Abstract
Cohesin is a ring‐shaped protein complex that plays a crucial role in sister chromatid cohesion and gene expression. The dynamic association of cohesin with chromatin is essential for these functions. However, the exact nature of cohesin dynamics, particularly cohesin translocation, remains unclear. We evaluated the dynamics of individual cohesin molecules on DNA and found that the cohesin core complex possesses an intrinsic ability to traverse DNA in an adenosine triphosphatase (ATPase)‐dependent manner. Translocation ability is suppressed in the presence of Wapl‐Pds5 and Sororin; this suppression is alleviated by the acetylation of cohesin and the action of mitotic kinases. In Xenopus laevis egg extracts, cohesin is translocated on unreplicated DNA in an ATPase‐ and Smc3 acetylation‐dependent manner. Cohesin movement changes from bidirectional to unidirectional when cohesin faces DNA replication; otherwise, it is incorporated into replicating DNA without being translocated or is dissociated from replicating DNA. This study provides insight into the nature of individual cohesin dynamics and the mechanisms by which cohesin achieves cohesion in different chromatin contexts.
Keywords: chromosome segregation, cohesin, DNA replication, post‐translational modification, single‐molecule TIRF microscopy
Subject Categories: Cell Cycle; Chromatin, Epigenetics, Genomics & Functional Genomics; DNA Replication, Repair & Recombination
Introduction
Cohesin, an evolutionarily conserved structural maintenance of chromosomes (SMC) family member, plays crucial roles in sister chromatid cohesion, chromatin structure organization, and gene expression in eukaryotes. Cohesin dysfunction in humans has been implicated in hereditary diseases, such as Cornelia de Lange syndrome (Liu & Krantz, 2009; Deardorff et al, 2012), Roberts syndrome (Vega et al, 2005), and cancer (Barber et al, 2008; Solomon et al, 2011; Welch et al, 2012; Kon et al, 2013). However, the pathogenicity mechanisms of these disorders remain unclear.
Since the discovery of the cohesin complex (Guacci et al, 1997; Michaelis et al, 1997; Losada et al, 1998), which consists of four core subunits, Smc1, Smc3, Scc1, and stromal antigen (SA/STAG) (Onn et al, 2008; Nasmyth & Haering, 2009), accumulating biochemical evidence has suggested that ring‐shaped cohesin complexes hold sister chromatids together by entrapping two DNA molecules (Anderson et al, 2002; Haering et al, 2002, 2008; Arumugam et al, 2003). Cohesin loading onto DNA depends on the cohesin loader complex Scc2‐Scc4 and the hydrolysis of adenosine triphosphate (ATP) by Smc1 and Smc3 (Ciosk et al, 2000; Arumugam et al, 2003; Weitzer et al, 2003). Several cohesin‐binding proteins and modification factors regulate sister chromatid cohesion. For example, to establish cohesion, the Smc3 subunit must be acetylated by the Eco1/Esco1/2 acetyltransferases (Ben‐Shahar et al, 2008; Unal et al, 2008; Zhang et al, 2008; Rowland et al, 2009), and Sororin, a vertebrate cohesin‐binding protein, is then recruited to chromatin‐bound cohesin following acetylation of Smc3 (Lafont et al, 2010; Nishiyama et al, 2010). The cohesin‐associated Wapl‐Pds5 heterodimer is required for cohesin removal from chromosome arms during mitotic prophase (Gandhi et al, 2006; Kueng et al, 2006; Shintomi & Hirano, 2009; Tedeschi et al, 2013). Cohesin dissociation also requires phosphorylation of the cohesin SA subunit (Losada et al, 2002; Sumara et al, 2002) and Sororin (Nishiyama et al, 2013). Wapl‐Pds5 and phosphorylation presumably act to open the “exit gate” (Smc3‐Scc1 interface) (Chan et al, 2012; Buheitel & Stemmann, 2013; Eichinger et al, 2013; Gligoris et al, 2014; Huis in ‘t Veld et al, 2014).
A previous study demonstrated that the depletion of Wapl stabilizes cohesin on DNA, resulting in the over‐cohesion of interphase chromatin, chromosome mis‐segregation, and altered gene expression (Tedeschi et al, 2013), suggesting that the dynamic interaction of cohesin with DNA is essential not only for faithful chromosome segregation, but also for proper chromosome architecture and gene expression. Previous studies have suggested two modes of cohesin dynamics: cohesin–chromatin “association/dissociation” and cohesin “translocation” along DNA. The former has been studied by analyzing the chromatin residence times of cohesin (Gerlich et al, 2006; Gause et al, 2010; Chan et al, 2012; Ladurner et al, 2014). However, the latter “translocation” mode is not well‐understood, although it is thought to occur in budding yeast (Lengronne et al, 2004; Hu et al, 2011). Recent single‐molecule studies revealed that cohesin is translocated on DNA by one‐dimensional diffusion and cohesin can be pushed by DNA motor proteins (Davidson et al, 2016; Stigler et al, 2016). It has also been suggested that cohesin slides along budding yeast chromatin via the pushing force of the transcription machinery (Ocampo‐Hafalla et al, 2016). However, the regulatory mechanisms of cohesin translocation are not understood.
Single‐molecule techniques such as optical/magnetic tweezers, atomic force microscopy, tethered particle microscopy, and fluorescence spectroscopy have been developed. Among them, the fluorescence imaging‐based flow‐stretching method enables high‐throughput analysis of single‐molecule dynamics on DNA. This technique has been used to show that Rad51 and lac repressor (LacI), as well as cohesin, diffuse on DNA (Graneli et al, 2006; Wang et al, 2006; Davidson et al, 2016; Stigler et al, 2016), and hOgg1, a DNA repair protein, slides along the DNA double helix (Blainey et al, 2006). Furthermore, recent studies showed that stretched DNA could be replicated by combining the flow‐stretching system with cell extracts (Yardimci et al, 2010, 2012b; Loveland et al, 2012). To determine the molecular mechanisms of cohesin translocation along DNA, we applied flow‐stretching method‐based single‐molecule techniques. After reconstituting the cohesin complex with its binding and modifying proteins and the loading complex, we analyzed the dynamics of single cohesin molecules on DNA or chromatin. These experiments provide novel insights into the regulatory mechanisms of cohesin translocation. The cohesin complex randomly translocates along DNA in an ATPase‐dependent manner. This translocation ability is attenuated by Wapl‐Pds5 and attenuation is alleviated by Smc3 acetylation, whereas Sororin further suppresses the translocation ability. Cohesin movement along the chromatin in Xenopus egg extracts depends on cohesin acetylation; translocation ability is influenced by DNA replication. Based on these results, we discuss the mechanisms of cohesin translocation along DNA and chromatin, as regulated by cohesin‐associated proteins, cell cycle‐dependent post‐translational modifications, and DNA replication.
Results
Cohesin translocates along DNA in an ATPase‐dependent manner
To investigate cohesin dynamics, we first examined single cohesin particles under physiological conditions. First, 48.5 kb of linear λ phage DNA (λ DNA) was biotinylated at both ends and tethered to a streptavidin‐coated coverslip that was assembled in a microfluidic flow cell attached to a syringe pump (Yardimci et al, 2012a) and then observed under a total internal reflection fluorescence (TIRF) microscope (Appendix Fig S1A). A human cohesin tetramer harboring Halo‐tagged Scc1 was expressed and purified from baculovirus‐infected insect cells and Scc1‐Halo was then covalently labeled with a fluorescent dye (Fig 1A). Based on the particle intensities and photobleaching results, particles with intensities similar to the peak intensity (0.5–0.8 × 103 EMCCD count; Appendix Fig S1B) were bleached in one step by continuous excitation (Appendix Fig S1C), suggesting that these particles were single molecules. After tethering the DNA at both ends to the coverslip, labeled cohesin was introduced to the flow cell in the presence of ATP. The cohesin tetramer alone could specifically bind to DNA under low‐salt conditions (30 mM NaCl, Fig EV1A). However, most cohesin particles were dissociated under high‐salt conditions (Fig EV1B), which is consistent with previous observations in vitro (Murayama & Uhlmann, 2014), indicating that these cohesin complexes were not topologically bound to DNA. In order to observe only cohesin particles that were topologically bound to DNA, cohesin was loaded onto DNA in the presence of the Scc2‐Scc4 complex (Fig EV1C and D) and washed with a high‐salt buffer. As expected, in the presence of Scc2‐Scc4, a significant number of cohesin particles remained on the DNA after washing with high‐salt buffer (Fig EV1B). If these DNA‐bound cohesins are topologically bound to DNA, cleavage of the cohesin ring should result in the dissociation of cohesin from DNA (Uhlmann et al, 1999, 2000). To test this, we utilized a cohesin complex in which the Scc1 subunit harbored tobacco etch virus (TEV) protease cleavage recognition sites (Fig 1A) to allow the artificial opening of cohesin rings by this enzyme. We found that most Scc1 was cleaved within 45 min of TEV protease treatment (Appendix Fig S1D). TEV‐cleavable cohesin was loaded onto DNA in the presence of Scc2‐Scc4, washed in high‐salt buffer, and then treated with TEV protease. TEV treatment significantly decreased the amount of cohesin on DNA (Fig 1B), indicating that cohesin was topologically bound to DNA.
Figure 1. Cohesin translocates along DNA in an ATPase‐dependent manner.
