Abstract
Hematopoietic stem cells (HSCs) are capable of giving rise to all blood cell lineages throughout adulthood, and the generation of engraftable HSCs from human pluripotent stem cells is a major goal for regenerative medicine. Here, we describe a functional genome‐wide RNAi screen to identify genes required for the differentiation of embryonic stem cell (ESC) into hematopoietic stem/progenitor cells (HSPCs) in vitro. We report the discovery of novel genes important for the endothelial‐to‐hematopoietic transition and subsequently for HSPC specification. High‐throughput sequencing and bioinformatic analyses identified twelve groups of genes, including a set of 351 novel genes required for HSPC specification. As in vivo proof of concept, four of these genes, Ap2a1, Mettl22, Lrsam1, and Hal, are selected for validation, confirmed to be essential for HSPC development in zebrafish and for maintenance of human HSCs. Taken together, our results not only identify a number of novel regulatory genes and pathways essential for HSPC development but also serve as valuable resource for directed differentiation of therapy grade HSPCs using human pluripotent stem cells.
Keywords: directed differentiation, embryonic stem cells, hematopoietic stem cells, RNAi
Subject Categories: Methods & Resources, Stem Cells
Introduction
Hematopoietic stem cells (HSCs) are a rare population of self‐renewing cells capable of replenishing all blood cell types throughout adulthood. HSC transplantation has become a standard treatment for hematologic and lymphoid cancers and is also used to treat other malignant and nonmalignant disorders 1, 2. However, the rarity and limited sources of HSCs [bone marrow (BM), peripheral blood, and umbilical cord blood] hamper their clinical application. Thus, a major goal for regenerative medicine is to find a method capable of generating engraftable hematopoietic stem/progenitor cells (HSPCs) from human pluripotent stem cells (hPSCs). Directed differentiation and transcription factor‐mediated conversion of HSCs provide new strategies to engineer therapy grade HSCs (reviewed in 3). Directed differentiation has been applied to differentiate mouse 4, 5 and human 6, 7, 8 ESCs into HSC lineages. Somatic cell reprogramming to induced pluripotent stem cells (iPSCs) 9, 10 provides new approaches to obtain autologous patient‐specific HSCs, if all the obstacles in ESCs to HSCs differentiations are solved. Recent work has shown that direct reprogramming of fibroblasts into hematopoietic lineage cells can be achieved by using transcription factors 11, 12, 13, 14. For example, infection of mouse fibroblasts with four transcription factors, Gata2, Gfi1b, cFos, and Etv6, induces hemogenic endothelial‐like cells that are able to differentiate into hematopoietic cells 13. Similarly, transduction of hPSC‐derived hematopoietic progenitors with HOXA9, ERG, RORA, SOX4, and MYB transcription factors induces their development to engraftable hematopoietic cells 15. Despite the remarkable progress made in the past few decades, efforts to generate transplantable hPSC‐derived HSCs have been largely elusive, due in part to insufficient long‐term self‐renewal capacity and an inadequate understanding of HSC ontogeny.
In the mouse, blood cells are thought to originate from the ventral floor of the dorsal aorta in the aorta‐gonad‐mesonephros region at embryonic day 10.5, and hematopoietic progenitors then sequentially colonize the fetal liver, spleen, and BM, where definitive hematopoiesis occurs 16. Although multipotent hematopoietic progenitors are widely accepted to arise from hemogenic endothelial cells 17, 18, 19, 20, 21, 22, 23, the molecular mechanisms regulating the endothelial–hematopoietic transition and the specification of definitive HSPCs from hematopoietic precursors remain poorly understood. Several studies have been performed with the goal of identifying the global regulatory networks controlling HSC hierarchy and differentiation 24, 25, the HSC niche 26, and HSC ontogeny 27. In addition, 159 long non‐coding RNAs (lncRNAs) were enriched in HSCs and knockdown of two HSC‐specific lncRNAs resulted in distinct effects on HSC self‐renewal and lineage commitment highlighting the role of another layer of RNA‐mediated control of HSC biology 28. Nevertheless, a genome‐wide functional study of HSC ontogeny is still lacking, making it difficult to generate a global picture of the gene networks governing definitive stage‐specific HSC development. Here, we report the results of a genome‐wide functional RNAi screen that identifies essential genes and networks regulating HSPC differentiation from ESCs in vitro.
Results
A genome‐wide functional genomics screen to identify regulators of HSPC development
The inducible ESC line iHoxB4 29 was infected with a pGIPZ lentiviral vector containing a mouse shRNA library of 57,600 shRNAs targeting 15,570 mouse genes 30, 31, and the cells were then cultured on an OP9 stromal cell layer to promote differentiation toward the hematopoietic lineage, as described previously 29 (Figs 1A and EV1). The pGIPZ vector also encoded GFP to identify successfully transduced ESCs and enable identification of the GFP+ population (Fig 1B and C). To dissect the development of HSPCs in vitro, we isolated five cell populations on days 6 and 20 of ESC differentiation: SSEA1−Flkl+CXCR4− (designated D6F) and SSEA1−/Flkl−CXCR4+ (D6C) cells on day 6; Lin−Sca1+c‐Kit− (D20LS), Lin−Sca1−c‐Kit+ (D20LK), and Lin−Sca1+c‐Kit+ (D20LSK) cells on day 20. mRNA microarray analysis and DNA‐seq were performed on ESCs and isolated cell populations from three independent experiments (Appendix Fig S1).
Figure 1. General outline of the experimental strategy.
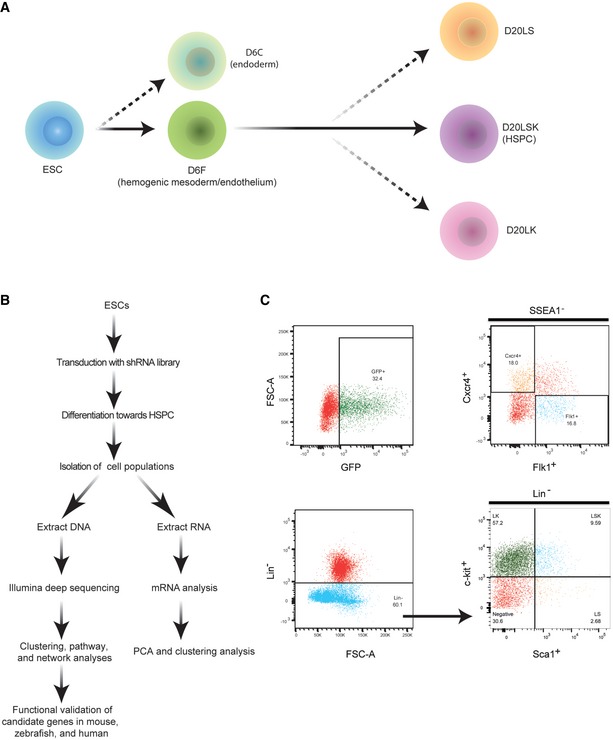
- Developmental stages of ESC differentiation to HSPCs, showing the populations isolated on days 6 and 20.
- Schematic overview of the experimental strategy.
- FACS plots from a representative experiment showing gates for purification of the cell populations. Upper left: GFP+ cells in the population of ESCs infected with pGIPZ‐shRNA library lentiviruses. Upper right: Isolation of mesodermal (SSEA1−Flk1+Cxcr4−) and endodermal (SSEA1−Flkl−Cxcr4+) cells. Lower panels: Isolation of D20LS (Lin−Sca1+c‐Kit−), D20LK (Lin−Sca1−c‐Kit+), and D20LSK (Lin−Sca1+c‐Kit+) cells.
Figure EV1. Morphology of embryoid bodies and differentiated cells.
EB formation and the morphology of cells during HSPC differentiation from ESCs. Scale bar = 100 μm.
SSEA1 is an embryonic stem cell marker and was used here to exclude ESCs from the differentiated populations, and CXCR4 is a marker for both primitive and definitive endoderm 32, 33. Flk1 is expressed by hemogenic mesodermal cells, which can differentiate into endothelial and hematopoietic cells in vitro and in vivo 20, 23, cardiomyocytic and mural cells in vitro 34, and functional blood vessels in vivo 35. Expression of CXCR4 by the D6C population identified them as endodermal. D6F cells expressed the highest levels of three endothelial marker genes: Cdh5 (vascular endothelial [VE]‐cadherin), CD31 (Pecam‐1), and Tie2 (Tek), and expressed the lowest level of the epithelial (E)‐cadherin (Cdh1) marker (Fig 2A), thus identifying the D6F fraction as hemogenic mesodermal/endothelial (HM/E) cells. Consistent with this, the hematopoietic markers, Scl (Tal1) 20 and CD41 (Itga2b) were expressed in D6F cells and in all day 20 cell populations, suggesting that the hematopoietic lineage was already established by day 6. The Lin−Sca1+c‐Kit+ (D20LSK) population has been defined as HSPCs in mouse BM and mouse ESC‐derived cells in vitro 36, whereas D20LK cells derived from ESCs in vitro and BM in vivo include hematopoietic, common myeloid, granulocyte/macrophage, and megakaryocyte/erythrocyte progenitors 36, 37. Thus, in this study, the D20LK population included the precursors and derivatives of HSPCs, whereas the D20LS population was heterogeneous and contained non‐hematopoietic and differentiated cells. The identity of the sorted day 20 populations was confirmed by qRT–PCR analysis of c‐Kit, Sca1, Gata1, Gata2, Lyz2, and Scl expression (Fig 2B and C). Finally, a large proportion of cells isolated on day 20 were confirmed by FACS analysis to be hematopoietic by their positive expression of CD41 and the pan‐hematopoietic antigen CD45 (Fig 2D).
