Abstract
Human malignancies overcome replicative senescence either by activating the reverse‐transcriptase telomerase or by utilizing a homologous recombination‐based mechanism, referred to as alternative lengthening of telomeres (ALT). In budding yeast, ALT exhibits features of break‐induced replication (BIR), a repair pathway for one‐ended DNA double‐strand breaks (DSBs) that requires the non‐essential subunit Pol32 of DNA polymerase delta and leads to conservative DNA replication. Here, we examined whether ALT in human cancers also exhibits features of BIR. A telomeric fluorescence in situ hybridization protocol involving three consecutive staining steps revealed the presence of conservatively replicated telomeric DNA in telomerase‐negative cancer cells. Furthermore, depletion of PolD3 or PolD4, two subunits of human DNA polymerase delta that are essential for BIR, reduced the frequency of conservatively replicated telomeric DNA ends and led to shorter telomeres and chromosome end‐to‐end fusions. Taken together, these results suggest that BIR is associated with conservative DNA replication in human cells and mediates ALT in cancer.
Keywords: alternative lengthening of telomeres, break‐induced replication, PolD3, PolD4, telomere length regulation
Subject Categories: DNA Replication, Repair & Recombination
Introduction
One of the factors limiting indefinite proliferation of somatic cells is telomere length 1, 2. Indeed, the inability to fully replicate both strands of a linear DNA molecule is expected to lead to gradual shortening of telomeres in cells that do not express telomerase. Telomere shortening may be even more severe, if the replication machinery fails to reach the telomeric end. Indeed, the highly repetitive primary structure of telomeres 3, the presence of G‐quadruplexes 4, DNA–RNA hybrids 5, 6, and T‐loops 7, as well as the extensive telomeric heterochromatinization 8, challenge the process of terminal DNA replication and make telomeres prone to fork collapse, similar to common fragile sites 9, 10. Fork collapse within a telomere is unlikely to be resolved by incoming forks or dormant forks, since human telomeres are thought to be devoid of replication origins. Instead, telomere replication is normally dependent on a single origin, located at the subtelomeric regions 11. In cancer cells, telomere replication poses an even greater challenge than in normal cells, due to the presence of oncogene‐induced DNA replication stress 12, 13, 14, 15, 16, 17, 18, 19.
Telomere shortening in cancer cells and immortalized cell lines is compensated either by expression of telomerase or by alternative lengthening of telomeres (ALT) 20, 21, 22, 23, 24, 25. ALT was originally described in the ever shorter telomeres (EST) yeast mutants 26 and in immortalized cancer cells lacking telomerase 23, 24, 25; it is relatively prevalent in aggressive mesenchymal tumors 2 and may contribute to the development of anti‐telomerase oncotherapy resistance 27. Thus, understanding ALT is expected to have important implications for the diagnosis and treatment of cancer.
From yeast to humans, ALT is characterized by extreme heterogeneity of telomere length and depends on homologous recombination for telomere extension 24, 25, 28, 29. As might be expected, the ALT pathway is best understood in yeast, where it has been shown to rely on break‐induced replication (BIR), a repair pathway for collapsed DNA replication forks and one‐ended DNA double‐strand breaks (DSBs) 30, 31, 32, 33. In BIR, the 3‐prime end of single‐stranded DNA invades a homologous double‐stranded DNA segment leading to the formation of a DNA displacement loop (D‐Loop); the invading strand then serves as a primer for initiation of DNA replication 34, 35, 36, 37. BIR requires Pol32, a non‐essential subunit of DNA polymerase delta 33, and, interestingly, leads to conservative DNA replication, in contrast to origin‐initiated replication, which is semiconservative 38, 39, 40, 41.
ALT in yeast has not yet been shown to involve conservative DNA replication, but is, nevertheless, considered to be mediated by BIR, because it is dependent on Pol32 33, 42. Yeast EST mutants demonstrate two discrete types of ALT: type I survivors, which are Rad51‐dependent and elongate their telomeres by amplifying repetitive subtelomeric sequences, and type II survivors, the most prevalent type, which do not require Rad51 and elongate their telomeres by expanding the telomeric repeats 31.
Most of the human ALT cell lines examined to date maintain their telomeres in a Rad51‐independent manner similar to yeast type II survivors 24, 25, 43, 44. However, whether BIR underlies ALT in human cells has not yet been established. The BIR pathway does exist in mammals and, as in yeast, it is involved in repair of collapsed DNA replication forks and one‐ended DNA DSBs 45. Furthermore, mammalian BIR is dependent on PolD3, the ortholog of yeast Pol32, as well as on PolD4, another subunit of DNA polymerase delta. One would assume that, as in yeast, DNA replication initiated by BIR in mammals would be conservative, but this remains to be demonstrated. Here, we examine whether ALT in human cancer cell lines is mediated by BIR. We demonstrate dependence of telomere length on PolD3 and PolD4, as well as the presence of conservatively replicated telomeric DNA. Together, our results reveal that ALT, in mammals, is mediated by BIR and involves conservative DNA replication.
