. 2014;31(5):1275–1291. doi: 10.1093/molbev/msu077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Painter HJ, Campbell TL, Llinás M. The Apicomplexan AP2 family: Integral factors regulating Plasmodium development. Mol Biochem Parasitol. 2011;176(1):1–7. doi: 10.1016/j.molbiopara.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin JT, et al. Plasmodium vivax isolates from Cambodia and Thailand show high genetic complexity and distinct patterns of P. vivax multidrug resistance gene 1 (pvmdr1) polymorphisms. Am J Trop Med Hyg. 2013;88(6):1116–1123. doi: 10.4269/ajtmh.12-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang H-H, et al. Genomic sequencing of Plasmodium falciparum malaria parasites from Senegal reveals the demographic history of the population. Mol Biol Evol. 2012;29(11):3427–3439. doi: 10.1093/molbev/mss161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mu J, et al. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nat Genet. 2010;42(3):268–271. doi: 10.1038/ng.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nwakanma DC, et al. Changes in malaria parasite drug resistance in an endemic population over a 25-year period with resulting genomic evidence of selection. J Infect Dis. 2014;209(7):1126–1135. doi: 10.1093/infdis/jit618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mobegi VA, et al. Genome-wide analysis of selection on the malaria parasite Plasmodium falciparum in West African populations of differing infection endemicity. Mol Biol Evol. 2014;31(6):1490–1499. doi: 10.1093/molbev/msu106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leber W, et al. A unique phosphatidylinositol 4-phosphate 5-kinase is activated by ADP-ribosylation factor in Plasmodium falciparum. Int J Parasitol. 2009;39(6):645–653. doi: 10.1016/j.ijpara.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 31.McNamara CW, et al. Targeting Plasmodium PI(4)K to eliminate malaria. Nature. 2013;504(7479):248–253. doi: 10.1038/nature12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amambua-Ngwa A, et al. SNP genotyping identifies new signatures of selection in a deep sample of West African Plasmodium falciparum malaria parasites. Mol Biol Evol. 2012;29(11):3249–3253. doi: 10.1093/molbev/mss151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheeseman IH, et al. Population structure shapes copy number variation in malaria parasites. Mol Biol Evol. 2016;33(3):603–620. doi: 10.1093/molbev/msv282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ocholla H, et al. Whole-genome scans provide evidence of adaptive evolution in Malawian Plasmodium falciparum isolates. J Infect Dis. 2014;210(12):1991–2000. doi: 10.1093/infdis/jiu349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park DJ, et al. Sequence-based association and selection scans identify drug resistance loci in the Plasmodium falciparum malaria parasite. Proc Natl Acad Sci USA. 2012;109(32):13052–13057. doi: 10.1073/pnas.1210585109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bozdech Z, et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci USA. 2008;105(42):16290–16295. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu L, et al. New insights into the Plasmodium vivax transcriptome using RNA-Seq. Sci Rep. 2016;6:20498. doi: 10.1038/srep20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoo R, et al. Integrated analysis of the Plasmodium species transcriptome. EBioMedicine. 2016;7:255–266. doi: 10.1016/j.ebiom.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bousema T, Drakeley C. Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin Microbiol Rev. 2011;24(2):377–410. doi: 10.1128/CMR.00051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuda M, Iwanaga S, Kaneko I, Kato T, Tomomi K. Global transcriptional repression: An initial and essential step for Plasmodium sexual development. Proc Natl Acad Sci USA. 2015;112(41):12824–12829. doi: 10.1073/pnas.1504389112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikadai H, et al. Transposon mutagenesis identifies genes essential for Plasmodium falciparum gametocytogenesis. Proc Natl Acad Sci USA. 2013;110(18):E1676–E1684. doi: 10.1073/pnas.1217712110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dembélé L, et al. Persistence and activation of malaria hypnozoites in long-term primary hepatocyte cultures. Nat Med. 2014;20(3):307–312. doi: 10.1038/nm.3461. [DOI] [PubMed] [Google Scholar]
- 43.Barnwell JW, Galinski MR. Malarial liver parasites awaken in culture. Nat Med. 2014;20(3):237–239. doi: 10.1038/nm.3498. [DOI] [PubMed] [Google Scholar]
