Significance
The progressive nature of neurodegenerative diseases is due to the spread of prions, misfolded infectious proteins, in the brain. In tauopathies, the protein tau misfolds, causing several diseases, including Alzheimer’s disease (AD) and chronic traumatic encephalopathy (CTE). Here we created a panel of mammalian cell lines expressing a fragment of tau fused to yellow fluorescent protein. Each cell line selectively detects tau prions that are misfolded into self-propagating conformations; such cells permit identification of minute differences among tauopathies. For example, tau prions in AD and CTE are distinct from prions in other tauopathies such as Pick’s disease and progressive supranuclear palsy. These insights are likely to contribute to the development of future therapeutics.
Keywords: argyrophilic grain disease, corticobasal degeneration, Pick’s disease, progressive supranuclear palsy, tauopathies
Abstract
Tau prions are thought to aggregate in the central nervous system, resulting in neurodegeneration. Among the tauopathies, Alzheimer’s disease (AD) is the most common, whereas argyrophilic grain disease (AGD), corticobasal degeneration (CBD), chronic traumatic encephalopathy (CTE), Pick’s disease (PiD), and progressive supranuclear palsy (PSP) are less prevalent. Brain extracts from deceased individuals with PiD, a neurodegenerative disorder characterized by three-repeat (3R) tau prions, were used to infect HEK293T cells expressing 3R tau fused to yellow fluorescent protein (YFP). Extracts from AGD, CBD, and PSP patient samples, which contain four-repeat (4R) tau prions, were transmitted to HEK293 cells expressing 4R tau fused to YFP. These studies demonstrated that prion propagation in HEK cells requires isoform pairing between the infecting prion and the recipient substrate. Interestingly, tau aggregates in AD and CTE, containing both 3R and 4R isoforms, were unable to robustly infect either 3R- or 4R-expressing cells. However, AD and CTE prions were able to replicate in HEK293T cells expressing both 3R and 4R tau. Unexpectedly, increasing the level of 4R isoform expression alone supported the propagation of both AD and CTE prions. These results allowed us to determine the levels of tau prions in AD and CTE brain extracts.
In the central nervous system (CNS), the soluble and unstructured protein tau binds to microtubules to stabilize and promote their polymerization in neurons (1). However, in the mid-1980s, brain samples from deceased Alzheimer’s disease (AD) patients containing neurofibrillary tangles (NFTs) were found to immunostain with antisera raised against tau proteins, indicating tau may play a deleterious role in neurodegeneration (2–5). Later, tau was identified in partially purified preparations from the brains of AD patients (6), and the resulting NFTs were subsequently correlated with cognitive impairment (7–9).
In addition to AD, tau was also linked to frontotemporal lobar degenerative diseases (FTLDs), including argyrophilic grain disease (AGD), corticobasal degeneration (CBD), Pick’s disease (PiD), and progressive supranuclear palsy (PSP), following the identification of mutations in patients with inherited forms of FTLD (10, 11). Although the identification of autosomal-dominant mutations in FTLD patients was pivotal in linking tau to FTLDs, the majority of these cases are sporadic, similar to AD (12). The presence of tau mutations in FTLD patients strikes a glaring dichotomy with familial AD (fAD) patients, where tau is not mutated; instead, mutations in the gene encoding amyloid precursor protein (APP), or the enzymes that cleave APP to generate β-amyloid (Aβ), have been identified (13). The difference in which gene is mutated results in the classification of FTLDs as primary tauopathies and AD as a secondary tauopathy, due to the additional presence of Aβ plaques in the brains of patients.
In FTLDs, tau acquires a β-sheet–rich structure that polymerizes into amyloid fibrils. Like other prions, tau prions multiply through a process of self-propagation, where the β-sheet acts as a template for the formation of nascent prions. As described here, different strains of tau prions are predicted to have unique conformations of misfolding that determine distinct tauopathies with different patterns of neuropathological lesions. These strain-dependent neuropathologies, including NFTs (2), Pick bodies, and globose tangles (14), are thought to arise from the progressive self-propagation of tau, spreading from one neuron to another, ultimately leading to widespread neurodegeneration.
The finding that tau, a single protein, gives rise to a wide array of neurological disorders arose from immunostaining studies conducted in conjunction with molecular-cloning investigations. Antibodies raised against the tau protein sequence demonstrated that diseases once thought to be unrelated, such as PiD and PSP, were in fact caused by the same protein (2–5, 14). Additionally, molecular cloning of tau cDNA resulted in the discovery that tau is expressed as six different isoforms (15, 16). These isoforms arise from alternative splicing of mRNA transcribed from the tau gene, microtubule-associated protein tau (MAPT), and are composed of two variable regions. In the N-terminal region, zero, one, or two insertions (0N, 1N, or 2N) arise from alternative splicing of exons 2 and 3, whereas in the C-terminal repeat domain (RD), exclusion or inclusion of exon 10 gives rise to either three repeats (3R) or four repeats (4R), respectively (17). As such, tau isoforms are identified by the number of inclusions present in the two regions (1N3R versus 2N4R, for example).
With this knowledge, a method emerged for categorizing the various tauopathies based on the tau isoforms present in the brains of patients who suffered from CNS dysfunction. For example, the Pick bodies seen in PiD patients typically consist of 3R tau isoforms (18), but the globose tangles seen in PSP and the astrocytic plaques seen in CBD are made up of 4R aggregates (19–21). Interestingly, although tau expression in a healthy adult brain is made up of an approximately equimolar ratio of the 3R and 4R isoforms, disease-promoting mutations first identified in 1998 were found to affect the splicing of tau mRNA, resulting in increased expression of the 4R tau isoforms over the 3R tau isoforms (22, 23). In addition to the 3R- and 4R-specific tauopathies, the combined tauopathies, AD and chronic traumatic encephalopathy (CTE), exhibit aggregation of both 3R and 4R tau isoforms (16, 22, 24).
The prion protein (PrP) was the first protein discovered to cause disease by protein-induced misfolding (25). Subsequent studies showed that distinct diseases caused by aggregation of the misfolded PrP scrapie conformation (PrPSc; as opposed to the cellular, soluble PrPC) were attributable to differences in prion strains (26, 27). For example, Creutzfeldt–Jakob disease (CJD) is thought to be the result of PrPSc misfolding into one conformation, whereas fatal familial insomnia (FFI) arises from PrPSc misfolding into a different conformation. This hypothesis is supported by the observation that a mutation at codon 178 in PrP, which results in an asparagine replacing an aspartic acid, will cause a patient to develop either CJD or FFI. A second polymorphism in the protein at codon 129 controls the difference between the PrPSc strains present in each disease, and thus the disease a patient will develop (28). Additional evidence comes from passaging studies demonstrating that transgenic (Tg) mice inoculated with brain homogenate from either CJD or FFI patients develop distinct PrPSc strains with unique properties (29). Similarly, it is posited that the unique diseases that arise from tau prions are also a result of differences between prion strains (30), but the factors that give rise to these differences are poorly understood.
Whereas categorizing tauopathies as 3R, 4R, or a combination of the two has been largely a descriptive exercise over the past two decades, we proposed that each tau isoform features in one or more specific tau prion strains. Previously, we and others used HEK293 cells expressing the RD of 4R tau, fused to yellow fluorescent protein (YFP), with the two familial mutations P301L and V337M [originally named TauRD(LM)–YFP cells but denoted here as Tau(4RD*LM)–YFP(1) cells] (Table S1) (31) to quantify tau prions isolated from PSP patient samples (32). To investigate the role of the RD in tau prion strains, we developed a panel of HEK293T cell lines expressing several variations of tau isoform fusion proteins: the RD of 3R tau, the RD of 4R tau, and the combination of both. Using tau prions isolated from several tauopathies, we found that 4R tau prions isolated from AGD, CBD, and PSP patient samples only infected cells expressing 4R tau. We also found that 3R tau prions isolated from PiD patient samples only infected cells expressing the 3R isoform. Although aggregates in the brains of AD and CTE patients are composed of both 3R and 4R tau, neither the AD nor the CTE brain extracts could infect HEK cells expressing either the 3R or 4R mutant tau fusion proteins.
Table S1.
Tau fusion protein constructs expressed in human embryonic kidney cells
| Tau expression construct | Alternative name | Repeat domains | Amino acid residues† | Mutations | Cell line | Fusion protein expression‡ |
| Tau(4RD*LM)–YFP(1) | TauRD(LM)–YFP§ | 4 | 243 to 375 | P301L,V337M (LM) | HEK293 | 1× |
| Tau(4RD*LM)–YFP(2) | 4 | 243 to 375 | P301L,V337M (LM) | HEK293T | 1.4× | |
| Tau(3RD*VM)–YFP | 3 | 243 to 274; 306 to 375 | L266V,V337M (VM) | HEK293T | n.d. | |
| Tau(3RD*VM,4RD*LM)–YFP | 3 | 243 to 274; 306 to 375 | L266V,V337M (VM) | HEK293T | n.d. | |
| 4 | 243 to 375 | P301L,V337M (LM) |
n.d., not determined.
Amino acid residues are based on the amino acid sequence of the longest tau isoform, 2N4R.
Protein expression was normalized to Tau(4RD*LM)–YFP(1) cells.
The cell line originally published in Sanders et al. (31).
To further investigate this finding, we modeled tau expression in the brains of AD and CTE patients by coexpressing the 3R and 4R fusion proteins in HEK293T cells. In the presence of both 3R and 4R tau fusion proteins, AD and CTE brain extracts templated de novo prion formation in mammalian cells. Furthermore, we also found that HEK293T cells expressing higher levels of the 4R tau fragment fused to YFP were sufficient to support propagation of AD and CTE prions. These findings demonstrate that tau prions feature in the pathogenesis of both AD and CTE. Importantly, the rapid and highly reproducible fusion protein bioassays described here have allowed us, and others, to begin unraveling the molecular pathogenesis of several neurodegenerative diseases (31, 32). Moreover, the insights gained from these assays make it possible to focus drug discovery efforts on the mechanism responsible for the propagation of these diseases, increasing the likelihood of developing effective therapeutics.
Results
Propagation of 4R Tauopathies in Cultured Cells Requires Expression of 4R Tau.
Previously, we described an enhanced cellular assay to quantify tau prions in PSP patient samples (32) based on initial studies using HEK293 cells expressing the Tau(4RD*LM)–YFP(1) fusion protein developed by Marc Diamond and colleagues. Sanders et al. (31) demonstrated that tau prions (both synthetic and patient-derived) induced protein aggregation in the cells after 12 d of incubation. We modified the Tau(4RD*LM)–YFP(1) cell assay to a 384-well-plate format and performed live-cell imaging after incubating the cells for 4 d with PSP patient samples (32). To improve the robustness of the assay and increase cell infection, we precipitated tau prions with sodium phosphotungstate (PTA), which selectively isolates aggregated proteins, including PrPSc, tau, and α-synuclein, from soluble proteins (32, 33). Adapting this automated, high-throughput approach increased the percentage of cells infected with aggregates from ∼4 to ∼61%, greatly enhancing the dynamic range of the assay.
Using this experimental design, we tested additional patient samples, including samples from control (n = 6), PiD (n = 6), AD (n = 7), CTE (n = 5), AGD (n = 2), CBD (n = 5), and PSP (n = 6) patients (Fig. 1 and Table S2). Control samples were from patients devoid of any detectable neuropathological lesions upon autopsy. Tauopathy patient samples were selected from the prototypical 3R tauopathy PiD; the 4R tauopathies AGD, CBD, and PSP; and two combined tauopathies, AD and CTE. The brain regions sampled from each patient are listed in Table S2. Western blot analysis of crude brain homogenate from 14 patient samples (2 from each patient group) using the total tau antibody, Tau12, showed tau is present in all of the samples tested, including the control patient samples (Fig. 1A).