Figure 2. Confirmation of the identity of the sorted cell populations.
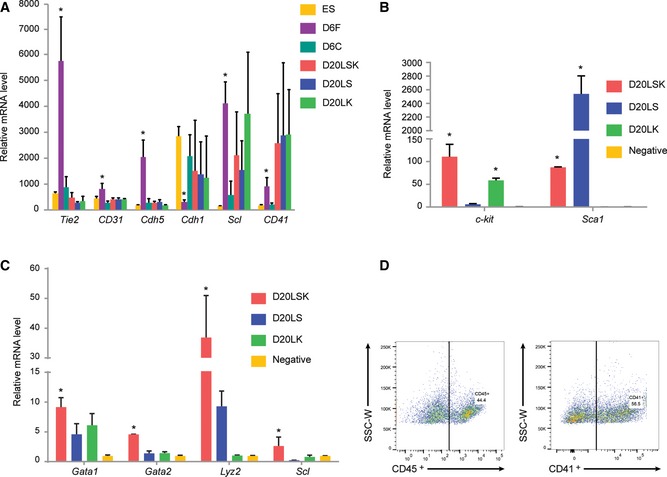
- mRNA analysis of expression of endothelial (Tie2, CD31, Cdh5), epithelial (Cdh1), and early hematopoietic (Scl, CD41) marker genes in the indicated sorted cell populations. Error bars represent standard deviation (SD), n = 3. Student's t‐test. *P < 0.05.
- Confirmation of the purity of sorted populations by qRT–PCR: c‐Kit is highly expressed only in the c‐Kit+ (D20LSK and D20LK) populations, and Sca1 is highly expressed only in the in Sca1+ (D20LSK and D20LS) populations. Error bars represent standard deviation (SD), n = 3. Student's t‐test. *P < 0.05.
- qRT–PCR analysis of Gata1, Gata2, Lyz2, and Scl expression in the sorted cell populations on day 20. Error bars represent standard deviation (SD), n = 3. Student's t‐test. *P < 0.05.
- FACS analysis of ESC‐derived cells confirming that D20LS, D20LK, and D20LSK cell populations express the early hematopoietic marker CD41 and the pan‐hematopoietic marker CD45.
PCA analysis of the mRNA microarray results showed clustering of the ESC, D6C, and D6F samples from three independent experiments, while the D20LS, D20LK, and D20LSK samples were more dispersed (Fig EV2). This was not surprising because long‐term cultures are expected to consist of a more heterogeneous cell population than the short‐term cultures, and the three surface markers used for selection are unlikely to be sufficient to distinguish cells with distinct gene expression patterns. Nevertheless, the D20LS, D20LK, and D20LSK cell populations may still have similar functions, as previously demonstrated 27, 36, 38.
Figure EV2. Gene expression analysis and validation.
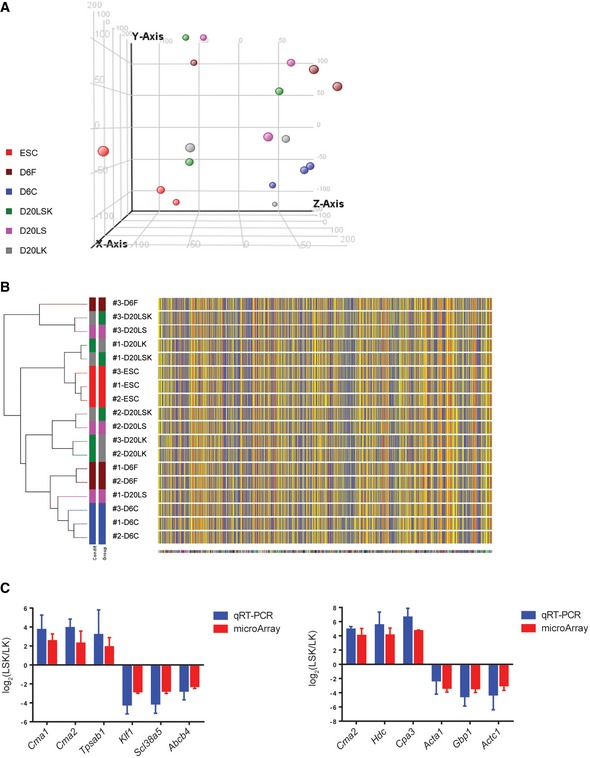
- PCA of mRNA microarray data.
- Clustering analysis of mRNA expression array data.
- qRT–PCR validation of several shRNA target genes with most increased or decreased expression in the LSK population compared with the LS and LK populations. Error bars represent standard deviation (SD), n=3. Student's t‐test shows no significance between the microarray and qRT‐PCR data.
Identification of genes involved in transitional stages of HSC development
We next performed DNA‐seq analysis to identify shRNA sequences that were enriched or depleted in the cell populations isolated on days 6 or 20 compared with the starting population of iHoxB4 ESCs. We reasoned that shRNAs targeted to genes that are essential for either cell survival or differentiation would cause either cell death or differentiation arrest and thus be underrepresented (depleted) in the cell populations isolated on day 6 or day 20 compared with ESCs. Conversely, shRNAs targeted to genes that are either not essential for cell survival or act as barriers to differentiation would be enriched in the day 6 and/or day 20 populations compared with ESCs. Thus, depletion of shRNAs in the five cell populations would identify genes that are essential for survival or differentiation. We conducted a clustering analysis using a k‐means algorithm based on log10 fold changes in shRNA reads in the D6C (endoderm), D6F (HM/E), D20LS, D20LK, and D20LSK (HSPC) populations compared with ESCs. For further analysis, shRNAs were assigned to ten groups based on the function of their target genes (Fig 3A).
Figure 3. Distinct groups of shRNA target genes are enriched or diminished in different cell populations.

- Clustering and enrichment analyses of shRNA reads in D6C (endoderm), D6F (HM/E), D20LS, D20LK, and D20LSK (HSPC) populations compared with ESCs. Specifically enriched/depleted shRNAs are grouped according to their target gene function (I–X). Color coding bar is shown below.
- Heat map of shRNA reads in D6C and D6F populations compared with ESCs. shRNAs specifically depleted in D6C and D6F are labeled as Groups XI and XII, respectively.
- GO pathway analysis showing the top 5 enriched biological processes specific to D6F (XII), D20LS (VI), D20LK (V), and D20LSK (X) populations. The enrichment analysis was conducted using the Metacore platform.
- GO analysis showing several key pathways, including Wnt signaling, cell adhesion, and cell cycle regulation, that are important at all stages of HSPC development.
- Heat map showing depletion of shRNAs targeting Cdc and Cdk genes in all day 20 samples.
Interestingly, only Group I shRNAs were significantly enriched in the day 20 populations, and those in Group VIII were essentially unchanged in all five populations compared with ESCs. Thus, the corresponding target genes of Groups I and VIII shRNAs are unlikely to play essential roles in HSPC development (Fig 3A). In contrast, shRNAs in the remaining groups were depleted to different extents in the three populations isolated at day 20. Groups V, VI, and X target genes were specifically depleted in the D20LK, D20LS, and D20LSK populations, respectively. Group II target genes appear to be essential for the development of the D20LK and D20LSK populations, whereas Group III genes were potentially required for differentiation to the D20LS and D20LSK stages. Although Group VII genes were not required for D20LSK differentiation, they appear to be critical for the development of both the D20LK and D20LS populations. To visualize the differentially enriched or depleted shRNAs in the D6C (endoderm) and D6F (HM/E) cell samples compared with ESCs, we performed clustering analysis using log2 fold changes in shRNA reads. We identified two groups of shRNAs that were specifically depleted in D6C cells and D6F cells, which we designated Groups XI and XII, respectively (Fig 3B).
We next performed gene ontogeny (GO) analysis to identify biological processes and pathways most highly represented by target genes in Groups XII, VI, V, and X, reflecting their requirement for the development of D6F (HM/E), D20LS, D20LK, and D20LSK (HSPC) populations, respectively (Fig 3C and Appendix Fig S2). We found that biological processes relevant to mesodermal development and endothelial specification, such as “cellular component organization or biogenesis”, “single‐organism cellular process”, and “single‐organism developmental process”, were highly enriched among the HM/E‐specific target genes, as expected (Fig 3C). The most enriched biological process networks among the D20LS‐specific target genes were “cell activation involved in immune response” and “leukocyte activation involved in immune response”. Several key activities related to phosphorylation of STAT protein were also enriched. Among the prominent D20LK‐specific processes were “negative regulation of neurological system process” and “neurological system process involved in regulation of systemic arterial blood pressure”. These processes are hallmarks of hematopoietic lineage precursors, suggesting that such cells were contained within the D20LK population. “Positive regulation of hematopoietic stem cell migration” was significantly enriched among the Group X target genes, confirming the HSPC identity of the D20LSK population. Interestingly, genes within the “phospholipase C‐activating angiotensin‐activated signaling pathway” were also enriched in D20LSK cells. In support of this finding, AGTR1 (angiotensin receptor II, type I) has been shown to positively regulate differentiation of BM granulocyte/macrophage progenitors (Lin−Sca‐1−c‐Kit+CD34+CD16/32+) and peripheral blood monocytes from HSCs 39, and blockade of AGTR1 leads to reduced number of BM and peripheral blood, as well as CD11b+ monocytes in bone marrow 40. The enrichment of target genes associated with “neuron projection morphogenesis” in this group also indicates that HSPC specification may involve novel pathways.
