Significance
The superfamily of α/β-hydrolase fold proteins consists of more than 60,000 different members that share a common Nucleophile-His-Acid catalytic triad at individual active sites. These enzymes function diversely in specific biochemical processes, many of which in fact remain poorly understood. In this study, we characterized a distinct α/β-hydrolase fold protein, TsrI, which exhibits the unprecedented dual activity for endopeptidyl hydrolysis and epoxide ring opening/macrocyclization. TsrI uses a same Ser-His-Asp catalytic triad to catalyze cascade C-N bond cleavage and formation, not only highlighting the versatility of α/β-hydrolase fold proteins, which apparently has not been fully appreciated thus far, but also providing insights into the biosynthesis of thiostrepton-type bicyclic thiopeptide members for side-ring system construction and molecular maturation.
Keywords: α/β-hydrolase fold protein, thiopeptide, dual activity, endopeptidyl hydrolysis, macrocyclization
Abstract
Thiostrepton (TSR), an archetypal bimacrocyclic thiopeptide antibiotic that arises from complex posttranslational modifications of a genetically encoded precursor peptide, possesses a quinaldic acid (QA) moiety within the side-ring system of a thiopeptide-characteristic framework. Focusing on selective engineering of the QA moiety, i.e., by fluorination or methylation, we have recently designed and biosynthesized biologically more active TSR analogs. Using these analogs as chemical probes, we uncovered an unusual indirect mechanism of TSR-type thiopeptides, which are able to act against intracellular pathogens through host autophagy induction in addition to direct targeting of bacterial ribosome. Herein, we report the accumulation of 6′-fluoro-7′, 8′-epoxy-TSR, a key intermediate in the preparation of the analog 6′-fluoro-TSR. This unexpected finding led to unveiling of the TSR maturation process, which involves an unusual dual activity of TsrI, an α/β-hydrolase fold protein, for cascade C-N bond cleavage and formation during side-ring system construction. These two functions of TsrI rely on the same catalytic triad, Ser72-His200-Asp191, which first mediates endopeptidyl hydrolysis that occurs selectively between the residues Met-1 and Ile1 for removal of the leader peptide and then triggers epoxide ring opening for closure of the QA-containing side-ring system in a regio- and stereo-specific manner. The former reaction likely requires the formation of an acyl-Ser72 enzyme intermediate; in contrast, the latter is independent of Ser72. Consequently, C-6′ fluorination of QA lowers the reactivity of the epoxide intermediate and, thereby, allows the dissection of the TsrI-associated enzymatic process that proceeds rapidly and typically is difficult to be realized during TSR biosynthesis.
Peptide natural products with ribosomal origins have become a focus that inspires the discovery of new chemical processes (1), because increasing evidence indicates that posttranslational modifications (PTMs) of genetically encoded precursor peptides are comparable to nonribosomal peptide synthetases (2) in terms of the creation of structurally complex molecules, e.g., thiopeptide antibiotics (3, 4). These antibiotics differ in amino acid composition and modification, but overall, they share a family-characteristic macrocyclic framework that contains a six-membered heterocycle domain central to multiple azoles and dehydroamino acids. Over the past several years, the biosynthetic gene clusters of a number of thiopeptides have been identified (5, 6), thereby setting the stage for diversification of their peptidyl skeletons, which, in fact, is a challenge to current chemical synthesis-based approaches, by genetic engineering of precursor-peptide sequences to improve the pharmaceutical properties for clinic use (7).
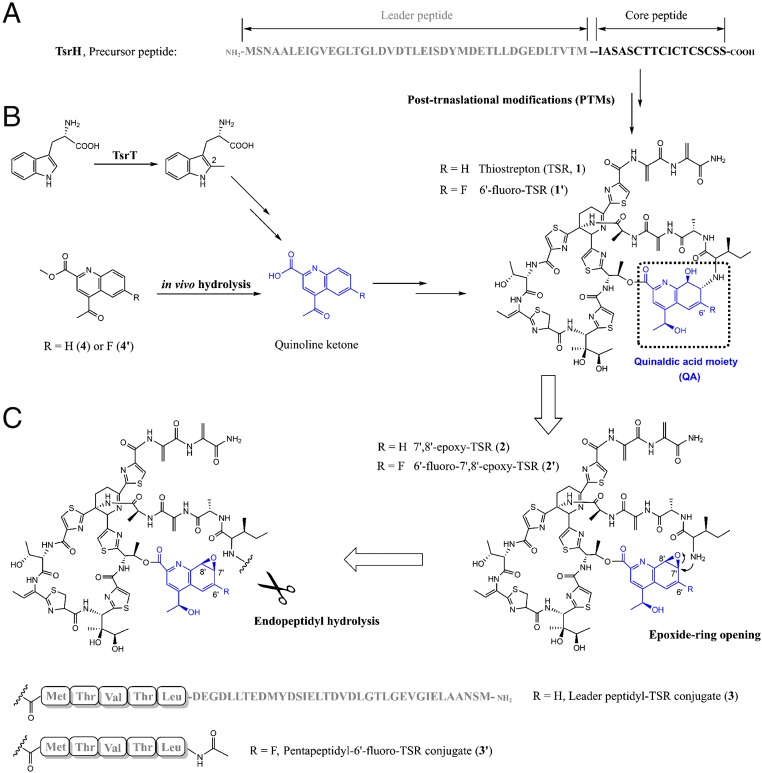