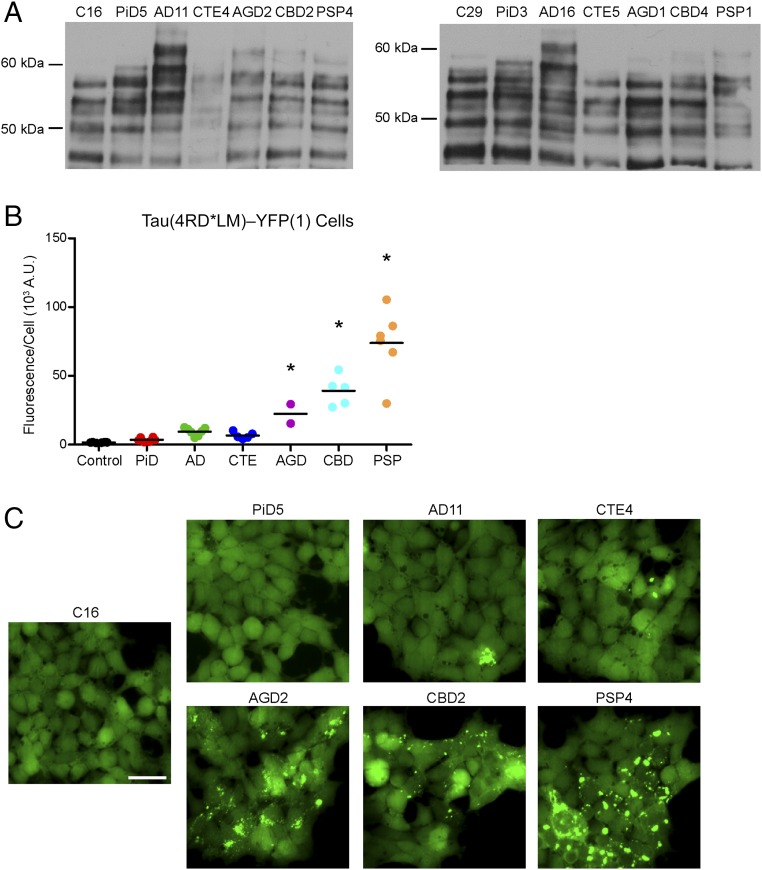
Fig. 1.
4R tau prions propagate in Tau(4RD*LM)–YFP(1) cells. (A) Crude brain homogenate from 14 patient samples (n = 2 for control, PiD, AD, CTE, AGD, CBD, and PSP) was analyzed for the presence of total tau by Western blot with the Tau12 antibody. (B and C) Tau prions were isolated from human brain homogenates by precipitating protein aggregates with sodium PTA. Protein aggregates were then incubated for 4 d with Tau(4RD*LM)–YFP(1) cells, which express the RD of 4R tau containing the mutations P301L and V337M. This protein fragment is fused to YFP and is expressed in HEK293 cells. (B) Quantification of cell infection using control (n = 6), PiD (n = 6), AD (n = 7), CTE (n = 5), AGD (n = 2), CBD (n = 5), and PSP (n = 6) patient samples. Prions isolated from the 4R tauopathies AGD (P < 0.05), CBD (P < 0.001), and PSP (P < 0.001) showed a significant increase in infectivity over the control samples, whereas PiD (P = 0.74), AD (P = 0.17), and CTE (P = 0.41) did not. Data are shown as the mean from five images per well in six wells. *P < 0.05. All values are shown in Table S2. (C) Representative images of Tau(4RD*LM)–YFP(1) cells infected with AGD, CBD, and PSP but not control, PiD, AD, and CTE patient samples. YFP is shown in green. (Scale bar, 50 μm.)
Table S2.
Infection of tau-expressing mammalian cells with tauopathy patient samples
| Patient sample | Brain region | Mean cell infection ± SD, fluorescence per cell, 103 A.U.† | |||
| Tau(4RD*LM)–YFP(1)‡ | Tau(3RD*VM)–YFP‡ | Tau(3RD*VM,4RD*LM)–YFP‡ | Tau(4RD*LM)–YFP(2)§ | ||
| C9 | Putamen | 2.0 ± 1.4 | 3.4 ± 1.8 | 6.2 ± 1.6 | 1.0 ± 0.4 |
| C10 | Putamen | 1.6 ± 0.3 | 1.8 ± 0.6 | 4.2 ± 1.7 | 2.5 ± 2.1 |
| C16 | Putamen | 1.3 ± 0.5 | 3.3 ± 1.2 | 4.9 ± 2.3 | 1.4 ± 1.2 |
| C17 | Putamen | 1.7 ± 0.5 | 5.0 ± 4.2 | 4.9 ± 2.4 | 1.2 ± 0.6 |
| C29 | Middle temporal gyrus | 1.8 ± 0.4 | 3.2 ± 2.0 | 6.8 ± 3.1 | 2.9 ± 3.0 |
| C30 | Middle temporal gyrus | 1.4 ± 0.3 | 2.6 ± 0.9 | 4.8 ± 2.1 | 3.2 ± 1.6 |
| PiD2 | Middle frontal gyrus | 2.0 ± 0.6 | 31 ± 15 | 15 ± 5.9 | 7.8 ± 4.4 |
| PiD3 | Angular gyrus | 3.1 ± 1.2 | 26 ± 6.1 | 9.9 ± 2.7 | 25 ± 26 |
| PiD4 | Angular gyrus | 5.5 ± 2.9 | 62 ± 5.0 | 19 ± 8.1 | 61 ± 87 |
| PiD5 | Inferior frontal gyrus | 2.2 ± 1.2 | 33 ± 13 | 15 ± 5.9 | 9.0 ± 3.3 |
| PiD6 | Inferior frontal gyrus | 5.3 ± 2.4 | 73 ± 20 | 19 ± 4.9 | 8.0 ± 14 |
| PiD7 | Inferior frontal gyrus | 3.5 ± 1.1 | 25 ± 11 | 11 ± 2.5 | 13 ± 13 |
| sAD11 | Middle temporal gyrus | 9.7 ± 5.2 | 3.0 ± 1.2 | 9.7 ± 1.3 | 51 ± 45 |
| sAD12 | Middle temporal gyrus | 12 ± 4.8 | 3.4 ± 1.6 | 13 ± 3.8 | 38 ± 12 |
| sAD13 | Middle temporal gyrus | 6.4 ± 2.6 | 3.0 ± 1.0 | 19 ± 6.2 | 35 ± 12 |
| sAD14 | Middle temporal gyrus | 13 ± 3.9 | 2.7 ± 0.6 | 12 ± 2.7 | 72 ± 110 |
| sAD15 | Middle temporal gyrus | 9.1 ± 4.8 | 4.0 ± 1.3 | 18 ± 4.4 | 27 ± 16 |
| fAD7 | Middle temporal gyrus | 11 ± 3.4 | 3.2 ± 1.5 | 20 ± 4.3 | 24 ± 17 |
| fAD16 | Middle temporal gyrus | 5.1 ± 3.5 | 1.6 ± 0.6 | 14 ± 6.9 | 12 ± 7.1 |
| CTE1 | Temporal pole | 5.2 ± 2.6 | 13 ± 4.4 | 17 ± 6.4 | 12 ± 7.0 |
| CTE1 | Frontal pole | n.d. | n.d. | n.d. | 11 ± 4.8 |
| CTE2 | Temporal pole | 5.7 ± 4.7 | 9.9 ± 4.3 | 48 ± 32 | 26 ± 16 |
| CTE2 | Frontal pole | n.d. | n.d. | n.d. | 2.9 ± 3.3 |
| CTE3 | Temporal pole | 7.9 ± 4.4 | 6.9 ± 5.9 | 58 ± 35 | 79 ± 56 |
| CTE3 | Frontal pole | n.d. | n.d. | n.d. | 13 ± 6.6 |
| CTE4 | Superior parietal cortex | 10 ± 5.1 | 6.0 ± 1.4 | 79 ± 22 | 90 ± 59 |
| CTE5 | Frontal pole | 4.1 ± 1.8 | 9.3 ± 4.8 | 30 ± 15 | 41 ± 10 |
| AGD1 | Middle frontal gyrus | 15 ± 7.7 | 2.5 ± 1.0 | 27 ± 7.0 | 47 ± 16 |
| AGD2 | Middle insula | 29 ± 21 | 1.2 ± 0.6 | 23 ± 4.4 | 240 ± 110 |
| CBD1 | Angular gyrus | 54 ± 16 | 4.7 ± 1.7 | 26 ± 8.2 | 24 ± 17 |
| CBD2 | Angular gyrus | 42 ± 20 | 2.9 ± 0.84 | 37 ± 11 | 62 ± 10 |
| CBD3 | Angular gyrus | 30 ± 9.0 | 5.0 ± 2.8 | 26 ± 5.6 | 80 ± 26 |
| CBD4 | Angular gyrus | 43 ± 12 | 8.9 ± 3.0 | 47 ± 7.4 | 37 ± 19 |
| CBD5 | Angular gyrus | 27 ± 3.7 | 4.8 ± 3.1 | 26 ± 5.7 | 72 ± 51 |
| PSP1 | Sensorimotor cortex | 67 ± 20 | 13 ± 5.7 | 35 ± 5.6 | 290 ± 16 |
| PSP4 | Inferior frontal gyrus | 86 ± 15 | 4.9 ± 2.2 | 21 ± 4.8 | 110 ± 21 |
| PSP5 | Inferior frontal gyrus | 76 ± 25 | 6.9 ± 2.5 | 40 ± 11 | 94 ± 19 |
| PSP6 | Angular gyrus | 80 ± 13 | 9.1 ± 5.3 | 23 ± 5.1 | 130 ± 31 |
| PSP7 | Inferior frontal gyrus | 110 ± 22 | 13 ± 7.2 | 32 ± 9.6 | 120 ± 24 |
| PSP8 | Inferior frontal gyrus | 30 ± 8.5 | 9.1 ± 4.6 | 20 ± 9.1 | 97 ± 101 |
n.d., not determined; sAD, sporadic AD.
Measurements were made from five images per well, n = 6 wells.
Phosphotungstic acid-precipitated samples were diluted in DPBS 1:40 [Tau(4RD*LM)–YFP(1) cells], 1:10 [Tau(3RD*VM)–YFP cells], or 1:4 [Tau(3RD*VM,4R*LM)–YFP cells] before testing.
Crude brain homogenate was diluted 1:40 in DPBS before incubation with Tau(4RD*LM)–YFP(2) cells.
A 10% (wt/vol) brain homogenate from each patient sample was prepared in Dulbecco’s PBS (DPBS) before digesting the sample in 2% (vol/vol) sarkosyl and 0.5% (vol/vol) benzonase. Importantly, benzonase digests all nucleic acids in the sample, leaving only protein, which we then incubated with 2% (vol/vol) PTA overnight before pelleting by centrifugation. The resulting pellets were diluted 1:40 in DPBS and incubated with Tau(4RD*LM)–YFP(1) cells for 4 d in the presence of Lipofectamine 2000 to increase the efficiency of protein uptake. The live cells were imaged using the IN Cell Analyzer 6000, collecting DAPI and FITC images from five distinct regions distributed across each of six technical replicate wells per sample. Images were then analyzed for the presence of YFP-positive aggregates. Previous quantification of infection measured the percentage of cells containing aggregates. However, to improve the window size of the assay, infection was measured by normalizing the total fluorescence of aggregates in each FITC image by the cell count [fluorescence density × area per cell reported in arbitrary units (A.U.)]. This measurement was calculated across all five images from each well; the average and SD were then determined for the six replicate wells. Finally, these values were multiplied by 10−3 for presentation.