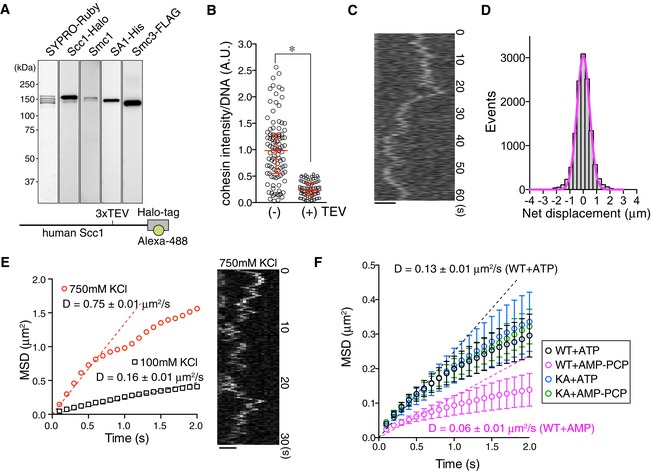
- Purification of the Halo‐tagged cohesin tetramer. Human cohesin tetramer tagged by Halo was purified from insect cells and Alexa 488 dye was conjugated to Halo‐tag (cohesinHalo488).
- CohesinHalo488 loaded onto DNA in the presence of Scc2‐Scc4 was incubated in the buffer with or without TEV protease. After high‐salt washing, cohesinHalo488 intensities on DNA were measured. Red bars denote the median, lower, and upper quartile values (n ≥ 98; *P < 0.0001, two‐tailed Mann–Whitney U‐test).
- Kymograph of a single DNA‐interacting cohesinHalo488 particle. CohesinHalo488 is loaded onto DNA in the presence of Scc2‐Scc4 and, after high‐salt washing, single cohesin particles were observed in the presence of ATP and 100 mM KCl. Scale bar, 2 μm.
- Histogram of the net displacement (x final − x initial) of cohesin particles treated as in (C). The magenta curve represents the Gaussian fit.
- CohesinHalo488 was loaded and washed in high‐salt as in (C), and the cohesin particles were observed in the presence of ATP and 100 mM or 750 mM KCl. Kymograph of DNA‐associated cohesin particles under high‐salt conditions (750 mM KCl, right) and the mean square displacement (MSD) vs. time (left) are shown. Dotted lines are regression lines used to estimate the diffusion coefficient (D). Scale bar, 2 μm.
- MSD vs. time for DNA‐bound cohesinWT‐Halo488 or cohesinKA‐Halo488 particles. CohesinWT‐Halo488 or cohesinKA‐Halo488 was loaded onto DNA in the presence of Scc2‐4 and ATP. After high‐salt washing, the cohesin particles on DNA were observed in the presence of 1 mM AMP‐PCP or ATP and 100 mM KCl. D indicates the diffusion coefficient (n = 45, mean ± s.e.m.).
Figure EV1. Proper loading of cohesin by Scc2‐Scc4 is required for translocation ability (related to Fig 1).
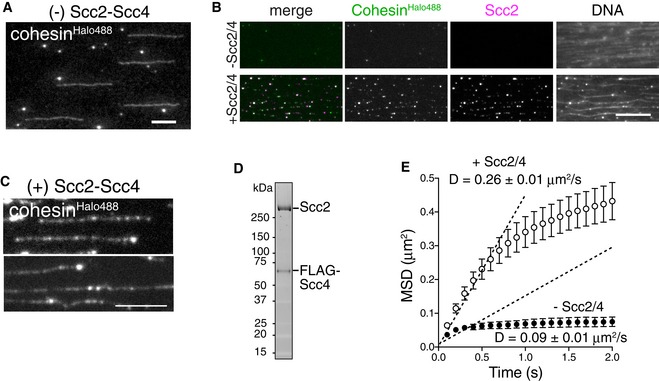
- CohesinHalo488 is specifically bound to DNA in the absence of Scc2‐Scc4. CohesinHalo488 (3.25 nM) was directly loaded onto DNA and DNA‐bound cohesinHalo488 was observed. CohesinHalo488 signals were detected all along DNA. Scale bar, 10 μm.
- CohesinHalo488 was loaded onto DNA in the presence or absence of Scc2‐Scc4 and washed in high‐salt buffer. Scc2 was immunostained by anti‐Scc2 antibody. DNA was counterstained with SYTOX. Scale bar, 10 μm.
- CohesinHalo488 specifically binds to DNA in the presence of Scc2‐Scc4. CohesinHalo488 (3.25 nM) was loaded onto DNA after Scc2‐Scc4 bound to DNA, and cohesinHalo488 was observed. Note that cohesinHalo488 signals were detected all along the DNA. Scale bar, 5 μm.
- SYPRO Ruby staining of the purified Scc2‐Scc4 complex after separation by SDS–PAGE.
- CohesinHalo488 was loaded onto DNA as in (B). After high‐salt wash, residual cohesin particles on DNA were observed in the presence of ATP and 100 mM KCl. MSD vs. time is shown. D indicates the diffusion coefficient (n = 45, mean ± s.e.m.).
Based on time‐lapse images, cohesin particles moved along λ DNA in the presence of ATP; the movement became more evident under higher salt conditions (≥ 100 mM KCl) (Fig 1C). A histogram of the net displacement of cohesin particles was fitted by a Gaussian curve (Fig 1D), suggesting that the movement was consistent with diffusion, rather than an active or a directional process. By plotting the mean square displacement (MSD) vs. time (Δt), we estimated the diffusion coefficient (D) of cohesin on DNA and found that it increased depending on the salt concentration and that cohesins were translocated along DNA even in the presence of 750 mM KCl (Fig 1E). As cohesin cannot be loaded onto DNA even in the presence of 100 mM NaCl (Appendix Fig S1E) (Murayama & Uhlmann, 2014), this result also indicates that cohesin was topologically bound to DNA, which is consistent with a previous report (Davidson et al, 2016; Stigler et al, 2016). Because the cohesin tetramer by itself may associate with DNA in the absence of Scc2‐Scc4 (Fig EV1A), we next evaluated whether cohesin loaded onto DNA independently of Scc2‐Scc4 could be translocated along DNA. When cohesin was directly loaded onto DNA, the DNA‐bound cohesin was much less mobile than that loaded by Scc2‐Scc4 (approximately threefold lower based on the diffusion coefficient: Fig EV1E), suggesting that the translocation ability of cohesin requires its proper loading via the Scc2‐Scc4 complex. To test whether this movement is non‐specific and is thus displayed by all DNA‐bound proteins, GFP‐tagged Tet repressor protein (TetR‐GFP) was mixed with a linear DNA fragment containing tet operator sequences (tetO), that is, TetR‐specific target sequences, at both ends. As expected, TetR‐GFP remained at both ends of tetO‐DNA (Appendix Fig S1F), demonstrating that proteins bound to specific sequences do not exhibit the random movement displayed by cohesin. We also tested the effect of aggregation of cohesin particles on translocation ability as cohesin frequently forms aggregates on DNA. The diffusion coefficients indicated that aggregated particles lost their mobility on DNA compared to non‐aggregated single particles (Appendix Fig S1G). Therefore, we selected particles with intensities corresponding to that of single molecule to avoid aggregation in subsequent experiments.
Next, we tested the requirement of cohesin ATPase activity for this movement. To rule out the possibility of secondary effects resulting from mis‐regulation in the cohesin loading step, cohesin was loaded onto DNA in the presence of Scc2‐Scc4 and ATP and then washed with high‐salt buffer. Cohesin movement was then observed in the presence of ATP or non‐hydrolyzable ATP analogues (AMP‐PCP). Although AMP‐PCP did not affect the amount of cohesin bound to DNA (Appendix Fig S1H), it significantly reduced the diffusion coefficient (Fig 1F). The translocation ability of cohesinKA, a cohesin mutant containing an Smc3 subunit harboring an ATP‐binding abolishing mutation (Smc3‐K38A) (Arumugam et al, 2006; Ladurner et al, 2014), was insensitive to AMP‐PCP (Fig 1F). Because the cohesin tetramer could be randomly translocated along DNA even in the absence of ATP (Davidson et al, 2016; Stigler et al, 2016), the physical disengagement of the Smc1/3 head domain after ATP hydrolysis may facilitate diffusion.
Cohesin acetylation antagonizes Wapl‐Pds5‐dependent attenuation of translocation ability
We next evaluated whether cohesin‐associated proteins and post‐translational modifications affect the translocation ability of cohesin. The Wapl‐Pds5 heterodimer is required for the dynamic interaction of cohesin to DNA in interphase as well as mitosis (Gandhi et al, 2006; Kueng et al, 2006; Shintomi & Hirano, 2009; Tedeschi et al, 2013). We first tested the ability of Wapl‐Pds5 to regulate translocation. The Wapl‐Pds5 heterodimer was expressed and purified as a stoichiometric complex from baculovirus‐infected insect cells (Appendix Fig S2A). When the Wapl‐Pds5 heterodimer was introduced into the flow cell after cohesin was loaded onto DNA by Scc2‐Scc4, Wapl‐Pds5 specifically bound to DNA‐bound cohesin (Appendix Fig S2B), which promoted cohesin dissociation from DNA after washing in high‐salt buffer (Appendix Fig S2C), confirming that Wapl‐Pds5 was functional in our flow cell system. We then examined whether Wapl‐Pds5 affects cohesin movement along DNA. Cohesin was topologically loaded by Scc2‐Scc4 followed by high‐salt washing and Wapl‐Pds5 was bound to the cohesin. When these cohesin particles were analyzed in the presence of ATP, we unexpectedly found that Wapl‐Pds5 significantly attenuated the translocation (Fig 2A and B). AMP‐PCP further decreased the translocation ability in the presence of Wapl‐Pds5 (Appendix Fig S2D), suggesting that Wapl‐Pds5 and ATPase activity independently affect translocation.
Figure 2. Acetylation of cohesin antagonizes Wapl‐Pds5‐dependent suppression of cohesin translocation along DNA.