Similar to many ribosomal peptide natural products, each thiopeptide antibiotic originates from a peptide precursor that consists of an N-terminal leader peptide and a C-terminal core peptide (Fig. 1A) (8). The latter undergoes a myriad of common and specific PTMs, which proceed in manners that either depend or are independent of the former, indicating how nature develops structural complexity and diversity from a Ser/Thr and Cys-rich peptide substrate (9). In general, the formation of a characteristic thiopeptide framework requires the activities of phosphorylation-based cyclodehydration that occurs on Cys/Ser residues and subsequent dehydrogenation to produce azoles (10), tRNAGlu-dependent glutamylation elimination for Ser/Thr-residue processing to yield dehydroamino acids (11), and an enzymatic intramolecular [4+2] cycloaddition to furnish the central heterocycle domain (12, 13). The production of thiostrepton (TSR, 1, Fig. 1B), a bicyclic member that is often referred to as the parent compound of the thiopeptide family, is not an exception (14, 15). In addition to the thiopeptide framework, TSR possesses a side-ring system in which a quinaldic acid (QA) moiety conjugates a four-residue peptide side chain. Specifically, QA is derived from a Trp residue that is independent of the precursor peptide of TSR, and the conversion from Trp relies on methyl transfer onto and rearrangement of its indole part to produce a quinoline ketone intermediate (Fig. 1B) (16). The mechanism by which QA is incorporated to construct the side-ring system of TSR remains poorly understood (17). Unlike current clinically used chemotherapeutics that target the bacterial ribosome (18), many thiopeptides, including TSR, display potent activity against various drug-resistant pathogens by perturbing translation factor binding and subsequent protein synthesis, largely upon the occupation of the common thiopeptide framework within a cleft located between the L11 protein and the 23S rRNA of the large ribosomal subunit (19). Although a total of only 14% of the surface buried by TSR on the ribosome is attributed to the side-ring system (20), the QA moiety approaches A1067 of the 23S rRNA, a key nucleobase that contributes to ligand interaction and mutation-based bacterial resistance (21).
Fig. 1.
The biosynthesis of TSR through PTMs of a genetically encoded precursor peptide. (A) TsrH, the precursor peptide of TSR that is composed of a leader peptide (gray) and a core peptide (black). (B) The (engineered) pathway of the (6′-fluorinated) QA moiety (blue) and its incorporation with the core peptide-derived thiopeptide framework to produce TSR (1) or 6′-fluoro-TSR (1′). (C) Retro-biosynthetic analysis of the construction of the side-ring system of 1 or 1′, showing here the necessity of the dual activity of TsrI for both C-N bond cleavage (endopeptidyl hydrolysis) and formation (epoxide ring opening).
Focusing on selective modification of the biologically relevant QA moiety to modulate molecular interaction with bacterial ribosome, we recently designed and effectively biosynthesized 5′- and 6′-fluoro-TSRs and 12′-methyl-STR, the unnatural analogs that have been demonstrated to be more potent than the parent compound in terms of antiinfection effects (22, 23). Using these analogs as chemical probes, we uncovered an unusual mode of action of TSR by inducing endoplasmic reticulum (ER) stress-mediated autophagy to enhance host cell defense (24). Compared with the previously known mechanism of TSR by targeting bacterial ribosome directly, ER stress-mediated host autophagy is an indirect but nonnegligible response and may inspire future changes in the treatment of intracellular pathogens. Following an unexpected result from the preparation of the analog 6′-fluoro-TSR, we here dissect the surprising dual activity of TsrI, an α/β-hydrolase fold protein that catalyzes both C-N bond cleavage and formation to construct the side-ring system of TSR. This study exemplifies the significance of engineering efforts, which, in addition to the production of the expected analogs either for mechanism probing or for antiinfective agent screening, allow the access into the biosynthetic processes that usually occur rapidly or are difficult to be realized and, thus, are able to considerably facilitate our understanding of thiopeptide biosynthesis by affecting the PTM capacity.
Results
Identification of the Side Ring-Open Product 6′-Fluoro-7′, 8′-Epoxy-TSR.