Results and Discussion
Human ALT telomeres display extended tracts of conservatively synthesized DNA
To distinguish between semiconservative and conservative DNA replication at human telomeres, we took advantage of the fact that the two telomeric DNA strands can be differentiated from each other using G‐strand‐ and C‐strand‐specific hybridization probes. Metaphase chromosomes, prepared from cells that had been exposed to BrdU and BrdC for one cell cycle, were subjected to three consecutive strand‐specific fluorescence in situ hybridization (FISH) staining steps (Fig 1). In the first step, the chromosomes were stained by conventional denaturing, telomere strand‐specific, dual color FISH. This allowed both telomeric strands to be labeled, providing a reference for the subsequent staining steps. After destaining, the chromosomes were stained by non‐denaturing, telomere strand‐specific, dual color chromosome orientation‐FISH (CO‐FISH). In this step, the nascent (newly synthesized) DNA strands are degraded and because the staining is performed under non‐denaturing conditions, only parental strands that have been replicated semiconservatively can hybridize with the probes (Fig 1A). In contrast, conservatively replicated telomeres would give no signal, because on one sister chromatid the nascent strands would be degraded and on the other sister chromatid the parental strands would anneal to each other and not be available to hybridize with the probes (Fig 1B). Thus, in this step, conservatively and semiconservatively replicated telomeres can be distinguished. After destaining, the chromosomes were stained for a third time by denaturing, telomere strand‐specific, dual color FISH. Following this third step, the staining pattern of semiconservatively replicated telomeres would not change (Fig 1A), but the previously annealed parental strands of conservatively replicated telomeres would be labeled, thereby ensuring that the absence of signal in the second step did not reflect a technical problem (Fig 1B). Notably, the triple‐FISH staining can also identify cases, where telomeres were partially semiconservatively replicated and partially conservatively replicated, as might occur if a replication fork collapses within the telomere and replication is completed by BIR (Fig 1C).
Figure 1. Triple‐FISH protocol to distinguish between telomeric semiconservative and conservative DNA replication.
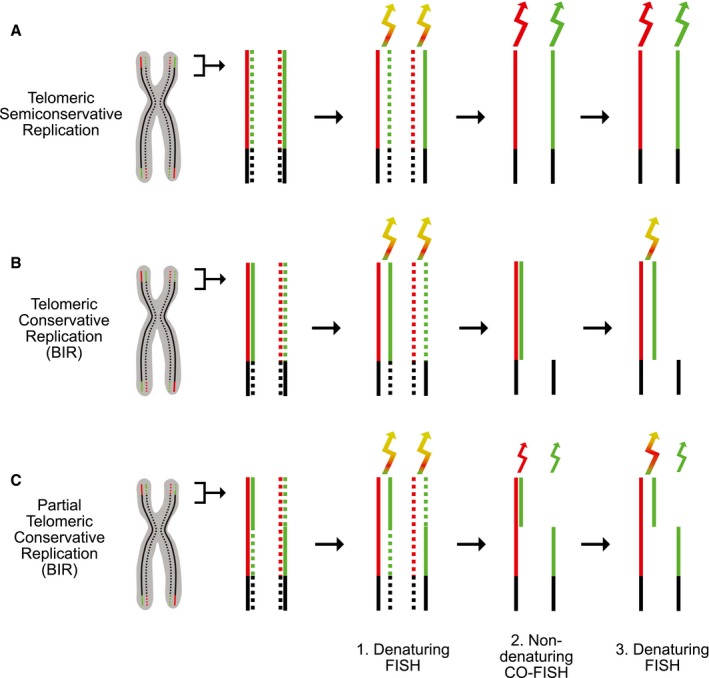
- Semiconservative replication. Diagram of a chromosome with the telomeric C‐rich and G‐rich strands colored red and green, respectively. Newly synthesized strands are indicated by dotted lines. The three steps of the protocol were strand‐specific: dual color, denaturing FISH (1), non‐denaturing chromosome orientation (CO)‐FISH (2), and denaturing FISH (3). In the second step, the nascent strands have been digested. In all steps, two sets of PNA primers specific for the G‐strand and C‐strand, respectively, were used to monitor the presence of both strands. The arrows indicate the color of the emitted light and its intensity (idealized).
- Conservative replication. Diagram showing that telomeric conservative replication (shown here to involve the entire length of the telomeres at the p arms) leads to distinct staining patterns, not observed with semiconservative replication. BIR, break‐induced replication.
