Abstract
Swi1 is required for programmed pausing of replication forks near the mat1 locus in the fission yeast Schizosaccharomyces pombe. This fork pausing is required to initiate a recombination event that switches mating type. Swi1 is also needed for the replication checkpoint that arrests division in response to fork arrest. How Swi1 accomplishes these tasks is unknown. Here we report that Swi1 copurifies with a 181-amino-acid protein encoded by swi3+. The Swi1-Swi3 complex is required for survival of fork arrest and for activation of the replication checkpoint kinase Cds1. Association of Swi1 and Swi3 with chromatin during DNA replication correlated with movement of the replication fork. swi1Δ and swi3Δ mutants accumulated Rad22 (Rad52 homolog) DNA repair foci during replication. These foci correlated with the Rad22-dependent appearance of Holliday junction (HJ)-like structures in cells lacking Mus81-Eme1 HJ resolvase. Rhp51 and Rhp54 homologous recombination proteins were not required for viability in swi1Δ or swi3Δ cells, indicating that the HJ-like structures arise from single-strand DNA gaps or rearranged forks instead of broken forks. We propose that Swi1 and Swi3 define a fork protection complex that coordinates leading- and lagging-strand synthesis and stabilizes stalled replication forks.
Accurate replication of the millions or billions of DNA base pairs in a eukaryotic genome is a remarkable achievement. This accomplishment is even more astonishing when one considers that the conditions for DNA synthesis are rarely ideal. Damaged templates, protein complexes bound to DNA, and inadequate supplies of deoxyribonucleotide triphosphates (dNTPs) are among the many obstacles that must be overcome to replicate a genome. All of these situations can stall replication forks. Stalled forks pose grave threats to genome integrity because they can rearrange, break, or collapse through disassembly of the replication complex (36).
Preserving genome integrity when forks stall is in large part the responsibility of the replication checkpoint (10, 44). The protein kinase Cds1 is a critical effector of the replication checkpoint in the fission yeast Schizosaccharomyces pombe (7, 31). One of its major functions is to prevent the onset of mitosis by regulating mitotic control proteins, but perhaps its most important activity is to stabilize replication forks (10). Cds1 is required to prevent fork breakage in cells treated with hydroxyurea (HU), a ribonucleotide reductase inhibitor that stalls replication by depleting dNTPs (41). In the budding yeast Saccharomyces cerevisiae, a failure to activate Rad53, a Cds1 homolog, is associated with collapse and regression of replication forks and gross chromosomal rearrangements in cells treated with HU (29, 33, 55, 58).
Replication checkpoint studies have typically used chemical agents or DNA replication mutants to stall replication, but there is abundant evidence of natural replication pause and termination sites (60). One of the best-characterized examples involves cell differentiation in fission yeast. Programmed fork pausing and termination events near the mating-type (mat1) locus in fission yeast are needed to create an imprint, probably a DNA strand discontinuity, that initiates a gene conversion event that switches mating type (13, 26). These events require swi1+ and swi3+. Swi1 has been identified (13) and shown to be required for proficient activation of Cds1 (41). Cds1 is not required for mating-type switching, thus Swi1 has both Cds1-dependent and -independent activities. A similar situation exists in budding yeast. Tof1, a Swi1 homolog, is involved in control of Rad53 (17). Tof1 travels with the replication fork and is needed to restrain fork progression when DNA synthesis is inhibited by HU (25). This function is shared with Mrc1, another protein involved in Rad53 activation (25, 43). Curiously, tof1Δ mutants are only weakly sensitive to HU (17), indicating that budding yeast readily tolerates uncoupling of replisome movement and DNA synthesis.
Swi1 is required for proficient DNA replication in the absence of agents that cause genotoxic stress (41). swi1Δ mutants accumulate foci containing the Rad22 DNA repair protein during S phase. Relatively fewer Rad22 foci appear in cds1Δ mutants, indicating that replication abnormalities in swi1Δ cells are not associated with an inability to activate Cds1. Mus81-Eme1, a DNA endonuclease implicated in cleavage of cruciform DNA structures such as Holliday junctions (HJs) and nicked HJs (8, 19, 45), is vital for survival in swi1Δ mutants (41). Mus81 is not required for viability in cds1Δ cells (9). These findings, coupled with evidence that Swi1 associates with chromatin specifically in S phase, led us to propose that Swi1 travels with the replisome and stabilizes stalled forks in a configuration that is recognized by the replication checkpoint (41).
Swi1 and Tof1 belong to a large protein family that was first defined by metazoan Tim1 (Timeless) (12, 13, 41). Drosophila melanogaster and mammalian Tim1s are implicated in circadian rhythmic oscillation (5), whereas the Caenorhabditis elegans Tim1 is required for proper chromosome cohesion and segregation (12). It is curious that members of the same protein family should appear to have such diverse functions. To better understand this protein family, we have identified proteins that interact with Swi1. Here we report that Swi1 has a tight association with a small conserved protein that we show is encoded by swi3+. Our studies suggest that mutants defective for the Swi1-Swi3 complex accumulate single-strand DNA (ssDNA) gaps during DNA replication that can recombine to form HJs. We propose that the Swi1-Swi3 heterodimer defines an evolutionarily conserved fork protection complex (FPC) that coordinates leading- and lagging-strand synthesis. This activity is required for accurate DNA replication, fork protection, and replication checkpoint signaling.
MATERIALS AND METHODS
General techniques.
Methods for genetic and biochemical analyses of fission yeast have been described elsewhere (2, 37). For immunoblotting, extracts from ∼1 × 108 cells were made by glass bead disruption in lysis buffer A (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 10% glycerol, 50 mM NaF, 1 mM Na3VO4, 5 mM EDTA, 5 mM N-methylmaleimide, 1 μM microcystin, 0.1 μM okadaic acid, and 1 mM dithiothreitol) supplemented with protease inhibitors (0.2 mM p-4-amidoinophenyl-methane sulfonyl fluoride hydrochloride monohydrate [p-APMSF] and Roche protease inhibitor cocktail). Protein extracts were clarified by centrifugation at 15,000 × g for 10 min at 4°C. For small-scale TAP tag protein precipitation, protein extracts were mixed with immunoglobulin G-Sepharose beads and incubated for 2 h at 4°C. The Sepharose beads were collected and washed three times in lysis buffer A. Proteins associated with the beads were analyzed by immunoblotting. Immunoblotting methods and Cds1 kinase assays have been described previously (31, 57). In situ chromatin binding assays were carried out as described elsewhere (27). To calculate a green fluorescent protein (GFP) signal value in live cells, we measured the total GFP signal intensity of an 81-pixel region in the nucleus by using IPlab software (Scanalytics Inc.). The total GFP signal intensity was divided by 81 to obtain a GFP signal value per pixel. Twenty-five nuclei from each strain were measured to obtain the average GFP signal value and standard deviation. Mating-type switching assays were performed as described elsewhere (21). For the UV sensitivity assay, cells were exposed to short-wavelength (254-nm) UV in a Stratalinker (Stratagene, La Jolla, Calif.). To visualize nuclear DNA and the cell wall, cells were fixed in 2.5% glutaraldehyde for 20 min on ice. Cells were washed in phosphate-buffered saline and stained with 4′,6-diamidino-2-phenylindole (DAPI).
Identification of Swi3.
Cells expressing Swi1-TAP at its own genomic locus were cultured in 8 liters of yeast extract supplemented (YES) medium until an optical density at 600 nm of 1.2 was reached, and cells were collected. Cell lysate was prepared by grinding a cell pellet that was frozen in lysis buffer B (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 10% glycerol, 50 mM NaF, 0.1 mM Na3VO4, 5 mM EDTA, 60 mM β-glycerophosphate, 15 mM p-nitrophenyl phosphate, and 1 mM dithiothreitol) supplemented with protease inhibitors (0.2 mM p-APMSF and Roche protease inhibitor cocktail) as described previously (8). Clarified cell extract was obtained by high-speed centrifugation at 30,000 rpm at 4°C in a Ti35 rotor for 1 h and filtration through a 0.45-μm-pore-size filter. Swi1-TAP and associated proteins were purified and trichloroacetic acid precipitated as described elsewhere (8, 50). The precipitated sample was analyzed by multidimensional protein identification technology (MudPIT) as described previously (8).
ChIP assay.
Chromatin immunoprecipitation (ChIP) was performed basically as described elsewhere (42). S. pombe cells (5 × 108) were fixed in 1% formaldehyde for 20 min at room temperature and quenched in 125 mM glycine for 5 min. Cells were washed in Tris-buffered saline and disrupted in lysis buffer (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, and 1% Triton X-100) supplemented with protease inhibitors (0.2 mM p-APMSF and Roche protease inhibitor cocktail). The broken cells were sonicated six times for 20 s each until chromatin DNA was sheared into 500- to 700-bp fragments. Cell lysate was clarified by two rounds of maximum-speed centrifugation in an Eppendorf 5415C microcentrifuge at 4°C. Immunoprecipitation was performed on the clarified lysate with anti-FLAG M2 agarose (Sigma) or anti-myc 9E10 antibody bound to protein G-Sepharose. PCR amplification and primers used in these studies have been described previously (42).
Branch migration experiments and 2D gel electrophoresis.