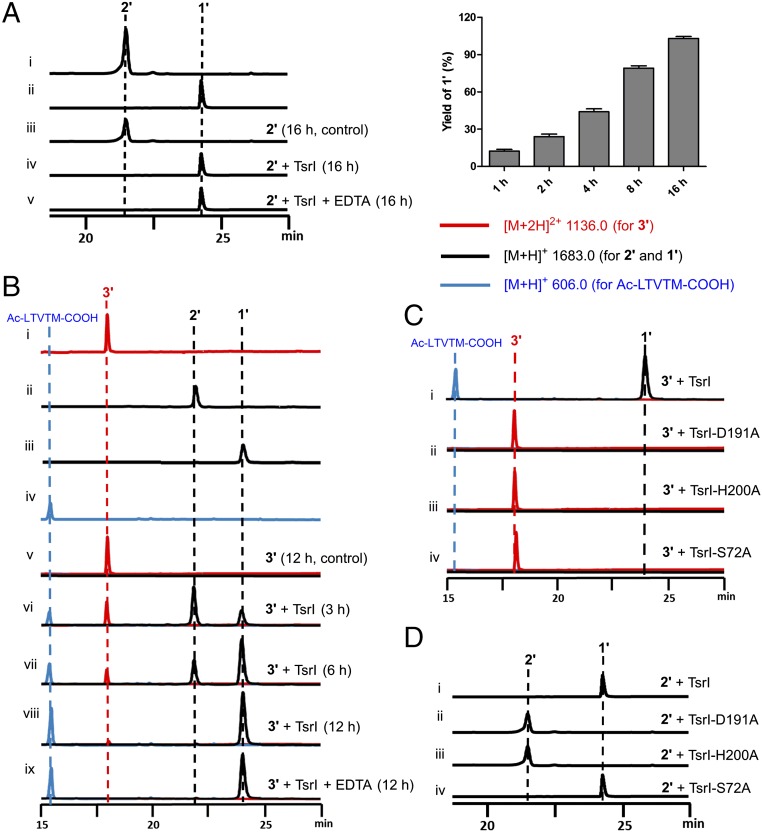
To produce 6′-fluoro-TSR (1′), we previously synthesized and fed the 6′-fluorinated ester analog (4′) of the quinoline ketone intermediate of QA into SL1102, the ΔtsrT Streptomyces laurentii mutant strain (Fig. 1B) (22). This mutant strain, in which the reaction for 2-methylation of Trp that initiates QA formation has been blocked, is incapable of producing the quinoline ketone intermediate and has thus lost the ability for TSR biosynthesis (16). As anticipated, the exogenous feeding of the fluorinated QA intermediate mimic led to robust production of the TSR-analog 1′, the yield of which in SL1102 was maximized on day 3.5. Unexpectedly, during the yield optimization process, we observed a distinct minor product (2′), which appeared with the major product 1′ on day 2 but disappeared over the following fermentation period (Fig. 2A). 1′ and 2′ are identical in molecular weight ([M + H]+ m/z: calculated 1,682.4902 for C72H85FN19O18S5, found 1,682.4906 and 1,682.4908, respectively) and highly similar in UV absorption (λmax 225, 280, and 350 nm, SI Appendix, Fig. S1), supporting the notion that they both are structurally related TSR-like thiopeptides. However, 2′ is relatively unstable and readily undergoes a spontaneous transformation in methanol to yield an adduct ([M + H]+ m/z: calculated 1,714.5164 for C73H89FN19O19S5, found 1,714.5169), indicating that 2′ is more reactive than 1′ in terms of nucleophillic attack by methanol (SI Appendix, Fig. S2).
Fig. 2.
Product examination in S. laurentii and its mutant strains. (A) Addition of ester analog into SL1102, the ΔtsrT S. laurentii mutant strain. i–iv, analysis of the product profiles on day 2 and day 3.5 in the presence of the 6′-fluorinated ester analog (4′) or the ester QA analog (4), respectively; and v and vi, the controls SL1102 and the S. laurentii wild-type strain. (B) Evaluation of the role of TsrI in TSR biosynthesis in vivo. i and ii, validation of the necessity for TSR biosynthesis by inactivation of tsrI and its in trans complementation into the resulting ΔtsrI S. laurentii mutant strain (SL6101); iii and iv, comparative analysis between SL1102 and SL6101 for transforming 6′-fluoro-7′, 8′-epoxy-TSR (2′) into 6′-fluoro-TSR (1′); and v and vi, analysis of TSR (1) or 1′ production in SL6101 in the presence of 4 or 4′. (C) Analysis of 1 production by selective mutation of the C-terminal residue Met-1 of the leader peptide into Ile (i), Lys (ii) and Asp (iii), respectively. λ = 254 nm.
We then scaled up the 4′-supplemented fermentation of SL1102 and accumulated a sufficient quantity of 2′ for structural elucidation. Consequently, 1D, 2D, and 19F NMR analyses of the extract from 500 L of the culture broth confirmed the identity of 2′ to 6′-fluoro-7′, 8′-epoxy-TSR (SI Appendix, Fig. S6), which possesses a thiopeptide framework that is identical to that of 1′ in amino acid composition and modification as well as 6′-fluoro-QA attachment (Fig. 1C). The only exception was found around the linkage positions between QA and Ile1, the first N-terminal residue arising from the TSR core peptide. Distinct from 1′, in which the QA moiety connects Ile1 via the C7′-N bond, 2′ is side ring open and its atoms C7′ and C8′ on QA conjugate an oxygen atom within an epoxide ring (Fig. 1C), which accounts for the aforementioned high reactivity of 2′ in methanol. The closure of the side-ring system appears to be necessary for antiinfection. As demonstrated by minimum inhibitory concentrations (MICs) assays, the activity of 2′ (MIC > 4 μg/mL) against Bacillus subtilis was more than 1,000-fold lower than that of the potent bicyclic product 1′ (MIC = 0.004 μg/mL).
TsrI Catalyzes Macrocyclization via C-N Bond Formation.