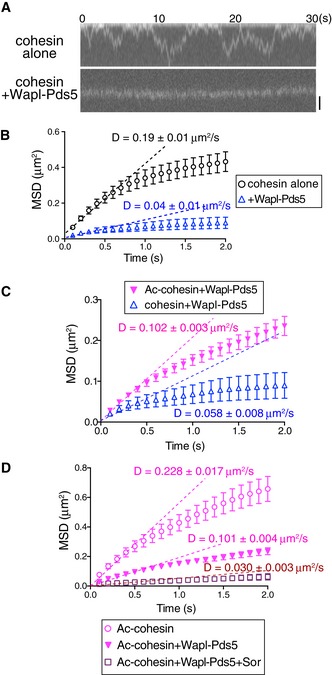
- Kymograph of cohesinHalo488 in the presence or absence of Wapl‐Pds5. CohesinHalo488 was loaded onto DNA in the presence of Scc2‐Scc4 and washed in high‐salt buffer. DNA‐bound cohesin was further incubated with or without Wapl‐Pds5, and then cohesinHalo488 particles were observed in the presence of ATP and 100 mM KCl. Scale bar, 1 μm.
- MSD vs. time of cohesin particles in (A) is shown. D indicates the diffusion coefficient (n = 45, mean ± s.e.m.).
- Acetylated or unacetylated cohesinHalo488 was loaded onto DNA in the presence of Scc2‐Scc4 and washed in high‐salt buffer. DNA‐bound cohesin was further incubated with Wapl‐Pds5, and then cohesinHalo488 particles were observed in the presence of ATP and 100 mM KCl. MSD vs. time is shown. D indicates the diffusion coefficient (n = 45, mean ± s.e.m.).
- Acetylated cohesinHalo488 was treated as in (C), the DNA‐bound cohesin was further incubated with buffer, Wapl‐Pds5, or Wapl‐Pds5 plus Sororin, and then cohesinHalo488 particles were observed in the presence of ATP and 100 mM KCl. MSD vs. time is shown. D indicates the diffusion coefficient (n = 45, mean ± s.e.m.).
Wapl and/or Pds5 exhibit anti‐establishment activity against Eco1/Esco1/2‐dependent cohesion (Gandhi et al, 2006; Ben‐Shahar et al, 2008; Unal et al, 2008; Rowland et al, 2009). Thus, we tested whether Smc3 acetylation affects the affinity of cohesin to DNA or cohesin translocation. In eukaryotes, the acetylation of two lysine residues in the Smc3 subunit, K105/K106 (human) and K112/K113 (budding yeast), by Eco1/Esco1/2 acetyltransferases is essential for the establishment of cohesion (Ben‐Shahar et al, 2008; Unal et al, 2008; Zhang et al, 2008; Rowland et al, 2009). Because of the low efficiency of the in vitro acetylation reaction, we purified the acetylated cohesin tetramer from insect cells co‐expressing human Esco1. Acetylated cohesin was loaded onto DNA in the presence of Scc2‐Scc4, washed with high‐salt buffer, and incubated with Wapl‐Pds5. Acetylation of cohesin was confirmed by immunofluorescence microscopy using an acetylated Smc3‐specific antibody; we estimated that ~50% of cohesin particles were acetylated (Fig EV2A). Intriguingly, the diffusion coefficient of acetylated cohesin was significantly higher than that of unacetylated cohesin, even in the presence of Wapl‐Pds5 (Fig 2C). This result suggests the diffusion coefficient increased because Wapl‐Pds5 is dissociated from acetylated cohesin. However, immunofluorescence microscopy revealed that Wapl was not dissociated from cohesin when cohesin was acetylated (Fig EV2B). Furthermore, acetylation itself facilitated translocation in the absence of Wapl‐Pds5 (Fig EV2C). Thus, acetylation directly affects the translocation ability of the cohesin core complex, irrespective of the presence of Wapl‐Pds5.
Figure EV2. Acetylation of Smc3 facilitates cohesin translocation (related to Fig 2).
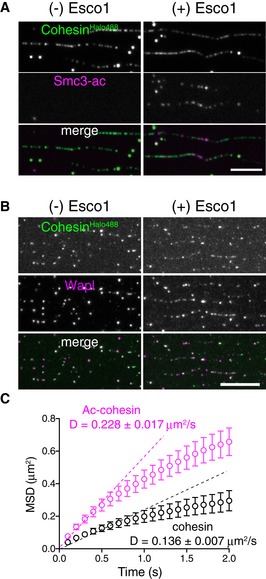
- Topologically loaded cohesinHalo488 (−Esco1) or ac‐cohesinHalo488 (+Esco1) was washed in high‐salt buffer. Acetylated Smc3 (Smc3‐ac) was detected by immunostaining. Scale bar, 5 μm.
- Topologically loaded cohesinHalo488 (−Esco1) or ac‐cohesinHalo488 (+Esco1) was washed in high‐salt buffer and treated with Wapl‐Pds5. Wapl was detected by immunostaining. Scale bar, 10 μm.
- CohesinHalo488 was treated as in (A) and cohesinHalo488 (cohesin) or acetylated cohesinHalo488 (Ac‐cohesin) particles were observed. MSD vs. time is shown. D indicates the diffusion coefficient (n = 45, mean ± s.e.m.).
We next assessed the effect of Sororin on cohesin translocation. Sororin is a cohesion establishment factor that associates with acetylated cohesin via Pds5 (Rankin et al, 2005; Schmitz et al, 2007; Nishiyama et al, 2010). Because Sororin associates with Pds5 and antagonizes its cohesin removal activity (Nishiyama et al, 2010), Sororin may also affect cohesin movement. His‐tagged recombinant human Sororin was expressed in and purified from Escherichia coli (Appendix Fig S2E) and incubated with acetylated cohesin in the presence of Wapl‐Pds5. We confirmed that Sororin particles were specifically associated with cohesin on DNA (Appendix Fig S2F) and stabilized cohesin on DNA in the presence of Wapl‐Pds5 (Appendix Fig S2G). Sororin further suppressed Wapl‐Pds5‐bound cohesin translocation (Fig 2D). These results indicate that the two cohesion establishment factors, that is, acetylation of Smc3 and Sororin, have different effects on cohesin translocation ability.
Plk1 and Aurora B facilitates translocation
Given the Sororin‐dependent stabilization of cohesin, we asked whether stabilized cohesin is relieved during mitosis. In living cells, mitotic phosphorylation of the cohesin SA/STAG subunit by polo‐like kinase 1 (Plk1) and cyclin‐dependent kinase 1 (CDK1)‐ and Aurora B‐dependent phosphorylation of Sororin are required for the mitotic dissociation of cohesin from chromosomes (Losada et al, 2002; Sumara et al, 2002; Hauf et al, 2005; Nishiyama et al, 2013). We therefore evaluated whether the mitotic kinases are responsible for the translocation of cohesin. The acetylated cohesin tetramer was loaded onto DNA in the presence of Scc2‐Scc4, incubated with Wapl‐Pds5 and Sororin after a high‐salt wash, and then treated with the mitotic kinases Plk1, Aurora B, or CDK1. Among these kinases, Plk1 and Aurora B significantly increased the diffusion coefficient of cohesin (Fig 3). When the cohesin complex was treated with recombinant Plk1 protein, phosphorylation was detected by the incorporation of [γ‐32P] ATP and mass spectrometry (Appendix Fig S3A and B). Mass spectrometry analysis indicated that Plk1 phosphorylated SA1 at multiple sites located in similar regions as the previously reported Plk1‐dependent mitotic phosphorylation sites of SA2 (Appendix Fig S3B) (Olsen et al, 2010; Hegemann et al, 2011). Cohesin was topologically loaded, treated with Plk1, and evaluated for the movement of cohesin particles. Plk1‐dependent phosphorylation did not affect the amount of DNA‐bound cohesin molecules over a range of salt concentrations (Appendix Fig S3C). DNA‐bound cohesin became more dynamic in the presence of Plk1, which was suppressed by BI4834, a Plk1 inhibitor (Fig 3A). However, immunofluorescence microscopy revealed that Plk1, as well as CDK1, significantly reduced the amount of Sororin, but not Wapl‐Pds5 on DNA (Fig 3B, and Appendix Fig S3D). Therefore, Plk1‐dependent promotion of translocation is presumably caused by the removal of Sororin from the cohesin–Wapl–Pds5 complex. Because phosphorylation of Sororin by Plk1 is not sufficient for removing Sororin from Wapl‐Pds5 (Nishiyama et al, 2013), the dissociation of Sororin observed in this study may be caused by the phosphorylation of cohesin itself or Wapl‐Pds5. In contrast, Aurora B reduced the levels of neither Sororin nor Wapl‐Pds5 on cohesin (Fig 3B and Appendix Fig S3D), even though Sororin was phosphorylated (Appendix Fig S3E). We previously showed that phosphorylation of Sororin by Aurora B inhibits Sororin binding to Wapl‐Pds5 in an in vitro binding assay (Nishiyama et al, 2013). This was not observed in our flow cell system presumably because (i) the flow cell was washed under mild conditions without any detergents or (ii) Aurora B‐dependent phosphorylations of the cohesin core complex or Wapl‐Pds5 (Hegemann et al, 2011) affect the affinity between Sororin and Wapl‐Pds5. Nevertheless, Aurora B significantly increased the diffusion coefficient of cohesin (Fig 3C), indicating that the Aurora B‐dependent phosphorylation of cohesin, Wapl‐Pds5, or Sororin counteracts the affinity between cohesin and DNA, presumably by inducing conformational changes or imparting a negative charge at the phosphorylation site to facilitate translocation. Interestingly, we found that Plk1 suppressed cohesin translocation in the absence of Wapl‐Pds5 and Sororin (Appendix Fig S3F), indicating that the effect of mitotic kinases on cohesin translocation is altered in the presence of Wapl‐Pds5 and Sororin.