Consistent with our previous findings, the control samples did not infect the Tau(4RD*LM)–YFP(1) cells (average fluorescence-per-cell measurement of 1.6 ± 0.2 × 103 A.U.), whereas tau prions isolated from PSP patient samples robustly induced aggregate formation (74 ± 25 × 103 A.U.; P < 0.001) (Fig. 1 B and C). The initial publication describing the Tau(4RD*LM)–YFP(1) cells found tau prions from PiD, AD, AGD, and CBD patient samples infected the cells (31). However, whereas we found that we could transmit AGD (22 ± 10 × 103 A.U.; P < 0.05) and CBD (39 ± 11 × 103 A.U.; P < 0.001) prions, neither PiD (3.6 ± 1.5 × 103 A.U.; P = 0.74) nor AD (9.5 ± 2.8 × 103 A.U.; P = 0.17) patient samples produced a substantive infection. In addition, incubation with CTE patient samples was also unable to yield a strong infection in the Tau(4RD*LM)–YFP(1) cells (6.7 ± 2.5 × 103 A.U.; P = 0.41).
Strikingly, of the samples tested here, only the 4R tauopathies yielded a robust infection in the 4R-expressing cells, suggesting that transmission of 4R tau prions is dependent on the presence of a 4R tau substrate. Visual comparison of the cells infected with AGD, CBD, or PSP shows distinct aggregate morphologies (Fig. 1C), consistent with previous reports (31). Whereas AGD-induced aggregates were diffuse, CBD aggregates presented as small puncta throughout the cytoplasm. In contrast, PSP induced large, bright puncta. These phenotypic differences between the diseases are reflected in the quantification of the infection; the larger and brighter the aggregates, the greater the fluorescence measurement (Fig. 1B).
Tau(3RD*VM)–YFP Cells Specifically Detect PiD Prions.
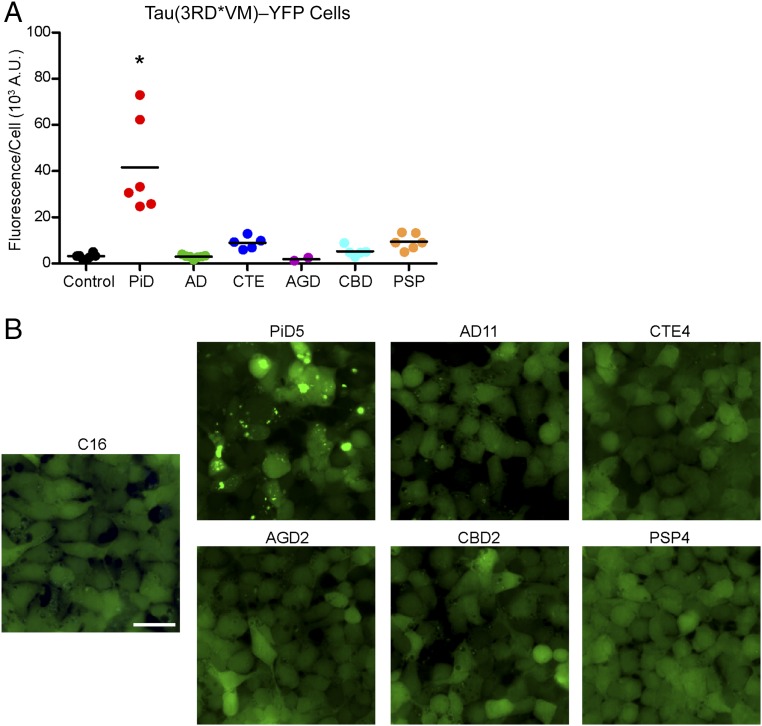
Our finding that only 4R-specific tauopathies propagate in cells expressing 4R tau led us to posit that the propagation of 3R tau prions requires the presence of 3R tau. To address this question, we developed HEK293T cells that express the RD of 3R tau with the familial L266V and V337M mutations fused to YFP [Tau(3RD*VM)–YFP]; the L266V mutation replaced the P301L mutation in this cell line because amino acid residue 301 is not present in the 3R isoforms of tau. We diluted the PTA-precipitated patient samples 1:10 in DPBS and incubated them with the 3R-expressing cell line for 4 d (Fig. 2). The control patient samples did not infect the Tau(3RD*VM)–YFP cells (average fluorescence-per-cell measurement of 3.2 ± 1.1 × 103 A.U.) (Fig. 2A and Table S2); however, tau prions isolated from the PiD patient samples transmitted to the cells (42 ± 21 × 103 A.U.; P < 0.001). Supporting the hypothesis that tau prions are isoform-specific, the AGD (1.9 ± 1.0 × 103 A.U.; P = 0.81), CBD (5.3 ± 2.2 × 103 A.U.; P = 0.67), and PSP (9.4 ± 3.4 A.U.; P = 0.14) patient samples, which infected the 4R tau cells, did not result in infection. Consistent with the Tau(4RD*LM)–YFP(1) results, neither the AD (3.0 ± 0.7 × 103 A.U.; P = 0.94) nor the CTE (9.0 ± 2.7 × 103 A.U.; P = 0.18) patient samples infected the cells. Importantly, these results indicate that the PiD-induced aggregates in the Tau(3RD*VM)–YFP cells, but not the other tauopathies (Fig. 2B), arise from the propagation of 3R tau-specific prions in PiD patient samples.
Fig. 2.
PiD prions infect Tau(3RD*VM)–YFP cells. Tau prions were precipitated from control, PiD, AD, CTE, AGD, CBD, and PSP patient samples using sodium PTA. The resulting protein pellets were incubated for 4 d with Tau(3RD*VM)–YFP cells expressing the repeat domain of 3R tau containing the L266V and V337M mutations and fused to YFP. (A) Quantification of cell infection using control (n = 6), PiD (n = 6), AD (n = 7), CTE (n = 5), AGD (n = 2), CBD (n = 5), and PSP (n = 6) patient samples was determined by summing the total fluorescence in each image and normalizing to the cell count. Prions isolated from PiD alone infected the Tau(3RD*VM)–YFP cells (P < 0.001) compared with controls. AD (P = 0.94), CTE (P = 0.18), AGD (P = 0.81), CBD (P = 0.67), and PSP (P = 0.14) showed no infectivity. Data are shown as the mean of five images per well in six wells. *P < 0.001. All values are shown in Table S2. (B) Representative images of HEK293T cells infected with PiD but not control, AD, CTE, AGD, CBD, or PSP patient samples. YFP is shown in green. (Scale bar, 50 μm.)
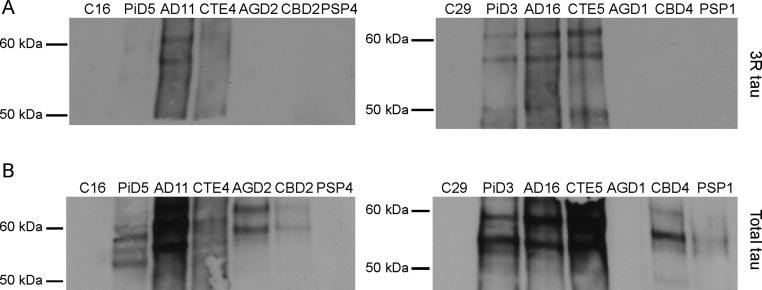
To confirm that these findings were the result of 3R prions in PiD and 4R prions in AGD, CBD, and PSP, we analyzed 14 of the PTA-precipitated samples by Western blot (Fig. S1). Using the 3R isoform-specific antibody, only 3R-containing aggregates were detected in the PiD, AD, and CTE patient samples (Fig. S1A). However, after probing with the Tau12 antibody, we found that the diseased patient samples contained tau aggregates, whereas the two control patient samples did not (Fig. S1B). These findings demonstrate the presence of 4R tau-containing aggregates in the AGD, CBD, and PSP patient samples, which were unable to infect the Tau(3RD*VM)–YFP cells.
Fig. S1.
Tau expression in human tissue samples. Tau prions were isolated from two representative patient samples from each patient group by PTA precipitation. The aggregated proteins were then analyzed by Western blot, probing with the 3R isoform-specific antibody (A) or the total tau antibody, Tau12 (B).
Identification of AD and CTE Prions.
Discovering that the propagation of 3R and 4R tauopathies in mammalian cells is isoform-specific, we posited that cells expressing both 3R and 4R tau isoforms could be infected by PiD samples, as well as by AGD, CBD, and PSP extracts. Testing this hypothesis, we developed HEK293T cells that express two tau–YFP constructs: the Tau(4RD*LM)–YFP construct expressed in the 4R cells, and the Tau(3RD*VM)–YFP construct expressed in the 3R cells [Tau(3RD*VM,4RD*LM)–YFP]. These fusion proteins are not expressed in tandem and, instead, are encoded by two separate plasmids. After isolating tau prions from the patient samples via PTA precipitation, the samples were diluted 1:4 in DPBS and incubated with the Tau(3RD*VM,4RD*LM)–YFP cells for 4 d (Fig. 3). Incubating the cells with the control patient samples yielded no infection (average fluorescence per cell of 5.3 ± 1.0 × 103 A.U.) (Fig. 3A and Table S2). However, because both the 3R and 4R tau isoforms were available as substrates for prion templating, not only did the PiD samples induce aggregates (15 ± 3.8 × 103 A.U.; P = 0.07), but the AGD (25 ± 2.2 × 103 A.U.; P < 0.01), CBD (32 ± 9.4 × 103 A.U.; P < 0.001), and PSP (28 ± 7.7 × 103 A.U.; P < 0.001) patient samples infected the cells as well. Interestingly, the PiD-induced tau aggregates are visibly smaller than the aggregates induced by the 4R tauopathies (Fig. 3B). Quantification of the smaller aggregates resulted in a lower fluorescence value compared with the 4R tauopathies. This is reflected in the quantification of infection (Fig. 3A), and likely the lack of statistical significance. Notably, the 95% confidence interval (CI) includes effect sizes from −0.9 to 20, suggesting additional samples could improve the precision of the estimate, which is impacted by intersample variability, to yield an effect that is statistically significant.
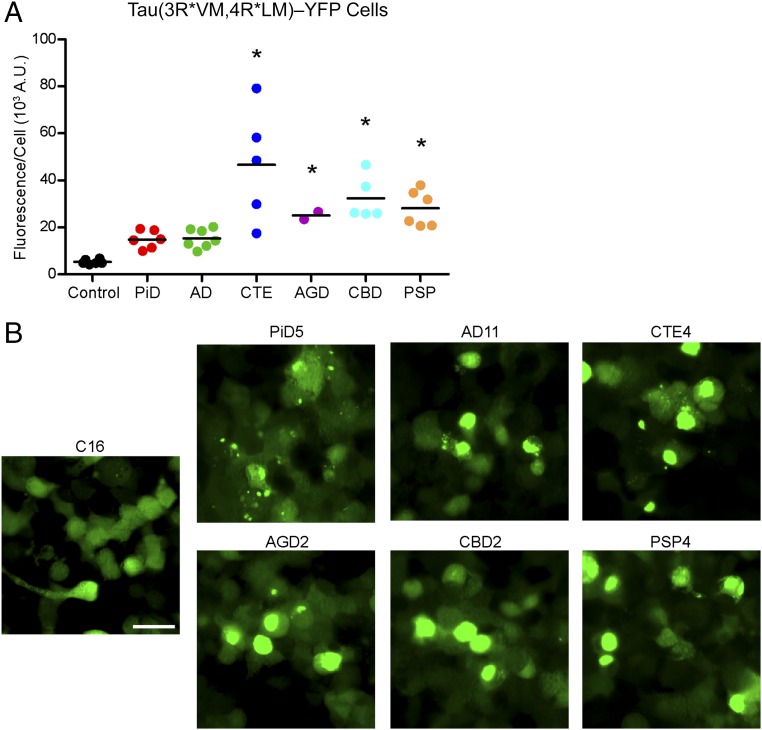
Fig. 3.