The identification of both 6′-fluoro-7′, 8′-epoxy-TSR (2′) and 6′-fluoro-TSR (1′) in SL1102 aroused the interesting query of whether 2′ could be cyclized through an epoxide ring-opening process to yield 1′. Most likely, this cyclization is an enzyme-dependent process because the incubation of the aqueous solution of 2′ at room temperature did not produce 1′ over a 16-h period (Fig. 3A). The conversion of 2′ to 1′ relies on the nucleophillic attack of the amino group of Ile1 onto the C7′ position of QA in a regio- and stereo-selective manner, mechanistically similar to the reactions catalyzed by various epoxide hydrolases that belong to the α/β-hydrolase superfamily (25, 26). These epoxide hydrolases usually share a Ser-His-Asp triad and catalyze epoxide ring opening by using H2O as the nucleophillic agent.
Fig. 3.
In vitro validation of the dual activity of TsrI by HPLC-MS. (A) Examination of the conversion of 6′-fluoro-7′, 8′-epoxy-TSR (2′) into 6’-fluoro-TSR (1′) over a 16-h incubation period (Left) and associated time-course analysis (Right). i and ii, standards 2′ and 1′; iii and v, in the absence and presence of TsrI, respectively; and v, in the presence of TsrI and EDTA. The error bars represent the SD from three different experiments. (B) Examination of the conversion of pentapeptidyl-TSR conjugate (3′) into 1′ through 2′. i–iv, standards 3′, 2′, 1′ and Ac-LTVTM-COOH; v, in the absence of TsrI for 12 h; vi–viii, in the presence of TsrI for 3, 6, and 12 h, respectively; and ix, in the presence of TsrI and EDTA for 12 h. (C and D) Examination of the catalytic triad for the conversions of 3′ and 2′, respectively. i–iv, TsrI and its mutant variants TsrI-D191A, TsrI-H200D, and TsrI-S72D. λ = 254 nm.
A search within the biosynthetic gene cluster of TSR revealed two genes coding for the α/β-hydrolase fold proteins TsrB and TsrI (15). In contrast to TsrB, a proven carboxylesterase that catalyzes a cryptic methyl-ester hydrolysis reaction for C-terminal tailoring in TSR biosynthesis (27), TsrI has not been functionally assigned. To correlate the role of TsrI with the construction of the side-ring system of TSR, we expressed and purified this protein from Escherichia coli and subsequently assayed its activity in vitro. In the presence of TsrI, the cyclization of 6′-fluoro-7′, 8′-epoxy-TSR (2′) proceeded in a time-dependent manner, completed over a 16-h incubation period, and yielded 6′-fluoro-TSR (1′) as the sole product (Fig. 3A). In contrast, no change was observed in the negative control reaction in which the inactivated enzyme was used as the catalyst. The addition of 2 mM ethylene diamine tetraacetic acid (EDTA) had no detectable effect on 1′ production (Fig. 3A). Consequently, TsrI exhibits activity that can trigger a regio- and stereo-specific process of epoxide ring opening and accelerate the closure of the side-ring system of 2′ for molecular maturation.
The Activity of TsrI Is Essential for TSR Biosynthesis.
To clarify the question of whether TSR biosynthesis involves the activity of TsrI, we inactivated the gene encoding TsrI in S. laurentii (15). Indeed, this inactivation completely abolished TSR production, which was then partially restored in the resulting mutant strain, SL6101, by in trans complementation of tsrI (Fig. 2B). Unambiguously, the activity of TsrI is necessary for TSR biosynthesis. Feeding exogenous 6′-fluoro-7′, 8′-epoxy-TSR (2′) into TSR nonproducing mutant strains revealed that SL6101, which lacks TsrI activity, was unable to transform 2′ and generate 6′-fluoro-TSR (1′); however, SL1102, the ΔtsrT mutant strain that has TsrI activity, had this ability (Fig. 2B). Clearly, in the presence of tsrI, 2′ is a transformable intermediate during the biosynthesis of 1′. Together, these findings suggest that the biosynthesis of TSR also involves the cyclization activity of TsrI for TSR maturation, which likely goes through 7′, 8′-epoxy-TSR (2), a nonfluorinated epoxide intermediate that is structurally related to 2′ (Fig. 1C). However, intermediate 2 was not observed in SL6101, even with the supplementation of the QA precursor 4 (Fig. 2B). Similar results were found following the supplementation of the precursor analog 4′ in SL6101, which resulted in no accumulation of any fluorinated product such as 2′. This inconsistency was unexpected and led to the hypothesis that the role of TsrI in the biosynthetic pathway is not confined to 7′, 8′-epoxy-TSR (2) or 6′-fluoro-7′, 8′-epoxy-TSR (2′) transformation.
TsrI Catalyzes Endopeptidyl Hydrolysis for C-N Bond Cleavage.