- Partially conservative and partially semiconservative telomeric replication. Diagram showing the staining patterns predicted for telomeres that are partially conservatively replicated (distal half) and partially semiconservatively replicated (proximal half), as might occur following fork collapse within a telomere (shown only for the telomeres at the p arms).
The triple‐FISH analysis described above was performed using a stably transfected U2OS cell clone that inducibly overexpresses cyclin E 12. In U2OS cells, telomere length is maintained by the ALT pathway. In the stably transfected clone that we used, cyclin E overexpression leads to DNA replication stress 12, 45. Since collapsed replication forks are repaired by BIR 45, cyclin E‐potentiated fork collapse within telomeres would be expected to lead to enhanced frequencies of BIR and telomeres exhibiting conservative DNA replication. We therefore examined metaphases from cells expressing normal levels of cyclin E (NE) and from cells overexpressing cyclin E (OE). A total of 75 metaphase plates were examined per condition (25 per experiment; three replicates), corresponding to a total of 10,830 and 11,538 chromosome arms for NE and OE cells, respectively. Of these, about 5% exhibited high‐quality staining in all three steps of the triple‐FISH protocol described above (Fig 2A) and were assigned to the three phenotypes shown in Fig 1.
Figure 2. Conservative DNA replication at telomeres of human ALT cells.
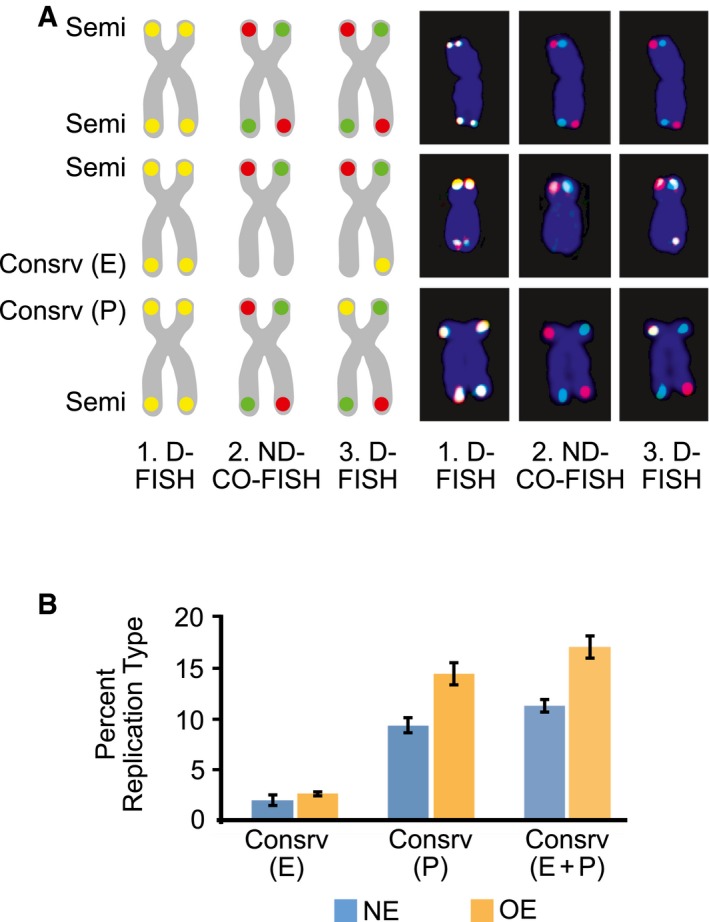
- Examples of chromosome arms exhibiting features of telomeric semiconservative (Semi) replication, conservative (Consrv) replication of the entire telomere (E) and conservative replication of part of the telomere (P). Idealized chromosome diagrams (left) and actual microscopy images (right) are shown. D‐FISH, denaturing FISH; ND‐CO‐FISH, non‐denaturing CO‐FISH.
- Percentages of chromosome arms exhibiting conservative (Consrv) replication of the entire telomere (E), conservative replication of part of the telomere (P), and conservative replication of either the entire telomere of part of it (E+P). NE, U2OS cells expressing normal levels of cyclin E; OE, U2OS cells overexpressing cyclin E for 4 days. Bars represent means and standard errors of the mean from three independent experiments. Cyclin E overexpression resulted in higher percentages of conservatively replicated telomeres: P < 0.025 for Consrv (P) and P < 0.05 for Consrv (E+P), as calculated using the paired t‐test.
As expected, the majority of chromosome arms exhibited staining indicative of semiconservative DNA replication of the telomeres. However, a significant fraction of the telomeres were replicated conservatively (Fig 2B). This fraction corresponded to about 11% of the telomeres in U2OS cells expressing normal levels of cyclin E and increased to about 17% after cyclin E overexpression. In about a fifth of the conservatively replicated telomeres, the entire telomere appeared conservatively replicated, while in the remaining four‐fifths only part of the telomere was conservatively replicated (Fig 2B).
