Significance
Filopodia are actin-based structures used by cells to sense chemical stimuli and promote adhesion to the extracellular environment during the development of multicellular organisms. Filopod formation in evolutionarily distant organisms requires MyTH4-FERM (myosin tail homology 4-band 4.1, ezrin, radixin, moesin; MF) myosins that consist of a motor domain paired with a tail domain that binds cytoskeletal and membrane proteins. Mutational analysis identified the minimal requirements for MF myosin function in filopod formation and revealed that the key features are conserved between amoebozoan and metazoan MF myosins. These findings have implications for understanding the fundamental principles of how filopodia form and how MF myosins function in phylogenetically distant organisms.
Keywords: myosin, actin, filopodia, cell motility, MyTH4-FERM
Abstract
The formation of filopodia in Metazoa and Amoebozoa requires the activity of myosin 10 (Myo10) in mammalian cells and of Dictyostelium unconventional myosin 7 (DdMyo7) in the social amoeba Dictyostelium. However, the exact roles of these MyTH4-FERM myosins (myosin tail homology 4-band 4.1, ezrin, radixin, moesin; MF) in the initiation and elongation of filopodia are not well defined and may reflect conserved functions among phylogenetically diverse MF myosins. Phylogenetic analysis of MF myosin domains suggests that a single ancestral MF myosin existed with a structure similar to DdMyo7, which has two MF domains, and that subsequent duplications in the metazoan lineage produced its functional homolog Myo10. The essential functional features of the DdMyo7 myosin were identified using quantitative live-cell imaging to characterize the ability of various mutants to rescue filopod formation in myo7-null cells. The two MF domains were found to function redundantly in filopod formation with the C-terminal FERM domain regulating both the number of filopodia and their elongation velocity. DdMyo7 mutants consisting solely of the motor plus a single MyTH4 domain were found to be capable of rescuing the formation of filopodia, establishing the minimal elements necessary for the function of this myosin. Interestingly, a chimeric myosin with the Myo10 MF domain fused to the DdMyo7 motor also was capable of rescuing filopod formation in the myo7-null mutant, supporting fundamental functional conservation between these two distant myosins. Together, these findings reveal that MF myosins have an ancient and conserved role in filopod formation.
Cells interact with their environment through protrusions such as filopodia that form in response to extracellular cues and mediate initial contact with the substrate. Filopodia are slender, actin-filled membrane projections that are highly dynamic, growing and shrinking from peripheral regions of cells, such as lamellipodia and the dorsal surface (1). A wide variety of cell types including amoebae such as Dictyostelium discoideum (2) and Acanthamoeba (3), as well as mammalian vascular endothelial cells (4) and developing neurons (5), extend filopodia. Filopodia are typically 1–10 µm long and 0.1–0.3 µm in diameter and have a core of 10–30 parallel actin filaments with a protein-rich complex at their tip (1, 6, 7). Modified forms of filopodia such as dendritic spines, cytonemes, and tunneling nanotubes promote intercellular communication during multicellular development (8–10). Defects in filopod formation alter cell spreading and adhesion (2, 11, 12), whereas overproduction of filopodia or filopodia-like protrusions is associated with increased invasiveness of metastatic cancer cells (13–15).
Filopod elongation is triggered by small GTPase activity (1, 16) and is driven by the activity of actin elongation factors including vasodilator-stimulated phosphoprotein (VASP) and formins, and the actin core is stabilized by actin cross-linking proteins (1). A MyTH4-FERM (myosin tail homology 4-band 4.1, ezrin, radixin, moesin; MF) myosin motor is also required. A broad survey of genomes reveals that these essential filopodial proteins are evolutionarily conserved between Holozoa (a group that includes Metazoa and their closest single-cell relatives such as the choanoflagellate Monosiga brevicollis) and Amoebozoa (17). The existence of shared core filopodial machinery suggests that diverse organisms may use fundamentally conserved means of generating filopodia.
Organisms in a range of species have been reported to make filopodia or filopodia-like protrusions, but little is known about how these structures are generated by nonmetazoan eukaryotes (17). Amoebozoa share a common ancestor with animals and fungi (18), thus making Dictyostelium an excellent model system to test the mechanistic conservation and diversity of these structures over evolutionary time (nearly a billion years of independent evolution). A number of proteins important for filopod formation in Metazoa are conserved with similar roles in Dictyostelium. The widely conserved actin regulator VASP and the Dictyostelium unconventional MF myosin DdMyo7 have critical roles in filopod initiation, and the formin dDia2 is required for filopod elongation (2, 11, 19). Although Dictyostelium lacks the key metazoan small GTPase Cdc42, the related GTPase Rac1a plays an analogous role in stimulating filopod formation (16). Instead of the metazoan actin cross-linking protein fascin (20) the actin-binding proteins ABP-34 and EF-1α are believed to play a role in actin cross-linking in Dictyostelium filopodia (21). The different actin cross-linking proteins used in Dictyostelium likely give rise to the less organized, shorter actin filaments in their filopodia (6). Despite these differences, the abundance of structural and genetic similarities argues that filopod formation is a conserved cell biological process in Metazoa and Amoebozoa.
Two different MF myosins have been found to be essential for filopodia formation in widely divergent organisms: myosin 10 (Myo10) in vertebrates and DdMyo7 in the social amoeba Dictyostelium (2, 22). These two phylogenetically distant MF myosins have several features in common (Fig. 1B). They both have a number of light-chain–binding sites (IQ motifs), a single α-helix (SAH) domain (23), and a C-terminal MF domain. Like several MF myosins, such as the Myo7 from humans or flies, DdMyo7 differs from Myo10 in having a second MF domain and an Src homology 3 (SH3) domain inserted before the C-terminal MF domain that is characteristic of this group of myosins. In contrast to DdMyo7, the tail of Myo10 (Fig. 1B) has pleckstrin homology (PH) domains in place of the internal MF and SH3 domains (24). Despite these differences in overall structure, the myo7-null Dictyostelium cells and HeLa or breast cancer cells with reduced Myo10 expression both exhibit a striking lack of filopodia (2, 22). The findings suggest that these two distinct MF myosins have functionally equivalent roles in filopod formation, one that has been conserved through almost a billion years of independent evolution. However, the phylogenetic relationship between Myo10 and DdMyo7 is unresolved, and it is unclear how these two motors cooperate with their respective cohorts of filopodial proteins to build filopodia. DdMyo7 has been classified as Myo7, Myo22, Myo25, or could not be classified (2, 25–27) (see also www.cymobase.org/cymobase). The uncertainty about the relationship between the metazoan and amoebozoan MF myosins and the differences in actin organization seen in Dictyostelium and mammalian filopodia raise questions about whether metazoan Myo10 and amoebozoan DdMyo7 myosins contribute to filopod formation in a similar way and whether their activities are conserved or are specific to each organism. In other words, do the filopodial MF myosins DdMyo7 and Myo10 possess convergent but distinct functions in filopodia, or do they share a conserved role in filopod formation despite their structural and phylogenetic differences? A functional dissection of the Dictyostelium MF myosin, DdMyo7, has been undertaken to begin to address this question.
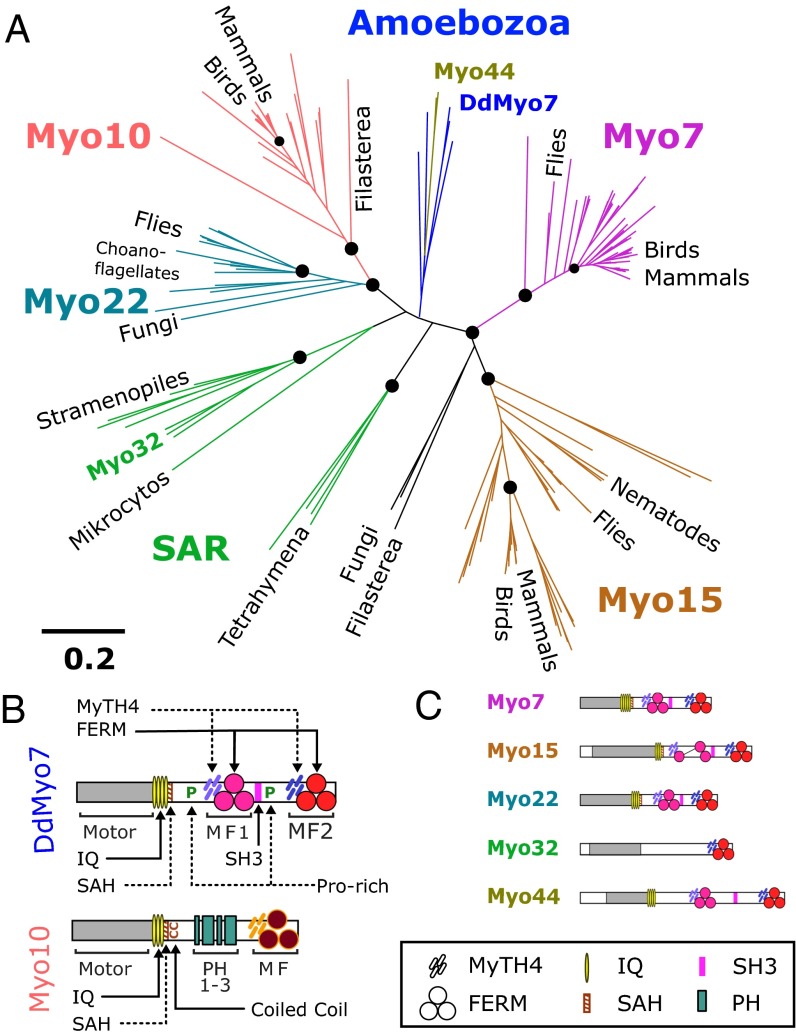
Fig. 1.
Evolutionary relationships of MyTH4-FERM myosins. (A) Phylogenetic analysis of the MF domains of myosins from representative organisms as determined by SATé (30). Likelihood values at major branch points (>95%) are indicated by circles, and the scale is as defined by FastTree (54). (B) Domain organization of DdMyo7 and Myo10, two MF myosins involved in filopod formation. Domains of DdMyo7 include the motor (myosin catalytic domain), IQ (light-chain–binding motifs 1–4); SAH (single α-helix); proline-rich regions; MyTH4 (myosin tail homology 4); FERM (band 4.1, ezrin, radixin, moesin); and SH3 (Src homology 3). Myo10 contains three PH (pleckstrin homology) domains. (C) Domain organization of other major MF myosin families. See the symbol legend at bottom for identification of domains.
Results
Deeper understanding of the role of DdMyo7 during filopod formation is needed to uncover functionally conserved and specific roles of MF myosins, particularly in comparison with Myo10’s role in filopod formation. Thus, a detailed characterization of DdMyo7’s function in vivo was undertaken. A phylogenetic reassessment of MF myosins also was conducted to determine if the amoebozoan Myo7s are indeed direct orthologs of any of the metazoan MF myosins or are a functionally distinct class of MF myosins that arose from a common ancestor.
Phylogenetic Relationship Between DdMyo7 and Metazoan Myosins.
The evolutionary time (∼600 My) (28) between Amoebozoa and Metazoa raises the question of how DdMyo7 differs from metazoan Myo10, given their common role in filopod formation. The relationship between these MF myosins was unclear in previous studies of myosin diversity based on a comparison of motor domain sequences (26, 27; see also www.cymobase.org/cymobase). A phylogenetic analysis of the MF myosins using their defining feature, the C-terminal MyTH4-FERM domain, was conducted to resolve their relationships better. Full-length MF myosin sequences were gathered from representative organisms across all eukaryotes known to possess MF myosins. A total of 162 sequences from 58 distinct species were used for this analysis (Dataset S1), including the metazoan MF myosins (Myo7, -10, --15, and -22), fungal and choanoflagellate Myo22, and amoebozoan MF myosins. Sequences for additional MF myosins including Myo32 were gathered from the Stramenopiles, Alveolates, and Rhizaria (SAR) clade, a branch of unicellular eukaryotes that includes ciliates such as Tetrahymena and oomycetes such as the pathogenic water mold Phytophthora (29). Phylogenetic relationships between the 162 C-terminal MF domain sequences were derived using simultaneous alignment and tree estimation (SATé) (30).
The resulting phylogenetic tree (Fig. 1A and Datasets S2 and S3) shows that the amoebozoan MF myosins form a single family with two myosin classes. The amoebozoan Myo7 (named for its structural resemblance to the animal Myo7) is found in social amoebae as well as in solitary species such as Acanthamoeba. DdMyo7 (also known as “MyoI”) is the best-characterized example of this class. A second class of myosin present in social amoebae, Myo44 (Dictyostelium MyoG), does not play a role in filopodia but has novel functions in chemotactic signaling (31). The amoebozoan MF myosins are distinguished with high confidence from the holozoan Myo7, Myo10, and Myo22 protein families and branch in a poorly defined region near the center of the tree, close to the presumptive root in the SAR group (Fig. 1A). The topology of the tree suggests that the common ancestor of these organisms contained a single MF myosin. Comparison of the domain organization of the myosins on each branch (Fig. 1 B and C) reveals a common structure for the DdMyo7, Myo22, and holozoan Myo7 proteins, each possessing a motor domain and a tail region that contains three domains (two MF domains and one SH3 domain) but lacking PH domains. The tree suggests that the ancestral MF myosin underwent duplication subsequent to the emergence of the Amoebozoa, resulting in the proteins that established the Myo10/22 and Myo7/15 branches (Fig. 1A). The striking similarity in tail structure in DdMyo7, Myo22, and Myo7 suggests that these proteins have the ancestral tail structure. Amoebozoan MF myosins are exemplified by DdMyo7, which is structurally dissimilar to Myo10 but is similar in function, suggesting that core molecular features needed for filopod formation will be present in both DdMyo7 and Myo10.
DdMyo7 Is Present in Filopodia and Is Required for Their Formation.