Figure 3. Plk1‐ and Aurora B‐dependent phosphorylation facilitates cohesin translocation.
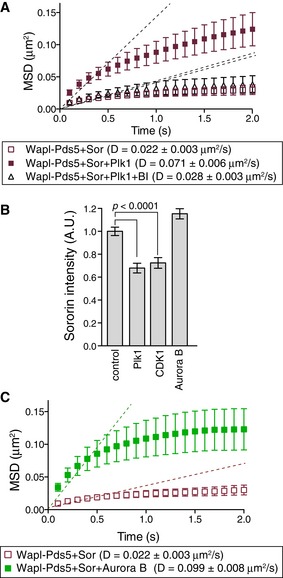
- Acetylated cohesinHalo488 was loaded onto DNA in the presence of Scc2‐Scc4 and washed in high‐salt buffer. DNA‐bound cohesin was further incubated with Wapl‐Pds5 plus Sororin and then treated with buffer, Plk1, or Plk1 plus BI4834. CohesinHalo488 particles were observed in the presence of ATP and 100 mM KCl. MSD vs. time is shown. D indicates the diffusion coefficient (n = 45, mean ± s.e.m.).
- Acetylated cohesinHalo488 was treated as in (A), or treated with CDK1 or Aurora B rather than Plk1. Sororin intensity on DNA was measured after immunofluorescence microscopy with anti‐Sororin antibody (n > 100, P < 0.0001, two‐tailed Mann–Whitney U‐test).
- Acetylated cohesinHalo488 was treated as in (A) but treated with Aurora B rather than Plk1. CohesinHalo488 particles were observed in the presence of ATP and 100 mM KCl. MSD vs. time is shown. D indicates the diffusion coefficient (n = 45, mean ± s.e.m.).
Cohesin preferentially accumulates in the nucleosome‐poor region of chromatin in Xenopus egg extract
To investigate the properties of single cohesin molecules under conditions mimicking those in vivo, we observed the dynamics of cohesin on DNA in interphase Xenopus egg extracts. In the egg extracts, cohesin loading onto the chromatin was dependent on the formation of a pre‐replication complex (pre‐RC) consisting of four factors: ORC, Cdc6, Cdt1, and Mcm2–7. The Cdt1 inhibitor geminin inhibits this loading (Gillespie & Hirano, 2004; Takahashi et al, 2004). Consistent with these characteristics, endogenous Xenopus cohesin was bound to tethered λ DNA in the high‐speed supernatant (HSS) of Xenopus egg extracts, whereas both the formation of the pre‐RC and loading of cohesin were abolished in the presence of geminin (Fig 4A and B). We next loaded fluorescently labeled human cohesin complexes onto DNA in HSS in the presence of the purified human Scc2‐Scc4 heterodimer. As expected, Scc2‐Scc4‐dependent cohesin loading was diminished with the ATP‐binding‐deficient cohesinKA mutant (Appendix Fig S4A and B). We also tested whether nucleosome density affected the cohesin–DNA association under these conditions. A previous study revealed that, in budding yeast, Scc2‐Scc4 recruits cohesins to a nucleosome‐free chromatin region, cooperating with the RSC chromatin remodeling complex (Lopez‐Serra et al, 2014). In contrast to budding yeast, where Scc2‐Scc4 recruitment is correlated with transcription (Lopez‐Serra et al, 2014), little transcriptional activity was observed in early Xenopus embryos and was nearly entirely absent in the HSS, where Scc2‐Scc4 recruitment depended on pre‐RC formation. Therefore, cohesin loading machinery in the two systems may differ. Taking advantage of the HSS system, where small circular DNA can form nucleosomes (Laskey et al, 1977), we examined cohesin loading onto chromatin. Nucleosome formation on loosely stretched DNA was initiated in the HSS supplemented with geminin, which was added to inhibit cohesin loading before nucleosome formation. Fluorescently labeled cohesin pre‐incubated with Scc2‐Scc4 was then added, and the mixture was introduced into the flow cell. Nucleosomes were detected by immunostaining of histone H3 and H2A.X‐F, an embryonic histone variant. Histone H3 signals were frequently co‐localized with H2A.X‐F, and a lack of co‐localization in some cases was presumably attributed to other H2A variants (Appendix Fig S4C), suggesting that most of the detected particles formed nucleosomes. When the relative positions of the nucleosomes and cohesin molecules were compared, we found that 38% of the cohesin particles co‐localized with nucleosomes, while 62% did not. Most non‐co‐localizing molecules were adjacent to nucleosomes (≤ 1 μm distance; Fig 4C and D). Remarkably, when we assayed nucleosome‐rich chromatin, the number of chromatin‐bound cohesin molecules was significantly reduced compared with that observed for nucleosome‐poor regions (Fig 4C and E). Most of those cohesin molecules resided in gaps between nucleosome‐rich regions (Fig 4C), suggesting that in our replication‐dependent system, cohesin preferentially accumulated in nucleosome‐poor regions on chromatin, as observed in budding yeast. If pre‐RC itself forms in a nucleosome‐poor region, this could explain the cohesin localization. However, as shown in Fig 4F, pre‐RC formation (i.e., localization of Mcm2) did not exhibit a preference for nucleosome‐poor region, suggesting that cohesin rather than pre‐RC is preferentially localized in nucleosome‐poor regions.
Figure 4. Cohesin preferentially accumulates in nucleosome‐poor regions on chromatin in Xenopus egg extracts.
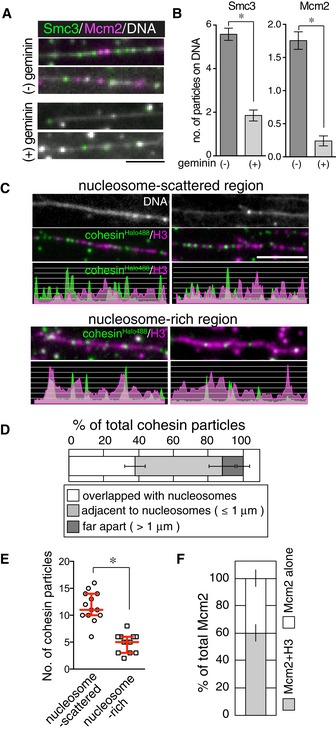
- Stretched DNA was incubated in the HSS in the presence or absence of geminin. After washing the HSS, endogenous Smc3 (green) and Mcm2 (magenta) were detected by immunofluorescence staining. DNA was counterstained with SYTOX (gray). Scale bar, 2 μm.
- Numbers of Smc3 and Mcm2 signals were counted on 10 μm DNA shown in (A). (22 DNAs per condition, n = 3, mean ± s.e.m., *P < 0.0001, two‐tailed Mann–Whitney U‐test).
- Chromatin formed in the HSS in the presence of geminin, and cohesinHalo488 (green) was loaded onto the chromatin with Scc2‐Scc4. Chromatin was detected by immunostaining of histone H3 (magenta). DNA was counterstained with SYTOX (gray). The distributions of cohesin particles and histone H3 are shown as line scan plots (bottom). Scale bar, 5 μm.
- Cohesin particles observed in (C) were categorized as indicated (> 37 particles per condition, n = 6, mean ± s.e.m.).
- Cohesin particle numbers for each condition. Red bars denote the median, lower, and upper quartile values (n ≥ 12, *P < 0.0004, unpaired t‐test).
- Co‐localization of Mcm2 with Smc3 was quantified (50 Mcm2 particles per condition, n = 3, mean ± s.e.m.)
Cohesin acetylation is required for the dynamic translocation of cohesin along unreplicated chromatin in Xenopus egg extracts
Next, we examined the cohesin translocation ability in Xenopus egg extracts. Immunofluorescence microscopy analyses revealed that Mcm helicases did not always co‐localize with cohesins (Figs 4 and 5A), despite the fact that cohesin loading requires pre‐RC formation. This indicates that cohesins and Mcm helicases are uncoupled after loading. Indeed, we observed that the fluorescently labeled Xenopus cohesin complex (xCohesinHaloTMR, Appendix Fig S5A) moved extensively along DNA in the HSS (Fig 5B, HSS). Consistent with recent in vitro observations (Davidson et al, 2016; Stigler et al, 2016), motion was lagging around the nucleosome‐rich region (Fig 5C), indicating that nucleosomes act as semipenetrable barriers under physiological conditions. When cohesin was pre‐loaded onto DNA in the HSS and washed with HSS supplemented with AMP‐PNP, a non‐hydrolyzable ATP analogue, we observed that cohesin particles transiently associated with DNA with a maximum residence time of 9 s (Fig 5B, HSSAMP‐PNP “transient”). This result is consistent with the fact that cohesin requires ATPase activity for stable loading onto DNA (Appendix Fig S4A and B) (Arumugam et al, 2003; Weitzer et al, 2003; Lopez‐Serra et al, 2014). In addition to the transient fraction, we frequently observed immobile cohesin particles in the presence of AMP‐PNP (Fig 5B, HSSAMP‐PNP “stable”). Considering that ATPase activity is required for cohesin acetylation (Ladurner et al, 2014) and that acetylation facilitates cohesin translocation (Figs 2C and EV2C), the stable association of cohesin in the presence of AMP‐PNP may be due to inefficient acetylation of cohesin. However, we did not detect a significant decrease in cohesin acetylation in the presence of AMP‐PNP compared to ATP (Appendix Fig S5B). This may be because: (i) ATPase activity does not affect cohesin acetylation in Xenopus egg extracts or (ii) acetylation is less stable on Xenopus chromatin because cohesin is translocated from its loading sites (pre‐RC), where XEco2, a major acetyltransferase responsible for Smc3 acetylation and cohesion establishment in Xenopus egg extracts, is loaded onto chromatin (Higashi et al, 2012).
