Abstract
We previously identified a novel cellular protein, RPB5-mediating protein (RMP), that retains corepressor activity and functionally antagonizes transcriptional modulation via hepatitis B virus X protein. The subcellular localization of RMP was examined using green fluorescent protein-fused protein forms. We found that a nuclear localization signal (NLS) and a coiled-coil (CC) domain functioning as a cytoplasmic localization signal (CLS) are important for the subcellular localization of RMP. The CLS apparently acts dominantly, since RMP was mostly localized in the cytoplasm with weak and diffuse signals in the nucleus, and the NLS was indispensable for the nuclear localization of RMP only in the absence of the CLS. Using a yeast two-hybrid method, we isolated a putative corepressor, DNA methyltransferase 1-associating protein (DMAP1), which was found to bind to the CC domain of RMP. DMAP1 facilitated the nuclear localization of RMP and the corepressor activity of RMP in a dose-dependent manner by interacting with the CC domain of RMP. These results are discussed in light of a recent paper showing a novel evolutionarily conserved role of URI in the TOR signaling pathway.
In eukaryotic cells, DNA-dependent RNA polymerases I, II, and III are highly conserved multisubunit enzymes responsible for the regulated expression of all genes (17, 24). The eukaryotic heteromeric enzyme RNA polymerase II consists of 12 different subunits (3, 28). Along with the well-documented regulatory role of the C-terminal domain of the largest subunit of RNA polymerase II (7, 8, 16), it is becoming increasingly apparent that other RNA polymerase II subunits are also targets for transcriptional regulation, determining interactions with either mediators-coactivators or transcriptional activators (2, 9, 12, 18, 19, 23, 25, 29).
Hepatitis B virus (HBV) X protein (HBx), a multifunctional viral regulator protein of HBV (13), directly interacts with RNA polymerase II subunit 5 (RPB5) and modulates activated transcription as a coactivator in vivo and in vitro (1). This observation suggested the notion that RPB5 is a communicating subunit of RNA polymerase II and that it interacts with transcriptional regulators (10, 11, 27). Consistent with this notion, a novel cellular protein, RPB5-mediating protein (RMP), was identified and selected by the far-Western cloning method (4). The specific binding of RMP and RPB5 requires the RPB5-binding region of RMP (amino acids [aa] 151 to 231) and the central part (or rather, the C terminus of the exposed domain) of RPB5, which overlaps with the HBx-binding region. RMP negatively modulates RNA polymerase II function in the absence of HBx and antagonizes the coactivator function of HBx by competitive binding to RPB5 (4, 11). RMP is composed of 508 aa residues. However, its cDNA might be reconstructed, since the 5′ sequence covering the first 25 aa is not from the sequence at the locus of the gene in human chromosome 19, and probably during cloning using a 5′ and 3′ rapid amplification of cDNA ends method due to the template switching.
Recently, a novel aspect of RMP suggested that RPB5 and RMP/URI, (unconventional prefoldin RPB5 interactor) are involved in a mammalian TOR (target of rapamycin) kinase signaling pathway that coordinates the regulation of nutrient availability with gene expression by interacting with STAP1 (SKP2-associating α-class prefoldin 1) (5). A version of RMP truncated at the N-terminal 50 aa has been reported to be the cytoplasmic protein NNX3 (26). We examined the subcellular localization of RMP using green fluorescent protein (GFP) and other tagging methods. Here, we report that RMP is mainly localized in the cytoplasm, with weak and diffuse signals in the nucleus, and that two localization signals, a nuclear localization signal (NLS) and a cytoplasmic localization signal (CLS), are important for the subcellular localization of RMP. Also, we report on the RMP-interacting partner, DNA methyltransferase 1 (DNMT1)-associating protein (DMAP1), a putative corepressor, which facilitates the exclusive nuclear localization of RMP and augments its corepressor activity.
MATERIALS AND METHODS
Plasmid construction.