The percentages of chromosome arms exhibiting conservative DNA replication were highly reproducible across biological replicates, but were nevertheless dependent on scoring only about 5% of all chromosome arms. This was because we scored only chromosome arms that exhibited excellent technical quality in all three steps of the triple‐FISH protocol. In addition, we excluded from our analysis telomeres that exhibited telomeric SCEs (T‐SCEs) 46. Telomeres with T‐SCEs hybridized with both the G‐strand and C‐strand probes in the second labeling step of the triple‐FISH protocol and, therefore, it was not possible to determine whether they were conservatively or semiconservatively replicated.
It is also worth noting that the minimum length of telomeric DNA that can be detected by PNA‐FISH is estimated to be about 200 base pairs 47. The sensitivity of detection is, of course, likely to decrease in the second and third steps of our triple‐FISH protocol. Thus, we estimate that the stretches of cytologically visible conservative DNA replication span from a few hundred to a few thousand base pairs.
Depletion of PolD3 or PolD4 suppresses telomeric conservative DNA replication and telomere function
The presence of conservatively replicated DNA at telomeres of ALT cells raises the possibility that ALT is mediated by BIR. To explore this possibility further, we examined whether depleting PolD3 or PolD4 by siRNA affected the frequency of telomeric conservative DNA replication, since PolD3 and PolD4 are required for BIR 45. The experiment was performed with U2OS cells expressing normal levels of cyclin E or overexpressing cyclin E. In one experiment, a total of 25 metaphase plates were examined for each data point, while, in a second experiment, a total of 10 metaphase plates were examined for each data point.
The transient depletion of PolD3 or PolD4 did not affect the frequencies of overall informative chromosome arms. However, the frequency of conservatively replicated telomeres decreased in both the NE and OE cells (Fig 3A). PolD4 depletion had a stronger effect than PolD3 depletion. This is also consistent with our prior experience depleting PolD3 and PolD4 on BIR and with the inability to completely deplete PolD3 protein levels by siRNA 45.
Figure 3. Conservative DNA replication at telomeres and telomere function in ALT cells are dependent on PolD3 and PolD4.
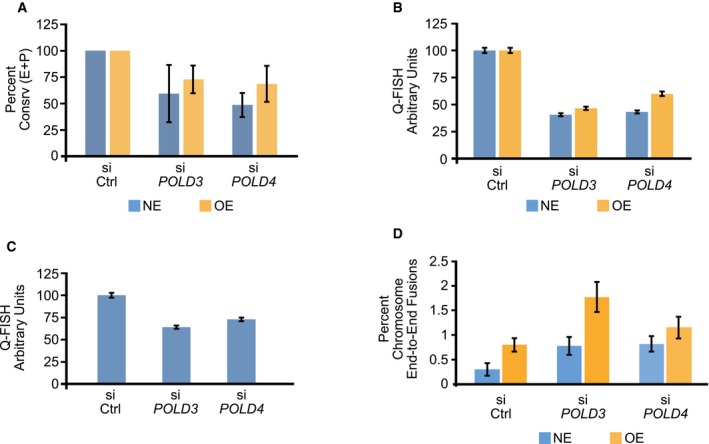
- Decrease in the fraction of chromosome arms exhibiting conservative (Consrv) DNA replication of the entire telomere length or part of the telomere (E+P) in U2OS cells after depletion of POLD3 or POLD4 by siRNA. siCtrl, control siRNA; NE, cells expressing normal levels of cyclin E; OE, cells overexpressing cyclin E for 4 days. Bars represent means and standard errors of the mean from two independent experiments. For statistical comparisons, the effects of PolD3 and Pold4 depletion were examined after grouping together the data from the NE and OE cells. PolD3 depletion reduced the fraction of conservatively replicated telomeres at a level of P = 0.072, whereas for PolD4 depletion the decrease was significant at a level of P < 0.025, as calculated using the paired t‐test.
- Decreased telomere length following PolD3 or PolD4 depletion in U2OS cells expressing normal levels of cyclin E (NE) or overexpressing cyclin E (OE). The cells were transfected with control siRNA (siCtrl) or siRNAs targeting POLD3 or POLD4 and, 3 days later, fixed and examined for telomere length by Q‐FISH. Bars indicate means and standard error of the mean from more than 2,600 telomeres examined per condition. The differences between control and PolD3‐ or PolD4‐depleted cells were significant (P < 10−6) for both NE and OE cells, as determined by unpaired t‐tests.