- 44.Fernández-Becerra C, et al. Increased expression levels of the pvcrt-o and pvmdr1 genes in a patient with severe Plasmodium vivax malaria. Malar J. 2009;8(1):55. doi: 10.1186/1475-2875-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melo GC, et al. Expression levels of pvcrt-o and pvmdr-1 are associated with chloroquine resistance and severe Plasmodium vivax malaria in patients of the Brazilian Amazon. PLoS One. 2014;9(8):e105922. doi: 10.1371/journal.pone.0105922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pava Z, et al. Expression of Plasmodium vivax crt-o is related to parasite stage but not ex vivo chloroquine susceptibility. Antimicrob Agents Chemother. 2015;60(1):361–367. doi: 10.1128/AAC.02207-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dharia NV, et al. Whole-genome sequencing and microarray analysis of ex vivo Plasmodium vivax reveal selective pressure on putative drug resistance genes. Proc Natl Acad Sci USA. 2010;107(46):20045–20050. doi: 10.1073/pnas.1003776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kryazhimskiy S, Plotkin JB, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4(12):e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duffy CW, et al. Comparison of genomic signatures of selection on Plasmodium falciparum between different regions of a country with high malaria endemicity. BMC Genomics. 2015;16:527. doi: 10.1186/s12864-015-1746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daniels RF, et al. Modeling malaria genomics reveals transmission decline and rebound in Senegal. Proc Natl Acad Sci USA. 2015;112(22):7067–7072. doi: 10.1073/pnas.1505691112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mok S, et al. Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science. 2015;347(6220):431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeeman A-M, der Wel AV, Kocken CHM. Ex vivo culture of Plasmodium vivax and Plasmodium cynomolgi and in vitro culture of Plasmodium knowlesi blood stages. Methods Mol Biol. 2013;923:35–49. doi: 10.1007/978-1-62703-026-7_4. [DOI] [PubMed] [Google Scholar]
- 53.Vaughan AM, Kappe SHI, Ploss A, Mikolajczak SA. Development of humanized mouse models to study human malaria parasite infection. Future Microbiol. 2012;7(5):657–665. doi: 10.2217/fmb.12.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mikolajczak SA, et al. Plasmodium vivax liver stage development and hypnozoite persistence in human liver-chimeric mice. Cell Host Microbe. 17(4):526–35. doi: 10.1016/j.chom.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joyner C, Barnwell JW, Galinski MR. No more monkeying around: Primate malaria model systems are key to understanding Plasmodium vivax liver-stage biology, hypnozoites, and relapses. Front Microbiol. 2015;6:145. doi: 10.3389/fmicb.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moraes Barros RR, et al. Editing the Plasmodium vivax genome, using zinc-finger nucleases. J Infect Dis. 2015;211(1):125–129. doi: 10.1093/infdis/jiu423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mok S, et al. Structural polymorphism in the promoter of pfmrp2 confers Plasmodium falciparum tolerance to quinoline drugs. Mol Microbiol. 2014;91(5):918–934. doi: 10.1111/mmi.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonzales JM, et al. Regulatory hotspots in the malaria parasite genome dictate transcriptional variation. PLoS Biol. 2008;6(9):e238. doi: 10.1371/journal.pbio.0060238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mideo N, et al. A deep sequencing tool for partitioning clearance rates following antimalarial treatment in polyclonal infections. Evol Med Public Health. 2016;2016(1):21–36. doi: 10.1093/emph/eov036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin JT, et al. Using amplicon deep sequencing to detect genetic signatures of Plasmodium vivax relapse. J Infect Dis. 2015;212(6):999–1008. doi: 10.1093/infdis/jiv142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Friedrich LR, et al. Complexity of infection and genetic diversity in Cambodian Plasmodium vivax. PLoS Negl Trop Dis. 2016;10(3):e0004526. doi: 10.1371/journal.pntd.0004526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nkhoma SC, et al. Population genetic correlates of declining transmission in a human pathogen. Mol Ecol. 2013;22(2):273–285. doi: 10.1111/mec.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hupalo DN, et al. Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet. 2016;48(8):953–958. doi: 10.1038/ng.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Molina-Cruz A, et al. The human malaria parasite Pfs47 gene mediates evasion of the mosquito immune system. Science. 2013;340(6135):984–987. doi: 10.1126/science.1235264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Durnez L, et al. Outdoor malaria transmission in forested villages of Cambodia. Malar J. 2013;12:329. doi: 10.1186/1475-2875-12-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinka ME, et al. The dominant Anopheles vectors of human malaria in the Asia-Pacific region: Occurrence data, distribution maps and bionomic précis. Parasit Vectors. 2011;4(1):89. doi: 10.1186/1756-3305-4-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson TJ, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17(10):1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 68.Leclerc MC, et al. Meager genetic variability of the human malaria agent Plasmodium vivax. Proc Natl Acad Sci USA. 2004;101(40):14455–14460. doi: 10.1073/pnas.0405186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lim CS, Tazi L, Ayala FJ. Plasmodium vivax: Recent world expansion and genetic identity to Plasmodium simium. Proc Natl Acad Sci USA. 2005;102(43):15523–15528. doi: 10.1073/pnas.0507413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prugnolle F, et al. Diversity, host switching and evolution of Plasmodium vivax infecting African great apes. Proc Natl Acad Sci USA. 2013;110(20):8123–8128. doi: 10.1073/pnas.1306004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu W, et al. African origin of the malaria parasite Plasmodium vivax. Nat Commun. 2014;5:3346. doi: 10.1038/ncomms4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sundararaman SA, et al. Genomes of cryptic chimpanzee Plasmodium species reveal key evolutionary events leading to human malaria. Nat Commun. 2016;7:11078. doi: 10.1038/ncomms11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nair S, et al. A selective sweep driven by pyrimethamine treatment in southeast asian malaria parasites. Mol Biol Evol. 2003;20(9):1526–1536. doi: 10.1093/molbev/msg162. [DOI] [PubMed] [Google Scholar]