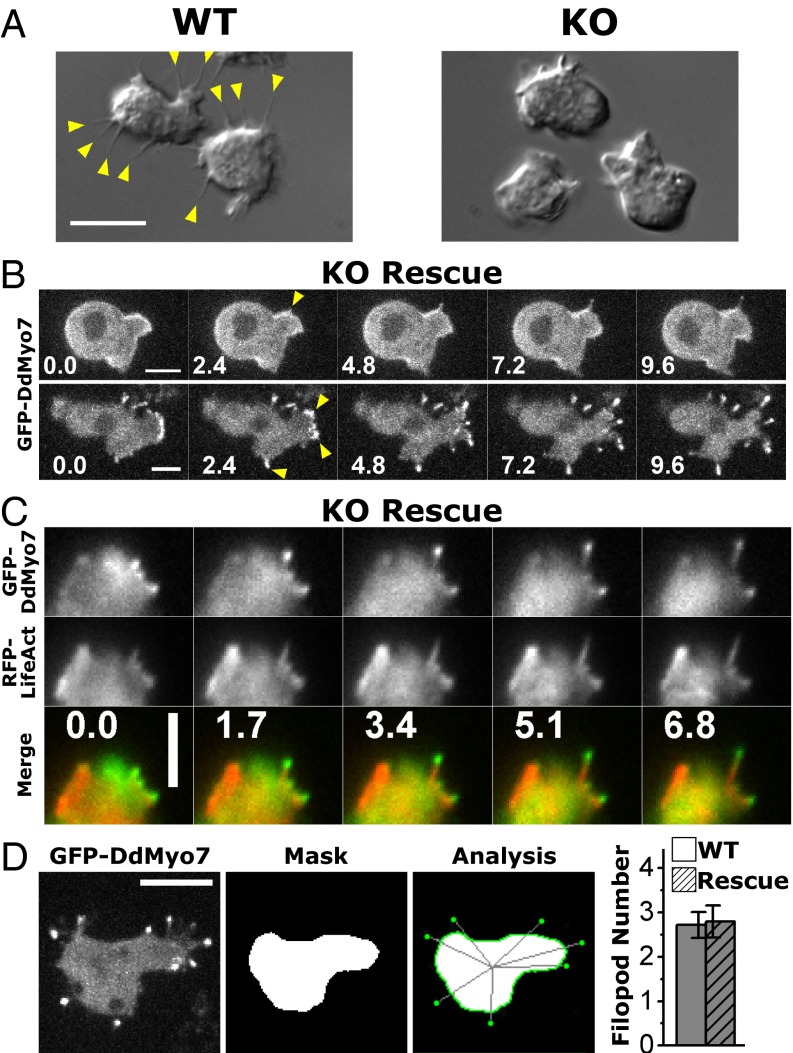
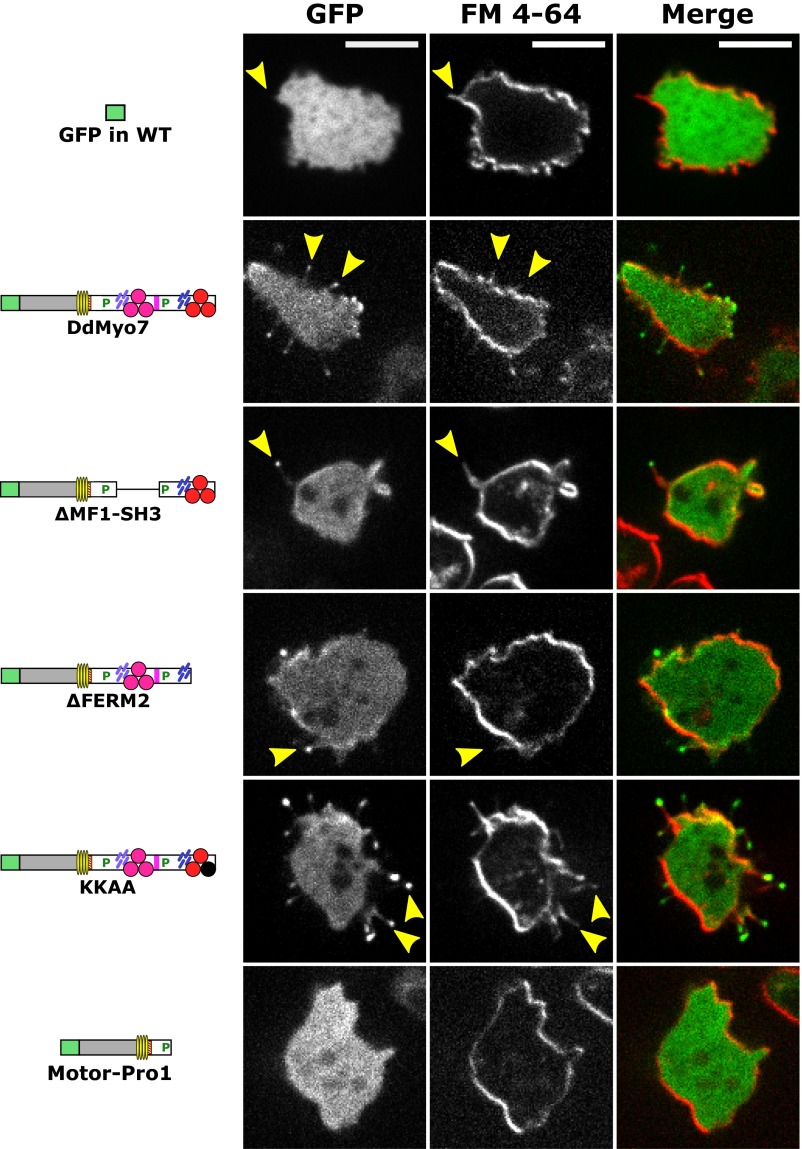
Filopodia are a common feature of Dictyostelium cells from the vegetative phase through early development (2). Wild-type Dictyostelium frequently display multiple filopodia (Fig. 2A), which appear as slender membrane projections that actively extend from the cell body close to the substrate, whereas myo7-null cells display virtually none (Fig. 2A). Filopod formation was rescued by expression of full-length GFP-tagged DdMyo7 (Fig. S1; referred to as “DdMyo7” hereafter). Spinning-disk confocal microscopy showed DdMyo7 localized to the actin cortex of an active leading edge (Fig. 2B), in filopodia tips, and in the cytosol, as previously reported (2, 32).
Fig. 2.
DdMyo7 is present in filopodia from their initiation and at filopod tips during elongation. (A) DIC images of wild-type and myo7-null (KO) cells illustrating the absence of filopodia in cells lacking DdMyo7. (Scale bar: 10 μm.) (B) Filopod initiation in two KO cells expressing GFP-DdMyo7 over 10 s. The arrowheads point to emerging and extending filopodia that show the myosin concentrating near the distal tips. (Scale bars: 5 μm.) (C) Time series showing formation of a filopodium at the edge of an actin-rich pseudopod in a myo7-null cell expressing GFP-DdMyo7 and RFP-LifeAct over time (seconds). (Scale bar: 4 μm.) (D) Automated analysis of filopod number in maximum intensity projection confocal images. Shown is a representative cell expressing GFP-DdMyo7. (Scale bar: 10 μm.) The cell body is masked (Mask), and filopod tips are located by GFP intensity before registering them to the cell (Analysis). Quantification of multiple images from either wild-type or myo7-null cells expressing GFP-DdMyo7 yields an average of 2.8 filopodia per cell, a baseline for further assays (see also Table 1).
Fig. S1.
Western blot analysis of GFP-DdMyo7 fusions. Western blotting of total cell lysates to validate the expression of GFP fusions. (A) Wild-type and myo7-null lysates probed with anti-Myo7 antibody showing the presence of the ∼270-kDa heavy chain in wild-type cells (Ax2) that is missing from the myo7-null cells. (B and C) Blots of wild-type (B) or myo7-null (C) cells expressing the DdMyo7 fusions probed with anti-GFP to detect the expressed heavy chain. The asterisk indicates the position of the full-length GFP fusion. All samples shown were also blotted for the 125-kDa MyoB heavy chain that is used as a loading control. Note that lanes from several independent blots are shown.
Filopod initiation events were monitored in cells coexpressing DdMyo7 and RFP-LifeAct to visualize actin filaments (33). DdMyo7 is present in the cytosol, and in a typical initiation event it was seen to become concentrated at the cortex (Fig. 2C). Then, a bright spot of myosin appeared, projecting from an actin-rich pseudopod. Filopodia elongated several micrometers within 7 s, with actin present along the length and DdMyo7 concentrated close to the filopod tip throughout the elongation process (Fig. 2C). These results show that filopod initiation is a highly dynamic and rapid process.
Filopod formation activity was measured in a live-cell assay by counting the number of filopodia per cell following 1 h of starvation, a condition that consistently stimulates the production of filopodia in wild-type Dictyostelium. Arbitrary fields of view were selected for imaging by differential interference contrast (DIC) and confocal GFP fluorescence. Blinded manual analysis demonstrated a low frequency of filopodia-like protrusions in myo7-null cells (Fig. S2 and Table S1) and that expression of DdMyo7 rescues filopod formation. DdMyo7 fluorescence was distinctly observed at the tips of protrusions, whereas GFP alone did not label protrusions (Fig. S3). Thus, although DIC imaging could not resolve filopodia from other types of protrusions such as ruffles or retraction fibers, DdMyo7 localization at the tip identified the protrusions as filopodia. Automated image analysis was used to measure the number and length of filopodia (Fig. 2D). The number of filopodia in cells expressing DdMyo7 was not significantly different in wild-type and rescued myo7-null cells (2.8 ± 0.4 filopodia per cell). The establishment of a quantitative assay for DdMyo7-based filopod formation allowed in-depth examination of the molecular requirements for this MF myosin in filopod formation.
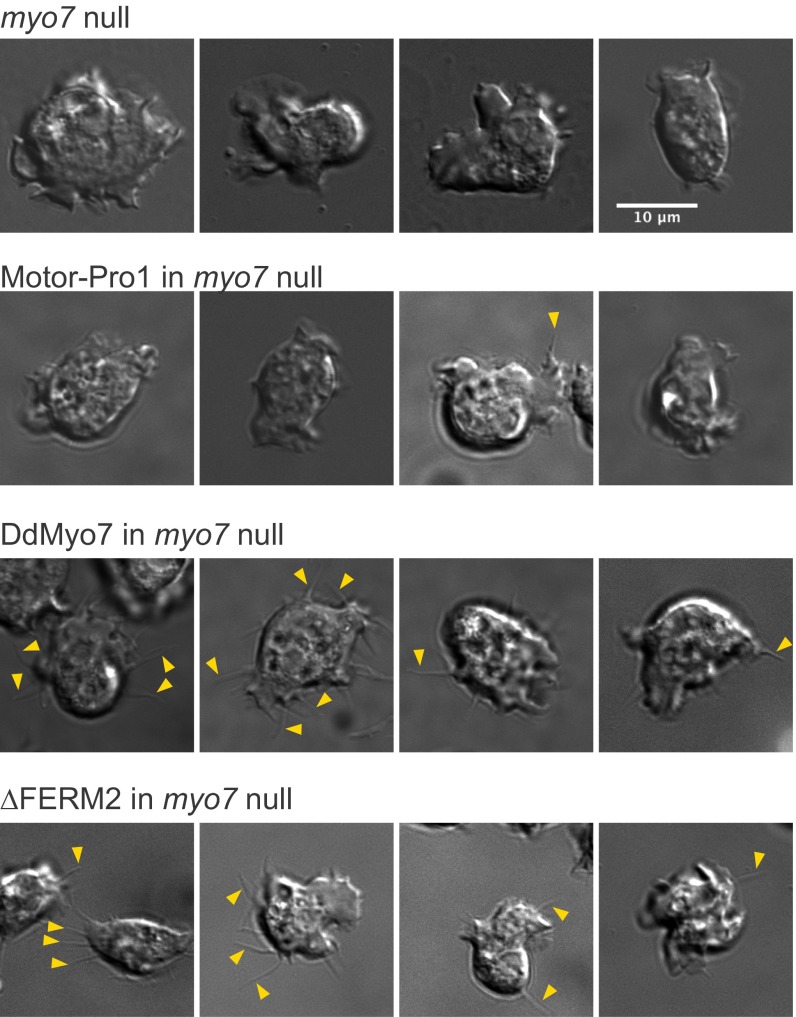
Fig. S2.
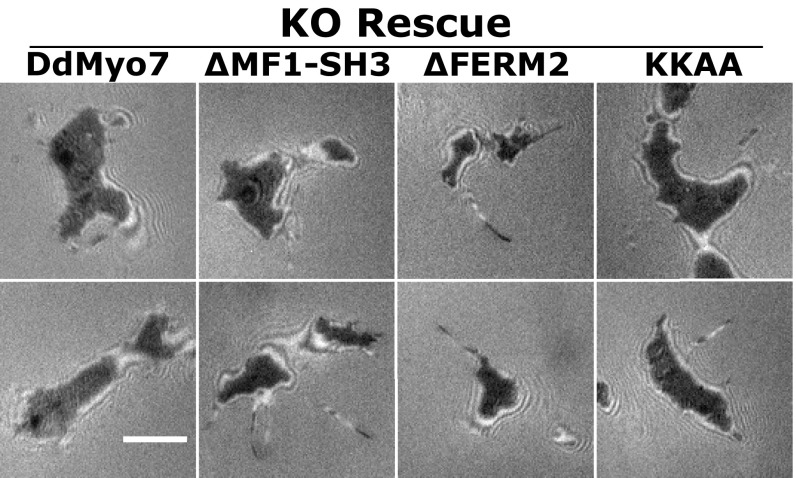
DIC images of Dictyostelium cells under optimal conditions for filopodia formation. Randomly selected images of cells imaged by DIC microscopy illustrating the scoring of filopodia for comparison with scoring by filopodial tip GFP fluorescence. Shown are the myo7-null cell line that does not extend filopodia, a nonrescued cell line (motor-Pro1), and two rescued lines (DdMyo7 and ∆FERM2). Examples of filopodial extensions are indicated by arrowheads. Nonrescued cell lines show very rare protrusions compared with rescued cell lines (also see Table S1). (Scale bar: 10 µm.)
Table S1.