The bacterial expression vectors pGENKS and pYFLAG and the mammalian expression vectors pNKFLAG and pNKGST were previously reported (1, 11). To generate pSG5UTPL-GFP, which drives the expression of GFP-fused protein forms in mammalian cells, GFP cDNA was prepared by PCR with phGFPS65T (Clontech Co. Ltd) as a template and was inserted into the NotI and EcoRI sites of pSG5UTPL (1). Sequences encoding full-length or truncated fragments of RMP were inserted into SacI or EcoRI sites and the BamHI sites of pSG5UTPL-GFP, pNKFLAG, pNKGST, and pGENKS. To construct internally deleted mutants of GFP-RMP, we used splicing PCR as previously described (4). For the construction of truncated RMP fused to TFIIB in GFP-fused forms (pGFP-RMP1-118-TFIIB and pGFP-RMP88-118-TFIIB), cDNA encoding full-length TFIIB was prepared by PCR with the forward primer 5′-TTGCTAGCCATGGCGTCTACCAGCCG-3′, containing an artificial NheI site, and the reverse primer 5′-GCGGATCCTTATAGCTGTGGTAGTTTGTC-3′, containing an artificial BglII site. The resultant fragment was inserted into the NheI and BglII sites of pGFP-RMP to generate pGFP-RMP1-118-TFIIB. Next, the EcoRI-NheI fragment of pGFP-RMP1-118-TFIIB was replaced by a DNA fragment encoding aa 88 to 118 with artificial EcoRI and NheI sites at the 5′ and 3′ ends to generate pGFP-RMP88-118-TFIIB. Full-length human DMAP1 cDNA was obtained by nested PCR using the Marathon liver cDNA library (Clontech Co. Ltd). The following primers were used: outside forward primer, 5′-ATCTCTCAGGCGCGATGGC-3′; outside reverse primer, 5′-ATAAGCTTATGGGATCGGCCATTG-3′; inside forward primer with artificial EcoRI and NcoI sites, 5′-ATGAATTCCATGGCTACGGGCGCGGAT-3′; inside reverse primer with artificial BamHI site, 5′-ATGGATCCCGGCTTCTTGGCTTTC-3′. The PCR product was cloned into the EcoRI and BamHI sites of the pNKGST and pGENKS plasmids. The luciferase reporter vector pFR-Luc was created by cloning the reporter gene sequence downstream of a basic promoter element (TATA box) and joining it to five tandem repeats of the GAL4 binding element (Stratagene). All constructs used in this study were sequenced by the dideoxy method using Taq sequencing kits and a DNA sequencer (catalog no. 370A; Applied Biosystems Inc.).
Yeast two-hybrid assays.
Yeast transformation and two-hybrid screening were performed as previously described (6, 30). Briefly, Saccharomyces cerevisiae strain PJ69-4A (GAL7-LacZ GAL1-HIS3 GAL2-ADE2) was used for yeast two-hybrid screening. Gal4 activation domain-fused cDNAs from a pACT library of human lymphocytes were used. A plasmid expressing RMP fused to the Gal4 DNA-binding domain was prepared by inserting full-length RMP cDNA into the yeast expression vector pAS1. The transformants were screened for growth on plates lacking tryptophan, leucine, and histidine and supplemented with 40 mM 3-amino-1,2,4-triazole at 27°C for 3 to 6 days. Histidine-positive colonies were further screened by β-galactosidase assays for positive interaction. Plasmids harboring cDNA were isolated from positive yeast colonies and transformed into Escherichia coli. The cDNAs of positive library plasmids were sequenced and compared to existing databases by using the BLAST algorithm.
Expression and purification of recombinant proteins.
Glutathione S-transferase (GST)-fused and FLAG-tagged proteins were expressed and purified as previously described (1, 10). Briefly, transformed bacterial cells (BL21) were harvested by centrifugation and suspended in PBST buffer A {phosphate-buffered saline without CaCl2 and MgCl2 [PBS(−)] containing 0.5% Triton X and 1 mM dithiothreitol (DTT)}. After centrifugation of the sonicated lysate, the supernatant was passed through DEAE-Sepharose, and GST-fused proteins were recovered with glutathione-Sepharose 4B beads (Amersham Biosciences, Inc.), while FLAG-tagged proteins were bound to anti-FLAG M2 resin (Sigma). The resin was washed, and the GST-fused or FLAG-tagged proteins were then eluted with glutathione or FLAG peptide, respectively. The eluted solution was dialyzed against buffer containing 100 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 1 mM DTT.