A survey of TsrI homologs in protein databases revealed SCO7095 from S. coelicolor A3 (2), an α/β-hydrolase fold protein that has been proven to exhibit endopeptidase activity (28). In fact, the hydrolysis of C-O, C-N, or C-C bond that follows a canonical esterase-like mechanism is most common among the reactions that are associated with the α/β-hydrolase superfamily (25, 26). Considering that TSR biosynthesis essentially is a peptide-transformation process, we propose that TsrI shares a similar activity with the precedent SCO7095 and is likely involved in the treatment of a leader peptide-coupled thiopeptide intermediate (Fig. 1C). This specific activity, which could be before the aforementioned cyclization activity of TsrI that closes the side-ring system for molecular maturation, would lead to the removal of the leader peptide part and the release of the amino group of Ile1 to generate 7′, 8′-epoxy-TSR (2). To validate this hypothesis, we synthesized Ac-LTVTM-COOH, an N-acetyl-protected pentapeptide that is composed of five residues at C-terminus of the leader peptide. This peptide was then ligated with 6′-fluoro-7′, 8′-epoxy-TSR (2′) to yield the conjugate 3′ ([M + Na]+ m/z: calculated 2291.7710 for C98H129FN24O26S6Na, found 2291.7714), which mimics the untreated, leader peptide-coupled thiopeptide intermediate (3, Fig. 1C). 3′ appeared to be stable in the aqueous solution because no transformation was observed at room temperature over a 12-h incubation period (Fig. 3B). In the presence of TsrI, this conjugate underwent a hydrolysis that occurs between the residues Met-1 at the C-terminus of the pentapeptide, which is a leader peptide mimic, and Ile1 at the N-terminus of the thiopeptide sequence, which resides in the core peptide part, to produce Ac-LTVTM-COOH and 2′ in a time-dependent manner (Fig. 3B). As expected, TsrI exhibited endopeptidase activity for specific C-N bond cleavage. The subsequent transformation of 3′ was completed within a 12-h period and eventually yielded 6′-fluoro-TSR (1′) as the end thiopeptide molecule (Fig. 3B).
Recently, we evaluated the overall capacity of TSR biosynthesis-related PTMs in regard to variations of Ile1 (29), the first N-terminal residue of the core peptide part, via saturation mutagenesis by using a newly developed, single “base”-based mutagenesis approach (30). The changes of Ile1 to Val, Leu, Ala, Phe, Met, Trp, and Pro led to the production (with various yields) of TSR analogs that bear different side chains at the first residue in the individually engineered S. laurentii strains; however, the changes of Ile1 to polar, acidic, or basic residues did not result in any products. According to this evaluation, the endopeptidase activity of TsrI is promiscuous and tolerates the substitution of Ile1 with other nonpolar amino acids. Using the same in vivo engineering approach, we here examined the variability of Met-1, the conserved residue that is upstream of Ile1 in the precursor peptide TsrH (specifically, Met-1 is located at the C-terminus of the lead peptide part). The mutations of Met-1 to Lys and Asp completely abolished TSR production, indicating that neither basic nor acidic amino acids were acceptable (Fig. 2C). In contrast, TSR production, albeit in a lower yield, was still observed when Met-1 was mutated to Ile (Fig. 2C), therefore supporting the notion that TsrI selectively exhibits endopeptidase activity and prefers amide bond hydrolysis between a nonpolar residue pair.
Examination of the Catalytic Triad for the Dual Activity of TsrI.
The sequence alignment of TsrI and its homologs revealed the highly conserved residues Ser72, His200, and Asp191, which are proposed to constitute a catalytic triad within the active site of TsrI on the α/β-hydrolase fold (SI Appendix, Fig. S4). To examine the necessity of these residues for the dual activity of the protein, we mutated them individually to Ala for in vitro assays. None of the mutant proteins, i.e., TsrI-S72A, TsrI-H200A, and TsrI-D191A, were capable of transforming the conjugate 3′ to 6′-fluoro-7′, 8′-epoxy-TSR (2′, Fig. 3C). Therefore, TsrI could be a serine protease that acts in a manner similar to typical α/β-hydrolase fold proteases, esterases, thioesterases, and lipases for selective hydrolysis between Ile1 and Met-1. The mutant proteins TsrI-H200A and TsrI-D191A were unable to convert 2′ to 6′-fluoro-TSR (1′, Fig. 3D); clearly, the residues His200 and Asp191 are indispensable for the cyclization activity of TsrI. However, the mutant protein TsrI-S72A catalyzed this conversion with an activity comparable to that of the native protein (Fig. 3D), indicating that the residue Ser72 is dispensable during closure of the side ring system. Combined with the aforementioned in vivo results regarding TsrI-related transformations, these assays validated the dual activity of TsrI for C-N bond cleavage and formation, both of which likely are associated with the variable functions of the catalytic triad Ser-His-Asp.
Discussion
The proteins that share the α/β-hydrolase fold as a catalytic domain constitute one of the largest (more than 60,000 individual members) and most important families of enzymes in living organisms (25, 26). This protein fold features a central β-sheet of eight α-helical interconnected strands, in which the second β-strand runs antiparallel to the others, to position a conserved Nucleophile-His-Acid triad (e.g., most commonly Ser-His-Asp) at the active site (31). Related enzymes have highly diverse catalytic functions in numerous biochemical processes, many of which remain to be characterized. In general, the functional diversity arises from the variation in substrate and associated transformation requirement and occasionally incorporates additional chemistry at the active site (26). However, to our knowledge, the utilization of the same catalytic triad within an α/β-hydrolase fold protein for two different decomposable reactions, as exemplified here by TsrI in the biosynthetic pathway of the thiopeptide antibiotic TSR for cascade C-N bond cleavage and formation during the construction of its side-ring system, is noncanonical.