- 74.Volkman SK, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39(1):113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 75.Snounou G, et al. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93(4):369–374. doi: 10.1016/s0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- 76.Paul RE, et al. Transmission intensity and Plasmodium falciparum diversity on the northwestern border of Thailand. Am J Trop Med Hyg. 1998;58(2):195–203. doi: 10.4269/ajtmh.1998.58.195. [DOI] [PubMed] [Google Scholar]
- 77.Jongwutiwes S, Putaporntip C, Hughes AL. Bottleneck effects on vaccine-candidate antigen diversity of malaria parasites in Thailand. Vaccine. 2010;28(18):3112–3117. doi: 10.1016/j.vaccine.2010.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith RC, Vega-Rodríguez J, Jacobs-Lorena M. The Plasmodium bottleneck: Malaria parasite losses in the mosquito vector. Mem Inst Oswaldo Cruz. 2014;109(5):644–661. doi: 10.1590/0074-0276130597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chang H-H, Hartl DL. Recurrent bottlenecks in the malaria life cycle obscure signals of positive selection. Parasitology. 2015;142(Suppl 1):S98–S107. doi: 10.1017/S0031182014000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Neafsey DE, et al. The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet. 2012;44(9):1046–1050. doi: 10.1038/ng.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hester J, et al. De novo assembly of a field isolate genome reveals novel Plasmodium vivax erythrocyte invasion genes. PLoS Negl Trop Dis. 2013;7(12):e2569. doi: 10.1371/journal.pntd.0002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beshir KB, et al. Measuring the efficacy of anti-malarial drugs in vivo: Quantitative PCR measurement of parasite clearance. Malar J. 2010;9:312. doi: 10.1186/1475-2875-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li H. 2013 Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv.org. Available at https://arxiv.org/abs/1303.3997. Accessed September 30, 2016.
- 84.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McKenna A, et al. The Genome Analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Auburn S, et al. Characterization of within-host Plasmodium falciparum diversity using next-generation sequence data. PLoS One. 2012;7(2):e32891. doi: 10.1371/journal.pone.0032891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Manske M, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487(7407):375–379. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Staab PR, Metzler D. Coala: An R framework for coalescent simulation. Bioinformatics. 2016;32(12):1903–1904. doi: 10.1093/bioinformatics/btw098. [DOI] [PubMed] [Google Scholar]
- 89.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18(2):337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 90.Schlamp F, et al. Evaluating the performance of selection scans to detect selective sweeps in domestic dogs. Mol Ecol. 2016;25(1):342–356. doi: 10.1111/mec.13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160(3):1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Szpiech ZA, Hernandez RD. selscan: An efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol. 2014;31(10):2824–2827. doi: 10.1093/molbev/msu211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: A probabilistic framework for structural variant discovery. Genome Biol. 2014;15(6):R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zimmerman PA, Mehlotra RK, Kasehagen LJ, Kazura JW. Why do we need to know more about mixed Plasmodium species infections in humans? Trends Parasitol. 2004;20(9):440–447. doi: 10.1016/j.pt.2004.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Köster J, Rahmann S. Snakemake--a scalable bioinformatics workflow engine. Bioinformatics. 2012;28(19):2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- 98.Robinson JD, Coffman AJ, Hickerson MJ, Gutenkunst RN. Sampling strategies for frequency spectrum-based population genomic inference. BMC Evol Biol. 2014;14:254. doi: 10.1186/s12862-014-0254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chang H-H, et al. Malaria life cycle intensifies both natural selection and random genetic drift. Proc Natl Acad Sci USA. 2013;110(50):20129–20134. doi: 10.1073/pnas.1319857110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Menard D, et al. Whole genome sequencing of field isolates reveals a common duplication of the Duffy binding protein gene in Malagasy Plasmodium vivax strains. PLoS Negl Trop Dis. 2013;7(11):e2489. doi: 10.1371/journal.pntd.0002489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Trenholme KR, et al. clag9: A cytoadherence gene in Plasmodium falciparum essential for binding of parasitized erythrocytes to CD36. Proc Natl Acad Sci USA. 2000;97(8):4029–4033. doi: 10.1073/pnas.040561197. [DOI] [PMC free article] [PubMed] [Google Scholar]