Two-dimensional (2D) gel electrophoresis was performed as described elsewhere (11, 28). Cells grown in YES medium were arrested with 0.1% sodium azide and chilled on ice for 5 min. Approximately 2 × 109 cells were harvested and washed with ice-cold water before storage at −80°C. Genomic DNA was purified as described elsewhere (41, 53). Samples of 5 μg of DNA were digested with 30 U each of HindIII and KpnI. Precipitated DNA was run on a 0.4% agarose gel in the first dimension. The gel slice of the first-dimension electrophoresis was incubated in branch migration buffer (10 mM Tris-HCl [pH 8.0], 0.1 M NaCl, and 0.1 mM EDTA) as described elsewhere (6, 19, 53, 70) at 4 or 65°C for 5 h. The gel slice was then processed for the second dimension of electrophoresis in a 1% gel. After electrophoresis, the DNA was transferred to a Hybond N+ membrane (Amersham Biosciences). Hybridization was carried out with the 3-kb HindIII-KpnI radiolabeled ribosomal DNA (rDNA) fragments (68). Radioactive signals were detected using a Molecular Dynamics Storm 840 instrument.
Gene cloning, plasmids, and S. pombe strain construction.
The 1.2-kb swi3+ genomic fragment was amplified by KOD-XL polymerase (Novagen) and introduced into the BamHI/SmaI site of pAL-sk, resulting in pAL-swi3. Plasmids pAL-cds1 and pAL-swi1 have been described elsewhere (41). swi3 cDNA was amplified from the S. pombe cDNA library with EXtaq polymerase (TaKaRa) and subcloned into the pBluescript II vector. Sequencing analysis of swi3 cDNA revealed that swi3+ has an additional exon in the 5′ region of the annotated open reading frame, SPBC30D10.04. Swi3 nucleotide and protein sequence data are available in the DDBJ, EMBL, and GenBank databases, with the accession number AY498547. swi1Δ (swi1::Kanr) was generated by a two-step PCR-based method (30) using primers P73, P74, P77, and P78. swi1-TAP (swi1-TAP::Kanr), swi1-13myc (swi1-13myc::Kanr), swi1-GFP (swi1-GFP::Kanr), and swi1-3FLAG (swi1-3FLAG::Kanr) were generated by a two-step PCR method (30) using primers P75, P76, P77, and P78 to place epitope tags at the C terminus of swi1. swi3Δ (swi3::Kanr) was generated by a two-step PCR method (30) using primers P280, P285, P287, and P282. swi3-TAP (swi3-TAP::Kanr), swi3-13myc (swi3-13myc::Kanr), swi3-GFP (swi3-GFP::Kanr), and swi3-3FLAG (swi3-3FLAG::Kanr) were generated by a two-step PCR-based method (30) using primers P281, P286, P287, and P282 to place epitope tags at the C terminus of swi3. rad11-3FLAG (rad11-3FLAG:Kanr) was generated by a one-step PCR method (4) using primers rpa1-1 and rpa1-2. Plasmids used as templates for PCR-based gene disruption and gene tagging have previously been described for pFA6a-KanMX6 (4), pFA6a-TAP-KanMX6 (8), pFA6a-13myc-KanMX6 (4), and pFA6a-GFP-KanMX6 (4). pFA6a-3FLAG-KanMX6 was constructed by replacing the PacI-AscI GFP fragment in pFA6a-GFP-KanMX6 with a FLAG3 fragment. Primers used for PCR-based gene disruption and gene tagging are available on request.
Mutations and epitope-tagged genes have previously been described for cdc6-23 (23), cdc20-M10 (40), cdc25-22 (16), swi3-1 (22), cds1Δ (cds1::ura4+) (7), chk1Δ (chk1::ura4+) (3), rad26Δ (rad26::ura4+) (3), mus81Δ (mus81::Kanr) (9), rad13Δ (rad13::ura4+) (69), uve1Δ (uve1::LEU2) (69), rad22Δ (rad22::ura4+) (46), rhp51Δ (rhp51::ura4+) (38), rhp54Δ (rhp54::ura4+) (39), rqh1Δ (rqh1::ura4+) (56), and rad22-YFP (rad22-YFP:Kanr) (15, 41).
S. pombe strains.
S. pombe strains used in this study were the following: AL2229 (h+ cdc6-23 ade6-210), BF2115 (h+ cds1::ura4+ chk1::ura4+), EN3180 (h− rad26::ura4+), EN3181 (h+ chk1::ura4+), EN3182 (h− swi1::Kanr), EN3190 (h+ mus81::Kanr), EN3191 (h+ swi1-GFP:Kanr), EN3192 (h− swi1::Kanr mus81::Kanr), EN3197 (h90 swi1::Kanr), EN3222 (h− rad22-YFP:Kanr), EN3286 (rad13::ura4+ uve1::LEU2), EN3363 (h− swi1-TAP:Kanr), EN3364 (h+ swi3::Kanr), EN3365 (h90 swi3::Kanr), EN3366 (h− swi3::Kanr), EN3367 (h90 swi3-1), EN3368 (h− swi1-TAP:Kanr swi3-13myc:Kanr), EN3369 (h− swi3-13myc:Kanr), EN3370 (h− swi3-TAP:Kanr swi1-13myc:Kanr), EN3371 (h− swi1-13myc:Kanr), EN3372 (h− swi1::Kanr swi3::Kanr), EN3373 (h− swi3::Kanr cds1::ura4+), EN3374 (h− swi3::Kanr chk1::ura4+), EN3375 (swi3::Kanr rad13::ura4+ uve1::LEU2), EN3376 (h− rad22-YFP:Kanr swi3::Kanr), EN3377 (h− swi1-3FLAG:Kanr cdc25-22), EN3378 (h+ swi3-3FLAG:Kanr cdc25-22), EN3379 (h− swi1-3FLAG:Kanr swi3::Kanr), EN3380 (h− swi3-3FLAG:Kanr swi1::Kanr), EN3381 (h− swi1-3FLAG:Kanr), EN3382 (h− swi3-3FLAG:Kanr), EN3383 (h− swi1-GFP:Kanr swi3::Kanr), EN3384 (h+ swi3-GFP:Kanr), EN3385 (h− swi3-GFP:Kanr swi1::Kanr), EN3410 (h+ swi1::Kanr), EN3455 (h− smt0 swi1::Kanr rad22::ura4+), EN3456 (h− smt0 swi3::Kanr rad22::ura4+), EN3457 (mus81::Kanr rad22::ura4+), EN3458 (swi1::Kanr mus81::Kanr rad22::ura4+), EN3459 (swi1-13myc:Kanr rad11-3FLAG:Kanr cdc25-22), JL1307 (h+ cdc20-M10 ade6-210 his7-366), KT2751 (h− cds1::ura4+), PR109 (h−), PS2382 (h− smt0 rhp54::ura4+), PS2383 (h− smt0 rhp51::ura4+), PS2384 (h− smt0 rad22::ura4+), and PS2375 (h− rqh1::ura4+). All strains are leu1-32 and ura4-D18.
RESULTS
Identification of the Swi1-Swi3 complex.
We used MudPIT to identify proteins that associated with the Swi1-TAP fusion protein expressed at endogenous levels from the swi1+ promoter. TAP consists of protein A and calmodulin binding domains separated by a tobacco etch virus protease cleavage site, allowing efficient two-step affinity purification of TAP-tagged proteins (50). MudPIT combines multidimensional liquid chromatography with electrospray ionization tandem mass spectrometry and proteomic methods to identify proteins in complex mixtures (65). The swi1-TAP strain used for these studies was not abnormally sensitive to genotoxic agents such as HU, indicating that Swi1-TAP was functional and retained physiologically significant protein interactions.
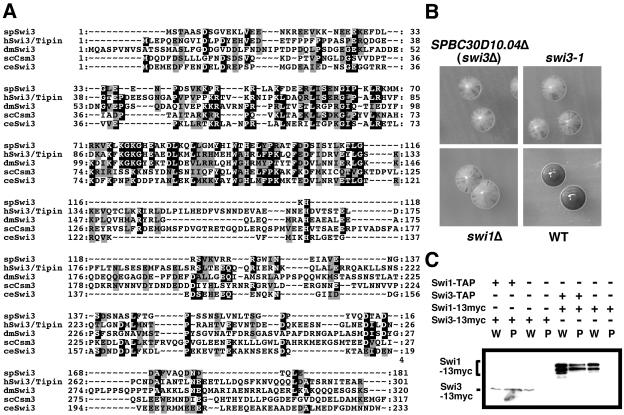
MudPIT analysis of Swi1-TAP identified Swi1 itself and a reproducible set of background proteins that are routinely identified in TAP fusion protein samples, together with a small number of potential interacting proteins. In the latter class, the highest coverage was of a predicted 148-amino-acid protein encoded by the SPBC30D10.04 open reading frame. Fourteen peptides from SPBC30D10.04 were identified. Sequence analysis of the corresponding cDNA revealed that this protein was actually derived from two exons that when spliced encoded a 181-amino-acid protein (Fig. 1A). This protein (which proved to be Swi3 [see the next paragraph]) has no evident protein motif, but it does have significant sequence similarity, especially between amino acids 38 and 116, to related proteins in budding yeast, humans, and other eukaryotes (Fig. 1A).
FIG. 1.
Identification of Swi3 as a Swi1-interacting protein. (A) Swi3 is conserved across evolution. Multiple alignments of Swi3 homologs from S. pombe (spSwi3; GenBank accession no. AY498547), humans (hSwi3/Tipin; GenBank accession no. NP_060328), Drosophila (dmSwi3; GenBank accession no. NP_609895), S. cerevisiae (scCsm3), and C. elegans (ceSwi3; GenBank accession no. NP_490977) are shown. (B) Deletion of SPBC30D10.04 (swi3Δ), swi3-1, and swi1Δ showed inefficient mating-type switching. Homothallic h90 strains with the indicated genotypes were incubated on sporulation medium for 6 to 7 days at 25°C. Plates were exposed to iodine vapors. Colonies that have efficient mating-type switching stain darkly with iodine vapors, whereas inefficient strains show mottled staining. (C) Swi1 and Swi3 form a protein complex in S. pombe. Cells expressing Swi1 or Swi3 as TAP or 13myc fusion proteins were used for coimmunoprecipitation studies. TAP proteins were precipitated and probed with anti-myc antibodies. All proteins were expressed at endogenous levels. W, whole-cell extract; P, precipitated fraction.