Blinded scoring of filopodial protrusions by GFP fluorescence and DIC
| GFP-tagged protein in KO cells | Total no. of cells | No. of GFP+ cells | Protrusions per cell, GFP | No. of protrusions, GFP | No. of protrusions, DIC | % cells with protrusions, GFP | % cells with protrusions, DIC |
| None | 128 | — | — | — | 15 | — | 6.3 |
| Rescue | |||||||
| DdMyo7 | 113 | 83 | 1.51 | 125 | 116 | 51.8 | 48.2 |
| ∆MF1-SH3 | 137 | 82 | 1.27 | 104 | 86 | 43.9 | 36.3 |
| ∆FERM2 | 82 | 39 | 1.33 | 52 | 52 | 35.9 | 35.9 |
| KKAA | 81 | 32 | 5.03 | 161 | 91 | 81.3 | 62.5 |
| No rescue | |||||||
| Motor-SAH | 138 | 74 | 0.03 | 2 | 1 | 2.7 | 1.4 |
| Motor-Pro1 | 266 | 126 | 0.01 | 1 | 11 | 0.8 | 2.4 |
| Tail | 1,028 | 58 | 0.00 | 0 | 0 | 0.0 | 0.0 |
| Chimeras | |||||||
| Motor-Pro1-HsM10M4 | 691 | 400 | 0.05 | 18 | 15 | 3.5 | 2.3 |
| Motor-Pro1-HsM10MF | 542 | 255 | 0.09 | 22 | 71 | 5.9 | 11.8 |
| Motor-Pro1-M44MF2 | 318 | 152 | 0.06 | 9 | 15 | 3.9 | 4.6 |
Paired DIC and GFP still images were manually scored for filopodia in several myo7-null (KO) cell lines expressing GFP-Myo7 fusions found to rescue or not rescue filopod extension. DIC images were first analyzed for cells extending slender projections, and the number of projections per cell was scored. The corresponding fluorescence image then was analyzed for GFP-expressing cells, and the number of GFP-tipped projections with a clear connection to the cell body was counted for each cell. The values shown in the table are the values obtained for the number of filopodia per cell for the GFP-expressing cells as observed both by DIC and fluorescence and the percent of GFP-expressing cells that were observed to extend protrusions in DIC. Note the strong correspondence between filopodia scored by GFP fluorescence and protrusions seen in DIC.
Fig. S3.
Filopodia visualized by GFP-DdMyo7 and the fluorescent membrane marker FM 4-64. Dictyostelium myo7-null cells expressing full-length or mutant DdMyo7 proteins or GFP alone were starved for 1 h and incubated with 0.25 µg/mL FM 4-64 membrane dye for 1–5 min before confocal imaging. A single image plane is shown for the GFP (Left) and red FM 4-64 (Center) channels; the merge of the two channels is shown on the right. DdMyo7 is enriched in filopod tips, and the filopodia are visible with FM 4-64 as well as GFP staining along their length (yellow arrows). Note the cytosolic localization of GFP expressed in wild-type cells (Top Row) contrasted with the localization of GFP-DdMyo7 at the filopod tip in the myo7-null cells.
Complementary Roles of the DdMyo7 Head and Tail in Filopod Initiation.
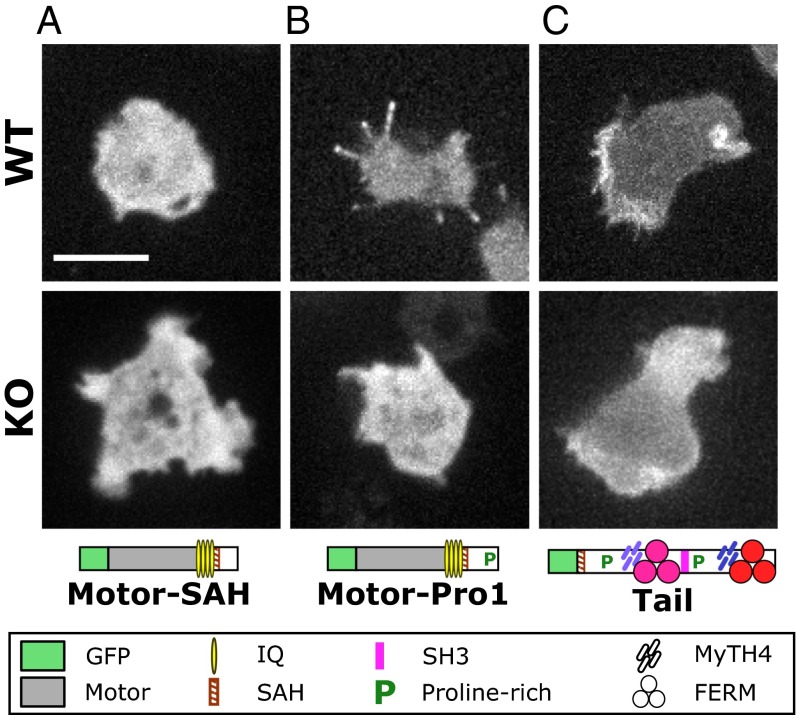
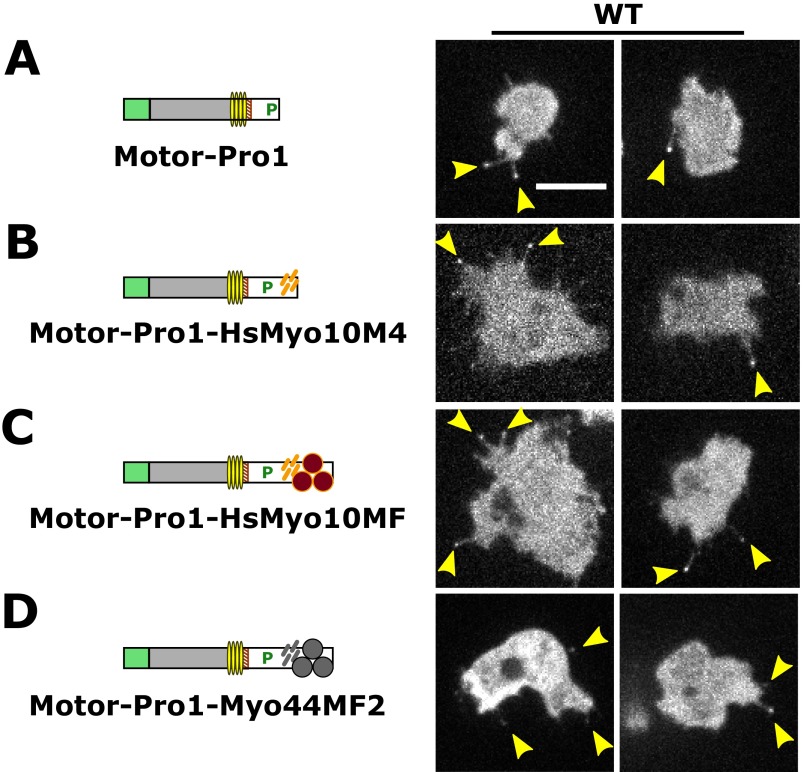
The motor activity of Myo10 may be sufficient for filopod formation because expression of truncated dimeric Myo10 motors was reported to induce filopodia in mammalian cells (34). Whether the DdMyo7 motor domain is necessary or sufficient for filopod initiation was tested with a series of mutants. Expression plasmids encoding DdMyo7 mutant proteins were transformed into wild-type cells as well as into myo7-null cells (to assay their effect on filopod formation). Transformants were screened for GFP fluorescence, and expression was confirmed by Western blot (Fig. S1). The role of the motor domain was tested with two tailless mutants encompassing the motor domain and a putative lever arm region consisting of four light-chain–binding IQ motifs, an SAH domain, and an uncharacterized sequence potentially extending the lever arm (motor-SAH; amino acids 1–1020) or this region plus the first proline-rich region (motor-Pro1; amino acids 1–1115) (Fig. 3A). Tailless mutants displayed diffuse cytosolic localization in both wild-type and myo7-null cells (Fig. 3 A and B), did not associate with pseudopodia, and did not rescue filopod formation in myo7-null cells. The longer motor-Pro1, in contrast to motor-SAH, did localize to filopod tips in wild-type cells, suggesting that the Pro1 region may aid the motor processivity necessary for DdMyo7 to reach filopod tips. A headless mutant comprised of the SAH and the full tail region was strongly enriched in pseudopodia of both wild-type and myo7-null cells but failed to rescue filopodia in myo7-null cells (Fig. 3C). Localization of the headless mutant to filopod tips was variable in wild-type cells, with the DdMyo7 tail either not reaching the tip (Fig. 3C) or being weakly enriched there (Fig. S4B). These observations establish that the tail region is required for localization of DdMyo7 to the cortical region, possibly through binding to a cortical or membrane-associated partner, and that the motor domain is required to target DdMyo7 to filopod tips and/or retain DdMyo7 at the tip of an elongating filopod. Thus, both the motor and tail are required for filopod formation.
Fig. 3.
The head and tail of DdMyo7 are required for filopod formation. (A) The DdMyo7 motor-SAH domains do not localize to filopodia when expressed in wild-type cells and do not rescue filopod formation in the myo7-null (KO) cells. (Scale bar: 10 μm.) (B) The motor-Pro1 region localizes to filopodia in wild-type cells but does not rescue filopod formation in myo7-null cells. (C) The DdMyo7 tail localizes to the cell leading edge in both wild-type and myo7-null cells and weakly localizes to filopodia in wild-type cells but does not rescue filopod formation in myo7-null cells.
Fig. S4.
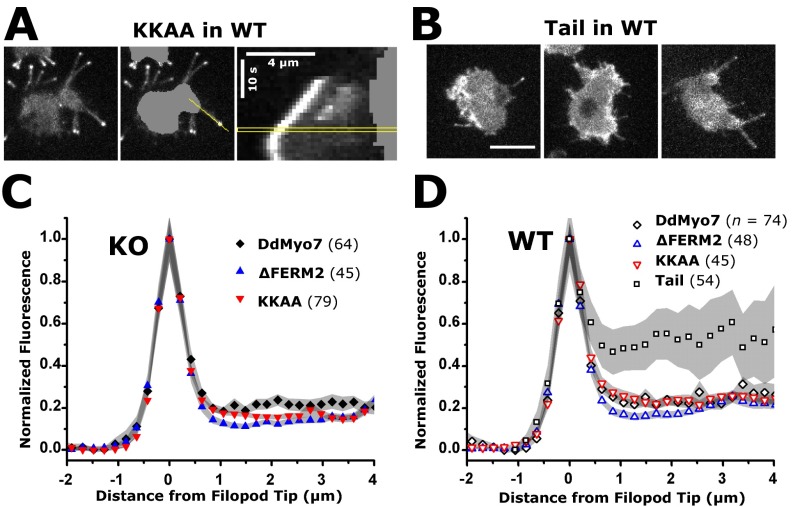
Deletion or mutation of the C-terminal FERM2 domain does not alter the distribution of GFP-DdMyo7 along the length of the filopodium. The fluorescence intensity of GFP-DdMyo7 was measured using the peak intensity in filopodia tips to align filopodia on the x axis (positive values are toward the cell body) and to localize DdMyo7 mutants in filopodia using the mean intensity along the length. (A, Left) A representative wild-type (WT) cell expressing the KKAA mutant protein. (Center) Images were processed by segmentation of the cell body followed by an intensity line scan when the filopod was near its maximum length. (Right) Kymograph with the time point used for the line scan outlined in yellow. (Scale bars: 4 μm × 10 s.) (B) The tail expressed in wild-type cells is occasionally localized to filopodia tips and along the length of the filopodium. (Scale bar: 10 μm.) (C) Mean GFP intensity along the length of filopodia extended by myo-7 null (KO) cells expressing full-length DdMyo7, the C-terminal FERM2 deletion (∆FERM2), or the double point mutant (KKAA). (D) Mean GFP intensity along the length of filopodia extended by wild-type cells expressing full-length or mutant DdMyo7 or the DdMyo7 tail. Note that a small portion of DdMyo7 is located uniformly along the actin-rich shaft in cells expressing the wild-type or deletion mutants. In contrast, higher relative levels of the tail are observed along the shaft. The tail does not rescue filopod formation and therefore is not shown in KO cells. The peak value (x = 0) is the normalized mean intensity of n filopodia (indicated for each condition). Shading indicates SEM of the number of filopodia averaged at each x value.
Functional Redundancy of DdMyo7 FERM Domains.
The role of the dual MF domains in DdMyo7 was examined by analysis of the tail region of this myosin. The MyTH4 and FERM domains form a compact supramodule, and in mammalian Myo7A the SH3 domain is coupled to the FERM domain of MF1 (35, 36). FERM domains interact with MF myosin cargo proteins, including adhesion and signaling receptors, and also can mediate autoinhibition of the motor (24). Therefore, removal of an MF domain was predicted to affect DdMyo7 function. As with the headless and tailless mutants, DdMyo7 deletion mutants (Fig. 4A) were expressed in wild-type and myo7-null cells. The number of filopodia per cell and the length and elongation velocity of the filopodia were measured in mutants that generate filopodia to determine which aspect of filopod formation was altered by domain deletion.
Fig. 4.
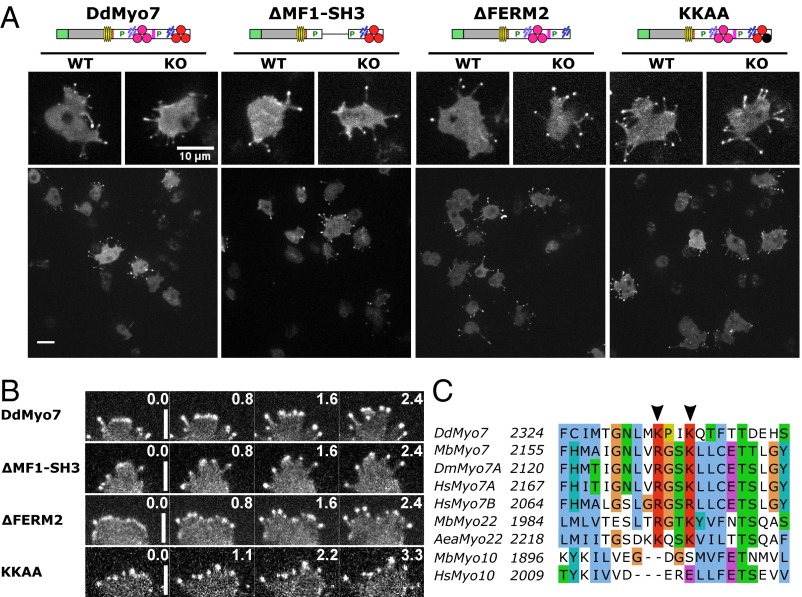
Rescue of filopod formation by mutant DdMyo7. (A) Domain organization of DdMyo7 mutants, with GFP fused to the N termini, is shown (see Fig. 1 for symbol key) with representative confocal fluorescence images of full-length DdMyo7, deletion mutants lacking the MF1-SH3 or C-terminal FERM2 domain, and the KKAA mutant with changes in conserved basic residues in the FERM2 domain. The large panels show representative fields used in analysis of each DdMyo7 expressed in myo7-null (KO) cells. (Scale bar: 10 μm.) (B) Detail of filopodia in extending pseudopods of KO cells expressing wild-type and mutant DdMyo7 proteins. Note the localization of DdMyo7 to the leading edge and subsequently to filopod tips during elongation. The time lapse is indicated in seconds. (Scale bar: 4 μm.) (C) Sequence alignment showing a motif (K/RxxK/R) that is conserved in the FERM2 domain of MF myosins including DdMyo7, animal and choanoflagellate Myo7, and Myo22 but excluding Myo10. Arrowheads indicate conserved basic residues. Aea, Aedes aegyptii; Dd, Dictyostelium discoideum; Dm, Drosophila melanogaster; Hs, Homo sapiens; Mb, Monosiga brevicollis. Sequence positions are indicated on the left, and conserved residues are highlighted: blue, hydrophobic; red, basic; magenta, acidic; green, Asn/Thr/Ser; cyan, His/Tyr; orange, Gly; yellow, Pro.