Tau prions in AD and CTE contain both 3R and 4R tau isoforms. Tau prions were isolated using sodium PTA from control, PiD, AD, CTE, AGD, CBD, and PSP patient samples and were then incubated with Tau(3RD*VM,4RD*LM)–YFP cells for 4 d. Tau(3RD*VM,4RD*LM)–YFP cells express both the 3R and 4R repeat domains of tau with mutations L266V and V337M (3RD) and P301L and V337M (4RD). (A) Quantification of cell infection using control (n = 6), PiD (n = 6), AD (n = 7), CTE (n = 5), AGD (n = 2), CBD (n = 5), and PSP (n = 6) patient samples was determined by standardizing the total fluorescence in each image to the total cell count. Prions from all six tauopathies, including AD and CTE, infected the Tau(3RD*VM,4RD*LM)–YFP cells, whereas the control samples showed no infection (PiD, P = 0.07; AD, P = 0.05; CTE, P < 0.001; AGD, P < 0.01; CBD, P < 0.001; PSP, P < 0.001). *P < 0.01. Data are shown as the mean from five images per well in six wells. All values are shown in Table S2. (B) Representative images of HEK293T cells infected with PiD, AD, CTE, AGD, CBD, and PSP but not control patient samples. YFP is shown in green. (Scale bar, 50 μm.)
We also proposed that coexpressing both the 3R and 4R isoforms in the cells would support infection by the combined tauopathies (AD and CTE). When we incubated the Tau(3RD*VM,4RD*LM)–YFP cells with AD (15 ± 4.0 × 103 A.U.; P = 0.05) and CTE (47 ± 24 × 103 A.U.; P < 0.001) patient samples, we found that the samples induced tau aggregates after 4 d (Fig. 3). Similar to results obtained with PiD, the effect size of infection with AD prions was not statistically significant, with a 95% CI of −0.49 to 20. The addition of more AD samples to our analysis would improve the precision of our estimate, overcoming the intersample variability, and likely result in a statistically significant effect. Remarkably, infection of Tau(3RD*VM,4RD*LM)–YFP cells across the five CTE patient samples was more variable than the five other tauopathies tested. All five CTE patients were diagnosed with CTE stage IV following neuropathological assessment, suggesting the variability seen here may be attributable to differences in sampling from patients with a disease that is highly focal in nature (34).
Cell Assay Sensitivity Is Enhanced by Overexpression of the 4R Tau Fusion Protein.
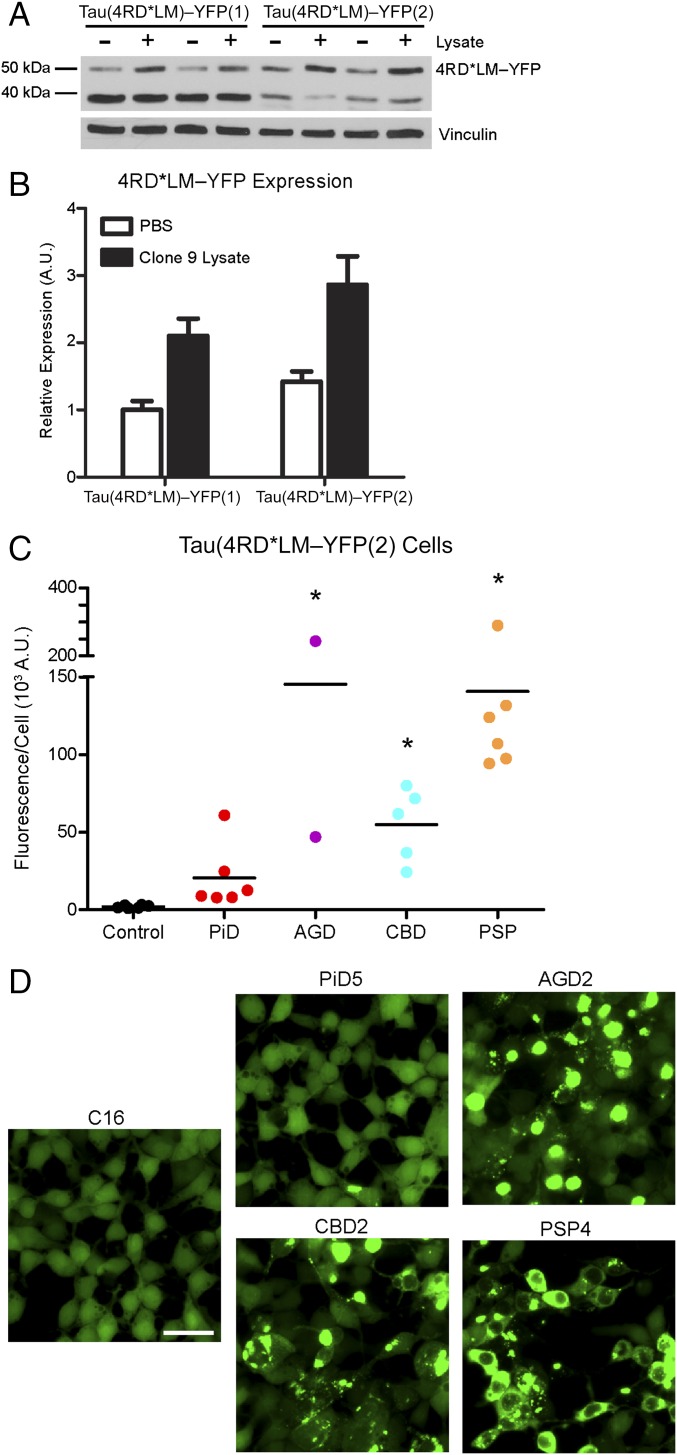
Quantification of tau prion infection in the Tau(3RD*VM,4RD*LM)–YFP cells overall showed a reduced window size, and therefore decreased infectivity of the isoform-specific tauopathies. Positing that reduced expression of the individual isoforms, compared with the Tau(4RD*LM)–YFP(1) and Tau(3RD*VM)–YFP cells, was responsible for decreasing susceptibility to infection, we tested the relationship between protein expression and sensitivity by developing a new HEK293T cell line with increased expression of the 4R RD, again containing the P301L and V337M mutations [Tau(4RD*LM)–YFP(2)]. Comparing the Tau(4RD*LM)–YFP(1) cells with the new Tau(4RD*LM)–YFP(2) cells, we examined expression of the fusion protein in the presence and absence of infection (Fig. 4 A and B). Lysate from clone 9 cells, which are Tau(4RD*LM)–YFP(1) cells that stably propagate infection with synthetic tau prions, has previously been shown to induce aggregate formation in Tau(4RD*LM)–YFP(1) cells (31). Here we found that in both the presence and absence of infection with clone 9 lysate, expression of the fusion protein is higher in the new Tau(4RD*LM)–YFP(2) cells compared with the original Tau(4RD*LM)–YFP(1) cells, as visualized by Western blot analysis (Fig. 4A) and quantification with ImageJ software (Fig. 4B).
Fig. 4.
Increased fusion protein expression improves assay sensitivity for 4R tau prions. HEK293T cells with a higher expression of the same fusion protein as the Tau(4RD*LM)–YFP(1) cells were created [Tau(4RD*LM)–YFP(2) cells]. (A) Western blot analysis of Tau(4RD*LM)–YFP(1) and Tau(4RD*LM)–YFP(2) cells in the absence and presence of infection with lysate from clone 9 cells, which stably propagate synthetic tau prions. The 4RD*LM–YFP construct was probed with anti-GFP (green fluorescent protein) antibody (Top). The membrane was reprobed for vinculin as a loading control (Bottom). (B) Quantification of protein expression in the Western blot was performed using ImageJ software (NIH). Expression levels were normalized to basal 4RD*LM–YFP expression in HEK293 cells. Data shown as mean ± SD (n = 2). (C and D) Crude brain homogenates from control (n = 6), PiD (n = 6), AGD (n = 2), CBD (n = 5), and PSP (n = 6) patient samples were diluted in DPBS and incubated with Tau(4RD*LM)–YFP(2) cells for 4 d. (C) Quantification of cell infection after incubation was determined by dividing the total fluorescence in each image by the total cell count. Tau prions from the 4R tauopathies were all capable of infecting the Tau(4RD*LM)–YFP(2) cells (AGD, P < 0.001; CBD, P < 0.05; PSP, P < 0.001), whereas the control samples did not. Of the six PiD samples, two infected the Tau(4RD*LM)–YFP(2) cells (PiD3 and PiD4) and the other four did not (P = 0.41). *P < 0.05. Data are shown as the mean of five images per well in six wells. All values are shown in Table S2. (D) Representative images of HEK293T cells infected with AGD, CBD, and PSP but not control or PiD patient samples. YFP is shown in green. (Scale bar, 50 μm.)
We then tested the susceptibility of the Tau(4RD*LM)–YFP(2) cells to infection with PiD, AGD, CBD, and PSP prions (Fig. 4 C and D). Although we previously used PTA to precipitate and concentrate tau prions before infecting the Tau(4RD*LM)–YFP(1) cells, the increased protein expression allowed us to instead incubate crude brain homogenate diluted 1:40 in DPBS with the cells for 4 d. Surprisingly, we found that crude homogenate alone was capable of transmitting AGD (150 ± 140 × 103 A.U.; P < 0.001), CBD (55 ± 24 × 103 A.U.; P < 0.05), and PSP (140 ± 74 × 103 A.U.; P < 0.001) prions to the new cell line, whereas the control samples showed no infection (2.0 ± 0.9 × 103 A.U.) (Fig. 4C and Table S2). Visual assessment of the infected cells (Fig. 4D) shows a similar result as seen in the Tau(4RD*LM)–YFP(1) cells (Fig. 1B); infection with AGD, CBD, and PSP samples induced cellular aggregates with distinct phenotypes. AGD-induced aggregates were large and round, whereas PSP prions induced tau aggregation throughout the entire cell with the noticeable exception of the nucleus. Compared with these larger aggregates, infection with CBD yielded a mixture of phenotypes, but the aggregates were predominantly smaller and emitted less fluorescence. This phenotypic difference ultimately contributes to variations in fluorescence values measured from the different patient groups, as seen in the quantification of tau prion infection (Fig. 4C). Notably, the distinct phenotypes induced by each 4R tauopathy in the Tau(4RD*LM)–YFP(1) cells (Fig. 1B) are unique from the phenotypes they induce in the Tau(4RD*LM)–YFP(2) cells (Fig. 4D).