We were intrigued to find that SPBC30D10.04 is located between spo6 and ade1 on chromosome II in a region where swi3 has been genetically mapped (2). We therefore investigated whether SPBC30D10.04 was allelic to swi3. We first examined mating-type switching efficiency. Mating-type switching within a single colony leads to mating and formation of spores that stain brown when exposed to iodine vapor. The strain with SPBC30D10.04Δ displayed a severe switching defect that was comparable to the defect in swi1Δ and swi3-1 mutants (Fig. 1B). We then cloned the SPBC30D10.04 genes from wild-type and swi3-1 strains and expressed them in the swi3-1 mutant. Only the SPBC30D10.04 clone from the wild-type strain rescued the mating-type switching defect of swi3-1 cells. Likewise, only the SPBC30D10.04 clone from the wild-type strain complemented the sensitivity that swi3-1 cells displayed to the genotoxic drugs HU and camptothecin (CPT). Finally, we sequenced the SPBC30D10.04 clone from the swi3-1 strain and found that it had an extra adenine at the 126th nucleotide. This mutation caused a frameshift that truncated the protein at amino acid 44. These results established that SPBC30D10.04 was allelic to swi3+.
To confirm the interaction of Swi1 and Swi3 in S. pombe, the strain expressing Swi1-TAP was engineered to express Swi3-13myc from the swi3 genomic locus. As expected, Swi3-13myc coprecipitated with Swi1-TAP (Fig. 1C). We also constructed a strain that expressed Swi3-TAP and Swi1-13myc from their own genomic loci and found that Swi1-13myc copurified with Swi3-TAP (Fig. 1C). The immunoblot signal of Swi1-13myc was much stronger than that of Swi3-13myc (Fig. 1C), but this difference probably reflected poor binding of Swi3-13myc to the blotting membrane. This conclusion is based on the microscopic analysis of Swi1-GFP and Swi3-GFP expressed from their respective genomic loci in live cells (see Fig. 4A, below). We measured the intensity of the GFP signals as described in Materials and Methods and found that the GFP signal value was 631 ± 81/pixel for Swi1-GFP and 640 ± 113/pixel for Swi3-GFP, indicating that both proteins were equally abundant in live cells (see Fig. 4A). Coupled with the evidence that swi1 and swi3 mutants present similar phenotypes in mating-type switching assays, these data strongly suggested that Swi1 and Swi3 function in a codependent manner as a stable heterodimer.
FIG. 4.
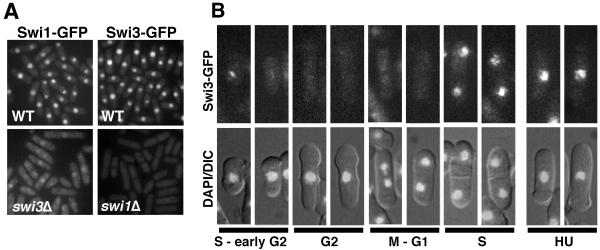
Recruitment of Swi3-GFP to chromatin in S phase. (A) Swi1-GFP and Swi3-GFP are nuclear proteins. Swi1-GFP delocalized from the nucleus in swi3Δ cells. Swi3-GFP was not detectable in the absence of Swi1. Live cells were analyzed for Swi1-GFP or Swi3-GFP fluorescence. (B) In situ chromatin binding assay of Swi3-GFP. Spheroplasts were extracted with Triton X-100 to remove soluble nuclear protein and then fixed for microscopic analysis (41). Representative patterns of fluorescence are shown. Swi3-GFP was detected predominantly in septated cells and unseptated small cells, which are in S phase or possibly early G2 phase. Representative photos of HU-treated cells are shown.
Swi3 is required for survival of fork arrest and for the replication checkpoint.
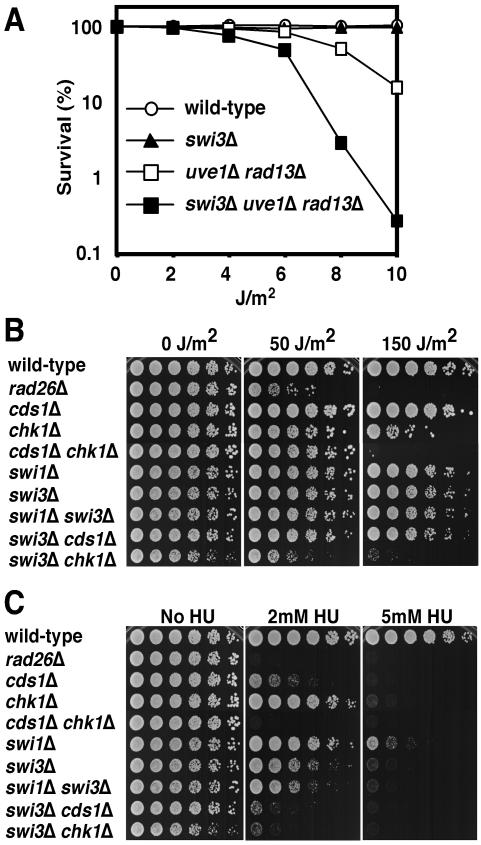
We carried out a series of studies to determine whether Swi3 is involved in tolerance of fork arrest. These studies were modeled on previous investigations of Swi1 (41). We first investigated whether Swi3 contributed to survival of DNA lesions that block replication forks. For these studies we used UV irradiation. UV light creates cyclobutane dimers and other lesions that arrest replication forks. These studies were carried out in strains that were defective for nucleotide excision repair (rad13Δ) and UV damage excision repair (uve1Δ). Nucleotide excision repair and UV damage excision repair account for all detectable UV damage repair in fission yeast (69). Strains defective for both pathways are acutely sensitive to UV. Their UV survival depends on homologous recombination or translesion DNA polymerase pathways that bypass UV lesions during DNA replication (9, 18, 67). swi3Δ cells were not significantly sensitive to low doses of UV, but the swi3Δ mutation substantially enhanced UV sensitivity in the rad13Δ uve1Δ double mutant background (Fig. 2A), indicating that Swi3 is vital for viability when UV damage in DNA is not repaired.
FIG. 2.
Swi3 is required for survival of fork arrest. (A) Swi3 contributes to survival with UV irradiation in a rad13Δ uve1Δ strain that cannot excise UV damage. Cells of the indicated genotypes were spread on YES agar medium and exposed to the indicated dose of UV. Agar plates were incubated for 3 days at 30°C to measure UV survival. (B) Interaction of swi3Δ and chk1Δ mutations in UV survival assays indicates that Swi1 is required for tolerance of UV damage during DNA replication. Fivefold serial dilutions of cells were plated on YES agar medium and exposed to the indicated dose of UV. Agar plates were incubated for 2 to 5 days at 30°C. (C) Swi3 has a role in tolerating HU-induced fork arrest. swi3Δ cells were highly sensitive to HU. Synergistic interactions of swi3Δ with cds1Δ and chk1Δ mutations indicate that Swi3 has an HU survival function that is at least partially independent of Cds1 and Chk1. Fivefold serial dilutions of cells were incubated on YES agar medium supplemented with the indicated amounts of HU for 2 to 5 days at 30°C.
We performed experiments to detect interactions between swi3Δ and mutations in checkpoint kinases in UV survival assays. The chk1Δ swi3Δ cells were substantially more sensitive than either single mutant (Fig. 2B). In contrast, there was no genetic interaction between swi3Δ and cds1Δ in UV survival assays. Chk1 is the effector kinase of the G2-M DNA damage checkpoint. The interaction of chk1Δ and swi3Δ mutations in UV survival assays may be explained by one or a combination of two mechanisms. The chk1Δ mutation allows cells irradiated in G2 phase to undergo mitosis and initiate a new round of DNA synthesis with unrepaired UV lesions. These lesions will obstruct replication forks and create an increased demand for Swi3's function in stabilizing stalled forks. In addition, the absence of Swi3 during S phase in cells containing UV lesions in DNA may lead to fork breakage or other abnormal DNA structures, creating an increased requirement for Chk1's function in the G2-M checkpoint. In either case the data support the idea that Swi3 has an important role in tolerance of DNA damage caused by UV.
We also examined whether Swi3 is required to survive exposure to HU. swi3Δ cells displayed strong HU sensitivity (Fig. 2C). Interestingly, swi3Δ and swi1Δ swi3Δ cells were more HU sensitive than swi1Δ cells, indicating that, in addition to its functions in partnership with Swi1, Swi3 might have additional functions that are independent of Swi1 or that Swi3 retains a partial function in the absence of Swi1. There was an additive interaction between swi3Δ and cds1Δ (Fig. 2C), showing that Swi3 has a role in tolerating HU-induced replication arrest that is partially independent of Cds1.
To investigate Swi3's role in HU-induced checkpoint arrest, we examined the effect of inactivating Swi3 in swi1Δ, cds1Δ, and chk1Δ backgrounds. swi3Δ and swi3Δ cds1Δ cells arrested division (Fig. 3A), but swi3Δ chk1Δ cells failed to arrest (Fig. 3A). The latter observation was consistent with the strong interaction between swi3Δ and chk1Δ mutations in HU survival assays (Fig. 2C). Cds1 and Chk1 define redundant pathways of HU-induced checkpoint arrest (10); thus, the checkpoint defect of swi3Δ chk1Δ cells suggested that Swi3 is required for proficient function of Cds1. In support of this conclusion, Cds1 activation by HU was strongly diminished in swi3Δ cells (Fig. 3B). A similar deficiency in Cds1 activation was observed in swi1Δ cells (Fig. 3B).