To identify pathways involved at all stages of differentiation, we examined target genes that were enriched in the five day 6 and day 20 cell populations (Fig 3D). The top four pathways included Wnt signaling, cell adhesion, cell cycle, and reproduction/differentiation. Among the common pathways was Wnt signaling, which is recognized to play crucial roles in the metabolism and differentiation of many cell lineages 41, 42 and is known to be a critical regulatory pathway for HSPC development 43, 44, 45. Wnt signaling mechanisms are often known as the canonical or Wnt/ß‐catenin and the non‐canonical pathways (reviewed in 46). High Wnt signals in the canonical pathway can lead to loss of stemness and enhance HSC differentiation 47. In addition, a switch in canonical to non‐canonical Wnt signaling could play important roles in HSC aging 48. As expected, pathways involved in cell adhesion 49 and cell cycle 50 regulation were also represented in all cell populations. Hedgehog signaling was also a prominent pathway in our GO analysis. Although several studies have shown that Hedgehog signaling is dispensable for HSC self‐renewal and function 51, 52, it is involved in the embryonic endothelial–hematopoietic transition 53. These results demonstrate that our experimental system faithfully captures the dynamic transitional stages of HSC development. To confirm our GO analysis findings, we analyzed reads of shRNAs targeted to > 50 cell cycle‐related genes, including many Cdc and Cdk genes. Indeed, most of the shRNAs targeting these genes were significantly decreased in D20LS, D20LK, and D20LSK cells, consistent with the findings of the GO analysis (Fig 3E).
Functional validation of genes required for differentiation of ESCs toward HSPCs
To validate the gene identified from our primary RNAi screen, we selected 3–4 genes from Groups XI, XII, IV, and X and individually analyzed their role in differentiation of mESCs toward HSPCs in vitro. Group XI genes, Atp5g3, Bub1b, Eci1, and Dis3l, and Group XII genes, Esrra, S100a8, and Hcfc2, were selected to examine their role in endoderm and mesoderm/endothelium specification. Silencing of Esrra, S100a8, and Hcfc2 in mESCs (Fig EV3A) significantly reduced the Flk1+Cxcr4− cells in EBs at day 6 (Fig 4A and B), confirming that these genes are required for generation of Flk1+Cxcr4− cell during differentiation. In contrast, Group XI genes Atp5g3, Bub1b, Eci1, and Dis3l knockdown (Fig EV3B) clearly diminished Cxcr4+Flk1− cells (Fig 4C and D), supporting that Group VI genes are essential for endoderm cell fate determination. Given that shRNAs in Group IV were largely decreased or depleted in D20 samples, the genes targeted by these shRNAs are most likely required for generation D20LK, D20LS, and D20LSK population. As expected, knockdown of Group IV genes Wbp5, Rbm26, Gdpd4, and Nrxn1 (Fig EV3C) caused reduced number of D20LK, D20LS, and D20LSK cells (Fig 4E and F). Interestingly, blockage of these genes increased Cxcr4+ cells while decreased Flk1+ cells at EB6 (Fig EV3D–F), suggesting their role on HSPC development may occur at earlier stage. Next, we want to examine whether genes identified in Group X are required for D20LSK population generation. We selected four genes Ap2a1, Mettl22, Lrsam1, or Hal which are conserved across Danio rerio, Mus musculus, and Homo sapiens. As we expected, the knockdown of these genes (Fig EV3G) remarkably diminished D20LSK population (Fig 4G and H) without affecting endoderm and mesoderm/endothelium cells on day 6 (Fig EV3H–J). Taken together, these data demonstrate that our primary screen was able to identify genes required for specific cell populations during differentiation.
Figure EV3. Knockdown of Group XII, XI, IV, and X genes and analysis of their function in differentiation.
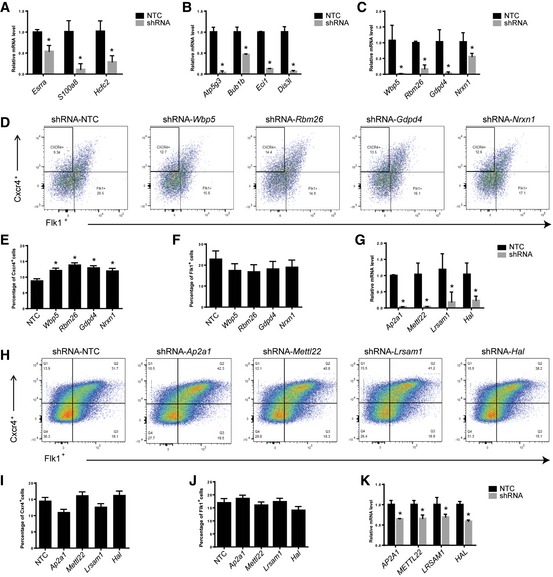
- qRT–PCR analysis showing the knockdown of Group XII genes Esrra, S100a8, and Hcfc2 in mESCs. Error bars represent standard deviation (SD) of technical triplicates. Student's t‐test, *P < 0.05.
- qRT–PCR analysis showing the knockdown of Group XI genes Atp5g3, Bub1b, Eci1, and Dis3l in mESCs. Error bars represent standard deviation (SD) of technical triplicates. Student's t‐test, *P < 0.05.
- qRT–PCR analysis showing the knockdown of Group IV genes Wbp5, Rbm26, Gdpd4, and Nrxn1 in mESCs. Error bars represent standard deviation (SD) of technical triplicates. Student's t‐test, *P < 0.05.
- Flow cytometric analysis of endoderm and mesoderm/endothelium cells in EBs on day 6 with knockdown of Wbp5, Rbm26, Gdpd4, and Nrxn1, respectively.
- Percentage of CXCR4+ cells in EBs on day 6 with knockdown of Wbp5, Rbm26, Gdpd4, and Nrxn1. Error bars represent standard deviation (SD), n = 2. Student's t‐test, *P < 0.05.
- Percentage of Flk1+ cells in EBs on day 6 with knockdown of Wbp5, Rbm26, Gdpd4, and Nrxn1. Error bars represent standard deviation (SD), n = 2. Student's t‐test shows no significance.
- qRT–PCR analysis showing the knockdown of Group X genes Ap2a1, Mettl22, Lrsam1, and Hal in mESCs. Error bars represent standard deviation (SD) of technical triplicates. Student's t‐test, *P < 0.05.
- Flow cytometric analysis of endoderm and mesoderm/endothelium cells in EBs on day 6 with knockdown of Ap2a1, Mettl22, Lrsam1, and Hal.
- Percentage of CXCR4+ cells in EBs on day 6 with knockdown of Ap2a1, Mettl22, Lrsam1, and Hal. Error bars represent standard deviation (SD), n = 3. Student's t‐test shows no significance.
- Percentage of Flk1+ cells at day 6 with knockdown of Ap2a1, Mettl22, Lrsam1, and Hal. Error bars represent standard deviation (SD), n = 3. Student's t‐test shows no significance.
- qRT–PCR analysis showing the knockdown AP2A1, METTL22, LRSAM1, and HAL in human CD34+ cells. Error bars represent standard deviation (SD) of technical triplicates. Student's t‐test, *P < 0.05.
Figure 4. Functional validation of candidate genes in Groups IV, X, XI, and XII required for differentiation of ESCs toward HSPCs.
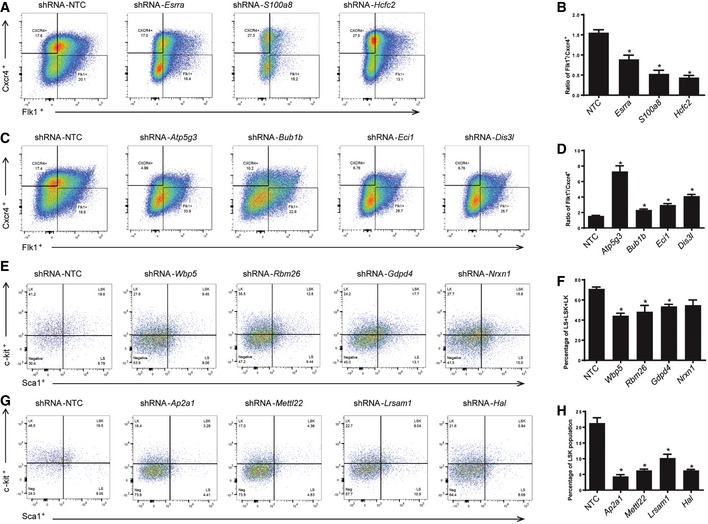
- The E14tg2a ESCs were transduced with lentiviral shRNAs targeting genes Esrra, S100a8, or Hcfc2 and induced to form EBs. The day 6 EB cells were analyzed by flow cytometry to measure the mesoderm/endothelium cells (Flk1+/Cxcr4−) and endoderm cells (Cxcr4+/Flk1−). The percentages of mesoderm/endothelium cells (Flk1+/Cxcr4−) were decreased on day 6 with shRNA knockdown of Group XII genes Esrra, S100a8, and Hcfc2 compared to control as quantified by flow cytometric analysis.
- Knockdown of Esrra, S100a8, and Hcfc2 showed that the ratios of percentage of mesoderm/endothelium population vs. endoderm population were decreased as measured by Flk1+/Cxcr4− vs. Flk1−/Cxcr4+ on day 6. The data were generated based on the quantification of flow cytometric analyses. Error bars represent standard deviation (SD), n = 2. Student's t‐test, *P < 0.05.