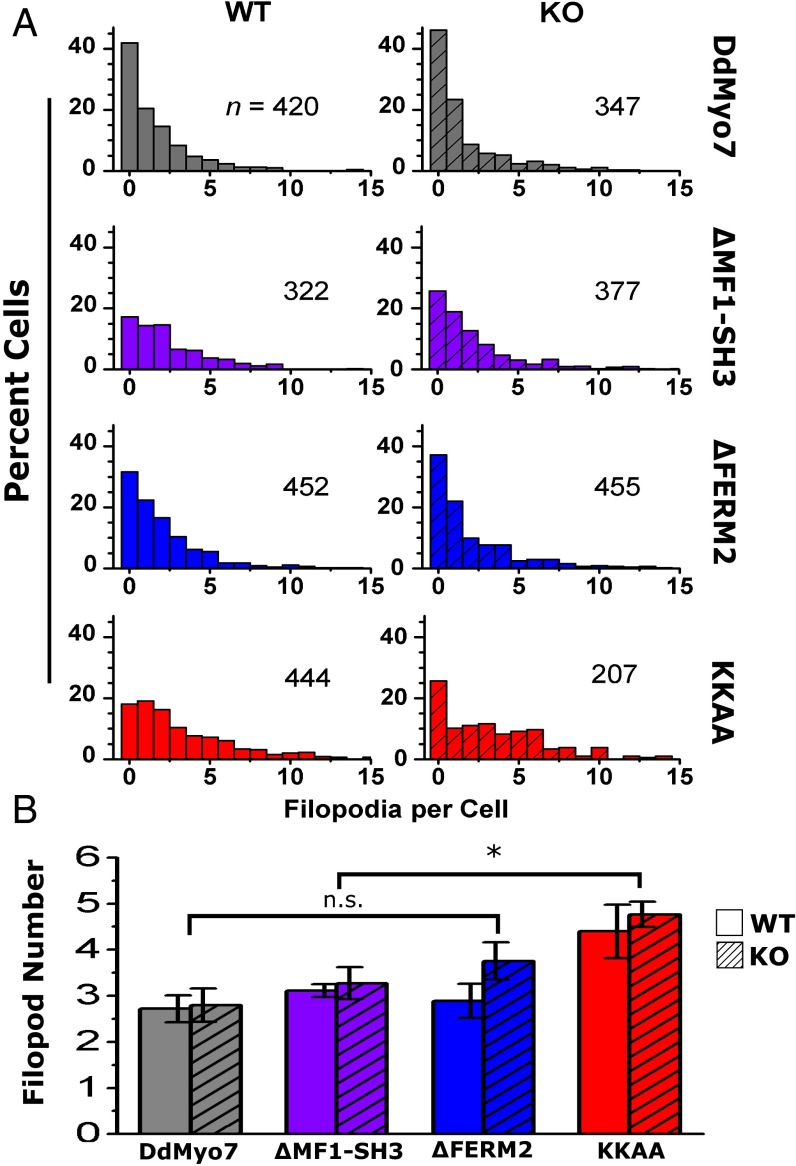
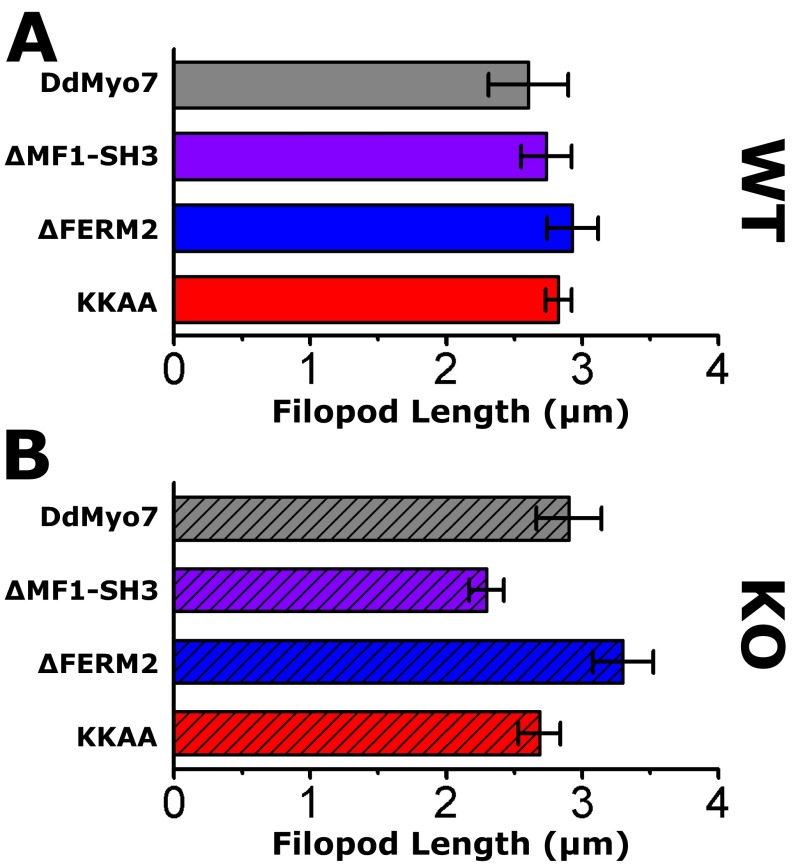
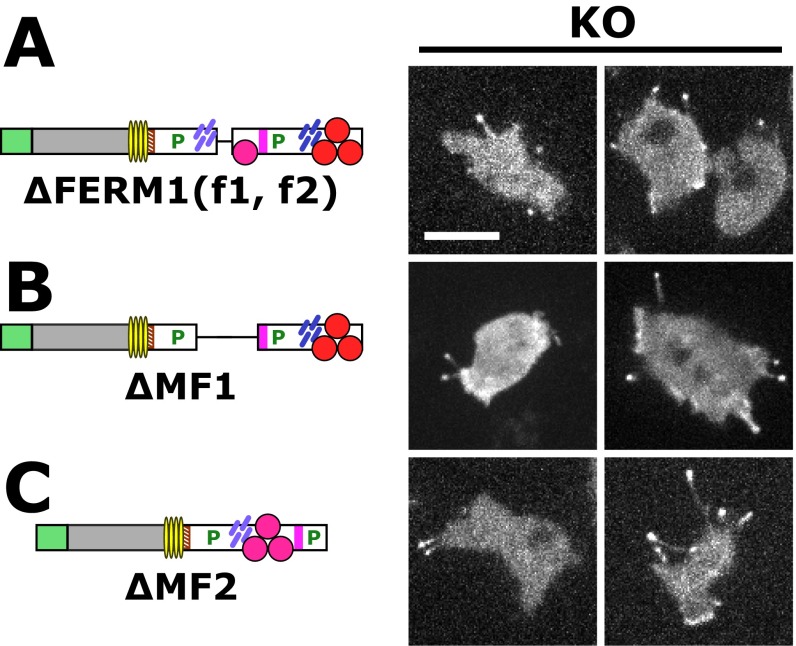
The contribution of the internal MF domain was examined with a DdMyo7 ∆MF1-SH3 mutant that is stably expressed and rescues filopod formation in myo7-null cells (Fig. 4A and Fig. S1). The ∆MF1-SH3 mutant localizes to both the leading edge and the tips of extending filopodia in wild-type and myo7-null cells (Fig. 4 A and B). Wild-type and myo7-null cells expressing either the ∆MF1-SH3 mutant or full-length DdMyo7 produce similar numbers of filopodia (Fig. 5 and Table 1), although the average length of filopodia in myo7-null cells expressing ∆MF1-SH3 was reduced by 20% (see Fig. S7 and Table 1). Smaller deletions encompassing only the MF1 or FERM1 domains [∆MF1 and ∆FERM1(f1, f2)] also did not affect the ability of DdMyo7 to promote filopod formation (Fig. S5). Similarly, a deletion mutant targeting MF2 also rescued filopod formation in myo7-null cells (Fig. S5). Thus, the MF domains appear to function redundantly with a single MF domain, either MF1 or MF2, being essential for the filopod-formation activity of this myosin.
Fig. 5.
Filopod formation by DdMyo7 is regulated by the C-terminal FERM2 domain. Filopodia were counted in Dictyostelium cells expressing DdMyo7. (A) Filopod number in cells expressing full-length DdMyo7, in DdMyo7 with MF1-SH3 or FERM2 domain deletions (ΔMF1-SH3, ΔFERM2), and in DdMyo7 with a double point mutation in the FERM2 domain (KKAA). Data are shown for wild-type (solid-colored bars) and myo7 null (KO) (hatched bars). The number of GFP+ cells (n) includes cells with and without filopodia. (B) Average number of filopodia per cell. Filopod number was defined as the total filopodia divided by the number of cells with filopodia. Cells expressing the KKAA mutant significantly increased filopod number by 66% compared with DdMyo7 (*P = 0.00008), by 43% relative to ΔMF1-SH3 (P = 0.002), and by 36% relative to ΔFERM2 (P = 0.003, multiple ANOVA with contrasts). Differences among other mutants were not significant (n.s.); error bars indicate the SEM; see Table 1.
Table 1.
Filopodia number, length, and elongation velocity in DdMyo7-expressing cells
| Back-ground | Protein | Filopod number ± SEM* | % change (95% confidence interval) | P | Filopod length ± SEM*, µm | % change (95% confidence interval) | P | Elongation velocity ± SEM†, µm/s | % change (95% confidence interval) | P | N |
| Wild type | DdMyo7 | 2.7 ± 0.3 | — | — | 2.6 ± 0.3 | — | — | N.d. | — | — | 4 |
| ∆MF1-SH3 | 3.1 ± 0.1 | 15 (−31, 61) | 0.51 | 2.7 ± 0.2 | 5 (−21, 31) | 0.70 | N.d. | — | — | 4 | |
| ∆FERM2 | 2.9 ± 0.4 | 6 (40, 52) | 0.78 | 2.9 ± 0.2 | 12 (−13, 39) | 0.34 | N.d. | — | — | 4 | |
| KKAA | 4.4 ± 0.6 | 62 (16, 108) | 0.01 | 2.8 ± 0.1 | 8 (−18, 35) | 0.52 | N.d. | — | — | 4 | |
| KO | DdMyo7 | 2.8 ± 0.4 | — | — | 2.9 ± 0.2 | — | — | 0.40 ± 0.03 (26) | — | — | 6 |
| ∆MF1-SH3 | 3.3 ± 0.3 | 17 (−20, 53) | 0.36 | 2.3 ± 0.1 | −20 (−2, −39) | 0.02 | 0.37 ± 0.02 (48) | −7 (−28, 13) | 0.77 | 6 | |
| ∆FERM2 | 3.8 ± 0.4 | 35 (1, 70) | 0.04 | 3.3 ± 0.2 | 14 (−4, 32) | 0.12 | 0.29 ± 0.01 (60) | −28 (−48, −8) | 0.002 | 8 | |
| KKAA | 4.8 ± 0.3 | 69 (31, 108) | 0.0008 | 2.7 ± 0.2 | −8 (−26, 11) | 0.41 | 0.38 ± 0.03 (31) | −4 (−26, 18) | 0.97 | 5 |
Proteins were expressed in wild-type or myo7-null (KO) Dictyostelium cells and filopodia parameters were assayed; % change is relative to DdMyo7 in the same background (wild-type or KO). N.d., no data.
SEM of N independent assays.
SEM of the no. of events (n).
Fig. S7.
Filopodia length in myo7-null cells expressing DdMyo7 is decreased with the deletion of the MF1-SH3 but not with the deletion of the FERM2 domain. Filopodia length was measured in cells expressing full-length or mutant (ΔMF1-SH3, ΔFERM2, or KKAA) DdMyo7. Average filopodia length is shown for (A) wild-type (WT) (solid bars) and (B) myo7-null (KO) cells (hatched bars). Error bars indicate the SEM; also see Table 1.
Fig. S5.
Deletion of the MyTH4-FERM domains does not impair filopod formation by DdMyo7. DdMyo7 mutants expressed in myo7-null (KO) cells were imaged by confocal microscopy. The domain organization of DdMyo7 mutants is shown on the left, and fluorescence images are shown on the right. (A) The ΔFERM1(f1, f2) mutant lacks the f1 and f2 subdomains of the internal FERM domain but not f3 or the associated MyTH4 domain. (B and C) Additional deletions remove the MF1 domain (located in the proximal tail region) (B) or the MF2 domain (the common MF domain at the C terminus) (C). All mutants shown here localize to tips of filopodia and rescue filopod formation. (Scale bar, 10 μm.)
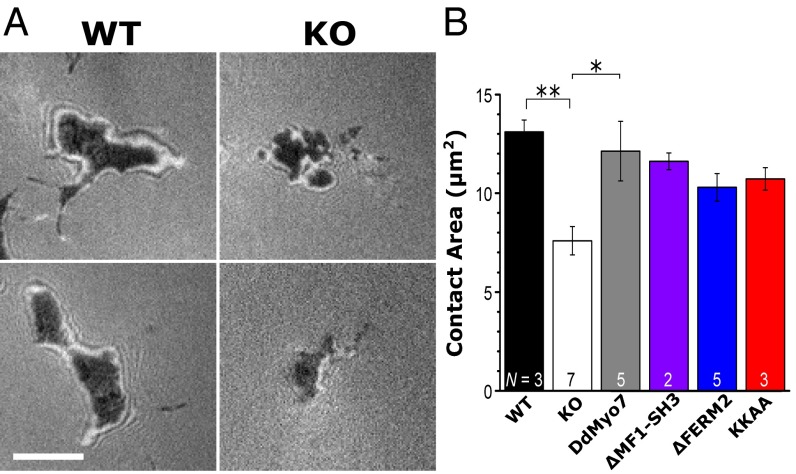
The loss of filopodia is correlated with a reduction in cell substrate adhesion in the myo7-null cells and in HeLa cells that have reduced levels of Myo10 (2, 22). The ability of the DdMyo7 FERM deletion mutants to rescue the myo7-null adhesion phenotype was assessed in polarized migrating cells by interference reflection microscopy (IRM) (37). Null cells expressing DdMyo7 exhibited adhesion to the substrate identical to that of wild-type control cells, and the ∆MF1-SH3, ∆FERM2, and K2333A/K2336A (KKAA) mutants also showed evidence of rescued adhesion (Fig. 6). Although the adhesion measured for the DdMyo7 mutants is slightly reduced compared with that of rescued wild-type cells, the differences are not significant. These results are consistent with the finding that the two MF domains are functionally redundant.