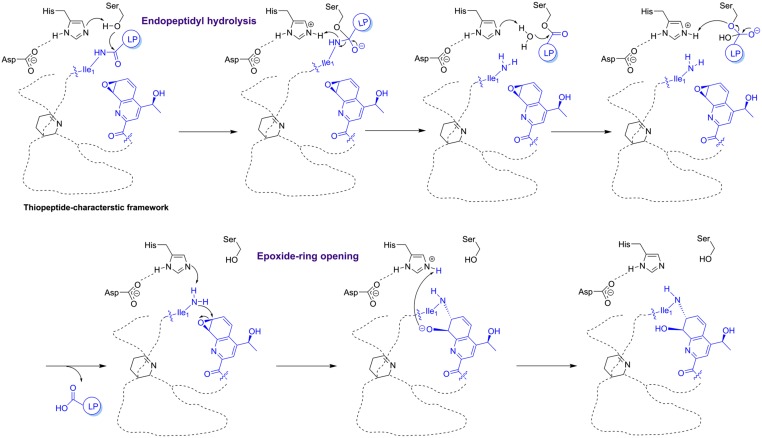
First, TsrI selectively catalyzes peptidyl bond cleavage through an esterase-like mechanism (Fig. 4), which typically requires the assistance of an oxyanion hole at the active site and the formation of an acyl-serine enzyme intermediate. In the presence of Asp191, His200 may act as a base to deprotonate Ser72, and its subsequent attack onto amide carbonyl followed by amine release yields the acyl-Ser72 enzyme intermediate. Then, His200 deprotonates water and enables it to replace Ser72 for leader peptide carboxylate release from the enzyme. Next, using a nascent primary amine as the nucleophillic agent, TsrI catalyzes epoxide ring opening for C-N bond formation, during which the catalytic residue Ser72 is unnecessary (Fig. 4). The residue His200 of TsrI may deprotonate primary amine with the assistance of Asp191, allowing the occurrence of intramolecular SN2 nucleophillic displacement at C7′ for the formation of a C-N bond. With this dual activity of TsrI, the native primary amine intermediate (7′, 8′-epoxy-TSR, 2) that results from peptidyl hydrolysis could be present temporarily and rapidly undergo the following cyclization reaction for TSR maturation. However, the potent electron-withdrawing effect that arises from the adjacent C6′-fluorination could lower the electrophilicity and reactivity of C7′. During 6′-fluoro-TSR (1′) biosynthesis in S. laurentii, the epoxide ring-opening reaction might accordingly slow down, and the intermediate 6′-fluoro-7′, 8′-epoxy-TSR (2′) would thus be able to escape from the active site of TsrI and become detectable at the early fermentation stage.
Fig. 4.
Proposed mechanism of TsrI in the construction of the side-ring system of TSR. LP, leader peptide. The key residue Ile1 and moiety QA for side-ring closure are indicated in blue.
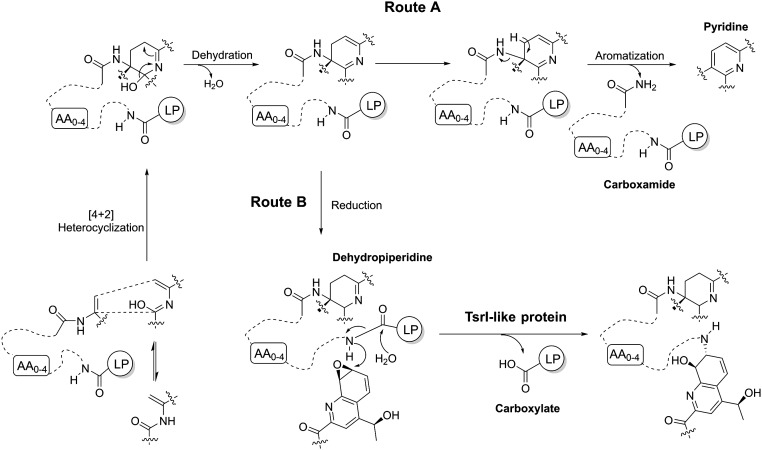
The dissection of the dual activity of TsrI led to unveiling of a unique thiopeptide-maturation process, which, however, could be common for all QA-containing, TSR-type bicyclic members. TsrI-mediated side-ring construction, which likely serves as the last step in the biosynthetic pathway of TSR, is responsible for converting a leader peptide-coupled inactive intermediate into an active mature molecule. Although the hydrolytic removal of the leader peptide part occurs selectively between the two nonpolar residues Met-1 and Ile1, TsrI appears to exhibit its activity largely according to the formation and functionalization of the thiopeptide framework (Fig. 1C), which involve a number of PTMs, including those common for all thiopeptides and those unique for TSR-type members. Specifically, whether a thiopeptide biosynthetic pathway involves TsrI-like dual activity depends on the paradigms for processing the central heterocycle domain that results from a common intramolecular [4+2] cycloaddition and subsequent dehydration (Fig. 5) (32). Heterocycle aromatization would undergo a protease activity-independent elimination process to produce a leader peptide carboxamide and a trisubstituted pyridine (e.g., for monocyclic members thiocillins) or hydroxypyridine (e.g., for bicyclic member nosiheptide) by an additional hydroxylation (route A) (33–38). Alternatively, reduction(s) can proceed to generate a tetrasubstituted piperidine or dehydropiperidine or imidazopiperidine via further modifications for TSR-type bicyclic thiopeptide members that bear a QA-containing side-ring system (route B) (9). During this process, a TsrI-like dual activity would be involved in the specific peptidyl cleavage to produce a leader peptide carboxylate and to release the N-terminal amino group of the core peptide for subsequent side-ring closure.