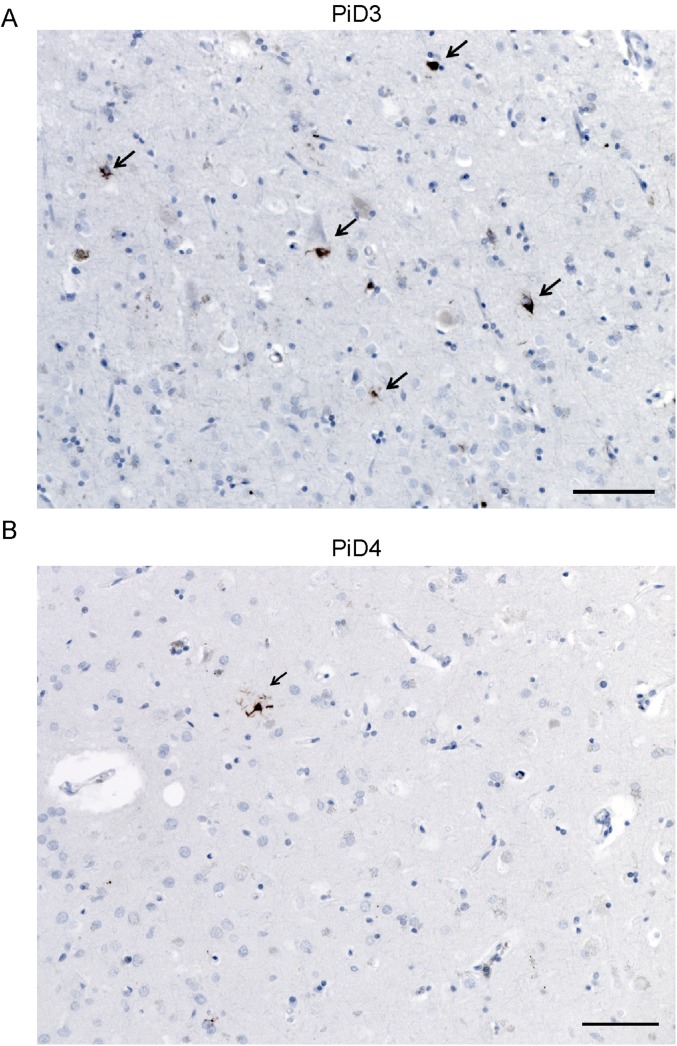
Intriguingly, when we incubated the Tau(4RD*LM)–YFP(2) cells with the PiD patient samples, we found one patient sample induced robust aggregate formation, one sample induced weak infection, and the other four showed very low infectivity (20 ± 21 × 103 A.U.; P = 0.41) (Fig. 4C and Table S2). It is important to note that although PiD has traditionally been classified as a 3R tauopathy, based on the initial discovery of 3R tau only in Pick’s bodies (18), some patient samples show conspicuous 4R tau inclusions in astroglia (35). To determine whether these results were due to astrocytic 4R tau, immunostaining using the 4R tau-specific antibody was performed on formalin-fixed sections from patients PiD3 and PiD4 (Fig. S2). Examining the angular gyrus from both patients, the same region tested in the cell assay, 4R tau astrocytic inclusions were detected, suggesting that the increased sensitivity of the Tau(4RD*LM)–YFP(2) cells enables detection of these less-prevalent lesions in PiD, compared with the Tau(4RD*LM)–YFP(1) cells.
Fig. S2.
4R tau inclusions in PiD patient samples. Immunostaining with the 4R isoform-specific tau antibody was performed on fixed tissue from the angular gyrus from PiD patients 3 (A) and 4 (B). Both patient samples contained 4R tau-positive lesions (arrows). (Scale bars, 30 µm.)
Overexpression of 4R Tau Supports Propagation of AD and CTE Prions.
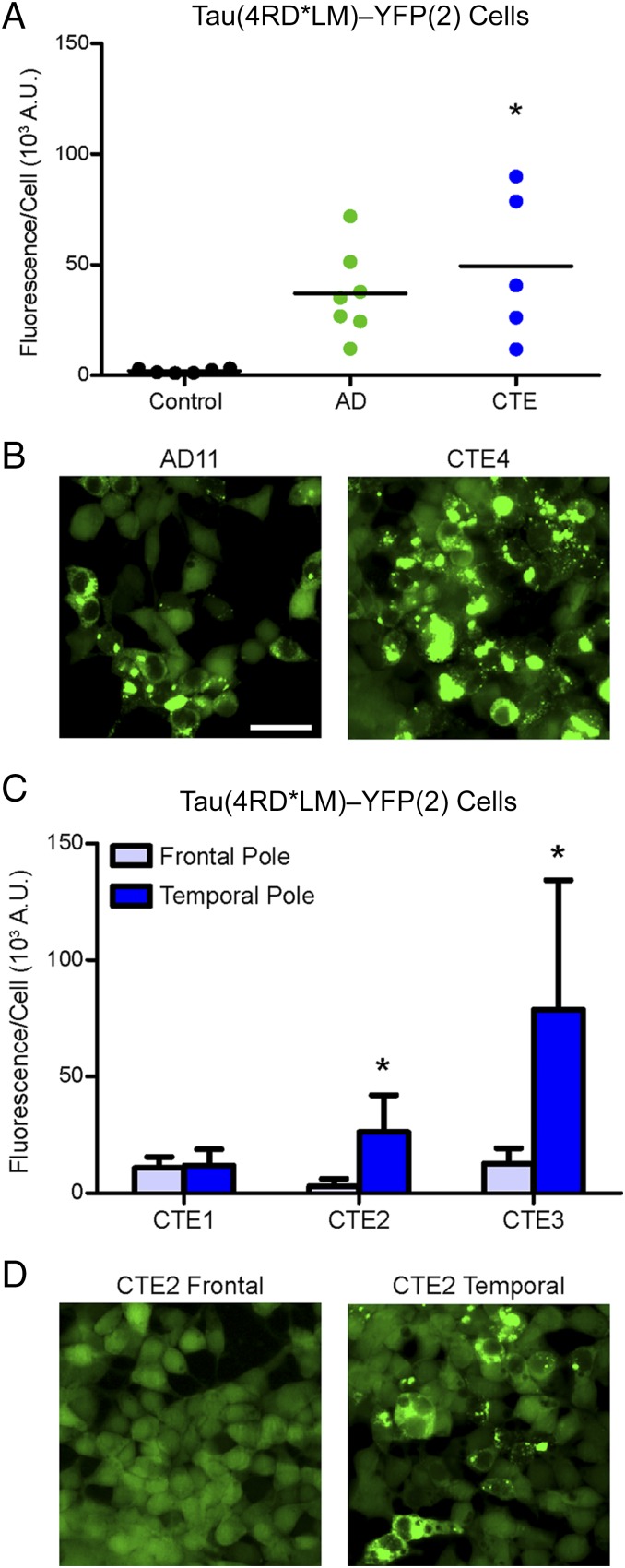
Following our observation that the Tau(4RD*LM)–YFP(2) cells enabled detection of 4R tau in two of the PiD patient samples tested, which previously did not induce aggregates, we decided to test whether or not overexpression of 4R tau also facilitates propagation of AD and CTE prions. After incubating crude brain homogenate from AD and CTE patients diluted 1:40 in DPBS with Tau(4RD*LM)–YFP(2) cells for 4 d, the cells were imaged and analyzed for infection. Remarkably, the increased sensitivity of the new HEK293T cells enabled infection with both AD (37 ± 20 × 103 A.U.; P = 0.1) and CTE (49 ± 34 × 103 A.U.; P < 0.005) patient samples, demonstrating the presence of tau prions in AD and CTE patients (Fig. 5 A and B and Table S2). As seen previously, the intersample variability of tau prions quantified from the AD patient samples yields an imprecise estimate for the number of samples tested in our statistical model. However, AD tau prions are estimated to increase infection in Tau(4RD*LM)–YFP(2) cells by 35 × 103 A.U. over control, which is a substantial increase over the estimated effect size for both Tau(4RD*LM)–YFP(1) (7.8 × 103 A.U.) and Tau(3RD*VM)–YFP cells (–0.4 × 103 A.U.), suggesting tau prions from AD patients have a large effect that is too imprecisely estimated given sample availability to conclude statistical significance.
Fig. 5.
Overexpression of 4R tau supports propagation of AD and CTE prions. Crude brain homogenates from control, AD, and CTE patient samples were diluted in DPBS and incubated for 4 d with Tau(4RD*LM)–YFP(2) cells. (A) Quantification of cell infection with control (n = 6; data also shown in Fig. 4C), AD (n = 7), and CTE (n = 5) patient samples. Both AD (P = 0.1) and CTE (P < 0.05) patient samples infected the Tau(4RD*LM)–YFP(2) cells. *P < 0.05. Data are shown as the mean of five images per well in six wells. All values are shown in Table S2. (B) Representative images of HEK293T cells infected with AD and CTE. YFP is shown in green. (Scale bar, 50 μm.) (C) Quantification of cell infection using two brain regions from three CTE patient samples. The frontal pole and temporal pole contain a significantly different concentration of tau prions in two of the CTE patient samples tested. Data are shown as the mean ± SD measured from five images per well in six wells. All values are shown in Table S2. *P < 0.05. (D) Representative images of HEK293T cells infected with samples from the frontal and temporal poles from patient CTE2. YFP is shown in green. The scale is the same as shown in B.
To assess the specificity of these findings, we tested samples from two different brain regions isolated from three CTE patients in the Tau(4RD*LM)–YFP(2) cells. In 2016, the results of the first consensus report on CTE concluded that the key neuropathological features of CTE include perivascular tau inclusions in neurons and astrocytes, particularly at the depths of the sulci in an irregular pattern (36). These focal deposits then spread from the cortical sulci to other brain structures (34). In three of the five CTE patients (CTE1, CTE2, and CTE3), we sampled both the temporal and frontal poles (Fig. 5 C and D and Table S2). Although infectivity of both brain regions for patient CTE1 was poor, the sulcus from the temporal pole tested from CTE patients 2 and 3 contained significantly more tau prions than the sulcus from the frontal pole that was tested (P < 0.05). Postmortem analysis found accumulation of NFTs in both the frontal and temporal poles from all three patients; however, consistent with our findings, tangle density was more robust in the temporal pole.
Discussion
Our findings show that specific tau isoforms must be available as substrates to transmit particular tauopathies, suggesting these diseases arise from distinct prion strains. AGD, CBD, and PSP patient samples infect the 4R-containing cell lines Tau(4RD*LM)–YFP(1), Tau(3RD*VM,4RD*LM)–YFP, and Tau(4RD*LM)–YFP(2). Similarly, prions from PiD samples infect both the Tau(3RD*VM)–YFP and Tau(3RD*VM,4RD*LM)–YFP cell lines. Both AD and CTE patient samples, which are composed of 3R and 4R tau aggregates, were originally found only to propagate in the Tau(3RD*VM,4RD*LM)–YFP cell line, suggesting tau misfolding in these two diseases requires both the 3R and 4R isoforms. However, increasing the expression level of the 4R tau isoform in the Tau(4RD*LM)–YFP(2) cells facilitated propagation not only of AGD, CBD, and PSP but also of AD and CTE. The biological mechanism by which this occurs remains to be elucidated.
Similar to tau, PrPSc and multiple system atrophy (MSA) prions aggregate into fibrils that coalesce into amyloid plaques and glial cytoplasmic inclusions, respectively (37–39). Measuring the kinetics of PrPSc and MSA propagation following intracerebral injection into Tg mice has been important in discerning the underlying biology of these prions. However, although intracerebral injection of tau fibrils into Tg mice expressing tau transgenes resulted in tau neuropathology, such experiments were inconclusive with respect to assessing the kinetics and accumulation of tau prions (40–44). Instead, measurements made from cells expressing the tau–YFP fusion proteins (31, 32, 45) have been more informative.
As described by us and others (31), six of the known tauopathies are characterized by transmission and de novo replication of tau aggregates. Our findings suggest that groups of tau prion strains responsible for each disease reflect the tau isoforms present—namely 3R tau in PiD; 4R tau in AGD, CBD, and PSP; and a combination of both 3R and 4R in AD and CTE. Although the isoform specificity of these tauopathies was previously established (24, 46), the discovery that the isoforms play an integral role in infection by tau prion strains provides an explanation for the range of phenotypes seen in patients. This research shows that it is possible to use cells to discriminate among 3R, 4R, and the combined tauopathies.
Previous work with synthetic tau prions provides further support for the isoform-directed specificity of tau prion replication. Using the recombinant K18 and K19 constructs, which encode the 4R and 3R RDs of tau, respectively, Dinkel et al. (47) demonstrated that heparin-induced K19 3R aggregates could not seed filaments formed from K18 4R monomers. In a separate experiment, recombinant 1N3R or 1N4R human tau fibrils were used to test the cross-isoform prion replication barrier in the neuroblastoma cell line SH-SY5Y (48). Transient expression of either 1N3R or 1N4R tau was initiated 14 h before exposing the cells to tau fibrils. When attempts were made to infect the cells with the 1N4R fibrils, only the cells expressing 4R tau developed aggregates. Similarly, the 1N3R fibrils only propagated in the 3R-expressing cells, demonstrating isoform specificity in synthetic prions.