Figure 5. XEco2 is required for the dynamic translocation of cohesin along unreplicated DNA in Xenopus egg extracts.
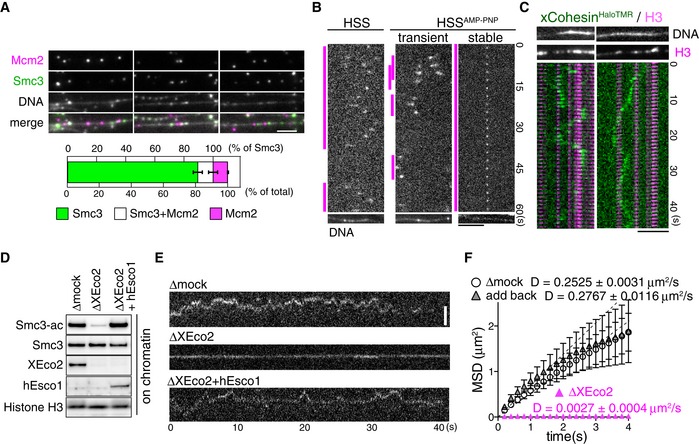
- Stretched DNA was incubated in the HSS. Endogenous Smc3 (green) and Mcm2 (magenta) were detected by immunofluorescence staining. DNA was counterstained with SYTOX (gray). Co‐localization of Smc3 and Mcm2 was quantified (bottom, n = 3, mean ± s.e.m.). Scale bar, 2 μm.
- Fluorescently labeled Xenopus cohesins (xCohesinHaloTMR) were loaded onto DNA in the HSS, and DNA‐bound cohesin particles were observed in the HSS with or without 5 mM AMP‐PNP supplementation. On the kymograph, the magenta lines indicate the duration of DNA binding of each cohesin particle. DNA was counterstained with SYTOX. Scale bar, 5 μm.
- Kymographs of DNA‐bound xCohesinHaloTMR (green) in HSS. After time‐lapse imaging of xCohesinHaloTMR, nucleosomes were immunostained by anti‐H3 antibody (magenta). Results of H3 immunostaining were merged with the kymograph. DNA was counterstained with SYTOX. Scale bar, 5 μm.
- HSS was subjected to mock or XEco2 immunodepletion and human Esco1‐His (hEsco1) was added back to XEco2‐depleted HSS. Sperm chromatin was incubated in these extracts and chromatin‐bound proteins were analyzed by immunoblotting.
- Kymographs of DNA‐bound xCohesinHaloTMR in mock‐, XEco2‐depleted, or hEsco1‐added XEco2‐depleted HSS. Scale bar, 2 μm.
- MSD vs. time of cohesin particle in the experiment in (E). D indicates the diffusion coefficient (n = 15, mean ± s.e.m.).
In order to directly test the effect of cohesin acetylation, we examined translocation ability in the absence of XEco2. After immunodepletion of endogenous XEco2 from HSS using anti‐XEco2 antibody beads (Fig 5D), minimal acetylated cohesin on chromatin was detected in XEco2‐depleted HSS (Fig 5D and Appendix Fig S5C). In mock‐depleted extracts, cohesin translocation was observed, as in untreated HSS (Fig 5E and F, Δmock). However, translocation was significantly diminished in XEco2‐depleted HSS (Fig 5E and F, ΔXEco2). The addition of purified human‐Esco1‐His protein (Appendix Fig S5D) to XEco2‐depleted HSS restored cohesin acetylation on chromatin to the same level as in mock‐depleted HSS (Fig 5D) and translocation ability was restored (Fig 5E and F, add back). Thus, acetylation is a prerequisite for sustaining the translocation ability of cohesin on unreplicated chromatin. Because we found that endogenous Wapl was bound to cohesin in both mock‐ and XEco2‐depleted HSS (Appendix Fig S5C), the diminished translocation in XEco2‐depleted HSS may have depended on Wapl‐Pds5. However, as the diffusion coefficient in XEco2‐depleted HSS was significantly lower than that in vitro, where cohesin bound to Wapl‐Pds5 (Figs 5F and 2C), there may be additional inhibitory mechanisms on chromatin in Xenopus egg extracts. Notably, not all cohesin molecules were acetylated, even in mock‐depleted HSS (Appendix Fig S5C), although both cohesin and XEco2 are recruited to chromatin in a pre‐RC formation‐dependent manner (Gillespie & Hirano, 2004; Takahashi et al, 2004; Higashi et al, 2012), suggesting that cohesin acetylation may be less stable on chromatin in HSS.
Cohesin dynamics on replicating chromatin in Xenopus egg extracts
To determine whether cohesin translocation is affected by DNA replication, we assessed cohesin dynamics in nucleoplasmic extracts (NPEs), in which linear DNA can be replicated (Yardimci et al, 2010; Loveland et al, 2012). Both‐end‐tethered DNA was first incubated in HSS in the presence of xCohesinHaloTMR to allow cohesin loading and subsequently incubated in NPE to start replication. DNA replication was monitored by incorporation of photoactivatable GFP (PAGFP)‐tagged Xenopus flap endonuclease 1 (xFen1)D179A (Fig 6A and Appendix Fig S6A; Loveland et al, 2012). On replicating DNA, we found that cohesin particles were (i) unidirectionally translocated, (ii) immobile, or (iii) dissociated. We further classified the cohesin motion into 5 groups (Fig 6B). A total of 14.7% of cohesin particles were located on the tip of the replication bubble and translocated together with the progression of replication (Fig 6B, group 1), whereas 23.5% of particles appeared to stop replication on the tip of the replication bubbles (Fig 6B, group 2), although they could be only arrested at the end of the DNA. In contrast, 32.4% of particles were incorporated into replicating DNA (Fig 6B, group 3) and 29.4% of particles were dissociated from chromatin during replication (Fig 6B, group 4) or when cohesin was faced replication sites (Fig 6B group 5). Interestingly, some cohesin particles, which were randomly translocated on unreplicated DNA, became immobile a few minutes before the initiation of replication and were then incorporated into replicating DNA (Fig 6B, group 3). Taking into account the function of cohesin, it must be present in replicated DNA and that it is indeed the case in our system (Appendix Fig S6B) as well as in vivo; thus, at least some populations of cohesin (~30% in our system) must be regulated such that they are not pushed away or dissociated from replicated DNA. Although we do not know how many cohesins properly entrap DNA on Xenopus chromatin, these remaining cohesins could be “cohesive cohesin”; otherwise, the replication machinery may displace cohesin molecules from the replication sites.
Figure 6. Cohesin behaviors on replicating chromatin in Xenopus egg extracts.
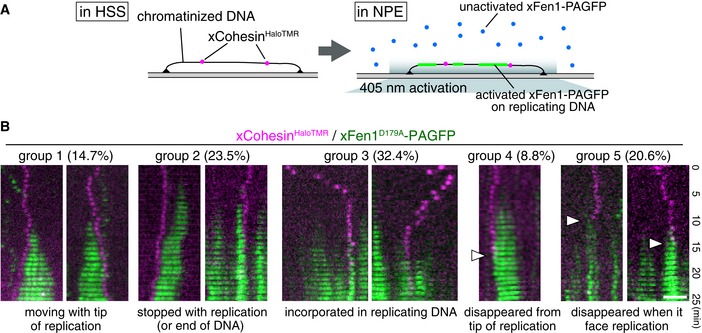
- Experimental design for monitoring replication and cohesin in Xenopus egg extracts.
- Kymographs of examples for cohesin motion during DNA replication. xCohesinHaloTMR (magenta) was loaded onto DNA in the HSS, and NPE was introduced to allow DNA replication. DNA replication was monitored by xFen1D179A‐PAGFP (green). Thirty‐four particles were categorized into five groups. Arrowheads indicate the time points when cohesin was dissociated from chromatin. Scale bar, 2 μm.
Discussion
The present study evaluated the dynamics and regulatory mechanisms of single cohesin molecules. We found that Scc2‐Scc4‐dependent topological loading and cohesin ATPase activity (disengagement of the head domain) are crucial for cohesin translocation along DNA. Consistent with this finding, the ATPase‐dependent translocation of cohesin in budding yeast was described in a previous study (Hu et al, 2011). Although Wapl‐Pds5 promotes the dissociation of cohesin from DNA as previously described (Gandhi et al, 2006; Kueng et al, 2006) (Appendix Fig S2C), we showed that Wapl‐Pds5 renders DNA‐associated cohesin immobile (Fig 2A and B). Considering that the engagement of Smc head domains restrains cohesin movement (Fig 1F), Wapl‐Pds5 may contribute to the tightening of the cohesin ring by associating with SA1, Scc1, and/or Smc3 (Shintomi & Hirano, 2009; Hara et al, 2014; Murayama & Uhlmann, 2015; Ouyang et al, 2016). Therefore, Wapl‐Pds5 may have dual activities: “anti‐establishment activity” and “anti‐translocation activity”.