Fig. 6.
Rescue of the myo7-null substrate adhesion defect in polarized cells. The cell–substrate contact area was measured by IRM. (A) IRM images of the cell contact area for polarized myo7-null (KO) and wild-type control cells. (Scale bar: 5 μm.) (B) Average contact area in myo7-null Dictyostelium is decreased significantly relative to wild-type control cells (**P = 0.008) and DdMyo7 KO rescue cells (*P = 0.01). Error bars indicate the SEM of the number of independent assays shown in each bar. See also Fig. S6.
Fig. S6.
Substrate adhesion in polarized myo7-null (KO) cells expressing DdMyo7 mutants. The cell–substrate contact area was imaged using IRM. Shown are IRM images of the cell contact area for two different myo7-null cells expressing full-length DdMyo7 and the ∆MF1-SH3, ∆FERM2, and KKAA mutant proteins in each column. (Scale bar: 5 μm.) See also Fig. 6.
DdMyo7 FERM2 Domain Regulates Filopod Formation and Elongation.
MF myosins have been shown to be autoinhibited, with the C-terminal MF domain serving to regulate the in vitro and in vivo activities of fly and human Myo7A as well as mammalian Myo10 (38–41). In Myo10, PH domains and the FERM domain are required for autoinhibition that is relieved by phospholipid binding to PH domains (40). In Myo7A, a basic motif (K/RxxK/R) in FERM2 is critical for autoinhibition (38, 41), consistent with an intramolecular head–tail interaction. This regulatory basic motif is highly conserved in holozoan Myo7 and Myo22 and is also present in DdMyo7 (Fig. 4C), suggesting that DdMyo7 also may be subject to autoinhibition.
The potential role of the DdMyo7 C-terminal FERM domain in controlling filopod formation was tested with two mutants of DdMyo7. The first mutant, ∆FERM2, deleted the C-terminal FERM domain implicated in the regulation of Myo7A and Myo10 and the interaction with signaling and adhesion receptors (12, 39, 40). The second mutant, KKAA, targeted two highly conserved basic residues in the FERM2 domain (Fig. 4C) implicated in the autoinhibition of Drosophila and human Myo7A (38, 41). The GFP-tagged mutant DdMyo7 proteins were expressed in cells (Fig. S1) and analyzed by confocal microscopy. Both mutants localized to filopodia tips in wild-type cells and rescued filopod formation in myo7-null cells (Fig. 4A) with a characteristic leading-edge concentration preceding filopod elongation (Fig. 4B). In the rescued cells, the DdMyo7 protein was highly concentrated at the tip and, although less abundant (about 20% relative to fluorescence at the tip), was distributed uniformly along the shaft of the filopod (Fig. S4 C and D). Wild-type and mutant DdMyo7 appeared locally enriched at the cortical actin/membrane interface visualized by the membrane stain FM 4-64 (Fig. S3), consistent with a subcellular fractionation study of DdMyo7 (42). Furthermore, cortical enrichment at nascent filopod tips during initiation appeared similar in ∆FERM2 and KKAA mutants and wild-type DdMyo7.
Quantification of filopodia formation in wild-type cells expressing the DdMyo7 ΔFERM2 mutant showed that the number of filopodia formed was not significantly different from those formed by wild-type cells expressing full-length DdMyo7 (Fig. 5 and Table 1). However, the number of filopodia was increased by 35% (P = 0.04) in myo7-null cells expressing DdMyo7 ΔFERM2 (3.8 ± 0.4 filopodia per cell), suggesting that the FERM2 domain negatively regulates filopod formation.
Expression of the DdMyo7 KKAA mutant resulted in a large increase in the number of filopodia formed per cell in wild-type or myo7-null cells, which had 4.4 ± 0.6 and 4.8 ± 0.3 filopodia per cell, respectively (Fig. 5 and Table 1). Overall, the KKAA mutant significantly increased filopod number by 66% with respect to full-length DdMyo7 (P = 0.00008) and 36% with respect to the ∆FERM2 mutant (P = 0.003) with similar results for wild-type and myo7-null cells (Table 1). The KKAA mutant exhibited localization similar to the wild-type DdMyo7 (Fig. 4A), suggesting that the conserved basic motif is not essential for activation or recruitment of DdMyo7 at the membrane. These data are consistent with the inhibition of DdMyo7 motor activity by the FERM2 domain, whereas relief of autoinhibition in the KKAA and ∆FERM2 mutants results in increased filopod formation activity. The increased activity of the KKAA mutant compared with ∆FERM2 and ∆MF1-SH3 mutants may indicate that both MF domains are needed for maximal filopod formation activity. The membrane recruitment of DdMyo7 likely depends upon additional, as-yet-unknown factors.
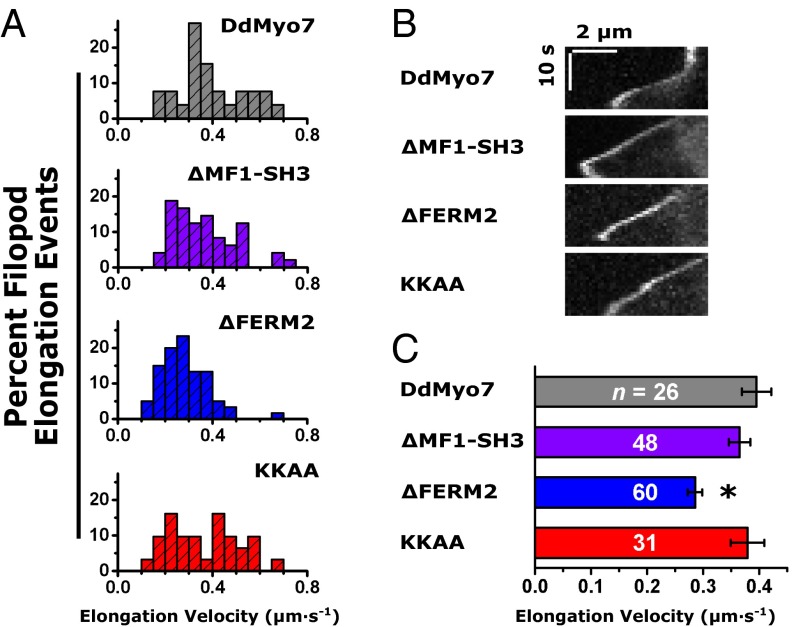
Overexpression of Myo10 in mammalian endothelial cells increases both filopod length and number (43), and it has been proposed that the Myo10 dimer plays a role in filopod initiation as well as elongation (24). To test whether the tail domains of DdMyo7 regulate filopod elongation, filopod length was measured in cells expressing DdMyo7 as described above (Fig. 2D). The average filopod length for wild-type cells expressing DdMyo7 was 2.6 ± 0.3 µm, similar to the length observed for myo7-null cells expressing the same protein (2.9 ± 0.2 µm) (Fig. S7). The average filopod length did not differ significantly between wild-type or myo7-null cells expressing full-length DdMyo7 and those expressing the ΔFERM2 or KKAA mutants (Table 1). Time-lapse kymograph analysis was performed to characterize filopod elongation further in cells expressing these mutant DdMyo7 proteins. Filopod elongation events were analyzed using DdMyo7 fluorescence at the filopod tip to determine elongation velocity (Fig. 7). A nearly constant slope was found in the kymographs (Fig. 7B), allowing the velocity to be measured with linear regression. The average velocity for myo7-null cells expressing DdMyo7 was 0.40 ± 0.03 μm/s (Fig. 7C and Table 1) and was not significantly altered in the ΔMF1-SH3 or KKAA mutants. Moreover, no apparent difference in filopod length was measured after expression of the KKAA mutant compared with DdMyo7, despite the observed increase in filopod number (Fig. 5B and Fig. S7B). In contrast, the velocity of filopod elongation in cells expressing the ΔFERM2 mutant decreased 28% relative to DdMyo7, to 0.28 ± 0.01 μm/s. The decrease in elongation velocity was significant compared with full-length DdMyo7 (P = 0.002; Tukey test). Taken together, these data support a role for the DdMyo7 FERM2 domain in promoting filopod elongation in a manner that is independent of the KKAA motif.
Fig. 7.
Filopod elongation velocity is reduced by deletion of the FERM2 domain. (A) Histograms of the elongation velocity during filopod formation events are shown for full-length DdMyo7 and the ∆MF1-SH3, ΔFERM2, and KKAA mutants. The y axis is the percent of total. (B) Representative kymographs of filopod elongation in myo7-null cells expressing DdMyo7 mutant proteins. (Scale bars: 2 μm × 10 s.) (C) Filopod elongation velocity is significantly decreased (by 28%) in the ∆FERM2 mutant (*P = 0.002) compared with full-length DdMyo7. The filopod elongation velocities in the ΔMF1-SH3 and KKAA mutants do not differ significantly from that of full-length DdMyo7. Error bars show the SEM of the number of events (n) indicated in each bar. (Also see Table 1.)
A Minimal Filopod Motor.
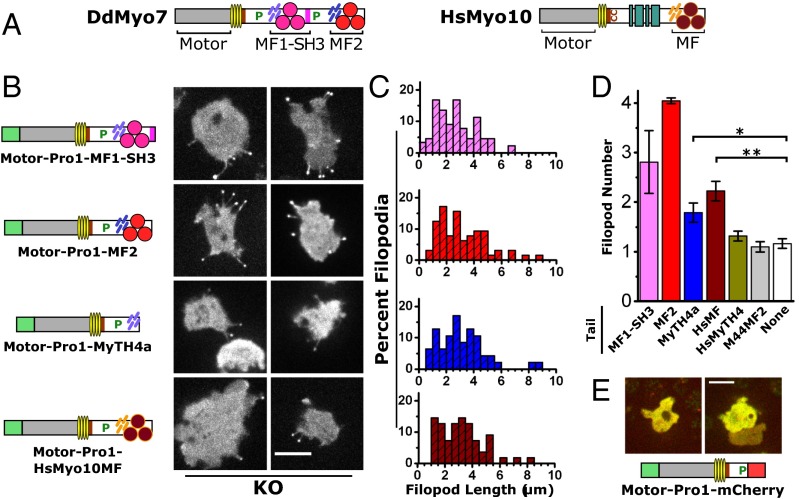
The expression of a minimal DdMyo7 motor region (motor-Pro1) produces virtually no filopodia in myo7-null cells, despite being localized at filopod tips in wild-type cells (Fig. 3B, Fig. S2, and Table S1). Thus, the motor-Pro1 was augmented with a MyTH4-FERM or MyTH4 domain added to the C terminus (Fig. 8B). Initial attempts to express a motor-Pro1-MF1 protein were unsuccessful, perhaps because the MF1-SH3 supramodule was disrupted (35). Instead, a motor-Pro1-MF1-SH3 protein (i.e., GFP-DdMyo7 truncated at the C terminus of the SH3 domain) was expressed in myo7-null cells (Fig. 8B and Fig. S1C). Filopodia were observed in GFP+ cells with characteristic tip localization of the GFP fusion protein. Filopod length was comparable to that seen for DdMyo7 (Fig. 8C and Table 1) with an average of 2.8 ± 0.6 filopodia per cell observed during time-lapse imaging. A fusion protein encoding motor-Pro1-MF2, with the MF2 domain taking the place of MF1-SH3, was tested with similar results (Fig. 8C). The large number of filopodia observed upon expression of motor-Pro1-MF1-SH3 and motor-Pro1-MF2 in myo7-null cells suggests that both the SH3 and the second proline-rich domain (amino acids 1685–1830) (Fig. 8A) are dispensable in filopod formation, whereas at least one MF domain is required.
Fig. 8.
Conservation of the requirement for a single MyTH4-FERM domain for MF myosin filopod-forming activity. (A) Domain structure of DdMyo7 and human Myo10. (B) Representative images of cells with filopodia marked by the indicated fusion proteins. All mutants shown rescued filopod formation and localized strongly to filopod tips when expressed in myo7-null (KO) cells. (Scale bar: 10 µm.) (C) Histograms of filopod length in measured in myo7-null cells expressing the indicated fusion proteins. The y axis is the percent of total (also see Table 2). (D) Filopod number in myo7-null cells expressing the motor-Pro1 fused to the indicated tail domains. Filopod number was defined as total filopodia during a 10-s time-lapse observation divided by the number of cells with filopodia. The weighted mean filopod number is shown with the SEM of three to nine independent experiments. Multiple ANOVA analysis (n = 10–50 cells) confirms significant increases in filopod formation for the MyTH4a (*P < 0.05) and HsMyo10MF (**P < 0.01) chimeras (see Table 2). MF1-SH3 and MF2 data were excluded from ANOVA because of their much larger variance. (E) Domain structure of the motor-Pro1-mCherry fusion protein and confocal fluorescence images showing merged GFP and mCherry channels (Scale bar: 10 μm.) The expressed protein is cytosolic, lacks membrane enrichment, and does not support filopod formation.
The ability to rescue filopod initiation conferred by the single MF motors strongly implies that the MF domain plays roles in targeting the DdMyo7 motor to the cortex and in the initiation of filopodia. The requirement for a FERM domain was tested further with motor-Pro1-MyTH4a protein, in which the tail contains only the internal MyTH4 domain. Filopod formation was rescued by motor-Pro1-MyTH4a despite the lack of a FERM domain (Fig. 8B). However, the number of filopodia was notably reduced compared with motor-Pro1-MF2 (Fig. 8D), although the average filopod length (Fig. 8C and Table S2) was again similar to that in the DdMyo7 rescue cells. Although myosin motor activity is critical for filopod initiation, the ability of motor-Pro1-MyTH4a to promote filopod formation could be explained by interactions of MyTH4a with yet-unidentified target proteins and/or conformational stability provided by the globular MyTH4 domain, favoring recruitment, clustering of myosin motors, or dimerization of the extended lever arm and post-lever arm region (44). Substitution of mCherry for MyTH4a did not support filopod formation (Fig. 8E), demonstrating that the mere presence of a globular domain (mCherry) at the C terminus is not sufficient for the assembly or recruitment of an active motor to promote filopodia at the plasma membrane. This analysis of DdMyo7 establishes that the minimal elements required for filopodial activity are a motor domain, a lever arm, and a MyTH4 domain. The supramodular MyTH4-FERM domain confers optimal function to the myosin, likely by allowing it to interact with partners that promote the growth of filopodia.