Preparation of cell extracts, coimmunoprecipitation, and Western blot analysis.
For in vivo immunoprecipitation assays, transfected COS1 cells were harvested and washed with PBS(−) and sonicated in lysis buffer (50 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 10 mM leupeptin, 10mM aprotinin, and 1 mM DTT). For each test tube, the total lysate derived from 2.5 × 105 cells was diluted 10-fold with lysis buffer containing 1% bovine serum albumin (BSA) and precleared by incubation with protein A-Sepharose resin (Amersham Pharmacia) for 1 h at 4°C. The precleared lysate was incubated with 10 μl of anti-FLAG M2 affinity resin for 2 h at 4°C. The resin was preblocked in lysis buffer containing 1% BSA. After an extensive wash with lysis buffer containing 300 mM NaCl, the bound proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes, and subjected to Western blot analysis with monoclonal anti-GST antibodies (Santa Cruz). For in vitro binding experiments, bacterially expressed and purified full-length FLAG-RMP or mammalian expressed and anti-FLAG M2 resin affinity-purified FLAG-RMP mutant proteins were mixed with bacterially expressed and affinity-purified GST-DMAP1. For one test tube, 200 ng of each purified protein was applied. The bound proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes, and subjected to Western blot analysis with anti-GST or anti-FLAG M2 antibodies. The proteins were visualized by enhanced chemiluminescence according to the manufacturer's instructions (Amersham Pharmacia Biotech).
Cell culture and transient transfection.
COS1 cells (a monkey kidney cell line), HepG2 cells, and HLE cells (a human hepatoma cell line) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 5% fetal calf serum (CELLect R GOLD) and 20 mg of antibiotics (ampicillin and kanamycin; Meigi Co., Ltd.)/ml and maintained in an incubator with 5% CO2 at 37°C. The cells were transfected by standard CaCl2-mediated transfection.
Immunofluorescence and confocal microscopy.
HLE cells grown on glass coverslips were washed once in PBS(−), fixed with 2% formaldehyde in PBS(−) for 30 min at room temperature (RT), permeabilized for 5 min with 100% cold methanol, and then dried at −25°C. GFP-fused proteins were detected after being counterstained with 0.0005% Evans Blue. For immunostaining, coverslips were blocked in 1.5% BSA in PBS(−) at RT for 1 h and incubated with primary antibodies in a humidifying chamber at RT for 1 h (1:300 monoclonal anti-GST antibody). The cells were washed five times with PBS(−) and incubated at RT for 1 h with secondary antibodies (1:30 goat anti-mouse immunoglobulin G [IgG]-Texas Red; Southern Biotechnology Associates, Inc.). Subsequently, the cells were washed five times in PBS(−) and mounted using Vectashield mounting medium. Immunofluorescent images were acquired using a confocal laser scanning microscope (LSM510; Carl Zeiss Co., Ltd).
Luciferase assay.
Approximately 105 HepG2 cells were plated in six-well tissue culture plates, and 1 day after being plated, they were cotransfected with plasmid cocktails. The total DNA concentration in the transfection cocktails was adjusted using plasmid pSG5UTPL. Forty-eight hours after transfection, luciferase and Renilla assays were performed using a Dual Luciferase Assay kit (Promega) according to the manufacturer's instructions. Luciferase activity was normalized against Renilla activity.
RESULTS
Subcellular localization of RMP and sequences important for its localization.