- The E14tg2a ESCs were transduced with lentiviral shRNAs targeting genes Atp5g3, Bub1b, Eci1, or Dis3l and induced to form EBs. The day 6 EB cells were analyzed by flow cytometry to measure the mesoderm/endothelium cells (Flk1+/Cxcr4−) and endoderm cells (Cxcr4+/Flk1−). Flow cytometric analysis showed that endoderm cells (Cxcr4+/Flk1−) were deceased while mesoderm/endothelium cells (Flk1+/Cxcr4−) were slightly increased in the cells with shRNA knockdown of Group XI genes Atp5g3, Bub1b, Eci1, and Dis3l compared to control.
- Knockdown of Atp5g3, Bub1b, Eci1, or Dis3l led to increase of mesoderm/endothelium/endoderm population ratio as quantified based on the flow cytometry data. Error bars represent standard deviation (SD), n = 3. Student's t‐test, *P < 0.05.
- The E14tg2a ESCs were transduced with lentiviral shRNAs targeting genes Wbp5, Rbm26, Gdpd4, and Nrxn1 and differentiated toward HSPCs. The differentiated cells on day 20 were analyzed by flow cytometry. Silencing of Group IV genes including Wbp5, Rbm26, Gdpd4, and Nrxn1 decreased overall LK, LS, and LSK population on day 20.
- Quantification of LK + LSK + LS cell fractions with Wbp5, Rbm26, Gdpd4, and Nrxn1 silencing based on flow cytometric analyses. Error bars represent standard deviation (SD), n = 2. Student's t‐test, *P < 0.05.
- The E14tg2a ESCs were transduced with lentiviral shRNAs targeting genes Ap2a1, Mettl22, Lrsam1, or Hal and differentiated toward HSPCs. The differentiated cells on day 20 were analyzed by flow cytometry. Flow cytometric analysis showed notable reduction of LSK population on day 20 with Group X genes Ap2a1, Mettl22, Lrsam1, or Hal knockdown.
- Quantification of flow cytometric analyses showed knockdown of Ap2a1, Mettl22, Lrsam1, and Hal significantly impaired generation of LSK population on day 20 in vitro compared to control. Error bars represent standard deviation (SD), n = 3. Student's t‐test, *P < 0.05.
Identification and functional validation of genes critical for HSPC specification
The largest group of shRNAs identified was Group IV, representing 7,222 genes. These shRNAs were significantly depleted in the day 20 populations, suggesting that the corresponding genes may be required for the HM/E–hematopoietic transition. The GO analysis of process networks revealed prominent enrichment of immune response and inflammation signaling pathways among the Group IV target genes, including IL‐6 signaling, amphoterin signaling, and IL‐2 signaling (Fig 5A). Notch signaling, progesterone signaling, ESR2 pathway, Wnt signaling, and platelet–endothelium–leukocyte interaction pathways were also significantly enriched among the Group IV genes, supporting the relevance of this group to the endothelial–hematopoietic transition. Interestingly, this group was enriched in genes associated with “male sex differentiation” and “spermatogenesis, motility, and copulation”, suggesting that these pathways may have previously unknown roles in hematopoiesis.
Figure 5. Identification of genes required for HSPC specification and validation in zebrafish.
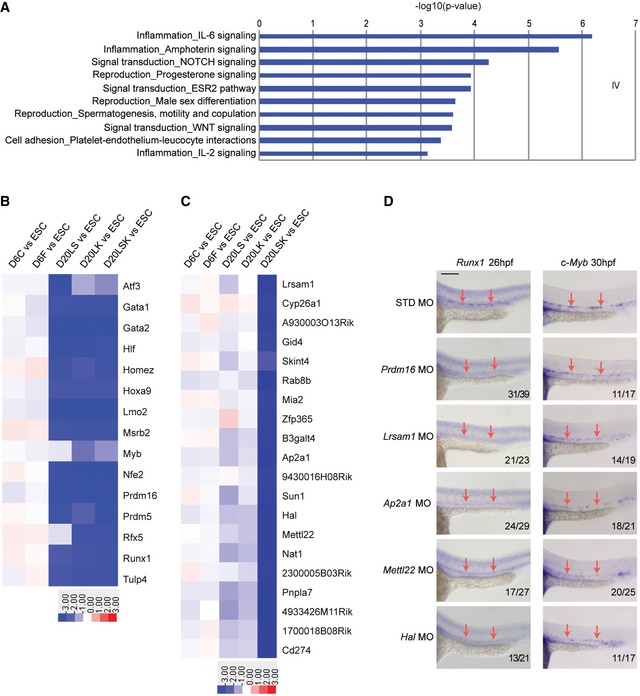
- GO analysis of biological processes among Group IV target genes, showing enrichment of networks in the endothelial–hematopoietic transition.
- Heat map showing bona fide HSC regulators in Group IV target genes.
- Heat map showing the most depleted shRNAs in pairwise comparisons between the D20LSK and D20LK populations.
- Validation of the involvement of Group X target genes Lrsam1, Ap2a1, Mettl22, and Hal in zebrafish hematopoiesis by MO‐mediated knockdown. Whole‐mount in situ hybridization of Runx1 and c‐Myb expression is shown at 26 hpf and 30 hpf, respectively. Prdm16 knockdown serves as a positive control. Note that nonspecific staining for Runx1 and c‐Myb was largely unaffected by targeted MO knockdown, whereas HSC markers Runx1 and c‐Myb staining were notably reduced in discrete cells in the ventral region of the dorsal aorta (arrows). Scale bar = 100 μm.
In contrast to Group IV, Group I shRNAs were significantly enriched in the populations isolated on day 20 relative to day 6, indicating that their target genes are potential barriers for the transition from HM/E to hematopoietic precursors. Interestingly, the most enriched pathway in Group I was related to cardiac development (P = 3.3 × 10−3). Although the Wnt signaling pathway plays a critical role in the HM/E–hematopoietic transition, Wnt antagonists induce cardiac specification from mesoderm and promote cardiomyocyte differentiation 54. Consistent with this, GO analysis showed high enrichment of the Wnt pathway among Group IV genes (P = 3.6 × 10−12) but not Group I genes (Figs 5A and EV4A). We also found that shRNAs targeting most of the well‐known HSC regulators, such as Hoxa9 55, Prdm16 27, Gata2, Runx1, Lmo2, and Gfi1b 56, were identified in Group IV and were significantly decreased in day 20 populations (Fig 5B). In contrast, some of the transcription factors represented in the enriched Group I target genes 57, 58 are known to be involved in cardiac lineage development (Fig EV4B).
Figure EV4. GO analysis (Groups IV and I genes) and heat map of Group I genes.
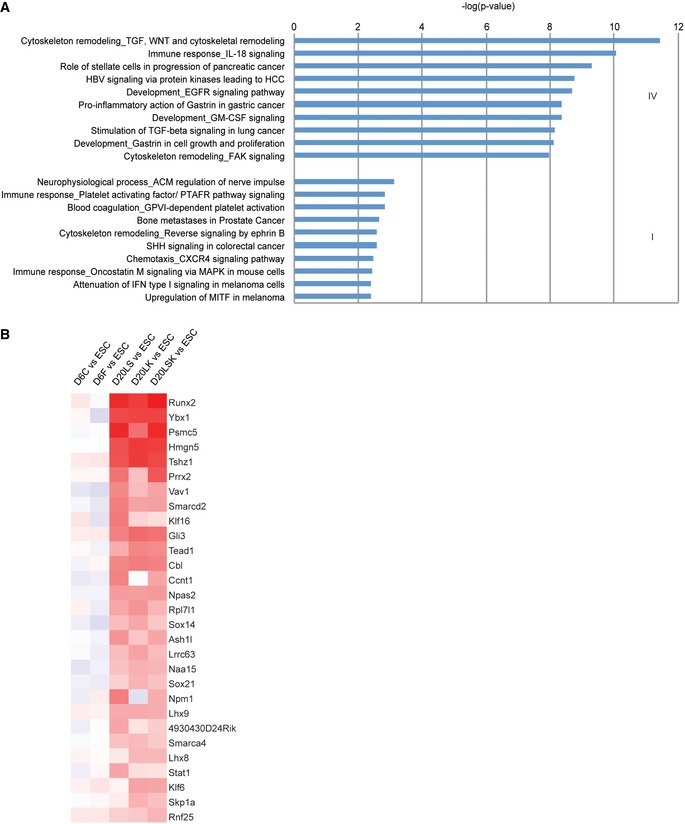
- GO analysis of Groups IV (upper) and I (lower) target genes showing the pathways enriched during the endothelial–hematopoietic transition.
- Transcription factors enriched in Group I genes are related to cardiomyocyte development.