FIG. 3.
Swi3 is required for the replication checkpoint. (A) Indicated strains were incubated in YES liquid medium supplemented with 0 or 12 mM HU for 6 h at 30°C and then stained with DAPI to visualize nuclear DNA as described in Materials and Methods. Wild-type, swi3Δ, swi3Δ swi1Δ, and swi3Δ cds1Δ cells treated with HU underwent checkpoint arrest, as indicated by the appearance of elongated, uninucleate cells without septa. In contrast, swi3Δ chk1Δ cells treated with HU failed to undergo cell cycle arrest and instead displayed aberrant mitosis, as indicated by a cut phenotype. The cut phenotype also appeared in ∼10% of septated swi3Δ chk1Δ cells grown in the absence of HU (indicated by arrow). In some of the HU-treated cells the nuclei were not stained efficiently with DAPI. (B) Cds1 kinase activation is defective in swi1Δ and swi3Δ cells. Cells of the indicated genotypes were incubated in YES liquid medium supplemented with 0 or 12 mM HU for 4 h at 30°C. Kinase activity of immunoprecipitated Cds1 was measured by using myelin basic protein as a substrate. (C) HU sensitivity of swi1Δ or swi3Δ cells was suppressed by a multicopy cds1+ plasmid. The swi1Δ or swi3Δ cells were transformed with the indicated plasmid and plated on YES medium containing 0 or 5 mM HU for 2 to 5 days at 30°C.
In view of these findings, we examined whether the HU sensitivity of swi1Δ and swi3Δ cells could be rescued by overexpressing Cds1. This analysis showed that a multicopy plasmid that contained cds1+ substantially rescued the HU sensitivity of swi1Δ and swi3Δ mutants (Fig. 3C). In contrast, Swi1 overexpression did not rescue the HU sensitivity of swi3Δ cells, nor did Swi3 overexpression rescue swi1Δ HU sensitivity (Fig. 3C).
The Swi1-Swi3 complex associates with chromatin in S phase.
To determine whether the Swi1-Swi3 complex functions at replication forks and to further understand the functional relationship between Swi1 and Swi3, we investigated Swi1 and Swi3 localization by fluorescence microscopy using GFP-tagged proteins expressed from their endogenous promoters. Swi3-GFP-expressing cells were resistant to HU and CPT and proficient for mating-type switching, indicating that Swi3-GFP was fully functional. Swi3-GFP localized in the nuclei of all cells in an asynchronous culture, showing that Swi3 is nuclear throughout the cell cycle (Fig. 4A). After Triton X-100 extraction removed nuclear soluble proteins, Swi3-GFP persisted predominantly in septated cells and short cells (Fig. 4B). Septated cells are in S phase, and short cells are in late S or early G2. Prior treatment with DNase I eliminated all Swi3-GFP (data not shown). Similar results were seen with the Swi1-GFP protein (41), indicating that the Swi1-Swi3 complex associates with chromatin only in S phase. These findings were consistent with the idea that the Swi1-Swi3 complex associates tightly with chromatin specifically during DNA replication.
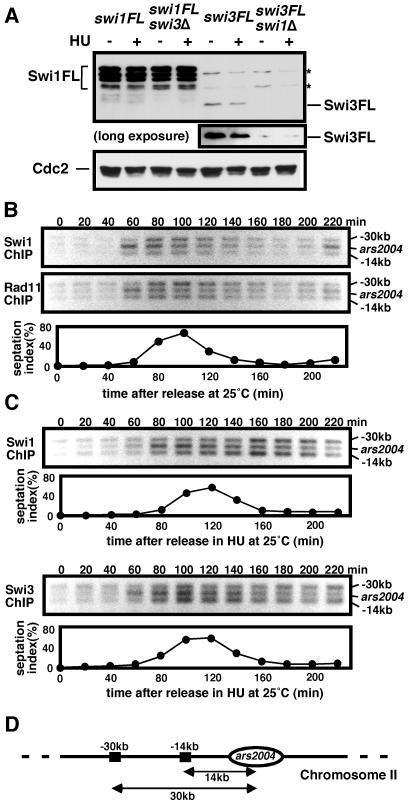
To further understand the functional relationship between Swi1 and Swi3, we examined Swi1-GFP localization in swi3Δ cells and Swi3-GFP localization in swi1Δ cells. Strikingly, the level of nuclear Swi1-GFP was strongly reduced in swi3Δ cells, and it was replaced by a strong cytoplasmic signal (Fig. 4A). The level of nuclear Swi3-GFP was strongly reduced in swi1Δ cells, but it was not replaced by an increase in the cytoplasmic signal of Swi3-GFP (Fig. 4A). Indeed, immunoblotting showed that Swi3 abundance was very low in swi1Δ cells (Fig. 5A), whereas Swi1 expression was unaffected in swi3Δ cells. The large reduction of Swi3 abundance in swi1Δ cells was curious in light of our studies showing that the phenotypes of swi3Δ cells were more severe than those seen in swi1Δ cells. These phenotypes suggested that residual amounts of Swi3 were present and presumably functional in swi1Δ cells. In fact, long exposures of the immunoblot showed that a small amount of Swi3 was detected in swi1Δ cells (Fig. 5A). These findings showed that Swi3's abundance in cells depends upon Swi1, and nuclear accumulation of Swi1 requires Swi3.
FIG. 5.
Swi1 associates with the origin in early S phase. (A) Immunoblot analysis of Swi1-3FLAG (Swi1-FL) and Swi3-3FLAG (Swi3-FL). Cells of the indicated strains were incubated in YES medium supplemented with 0 or 12 mM HU. Protein samples of the indicated cells were analyzed by immunoblotting with anti-FLAG antibody. A longer exposure of the immunoblot showed that a very low amount of Swi3-FL was detected in swi1Δ cells. Asterisks show proteins that were cross-reactive with anti-FLAG antibody. (B) ChIP assays of Swi1-13myc and Rad11-3FLAG were performed at ars2004 and sites located 14 or 30 kb away from ars2004 (42). The cdc25-22 cells were synchronized at the G2-M boundary by incubation at 36°C for 4 h and then released in fresh YES medium at 25°C. An increase in the septation index indicates the onset of S phase. (C) ChIP assays of Swi1-FL and Swi3-FL were performed as described above, except that cells were released into YES medium supplemented with 10 mM HU. (D) Diagram of region used in the ChIP assay. ars2004 and surrounding regions are shown.
The Swi1-Swi3 complex moves with the replication fork.
To address whether the S-phase-specific association of Swi1-Swi3 with chromatin reflects its localization at the replication fork, we performed ChIP with cells expressing FLAG- or myc-tagged proteins expressed from their endogenous promoters. The cdc25-22 allele was used to synchronize cells at the G2-M boundary. The cdc25-22 strain was engineered to express both Swi1-13myc and Rad11-3FLAG proteins. Rad11 is the large subunit of the replication protein A (RPA) complex that binds ssDNA at the fork. We monitored Swi1-13myc at the ars2004 replication origin and at two positions, 14 and 30 kb away from ars2004 (42). Upon release from the arrest, Swi1-13myc associated with the replication origin ars2004 at 60 min, and this association declined by 120 min (Fig. 5B). Swi1's association with ars2004 occurred as the septation index increased, which coincided with the onset of S phase. Swi1's association with a site located 14 kb away from ars2004 peaked at 60 to 100 min and declined by 140 min, shortly after its association with ars2004. Swi1's association with a site located 30 kb away from ars2004 peaked slightly later, at 80 to 120 min, and declined by 160 to 180 min. The ChIP pattern of Rad11-3FLAG closely matched that of Swi1-13myc (Fig. 5B), suggesting that Swi1 associated with the moving replication fork. To further confirm that Swi1 relocates on chromatin during DNA replication, cells were released into the presence of a low dose of HU to extend the duration of S phase (Fig. 5C, upper panels). Peak association of Swi1 at ars2004 occurred at 80 min and, thereafter, it slowly declined. The decrease in Swi1 at ars2004 coincided with its increased association at the sites 14 and 30 kb away, again consistent with Swi1 moving with the fork. We obtained similar results with the Swi3-FLAG protein (Fig. 5C, bottom panels), suggesting that the Swi1-Swi3 complex moves with the fork.
Replication abnormalities in swi3Δ cells.
Having established that Swi3 is required for tolerance of fork arrest and for checkpoint signaling to Cds1 and having found that it associates with chromatin in S phase, we then investigated Swi3's role in cells that have not been exposed to genotoxic agents. These studies were performed because several observations suggested that Swi3 was required for accurate DNA replication in the absence of genotoxic stress. In particular, we noticed that swi3Δ cells grown in the absence of genotoxic agents were moderately elongated in relation to wild type, whereas swi3Δ chk1Δ cells were not elongated (Fig. 3A). These phenotypes indicated that swi3Δ cells spontaneously accumulate unusual DNA structures that activate Chk1 and delay mitosis. Consistent with this possibility, we noted that swi3Δ chk1Δ cells frequently displayed a spontaneous “cut” phenotype. This phenotype was characterized by a DNA mass that was unequally distributed to daughter cells or bisected by the division plate (Fig. 3A). About 10% of the septated swi3Δ chk1Δ cells grown in liquid culture displayed a cut phenotype. Significantly, all of the swi3Δ chk1Δ phenotypes were also seen in swi1Δ chk1Δ cells but not in cds1Δ chk1Δ cells (41). The latter observation showed that the genetic interactions involving swi3Δ and chk1Δ were not solely attributable to Swi3's role in Cds1 activation.