- Decreased telomere length following PolD3 or PolD4 depletion in parental U2OS cells. The cells were transfected with control siRNA (siCtrl) or siRNAs targeting POLD3 or POLD4 and, 3 days later, fixed and examined for telomere length by Q‐FISH. Bars indicate means and standard error of the mean from more than 2,800 telomeres examined per condition. The differences between control and PolD3‐ or PolD4‐depleted cells were significant (P < 10−6), as determined by unpaired t‐tests.
- Increased frequency of chromosome end‐to‐end fusions following POLD3 or POLD4 depletion. U2OS cells expressing normal levels of cyclin E (NE) or overexpressing cyclin E for 4 days (OE) were transfected with control siRNA (siCtrl) os siRNAs targeting POLD3 or POLD4. For each condition, 25 metaphases were examined (representing about 1,900 high‐quality chromosome arms per condition); the percentage of chromosome fusions was determined for each metaphase and used to calculate means and standard errors of the mean. For statistical analysis, the numbers of chromosome end‐to‐end fusions in the various conditions were compared by chi‐square tests. Cyclin E overexpression led to a higher number of fusions (P < 0.0005); PolD3 and PolD4 depletion also led to a higher number of fusions (P < 0.001 and P < 0.025, respectively).
To further establish the link between ALT and BIR, we examined the effects of depleting PolD3 or PolD4 on telomere length and on the frequency of chromosome end‐to‐end fusions. Telomere length was quantified 3 days after siRNA transfection using a modified PNA Q‐FISH protocol 48 using cells expressing normal levels of cyclin E or overexpressing cyclin E (Fig 3B) or parental U2OS cells (Fig 3C). In all cases, depletion of PolD3 or PolD4 led to telomere shortening. Depletion of PolD3 or PolD4 led to an increased frequency of telomeric end‐to‐end fusions in cells expressing normal levels of cyclin E or overexpressing cyclin E (Fig 3D). Together these results argue that inhibiting BIR compromises telomere length and function.
Eroded telomeres will activate diverse types of DNA repair pathways, including homologous recombination and non‐homologous end joining 49, 50, 51, 52. However, these two major DSB repair pathways are not capable of synthesizing long tracts of telomeric DNA and are, therefore, not suited for telomere elongation. Here, we propose that ALT in human cells is mediated by BIR, a repair pathway that utilizes a one‐ended DNA DSB, such as the end of an eroded telomere, to initiate DNA replication.
A unique property of BIR is that it generates long tracts of conservatively replicated DNA 38, 39, 40, 41. This has been demonstrated in yeast by showing that newly synthesized strands are associated with the recipient, but not the donor chromosome. In higher eukaryotes, the type of DNA replication initiated by BIR has not been experimentally established, but it has been proposed to be conservative, as in yeast 45. The ability of PNA CO‐FISH to distinguish between parental and nascent strands 53 and the repetitive nature of human telomeres allowed us to establish a triple‐FISH protocol to demonstrate that in ALT cells a significant fraction of telomeres, ranging between about 11 and 17% depending on whether cyclin E was overexpressed or not, are conservatively replicated. To our knowledge, this is the first demonstration of the presence of long stretches of conservatively replicated DNA in human cells.
The dependence of conservative replication at telomeres on PolD3 and PolD4 further supports our conclusion that BIR underlies ALT, since PolD3 and PolD4 are required for BIR in human cells 45. Importantly, depletion of PolD3 or PolD4 also resulted in shorter telomere lengths and higher frequencies of end‐to‐end chromosome fusions, further indicating that BIR is important for telomere function in human ALT cells, as it is in telomerase‐deficient yeast cells.
Materials and Methods
Telomeric peptide nucleic acid (PNA) FISH
Telomeric PNA‐FISH was performed using established protocols 54, 55. Briefly, methanol/acetic acid‐fixed cell pellets were resuspended and dropped on wet slides that were left to age overnight. Chromosome preparations were rehydrated in phosphate‐buffered saline (PBS, pH 7.5) for 15 min at room temperature, treated with RNase A (100 μg/μl) (Roche) for 1 h at 37°C, fixed in 3.7% formaldehyde in 1× TBS for 2 min, and then washed in 1× TBS twice for 5 min each time. The chromosome preparations were then digested with pepsin (1 mg/ml) in 10 mM HCl (pH 2) at 37°C for 10 min, washed twice for 5 min in 1× TBS, and then dehydrated by serial incubations in 70, 85, and 96% cold ethanol and air‐dried. Telomere‐specific hybridizations were performed using Cy3‐labeled (TTAGGG)3 and FITC‐labeled (CCCTAA)3 PNA probes (BioSynthesis). After one wash in PBS and one wash in Wash solution (0.1 M Tris–HCl, 0.15 M NaCl, 0.08% Tween‐20, pH 7.5) at 65°C for 5 min, both slides and probe were prewarmed at 37°C for 5 min and then 10 μl of hybridization mixture containing 0.2–0.8 μM of PNA telomeric probes, 70% formamide, and 10 mM Tris, pH 7.2 (Cytocell), was placed on the spotted area of the slide. For denaturing FISH, the slides were heated for 5 min at 80°C; this step was omitted for non‐denaturing FISH. Then, for both denaturing and non‐denaturing FISH, the slides were incubated overnight at 37°C in humidity. The following day, the slides were washed once in PBS for 15 min, once in 0.5× SSC/0.1% SDS at 72°C for 2 min, once in 2× SSC, pH 7/0.05% Tween‐20 at room temperature for 30 min, and twice in PBS for 15 min each. Finally, the preparations were counterstained and mounted with Vectashield containing 4′,6‐diamidino‐2‐phenylindole‐dihydrochloride (DAPI 0.6 μg/ml, Vector Laboratories). Fluorescence images were acquired using an Axio Imager Z1 Zeiss microscope equipped with a 63× magnification lens and a charge‐coupled device (CCD) camera and analyzed using MetaSystems Isis software.