Table S2.
Filopodia length in cells expressing minimal and chimeric DdMyo7 motors
| Protein | Filopod length ± SEM, µm | n | N | P, vs. Myo10MF |
| Motor-Pro1-MF1-SH3 | 2.5 ± 0.2 | 89 | 5 | 0.83 |
| Motor-Pro1-MF2 | 3.1 ± 0.6 | 64 | 3 | 0.12 |
| Motor-Pro1-MyTH4a | 3.2 ± 0.4 | 47 | 4 | 0.76 |
| Motor-Pro1-HsMyo10MF | 3.6 ± 0.4 | 55 | 8 | — |
Filopodia length in myo7-null Dictyostelium cells expressing the indicated proteins. The mean of N independent assays is shown.
Multiple ANOVA of n filopodia (P = 0.34) contrasted with HsMyo10MF.
Functional Conservation of MF Domains Across Kingdoms.
MyTH4-FERM myosins are required for filopod formation in humans (22) and amoebae (2). The strong morphological and genetic parallels between these distantly related proteins suggest that divergent MF myosins have conserved roles in filopodia formation. Several attempts to express GFP-tagged bovine Myo10 (43) in Dictyostelium to test this possibility were unsuccessful, most likely because of the difficulty of cross-kingdom gene expression. However, the identification of MF2 as a minimal tail domain supporting robust filopod formation raised the question whether the common MF domain from the evolutionarily distant Myo10 might be able to substitute functionally for the DdMyo7 MF domain. This possibility was tested using a DdMyo7 motor-Pro1-HsMyo10 MF domain chimera (Fig. 8B) that localized to filopodia in wild-type cells (Fig. S8). The Myo10MF chimera rescued filopodia formation in myo7-null cells, averaging 2.2 ± 0.2 filopodia per cell (Fig. 8D and Table 2). However, the extent of rescue was less than that seen with motor-Pro1-MF2 (4.0 ± 0.1) but was comparable to the motor-Pro1-MyTH4a (1.8 ± 0.2 filopodia per cell). A chimeric protein with a Myo10 MyTH4 tail (motor-Pro1-HsMyo10MyTH4) localized to filopodia in wild-type cells (Fig. S8) but did not stimulate filopodia formation (Fig. 8D and Table 2). These results reveal that there is functional conservation between filopodial MF domains, most likely because of shared features of the FERM domains of the human Myo10 and DdMyo7 MF domains.
Fig. S8.
Localization of DdMyo7 mutants with chimeric tail domains in wild-type cells. DdMyo7 mutants expressed in wild-type cells were imaged by confocal microscopy. The domain organization of the DdMyo7 mutants is shown on the left, and fluorescence images are shown on the right. (A) GFP-tagged motor-Pro1 is shown. Note localization at filopodia tips (arrowheads). (B–D) The equivalent proteins fused at the C terminus with human Myo10 MyTH4 domain (HsMyo10M4) (B), the human Myo10 MF domain (HsMyo10MF) (C), or the Dictyostelium Myo44 MF2 domain (D). (Scale bar: 10 µm.) Also see Fig. 8 and Table 2.
Table 2.
Filopodia number in cells expressing minimal DdMyo7 motors
| Protein | Filopod number ± SEM | No. of cells analyzed | No. of independent assays |
| Motor-Pro1-MF1-SH3 | 2.8 ± 0.6 | 26 | 5 |
| Motor-Pro1-MF2 | 4.0 ± 0.1 | 42 | 3 |
| Motor-Pro1-MyTH4a | 1.8 ± 0.2 | 38 | 5 |
| Motor-Pro1-HsMyo10MF | 2.2 ± 0.2 | 18 | 7 |
| Motor-Pro1-HsMyo10MyTH4 | 1.3 ± 0.1 | 50 | 9 |
| Motor-Pro1-DdMyo44MF2 | 1.1 ± 0.1 | 10 | 4 |
| Motor-Pro1 | 1.2 ± 0.1 | 18 | 4 |
The indicated proteins were expressed in myo7-null Dictyostelium. Weighted mean of N independent assays is shown. SEM, standard error of the number of independent assays.
Only a subset of MF myosins has a role in filopodia formation. Others such as HsMyo7A and HsMyo7B function as transporters and anchors in stereocilia and microvilli, respectively (38, 45, 46), raising the question whether there are features specific to filopodial MF domains or if an MF domain from a nonfilopodial myosin can substitute functionally for the DdMyo7 MF domain. Dictyostelium expresses a second MF myosin with two MF domains, Myo44 (MyoG), that is essential for chemotactic signaling but not required for filopodia formation (31). The MF2 domain of Myo44 was fused to DdMyo7 motor-Pro1, and the resulting chimera was expressed in wild-type and myo7-null cells. Interestingly, the motor-Pro1-Myo44MF2 chimera expressed in myo7-null cells failed to rescue the filopodia defect (Table 2) and only weakly localized to filopodia in wild-type cells (Fig. S8), revealing that there are intrinsic, conserved features of the filopodial MF domain that are required for its role in filopodia formation.
Discussion
Filopodia or filopodia-like protrusions are produced by a range of cell types in a diverse array of eukaryotic organisms, raising the question of whether these protrusions are formed by a core conserved mechanism or whether independent pathways emerged to give rise to these structures throughout phylogeny (17). Comparison of filopodia formation in Metazoa and Amoebozoa, groups that have been evolving independently for hundreds of millions of years (28), reveals that filopodia formation requires the same shared core set of filopodial proteins in these two branches of the phylogenetic tree. Specifically, several mammalian proteins essential for filopod formation either have direct orthologs [VASP (11) and Dia2 (19)] or functional homologs [e.g., Rac1a (16)] in Dictyostelium; furthermore, both mammalian cells and Dictyostelium absolutely require the activity of a specialized MyTH4-FERM myosin (2, 22). Analysis of a series of DdMyo7 mutants and chimeras reveals that the important features of the DdMyo7 and Myo10 required for filopod formation are evolutionarily conserved. The results indicate that a functional filopodial MF myosin in Metazoa and Amoebozoa consists of its motor domain, an extended lever arm and a short post-lever arm region that may be involved in dimerization of the motor, and an MF domain (Fig. 8).
Evolution of MF Myosins.
The MF myosin responsible for filopod formation in vertebrates, Myo10, is phylogenetically distinct from the Dictyostelium MF myosin DdMyo7 (Fig. 1). Attempts to classify DdMyo7 in relation to other myosin motor domain sequences have yielded conflicting results (26, 27). The molecular phylogenetic analysis focusing on the common MF domain (Fig. 1) reveals that amoebozoan MF myosins are an evolutionarily distinct class of motor proteins. DdMyo7 fails to group with the animal MF myosin classes, most likely because the gene duplications that produced these proteins occurred after the split between the Amoebozoa and Holozoa, more than 600 Mya (28). The analysis suggests that the common ancestor of Amoebozoa, Fungi, and Holozoa contained a myosin that had two MF domains and a tail structure similar to that of DdMyo7. These myosins share many structural similarities (Fig. 1B), including a highly conserved basic motif at the C terminus of Myo7, Myo22, and amoebozoan Myo7 (Fig. 4C) essential for autoinhibition of Drosophila and human Myo7A (38, 41) and regulation of DdMyo7 activity (Fig. 5). These findings are consistent with DdMyo7 having retained core structural features of the last common ancestral MF myosin.
MF myosins play critical roles in the formation and organization of protrusions containing parallel actin bundles, such as filopodia (2, 22, 46), microvilli (45), and stereocilia (47, 48). These myosins may plausibly be seen as conserving a core ancestral function (17). However, the origins of this functional conservation are unclear. Although it is commonly assumed that gene duplications lead to novel functions (i.e., duplicated genes are maintained because one of the genes evolves a new adaptive function), a more frequent outcome is that gene duplications lead to the partitioning of ancestral functions between the duplicated genes (49, 50). This logic implies that the functions of the ancestral protein have been partitioned among the various MF myosin classes in Metazoa, such as filopodial versus nonfilopodial myosins. Amoebozoan MF myosins, exemplified by DdMyo7, represent a branch preceding the expansion of holozoan MF myosins on the Myo7/15 and Myo10/22 branches (Fig. 1A). In this view DdMyo7 is not an ortholog of any single animal MF myosin but potentially is a model for all animal MF myosins.
Basic Requirements for MF Myosin-Based Regulation of Filopod Formation.
Filopod formation in both animals and amoebae requires myosin motor activity as well as the MF tail (Fig. 3) (22). A DdMyo7 motor-lever arm mutant lacking domains following the SAH fails to localize to filopod tips in wild-type Dictyostelium (Fig. 3A), paralleling results with an Myo10 missing the tail region following the SAH domain that also is not observed in filopodia tips in COS7 cells (34). A longer DdMyo7 protein including a proline-rich domain (motor-Pro1; corresponding to ∆MF1-SH3-Pro2-MF2) localized to filopod tips in wild-type cells without rescuing filopod formation in myo7-null cells (Fig. 3B), again similar to the results in COS7 cells in which Myo10 ∆PH-MF or ∆MF mutants localized properly to basal filopodia but failed to promote the formation of dorsal filopodia (22). These results strongly imply that the ability of the motor to localize to filopodia is necessary but not sufficient for their formation. Although a tailless Myo10 was found to stimulate basal filopod formation in COS7 cells when long-lived Myo10 dimers were experimentally induced, these filopodia were short and unstable (34). The results seen with tailless DdMyo7 and Myo10 support the view that filopod initiation proceeds from mechanical reorganization of actin filaments by dimeric MF myosin motors targeted to the cortex, with elongation requiring both motor activity and the MF tail (either for cargo transport to the tip or to localize myosin activity to the tip).
Functional analysis of the MF tail domain implicates the FERM2 domain in regulating DdMyo7 activity. Deletion of the FERM2 domain results in increased numbers of filopodia that extend at a slower velocity (Figs. 4, 5, and 7). Mutation of a conserved motif in FERM2 (KKAA) also leads to an increase in filopod formation (Fig. 5) but does not affect either filopod length or elongation velocity (Fig. 7 and Fig. S7). The observed increase in the activity of the KKAA mutant is reminiscent of observations with human Myo7A, which is activated both in vitro and in vivo when the equivalent residues are mutated (38). Furthermore, although ectopic expression of Myo10 in COS7 cells increases filopod length, cells expressing a ∆FERM mutant produce shorter filopodia (43) that extend faster (51). This faster elongation likely results from the release of autoinhibition. Together, these results could be explained in at least two ways. FERM2 could interact with actin polymerization factors such as VASP or Dia2, or with regulators of these proteins, to control the velocity of filopod elongation, and thus the loss of this domain leads to a slower elongation. FERM2 also might control myosin activity in vivo by autoinhibition of the motor, as has been seen for the metazoan Myo7A and Myo10 (39–41). This possibility is consistent with a conserved mechanism of DdMyo7 activation by a binding partner, either in the cytosol, as with Myo7A (52), or at the plasma membrane. The contrasting effects of deleting the C-terminal FERM domains of DdMyo7 and mammalian Myo10 on filopod length and elongation velocity potentially might be explained by differences in their binding partners or by the redundancy of DdMyo7 FERM domains as opposed to the single FERM domain of Myo10. Together, these findings support conserved roles for the C-terminal FERM domain in regulating MF myosin activity as well as in promoting the filopod formation activity of DdMyo7 and Myo10.
Minimal and Conserved Features of MF Myosin Required for Filopod Formation.
DdMyo7 proved surprisingly robust to domain deletion, because it was able to rescue filopod formation with either one of the MF domains (Fig. 4). The motor plus the post-lever arm region (motor-Pro1) itself is not sufficient to generate filopodia (Fig. S2 and Table S1). However, the addition of either the MF1-SH3 or the MF2 domain to the motor imparted full functionality. Consistent with this finding, Myo10 lacking the MF domain does not promote the formation of dorsal filopodia in COS7 cells (22). A minimal DdMyo7 motor fused to the MyTH4 domain alone can promote filopodia formation, but this activity is significantly augmented by the presence of the adjacent FERM domain (Fig. 8D and Table 2). The apparent congruence of amoebozoan and metazoan MF myosins suggested the use of a chimeric approach to test their relationship. DdMyo7 motor-Pro1 fused to the human Myo10 MF domain (Fig. 8 B–D) does rescue filopodia formation in the myo7-null mutant, albeit with reduced efficiency, with localization and filopod length comparable to native DdMyo7 (Fig. 8, Table 2, and Table S2). This result suggests that the Myo10 MF domain retains enough homology with DdMyo7 to function as a tail domain for filopod formation and that the MF domains of human and amoeboid filopodial myosins interact with conserved or analogous binding partners that mediate their role in stimulating filopod formation.
The failure of the DdMyo7 motor-Pro1-Myo44 MF2 chimera to rescue filopodia formation establishes that MF domains are specific for the function of a particular MF myosin and that specific features not found in the Myo44 MF must be present in a MF domain for it to contribute to filopodia formation. In contrast, Myo10 and DdMyo7 MF appear to share such features, although the nature of these features is not yet clear. These two MF domains may be important for myosin activation or may recognize an as-yet-unknown common partner that promotes dimerization. The Myo10 MyTH4-FERM domain, but not MyTH4 alone, was able to substitute for DdMyo7 MF domains in filopod formation (Fig. 8 B–D). This lack of functionality may result from a loss of stability induced by deletion of the FERM domain from the MF supramodule. The supramodule is critical for coordinating how partners bind to the MF domain, and the structures of the Myo10 and DdMyo7 MyTH4 (36, 53) establish that they have divergent binding surfaces. The surface differences are consistent with low sequence conservation (24% identical and 40% similar) between the DdMyo7 and Myo10 MyTH4 domains. Thus, the absence of the FERM domain compromises MyTH4 function, and the Myo10 MyTH4 domain alone appears to be too divergent to confer minimal function. Future studies of specific surface mutations and their impact on the interaction with partners will be required to elucidate the role(s) of the MF supramodule in filopodia formation activity.