These findings suggested that swi3Δ cells suffer spontaneous DNA damage. This possibility was investigated further by monitoring the formation of Rad22 DNA repair foci in swi3Δ cells (15, 41). Fission yeast Rad22 (also known as Rad22A) is a homolog of budding yeast Rad52, a protein that binds to ssDNA during homologous recombination (47). If swi3Δ cells suffered broken forks or other forms of DNA damage as the result of replication abnormalities, these structures would be expected to recruit Rad22. Our studies were carried out with strains that expressed endogenous Rad22 tagged with yellow fluorescent protein (YFP). This analysis showed that there was a large increase in spontaneous Rad22-YFP foci in swi3Δ cells (Fig. 6A). In wild-type cells, 15.8% of nuclei contained a single Rad22-YFP focus and none showed multiple Rad22-YFP foci, whereas in swi3Δ cells 65.6% of the nuclei contained at least one Rad22-YFP focus and 48.9% had multiple foci (Fig. 6A).
FIG. 6.
Spontaneous DNA damage occurs during S phase in swi3Δ cells. (A) Multiple Rad22-YFP foci accumulated in swi3Δ cells. Log-phase cells were grown in Edinburgh minimal medium at 25°C. In wild-type cells, 15.8% of the nuclei contained a single Rad22-YFP focus and none showed multiple Rad22-YFP foci. In swi3Δ cells, 65.6% of the nuclei contained at least one Rad22-YFP focus and 48.9% had multiple foci. (B) Quantification of Rad22-YFP foci according to cell cycle stage estimated from cell and nuclear morphology. The percentages of nuclei that have at least one focus or two or more foci are shown. S-phase cells had the most Rad22-YFP foci.
We suspected that swi3Δ cells were suffering DNA damage or accumulating unusual DNA structures during DNA replication. This possibility was addressed by evaluating the cell cycle position of cells that contained Rad22-YFP foci. Cell cycle position in fission yeast can be estimated by noting cell length, nuclear morphology, and the appearance of a septum. S phase initiates very soon after nuclear division, continues during cell septation, and ends about when daughter cells finally detach. Cell cycle stage analysis revealed that swi3Δ cells with multiple Rad22-YFP foci were predominantly in S or early G2 (Fig. 6B), indicating that the foci arose from DNA replication abnormalities. A similar phenotype was observed with swi1Δ cells (41), whereas cds1Δ cells displayed only a small increase in Rad22-YFP foci (41). These findings provided evidence that the Swi1-Swi3 complex has Cds1-independent functions that help ensure faithful DNA replication.
Rhp51 and Rhp54 are not required for survival of swi3Δ cells.
Rad52 homologs have well-established roles in double-strand break (DSB) repair (47). We therefore hypothesized that the increased Rad22-YFP foci in swi3Δ cells were representative of DSBs created by fork collapse. A broken fork produces a DNA free end that is resected on one strand as a prelude to repair by homologous recombination (Fig. 7A). Recapture of broken forks is thought to proceed by strand invasion and DNA annealing steps that are dependent on Rad51 and Rad54 (36, 47). This process is thought to necessarily lead to formation of an HJ (Fig. 7A). Current evidence in fission yeast indicates that Rhp51 and Rhp54 are needed for strand invasion of DNA duplexes, while the Mus81-Eme1 complex is required for HJ resolution (8, 19, 45). Consistent with the fork recapture model, Rhp51, Rhp54, and Mus81-Eme1 are required for survival of fork breakage caused by CPT, a topoisomerase I inhibitor (14).
FIG. 7.
swi1Δ cells accumulate X-shaped DNA. (A) Model of the fork recapture mechanism. Fork recapture requires strand invasion catalyzed by Rhp51 and Rhp54 followed by HJ resolution by the Mus81-Eme1 complex. (B) X-shaped DNA accumulates in swi1Δ mus81Δ cells. Genomic DNA samples prepared from exponentially growing S. pombe cells with the indicated genotypes were analyzed by 2D gel electrophoresis with the ars3001 HindIII-KpnI fragment as a probe (41). The amount of X-shaped DNA expressed as the percentage of entire replication and recombination intermediates was quantified. The arrow points to X-shaped DNA in swi1Δ mus81Δ cells. (C) The X-shaped DNA branch migrates into linear DNA. Genomic DNA from swi1Δ mus81Δ cells was run in the first-dimension gel and gel slices were incubated in branch migration buffer at 4 or 65°C for 5 h, as described in Materials and Methods, and then DNA was electrophoresed in the 2D gel. The arrow indicates the spot corresponding to the linear DNA products derived from the X-shaped DNA molecules by branch migration. (D) Diagram of the migration pattern of replication and recombination intermediates that can be detected by 2D gel electrophoresis before (4°C) and after (65°C) the branch migration reaction. X-shaped DNA that has the property of HJs can be converted into a linear product that produces a distinct spot which migrates below the X-shaped DNA on the 1N line.
To address whether swi1Δ and swi3Δ cells experience spontaneous fork breakage, we carried out genetic crosses that introduced swi1Δ or swi3Δ mutations into rhp51Δ or rhp54Δ strains. Tetrad analysis revealed that rhp51Δ and rhp54Δ mutations displayed no obvious genetic interactions with swi1Δ or swi3Δ mutations (Table 1). Viable double mutants were recovered at the expected frequency, and they grew as well as the rhp51Δ and rhp54Δ single mutants. Given the vital role that Rhp51 and Rhp54 play in homologous recombination and survival of CPT treatment (14), these findings strongly indicated that swi1Δ and swi3Δ strains do not experience an elevated occurrence of fork breakage.
TABLE 1.
Genetic interactions involving swi1Δ and swi3Δ
| Genotype | Growth ratea |
|---|---|
| Wild type | ++++ |
| swi1Δ | +++ |
| swi3Δ | +++ |
| rhp51Δ | ++ |
| rhp54Δ | ++ |
| rad22Δ | ++ |
| mus81Δ | ++ |
| swi1Δ rhp51Δ | ++ |
| swi3Δ rhp51Δ | ++ |
| swi1Δ rhp54Δ | ++ |
| swi3Δ rhp54Δ | ++ |
| swi1Δ rad22Δ | ++ |
| swi3Δ rad22Δ | ++ |
| swi1Δ mus81Δ | −b |
| swi3Δ mus81Δ | −c |
| swi1Δ mus81Δ rhp51Δ | − |
| swi1Δ mus81Δ rhp54Δ | − |
| swi1Δ mus81Δ rad22Δ | + |
Doubling times: ++++, ∼2 h; +++, ∼2.5 h; ++, ∼3 h; +, ∼6 h; −, no growth.
Two out of 37 swi1Δ mus81Δ spores were viable.
None of the 33 swi3Δ mus81Δ spores was viable.
We also performed a genetic cross involving swi3Δ and mus81Δ mutations. In contrast to the situation with rhp51Δ and rhp54Δ mutations, swi3Δ had a synthetic lethal interaction with mus81Δ (Table 1). The swi3Δ mus81Δ spores germinated and formed microcolonies of ∼4 to 30 elongated or misshapen cells. No viable swi3Δ mus81Δ cells were recovered among 33 double mutant spores. A very similar genetic interaction was seen with swi1Δ and mus81Δ mutations (41). Tetrad analysis revealed that 35 of 37 swi1Δ mus81Δ spores germinated to produce inviable microcolonies. The other two swi1Δ mus81Δ spores propagated to form very slow growing colonies that contained many dead cells. Taken together, these studies provided strong evidence that swi1Δ and swi3Δ strains experience replication abnormalities that lead to HJ formation without fork breakage.
X-shaped DNA structures accumulate in swi1Δ mus81Δ cells.
The genetic interactions described above suggested that swi1Δ and swi3Δ mutants form abnormal DNA structures during DNA replication that recombine to form HJs by an Rhp51- and Rhp54-independent mechanism. We explored this possibility by performing 2D gel electrophoresis of chromosomal DNA to detect branched DNA structures. X-shaped DNA migrates in a characteristic spike in 2D gels (Fig. 7D). We monitored the ars3001 HindIII-KpnI region. This region contains the replication origin of the rDNA (41, 51). We analyzed wild-type, mus81Δ, swi1Δ, swi3Δ, and cds1Δ strains, as well as cds1Δ mus81Δ and swi1Δ mus81Δ double mutants (Fig. 7B). The latter strain was derived from one of the very slow growing swi1Δ mus81Δ strains mentioned above. The amount of X-shaped DNA was quantified as the percentage of replication and recombination intermediates (Fig. 7B). Wild-type, swi1Δ, swi3Δ, and cds1Δ strains contained approximately equal amounts of X-shaped DNA (14.2 to 19.6%, expressed as a percentage of the total replication and recombination intermediates), whereas X-shaped DNA was increased in the mus81Δ cells (26.7%). Importantly, there was substantially more X-shaped DNA in swi1Δ mus81Δ cells (38.6%). In contrast, the amount of X-shaped DNA in cds1Δ mus81Δ cells (29.1%) was not substantially increased relative to that in mus81Δ cells (Fig. 7B).
The structure of the X-shaped DNA in swi1Δ mus81Δ cells was investigated further by performing branch migration assays (6, 19, 53, 70). In these assays HJs branch migrate to form linear DNA fragments. The agarose gel slice from the first-dimension gel electrophoresis was incubated in branch migration buffer at 65°C for 5 h prior to 2D gel electrophoresis (Fig. 7C). A control gel slice was incubated at 4°C for 5 h. When incubated at 65°C, the amount of X-shaped DNA in swi1Δ mus81Δ cells was strongly reduced (Fig. 7C and D). The branch migration reaction produced a distinct spot that migrated directly below the X-shaped DNA at the level of linear fragment (Fig. 7C and D), indicating that the majority of X-shaped DNAs in swi1Δ mus81Δ cells were HJs, or possibly hemicatenanes that formed by the fusion of HJs (32, 66).
Rad22 inactivation suppresses swi1Δ mus81Δ synthetic lethality and formation of X-shaped DNA.