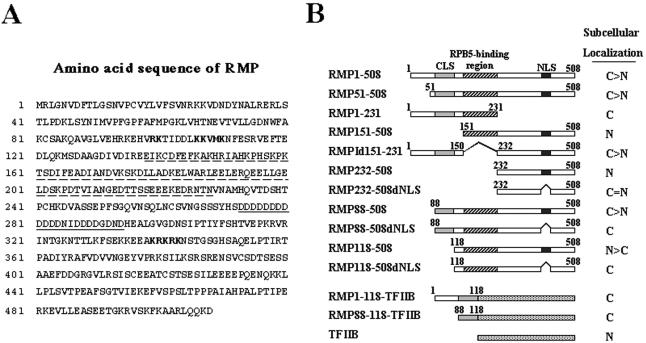
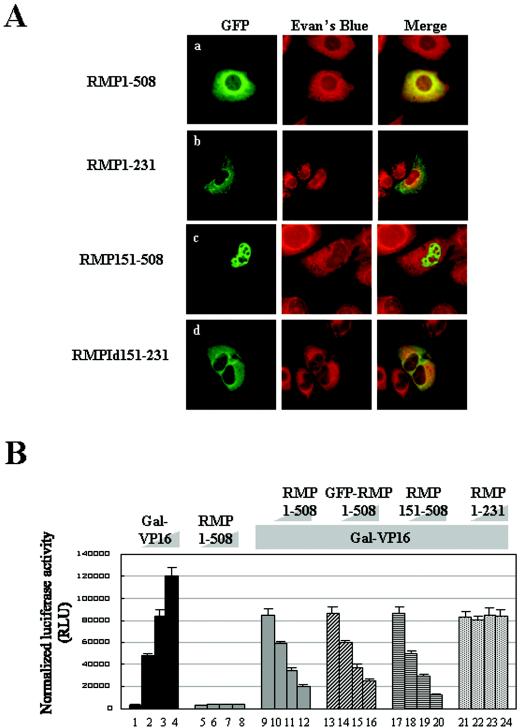
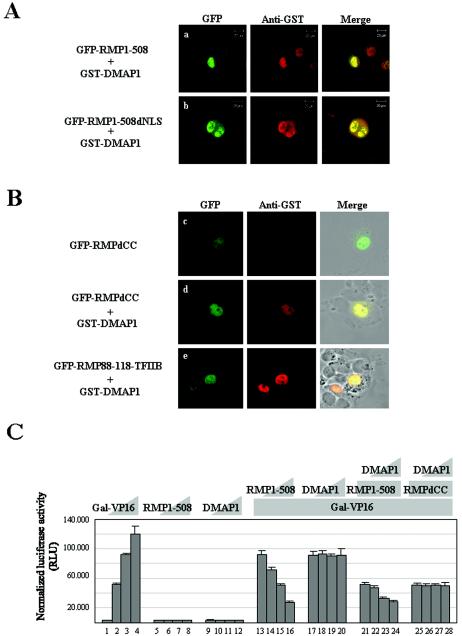
RMP harbors the RBP5-binding region (aa 151 to 231) and an Asp-rich sequence (aa 272 to 298), a coiled-coil (CC) domain (aa 88 to 188), and two different kinds of putative NLS sequences (aa 99 to 110 and 339 to 343) (Fig. 1A). To examine the subcellular localization of RMP, we constructed mammalian expression plasmids harboring different truncated versions of RMP in GFP-fused protein form (Fig. 1B). We also constructed, for comparison, RMP1-508 and RMP51-508, which is reported to be a shorter isoform of RMP (see Discussion). HLE cells were transiently transfected with GFP-RMP1-508 or a GFP-expressing plasmid, and the resulting GFP signals were observed by confocal microscopy. GFP-RMP1-508 signals were found mostly in the cytoplasm, but faint signals were reproducibly detected in the nucleus (Fig. 2A, row a). Similar distributions of GFP-RMP1-508 signals were observed in COS1 and Huh7 cells (data not shown).
FIG. 1.
Subcellular localization of RMP. (A) Amino acid sequence of RMP. The RPB5-binding region is underlined with a dashed line, the Asp-rich region is underlined, and two different putative NLSs are shown in boldface. (B) Schematic presentation of different truncated mutants of RMP in GFP-fused form. The subcellular localization of the fusion proteins elucidated by confocal microscopy is shown at the right (C, exclusively cytoplasmic; N, exclusively nuclear; >, mostly or strongly; =, equally). The numbers indicate the number of amino acids in a given construct.