In addition to these in vitro findings, several in vivo studies have demonstrated homotypic seeding of tau. Clavaguera and colleagues used two similar Tg mouse models, P301S (human 0N4R tau with the P301S mutation) and ALZ17 (wild-type human 2N4R tau), to demonstrate that inoculating diseased P301S mouse brain extracts into ALZ17 mice induced NFTs, neuropil threads, and coiled bodies by self-templating of 4R tau (49). Using the same P301S model, other groups found that brain extracts prepared from aged, symptomatic animals inoculated into 2-mo-old presymptomatic mice induced NFT pathology within 2 wk of inoculation, demonstrating rapid propagation of 4R tau prions (50). Similarly, recombinant human 2N4R tau with the P301S mutation, and K18 with the P301L mutation, were fibrillized in the presence of heparin and inoculated into young PS19 mice (human 1N4R tau with the P301S mutation) (51). Both of these 4R-containing fibrils induced tau neuropathology, whereas inoculation of α-synuclein fibrils had no effect, highlighting homotypic propagation of synthetic 4R tau prions in a third mouse model (40).
The aforementioned tau studies, as well as our own findings, are reminiscent of earlier studies in Tg mice expressing Syrian hamster (SHa) PrP (52). These animals expressed both mouse (MoPrPC) and SHaPrPC, and were therefore able to propagate either MoPrPSc or SHaPrPSc depending on the inoculum chosen; MoPrPSc prions stimulated the formation of nascent MoPrPSc prions, whereas inoculated SHaPrPSc prions initiated the formation of de novo SHaPrPSc prions. Although these studies demonstrate a transmission barrier between species, additional studies show that differences in the PrP amino acid sequence between the host and the inoculum can give rise to distinct prion strain properties within a species. Comparing the a and b alleles of the mouse PrP gene, which differ by two amino acids, inoculation of PrP-A prions into mice expressing PrP-B yielded inefficient disease transmission with extended incubation times compared with PrP-B prions inoculated into the PrP-B mice (53, 54). This mouse strain barrier can be overcome by increasing the expression level of PrP, as may be the case for tau prions here. Together, these PrP data demonstrate that homotypic interactions feature in prion replication whereas heterotypic templating is inefficient, both between and within species.
SDS/PAGE studies demonstrated tau isoform specificity in detergent-extracted samples obtained from AD, PiD, AGD, CBD, and PSP patient samples more than a decade ago (46), but the more recent development of a rapid tau prion bioassay using Tau(4RD*LM)–YFP(1) cells (31), followed by modifications to the bioassay conditions (32), has facilitated the rapid detection of tau prions. Using this powerful approach, we posited that homotypic interactions in tau prion replication were just as critical as those demonstrated in PrP Tg mice described above (52). In our current studies, new cell lines were created to measure tau prion infection based on the ability to template the RDs of 3R and/or 4R tau. Our findings, and those of others (31), demonstrate that the 4R tauopathies can readily initiate formation of 4R tau aggregates in HEK cells (Figs. 1 and 4). In contrast, the 3R tauopathy PiD initiates formation of 3R tau aggregates (Fig. 2). Whereas neither AD nor CTE patient samples initiated the formation of tau aggregates in the 4R or 3R cell lines (Figs. 1 and 2), expression of both fusion proteins in the Tau(3RD*VM,4RD*LM)–YFP cells supported the formation of tau aggregates in HEK cells incubated with either AD or CTE brain extracts (Fig. 3). Notably, these findings recapitulate the tau isoforms found in the brains of patients with these diseases (Fig. S1) (24, 46).
Our finding that both 3R and 4R tau are more efficient than either isoform alone in propagating AD and CTE prions in HEK293T cells suggests that tau misfolding in AD and CTE prion strains incorporates both tau isoforms. The subsequent discovery that increased expression of the RD of 4R tau in the Tau(4RD*LM)–YFP(2) cells facilitates transmission of AD and CTE prions seemingly contradicts this conclusion (Fig. 5), and instead suggests tau prions in AD and CTE comprise distinct 3R and 4R tau prions. One argument against this interpretation is the inability of the samples to infect the Tau(3RD*VM)–YFP and Tau(4RD*LM)–YFP(1) cells. If AD and CTE samples contained distinct 3R and 4R aggregates, those prions should propagate in the respective cell lines, as demonstrated with PiD, AGD, CBD, and PSP. A second explanation could arise from small amounts of isolated 4R aggregates along with the combined 3R/4R tau aggregates typically associated with the diseases. Due to its greater sensitivity, only the Tau(4RD*LM)–YFP(2) cells were able to propagate the minor 4R component of the total aggregated tau, compared with the Tau(4RD*LM)–YFP(1) cells. One possible source of distinct 4R aggregates in these samples is aging-related tau astrogliopathy (ARTAG), which is selectively immunostained with 4R tau antibodies, even in PiD patients (55). Consistent with this finding, two PiD patient samples containing 4R tau astrocytic lesions infected the more sensitive Tau(4RD*LM)–YFP(2) cells. Whereas ARTAG has been identified in AD patients, 4R astrocytic inclusions are a key component of CTE neuropathology. However, AD and CTE showed similar infectivity in the Tau(4RD*LM)–YFP(2) cells, which would be unexpected given the variability of ARTAG in AD patient samples.
A third explanation of our seemingly disparate findings may lie in the kinetic differences between the two RDs. In vitro analysis of the aggregation propensity of the six tau isoforms found the 4R isoforms have a higher rate of aggregation than the 3R isoforms; the 4R fibrils showed an increase in the rate of both tau fibril nucleation and fibril extension compared with 3R fibrils (56). This work suggests 4R tau is intrinsically more aggregation-prone than 3R tau. It is therefore plausible that the high expression of 4R tau in the Tau(4RD*LM)–YFP(2) cells is seeded by the 4R component of tau prions in AD and CTE. It is noteworthy that the [18F]AV-1451 PET tracer selectively binds to pathologic tau in AD patient samples compared with PSP and PiD patient samples (57–59), suggesting a conformation difference may exist between tau in AD versus tau in the isoform-specific tauopathies. Our conclusion, that AD prions contain both 3R and 4R tau isoforms, whereas PSP and PiD contain either 4R or 3R, respectively, provides one possible explanation for the results reported for [18F]AV-1451.
Finally, it is also possible that posttranslational modifications (PTMs) on tau facilitate protein misfolding into distinct prion strains. A large number of PTMs have been identified on tau isolated from both human and Tg mouse samples, including multiple sites for phosphorylation, acetylation, ubiquitination, and O-glycosylation (60–62). However, none of these differences have been consistently detected in distinct patient groups in a manner that would explain the differences observed here.
We, and others, have proposed that tau prions polymerize into paired-helical and straight filaments that form NFTs in AD patients. There is considerable evidence demonstrating that NFTs correlate with dementia, and the absence of NFTs is generally accompanied by normal cognition (7–9). Our studies establish that biologically active tau prions are found in AD patient samples; it is possible the replication of these tau prions may be responsible for the progressive dementia seen in patients. In contrast to AD, the tau prions identified in CTE patient samples likely arise as a result of repetitive mild traumatic brain injury (TBI). CTE, first defined as “punch drunk” (63), has been increasingly diagnosed in amateur and professional athletes in contact sports. In addition, many soldiers who suffer mild TBIs later develop posttraumatic stress disorders and progressive dementia. Some studies argue that these signature injuries of the recent conflicts in Afghanistan and Iraq are the result of exposure to blast waves from improvised explosive devices (34, 64–67). Similar to AD, biochemical analyses of NFTs isolated from CTE patients have shown that these aggregates consist of both 3R and 4R tau isoforms (24). Although tau pathology in AD patients is localized to neurons, astrocytic tangles can be prominent in CTE patients (68). Additionally, the location of NFTs in the cortical layers also differs between AD and CTE patients. AD patients often develop NFTs in both superficial and deeper cortical layers, with the highest density seen in layers V and VI. On the other hand, CTE patients typically develop NFTs in the more superficial layers II and III of the neocortex. Despite these regional differences, the hippocampus is similarly affected by both diseases (7) and may be a major contributor to the progressive dementia seen in these patients.
In summary, the findings presented here use HEK cells expressing mutant tau fragments fused to YFP to demonstrate that tau prions from patients with tauopathies, including AD and CTE, can infect mammalian cells. Notably, we used a panel of cell lines to show that the tau isoforms identified in the neuropathological lesions associated with each tauopathy play a critical role in the formation and transmission of tau prion strains. Although these results provide critical insight into the pathogenesis of each tauopathy, the involvement of 3R and 4R tau isoforms in the propagation of AD and CTE suggests a complex mechanism of prion replication, where heterodimers interact through a process, as yet undefined, to produce different strains of tau prions causing these two diseases. Most importantly, the identification of biologically active tau prions in all of the patient samples tested suggests developing successful anti-tau neurotherapeutics will require inhibiting the propagation of specific tau prion strains.
Materials and Methods
Human Tissue Samples.
Frozen brain tissue samples from neuropathologically confirmed cases of AD, AGD, CBD, PiD, and PSP were provided by the University of California, San Francisco (UCSF) Neurodegenerative Disease Brain Bank, as were two control samples. Frozen brain tissue samples from neuropathologically confirmed cases of CTE were provided by the Chronic Traumatic Encephalopathy Program at Boston University’s Alzheimer’s Disease Center. Four control samples were provided by Deborah Mash, University of Miami, Coral Gables, FL.
Cell Line Development.
Constructs encoding the RD of 4R tau (amino acids 243 to 375, corresponding to 2N4R tau) containing the mutations P301L and V337M, and the RD of 3R tau (amino acids 243 to 274 and 306 to 375) containing the mutations L266V and V337M, were fused with YFP at the C terminus with an 18-amino acid flexible linker (EFCSRRYRGPGIHRSPTA). These constructs were introduced into the pIRESpuro3 vector (Clontech) or the pIRESblaS vector in which the puromycin resistance gene of the pIRESpuro3 was replaced with the blasticidin resistance gene. All tau fusion protein constructs are listed with amino acid residues, mutations, and background cell line in Table S1.
HEK293T cells (ATCC) were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 50 units/mL penicillin, 50 μg/mL streptomycin, and 10% (vol/vol) FBS (Thermo Fisher). Cultures were maintained in a humidified atmosphere of 5% (vol/vol) CO2 at 37 °C. Cells plated in DMEM were transfected using Lipofectamine 2000 (Thermo Fisher). Stable cells were selected in DMEM containing 1 μg/mL puromycin or 10 μg/mL blasticidin S (Thermo Fisher). Monoclonal lines were generated by limiting dilution of polyclonal cell populations in 96- or 384-well plates.
SI Materials and Methods
Human Patient Neuropathology.
Two control samples, as well as the AD, AGD, CBD, PiD, and PSP samples, were received from patients enrolled in the UCSF Memory and Aging Center longitudinal clinical research programs. Fresh brains were cut into ∼1-cm coronal sections and alternately fixed in 10% neutral buffered formalin for 72 h or rapidly frozen. Neuropathological diagnoses were made in accordance with consensus diagnostic criteria (69) using histological and immunohistochemical methods previously described (70). Patient samples were selected by W.W.S. and L.T.G. Brain regions from each patient were selected based on the presence of confirmed tau neuropathology. The CTE patient samples were provided by the Chronic Traumatic Encephalopathy Program at Boston University’s Alzheimer’s Disease Center and the Veterans Affairs Boston Healthcare System. Neuropathological processing was carried out as described previously (71). Staining and neuropathological assessment of patient samples are described (68). Patient samples were selected by A.C.M.
Cell Aggregation Assay.