The aforementioned studies suggested that the absence of functional Swi1-Swi3 complex leads to the formation of ssDNA regions that are bound by Rad22 and are converted into HJs. If there was a causal connection between formation of Rad22 foci and the appearance of HJs in swi1Δ mus81Δ cells, we would expect that inactivation of Rad22 might ameliorate the genetic interactions involving swi1Δ and mus81Δ. Moreover, if HJs were formed without the participation of Rhp51 and Rhp54, as suggested by our studies, it would be expected that inactivation of Rhp51 and Rhp54 should not rescue the synthetic lethal interactions involving swi1Δ and mus81Δ. Indeed, genetic crosses showed that rhp51Δ and rhp54Δ mutations did not suppress swi1Δ mus81Δ synthetic lethality (Table 1). In contrast, swi1Δ mus81Δ rad22Δ cells were viable, although they grew slowly relative to each of the single mutants (Table 1).
These results suggested that the Rad22 foci detected in swi1Δ and swi3Δ mutants were precursors of HJs that were formed, apparently without fork breakage, in a reaction requiring DNA strand annealing promoted by Rad22. To test this possibility, we examined whether the increase in X-shaped DNA in swi1Δ mus81Δ cells required Rad22. The amount of X-shaped DNA in swi1Δ mus81Δ rad22Δ cells (13.1%) was approximately equivalent to that seen in swi1Δ (17.3%) and rad22Δ (14.0%) single mutants (Fig. 7B). These data suggested that accumulated ssDNA in the swi1Δ and swi3Δ mutants recombined to form HJs through a Rad22-dependent mechanism.
Interestingly, the amount of X-shaped DNA in rad22Δ cells and the wild type was approximately the same (Fig. 7B). The presence of this Rad22-independent X-shaped DNA seems to contradict earlier studies in S. pombe and S. cerevisiae (53, 70), in which formation of X-shaped DNA required Rad22/Rad52. However, recent studies of S. cerevisiae have shown that X-shaped DNA can form without Rad52 (32, 66). These studies used a modified method of preparing DNA that preserves X-shaped DNA structures. In this study, we have detected both Rad22-dependent and -independent X-shaped DNA. We suspect that the Rad22-dependent X-shaped DNA molecules are HJs, because they arose in the mus81Δ background and readily branch migrated. The Rad22-independent X-shaped DNA might be converged forks or hemicatenanes.
Specific genetic interactions involving the Swi1-Swi3 complex and DNA polymerases.
Previous studies showed that Swi1 and Swi3 are required for viability in a strain that has the swi7-1 (DNA polymerase α [Polα]) mutation that impairs mating-type switching (52). Polα is required to initiate both leading- and lagging-strand synthesis (63). To further explore the source of the abnormal DNA structures in swi1Δ and swi3Δ cells, we examined genetic interactions with cdc6-23 (Polδ) and cdc20-M10 (Polɛ) mutations. The exact roles of these polymerases are uncertain, but mutational analyses using exonuclease-deficient Polδ and Polɛ strains have shown a strand specificity of mutation rates and spectra, suggesting that the two polymerases operate preferentially on different sides of the fork (24, 48, 54). We found that swi1Δ and swi3Δ mutations were synthetically lethal with cdc6-23 but not with cdc20-M10. We cannot exclude the possibility that swi1Δ and swi3Δ might have genetic interactions with other alleles of cdc20, but the specific genetic interactions with DNA Polα and Polδ suggest that Swi1-Swi3 might be required to coordinate leading- and lagging-strand synthesis.
DISCUSSION
We have shown that Swi1 and Swi3 function together in a protein complex that helps to maintain genome integrity in fission yeast. This complex is recruited to chromatin in S phase, and it travels in close proximity to the replication fork. It is needed to pause the replication fork at specific sites near the mat1 locus, and it is required for proficient activation of the replication checkpoint in response to fork arrest. In the absence of the Swi1-Swi3 complex, abnormal DNA structures represented by Rad22 foci accumulate during S phase. These structures are unlikely to be broken forks, because rhp51Δ and rhp54Δ do not have genetic interactions with swi1Δ and swi3Δ mutations. Instead, we hypothesize that the Rad22 foci represent ssDNA gaps that can mature into HJs or related structures that are processed by Mus81-Eme1 resolvase. Both swi1Δ and swi3Δ have striking genetic interactions with mutations that impair DNA Polα and Polδ but not Polɛ. In view of these and earlier studies, we suggest that the Swi1-Swi3 heterodimer can be described as an FPC that is required to properly coordinate leading- and lagging-strand synthesis. We suggest that an inability to coordinate leading- and lagging-strand synthesis in FPC-deficient cells leads to formation of ssDNA gaps during S phase, as well as an inability to activate the replication checkpoint.
Swi1-Swi3 FPC.
Swi1 and Swi3 readily coprecipitate as components of a stable complex. We cannot formally exclude the possibility that there may be other proteins in this complex, but mass spectrometry and genetic studies have failed to identify additional candidates. In cells that express GFP-tagged Swi1 or Swi3 from the endogenous loci, the two proteins appear to be expressed at a 1:1 ratio and they colocalize in the nucleus. Swi3 appears to be very unstable in swi1Δ cells, whereas Swi1 is delocalized from the nucleus in swi3Δ cells. The swi1Δ and swi3Δ mutations impart nearly identical phenotypes in a wide range of assays, with the only exception being that swi3Δ cells appear to be slightly more sensitive to HU. Taken together, these data show that Swi1 and Swi3 function as an integrated, self-contained complex.
Our studies suggest that the Swi1-Swi3 FPC is closely associated with the replication fork, and it may in fact be an ancillary component of the larger replisome complex. FPC associates with chromatin specifically in S phase, and it moves away from a replication origin in synchrony with RPA. These findings are consistent with genome-wide studies of S. cerevisiae Tof1, the presumptive homolog of Swi1 (25). Exactly how FPC associates with the fork is unknown. Tof1 was shown to coprecipitate with the essential DNA replication protein Cdc45 specifically during S phase (25); thus, it is reasonable to speculate that Cdc45 (Sna41 in fission yeast) or an associated protein recruits FPC to replication forks. In such a position, FPC is well placed to stabilize stalled forks in a configuration that can be recognized by the replication checkpoint system.
Abnormal DNA structures in FPC mutants.
Genetic interactions involving swi1Δ and swi3Δ mutations with chk1Δ strongly suggest that FPC mutants accumulate abnormal DNA structures during S phase that activate the DNA damage checkpoint. Deciphering the nature of this damage and FPC's role in DNA replication necessitates an understanding of homologous recombination repair. Rad51, Rad52, and Rad54 homologs play central roles in mediating strand invasion of resected DSBs into intact duplex targets (47). They are thought to perform the same activities in recapture of broken forks. In the case of fork recapture, strand invasion necessarily leads to formation of a D-loop and an HJ or an HJ-like structure that must be resolved to allow chromosome segregation during mitosis (36). The Mus81-Eme1 complex is thought to resolve these structures (8, 19, 45). Consistent with these models, Rhp51, Rhp54, and Mus81-Eme1 endonuclease are all vital for survival of cells treated with CPT, a topoisomerase inhibitor that induces fork breakage (14). In contrast to the situation with CPT, Rhp51 and Rhp54 but not Mus81-Eme1 are required for survival of DSBs caused by ionizing radiation (9). These results suggest that DSBs in G2 phase are preferentially repaired by synthesis-dependent strand annealing or a related mechanism (47) that requires strand invasion but not HJ resolution. Consistent with this notion, crossing over is rarely associated with mitotic recombination in S. pombe (62). Thus, synthesis-dependent strand annealing appears to be the dominant recombination mechanism of repairing DSBs in mitotic S. pombe cells, as it is in S. cerevisiae (47).
Mus81-Eme1, Rhp51, and Rhp54 are all required to survive CPT-induced fork breakage, whereas only Rhp51 and Rhp54 are required to survive DSBs caused by ionizing radiation. In swi1Δ and swi3Δ mutants, a third situation exists in which Mus81-Eme1, but not Rhp51 and Rhp54, is required for viability. These relationships imply that swi1Δ mus81Δ and swi3Δ mus81Δ cells form HJs without breaking forks. The alternative possibility is that FPC mutants suffer broken forks that are recaptured by a mechanism that does not require Rhp51 and Rhp54. It is conceivable that in some instances broken forks might be repaired without Rhp51 and Rhp54, for example, in the rDNA repeats. However, if broken forks were constrained to the rDNA, we would not expect to see multiple Rad22 foci in FPC mutants. We therefore conclude that Rad22 foci detected in swi1Δ and swi3Δ mutants are unlikely to represent broken forks and, thus, are probably sites of ssDNA gaps left in the wake of a moving replication fork. The idea that FPC mutants were not suffering broken forks was further strengthened by the fact that the swi1Δ rad22Δ rhp51Δ triple mutant was still highly viable (data not shown). A causal connection between Rad22 foci and Rad22-dependent formation of X-shaped DNAs is strongly suggested by the rescue of swi1Δ mus81Δ by rad22Δ. These genetic interactions indicate that a Rhp51/Rhp54-independent strand annealing activity of Rad22 promotes formation of recombination structures in swi1Δ and swi3Δ cells that must be resolved by Mus81-Eme1. Such a Rad51/Rad54-independent activity has been described for Rad52 in budding yeast (47). This activity of Rad22 is not required for viability in FPC mutants (Table 1); thus, it is likely that the ssDNA gaps in rad22Δ swi1Δ mus81Δ cells are repaired by a gap-filling mechanism similar to that involved in nucleotide excision repair.