Although many known HSC regulators were found in Group IV, Group X target genes appeared to be critical for HSPC specification, as shRNAs in this group were specifically diminished in the D20LSK population. To identify key genes critical to the specification of HSPCs, we examined Group X shRNAs in more detail. These shRNAs were slightly depleted in D20LS and D20LK cells and markedly depleted in the D20LSK population (Figs 3A and 5C), suggesting that Group X target genes may play essential roles in LSK specification. To test this, we examined the in vivo functional importance in HSC development of four of the top ranked Group X target genes: Ap2a1, Lrsam1, Hal, and Mettl22 (Fig 5C). All four genes are conserved between the zebrafish Danio rerio and Homo sapiens, allowing us to validate their function in HSPC development in zebrafish. Ap2a1 is a subunit of the adaptor‐related protein complex 2 (AP2), which is important for cell polarization and is known to be involved in HSPC migration 59. Lrsam1 is an E3 ubiquitin ligase and may be involved in a novel regulatory pathway for HSC specification. Mettl22 (methyltransferase like 22) is a protein methyltransferase and acts on various chaperone complexes; for example, trimethylation of K135 of KIN/Kin17, K315 of VCP/p97, K561 of HSPA8/Hsc70, as well as lysine residues of other Hsp70 isoforms 60. To date, Mettl22 has not been reported to function in hematopoiesis. Similarly, while deficiency of the cytosolic catabolic enzyme HAL (histidine ammonia‐lyase) 61 causes histidinemia, the enzyme has not previously been implicated in HSC ontogeny 61. HAL catalyzes the deamination of L‐histidine to urocanic acid, which is involved in UV radiation‐induced immunosuppression 62. Although not yet reported, we hypothesized that histidine deamination may also play a role in hematopoiesis. Knockdown of the four genes was achieved by injecting splice‐blocking morpholino oligomers (MOs) into zebrafish embryos at the one‐cell stage. For each gene, MO‐induced abnormal splicing was confirmed by RT–PCR using three different primer sets, and overall mRNA knockdown efficiency was calculated by qRT–PCR (Appendix Fig S3A–E). To assess the effects of MOs on expression of the HSC differentiation‐associated genes, embryos were collected and fixed for in situ hybridization at approximately 26 h post‐fertilization (Runx1), 30 hpf (c‐Myb) and 54 hpf (c‐Myb). Knockdown of Prdm16 serves as a positive control because it has previously been demonstrated to be required for HSC development in zebrafish 27. Compared with negative control MO injections, knockdown of Lsram1, Ap2a1, Mettl22, Hal, and Prdm16 dramatically reduced staining of both c‐Myb and Runx1 in the aorta‐gonad‐mesonephros region at each of the three time points (Fig 5D and Appendix Fig S3F), demonstrating a requirement for these genes during HSC specification. Interestingly, knockdown of these genes had a similar or greater effect than the previously described Prdm16 gene. These results validate the RNAi‐based identification of Group X genes as essential for mammalian HSPC development and further testify to the high discovery rate of functionally relevant genes in our genome‐wide analysis.
Group X genes, AP2A1, METTL22, LRSAM1, and HAL, are crucial for maintaining human CD34+ HSCs
Given the conservation of group X genes in fish, mouse, and human, we sought to determine whether our identified genes play important roles in human HSC biology. Many genes required for HSC specification are also essential for HSC self‐renewal. Group X genes were identified to be required for HSPC specification; therefore, we reasoned that these genes may also be involved in maintaining self‐renewals of adult HSCs. To address this question, we tested the role of four Group X genes, AP2A1, METTL22, LRSAM1, and HAL, in human CD34+ cells. Knockdown of Ap2a1, Lrsam1, Hal, and Mettl22 in human cord blood CD34+ cells (Fig EV3K) caused significant decrease in CD34+ cells after 5 days post‐transduction (Fig 6A and B). This phenotype is much more dramatic in long‐term HSCs as measured by two human HSC markers CD34 and CD90, with nearly 90% loss of CD34+CD90+ cells (Fig 6C and D). These results demonstrate that Group X genes, AP2A1, METTL22, LRSAM1, and HAL, are critical for maintaining human HSPC self‐renewal, especially for long‐term HSCs. Overall, our data confirmed that Group X genes, AP2A1, METTL22, LRSAM1, and HAL, are crucial for HSPC specification and HSC self‐renewal. Notably, whereas most genes known to be important for HSC development are transcriptional regulators, we show here that proteins with diverse functions, including ligase, methyltransferase, transporter, and lyase activities, also play crucial roles in specification of HSPCs and that these functions are conserved from zebrafish to humans.
Figure 6. Novel genes from Group X are required for maintaining human HSCs.
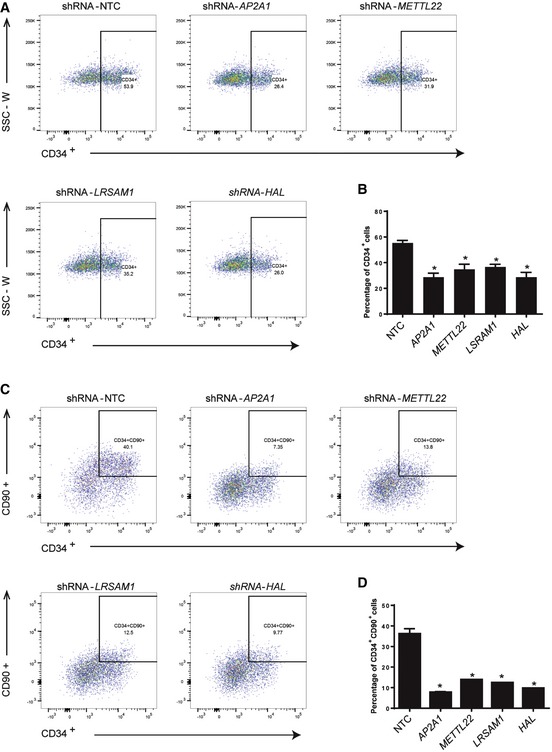
- Representative flow cytometric analysis showed that CD34+ cells were significantly decreased at 5 dpi with knockdown of AP2A1, METTL22, LRSAM1, or HAL. The human cord blood CD34+ cells were transduced with pLKO‐shRNAs targeting human AP2A1, METTL22, LRSAM1, or HAL and cultured for 5 days. Then, the flow cytometric analysis was performed to examine the percentage of CD34+ cells in culture.
- Quantification of CD34+ cells at 5 dpi with knockdown of AP2A1, METTL22, LRSAM1, and HAL showed significant decrease in CD34+ cells compared to control. Error bars represent standard deviation (SD) from duplicates, Student's t‐test, *P < 0.05.
- Flow cytometric analysis showed CD34+/CD90+ cells were strikingly decreased at 5 dpi with knockdown of AP2A1, METTL22, LRSAM1, and HAL.
- Quantification of CD34+CD90+ cells at 5 dpi with knockdown of AP2A1, METTL22, LRSAM1, and HAL showed significant decrease in CD34+CD90+ cells compared to control. Error bars represent standard deviation (SD) from duplicates, Student's t‐test, *P < 0.05.
Discussion
Our analysis of the population‐specific gene groups suggests the existence of a pre‐HSC stage prior to maturation of definitive HSPCs, as proposed in a developmental stage map of ESC to HSPC differentiation (Fig 7). To identify functional stage‐specific gene networks, we compared the target gene sets associated with the five differentiating cell populations (Fig 3A and B). For Stage I, which represents the differentiation of ESCs to HM/Es, we compared target genes in Group XII between HM/E (D6F) and endoderm (D6C) populations. For the HM/E to “pre‐HSC” transition (Stage II), we compared day 20 populations (D20LS, D20LK, and D20LSK) with HM/E (D6F) in Group IV target genes, which are required for the differentiation of all day 20 populations. For Stage III, representing the specification of HSPCs, a network was built by comparing genes present in Group X between D20LSK and D20LK populations. String 10 63 was used to build networks for the top 20 genes identified in each stage (Fig EV5 and Datasets EV1, EV2 and EV3). We found that the Stage I network is connected by Ppara‐Set‐Nup98‐Ddx39b gene sets; the Stage II network is mainly composed of Cct‐Cps1‐Cad‐Dpysl gene sets; and the Ap2a network is dominant at Stage III. At the HM/E transition (Stage I), the network is more broadly connected, as expected, whereas the networks at the pre‐HSPC transition and definitive HSPC specification stages (Stages II and III) become progressively more restricted to a few connected gene sets (Fig EV5).
Figure 7. Summary of key genes and networks identified for various stages of HSPC development.
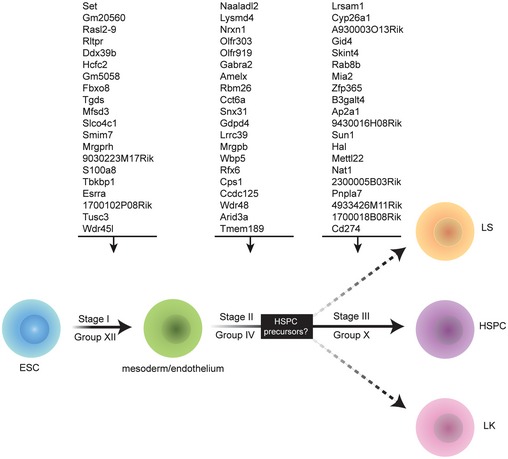
In the proposed roadmap of HSPC development, Stage I refers to the differentiation of ESCs into HM/E, which required Group XII genes. The transition from HM/E, which is dependent on Group IV genes, is denoted as Stage II. Group X genes are specifically required for HSPC specification, indicated as Stage III. See text for details.
Figure EV5. Identification of stage‐specific gene networks that are critical to HSPC development.