Conclusion
Evolutionarily distant MF myosins with essential roles in filopod formation have several core features in common, indicating that their activity is not an example of convergent evolution but rather reveals a high degree of functional conservation between these motor proteins. The motor domains and post-lever arm regions of the amoebozoan DdMyo7 and metazoan Myo10 are essential for in vivo activity, and the tail domains play critical roles in the activity of these myosins. Filopod formation activity minimally requires a MyTH4-FERM supramodule specific to filopodial MF myosins that imparts optimal activity as well as regulating filopod elongation (Fig. 8); however, the exact role of the MF domain in filopod formation remains to be clarified. FERM domains interact variously with membrane receptors, actin filaments, and other binding partners (36) that may work in concert with filopodial MF myosins to promote the formation of filopodia, but the specific interactions have not been identified. Additionally, the exact contribution of the motor during the different stages of filopod formation, especially the initiation step, remains to be clarified. It appears that the length and flexibility of the lever arms is critical (34), but how the motor might organize actin at the cortex and/or support transport along cortical actin to promote filopod formation is still unclear. Another intriguing question is what specific features confer filopod formation activity on an MF myosin. Although metazoan Myo7 and Dictyostelium Myo44 have all the essential domains required for filopod formation, including a C-terminal MF domain, they do not support filopod formation (31, 52). Future studies using Dictyostelium as an ancestral MF myosin model system should address these fundamental questions and provide further insight into the evolutionary conservation of MF myosin function.
Methods
Cell Culture.
Wild-type Dictyostelium Ax2 cells and wild-type control (G1-21) and myo7-null (HTD17-1) (2) Ax3 cells were grown at 21 °C on bacteriological plastic plates in HL5 glucose medium (Formedium) supplemented with 10 kU/mL penicillin G and 10 µg/mL streptomycin sulfate (Sigma). Null cells were periodically selected in 10 µg/mL blasticidin S (Calbiochem). Cells were transformed by electroporation and then were selected and maintained using 20 µg/mL G418 (for neomycin resistance) (Fisher Scientific) or 50 µg/mL hygromycin B (Gold Biotechnology).
Plasmid Design and Expression.
An integrating expression plasmid for the N-terminal GFP-tagged DdMyo7 full-length myosin, pDTi74 and the extrachromosomal GFP-Myo7 and GFP-Myo7 tail plasmids have been described previously (2, 42). Standard molecular biological methods were used to generate mutant expression plasmids in the integrating pDTi74 background. Enzymes were obtained from New England Biolabs, and the sequences of all PCR-generated DNAs were verified by sequencing at the University of Minnesota Genomics Center. RFP-tagged Lifeact in pDM358 (33) was a gift from Hanna Brzeska, Laboratory of Cell Biology, National Heart, Lung and Blood Institute, NIH, Bethesda, MD.
Microscopy.
Epifluorescence and IRM microscopy experiments were performed on a Zeiss Axiovert 200 microscope equipped with a 63× Ph3 Plan-Apochromat (Plan Apo) (NA 1.4) objective and Spot RT camera (SPOT Diagnostics). Confocal microscopy was performed with 63× and 100× Plan Apo oil-immersion objectives (NA 1.4) on a Marianas Spinning Disk Confocal imaging system based on a Zeiss Axiovert microscope equipped with a Yokogawa CSU-X1, a Photometrics Evolve 512 electron-multiplying (EM) CCD camera, a Photometrics HQ2 CCD camera, an ASI MS-2000 stage controller, and laser lines at 488 and 561 nm, all controlled by SlideBook 6.0 software (Intelligent Imaging Innovations). Total internal reflection fluorescence (TIRF) microscopy was performed on a Zeiss Axiovert microscope equipped with a Photometrics QuantEM 512SC EMCCD camera, 100× α-Plan Apo (NA 1.46) objective, and laser lines at 488 and 561 nm controlled by ZEN2 Blue software (Carl Zeiss).
Live-Cell Imaging.
Cells were plated in 35-mm glass observation dishes (MatTek or Bioptechs) at a density of ∼105 cells/cm2 and were allowed to adhere for 10 min. Cells were rinsed twice in starvation buffer (SB) (16.8 mM sodium/potassium phosphate adjusted to pH 6.4) and were placed in 1–2 mL of SB for imaging 45–75 min following the onset of starvation. Assays of vegetative cells used low-fluorescence medium (Formedium) in place of SB. For IRM experiments, cells were rinsed in twice in SB and were plated on bacteriological plastic at a density of 1.4 × 105 cells/cm overnight (12–15 h) at 11 °C to induce polarization (33). Cells then were resuspended, plated in observation dishes at room temperature, and imaged by IRM from 10 to 30 min after plating.
Data Analysis.
Statistical analyses were performed in Origin 9.0 (for ANOVA with Tukey post hoc tests) or SPSS Statistics version 20 (for multiple ANOVA with contrasts). Significance was accepted at the P < 0.05 level.
SI Methods
Plasmid Design and Expression.
An integrating expression plasmid for the N-terminal GFP-tagged DdMyo7 full-length myosin, pDTi74, and the extrachromosomal GFP-Myo7 and GFP-Myo7 tail plasmids, pDTi112 and pDTi35, have been described previously (2, 42). Standard molecular biological methods were used to generate mutant expression plasmids in the integrating pDTi74 background. Enzymes were obtained from New England Biolabs, and the sequences of all PCR-generated DNAs were verified by sequencing at the University of Minnesota Genomics Center. RFP-tagged Lifeact in pDM358 (33) was a gift from Hannah Brzeska, Laboratory of Cell Biology, National Heart, Lung and Blood Institute, NIH, Bethesda, MD. The ∆FERM2 deletion mutant was created using PCR-based methods to delete the C-terminal coding sequence from the myoI gene (GenBank accession number L35321.2) encoding the FERM domain (nucleotides 6551–7453; ends at amino acid 2056). The ∆MF1-SH3 deletion mutant (nucleotides 3725–5434; amino acids 1115–1684) and Motor-Pro1-MF1-SH3 mutant (∆Pro2-MF2 deletion at nucleotides 5453–7453; ends at amino acid 1690) were generated similarly. The mutant K2333A, K2336A (KKAA) was generated using the Q5 mutagenesis kit (New England Biolabs) to change the corresponding codons to GCA. DdMyo7 motor-SAH (amino acids 1–1020) was generated using PCR to obtain a genomic clone encoding the 5′ region of the myoI gene (nucleotides 49–3442) and ligating this to the extrachromosomal vector pTX-GFP (55). DdMyo7 motor-Pro1 (amino acids 1–1115) was generated by PCR (nucleotides 49–3727) followed by ligation into a pDXA-based GFP expression vector (56). Expression vectors for a motor-SAH construct (amino acids 1–879) in pTX-GFP or pDXA were tested also, but protein expression was poor and declined over time in culture. For tail domain fusions, the motor-Pro1 plasmid was digested with BstEII and XhoI, yielding a linearized vector for Gibson assembly. The 5′ fragment from the BstEII site through amino acid 1115 was amplified by PCR (forward: 5′-cattagaattagaaaattgg-GTTACCCAATCCGTCATAC-3′; reverse: 5′-tcgatcgg-TGAAACAGTTGGTTGTGG-3′). For motor-Pro1-MyTH4a, the 5′ fragment through amino acid 1316 was amplified (reverse: 5′-aatttttgttctaatgcatctcgagctagctta-ACGATTCTCTTTAATTGATTCTAATTC-3′). For motor-Pro1-MF2, the 5′ fragment through amino acid 1115 was amplified (reverse: 5′-ggtgatga-TGAAACAGTTGGTTGTGG-3′) along with a fragment encoding MF2 (amino acids 1811–2357; forward: 5′-ccaactgtttca-TCATCACCACCAAACTTTAG-3′; reverse: 5′-aatttttgttctaatgcatctcgagctagc-TTATTGAGAAGAATAAAATTGATAAACTG-3′). The motor-Pro1-HsMyo10 sequences were constructed as follows: The regions of the human MYO10 gene (GenBank accession no. NP_036466) encoding either the MyTH4 (amino acids 1501–1699) or MF domain (amino acids 1501–2058, omitting residues 1871–1906 as previously described (53), were amplified by PCR (forward: 5′-actgtttca-CCGATCGACACCCCCACC-3′; MyTH4 reverse: 5′-aatttttgttctaatgcatcgctagctta-TTCCTGCCTGTGGATCAGAG-3′; MF reverse: 5′-aatttttgttctaatgcatc-GCTAGCTCACCTGGAGCTG-3′). The linearized vector, the 5′ DdMyo7 fragment, and the 3′ Myo10-MF or Myo10-MyTH4 tail fragment were assembled using HiFi Assembly Master Mix (New England Biolabs), and the resulting plasmid verified by sequencing. DdMyo7 Motor-Pro1-Myo44 MF was generated in a similar fashion but instead the coding sequence for the DdMyo44 MF2 domain (amino acids 2906–3425) was amplified from a genomic clone (31); (accession number XM_638108.1), as was a linker region encoding the Pro1 region (forward: 5′-cattagaattagaaaattgg-GTTACCCAATCCGTCATAC-3′; reverse: 5′-ggttcacc-TGAAACAGTTGGTTGTGG-3′; forward: 5′-actgtttca-GGTGAACCACAATTTTCTAC-3′; reverse: 5′-aatttttgttctaatgcatctcgagtt-ATCTCTTTTCTTTTTCTTTTCTTTCTC-3′), and these then were assembled in frame into an integrating plasmid containing the GFP-DdMyo7 motor-Pro1 coding sequence and were verified by sequencing. DdMyo7 Motor-Pro1-mCherry was generated in a similar fashion but amplifying the mCherry coding sequence as a 3′ tail fragment (forward: 5′-actgtttca-ATGGTGAGCAAGGGCGAG-3′; reverse: 5′-aatttttgttctaatgcatcgctagc-CTACTTGTACAGCTCGTCCATG-3′) and assembled in frame. The ∆FERM1 (nucleotides 3734–5041; deleted for amino acids 1314–1553), ∆MF1 (nucleotides 3734–5230; deleted for amino acids 1118–1616) and ∆MF2 (nucleotides 5903–7453; ends at amino acid 1840) deletion mutants integrating expression plasmids were generated using PCR (2). The clone encoding bovine Myo10 was a gift from Richard E. Cheney, University of North Carolina at Chapel Hill, Chapel Hill, NC. The bovine Myo10 gene was amplified by PCR and cloned into the pDM358 expression plasmid.
Molecular Phylogenetics.
The majority of MF myosin heavy chain sequences were downloaded from www.cymobase.org/cymobase, a database of cytoskeletal protein sequences (57) curated by Martin Kollmar and the CyMoBase team at the Max Planck Institute for Biophysical Chemistry, Gottingen, Germany. Additional sequences were produced by the Joint Genome Institute (58) or from the Physarum polycephalum Genome Resources server (59) (www.physarum-blast.ovgu.de). A total of 162 sequences from 58 taxa were gathered, and an initial alignment of these sequences was performed using MUSCLE (60). Only sequences with intact motor and MF domains were retained, to exclude incomplete or pseudogenic sequences. Domain boundaries were set using the nominal position of human Myo7A MF2 (residues 1749–2206). When two MF domains were present, only the C-terminal domain was used. Simultaneous alignment and tree estimation (SATé II) was performed using Multiple Alignment using Fast Fourier Transform (MAFFT)/MUSCLE and FastTree (30). SATé performed an initial realignment followed by centroid decomposition and alignment using a Whelan and Goldman (WAG)+gamma substitution model. Maximum subproblem size was set to 50% (81 sequences).
Image Analysis.
Filopodia were imaged in the plane of contact between the cell and the coverslip where the fluorescence of filopodia tips was greatest. The focal plane was located by DIC or by single-frame confocal exposures. For counting assays, three to six planes were acquired at 63× magnification with a vertical step size of 0.3 µm repeated for 30–60 s. Assays of initiation and elongation used 100× magnification in two planes (1-µm step size) to increase the imaging rate. Automated image analysis generated a mask of the cell body and identified filopodia tips using GFP-DdMyo7 fluorescence. The use of DdMyo7 as a filopodial marker was validated by blinded manual analysis of paired DIC and GFP images in which a single DIC image was acquired a few seconds before the start of GFP imaging (Fig. S2 and Table S1). Membrane protrusions identified in DIC were strongly correlated in wild-type or rescue cells with GFP-DdMyo7 punctae at filopodia tips as previously reported (2, 61). The number and length (the distance between the DdMyo7 puncta and cell periphery) of filopodia then were measured (Fig. 2D). Two populations of cells were observed, those expressing DdMyo7 (showing cytosolic GFP fluorescence) and those not expressing the fusion protein (no fluorescence). An accurate count of cells expressing the protein but showing zero filopodia was not feasible with this method because of the difficulty in differentiating between cells not expressing or only weakly expressing DdMyo7. Consequently, filopod number was defined as the number of filopodia divided by number of cells with one or more filopodia (Fig. 2D). Filopodia number was determined by counting filopodia tips marked with GFP. Briefly, regions corresponding to the cell body were identified using a median filter and thresholding to create an image mask. Two thresholding methods were tried in Fiji/ImageJ (62): full-frame thresholding with moments and local thresholding with the Bernsen method (63). The former had reliable results for filopodia number but unreliable results for filopodia length based on small-scale manual analysis. Local thresholding required manual correction of the mask file to remove fluorescent debris and autofluorescent cells with low GFP expression. However, local thresholding produced reliable results for both number and length and therefore was used for all analyses.
Rescue of filopod formation by DdMyo7 mutants was classified in two categories. First, mutant proteins that produced numerous filopodia at nearly wild-type frequency were analyzed by automated counting of filopod tips in the first frame of fluorescence imaging. The second category included mutant proteins with minimal tail domains (MF1-SH3, MF2, M4a, HsMyo10MF, HsMyo10M4, and Myo44MF2) that produced filopodia more rarely and required manual analysis. For automated counting, filopodia tips were identified using the Find Maxima function in ImageJ. The radial intensity distribution around each candidate was analyzed in terms of the sample skewness, (64). Candidates with skewness less than 1 were considered to be fluorescent debris and were discarded. The remaining candidates were registered to the nearest cell center of area. Candidates >20 µm distant were discarded, and candidates >10 µm distant were manually reviewed for accuracy. Both the total number of cells and number of filopodia registered to each cell were recorded. Because the proportion of cells with filopodia varied, filopodia number was defined as number of filopodia divided by number of cells with one or more filopodia. Mutants with a low frequency of filopodia were reviewed manually to identify filopodia in each frame of the time-lapse image stack (typically 10–30 s, no longer than 80 s). Image stacks were analyzed by an operator blinded to the identity of the DdMyo7 mutant protein in each image. Filopodia tips were identified as fluorescent puncta neighboring a GFP+ cell that were observed in two or more frames and for which the linear extent of the filopod either was clearly visible by GFP fluorescence or was not visible in the confocal section but was inferred by the directed motion of the tip relative to the cell body. Regions of interest were acquired to indicate the locations of the filopodia identified by the first operator. For length analysis, a second operator manually reviewed the position of the region of interest in relation to the tip and the threshold mask of the cell body. For filopod number analysis, only filopodia appearing within 10 ± 1 s of the onset of GFP excitation were included to establish a uniform counting procedure for these low-frequency events.