Two models that illustrate how HJs might arise without fork breakage from a single-strand gap in the vicinity of a replication fork are shown in Fig. 8. These ssDNA gaps occur in the absence of FPC. These gaps may be generated when leading- and lagging-strand DNA synthesis are uncoordinated (see next paragraph). In model A, an ssDNA gap left in the wake of a replication fork becomes involved in strand exchange events that are promoted by Rad22. Coupled DNA synthesis leads to formation of double HJs that are cleaved by Mus81-Eme1. In principle, Mus81-Eme1 could cleave HJs or HJ precursors. We did not observe an increased amount of X-shaped DNA in swi1Δ or swi3Δ single mutants, suggesting that the HJs or HJ precursors can be readily resolved by Mus81-Eme1. An alternative possibility for formation of HJs without fork breakage is shown in Fig. 8B. In this model, problems with DNA synthesis in FPC mutants lead to fork regression and formation of a “chicken foot” structure that is similar to an HJ. The exposed nascent strand of DNA is bound by Rad22, which in turn promotes annealing to the template parental strand ahead of the fork. The activity of a replicative helicase might disassociate the parental strands ahead of the fork and thereby facilitate Rad22-dependent strand invasion.
FIG. 8.
Two models for Rad22-dependent generation of X-shaped DNA molecules and their resolution by Mus81-Eme1 in swi1Δ and swi3Δ cells. (A) The ssDNA gap left in the lagging strand participates in a recombination event catalyzed by Rad22. Strand invasion by the 3′ end of an Okazaki fragment and following DNA synthesis leads to formation of double HJs that are targeted by Mus81-Eme1. (B) An ssDNA gap may lead to fork regression, creating a chicken foot structure that is similar to an HJ. The exposed nascent strand of DNA may anneal to the template parental strand ahead of the fork, resulting in formation of double HJs.
Relationship between FPC and DNA Pols.
The relationships involving swi1Δ and swi3Δ with rhp51Δ, rhp54Δ, and mus81Δ are not completely unique. Mutations in swi7-1 (Polα) and cdc6-23 (Polδ) also display strong genetic interactions with mus81Δ but not with rhp51Δ or rhp54Δ (9). Polα is required for both lagging- and leading-strand synthesis, while Polδ and Polɛ are thought to operate on different sides of the fork. Using the same rationale described above, these genetic relationships suggest that swi7-1 and cdc6-23 mutants accumulate ssDNA gaps but not broken forks during S phase. One possible interpretation of these findings is that FPC works together with Polα and Polδ to promote effective DNA synthesis. This possibility is consistent with the synthetic lethal interactions involving swi1Δ and swi3Δ with swi7-1 and cdc6-23 mutations. Significantly, swi1Δ and swi3Δ did not display a synthetic lethal interaction with the cdc20-M10 mutation that encodes a defective form of Polɛ. In this context, it is worth noting the correlations of our investigations with earlier studies by Zou and Rothstein (70). Working with budding yeast, they showed that Polα and Polδ mutations, but not Polɛ mutations, led to formation of X-shaped DNAs in the rDNA repeats by a mechanism that required Rad52 but not Rad51 or Rad54.
Genetic studies have indicated that Polδ and Polɛ operate on different DNA strands (24, 48, 54), and in one study it was further suggested that Polδ might be specifically involved in lagging-strand synthesis, although definitive evidence is lacking. On the basis of these studies and our evidence that the Swi1-Swi3 complex is essential in cells that are partially defective for Polδ but not Polɛ, it is tempting to speculate that Swi1-Swi3 FPC is needed to stabilize forks when lagging-strand synthesis is blocked. FPC might directly participate in lagging-strand synthesis, or it might be required to slow down the fork when lagging-strand replication encounters difficulties. Vengrova and Dalgaard recently made a similar suggestion (61). These difficulties could be in the form of DNA damage that blocks lagging-strand synthesis. Tof1, the Swi1 homolog in budding yeast, is required to retard replisome movement when DNA synthesis is inhibited by HU (25). Perhaps Tof1 and Swi1 are subunits of a molecular brake that slows down or arrests the replisome when dNTPs are unavailable or when lagging-strand synthesis encounters barriers.
FPC's checkpoint function.
Swi1 and Swi3 have the properties expected of proteins that are specifically involved in the replication checkpoint. They are necessary for survival of genotoxic agents that arrest replication forks, they are required for HU-induced cell cycle arrest in a chk1Δ strain, and they are needed for proficient activation of Cds1. Moreover, the HU-sensitive phenotype of FPC mutants can be substantially rescued by overexpression of Cds1. Swi1 and Swi3 are not needed for the Chk1-dependent HU arrest that occurs in cds1Δ cells; thus, FPC is not required for Chk1 function. These findings show that FPC is specifically required for the replication checkpoint. The replication checkpoint properties of FPC closely match those of Mrc1, a protein that channels the replication checkpoint signal to Cds1 (1, 57). This specificity correlates with the fact that Mrc1 abundance is highly cyclical and specific for S phase (57) and with the observations that Swi1 and Swi3 associate with chromatin only in S phase.
While FPC, Mrc1, and Cds1 are all specifically required for the replication checkpoint, it is evident that swi1Δ and swi3Δ mutants differ from cds1Δ and mrc1Δ mutants. The mutations in cds1Δ and mrc1Δ do not impair mating-type switching, and they do not cause an increased incidence of aberrant mitosis in a chk1Δ background (41, 57). Inactivation of Cds1 causes only a small increase in Rad22 foci, and cds1Δ is not synthetic lethal with mus81Δ (9, 41). Unlike a number of DNA replication proteins that have been implicated in checkpoint signaling, FPC is not essential for DNA replication. Thus, the replication checkpoint defects in swi1Δ and swi3Δ cells cannot be readily ascribed to deficiencies in origin activation or DNA synthesis. These observations led us to conclude that FPC has a checkpoint-independent role in preserving replication forks, and this function stabilizes forks in a configuration that is required for efficient activation of the replication checkpoint kinase Cds1. These properties distinguish the Swi1-Swi3 FPC from previously described checkpoint proteins.
We previously showed that Swi1 associates with chromatin in S phase in a manner that is independent of other checkpoint proteins, such as Rad26, indicating that Swi1 has a role in DNA replication and checkpoint control that is independent of checkpoint activation (41). These finding suggested that FPC might be a component of a replisome whose primary role is to stabilize forks when they encounter obstacles. In this study, we showed that Swi1, as well as RPA, preferentially associates with a replication origin early in S phase and then associates with sites distal to the origin as replication proceeds. These findings suggest that Swi1 moves with the replication fork, consistent with recent studies of Tof1 (25). On the basis of these studies we propose that the Swi1-Swi3 complex moves with the fork and stabilizes it when it encounters a replication obstacle. In this model, FPC acts at a very early stage of fork stabilization and checkpoint signaling, whereas Cds1 acts at a later stage and is needed to maintain stalled replication forks in a stable state. Mrc1 might act as a bridge between the initial fork stabilization by FPC and maintenance of the stalled replication fork by Cds1.
FPC in other organisms.
Is FPC conserved in other eukaryotes? As discussed above, Swi1 and Tof1 are structurally related and share a number of phenotypes and activities, and thus they are likely to be true functional homologs. There are some significant phenotypic differences between swi1Δ and tof1Δ cells, for example, tof1Δ cells are not sensitive to HU (17), but these differences probably reflect organism differences in repair and checkpoint systems as opposed to differences in Swi1 and Tof1 functions. Interestingly, a large-scale yeast two-hybrid screen identified a putative interaction between budding yeast Tof1 and the Swi3-related protein Csm3 (59), suggesting conservation of FPC in the two highly divergent yeasts. The Tof1-Csm3 interaction was recently confirmed by coimmunoprecipitation (35). Budding yeast csm3Δ mutants appear to have a mild defect in meiotic chromosome segregation (49), and recent studies have uncovered a partial sister chromatid cohesion defect in tof1Δ and csm3Δ cells (35, 64). This phenotype is interesting in light of the role of C. elegans Tim1 (Tof1/Swi1 homolog) in chromosome cohesion and segregation (12). C. elegans has a Csm3/Swi3 homolog (Fig. 1A), and we speculate that it functions together with Tim1. There may be a causal connection between fork protection and chromosome cohesion. FPC-dependent fork protection may assist in establishing cohesion during S phase. Another possibility is that FPC-dependent activation of Cds1 homologs may be involved in establishing chromosome cohesion. This proposal is consistent with evidence that a mutation in C. elegans chk-2, a cds1 homolog, causes defects in meiotic chromosome segregation (34).
Drosophila and mammalian Timeless (Tim1) have been characterized as proteins that regulate circadian rhythms (5). At present we cannot offer a model that connects circadian rhythm control to fork protection, checkpoint signaling, or chromosome cohesion. However, a mouse Csm3/Swi3 homolog named TIPIN was recently detected in a two-hybrid screen with mouse Tim1 (20). Thus, it appears that the protein complexes related to Swi1-Swi3 and Tof1-Csm3 are broadly conserved among eukaryotes. It will be interesting to determine whether these complexes are involved in DNA replication and maintenance of genome integrity.
Acknowledgments
We thank V. Martin for providing the Rad11-3FLAG strain. Members of the Scripps Cell Cycle groups are thanked for their support and encouragement. We thank Joel Huberman for his helpful discussion.
E.N. was a fellow of the Human Frontier Science Program. E.N. was also supported by the UEHARA Memorial Foundation. This work was funded by MERK-MGRI-241 (W.H.M. and J.R.Y.), NIH EY1328801 (J.R.Y.), and NIH grant GM59447 awarded to P.R.
REFERENCES
- 1.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 2.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast. Cold Spring Harbor Laboraotry Press, Cold Spring Harbor, N.Y.