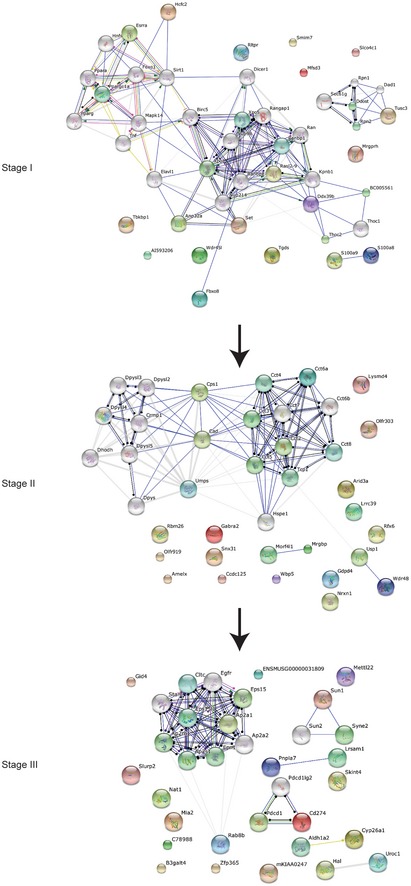
Stage‐specific gene networks during HSPC development. The positioning of Stages I–III is shown in Fig 7. The top 20 genes shown in Fig 7 were used to build the interaction network.
Collectively, these analyses identify stage‐specific gene networks that are critical to the differentiation of definitive HSPCs. Furthermore, we identified 351 genes in Group X that function in the definitive HSPC specification step that confers multipotency. Because RNAi functional screens can identify genes that are not detected by mRNA expression‐based datasets, we were able to discover new genes and pathways required for HSPC development. For example, comparison of our Group X target genes with those previously reported to play roles in HSPC differentiation 27 revealed that only 67 of the 351 genes in Group X were previously identified. The four candidate genes selected from Group X were successfully validated in zebrafish and human CD34+ cells (Figs 5D and 6), demonstrating that our RNAi screen was highly effective in identifying functionally essential and universally conserved genes for HSC differentiation.
Our proposal that a pre‐HSPC stage lies between the HM/E and definitive HSPC stages is based on the observation that most of the known HSC regulators were represented in Group IV, whereas most of the D20LSK‐specific Group X genes have not been previously reported. However, this hypothesis does not exclude the possibility that Group IV genes are required throughout HSPC specification. If our hypothesis is correct, the pre‐HSPC stage may serve as a checkpoint for cells entering the hematopoietic lineage rather than other HM/E‐related lineages such as cardiomyocytes and mural cells. Because we did not examine additional cell populations between days 6 and 20, further experiments will be required to determine whether the putative pre‐HSPC population exists in vitro or in vivo and, if so, to establish whether it is multipotent.
Materials and Methods
Mouse embryonic stem cell culture
The mouse ESC line iHoxB4 (kindly provided by Dr. Michael Kyba) and E14tg2a (a gift from Dr. Chuan He) were used in this study. Mouse ESCs were cultured on irradiated mouse embryonic fibroblasts (MEFs) in DMEM‐based medium supplemented with 15% ES screened fetal bovine serum (FBS; Hyclone Laboratories), 0.1 mM nonessential amino acids (NAEE, GIBCO), 2 mM glutamine (Gibco), 50 μg/ml penicillin and 75 μg/ml streptomycin (GIBCO), 0.00086% 1‐thioglycerol (Sigma), and 1,000 U/ml leukemia inhibitory factor (LIF; Life Technologies). The medium was changed every other day and the ES cells were normally passaged every 3 days.
In vitro differentiation of hematopoietic stem/progenitor cells
In vitro differentiation of HSPCs from mouse ESCs was described previously 29, 64. In brief, on day 0, ESCs were trypsinized and resuspended in EBD medium [IMDM containing 15% FBS, 2 mM glutamine, 50 μg/ml penicillin, 75 μg/ml streptomycin, 0.45 mM 1‐thioglycerol, 200 g/ml iron‐saturated transferrin (Sigma), and 50 g/ml ascorbic acid (Sigma)]. The cells were then cultured in hanging drops at 150 cells/15 μl in inverted 150 × 15 mm petri dishes. On day 2, the EBs were collected and placed in 10 ml EBD medium in a 100 mm ultra‐low attachment dish. The medium was exchanged on day 4. On day 6, EBs were dissociated by incubating for 20–30 min at 37°C in DMEM containing 10 mg/ml collagenase IV (Invitrogen, cat. no. 17104‐019), 20 mg/ml hyaluronidase (Sigma, cat. no. H2126), and 800 U/ml DNase (Sigma, cat. no. D4527), with occasional shaking. The dissociated cells were resuspended in IMDM differentiation medium (supplemented with 10% FBS, 2 mM glutamine, 50 μg/ml penicillin, 75 μg/ml streptomycin, 100 ng/ml stem cell factor, 40 ng/ml VEGF, 40 ng/ml thrombopoietin, and 100 ng/ml Flt‐3 ligand) and plated at 1 × 105 cells/ml on an OP9 stromal layer plated 1 day prior. Doxycycline was added to the medium at final concentration of 1 μg/ml to induce HoxB4 expression in iHoxB4 cells. For E14tg2a cells, the retrovirus expressing HoxB4 was used to infect EB6 cells to overexpress HoxB4 in differentiating cells. The cells were passaged and replated onto fresh OP9 cells when they became ~80% confluent.
Human CD34+ cell culture
Human cord blood CD34+ cells were cultured in StemSpan SFEM (Stem cell Technologies) containing 1× antibiotics and 100 ng/ml of recombinant human cytokines: stem cell factor (hSCF), Flt3 ligand (Flt3L), thrombopoietin (hTPO), interleukin‐6 (hIL6) (Peprotech), supplemented with 1 μM StemRegenin 1 (SR1). The cells were plated at 5 × 105 cells/ml as needed.
Lentivirus generation and transduction
The mouse pGIPZ‐shRNA library lentiviruses and pLKO shRNA lentiviruses were prepared as previously described 30, 31. Briefly, the lentiviral vectors were transfected into 293FT cells together with pPAXs and pMD2 vectors using Lipofectamine® LTX with Plus™ Reagent (ThermoFisher Scientific, #15338100). The viral supernatant was collected 48 h post‐infection and concentrated by ultracentrifugation. The concentrated viruses were divided into aliquots and snap frozen in liquid nitrogen before freezing at −80°C. Transduction of iHoxB4 cells was carried out as previously described 30. Transduction of human CD34+ cells were performed using RetroNectin reagent (TaKaRa).
FACS analysis and whole‐genome RNAi screening
Cells were collected and stained with fluorochrome‐conjugated antibodies in staining medium (PBS supplemented 0.5% BSA and 2 mM EDTA) according to manufacturers' instructions. UltraComp eBeads (eBioscience) was used for single staining samples. The flow cytometry FCS data were further analyzed in FlowJo software. Differentiated mesodermal cells (Ssea1−Flkl+Cxcr4−), endodermal cells (Ssea1−Flkl−Cxcr4+), D20LS (Lin−Sca1+c‐Kit−), D20LK (Lin−Sca1−c‐Kit+), and D20LSK (Lin−Sca1+c‐Kit+) cells were purified by FACS. Transduced iHoxB4 cells were analyzed as undifferentiated control cells. Detailed information on the antibodies used in this study is provided in Table EV1. Genomic DNA and total RNA were extracted from the sorted cells for mRNA microarray and Illumina Hi‐seq analysis, respectively. For DNA‐seq, the sequences corresponding to each shRNA were PCR‐amplified using universal primers, digested with XhoI and EcoRI restriction enzymes, and gel‐purified. The purified fragments were then used for preparation of a sequencing library according to the Illumina Hi‐seq kit instructions. For mRNA microarrays, total RNA was extracted with TRIzol and used directly for microarray analysis using the Illumina MouseWG‐6 v2.0 Expression BeadChip kit.
Quantitative RT–PCR
Total RNA was extracted from sorted cells or zebrafish using TRIzol (Life Technologies) and treated with TURBO DNA‐free DNase (Ambion) to remove residual genomic DNA. Samples of 400 ng DNase‐treated RNA served as templates for reverse transcription using oligo(dT)15 primers (Promega) and SuperScript III reverse transcriptase (Life Technologies). Diluted cDNA samples were mixed with 2× SYBR Green master mix (Bio‐Rad) and analyzed on a Roche Light‐Cycler 480. All qRT–PCR primers used in this study are listed in Table EV2.
Data analysis
FASTQ files of DNA‐seq data were mapped to reference shRNA libraries, and perfectly matched shRNAs were further analyzed. Clustering analysis of sorted populations was performed using Cluster 3.0 and visualized in Java TreeView. GO and enrichment analyses were carried out using the Metacore platform. Functional interactions of overrepresented genes were analyzed using the String database.
For DNA‐seq data analysis, a dataset of shRNA library sequences was generated from 57,600 22‐bp hairpin stem sequences. Next, we removed the low‐quality reads and converted the FASTQ file format to FASTA format. Blast analysis of the sequenced reads (100 bp) against the shRNA library was carried out, and perfectly matched shRNAs were used for normalization.
shRNAs targeting the same gene were totaled in each experiment and the average read numbers from the three independent experiments were used for further analysis. Analysis of variance (ANOVA) and post hoc analysis were performed in Partek Genomics Suite 6.6 to calculate the fold change of shRNA reads in the D6F, D6C, D20LS, D20LK, D20LSK populations compared with ESCs, and the corresponding P‐values. An shRNA read in ESCs of < 10 reads per million total reads was set as the cutoff value. For analysis of shRNA reads in the five sorted populations compared with ESCs, the fold change in log10 was employed for clustering analysis with a k‐means algorithm using Cluster 3.0. The clustered data were visualized as heat maps using Java TreeView. To better visualize the difference between D6F and D6C, clustering analysis was performed using the fold change in log2.