FIG. 2.
Subcellular localizations and corepressor activities of RMP and its truncated mutants. (A) HLE cells transfected with the indicated GFP-RMP constructs were stained with Evans Blue to visualize the cell structure and were observed by confocal microscopy. The expression of GFP-RMP proteins was detected by green fluorescence. (B) Corepressor activity was addressed by a dual luciferase assay as described in Materials and Methods. Various amounts of Gal-VP16 and RMP constructs were transfected together with 200 ng of reporter luciferase and 20 ng of control luciferase constructs, respectively, for each well. The cotransfection mixture contained the following constructs: bars 1 to 4, 0, 0.2, 0.4, and 0.8 ng of Gal-VP16, respectively; bars 5 to 8, 0, 0.1, 0.5, and 1.0 μg of RMP1-508, respectively; bars 9 to 12 0, 0.1, 0.5, and 1.0 μg of RMP1-508, respectively, plus 0.4 ng of Gal-VP16; bars 13 to 16, 0, 0.5, 1.0, and 2.0 μg of GFP-RMP1-508, respectively, plus 0.4 ng of Gal-VP16; bars 17 to 20, 0, 0.1, 0.5, and 1.0 μg of RMP151-508, respectively, plus 0.4 ng of Gal-VP16; and bars 21 to 24, 0, 0.1, 0.5, and 1.0 μg of RMP1-231, respectively, plus 0.4 ng of Gal-VP16. The error bars indicate standard deviations.
To determine the regions or sequences responsible for the subcellular localization of RMP, two truncation mutants, RMP151-508 and RMP1-231, were examined using GFP-fused protein forms. GFP-RMP1-231 was exclusively localized in the cytoplasm; in contrast, GFP-RMP151-508 had exclusive nuclear localization (Fig. 2A, rows b and c). This suggests that there are at least two sequences, one responsible for cytoplasmic localization within aa 1 to 151 and the other responsible for nuclear localization within aa 151 to 508. The putative NLS sequence within aa 99 to 110 and the RPB5-binding region cannot work as an NLS, since GFP-RMP1-231 was exclusively localized in the cytoplasm. The signals associated with GFP-RMPId151-231 lacking the RPB5-binding region (Fig. 2A, row d) were similar to that for GFP-RMP1-508, supporting the hypothesis that the RPB5-binding region does not contribute much to the subcellular localization of RMP.
We examined the correlation between subcellular localization and the corepressor ability of RMP using these truncation mutants. GFP-RMP1-508 exhibited a corepressor activity similar to that of FLAG-RMP1-508 in the presence of Gal-VP16 with a luciferase reporter under the control of a Gal4 DNA-binding repeat acting as an enhancer (Fig. 2B, bars 9 to 16). On the other hand, RMP151-508 exhibited significantly higher repression activity than RMP1-508 did, while RMP1-231 containing an RPB5-binding region failed to repress transactivation by GAL-VP16. This result indicates that the corepressor activity might be localized in the C terminus, that the RPB5-binding region does not contribute much to the corepressor function in the presence of Gal-VP16, and that the RPB5-binding region itself has no effect on corepressor activity (Fig. 2B, bars 17 to 24).
The NLS at the C terminus and the CC domain as a CLS.