It remains unclear how cohesin acetylation facilitates the translocation ability of cohesin irrespective of the presence or absence of Wapl‐Pds5. Although acetylation does not affect the ATPase activity of cohesin in vitro (Ladurner et al, 2014), it has been suggested to reduce DNA‐stimulated ATPase activity (Murayama & Uhlmann, 2015). If this is because of lower ATP binding to the cohesin's head domain, resulting in less frequent closure of the ring, this may explain why acetylation facilitates translocation ability. Another possibility is that if Wapl‐Pds5 tightens the cohesin ring, as discussed above, so that the cohesin ring cannot readily translocate along DNA, Smc3 acetylation may neutralize the tightening ability of Wapl‐Pds5 by changing the cohesin conformation to release Wapl‐Pds5‐dependent constraint or changing the affinity of the cohesin complex to DNA. In vertebrates, chromatin‐bound cohesin is acetylated by Esco1 during G1 phase, after cohesin is loaded onto DNA by Scc2‐Scc4 during telophase/G1 phase, whereas both Esco1 and Esco2 acetylate cohesin during S phase (Hou & Zou, 2005). Although cohesin acetylation in S phase is essential for cohesion establishment, the biological significance of cohesin acetylation in G1 phase is not well‐understood. Our findings revealed that the acetylation of cohesin facilitates its translocation irrespective of the presence or absence of Wapl‐Pds5. This translocation ability is presumably required to prevent the transcription machinery from stalling after encountering the cohesin complex (Davidson et al, 2016; Ocampo‐Hafalla et al, 2016). Indeed, acetylated cohesin preferentially accumulates at the 3′ end of active genes, whereas cohesin itself localizes equally on the 5′ and 3′ ends of genes (Deardorff et al, 2012), indicating that acetylated cohesin can translocate on active genes depending on transcription progress. Cohesin may also encounter the replication machinery during S phase. In Xenopus egg extracts, XEco2/Esco2‐dependent acetylation occurs in a pre‐RC‐dependent manner, that is, before the initiation of DNA replication (Higashi et al, 2012), indicating that XEco2‐dependent acetylation of Smc3 in S phase is also required to avoid collision between cohesin and the replication machinery (see below). In contrast to acetylation, Sororin negatively regulates cohesin translocation in the presence of Wapl‐Pds5 (Fig 2D). It has been shown that the establishment of Sororin‐dependent cohesion relies on the association of Sororin with acetylated cohesin and Pds5 (Nishiyama et al, 2010) and stabilization of the cohesin–chromatin interaction (Schmitz et al, 2007; Ladurner et al, 2016). Although at least one role of Sororin is to antagonize the cohesin dissociation activity of Wapl‐Pds5, Sororin may also protect immobility induced by Wapl‐Pds5 when cohesin regains translocation ability by Smc3 acetylation. Thus, cohesion establishment activity may be closely related to the mobility of the cohesin complex on chromatin. In this sense, it would be reasonable to infer that only mitotic kinases, particularly Aurora B and Plk1, could alleviate this immobility in the presence of Sororin.
We demonstrated that cohesins were indeed translocated along unreplicated DNA/chromatin in Xenopus egg extracts in an ATPase‐dependent manner. Consistent with this finding, cohesin particles and the Mcm complexes were detected at different loci (Fig 5A). Because Mcm helicases are not likely to be active in the HSS (Pacek & Walter, 2004), it appears likely that cohesin, but not Mcm, is translocated from its loading sites, as has been suggested for budding yeast (Lengronne et al, 2004; Hu et al, 2011; Ocampo‐Hafalla et al, 2016). As XEco2 (i.e., cohesin acetylation) is indeed required for cohesin translocation in HSS, it is plausible that the acetylation of cohesin is required to avoid collision with the DNA replication machinery in the presence of Wapl‐Pds5. In our replication system, we found that ~15% of cohesin particles were translocated together with the progression of replication (Fig 6B, group 1). This replication‐associated translocation may require cohesin acetylation. More importantly, approximately one‐third of cohesin particles were incorporated into replicated DNA without being translocated (Fig 6B, group 3). These particles could be “cohesive cohesins”, which contribute to sister chromatid cohesion, and Sororin may be important for this immobile status of cohesin, which should allow the progression of DNA replication. If this process is misregulated and cohesin becomes improperly immobile, DNA replication can be arrested, as shown in Fig 6B (group 2). It remains unclear whether there are any regulatory mechanisms by which cohesin is incorporated into replicating DNA or if replication machinery is simply passing through inside of the cohesin ring. Further studies are necessary to clarify this point.
Materials and Methods
Microscopy and flow cell assembly
The TIRFM system was built based on a Nikon Ti inverted microscope with a 100× objective (NA1.49, oil immersion, Nikon) and an EMCCD camera iXon DU888E (Andor) or Evolve (Roper). Illumination was provided by sapphire solid‐state lasers (20 mW, 488 nm and 20 mW, 561 nm; Coherent) and diode lasers (100 mW, 405 nm and 40 mW, 640 nm; Coherent) and passed through an excitation filter (TRF89902‐ET quad band set, Chroma). All images were taken through the 1.5× intermediate magnification module. Assembly of the flow cell and preparation of λDNA were performed based on the method previously described (Yardimci et al, 2012a). In several experiments, PDMS (2 mm × 100 nm) flow cells were used. The flow cell was attached to the syringe pump (Pump 11 Elite, Harvard Apparatus) via a 3‐ml syringe.
Preparation of Xenopus egg extracts and XEco2 immunodepletion
Low‐speed interphase Xenopus egg extracts were prepared as described (Nishiyama et al, 2010). To prepare HSS, low‐speed extracts were centrifuged at 200,000 g for 90 min, and the supernatant was collected. NPE was prepared as previously described (Lebofsky et al, 2009). To deplete XEco2 from the HSS, 1 vol of XEco2 antiserum (Higashi et al, 2012) was bound to 1 vol of Affi‐Prep beads. Then, 0.4 or 0.6 vol of the antibody‐coupled beads was incubated in 1 vol of HSS for 30 min on ice. Retrieved HSS was used for TIRFM observations. hEsco1 was added to the XEco2‐depleted HSS to a final concentration of 17.5 nM.
Chromatin isolation and immunoblotting
Preparation of Xenopus chromatin fraction was performed as described previously (Nishiyama et al, 2010) with some modifications. Sperm nuclei were incubated in HSS at a concentration of 8,000 nuclei/μl for 30 min at 22°C. 50 μl of HSS was diluted by 450 μl of ice‐cold extract buffer (EB) [5 mM MgCl2, 100 mM KCl, HEPES–KOH pH 7.5] containing 0.25% Triton X‐100 and 0.25 M sucrose, and layered over EB containing 0.5 M sucrose. After centrifugation at 20,000 g for 5 min at 4°C, the chromatin pellets were washed with EB containing 0.25% Triton X‐100 and 0.25 M sucrose, and suspended in SDS loading sample buffer. Chromatin‐bound proteins were analyzed by immunoblotting using anti‐Smc3 polyclonal antibody (A300‐060A, Bethyl Laboratories), anti‐Smc3‐ac monoclonal antibody (Nishiyama et al, 2010), anti‐His monoclonal antibody (9c11, Wako), anti‐XEco2 polyclonal antibody (Higashi et al, 2012), and anti‐histone H3 polyclonal antibody (9715L, Cell Signaling).
TIRFM observation of cohesin and the particle analysis
Cohesin loading reaction was performed as described in a previous study (Murayama & Uhlmann, 2014) with some modifications. Scc2‐Scc4 complex (7.5 nM) in CL buffer [35 mM Tris–HCl (pH 7.4), 30 mM NaCl, 1 mM TCEP, 1 mM MgCl2, 15% (v/v) glycerol, 0.003% Tween‐20] was introduced into a DNA‐tethered flow cell and incubated for 15 min. Then, cohesinHalo488 (3.25 nM) in CL buffer containing 0.5 mM ATP was introduced and incubated for 30 min. After incubation, the flow cell was washed by TW buffer [35 mM Tris–HCl (pH 7.4), 300 mM NaCl, 0.5 mM TCEP, 0.25 mM ATP, 0.1% Triton X‐100] followed by TC buffer [35 mM Tris–HCl (pH 7.4), 100 mM NaCl, 0.5 mM TCEP, 1 mM MgCl2, 0.25 mM ATP or AMP‐PCP, 0.05% Tween‐20]. Then, SB buffer [40 mM Tris–HCl (pH 7.4), 100 mM KCl, 1 mM DTT, 1 mM MgCl2, 1 mM ATP or AMP‐PCP, 1 mg/ml BSA] was flowed into the flow cell and cohesin particles were observed under 100, 200, or 400 mM KCl conditions.
To observe effects of Wapl‐Pds5, Sororin, and mitotic kinases on cohesin movements, cohesin complexes were loaded onto DNA and washed by TW buffer, and then TC buffer. After washing by SB buffer, Wapl‐Pds5 complex (15 nM) in SB buffer (100 mM KCl) was introduced into the flow cell and incubated for 30 min. After incubation, SB buffer (100 mM KCl) was flowed and cohesin movement was observed in SB buffer (100, 200, or 400 mM KCl).
Sororin (15 nM) was introduced into the flow cell after incubation of cohesin with Wapl‐Pds5 for 20 min. Sororin was introduced and incubated for 20 min in SB buffer (100 mM KCl). After washing by SB buffer (100 mM KCl), cohesin particles were observed under 100, 200, or 400 mM KCl conditions.
For kinase reactions, the flow cell was washed by kinase buffer [40 mM Tris–HCl (pH 7.4), 100 mM KCl, 1 mM DTT, 5 mM MgCl2, 1 mM ATP] after the incubation of cohesin with Wapl‐Pds5 and Sororin. Then, 2 μl of CDK (0.1 mg/ml, 14 nmol/min/mg; SignalChem), 1.5 μl of Aurora B (0.1 mg/ml, 190 nmol/min/mg; Cell Signaling), or 1 μl of Plk1 (0.1 mg/ml, 17 nmol/min/mg; Sigma‐Aldrich) in a total 30 μl of kinase buffer was introduced into the flow cell and incubated for 30 min at 22°C for Plk1, or 26°C for CDK and Aurora B. To inhibit Plk1 activity, BI (final 10 μM) was added to the kinase reaction mix. After incubation, the flow cell was washed by SB buffer (100 mM KCl) and cohesin particles were observed under 100, 200, or 400 mM KCl conditions. In all experiments, DNA was visualized by SYTOX Orange (25 nM) or SYTOX Blue (125 nM).