For GO and enrichment analysis, the ANOVA file was uploaded to the Metacore server. The cutoff values for the fold change and P‐value were set as 2 and 0.05, respectively. The enriched genes, networks, and pathways were identified using the Metacore platform. To analyze the enriched genes and networks in Stage I, the shRNAs from Group XII were used to calculate the fold change in D6F compared with D6C populations. For Stage II, the shRNAs from Group IV were used to calculate the fold change in the day 20 populations (average of shRNAs in D20LS, D20LK, and D20LSK) compared with the D6F population. For Stage III, Group X shRNAs were used to calculate the fold change in D20LSK compared with D20LK populations. To exclude variation resulting from zero‐read shRNAs, 1 was added to all reads of < 1 per million total reads. For genetic network construction, the top 20 target genes of depleted shRNAs at Stages I (Dataset EV1), II (Dataset EV2), and III (Dataset EV3) were selected for String analysis with action view.
For microarray analysis, the sample probe files of all samples were collected using Illumina's GenomeStudio software after expression intensities calculation and quality control which was carried out based on the cutoff of detection P‐value < 0.05 for each gene probed on the array. Then, quantile normalization, log transformation, and statistical analysis were performed using Agilent's GeneSpring GX 11.5 software. PCA analysis (Fig EV2A) and hierarchical clustering with the “average” linkage algorithm were performed in Partek Genomics Suite 6.6 (Fig EV2B).
Zebrafish studies
General maintenance, collection, and staging were performed as previously described 65. All animal work was approved by the Institutional Review Board at the University of California, San Diego, and was performed in accordance with Institutional Animal Care and Use Committee guidelines. To prepare the Runx1 antisense RNA probe, a fragment containing the full‐length cDNA and the T7 promoter sequence was PCR‐amplified from the pCS2‐Runx1 vector using Runx1‐probe‐F (5′‐CATACCAAATGGTTTTTCTTTGGGACGCC3′) and T7‐Runx1‐R (5′‐TAATACGACTCACTATAGGTTCTAGACTGTCCCT‐3′) primers. For the c‐Myb antisense RNA probe, c‐Myb‐probe‐F (5′‐GCAACACAAACAGCCCAAATA‐3′) and T7‐c‐Myb‐R (5′‐CGACGGCCAGTGAATTGTAATA‐3′) primers were used to amplify a partial cDNA sequence of c‐Myb and the T7 promoter sequence from the pBK‐c‐Myb vector. The PCR products were digested with DpnI and purified, and the fragments were used as a template for reverse transcription using DIG RNA Labeling Mix (T7) (Roche Diagnostics). The RNA probes were divided into aliquots, snap frozen, and placed at −80°C.
Splice‐blocking morpholinos (MOs) targeting Prdm16, Ap2a1, Lrsam1, Mettl22, and Hal were designed and supplied by Gene Tools, LLC. The MOs, including a standard control oligo CCTCTTACCTCAGTTACAATTTATA, were injected into embryos at the one‐cell stage and larvae were collected for analysis of Runx1 (26 hpf) and c‐Myb (30 hpf and 54 hpf) expression, respectively. Three primer sets were designed (Table EV3) to confirm splicing alterations of MO‐targeted exons using zebrafish RNA (Appendix Fig S3A–E). The MO knockdown efficiency was quantified by qRT–PCR using primer set c for Prdm16, Lrsam1, Ap2a1, Mettl22, and Hal as previously described 27, 43, 66, 67 (Appendix Fig S3A–E). For whole‐mount in situ hybridization, the fish were fixed with 4% paraformaldehyde in PBS at 4°C overnight, and in situ hybridization was performed as previously described 27, 43, 66, 67.
Accession numbers
The GEO accession numbers for the DNA‐seq and RNA microarray data are GSE86898 and GSE86853, respectively.
Author contributions
TH designed and performed the experiments, analyzed the data, and wrote the manuscript. C‐SY and K‐YC analyzed the data. DZ performed the zebrafish experiments. FBI designed the zebrafish functional studies and analyzed the data. TMR designed the overall study and experiments, analyzed the data, wrote the manuscript, and provided financial support.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Table EV1
Table EV2
Table EV3
Review Process File
Acknowledgements
We thank Dr. Michael Kyba (University of Minnesota) for kindly providing the iHoxB4 cell line and vectors and for valuable suggestions on hematopoietic differentiation of mouse ESCs. We also thank Dr. David Traver (UCSD) for kindly providing the pBK‐c‐Myb and pCS2‐Runx1 probe vectors and Runx1 comparison probe for zebrafish in situ staining; Dr. Chuan He (University of Chicago) for kindly providing E14tg2a ES cell line; and Dr. Giulio Cattarossi for his help with the animal experiments. We gratefully acknowledge the staff of the core facilities at the Sanford Burnham Prebys Medical Discovery Institute, UCSD, and The Scripps Research Institute. At SBP, Yoav Altman and Amy Cortez of the Flow Cytometry Core, Alexey Eroshkin and Stacy Huang of the Bioinformatics and Data Management Core, and the staff of the Animal Facility; at UCSD, Neal Sekiy and Tara Rambaldo of the Flow Cytometry Research Core facility (funded by CFAR at the VA Hospital, UCSD), and staff of the Animal Care Program; and at TSRI, Steve Head and the staff of the Next Generation Sequencing Core Facility for help with the HT‐seq. We thank Jennifer Klabis and Jason Dang for their help with preparation of the artwork and figures, and members of the Rana laboratory for helpful discussions and advice, especially Drs. Zhonghan Li and Nianwe Lin. Funded by the National Institute of Allergy and Infectious Diseases (NIAID) (AI43198, AI41404) and the National Institute on Drug Abuse (NIDA) (DA039562).
EMBO Reports (2016) 17: 1814–1828
References
- 1. Copelan EA (2006) Hematopoietic stem‐cell transplantation. N Engl J Med 354: 1813–1826 [DOI] [PubMed] [Google Scholar]
- 2. Gratwohl A, Baldomero H, Aljurf M, Pasquini MC, Bouzas LF, Yoshimi A, Szer J, Lipton J, Schwendener A, Gratwohl M, et al (2010) Hematopoietic stem cell transplantation: a global perspective. JAMA 303: 1617–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rowe RG, Mandelbaum J, Zon LI, Daley GQ (2016) Engineering hematopoietic stem cells: lessons from development. Cell Stem Cell 18: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schmitt RM, Bruyns E, Snodgrass HR (1991) Hematopoietic development of embryonic stem cells in vitro: cytokine and receptor gene expression. Genes Dev 5: 728–740 [DOI] [PubMed] [Google Scholar]
- 5. Wiles MV, Keller G (1991) Multiple hematopoietic lineages develop from embryonic stem (ES) cells in culture. Development 111: 259–267 [DOI] [PubMed] [Google Scholar]
- 6. Chadwick K, Wang L, Li L, Menendez P, Murdoch B, Rouleau A, Bhatia M (2003) Cytokines and BMP‐4 promote hematopoietic differentiation of human embryonic stem cells. Blood 102: 906–915 [DOI] [PubMed] [Google Scholar]
- 7. Kaufman DS, Hanson ET, Lewis RL, Auerbach R, Thomson JA (2001) Hematopoietic colony‐forming cells derived from human embryonic stem cells. Proc Natl Acad Sci USA 98: 10716–10721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vodyanik MA, Bork JA, Thomson JA, Slukvin II (2005) Human embryonic stem cell‐derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood 105: 617–626 [DOI] [PubMed] [Google Scholar]
- 9. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872 [DOI] [PubMed] [Google Scholar]
- 10. Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- 11. Batta K, Florkowska M, Kouskoff V, Lacaud G (2014) Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Rep 9: 1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elcheva I, Brok‐Volchanskaya V, Kumar A, Liu P, Lee JH, Tong L, Vodyanik M, Swanson S, Stewart R, Kyba M, et al (2014) Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat Commun 5: 4372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pereira CF, Chang B, Qiu J, Niu X, Papatsenko D, Hendry CE, Clark NR, Nomura‐Kitabayashi A, Kovacic JC, Ma'ayan A, et al (2013) Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell 13: 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Riddell J, Gazit R, Garrison BS, Guo G, Saadatpour A, Mandal PK, Ebina W, Volchkov P, Yuan GC, Orkin SH, et al (2014) Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell 157: 549–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doulatov S, Vo LT, Chou SS, Kim PG, Arora N, Li H, Hadland BK, Bernstein ID, Collins JJ, Zon LI, et al (2013) Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage‐restricted precursors. Cell Stem Cell 13: 459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Medvinsky A, Rybtsov S, Taoudi S (2011) Embryonic origin of the adult hematopoietic system: advances and questions. Development 138: 1017–1031 [DOI] [PubMed] [Google Scholar]
- 17. Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D (2010) Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464: 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boisset JC, van Cappellen W, Andrieu‐Soler C, Galjart N, Dzierzak E, Robin C (2010) In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464: 116–120 [DOI] [PubMed] [Google Scholar]
- 19. Lam EY, Hall CJ, Crosier PS, Crosier KE, Flores MV (2010) Live imaging of Runx1 expression in the dorsal aorta tracks the emergence of blood progenitors from endothelial cells. Blood 116: 909–914 [DOI] [PubMed] [Google Scholar]
- 20. Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G (2009) The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457: 892–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA (2009) Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457: 887–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kissa K, Herbomel P (2010) Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464: 112–115 [DOI] [PubMed] [Google Scholar]