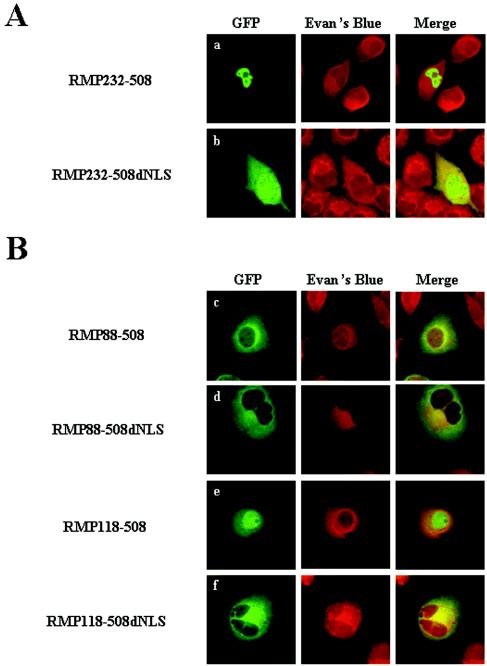
Since GFP-RMP151-508 was exclusively localized in the nucleus (Fig. 2A, row c), the contribution of the putative NLS located between aa 339 and 343 was evaluated by amino acid deletion and substitution. GFP-RMP232-508 was exclusively localized in the nucleus, but GFP-RMP232-508dNLS was distributed in the cytoplasm and nucleus (Fig. 3A, rows a and b). When the NLS was replaced with alanines, the same results as for GFP-RMP232-508dNLS were observed (data not shown). This indicates that aa 339 to 343 function as an NLS, although apparently it does not contribute much to the subcellular localization of RMP.
FIG. 3.
Mapping of CLS in an N-terminal region and the functional role of the NLS in the C terminus of RMP. HLE cells transfected with the indicated GFP-RMP constructs were fixed and counterstained with Evans Blue to visualize the cell structure and were observed by confocal microscopy. The expression of GFP-RMP proteins was detected by green fluorescence.
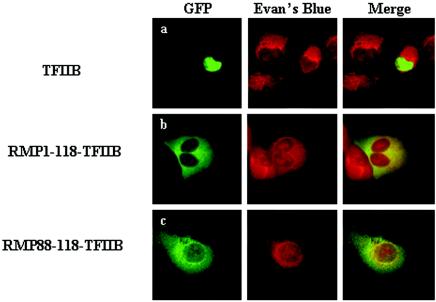
Truncation mutants were constructed to map the sequence(s) responsible for the cytoplasmic localization of RMP. GFP-RMP118-508, but not GFP-RMP88-508, resulted in mostly nuclear localization (Fig. 3B, rows c and e), and an NLS was required for nuclear localization in the latter construct (Fig. 3B, rows d and f). Two possibilities may explain the apparent masking effect of the N-terminal region, the region likely spanning aa 88 to 118: the region is necessary to mask the function of the NLS of RMP, or it is a strong CLS by itself. To differentiate between these possibilities, GFP-RMP1-118 or GFP-RMP88-118 was fused to TFIIB, a general transcription factor that is a nuclear protein (Fig. 4A, row a). The GFP signals were dramatically changed from the nucleus to the cytoplasm by inserting aa 1 to 118 or 88 to 118 into GFP-TFIIB (Fig. 4A, rows b and c). This clearly indicates that a small region covering aa 88 to 118 itself has the ability to inhibit the nuclear localization of TFIIB. The CLS almost overlapped the CC domain of RMP, indicating that the domain acts as a CLS.
FIG. 4.
The CC domain of RMP inhibits the nuclear localization of TFIIB. HLE cells were transfected with plasmids expressing GFP-TFIIB (a) and its chimeras fused to aa 1 to 118 (b) or 88 to 118 (c) of RMP. The fixed cells were stained with Evans Blue to visualize the cell structure and were observed by confocal microscopy. The expression of GFP-RMP proteins was detected by green fluorescence.
Identification of DMAP1 as an RMP-interacting protein.