Cells were plated in 384-well plates with black polystyrene walls (Greiner) with 0.012 μg Hoechst 33342 (Thermo Fisher) at a density of 1,000 cells per well [Tau(4RD*LM)–YFP(1)], 3,000 cells per well [Tau(3RD*VM)–YFP and Tau(4RD*LM)–YFP(2)], or 4,000 cells per well [Tau(3RD*VM,4RD*LM)–YFP]. Cells were incubated at 37 °C for 2 to 4 h to allow adherence to the plate. Lipofectamine 2000 (1.5% final volume; Thermo Fisher) and OptiMEM (78.5% final volume; Thermo Fisher) were premixed and added to each patient sample before incubation at room temperature for 2 h. Patient samples were plated in six replicate wells. Plates were then incubated at 37 °C in a humidified atmosphere of 5% (vol/vol) CO2 for 4 d before imaging on the IN Cell Analyzer 6000 (GE Healthcare). Images of both the DAPI and FITC channels were collected from five different regions in each well. The images were analyzed using IN Cell Developer software with algorithms developed to identify intracellular aggregates only in live cells.
Cell Assay Sample Preparation.
Patient tissue samples were homogenized to 10% (wt/vol) in calcium- and magnesium-free DPBS using an Omni Tip (Omni International) and PowerGen homogenizer (Fisher Scientific). Samples were aliquoted and stored at −80 °C.
PTA precipitation was performed as described (33). Briefly, 10% (wt/vol) brain homogenate was incubated in final concentrations of 2% (vol/vol) sarkosyl (Sigma) and 0.5% (vol/vol) benzonase (Sigma) at 37 °C with constant agitation in an orbital shaker for 2 h. Sodium PTA (Sigma) was dissolved in ddH2O, and the pH was adjusted to 7.0. PTA was added to the samples to a final concentration of 2% (vol/vol), which was then incubated overnight in the same conditions. The sample was centrifuged at 16,000 × g for 30 min at room temperature, and the supernatant was removed. The resulting pellet was resuspended in 2% (vol/vol) sarkosyl in DPBS and 2% (vol/vol) PTA in ddH2O (pH 7.0). The sample was again incubated for at least 1 h before a second centrifugation as above. The supernatant was again removed, and the pellet was resuspended in DPBS using 10% of the initial starting volume. This suspension was diluted in DPBS [1:40 for Tau(4RD*LM)–YFP(1) cells, 1:10 for Tau(3RD*VM)–YFP cells, and 1:4 for Tau(3RD*VM,4RD*LM)–YFP cells] before incubation with Lipofectamine 2000 and OptiMEM.
Crude brain homogenate was diluted 1:40 in DPBS [for Tau(4RD*LM)–YFP(2) cells] before incubation with Lipofectamine 2000 and OptiMEM.
Immunohistochemistry.
Immunostaining of the angular gyrus from the PiD patient samples was performed using formalin-fixed tissue. Blocks were embedded in paraffin wax, cut into 8-µm-thick sections, and stained with the four-repeat tau primary antibody (RD4; 1:250; Millipore). Primary antibody was incubated overnight at 4 °C, and mouse-specific biotinylated secondary antibody was subsequently incubated for 1 h at room temperature. Slides were pretreated for antigen retrieval by immersion in citrate (10 mM citric acid, 0.05% Tween 20, pH 6.0) in an autoclave at 121 °C for 5 min. Immunoperoxidase staining was performed using an avidin–biotin complex detection system (Vectastain ABC Kit; Vector Laboratories) with 3,3-diaminobenzidine as the chromogen.
Clone 9 Infection Assay.
Tau(4RD*LM)–YFP(1) and Tau(4RD*LM)–YFP(2) cells were plated at a density of 4 × 105 cells per well in a six-well plate (Costar). Cells were incubated at 37 °C for 2 to 4 h to allow adherence to the plate. Clone 9 lysate was diluted in DPBS (1.5 μg/µL) and incubated in Lipofectamine 2000 [final concentration of 2% for Tau(4RD*LM)–YFP(1) cells and 1.2% for Tau(4RD*LM)–YFP(2) cells] for 1.5 h. OptiMEM was added to the samples [78% of final concentration for Tau(4RD*LM)–YFP(1) cells and 78.8% of final concentration for Tau(4RD*LM)–YFP(2) cells]. Plates were then incubated at 37 °C in a humidified atmosphere of 5% (vol/vol) CO2 for 3 d before collecting lysates.
Production of Cell Lysate.
Cell lysates were prepared by incubating washed, confluent cells in cold radioimmunoprecipitation assay buffer (50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing cOmplete EDTA-free protease inhibitor mixture (Roche) for 10 min on ice. Cells were then collected using a cell scraper (Celltreat). Using a 22-gauge needle and syringe (BD), lysate was produced by drawing and expelling the collected cells through the needle 10 times. The lysate was centrifuged at 3,000 × g for 10 min. The supernatant was collected, and the protein concentration was determined using the bicinchoninic acid assay (Pierce).
Immunoblotting.
Cell lysates were diluted to a concentration of 1 mg/mL in DPBS and then combined with 4× NuPAGE loading buffer (final concentration 1×; Thermo Fisher) and 10× reducing agent (final concentration 1×; Thermo Fisher), boiled for 10 min, and loaded onto a 4 to 12% Novex bis-Tris gel (Thermo Fisher). Crude brain homogenates (10% wt/vol) from control and tauopathy patient samples were combined with 2× NuPAGE loading buffer (final concentration 1.8×), boiled for 10 min, and loaded onto an 8% Bolt bis-Tris gel (Thermo Fisher). PTA-precipitated brain homogenates were diluted 1:2 in DPBS with 1× NuPAGE loading buffer and 1× NuPAGE reducing agent (Thermo Fisher). Samples were sonicated for 10 min and boiled for 20 min before loading onto an 8% Bolt bis-Tris gel (Thermo Fisher).
SDS/PAGE was performed using MES buffer (Thermo Fisher). Gels were transferred to a PVDF membrane (Thermo Fisher) using a wet transfer system. The membrane was blocked for 30 min in blocking buffer [5% (wt/vol) nonfat milk in 1× Tris-buffered saline containing 0.05% (vol/vol) Tween 20 (TBST)] and incubated with primary antibody overnight at 4 °C. With the exception of the GFP antibody used to probe the cell lysates, membranes were washed three times with 1× TBST before incubating with a secondary antibody conjugated to horseradish peroxidase for 1 h. Membranes were then washed a final time in 1× TBST before developing with enhanced chemiluminescent detection (GE Healthcare) and exposing to X-ray film. The GFP antibody used was conjugated to horseradish peroxidase, so membranes were developed immediately after the first wash step. When membranes were stripped and reprobed, Restore Western Blot Stripping Buffer (Thermo Fisher) was used before blocking the membranes for 30 min in blocking buffer and repeating the methods described above. Primary antibodies used included GFP (1:1,000; Santa Cruz), vinculin (1:10,000; Abcam), Tau12 (1:10,000), and the 3R isoform-specific antibody (clone 8E6/C11; 1:1,000; Millipore). Production and isolation of Tau12 were performed as described (32). Horseradish peroxidase-conjugated secondary antibodies used included goat anti-rabbit and goat anti-mouse (both 1:10,000; Bio-Rad). Quantification of Western blot band intensity was done with ImageJ software (NIH).
Statistical Analysis.
Cell infection data are presented as mean ± SD. Values represent averages of five images collected from each well of a 384-well plate. Technical replicates for each patient sample were averaged across six wells. Statistical comparisons between control and diseased patient samples were performed using a mixed-effects model on data in their original scale. A mixed-effects model was used to incorporate correlation among fluorescence-per-cell measurements collected from the same patient sample (technical replicates), as opposed to other statistical models that assume technical replicates are independent measures. Data were kept in the original scale because the data are distributed approximately normal for most measures, in contrast to other distributional assumptions that would require a data transformation, such as a log-normal distribution. Statistical comparisons between the two brain regions in the three CTE patient samples were done using a Student’s t test with unequal variance. Statistical significance for all tests was determined with a P value < 0.05.
Acknowledgments
We thank George A. Carlson for his thoughtful revisions and comments, and Brittany N. Dugger for microdissecting the CTE patient samples. This work was supported by NIH Grants AG002132 and AG031220, Daiichi Sankyo, Dana Foundation, Glenn Foundation, Sherman Fairchild Foundation, and a gift from the Rainwater Charitable Foundation. Control, AD, AGD, CBD, PiD, and PSP tissue samples were provided by the University of California, San Francisco, Neurodegenerative Disease Brain Bank, which is supported by NIH Grants AG023501 and AG19724 (to W.W.S.), Tau Consortium, and Consortium for Frontotemporal Dementia Research. CTE tissue samples were provided by the Boston University Alzheimer’s Disease Center Chronic Traumatic Encephalopathy Center, which is supported by the Department of Veterans Affairs, National Institute of Neurological Disorders and Stroke, National Institute of Biomedical Imaging and Bioengineering (NS086559), National Institute on Aging, Boston University Alzheimer’s Disease Center (AG13846; Supplement 0572063345-5), Concussion Legacy Foundation, Andlinger Foundation, and World Wrestling Entertainment, Inc. Additional control samples were provided by the University of Miami Brain Endowment Bank.
Footnotes
Conflict of interest statement: A provisional patent application has been submitted in connection with this work. Inventors include A.L.W., A.A., S.P., S.A.K., S.H.O., and S.B.P.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616344113/-/DCSupplemental.