In each condition, 45 cohesin particles were filmed in every 100 ms for 30 s, and the particle tracking was performed by NIS‐Elements (Nikon). Based on the experimentally obtained trajectory of 45 particles, we determined the mean square displacement (MSD) and diffusion coefficient (D) (Tafvizi et al, 2008). The MSD was calculated as
where N is the total number of frames (N = 300) and Δt is time interval (Δt = 0.1 s). MSD was plotted as a function of time window nΔt and D was estimated from the initial slope of the MSD vs. nΔt curve (i.e., by fitting a straight line to the MSD calculated at Δt1, Δt2, Δt3, Δt4, and Δt5; from 0 s to 0.5 s). Line scan analysis was performed by NIS‐Elements (Nikon) and other measurements/analyses/processing was performed by ImageJ software.
Immunofluorescence microscopy
For immunofluorescence microscopy, following primary antibodies are used: anti‐Smc3 polyclonal antibody (A300‐060A, Bethyl Laboratories), anti‐histone H3 monoclonal antibody (304‐34781, MBL), anti‐histone H2A.X‐F1 polyclonal antibody (gift from M. Iwabuchi and K. Ohsumi), anti‐Mcm2 monoclonal antibody (BM28, BD Biosciences), anti‐NIPBL (Scc2) monoclonal antibody ab5 (gift from L. Strom), anti‐hSororin polyclonal antibody (Schmitz et al, 2007), anti‐xWapl polyclonal antibody (Nishiyama et al, 2010), anti‐Smc3‐ac monoclonal antibody (Nishiyama et al, 2010), and anti‐hWapl monoclonal antibody (ab109537, Abcam). Primary antibodies were diluted in ELB [10 mM HEPES–KOH (pH 7.7), 2.5 mM MgCl2, 50 mM KCl] (1 μg/ml in final concentration or 1/100 for anti‐hWapl) containing 40 μg/ml κ‐casein and introduced into the flow cell, then incubated for 7.5–10 min (Smc3, H3, H2A.X‐F1, Mcm2, Scc2, hSororin, and hWapl) or 30 min (xWapl and Smc3‐ac). After washing by ELB containing 1 mg/ml BSA (ELB++), 2 μg/ml of secondary antibodies (anti‐rabbit, anti‐mouse and anti‐guinea pig Alexa Fluor 568, anti‐rabbit and anti‐mouse Alexa Fluor 647, and anti‐rabbit and anti‐mouse Alexa Fluor 488; Invitrogen) diluted in ELB containing 40 μg/ml κ‐casein was flowed for 2.5 min, washed by ELB++, and fluorescent signals were observed by TIRFM.
Chromatin formation, cohesin loading, and DNA replication in Xenopus egg extracts
High‐speed supernatant supplemented with 2 mM ATP (final 4 mM; HSS was supplemented with 2 mM ATP during the preparation) or 5 mM AMP‐PNP, 20 mM phosphocreatine, 5 μg/ml creatine kinase, 20 μg/ml aprotinin, 20 μg/ml leupeptin, and 15 μg/ml nocodazole was centrifuged at 20,000 g for 5 min at 22°C to remove debris. For chromatin formation in HSS, a DNA‐tethered flow cell was washed by ELB++, and HSS was introduced into the flow cell in the presence or absence of geminin. Mcm2 and Smc3 were detected by immunostaining. For fluorescence‐labeled cohesin loading, HSS was incubated with human cohesinHalo488 or Xenopus cohesinHaloTMR (61.4–210 nM) and 60 ng/μl of oligo‐duplex for 5 min at RT, and the exogenous cohesin‐containing HSS was flowed into the flow cell and incubated for 30 min. Then, HSS containing 10 nM SYTOX Green and HSS diluted to 1/2 by ELB containing 5 nM SYTOX Green, without cohesinHaloTMR, were introduced and cohesin particles were observed under TIRFM.
Replication reaction in a flow cell was performed as described previously (Loveland et al, 2012; Yardimci et al, 2012a) with some modifications. For replication reaction, HSS was incubated with cohesinHaloTMR (210–450 nM) and 60 ng/μl of oligo‐duplex for 5–10 min at 22°C and then introduced into the DNA‐tethered flow cell. To prepare NPE‐HSS mix, 20 μl of HSS was incubated with 1.2 μl of 15.6 μM geminin, and 20 μl of NPE was incubated with 1.2 μl of ATP regeneration mix [66.7 μM ATP, 667 μM phosphocreatine, 166 μg/ml creatine kinase], 0.6 μl of aprotinin/leupeptin mix [10 mg/ml aprotinin, 10 mg/ml leupeptin], and 0.4 μl of 1 M DTT for 5 min at 22°C, respectively. Then, HSS and NPE were mixed and incubated at 22°C for 5 min. After incubation, 0.78 μl of 1 mM DIG‐11‐dUTP (Roche), 1.65 μl of 530 ng/μl carrier plasmid (pBS(−)SK+), and 5.7 μl of 42.3 μM xFen1D179A‐PAGFP (final 4 μM) were added and incubated for 5 min at 22°C. Then, 9.7 μl of ELB supplemented with 15 μg/ml nocodazole, 4 mM ATP, 40 mM phosphocreatine, and 10 μg/ml creatine kinase was added and the HSS–NPE mix was flowed into the flow cell. Progression of DNA replication was monitored by photoactivation of xFen1D179A‐PAGFP, and cohesin movements during DNA replication were observed by TIRFM. DIG‐dUTP incorporation on DNA was visualized by staining with anti‐DIG‐Rhodamine, Fab fragments (Roche).
Author contributions
TN designed experiments. MK and TN performed TIRFM experiments and analyzed and interpreted the data. ET purified proteins and performed particle tracking. PJHV produced human cohesin complex‐expressing MultiBac system. MK and TN prepared the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We are grateful to G. Goshima and the members of his laboratory for sharing reagents, equipment, and the TIRF microscope system as well as for valuable discussion. We also thank I. F. Davidson, R. Ladurner, V. Bhaskara, and J.‐M. Peters for sharing unpublished results and for discussions and comments; T. S. Takahashi for the Xenopus cohesin constructs, baculoviruses, instructions for NPE preparation, and anti‐XEco2 antibody; K. Shirahige for anti‐Smc3‐ac antibody; L. Strom for anti‐NIPBL antibody; K. Kuwata and Y. Matsubayashi for mass spectrometry analysis; M. Iwabuchi and K. Ohsumi for the anti‐Xenopus histone H2A.X‐F1; S. Westermann for the tetO‐TetR system; A. Tomioka, E. Teruya, and S. Machida for their technical assistance; Y. Matsuda for discussions and comments regarding the single‐molecule analysis; and J. Hutchins for crucial comments on the manuscript. M.K. is supported by the Japan Society for the Promotion of Science (JSPS). T.N. is supported by a JSPS (KAKENHI #25711002), JST‐PRESTO, the JST Tenure Track Program, the Uehara Memorial Foundation, Daiko Foundation, the Naito Foundation, and the Sumitomo Foundation.