Quantitative telomeric PNA‐FISH
PNA‐FISH was performed as described above. For the evaluation of telomere length by Q‐FISH, we utilized the MetaSystems Isis/Telomere software (MetaSystem Hardware and Software) to normalize the signals from each metaphase spread according to a particular chromosomal reference fluorescence signal 48. The signal from the centromere of chromosome 2 served as the internal reference control. Human centromere 2‐specific PNA probes labeled with Cy3 or FITC were kindly provided as a gift from DAKOCytomation, Glostrup, Denmark.
Strand‐specific CO‐FISH
Metaphase spreads were prepared by conventional cytogenetic methods. Chromosome preparations were stained with Hoechst 33258 (0.5 μg/ml) (Sigma), incubated in 2× SSC (Invitrogen) for 15 min at RT, and exposed to 365‐nm UV light (Stratalinker 1800 UV irradiator) for 45 min. The 5′‐bromo‐2′‐deoxyuridine‐ and 5′‐bromo‐2′‐deoxycytosine‐substituted DNA was digested with exonuclease III (Promega) in a buffer supplied by the manufacturer (5 mM DTT, 5 mM MgCl2, and 50 mM Tris–HCl, pH 8.0) for 15 min at 37°C. The slides were then dehydrated through a cold ethanol series (70, 85, and 100%) and air‐dried. PNA‐FISH was performed with probes specific for C‐ and G‐rich telomere repeats [FITC‐(CCCTAA)3 and Cy3‐(TTAGGG)3] (BioSynthesis). The slides were incubated in hybridization mix (10 mM Tris–HCl, pH 7.2, 70% formamide, 0.5% blocking solution) for 10 min at RT, hybridized with the first probe 0.5 μM TelG‐Cy3 for 1 h in humidity, rinsed with Wash I solution (70% formamide, 10 mM Tris–HCl, pH 7.2, and 0.1% BSA) for 5 min and hybridized with the second probe 0.8 μM TelC‐FITC for 1 h in humidity; both probes were diluted in hybridization mix. The slides were washed twice with solution Wash I for 15 min each and three times with solution Wash II (0.1 M Tris–HCl, pH 7.4, 0.15 M NaCl, 0.08% Tween‐20) for 5 min each and dehydrated through a cold ethanol series (70, 95, and 100%). All preparations were mounted and counterstained with Vectashield antifade medium (Vector), containing 0.1 μg/ml DAPI (Vector). Digital images were captured and analyzed as described above.
Triple‐FISH for visualization of conservative DNA replication
Human ALT osteosarcoma U2OS cells were synchronized by mitotic shake‐off, exposed for one round of DNA replication to BrdU/BrdC, arrested in mitosis by colcemid, and harvested under hypotonic treatment to obtain chromosome preparations. Chromosome specimens mounted onto microscope slides were stained by conventional denaturing telomere strand‐specific dual color PNA‐FISH. Selected “complete and good‐quality” metaphase spreads were photographed under a fluorescent microscope in three color channels, slide coordinates were recorded, and the specimen was destained by heat (72°C) in 2× SSC solution for 30 min. Then, the same microscope slide was exposed to the strand‐specific dual color non‐denaturing telomeric PNA CO‐FISH. Preselected metaphase plates were retrieved and recaptured, and the slide was destained again as above. The third subsequent dual color telomere strand‐specific PNA‐FISH was performed under denaturing conditions. Retrieved metaphase plates were recaptured, and the staining patterns obtained in the three consecutive FISH steps were used to identify the type of DNA replication using the criteria shown in Fig 1.