References
- 1.Iqbal K, et al. Defective brain microtubule assembly in Alzheimer’s disease. Lancet. 1986;2(8504):421–426. doi: 10.1016/s0140-6736(86)92134-3. [DOI] [PubMed] [Google Scholar]
- 2.Brion J-P, Passareiro H, Nunez J, Flament-Durand J. Mise en évidence immunologique de la protéine tau au niveau des lésions de dégénérescence neurofibrillaire de la maladie d’Alzheimer. Arch Biol (Liege) 1985;95:229–235. French. [Google Scholar]
- 3.Grundke-Iqbal I, et al. Abnormal phosphorylation of the microtubule-associated protein τ (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci USA. 1986;83(13):4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosik KS, Joachim CL, Selkoe DJ. Microtubule-associated protein tau (τ) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc Natl Acad Sci USA. 1986;83(11):4044–4048. doi: 10.1073/pnas.83.11.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood JG, Mirra SS, Pollock NJ, Binder LI. Neurofibrillary tangles of Alzheimer disease share antigenic determinants with the axonal microtubule-associated protein tau (τ) Proc Natl Acad Sci USA. 1986;83(11):4040–4043. doi: 10.1073/pnas.83.11.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wischik CM, et al. Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc Natl Acad Sci USA. 1988;85(12):4506–4510. doi: 10.1073/pnas.85.12.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hof PR, et al. Differential distribution of neurofibrillary tangles in the cerebral cortex of dementia pugilistica and Alzheimer’s disease cases. Acta Neuropathol. 1992;85(1):23–30. doi: 10.1007/BF00304630. [DOI] [PubMed] [Google Scholar]
- 8.Jansen WJ, et al. Amyloid Biomarker Study Group Prevalence of cerebral amyloid pathology in persons without dementia: A meta-analysis. JAMA. 2015;313(19):1924–1938. doi: 10.1001/jama.2015.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serrano-Pozo A, et al. Thal amyloid stages do not significantly impact the correlation between neuropathological change and cognition in the Alzheimer disease continuum. J Neuropathol Exp Neurol. 2016;75(6):516–526. doi: 10.1093/jnen/nlw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutton M, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393(6686):702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 11.Poorkaj P, et al. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol. 1998;43(6):815–825. doi: 10.1002/ana.410430617. [DOI] [PubMed] [Google Scholar]
- 12.Stanford PM, et al. Frequency of tau mutations in familial and sporadic frontotemporal dementia and other tauopathies. J Neurol. 2004;251(9):1098–1104. doi: 10.1007/s00415-004-0489-x. [DOI] [PubMed] [Google Scholar]
- 13.Goate A, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349(6311):704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 14.Pollock NJ, Mirra SS, Binder LI, Hansen LA, Wood JG. Filamentous aggregates in Pick’s disease, progressive supranuclear palsy, and Alzheimer’s disease share antigenic determinants with microtubule-associated protein, tau. Lancet. 1986;2(8517):1211. doi: 10.1016/s0140-6736(86)92212-9. [DOI] [PubMed] [Google Scholar]
- 15.Goedert M, Spillantini MG, Potier MC, Ulrich J, Crowther RA. Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: Differential expression of tau protein mRNAs in human brain. EMBO J. 1989;8(2):393–399. doi: 10.1002/j.1460-2075.1989.tb03390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goedert M, Spillantini MG, Cairns NJ, Crowther RA. Tau proteins of Alzheimer paired helical filaments: Abnormal phosphorylation of all six brain isoforms. Neuron. 1992;8(1):159–168. doi: 10.1016/0896-6273(92)90117-v. [DOI] [PubMed] [Google Scholar]
- 17.Andreadis A, Brown WM, Kosik KS. Structure and novel exons of the human τ gene. Biochemistry. 1992;31(43):10626–10633. doi: 10.1021/bi00158a027. [DOI] [PubMed] [Google Scholar]
- 18.Delacourte A, et al. Specific pathological Tau protein variants characterize Pick’s disease. J Neuropathol Exp Neurol. 1996;55(2):159–168. doi: 10.1097/00005072-199602000-00004. [DOI] [PubMed] [Google Scholar]
- 19.Ksiezak-Reding H, et al. Ultrastructure and biochemical composition of paired helical filaments in corticobasal degeneration. Am J Pathol. 1994;145(6):1496–1508. [PMC free article] [PubMed] [Google Scholar]
- 20.Flament S, Delacourte A, Verny M, Hauw J-J, Javoy-Agid F. Abnormal Tau proteins in progressive supranuclear palsy. Similarities and differences with the neurofibrillary degeneration of the Alzheimer type. Acta Neuropathol. 1991;81(6):591–596. doi: 10.1007/BF00296367. [DOI] [PubMed] [Google Scholar]
- 21.Lee VM-Y, Trojanowski JQ. Neurodegenerative tauopathies: Human disease and transgenic mouse models. Neuron. 1999;24(3):507–510. doi: 10.1016/s0896-6273(00)81106-x. [DOI] [PubMed] [Google Scholar]
- 22.Hong M, et al. Mutation-specific functional impairments in distinct tau isoforms of hereditary FTDP-17. Science. 1998;282(5395):1914–1917. doi: 10.1126/science.282.5395.1914. [DOI] [PubMed] [Google Scholar]
- 23.D’Souza I, et al. Missense and silent tau gene mutations cause frontotemporal dementia with parkinsonism-chromosome 17 type, by affecting multiple alternative RNA splicing regulatory elements. Proc Natl Acad Sci USA. 1999;96(10):5598–5603. doi: 10.1073/pnas.96.10.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt ML, Zhukareva V, Newell KL, Lee VM, Trojanowski JQ. Tau isoform profile and phosphorylation state in dementia pugilistica recapitulate Alzheimer’s disease. Acta Neuropathol. 2001;101(5):518–524. doi: 10.1007/s004010000330. [DOI] [PubMed] [Google Scholar]
- 25.Pan K-M, et al. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci USA. 1993;90(23):10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser H, Dickinson AG. Scrapie in mice. Agent-strain differences in the distribution and intensity of grey matter vacuolation. J Comp Pathol. 1973;83(1):29–40. doi: 10.1016/0021-9975(73)90024-8. [DOI] [PubMed] [Google Scholar]
- 27.Peretz D, et al. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci. 2001;10(4):854–863. doi: 10.1110/ps.39201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldfarb LG, et al. Fatal familial insomnia and familial Creutzfeldt-Jakob disease: Disease phenotype determined by a DNA polymorphism. Science. 1992;258(5083):806–808. doi: 10.1126/science.1439789. [DOI] [PubMed] [Google Scholar]
- 29.Telling GC, et al. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274(5295):2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 30.Prusiner SB. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet. 2013;47:601–623. doi: 10.1146/annurev-genet-110711-155524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanders DW, et al. Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron. 2014;82(6):1271–1288. doi: 10.1016/j.neuron.2014.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woerman AL, et al. Propagation of prions causing synucleinopathies in cultured cells. Proc Natl Acad Sci USA. 2015;112(35):E4949–E4958. doi: 10.1073/pnas.1513426112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Safar J, et al. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med. 1998;4(10):1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 34.Stein TD, Alvarez VE, McKee AC. Chronic traumatic encephalopathy: A spectrum of neuropathological changes following repetitive brain trauma in athletes and military personnel. Alzheimers Res Ther. 2014;6(1):4. doi: 10.1186/alzrt234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhukareva V, et al. Sporadic Pick’s disease: A tauopathy characterized by a spectrum of pathological τ isoforms in gray and white matter. Ann Neurol. 2002;51(6):730–739. doi: 10.1002/ana.10222. [DOI] [PubMed] [Google Scholar]
- 36.McKee AC, et al. TBI/CTE Group The first NINDS/NIBIB consensus meeting to define neuropathological criteria for the diagnosis of chronic traumatic encephalopathy. Acta Neuropathol. 2016;131(1):75–86. doi: 10.1007/s00401-015-1515-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kidd M. Paired helical filaments in electron microscopy of Alzheimer’s disease. Nature. 1963;197:192–193. doi: 10.1038/197192b0. [DOI] [PubMed] [Google Scholar]
- 38.Prusiner SB, et al. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell. 1983;35(2 Pt 1):349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 39.Prusiner SB, et al. Evidence for α-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc Natl Acad Sci USA. 2015;112(38):E5308–E5317. doi: 10.1073/pnas.1514475112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iba M, et al. Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer’s-like tauopathy. J Neurosci. 2013;33(3):1024–1037. doi: 10.1523/JNEUROSCI.2642-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iba M, et al. Tau pathology spread in PS19 tau transgenic mice following locus coeruleus (LC) injections of synthetic tau fibrils is determined by the LC’s afferent and efferent connections. Acta Neuropathol. 2015;130(3):349–362. doi: 10.1007/s00401-015-1458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clavaguera F, et al. Peripheral administration of tau aggregates triggers intracerebral tauopathy in transgenic mice. Acta Neuropathol. 2014;127(2):299–301. doi: 10.1007/s00401-013-1231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goedert M. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science. 2015;349(6248):1255555. doi: 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- 44.Clavaguera F, et al. Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc Natl Acad Sci USA. 2013;110(23):9535–9540. doi: 10.1073/pnas.1301175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kfoury N, Holmes BB, Jiang H, Holtzman DM, Diamond MI. Trans-cellular propagation of Tau aggregation by fibrillar species. J Biol Chem. 2012;287(23):19440–19451. doi: 10.1074/jbc.M112.346072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buée L, Bussière T, Buée-Scherrer V, Delacourte A, Hof PR. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Brain Res Rev. 2000;33(1):95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 47.Dinkel PD, Siddiqua A, Huynh H, Shah M, Margittai M. Variations in filament conformation dictate seeding barrier between three- and four-repeat tau. Biochemistry. 2011;50(20):4330–4336. doi: 10.1021/bi2004685. [DOI] [PubMed] [Google Scholar]
- 48.Nonaka T, Watanabe ST, Iwatsubo T, Hasegawa M. Seeded aggregation and toxicity of α-synuclein and tau: Cellular models of neurodegenerative diseases. J Biol Chem. 2010;285(45):34885–34898. doi: 10.1074/jbc.M110.148460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clavaguera F, et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 2009;11(7):909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahmed Z, et al. A novel in vivo model of tau propagation with rapid and progressive neurofibrillary tangle pathology: The pattern of spread is determined by connectivity, not proximity. Acta Neuropathol. 2014;127(5):667–683. doi: 10.1007/s00401-014-1254-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshiyama Y, et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53(3):337–351. doi: 10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 52.Prusiner SB, et al. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63(4):673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 53.Carlson GA, Westaway D, DeArmond SJ, Peterson-Torchia M, Prusiner SB. Primary structure of prion protein may modify scrapie isolate properties. Proc Natl Acad Sci USA. 1989;86(19):7475–7479. doi: 10.1073/pnas.86.19.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carlson GA, et al. Prion isolate specified allotypic interactions between the cellular and scrapie prion proteins in congenic and transgenic mice. Proc Natl Acad Sci USA. 1994;91(12):5690–5694. doi: 10.1073/pnas.91.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kovacs GG, et al. Aging-related tau astrogliopathy (ARTAG): Harmonized evaluation strategy. Acta Neuropathol. 2016;131(1):87–102. doi: 10.1007/s00401-015-1509-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong Q, Congdon EE, Nagaraja HN, Kuret J. Tau isoform composition influences rate and extent of filament formation. J Biol Chem. 2012;287(24):20711–20719. doi: 10.1074/jbc.M112.364067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marquié M, et al. Validating novel tau positron emission tomography tracer [F-18]-AV-1451 (T807) on postmortem brain tissue. Ann Neurol. 2015;78(5):787–800. doi: 10.1002/ana.24517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harada R, et al. Characteristics of tau and its ligands in PET imaging. Biomolecules. 2016;6(1):7. doi: 10.3390/biom6010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sander K, et al. Characterization of tau positron emission tomography tracer [(18)F]AV-1451 binding to postmortem tissue in Alzheimer’s disease, primary tauopathies, and other dementias. Alzheimers Dement. February 15, 2016 doi: 10.1016/j.jalz.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 60.Thomas SN, et al. Dual modification of Alzheimer’s disease PHF-tau protein by lysine methylation and ubiquitylation: A mass spectrometry approach. Acta Neuropathol. 2012;123(1):105–117. doi: 10.1007/s00401-011-0893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morris M, et al. Tau post-translational modifications in wild-type and human amyloid precursor protein transgenic mice. Nat Neurosci. 2015;18(8):1183–1189. doi: 10.1038/nn.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mair W, et al. FLEXITau: Quantifying post-translational modifications of tau protein in vitro and in human disease. Anal Chem. 2016;88(7):3704–3714. doi: 10.1021/acs.analchem.5b04509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martland HS. Punch drunk. J Am Med Assoc. 1928;91:1103–1107. [Google Scholar]
- 64.Owens BD, et al. Combat wounds in operation Iraqi Freedom and operation Enduring Freedom. J Trauma. 2008;64(2):295–299. doi: 10.1097/TA.0b013e318163b875. [DOI] [PubMed] [Google Scholar]
- 65.Hoge CW, et al. Mild traumatic brain injury in U.S. soldiers returning from Iraq. N Engl J Med. 2008;358(5):453–463. doi: 10.1056/NEJMoa072972. [DOI] [PubMed] [Google Scholar]
- 66.Terrio H, et al. Traumatic brain injury screening: Preliminary findings in a US Army Brigade Combat Team. J Head Trauma Rehabil. 2009;24(1):14–23. doi: 10.1097/HTR.0b013e31819581d8. [DOI] [PubMed] [Google Scholar]
- 67.Shively SB, et al. Characterisation of interface astroglial scarring in the human brain after blast exposure: A post-mortem case series. Lancet Neurol. 2016;15(9):944–953. doi: 10.1016/S1474-4422(16)30057-6. [DOI] [PubMed] [Google Scholar]
- 68.McKee AC, et al. The spectrum of disease in chronic traumatic encephalopathy. Brain. 2013;136(Pt 1):43–64. doi: 10.1093/brain/aws307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mackenzie IR, et al. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: An update. Acta Neuropathol. 2010;119(1):1–4. doi: 10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim E-J, et al. Selective frontoinsular von Economo neuron and fork cell loss in early behavioral variant frontotemporal dementia. Cereb Cortex. 2012;22(2):251–259. doi: 10.1093/cercor/bhr004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vonsattel JPG, et al. An improved approach to prepare human brains for research. J Neuropathol Exp Neurol. 1995;54(1):42–56. doi: 10.1097/00005072-199501000-00006. [DOI] [PubMed] [Google Scholar]