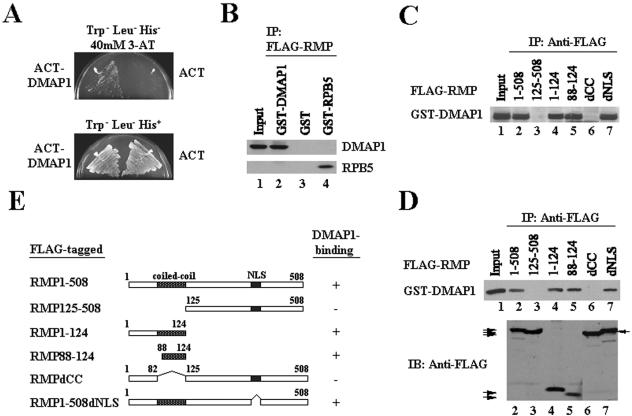
Since the CC domain is well known for homotypic or heteromeric interactions, the RMP domain may associate with a cytoplasmic factor(s) or facilitate the nuclear export signal function. Therefore, we searched for an RMP-interacting partner(s) using yeast two-hybrid screening (see Materials and Methods). Among the clones isolated (228 cDNA clones), more than half shared the middle portion of DMAP1 cDNA and ∼40% of the clones possessed the common N-terminal portion of RMP itself. DMAP1 is a putative corepressor and an associating factor with DNMT1, which is involved in gene silencing (20, 21, 22). The interaction between DMAP1 and RMP was confirmed in yeast cells expressing RMP and DMAP1 in the presence of 40 mM 3-amino triazole (Fig. 5A). The specific interaction was also demonstrated in mammalian cells transiently coexpressing differently tagged proteins (Fig. 5B, lanes 2 and 4). Consistent with the results in vivo, RMP immunoprecipitated DMAP1, demonstrating direct binding between RMP and DMAP1 in vitro with partially purified GST-DMAP1 and FLAG-RMP proteins (Fig. 5D, lane 2). These results are consistent with specific and direct interaction between RMP and DMAP1.
FIG. 5.
Interaction of RMP and DMAP1. (A) RMP interacts with DMAP1 in a yeast two-hybrid system. Transformed cells were plated on Trp− Leu− His− plates containing 40 mM 3-amino triazole (3-AT) or on Trp− Leu− His+ plates. (B) Interaction of RMP and DMAP1 in COS1 cells. The cells were cotransfected with plasmids expressing FLAG-RMP and GST-DMAP1. GST-RPB5 and GST were used as positive and negative controls, respectively. Total cell lysate was immunoprecipitated (IP) with anti-FLAG M2 antibody-bound resin, and the precipitated proteins were detected using anti-GST antibody. (C and D) Mapping the RMP region responsible for interaction with DMAP1 in vivo and in vitro. (C) A construct harboring GST-DMAP1 was cotransfected along with constructs of different FLAG-tagged truncation mutants of RMP, as indicated. COS1 cell lysate was immunoprecipitated using anti-FLAG M2 affinity resin. (D) Bacterially expressed and purified full-length FLAG-RMP or mammalian expressed and anti-FLAG M2 resin affinity-bound RMP mutants were mixed with bacterially expressed and affinity-purified GST-DMAP1. The lower part of the panel shows a comparable level of FLAG-RMP proteins applied in the binding assay (the arrows indicate the positions of the mutant proteins). (E) Schematic presentation of FLAG-RMP truncation mutants used for binding assays. The DMAP1-binding ability of the indicated protein is shown on the right. − and +, no binding and ability to bind to DMAP1, respectively.
Mapping of the RMP region responsible for interaction with DMAP1.
To further confirm the specific binding of DMAP1, the DMAP1-binding site in RMP was mapped using externally and internally truncated RMP proteins transiently coexpressed in the presence of GST-DMAP1 in COS1 cells. GST-DMAP1 was recovered only when RMP mutants harbored the CC domain (Fig. 5C, lanes 2, 4, 5, and 7), indicating that the CC domain is responsible for DMAP1 binding in vivo. A pull-down assay with affinity-purified FLAG-RMP mutant proteins expressed in mammalian cells and purified GST-DMAP1 confirmed that the CC domain of RMP is required for DMAP1 binding in vitro (Fig. 5D).
Homotypic interaction of RMP.