Author contributions
F‐MR, SKS, VK, and MC performed the experiments and counted the various phenotypes. TDH and SG conceived the project, supervised the experiments, and wrote the manuscript. All authors contributed to the planning of the experiments and commented on the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Review Process File
Acknowledgements
This study was supported by funds from the Swiss National Science Foundation and the European Commission (ERC grant: ONIDDAC) to TDH and BRFAA Intramural Funding to SG.
EMBO Reports (2016) 17: 1731–1737
Contributor Information
Thanos D Halazonetis, Email: thanos.halazonetis@unige.ch.
Sarantis Gagos, Email: sgagos@bioacademy.gr.
References
- 1. Harley CB, Futcher AB, Greider CW (1990) Telomeres shorten during ageing of human fibroblasts. Nature 345: 458–460 [DOI] [PubMed] [Google Scholar]
- 2. Shay JW, Bacchetti S (1997) A survey of telomerase activity in human cancer. Eur J Cancer 33: 787–791 [DOI] [PubMed] [Google Scholar]
- 3. Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR (1988) A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA 85: 6622–6626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D (2006) Human telomeric sequence forms a hybrid‐type intramolecular G‐quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res 34: 2723–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Azzalin CM, Lingner J (2015) Telomere functions grounding on TERRA firma. Trends Cell Biol 25: 29–36 [DOI] [PubMed] [Google Scholar]
- 6. Arora R, Azzalin CM (2015) Telomere elongation chooses TERRA ALTernatives. RNA Biol 12: 938–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Lange T (2004) T‐loops and the origin of telomeres. Nat Rev Mol Cell Biol 5: 323–329 [DOI] [PubMed] [Google Scholar]
- 8. Blasco MA (2007) The epigenetic regulation of mammalian telomeres. Nat Rev Genet 8: 299–309 [DOI] [PubMed] [Google Scholar]
- 9. Arlt MF, Wilson TE, Glover TW (2012) Replication stress and mechanisms of CNV formation. Curr Opin Genet Dev 22: 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drosopoulos WC, Kosiyatrakul ST, Yan Z, Calderano SG, Schildkraut CL (2012) Human telomeres replicate using chromosome‐specific, rather than universal, replication programs. J Cell Biol 197: 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C et al (2005) DNA damage response as a candidate anti‐cancer barrier in early human tumorigenesis. Nature 434: 864–870 [DOI] [PubMed] [Google Scholar]
- 13. Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B et al (2005) Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434: 907–913 [DOI] [PubMed] [Google Scholar]
- 14. Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC et al (2006) Oncogene‐induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637 [DOI] [PubMed] [Google Scholar]
- 15. Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene‐induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- 16. Hills SA, Diffley JF (2014) DNA replication and oncogene‐induced replicative stress. Curr Biol 24: r435–r444 [DOI] [PubMed] [Google Scholar]
- 17. Macheret M, Halazonetis TD (2015) DNA replication stress as a hallmark of cancer. Annu Rev Pathol 10: 425–448 [DOI] [PubMed] [Google Scholar]
- 18. Munoz S, Mendez J (2016) DNA replication stress: from molecular mechanisms to human disease. Chromosoma doi:10.1007/s00412‐016‐0573‐x [DOI] [PubMed] [Google Scholar]
- 19. Suram A, Kaplunov J, Patel PL, Ruan H, Cerutti A, Boccardi V, Fumagalli M, Di Micco R, Mirani N, Gurung RL et al (2012) Oncogene‐induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J 31: 2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S (1992) Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J 11: 1921–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015 [DOI] [PubMed] [Google Scholar]
- 22. Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE (1998) Extension of life‐span by introduction of telomerase into normal human cells. Science 279: 349–352 [DOI] [PubMed] [Google Scholar]
- 23. Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR (1995) Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J 14: 4240–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cesare AJ, Reddel RR (2010) Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 11: 319–330 [DOI] [PubMed] [Google Scholar]
- 25. Pickett HA, Reddel RR (2015) Molecular mechanisms of activity and derepression of alternative lengthening of telomeres. Nat Struct Mol Biol 22: 875–880 [DOI] [PubMed] [Google Scholar]
- 26. Lundblad V, Blackburn EH (1993) An alternative pathway for yeast telomere maintenance rescues est1‐ senescence. Cell 73: 347–360 [DOI] [PubMed] [Google Scholar]
- 27. Bechter OE, Zou Y, Walker W, Wright WE, Shay JW (2004) Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res 64: 3444–3451 [DOI] [PubMed] [Google Scholar]
- 28. Nabetani A, Ishikawa F (2011) Alternative lengthening of telomeres pathway: recombination‐mediated telomere maintenance mechanism in human cells. J Biochem 149: 5–14 [DOI] [PubMed] [Google Scholar]