The EMBO Journal (2016) 35: 2686–2698
References
- Anderson DE, Losada A, Erickson HP, Hirano T (2002) Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol 156: 419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K (2003) ATP hydrolysis is required for cohesin's association with chromosomes. Curr Biol 13: 1941–1953 [DOI] [PubMed] [Google Scholar]
- Arumugam P, Nishino T, Haering CH, Gruber S, Nasmyth K (2006) Cohesin's ATPase activity is stimulated by the C‐terminal Winged‐Helix domain of its kleisin subunit. Curr Biol 16: 1998–2008 [DOI] [PubMed] [Google Scholar]
- Barber TD, McManus K, Yuen KW, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, Velculescu VE, Kinzler KW, Vogelstein B, Lengauer C, Hieter P (2008) Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci U S A 105: 3443–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F (2008) Eco1‐dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321: 563–566 [DOI] [PubMed] [Google Scholar]
- Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS (2006) A base‐excision DNA‐repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci U S A 103: 5752–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buheitel J, Stemmann O (2013) Prophase pathway‐dependent removal of cohesin from human chromosomes requires opening of the Smc3‐Scc1 gate. EMBO J 32: 666–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Roig MB, Hu B, Beckouet F, Metson J, Nasmyth K (2012) Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell 150: 961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5: 243–254 [DOI] [PubMed] [Google Scholar]
- Davidson IF, Goetz D, Zaczek M, Molodtsov M, Huis in ‘t Veld PJ, Weissmann F, Litos G, Cisneros D, Ocampo‐Hafalla M, Ladurner R, Uhlmann F, Vaziri A, Peters JM (2016) Rapid movement and transcriptional re‐localization of human cohesin on DNA. EMBO J doi:10.15252/embj.201695402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Bando M, Nakato R, Watrin E, Itoh T, Minamino M, Saitoh K, Komata M, Katou Y, Clark D, Cole KE, De Baere E, Decroos C, Di Donato N, Ernst S, Francey LJ, Gyftodimou Y, Hirashima K, Hullings M, Ishikawa Y et al (2012) HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 489: 313–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger CS, Kurze A, Oliveira RA, Nasmyth K (2013) Disengaging the Smc3/kleisin interface releases cohesin from Drosophila chromosomes during interphase and mitosis. EMBO J 32: 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T (2006) Human Wapl is a cohesin‐binding protein that promotes sister‐chromatid resolution in mitotic prophase. Curr Biol 16: 2406–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Misulovin Z, Bilyeu A, Dorsett D (2010) Dosage‐sensitive regulation of cohesin chromosome binding and dynamics by Nipped‐B, Pds5, and Wapl. Mol Cell Biol 30: 4940–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J (2006) Live‐cell imaging reveals a stable cohesin‐chromatin interaction after but not before DNA replication. Curr Biol 16: 1571–1578 [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Hirano T (2004) Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol 14: 1598–1603 [DOI] [PubMed] [Google Scholar]
- Gligoris TG, Scheinost JC, Burmann F, Petela N, Chan KL, Uluocak P, Beckouet F, Gruber S, Nasmyth K, Lowe J (2014) Closing the cohesin ring: structure and function of its Smc3‐kleisin interface. Science 346: 963–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graneli A, Yeykal CC, Robertson RB, Greene EC (2006) Long‐distance lateral diffusion of human Rad51 on double‐stranded DNA. Proc Natl Acad Sci USA 103: 1221–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A (1997) A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Lowe J, Hochwagen A, Nasmyth K (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9: 773–788 [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K (2008) The cohesin ring concatenates sister DNA molecules. Nature 454: 297–301 [DOI] [PubMed] [Google Scholar]
- Hara K, Zheng G, Qu Q, Liu H, Ouyang Z, Chen Z, Tomchick DR, Yu H (2014) Structure of cohesin subcomplex pinpoints direct shugoshin‐Wapl antagonism in centromeric cohesion. Nat Struct Mol Biol 21: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM (2005) Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol 3: e69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann B, Hutchins JR, Hudecz O, Novatchkova M, Rameseder J, Sykora MM, Liu S, Mazanek M, Lenart P, Heriche JK, Poser I, Kraut N, Hyman AA, Yaffe MB, Mechtler K, Peters JM (2011) Systematic phosphorylation analysis of human mitotic protein complexes. Sci Signal 4: rs12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi TL, Ikeda M, Tanaka H, Nakagawa T, Bando M, Shirahige K, Kubota Y, Takisawa H, Masukata H, Takahashi TS (2012) The prereplication complex recruits XEco2 to chromatin to promote cohesin acetylation in Xenopus egg extracts. Curr Biol 22: 977–988 [DOI] [PubMed] [Google Scholar]
- Hou F, Zou H (2005) Two human orthologues of Eco1/Ctf7 acetyltransferases are both required for proper sister‐chromatid cohesion. Mol Biol Cell 16: 3908–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Itoh T, Mishra A, Katoh Y, Chan KL, Upcher W, Godlee C, Roig MB, Shirahige K, Nasmyth K (2011) ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr Biol 21: 12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis in ‘t Veld PJ, Herzog F, Ladurner R, Davidson IF, Piric S, Kreidl E, Bhaskara V, Aebersold R, Peters JM (2014) Characterization of a DNA exit gate in the human cohesin ring. Science 346: 968–972 [DOI] [PubMed] [Google Scholar]
- Kon A, Shih LY, Minamino M, Sanada M, Shiraishi Y, Nagata Y, Yoshida K, Okuno Y, Bando M, Nakato R, Ishikawa S, Sato‐Otsubo A, Nagae G, Nishimoto A, Haferlach C, Nowak D, Sato Y, Alpermann T, Nagasaki M, Shimamura T et al (2013) Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet 45: 1232–1237 [DOI] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM (2006) Wapl controls the dynamic association of cohesin with chromatin. Cell 127: 955–967 [DOI] [PubMed] [Google Scholar]
- Ladurner R, Bhaskara V, Huis in ‘t Veld PJ, Davidson IF, Kreidl E, Petzold G, Peters JM (2014) Cohesin's ATPase activity couples cohesin loading onto DNA with Smc3 acetylation. Curr Biol 24: 2228–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladurner R, Kreidl E, Ivanov MP, Ekker H, Idarraga‐Amado MH, Busslinger GA, Wutz G, Cisneros DA, Peters JM (2016) Sororin actively maintains sister chromatid cohesion. EMBO J 35: 635–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont AL, Song J, Rankin S (2010) Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication‐dependent sister chromatid cohesion. Proc Natl Acad Sci U S A 107: 20364–20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey RA, Mills AD, Morris NR (1977) Assembly of SV40 chromatin in a cell‐free system from Xenopus eggs. Cell 10: 237–243 [DOI] [PubMed] [Google Scholar]
- Lebofsky R, Takahashi T, Walter JC (2009) DNA replication in nucleus‐free Xenopus egg extracts. Methods Mol Biol 521: 229–252 [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430: 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Krantz ID (2009) Cornelia de Lange syndrome, cohesin, and beyond. Clin Genet 76: 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Serra L, Kelly G, Patel H, Stewart A, Uhlmann F (2014) The Scc2‐Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome‐free regions. Nat Genet 46: 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T (1998) Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 12: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T (2002) Cohesin release is required for sister chromatid resolution, but not for condensin‐mediated compaction, at the onset of mitosis. Genes Dev 16: 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland AB, Habuchi S, Walter JC, van Oijen AM (2012) A general approach to break the concentration barrier in single‐molecule imaging. Nat Methods 9: 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45 [DOI] [PubMed] [Google Scholar]
- Murayama Y, Uhlmann F (2014) Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 505: 367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama Y, Uhlmann F (2015) DNA Entry into and Exit out of the Cohesin Ring by an Interlocking Gate Mechanism. Cell 163: 1628–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2009) Cohesin: its roles and mechanisms. Annu Rev Genet 43: 525–558 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, Peters JM (2010) Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 143: 737–749 [DOI] [PubMed] [Google Scholar]
- Nishiyama T, Sykora MM, Huis in ‘t Veld PJ, Mechtler K, Peters JM (2013) Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc Natl Acad Sci USA 110: 13404–13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo‐Hafalla M, Munoz S, Samora CP, Uhlmann F (2016) Evidence for cohesin sliding along budding yeast chromosomes. Open Biol 6: 150178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 3: ra3 [DOI] [PubMed] [Google Scholar]
- Onn I, Heidinger‐Pauli JM, Guacci V, Unal E, Koshland DE (2008) Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol 24: 105–129 [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Zheng G, Tomchick DR, Luo X, Yu H (2016) Structural basis and IP requirement for Pds5‐dependent cohesin dynamics. Mol Cell 62: 248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Walter JC (2004) A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J 23: 3667–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin S, Ayad NG, Kirschner MW (2005) Sororin, a substrate of the anaphase‐promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell 18: 185–200 [DOI] [PubMed] [Google Scholar]
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, Mechtler K, Katis VL, Nasmyth K (2009) Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell 33: 763–774 [DOI] [PubMed] [Google Scholar]
- Schmitz J, Watrin E, Lenart P, Mechtler K, Peters JM (2007) Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr Biol 17: 630–636 [DOI] [PubMed] [Google Scholar]
- Shintomi K, Hirano T (2009) Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl‐Pds5 and Sgo1. Genes Dev 23: 2224–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz‐Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, Samuels Y, James CD, Yu H, Kim JS, Waldman T (2011) Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333: 1039–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigler J, Camdere GO, Koshland DE, Greene EC (2016) Single‐Molecule Imaging Reveals a Collapsed Conformational State for DNA‐Bound Cohesin. Cell Rep 15: 988–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters JM (2002) The dissociation of cohesin from chromosomes in prophase is regulated by Polo‐like kinase. Mol Cell 9: 515–525 [DOI] [PubMed] [Google Scholar]
- Tafvizi A, Huang F, Leith JS, Fersht AR, Mirny LA, van Oijen AM (2008) Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys J 95: L01–L03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC (2004) Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre‐replication complex. Nat Cell Biol 6: 991–996 [DOI] [PubMed] [Google Scholar]
- Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, Nishiyama T, Vaziri A, Wutz A, Ellenberg J, Peters JM (2013) Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501: 564–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K (1999) Sister‐chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400: 37–42 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K (2000) Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103: 375–386 [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger‐Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE (2008) A molecular determinant for the establishment of sister chromatid cohesion. Science 321: 566–569 [DOI] [PubMed] [Google Scholar]
- Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K, Jabs EW, Inui K, Joenje H (2005) Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet 37: 468–470 [DOI] [PubMed] [Google Scholar]
- Wang YM, Austin RH, Cox EC (2006) Single molecule measurements of repressor protein 1D diffusion on DNA. Phys Rev Lett 97: 048302 [DOI] [PubMed] [Google Scholar]
- Weitzer S, Lehane C, Uhlmann F (2003) A model for ATP hydrolysis‐dependent binding of cohesin to DNA. Curr Biol 13: 1930–1940 [DOI] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, Kandoth C, Fulton RS, McLellan MD, Dooling DJ, Wallis JW, Chen K, Harris CC, Schmidt HK, Kalicki‐Veizer JM, Lu C et al (2012) The origin and evolution of mutations in acute myeloid leukemia. Cell 150: 264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC (2010) Uncoupling of sister replisomes during eukaryotic DNA replication. Mol Cell 40: 834–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci H, Loveland AB, van Oijen AM, Walter JC (2012a) Single‐molecule analysis of DNA replication in Xenopus egg extracts. Methods 57: 179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci H, Wang X, Loveland AB, Tappin I, Rudner DZ, Hurwitz J, van Oijen AM, Walter JC (2012b) Bypass of a protein barrier by a replicative DNA helicase. Nature 492: 205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, Zhang P, Kim ST, Pan X, Qin J (2008) Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell 31: 143–151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