Fig. 5.
Differences in processing an established common central domain and eliminating leader peptide (LP) in the biosynthesis of thiopeptide antibiotics. Route A, for monocyclic and nosiheptide-type bicyclic thiopeptide members that contain a trisubstituted pyridine-derived domain; and route B, for TSR-type bicyclic members that share a tetrasubstituted piperidine-derived domain.
Conclusions
Using fluorine incorporation to slow down the biochemical process, we have dissected the unexpected dual activity of TsrI, an α/β-hydrolase fold protein that employs the same Ser-His-Asp catalytic triad for two different mechanisms to catalyze cascade C-N bond cleavage and formation. The findings presented here highlight the versatility of α/β-hydrolase fold proteins that results from the variable functions of the conserved catalytic triad Ser-His-Asp. Characterization of these functions may aid in accessing the biological processes that have previously not been characterized. As exemplified in this study, TSR biosynthesis involves the unusual dual activity of TsrI to construct a pharmaceutically important side-ring system through the conjugation of a precursor peptide-derived thiopeptide framework with the QA moiety, which, biosynthetically, is independent of the precursor peptide.
Materials and Methods
Materials and methods are summarized in SI Appendix, SI Materials and Methods. Bacteria strains and plasmids used in this study are listed in SI Appendix, Table S1. Primers used in this study are summarized in SI Appendix, Table S2. Chemical characterization is also described in Supplementary Results of SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Heinz G. Floss (University of Washington) for providing the TSR-producing strain S. laurentii ATCC 31255. This work was supported in part by National Natural Science Foundation of China Grants 31430005, 21520102004, and 21472231; Chinese Academy of Sciences Grant XDB20020200; Science and Technology Commission of Shanghai Municipality Grants 14JC1407700 and 15JC1400400; and Chang-Jiang Scholars Program of China.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612607113/-/DCSupplemental.
References
- 1.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30(1):108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marahiel MA. Working outside the protein-synthesis rules: Insights into non-ribosomal peptide synthesis. J Pept Sci. 2009;15(12):799–807. doi: 10.1002/psc.1183. [DOI] [PubMed] [Google Scholar]
- 3.Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem Rev. 2005;105(2):685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 4.Walsh CT, Acker MG, Bowers AA. Thiazolyl peptide antibiotic biosynthesis: A cascade of post-translational modifications on ribosomal nascent proteins. J Biol Chem. 2010;285(36):27525–27531. doi: 10.1074/jbc.R110.135970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C, Kelly WL. Recent advances in thiopeptide antibiotic biosynthesis. Nat Prod Rep. 2010;27(2):153–164. doi: 10.1039/b922434c. [DOI] [PubMed] [Google Scholar]
- 6.Li J, et al. ThioFinder: A web-based tool for the identification of thiopeptide gene clusters in DNA sequences. PLoS One. 2012;7(9):e45878. doi: 10.1371/journal.pone.0045878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Liu W. Biosynthesis of thiopeptide antibiotics and their pathway engineering. Nat Prod Rep. 2013;30(2):218–226. doi: 10.1039/c2np20107k. [DOI] [PubMed] [Google Scholar]
- 8.Ortega MA, van der Donk WA. New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products. Cell Chem Biol. 2016;23(1):31–44. doi: 10.1016/j.chembiol.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S, Zhou S, Liu W. Opportunities and challenges from current investigations into the biosynthetic logic of nosiheptide-represented thiopeptide antibiotics. Curr Opin Chem Biol. 2013;17(4):626–634. doi: 10.1016/j.cbpa.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 10.Melby JO, Nard NJ, Mitchell DA. Thiazole/oxazole-modified microcins: Complex natural products from ribosomal templates. Curr Opin Chem Biol. 2011;15(3):369–378. doi: 10.1016/j.cbpa.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ortega MA, et al. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature. 2015;517(7535):509–512. doi: 10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wever WJ, et al. Chemoenzymatic synthesis of thiazolyl peptide natural products featuring an enzyme-catalyzed formal [4 + 2] cycloaddition. J Am Chem Soc. 2015;137(10):3494–3497. doi: 10.1021/jacs.5b00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hudson GA, Zhang Z, Tietz JI, Mitchell DA, van der Donk WA. In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin. J Am Chem Soc. 2015;137(51):16012–16015. doi: 10.1021/jacs.5b10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly WL, Pan L, Li C. Thiostrepton biosynthesis: Prototype for a new family of bacteriocins. J Am Chem Soc. 2009;131(12):4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 15.Liao R, et al. Thiopeptide biosynthesis featuring ribosomally synthesized precursor peptides and conserved posttranslational modifications. Chem Biol. 2009;16(2):141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan L, Wang S, Liao R, Liu W. Insights into quinaldic acid moiety formation in thiostrepton biosynthesis facilitating fluorinated thiopeptide generation. Chem Biol. 2012;19(4):443–448. doi: 10.1016/j.chembiol.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Zheng Q, Wang S, Liao R, Liu W. Precursor-directed mutational biosynthesis facilitates the functional assignment of two cytochromes P450 in thiostrepton biosynthesis. ACS Chem Biol. 2016;11(10):2673–2678. doi: 10.1021/acschembio.6b00419. [DOI] [PubMed] [Google Scholar]