Filopodia length was estimated as the distance to the edge of the cell body on a line from the tip to the cell center. This estimate was validated by small-scale manual analysis. Kymographs and line scans were performed using manual tracing to define regions of interest. Filopod line scans used for fluorescence intensity analysis were aligned either automatically at the peak fluorescence intensity or, in the case of the DdMyo7 tail, manually at the distal ends of filopodia when tip enrichment was not observed. Kymographs used the maximum intensity from a 1.3-µm-wide perpendicular region at each time point. The resulting image was processed with edge detection and thresholding to produce a plot of tip position in which velocity was equal to the slope. Linear regression was performed (minimum of five points, ≤0.8 s per point, 133-nm pixel width), and an elongation velocity event was recorded using the regression slope of the longest segment in which the squared correlation coefficient exceeded 0.98.
IRM images were processed by correction for flat-field illumination (using the averaged image intensity for each assay followed by a 2-µm median filter) and thresholding as before using the same filter radius. The average contact area was calculated using the ImageJ Analyze Particles function with a circularity range of 0.10–1.0 to remove thresholding artifacts and with a size range of 1–50 µm2 to exclude both small debris and cells with a large contact area (i.e., nonmigrating cells).
Western Blotting.
Expression of all of the DdMyo7 fusions was evaluated by Western blotting (Fig. S1) using a mouse monoclonal anti-GFP antibody (MMS-118R; Covance Inc.), a rabbit polyclonal antibody specific for the DdMyo7 heavy chain (42), and the heavy chain of the class I myosin 1B (MyoB) as a control (2, 32). Washed blots were incubated with Alexa Fluor 680- or 800-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies (Molecular Probes, Invitrogen). Detection was performed using an Odyssey infrared imaging system (LI-COR Biosciences).
Supplementary Material
Acknowledgments
We thank Hilary Bauer, Zoé Henrot, and Alex McQuown and Drs. Guillermo Marqués and Mark Sanders (University of Minnesota Imaging Centers) for expert technical assistance; Drs. Naomi Courtemanche, Melissa Gardner, Sivaraj Sivaramakrishnan, and Gaku Ashiba for many valuable discussions; Dr. Martin Kollmar for developing and maintaining CyMoBase (cymobase.org); Dr. Tandy Warnow (University of Illinois) for introducing us to SATé; and the Minnesota Supercomputing Institute for providing excellent computational resources. K.J.P. was supported by NIH Training Program in Muscle Research Grant AR007612 and a University of Minnesota Doctoral Dissertation Fellowship; H.V.G. was supported by National Science Foundation (NSF) Award MCB-1244593; G.W.G.L. was supported by NIH Awards R42DA037622 and R21NS095109-01; A.H. was supported by grants from the ANR-13-BSV8-0019-01, Ligue Nationale Contre le Cancer, and Association pour la Recherche sur le Cancer Subvention Fixe; and M.A.T. was supported by NSF Grant MCB-124423.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615392113/-/DCSupplemental.
References
- 1.Mattila PK, Lappalainen P. Filopodia: Molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9(6):446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 2.Tuxworth RI, et al. A role for myosin VII in dynamic cell adhesion. Curr Biol. 2001;11(5):318–329. doi: 10.1016/s0960-9822(01)00097-5. [DOI] [PubMed] [Google Scholar]
- 3.Ostap EM, et al. Dynamic localization of myosin-I to endocytic structures in Acanthamoeba. Cell Motil Cytoskeleton. 2003;54(1):29–40. doi: 10.1002/cm.10081. [DOI] [PubMed] [Google Scholar]
- 4.Eilken HM, Adams RH. Dynamics of endothelial cell behavior in sprouting angiogenesis. Curr Opin Cell Biol. 2010;22(5):617–625. doi: 10.1016/j.ceb.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Mortimer D, Fothergill T, Pujic Z, Richards LJ, Goodhill GJ. Growth cone chemotaxis. Trends Neurosci. 2008;31(2):90–98. doi: 10.1016/j.tins.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Medalia O, et al. Organization of actin networks in intact filopodia. Curr Biol. 2007;17(1):79–84. doi: 10.1016/j.cub.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 7.Mellor H. The role of formins in filopodia formation. Biochim Biophys Acta. 2010;1803(2):191–200. doi: 10.1016/j.bbamcr.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 8.Gerdes H-H, Rustom A, Wang X. Tunneling nanotubes, an emerging intercellular communication route in development. Mech Dev. 2013;130(6-8):381–387. doi: 10.1016/j.mod.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Kornberg TB, Roy S. Cytonemes as specialized signaling filopodia. Development. 2014;141(4):729–736. doi: 10.1242/dev.086223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ziv NE, Smith SJ. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron. 1996;17(1):91–102. doi: 10.1016/s0896-6273(00)80283-4. [DOI] [PubMed] [Google Scholar]
- 11.Han Y-H, et al. Requirement of a vasodilator-stimulated phosphoprotein family member for cell adhesion, the formation of filopodia, and chemotaxis in Dictyostelium. J Biol Chem. 2002;277(51):49877–49887. doi: 10.1074/jbc.M209107200. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, et al. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6(6):523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- 13.Arjonen A, et al. Mutant p53-associated myosin-X upregulation promotes breast cancer invasion and metastasis. J Clin Invest. 2014;124(3):1069–1082. doi: 10.1172/JCI67280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao R, et al. Elevated expression of myosin X in tumours contributes to breast cancer aggressiveness and metastasis. Br J Cancer. 2014;111(3):539–550. doi: 10.1038/bjc.2014.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibue T, Brooks MW, Inan MF, Reinhardt F, Weinberg RA. The outgrowth of micrometastases is enabled by the formation of filopodium-like protrusions. Cancer Discov. 2012;2(8):706–721. doi: 10.1158/2159-8290.CD-11-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumontier M, Höcht P, Mintert U, Faix J. Rac1 GTPases control filopodia formation, cell motility, endocytosis, cytokinesis and development in Dictyostelium. J Cell Sci. 2000;113(Pt 12):2253–2265. doi: 10.1242/jcs.113.12.2253. [DOI] [PubMed] [Google Scholar]
- 17.Sebé-Pedrós A, et al. Insights into the origin of metazoan filopodia and microvilli. Mol Biol Evol. 2013;30(9):2013–2023. doi: 10.1093/molbev/mst110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavalier-Smith T, et al. Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa. Mol Phylogenet Evol. 2014;81:71–85. doi: 10.1016/j.ympev.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol. 2005;7(6):619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- 20.Vignjevic D, et al. Role of fascin in filopodial protrusion. J Cell Biol. 2006;174(6):863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furukawa R, Fechheimer M. The structure, function, and assembly of actin filament bundles. Int Rev Cytol. 1997;175:29–90. doi: 10.1016/s0074-7696(08)62125-7. [DOI] [PubMed] [Google Scholar]
- 22.Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA. 2006;103(33):12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight PJ, et al. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J Biol Chem. 2005;280(41):34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 24.Kerber ML, Cheney RE. Myosin-X: A MyTH-FERM myosin at the tips of filopodia. J Cell Sci. 2011;124(Pt 22):3733–3741. doi: 10.1242/jcs.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci USA. 2006;103(10):3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8(9):R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sebé-Pedrós A, Grau-Bové X, Richards TA, Ruiz-Trillo I. Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome Biol Evol. 2014;6(2):290–305. doi: 10.1093/gbe/evu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heidel AJ, et al. Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Res. 2011;21(11):1882–1891. doi: 10.1101/gr.121137.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adl SM, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59(5):429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu K, et al. SATé-II: Very fast and accurate simultaneous estimation of multiple sequence alignments and phylogenetic trees. Syst Biol. 2012;61(1):90–106. doi: 10.1093/sysbio/syr095. [DOI] [PubMed] [Google Scholar]
- 31.Breshears LM, Wessels D, Soll DR, Titus MA. An unconventional myosin required for cell polarization and chemotaxis. Proc Natl Acad Sci USA. 2010;107(15):6918–6923. doi: 10.1073/pnas.0909796107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galdeen SA, Stephens S, Thomas DD, Titus MA. Talin influences the dynamics of the myosin VII-membrane interaction. Mol Biol Cell. 2007;18(10):4074–4084. doi: 10.1091/mbc.E06-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brzeska H, Pridham K, Chery G, Titus MA, Korn ED. The association of myosin IB with actin waves in Dictyostelium requires both the plasma membrane-binding site and actin-binding region in the myosin tail. PLoS One. 2014;9(4):e94306. doi: 10.1371/journal.pone.0094306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tokuo H, Mabuchi K, Ikebe M. The motor activity of myosin-X promotes actin fiber convergence at the cell periphery to initiate filopodia formation. J Cell Biol. 2007;179(2):229–238. doi: 10.1083/jcb.200703178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu L, Pan L, Wei Z, Zhang M. Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo. Science. 2011;331(6018):757–760. doi: 10.1126/science.1198848. [DOI] [PubMed] [Google Scholar]
- 36.Planelles-Herrero VJ, et al. Myosin MyTH4-FERM structures highlight important principles of convergent evolution. Proc Natl Acad Sci USA. 2016;113(21):E2906–E2915. doi: 10.1073/pnas.1600736113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber I, Wallraff E, Albrecht R, Gerisch G. Motility and substratum adhesion of Dictyostelium wild-type and cytoskeletal mutant cells: A study by RICM/bright-field double-view image analysis. J Cell Sci. 1995;108(Pt 4):1519–1530. doi: 10.1242/jcs.108.4.1519. [DOI] [PubMed] [Google Scholar]
- 38.Sakai T, et al. Structure and regulation of the movement of human myosin VIIA. J Biol Chem. 2015;290(28):17587–17598. doi: 10.1074/jbc.M114.599365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umeki N, et al. The tail binds to the head-neck domain, inhibiting ATPase activity of myosin VIIA. Proc Natl Acad Sci USA. 2009;106(21):8483–8488. doi: 10.1073/pnas.0812930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Umeki N, et al. Phospholipid-dependent regulation of the motor activity of myosin X. Nat Struct Mol Biol. 2011;18(7):783–788. doi: 10.1038/nsmb.2065. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y, et al. A FERM domain autoregulates Drosophila myosin 7a activity. Proc Natl Acad Sci USA. 2009;106(11):4189–4194. doi: 10.1073/pnas.0808682106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuxworth RI, Stephens S, Ryan ZC, Titus MA. Identification of a myosin VII-talin complex. J Biol Chem. 2005;280(28):26557–26564. doi: 10.1074/jbc.M503699200. [DOI] [PubMed] [Google Scholar]
- 43.Berg JS, Cheney RE. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat Cell Biol. 2002;4(3):246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 44.Lu Q, Ye F, Wei Z, Wen Z, Zhang M. Antiparallel coiled-coil-mediated dimerization of myosin X. Proc Natl Acad Sci USA. 2012;109(43):17388–17393. doi: 10.1073/pnas.1208642109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crawley SW, et al. Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell. 2014;157(2):433–446. doi: 10.1016/j.cell.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Amraoui A, Petit C. Usher I syndrome: Unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J Cell Sci. 2005;118(Pt 20):4593–4603. doi: 10.1242/jcs.02636. [DOI] [PubMed] [Google Scholar]
- 47.Belyantseva IA, et al. Myosin-XVa is required for tip localization of whirlin and differential elongation of hair-cell stereocilia. Nat Cell Biol. 2005;7(2):148–156. doi: 10.1038/ncb1219. [DOI] [PubMed] [Google Scholar]
- 48.Probst FJ, et al. Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science. 1998;280(5368):1444–1447. doi: 10.1126/science.280.5368.1444. [DOI] [PubMed] [Google Scholar]
- 49.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151(4):1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prince VE, Pickett FB. Splitting pairs: The diverging fates of duplicated genes. Nat Rev Genet. 2002;3(11):827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe TM, Tokuo H, Gonda K, Higuchi H, Ikebe M. Myosin-X induces filopodia by multiple elongation mechanism. J Biol Chem. 2010;285(25):19605–19614. doi: 10.1074/jbc.M109.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakai T, Umeki N, Ikebe R, Ikebe M. Cargo binding activates myosin VIIA motor function in cells. Proc Natl Acad Sci USA. 2011;108(17):7028–7033. doi: 10.1073/pnas.1009188108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei Z, Yan J, Lu Q, Pan L, Zhang M. Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain. Proc Natl Acad Sci USA. 2011;108(9):3572–3577. doi: 10.1073/pnas.1016567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levi S, Polyakov M, Egelhoff TT. Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid. 2000;44(3):231–238. doi: 10.1006/plas.2000.1487. [DOI] [PubMed] [Google Scholar]
- 56.Knetsch MLW, Tsiavaliaris G, Zimmermann S, Rühl U, Manstein DJ. Expression vectors for studying cytoskeletal proteins in Dictyostelium discoideum. J Muscle Res Cell Motil. 2002;23(7-8):605–611. doi: 10.1023/a:1024498805771. [DOI] [PubMed] [Google Scholar]
- 57.Odronitz F, Kollmar M. Pfarao: A web application for protein family analysis customized for cytoskeletal and motor proteins (CyMoBase) BMC Genomics. 2006;7(1):300. doi: 10.1186/1471-2164-7-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nordberg H, et al. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014;42(Database issue):D26–D31. doi: 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaap P, et al. The Physarum polycephalum genome reveals extensive use of prokaryotic two-component and Metazoan-type tyrosine kinase signaling. Genome Biol Evol. 2015;8(1):109–125. doi: 10.1093/gbe/evv237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Faix J, Breitsprecher D, Stradal TEB, Rottner K. Filopodia: Complex models for simple rods. Int J Biochem Cell Biol. 2009;41(8-9):1656–1664. doi: 10.1016/j.biocel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 62.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sezgin M, Sankur B. Survey over image thresholding techniques and quantitative performance evaluation. J Electron Imaging. 2004;13(1):146–168. [Google Scholar]
- 64.Joanes DN, Gill CA. Comparing measures of sample skewness and kurtosis. J R Stat Soc Ser Stat. 1998;47(1):183–189. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.