- 29. Doksani Y, de Lange T (2014) The role of double‐strand break repair pathways at functional and dysfunctional telomeres. Cold Spring Harb Perspect Biol 6: a016576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bosco G, Haber JE (1998) Chromosome break‐induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150: 1037–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Le S, Moore JK, Haber JE, Greider CW (1999) RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152: 143–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McEachern MJ, Haber JE (2006) Break‐induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem 75: 111–135 [DOI] [PubMed] [Google Scholar]
- 33. Lydeard JR, Jain S, Yamaguchi M, Haber JE (2007) Break‐induced replication and telomerase‐independent telomere maintenance require Pol32. Nature 448: 820–823 [DOI] [PubMed] [Google Scholar]
- 34. Davis AP, Symington LS (2004) RAD51‐dependent break‐induced replication in yeast. Mol Cell Biol 24: 2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Llorente B, Smith CE, Symington LS (2008) Break‐induced replication: what is it and what is it for? Cell Cycle 7: 859–864 [DOI] [PubMed] [Google Scholar]
- 36. Anand RP, Lovett ST, Haber JE (2013) Break‐induced DNA replication. Cold Spring Harb Perspect Biol 5: a010397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Malkova A, Ira G (2013) Break‐induced replication: functions and molecular mechanism. Curr Opin Genet Dev 23: 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith CE, Llorente B, Symington LS (2007) Template switching during break‐induced replication. Nature 447: 102–105 [DOI] [PubMed] [Google Scholar]
- 39. Donnianni RA, Symington LS (2013) Break‐induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci USA 110: 13475–13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A (2013) Migrating bubble during break‐induced replication drives conservative DNA synthesis. Nature 502: 389–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P et al (2013) Pif1 helicase and Poldelta promote recombination‐coupled DNA synthesis via bubble migration. Nature 502: 393–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hu Y, Tang HB, Liu NN, Tong XJ, Dang W, Duan YM, Fu XH, Zhang Y, Peng J, Meng FL et al (2013) Telomerase‐null survivor screening identifies novel telomere recombination regulators. PLoS Genet 9: e1003208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Potts PR, Yu H (2007) The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere‐binding proteins. Nat Struct Mol Biol 14: 581–590 [DOI] [PubMed] [Google Scholar]
- 44. Verma P, Greenberg RA (2016) Noncanonical views of homology‐directed DNA repair. Genes Dev 30: 1138–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD (2014) Break‐induced replication repair of damaged forks induces genomic duplications in human cells. Science 343: 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Londoño‐Vallejo JA, Der‐Sarkissian H, Cazes L, Bacchetti S, Reddel RR (2004) Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res 64: 2324–2327 [DOI] [PubMed] [Google Scholar]
- 47. Martens UM, Zijlmans JM, Poon SS, Dragowska W, Yui J, Chavez EA, Ward RK, Lansdorp PM (1998) Short telomeres on human chromosome 17p. Nat Genet 18: 76–80 [DOI] [PubMed] [Google Scholar]
- 48. Perner S, Brüderlein S, Hasel C, Waibel I, Holdenried A, Ciloglu N, Chopurian H, Nielsen KV, Plesch A, Högel J et al (2003) Quantifying telomere lengths of human individual chromosome arms by centromere‐calibrated fluorescence in situ hybridization and digital imaging. Am J Pathol 163: 1751–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. d'Adda di Fagagna F, Reaper PM, Clay‐Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP (2003) A DNA damage checkpoint response in telomere‐initiated senescence. Nature 426: 194–198 [DOI] [PubMed] [Google Scholar]
- 50. Sabatier L, Ricoul M, Pottier G, Murnane JP (2005) The loss of a single telomere can result in instability of multiple chromosomes in a human tumor cell line. Mol Cancer Res 3: 139–150 [DOI] [PubMed] [Google Scholar]
- 51. Jackson SP, Bartek J (2009) The DNA‐damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O'Sullivan RJ, Karlseder J (2010) Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 11: 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bailey SM, Williams ES, Cornforth MN, Goodwin EH (2010) Chromosome orientation fluorescence in situ hybridization or strand‐specific FISH. Methods Mol Biol 659: 173–183 [DOI] [PubMed] [Google Scholar]
- 54. Cukusic Kalajzic A, Vidacek NS, Huzak M, Ivankovic M, Rubelj I (2014) Telomere Q‐PNA‐FISH–reliable results from stochastic signals. PLoS ONE 9: e92559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sakellariou D, Chiourea M, Raftopoulou C, Gagos S (2013) Alternative lengthening of telomeres: recurrent cytogenetic aberrations and chromosome stability under extreme telomere dysfunction. Neoplasia 15: 1301–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Review Process File