- 18.Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12(1):35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 19.Bausch SL, Poliakova E, Draper DE. Interactions of the N-terminal domain of ribosomal protein L11 with thiostrepton and rRNA. J Biol Chem. 2005;280(33):29956–29963. doi: 10.1074/jbc.M504182200. [DOI] [PubMed] [Google Scholar]
- 20.Harms JM, et al. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell. 2008;30(1):26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Baumann S, et al. Molecular determinants of microbial resistance to thiopeptide antibiotics. J Am Chem Soc. 2010;132(20):6973–6981. doi: 10.1021/ja909317n. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, et al. Target-oriented design and biosynthesis of thiostrepton-derived thiopeptide antibiotics with improved pharmaceutical properties. Org Chem Front. 2015;2(2):106–109. [Google Scholar]
- 23.Wang S, et al. Concurrent modifications of the C-terminus and side ring of thiostrepton and their synergistic effects with respect to improving antibacterial activities. Org Chem Front. 2016;3(4):496–500. [Google Scholar]
- 24.Zheng Q, et al. Thiopeptide antibiotics exhibit a dual mode of action against intracellular pathogens by affecting both host and microbe. Chem Biol. 2015;22(8):1002–1007. doi: 10.1016/j.chembiol.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 25.Holmquist M. Alpha/Beta-hydrolase fold enzymes: Structures, functions and mechanisms. Curr Protein Pept Sci. 2000;1(2):209–235. doi: 10.2174/1389203003381405. [DOI] [PubMed] [Google Scholar]
- 26.Rauwerdink A, Kazlauskas RJ. How the same core catalytic machinery catalyzes 17 different reactions: The serine-histidine-aspartate catalytic triad of α/β-hydrolase fold enzymes. ACS Catal. 2015;5(10):6153–6176. doi: 10.1021/acscatal.5b01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao R, Liu W. Thiostrepton maturation involving a deesterification-amidation way to process the C-terminally methylated peptide backbone. J Am Chem Soc. 2011;133(9):2852–2855. doi: 10.1021/ja1111173. [DOI] [PubMed] [Google Scholar]
- 28.Nagy I, et al. Characterization of a novel intracellular endopeptidase of the alpha/beta hydrolase family from Streptomyces coelicolor A3(2) J Bacteriol. 2003;185(2):496–503. doi: 10.1128/JB.185.2.496-503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan P, et al. Molecular engineering of thiostrepton via single “base”-based mutagenesis to generate side ring-derived variants. Org Chem Front. 2016;3(10):1254–1258. [Google Scholar]
- 30.Guo H, et al. Insight into bicyclic thiopeptide biosynthesis benefited from development of a uniform approach for molecular engineering and production improvement. Chem Sci (Camb) 2014;5(1):240–246. [Google Scholar]
- 31.Nardini M, Dijkstra BW. Alpha/beta hydrolase fold enzymes: The family keeps growing. Curr Opin Struct Biol. 1999;9(6):732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 32.Chen M, Liu J, Li M, Duan P, Liu W. Biosynthesis and molecular engineering of template natural products. Natl Sci Rev. 2016 doi: 10.1093/nsr/nww045. [DOI] [Google Scholar]
- 33.Bowers AA, Walsh CT, Acker MG. Genetic interception and structural characterization of thiopeptide cyclization precursors from Bacillus cereus. J Am Chem Soc. 2010;132(35):12182–12184. doi: 10.1021/ja104524q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malcolmson SJ, Young TS, Ruby JG, Skewes-Cox P, Walsh CT. The posttranslational modification cascade to the thiopeptide berninamycin generates linear forms and altered macrocyclic scaffolds. Proc Natl Acad Sci USA. 2013;110(21):8483–8488. doi: 10.1073/pnas.1307111110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tocchetti A, et al. Capturing linear intermediates and C-terminal variants during maturation of the thiopeptide GE2270. Chem Biol. 2013;20(8):1067–1077. doi: 10.1016/j.chembiol.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Hayashi S, et al. Genome mining reveals a minimum gene set for the biosynthesis of 32-membered macrocyclic thiopeptides lactazoles. Chem Biol. 2014;21(5):679–688. doi: 10.1016/j.chembiol.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Yu Y, et al. Nosiheptide biosynthesis featuring a unique indole side ring formation on the characteristic thiopeptide framework. ACS Chem Biol. 2009;4(10):855–864. doi: 10.1021/cb900133x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding Y, et al. Moving posttranslational modifications forward to biosynthesize the glycosylated thiopeptide nocathiacin I in Nocardia sp. ATCC202099. Mol Biosyst. 2010;6(7):1180–1185. doi: 10.1039/c005121g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.