- 3.al-Khodairy, F., E. Fotou, K. S. Sheldrick, D. J. Griffiths, A. R. Lehmann, and A. M. Carr. 1994. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol. Biol. Cell 5:147-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 5.Barnes, J. W., S. A. Tischkau, J. A. Barnes, J. W. Mitchell, P. W. Burgoon, J. R. Hickok, and M. U. Gillette. 2003. Requirement of mammalian Timeless for circadian rhythmicity. Science 302:439-442. [DOI] [PubMed] [Google Scholar]
- 6.Benard, M., C. Maric, and G. Pierron. 2001. DNA replication-dependent formation of joint DNA molecules in Physarum polycephalum. Mol. Cell 7:971-980. [DOI] [PubMed] [Google Scholar]
- 7.Boddy, M. N., B. Furnari, O. Mondesert, and P. Russell. 1998. Replication checkpoint enforced by kinases Cds1 and Chk1. Science 280:909-912. [DOI] [PubMed] [Google Scholar]
- 8.Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates III, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107:537-548. [DOI] [PubMed] [Google Scholar]
- 9.Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W. D. Heyer, and P. Russell. 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20:8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boddy, M. N., and P. Russell. 2001. DNA replication checkpoint. Curr. Biol. 11:R953-R956. [DOI] [PubMed] [Google Scholar]
- 11.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 12.Chan, R. C., A. Chan, M. Jeon, T. F. Wu, D. Pasqualone, A. E. Rougvie, and B. J. Meyer. 2003. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 424:1002-1009. [DOI] [PubMed] [Google Scholar]
- 13.Dalgaard, J. Z., and A. J. Klar. 2000. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102:745-751. [DOI] [PubMed] [Google Scholar]
- 14.Doe, C. L., J. S. Ahn, J. Dixon, and M. C. Whitby. 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277:32753-32759. [DOI] [PubMed] [Google Scholar]
- 15.Du, L.-L., T. Nakamura, B. A. Moser, and P. Russell. 2003. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell. Biol. 23:6150-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fantes, P. 1979. Epistatic gene interactions in the control of division in fission yeast. Nature 279:428-430. [DOI] [PubMed] [Google Scholar]
- 17.Foss, E. J. 2001. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157:567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 19.Gaillard, P. H., E. Noguchi, P. Shanahan, and P. Russell. 2003. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell 12:747-759. [DOI] [PubMed] [Google Scholar]
- 20.Gotter, A. L. 2003. Tipin, a novel timeless-interacting protein, is developmentally co-expressed with timeless and disrupts its self-association. J. Mol. Biol. 331:167-176. [DOI] [PubMed] [Google Scholar]
- 21.Grewal, S. I., and A. J. Klar. 1997. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146:1221-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutz, H., and H. Schmidt. 1985. Switching genes in Schizosaccharomyces pombe. Curr. Genet. 9:325-331. [DOI] [PubMed] [Google Scholar]
- 23.Iino, Y., and M. Yamamoto. 1997. The Schizosaccharomyces pombe cdc6 gene encodes the catalytic subunit of DNA polymerase delta. Mol. Gen. Genet. 254:93-97. [DOI] [PubMed] [Google Scholar]
- 24.Karthikeyan, R., E. J. Vonarx, A. F. Straffon, M. Simon, G. Faye, and B. A. Kunz. 2000. Evidence from mutational specificity studies that yeast DNA polymerases delta and epsilon replicate different DNA strands at an intracellular replication fork. J. Mol. Biol. 299:405-419. [DOI] [PubMed] [Google Scholar]
- 25.Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1078-1083. [DOI] [PubMed] [Google Scholar]
- 26.Kaykov, A., A. M. Holmes, and B. Arcangioli. 2004. Formation, maintenance and consequences of the imprint at the mating-type locus in fission yeast. EMBO J. 23:930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kearsey, S. E., S. Montgomery, K. Labib, and K. Lindner. 2000. Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 19:1681-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, S. M., and J. A. Huberman. 2001. Regulation of replication timing in fission yeast. EMBO J. 20:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 30.Krawchuk, M. D., and W. P. Wahls. 1999. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 15:1419-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindsay, H. D., D. J. Griffiths, R. J. Edwards, P. U. Christensen, J. M. Murray, F. Osman, N. Walworth, and A. M. Carr. 1998. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12:382-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopes, M., C. Cotta-Ramusino, G. Liberi, and M. Foiani. 2003. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol. Cell 12:1499-1510. [DOI] [PubMed] [Google Scholar]
- 33.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 34.MacQueen, A. J., and A. M. Villeneuve. 2001. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 15:1674-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayer, M. L., I. Pot, M. Chang, H. Xu, V. Aneliunas, T. Kwok, R. Newitt, R. Aebersold, C. Boone, G. W. Brown, and P. Hieter. 2004. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell 15:1736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGlynn, P., and R. G. Lloyd. 2002. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 3:859-870. [DOI] [PubMed] [Google Scholar]
- 37.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 38.Muris, D. F., K. Vreeken, A. M. Carr, B. C. Broughton, A. R. Lehmann, P. H. Lohman, and A. Pastink. 1993. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 21:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muris, D. F., K. Vreeken, A. M. Carr, J. M. Murray, C. Smit, P. H. Lohman, and A. Pastink. 1996. Isolation of the Schizosaccharomyces pombe RAD54 homologue, rhp54+, a gene involved in the repair of radiation damage and replication fidelity. J. Cell Sci. 109:73-81. [DOI] [PubMed] [Google Scholar]
- 40.Nasmyth, K., and P. Nurse. 1981. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 182:119-124. [DOI] [PubMed] [Google Scholar]
- 41.Noguchi, E., C. Noguchi, L. L. Du, and P. Russell. 2003. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol. Cell. Biol. 23:7861-7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogawa, Y., T. Takahashi, and H. Masukata. 1999. Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol. 19:7228-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osborn, A. J., and S. J. Elledge. 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17:1755-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osborn, A. J., S. J. Elledge, and L. Zou. 2002. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12:509-516. [DOI] [PubMed] [Google Scholar]
- 45.Osman, F., J. Dixon, C. L. Doe, and M. C. Whitby. 2003. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12:761-774. [DOI] [PubMed] [Google Scholar]
- 46.Ostermann, K., A. Lorentz, and H. Schmidt. 1993. The fission yeast rad22 gene, having a function in mating-type switching and repair of DNA damages, encodes a protein homolog to Rad52 of Saccharomyces cerevisiae. Nucleic Acids Res. 21:5940-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paques, F., and J. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pavlov, Y. I., C. S. Newlon, and T. A. Kunkel. 2002. Yeast origins establish a strand bias for replicational mutagenesis. Mol. Cell 10:207-213. [DOI] [PubMed] [Google Scholar]
- 49.Rabitsch, K. P., A. Toth, M. Galova, A. Schleiffer, G. Schaffner, E. Aigner, C. Rupp, A. M. Penkner, A. C. Moreno-Borchart, M. Primig, R. E. Esposito, F. Klein, M. Knop, and K. Nasmyth. 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 11:1001-1009. [DOI] [PubMed] [Google Scholar]
- 50.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 51.Sanchez, J. A., S. M. Kim, and J. A. Huberman. 1998. Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp. Cell Res. 238:220-230. [DOI] [PubMed] [Google Scholar]
- 52.Schmidt, H., P. Kapitza, and H. Gutz. 1987. Switching genes in Schizosaccharomyces pombe: their influence on cell viability and recombination. Curr. Genet. 11:303-308. [DOI] [PubMed] [Google Scholar]
- 53.Segurado, M., M. Gomez, and F. Antequera. 2002. Increased recombination intermediates and homologous integration hot spots at DNA replication origins. Mol. Cell 10:907-916. [DOI] [PubMed] [Google Scholar]
- 54.Shcherbakova, P. V., and Y. I. Pavlov. 1996. 3′→5′ exonucleases of DNA polymerases epsilon and delta correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics 142:717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 56.Stewart, E., C. R. Chapman, F. Al-Khodairy, A. M. Carr, and T. Enoch. 1997. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16:2682-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanaka, K., and P. Russell. 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3:966-972. [DOI] [PubMed] [Google Scholar]
- 58.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 59.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 60.Vengrova, S., S. Codlin, and J. Z. Dalgaard. 2002. RTS1—an eukaryotic terminator of replication. Int. J. Biochem. Cell Biol. 34:1031-1034. [DOI] [PubMed] [Google Scholar]
- 61.Vengrova, S., and J. Z. Dalgaard. 2004. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev. 18:794-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Virgin, J. B., J. P. Bailey, F. Hasteh, J. Neville, A. Cole, and G. Tromp. 2001. Crossing over is rarely associated with mitotic intragenic recombination in Schizosaccharomyces pombe. Genetics 157:63-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 64.Warren, C. D., D. M. Eckley, M. S. Lee, J. S. Hanna, A. Hughes, B. Peyser, C. Jie, R. Irizarry, and F. A. Spencer. 2004. S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol. Biol. Cell 15:1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Washburn, M. P., D. Wolters, and J. R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242-247. [DOI] [PubMed] [Google Scholar]
- 66.Wellinger, R. E., P. Schar, and J. M. Sogo. 2003. Rad52-independent accumulation of joint circular minichromosomes during S phase in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:6363-6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woodgate, R. 1999. A plethora of lesion-replicating DNA polymerases. Genes Dev. 13:2191-2195. [DOI] [PubMed] [Google Scholar]
- 68.Yanow, S. K., Z. Lygerou, and P. Nurse. 2001. Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J. 20:4648-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yonemasu, R., S. J. McCready, J. M. Murray, F. Osman, M. Takao, K. Yamamoto, A. R. Lehmann, and A. Yasui. 1997. Characterization of the alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe. Nucleic Acids Res. 25:1553-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90:87-96. [DOI] [PubMed] [Google Scholar]