- 23. Eilken HM, Nishikawa S, Schroeder T (2009) Continuous single‐cell imaging of blood generation from haemogenic endothelium. Nature 457: 896–900 [DOI] [PubMed] [Google Scholar]
- 24. Cabezas‐Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, Weichenhan D, Lier A, von Paleske L, Renders S, et al (2014) Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 15: 507–522 [DOI] [PubMed] [Google Scholar]
- 25. Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al (2011) Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144: 296–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Charbord P, Pouget C, Binder H, Dumont F, Stik G, Levy P, Allain F, Marchal C, Richter J, Uzan B, et al (2014) A systems biology approach for defining the molecular framework of the hematopoietic stem cell niche. Cell Stem Cell 15: 376–391 [DOI] [PubMed] [Google Scholar]
- 27. McKinney‐Freeman S, Cahan P, Li H, Lacadie SA, Huang HT, Curran M, Loewer S, Naveiras O, Kathrein KL, Konantz M, et al (2012) The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell 11: 701–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luo M, Jeong M, Sun D, Park HJ, Rodriguez BA, Xia Z, Yang L, Zhang X, Sheng K, Darlington GJ, et al (2015) Long non‐coding RNAs control hematopoietic stem cell function. Cell Stem Cell 16: 426–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kyba M, Perlingeiro RC, Daley GQ (2002) HoxB4 confers definitive lymphoid‐myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell 109: 29–37 [DOI] [PubMed] [Google Scholar]
- 30. Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al (2005) Second‐generation shRNA libraries covering the mouse and human genomes. Nat Genet 37: 1281–1288 [DOI] [PubMed] [Google Scholar]
- 31. Yang CS, Chang KY, Rana TM (2014) Genome‐wide functional analysis reveals factors needed at the transition steps of induced reprogramming. Cell Rep 8: 327–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Drukker M, Tang C, Ardehali R, Rinkevich Y, Seita J, Lee AS, Mosley AR, Weissman IL, Soen Y (2012) Isolation of primitive endoderm, mesoderm, vascular endothelial and trophoblast progenitors from human pluripotent stem cells. Nat Biotechnol 30: 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE (2005) Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23: 1534–1541 [DOI] [PubMed] [Google Scholar]
- 34. Schroeder T, Meier‐Stiegen F, Schwanbeck R, Eilken H, Nishikawa S, Hasler R, Schreiber S, Bornkamm GW, Nishikawa S, Just U (2006) Activated Notch1 alters differentiation of embryonic stem cells into mesodermal cell lineages at multiple stages of development. Mech Dev 123: 570–579 [DOI] [PubMed] [Google Scholar]
- 35. Yamashita J, Itoh H, Hirashima M, Ogawa M, Nishikawa S, Yurugi T, Naito M, Nakao K, Nishikawa S (2000) Flk1‐positive cells derived from embryonic stem cells serve as vascular progenitors. Nature 408: 92–96 [DOI] [PubMed] [Google Scholar]
- 36. Kitajima K, Minehata K, Sakimura K, Nakano T, Hara T (2011) In vitro generation of HSC‐like cells from murine ESCs/iPSCs by enforced expression of LIM‐homeobox transcription factor Lhx2. Blood 117: 3748–3758 [DOI] [PubMed] [Google Scholar]
- 37. Akashi K, Traver D, Miyamoto T, Weissman IL (2000) A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404: 193–197 [DOI] [PubMed] [Google Scholar]
- 38. McKinney‐Freeman SL, Naveiras O, Yates F, Loewer S, Philitas M, Curran M, Park PJ, Daley GQ (2009) Surface antigen phenotypes of hematopoietic stem cells from embryos and murine embryonic stem cells. Blood 114: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsubakimoto Y, Yamada H, Yokoi H, Kishida S, Takata H, Kawahito H, Matsui A, Urao N, Nozawa Y, Hirai H, et al (2009) Bone marrow angiotensin AT1 receptor regulates differentiation of monocyte lineage progenitors from hematopoietic stem cells. Arterioscler Thromb Vasc Biol 29: 1529–1536 [DOI] [PubMed] [Google Scholar]
- 40. Strawn WB, Ferrario CM (2008) Angiotensin II AT1 receptor blockade normalizes CD11b+ monocyte production in bone marrow of hypercholesterolemic monkeys. Atherosclerosis 196: 624–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Logan CY, Nusse R (2004) The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20: 781–810 [DOI] [PubMed] [Google Scholar]
- 42. Clevers H (2006) Wnt/beta‐catenin signaling in development and disease. Cell 127: 469–480 [DOI] [PubMed] [Google Scholar]
- 43. Clements WK, Kim AD, Ong KG, Moore JC, Lawson ND, Traver D (2011) A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature 474: 220–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL (2003) A role for Wnt signalling in self‐renewal of haematopoietic stem cells. Nature 423: 409–414 [DOI] [PubMed] [Google Scholar]
- 45. Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, et al (2005) Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol 6: 314–322 [DOI] [PubMed] [Google Scholar]
- 46. Staal FJ, Luis TC, Tiemessen MM (2008) WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol 8: 581–593 [DOI] [PubMed] [Google Scholar]
- 47. Famili F, Brugman MH, Taskesen E, Naber BE, Fodde R, Staal FJ (2016) High levels of canonical Wnt signaling lead to loss of stemness and increased differentiation in hematopoietic stem cells. Stem Cell Reports 6: 652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Florian MC, Nattamai KJ, Dorr K, Marka G, Uberle B, Vas V, Eckl C, Andra I, Schiemann M, Oostendorp RA, et al (2013) A canonical to non‐canonical Wnt signalling switch in haematopoietic stem‐cell ageing. Nature 503: 392–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Halbleib JM, Nelson WJ (2006) Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev 20: 3199–3214 [DOI] [PubMed] [Google Scholar]
- 50. Pietras EM, Warr MR, Passegue E (2011) Cell cycle regulation in hematopoietic stem cells. J Cell Biol 195: 709–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hofmann I, Stover EH, Cullen DE, Mao J, Morgan KJ, Lee BH, Kharas MG, Miller PG, Cornejo MG, Okabe R, et al (2009) Hedgehog signaling is dispensable for adult murine hematopoietic stem cell function and hematopoiesis. Cell Stem Cell 4: 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gao J, Graves S, Koch U, Liu S, Jankovic V, Buonamici S, El Andaloussi A, Nimer SD, Kee BL, Taichman R, et al (2009) Hedgehog signaling is dispensable for adult hematopoietic stem cell function. Cell Stem Cell 4: 548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim PG, Albacker CE, Lu YF, Jang IH, Lim Y, Heffner GC, Arora N, Bowman TV, Lin MI, Lensch MW, et al (2013) Signaling axis involving Hedgehog, Notch, and Scl promotes the embryonic endothelial‐to‐hematopoietic transition. Proc Natl Acad Sci USA 110: E141–E150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Van Handel B, Montel‐Hagen A, Sasidharan R, Nakano H, Ferrari R, Boogerd CJ, Schredelseker J, Wang Y, Hunter S, Org T, et al (2012) Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium. Cell 150: 590–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Humphries RK, Largman C (2005) Loss of expression of the Hoxa‐9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood 106: 3988–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, Chilarska PM, Kinston S, Ouwehand WH, Dzierzak E, et al (2010) Combinatorial transcriptional control in blood stem/progenitor cells: genome‐wide analysis of ten major transcriptional regulators. Cell Stem Cell 7: 532–544 [DOI] [PubMed] [Google Scholar]
- 57. Gitler AD, Lu MM, Jiang YQ, Epstein JA, Gruber PJ (2003) Molecular markers of cardiac endocardial cushion development. Dev Dyn 228: 643–650 [DOI] [PubMed] [Google Scholar]
- 58. Lu ZH, Books JT, Ley TJ (2005) YB‐1 is important for late‐stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Mol Cell Biol 25: 4625–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fonseca AV, Corbeil D (2011) The hematopoietic stem cell polarization and migration: a dynamic link between RhoA signaling pathway, microtubule network and ganglioside‐based membrane microdomains. Commun Integr Biol 4: 201–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cloutier P, Lavallee‐Adam M, Faubert D, Blanchette M, Coulombe B (2013) A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet 9: e1003210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Taylor RG, Levy HL, McInnes RR (1991) Histidase and histidinemia. Clinical and molecular considerations. Mol Biol Med 8: 101–116 [PubMed] [Google Scholar]
- 62. Welsh MM, Karagas MR, Applebaum KM, Spencer SK, Perry AE, Nelson HH (2008) A role for ultraviolet radiation immunosuppression in non‐melanoma skin cancer as evidenced by gene‐environment interactions. Carcinogenesis 29: 1950–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta‐Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al (2015) STRING v10: protein‐protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McKinney‐Freeman SL, Naveiras O, Daley GQ (2008) Isolation of hematopoietic stem cells from mouse embryonic stem cells. Curr Protoc Stem Cell Biol Chapter 1: Unit 1F 3 [DOI] [PubMed] [Google Scholar]
- 65. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310 [DOI] [PubMed] [Google Scholar]
- 66. Thisse C, Thisse B (2008) High‐resolution in situ hybridization to whole‐mount zebrafish embryos. Nat Protoc 3: 59–69 [DOI] [PubMed] [Google Scholar]
- 67. Woods IG, Imam FB (2015) Transcriptome analysis of severe hypoxic stress during development in zebrafish. Genom Data 6: 83–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Table EV1
Table EV2
Table EV3
Review Process File