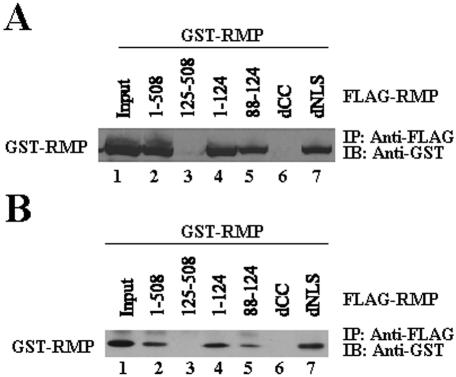
To confirm the homotypic interaction of RMP in mammalian cells, immunoprecipitation experiments were carried out with the lysates of COS1 cells transiently overexpressing FLAG-RMP and GST-RMP. The homotypic interaction of RMP demonstrated in vivo was consistent with the cloning results of the two-hybrid system (Fig. 6A). Homotypic interaction was demonstrated in vitro with partially purified FLAG-RMP and GST-RMP (Fig. 6B), indicating that the CC domain is responsible for the homotypic interaction of RMP in vivo and in vitro (Fig. 6).
FIG. 6.
RMP region responsible for homotypic interaction in vivo and in vitro. (A) A plasmid expressing GST-RMP was cotransfected with constructs of different FLAG-tagged truncation mutants of RMP, as indicated. Cell lysate was immunoprecipitated (IP) using anti-FLAG M2 affinity resin, and the bound GST-RMP was detected by anti-GST antibodies. IB, immunoblotting. (B) Bacterially expressed full-length FLAG-RMP or mammalian expressed and partially purified FLAG-RMP mutant proteins were mixed with bacterially expressed and affinity-purified GST-RMP. The protein complexes were precipitated by anti-FLAG M2 affinity resin, and bound proteins were detected using anti-GST antibodies.
DMAP1 facilitates the nuclear localization of RMP and augments the corepressor activity of RMP.
We investigated whether DMAP1 affects the subcellular localization and corepressor activity of RMP due to the specific interaction. The coexpression of DMAP1 changed the state of subcellular localization of RMP1-508 from mostly cytoplasmic to exclusively nuclear (Fig. 2A and Fig. 7A), Nuclear localization of GFP-DMAP1 was confirmed (data not shown) as previously reported (22). To further understand the role of DMAP1 in the subcellular localization of RMP, several mutant RMPs in GFP-fused forms were examined in the presence or absence of DMAP1. The signals of GFP-RMP1-508dNLS, lacking NLS, and those of GFP-RMP88-118-TFIIB, harboring the CC domain of RMP, were mostly localized in the nucleus when GST-DMAP1 was coexpressed (Fig. 7A and B). These GFP-fused RMP mutants were exclusively localized in the cytoplasm, as shown Fig. 4 (data not shown). In contrast, GST-DMAP1 could not significantly affect the subcellular distribution of GFP-RMPdCC, lacking the CC domain, which is exclusively nuclear even in the absence of GST-DMAP1 (Fig, 7B). Taken together, the data show that the CC domain by itself is responsible for the mostly cytoplasmic localization of RMP, and NLS functions if the CC domain is missing or occupied by DMAP1. This result strongly argues that the nuclear localization of RMP in the presence of DMAP1 is due in part to the CC domain and that the nuclear localization mechanism of DMAP1 might function when complexed with the CC domain of RMP.
FIG. 7.
DMAP1 facilitates the nuclear localization of RMP and augments corepressor activity of RMP. (A and B) HLE cells were transiently cotransfected with a plasmid expressing GST-DMAP1, along with constructs containing cDNAs of full-length GFP-RMP1-508 (a), GFP-RMP1-508dNLS mutants (b), GFP-RMPdCC (d), and GFP-RMP88-118-TFIIB (e), or HLE cells were transiently transfected with a plasmid expressing GFP-RMPdCC (c). The cells were stained with monoclonal anti-GST antibodies and visualized by Texas Red-linked goat-anti mouse IgGs. The expression of GFP-fused proteins was detected by green fluorescence. (C) A luciferase assay was performed as for Fig. 2B. The cotransfection mixture contained the following constructs: bars 1 to 4, 0, 0.2, 0.4, and 0.8 ng of Gal-VP16, respectively; bars 5 to 8, 0, 0.1, 0.5, and 1.0 μg of RMP1-508, respectively; bars 9 to 12, 0, 0.5, 1.0, and 2.0 μg of DMAP1, respectively; bars 13 to 16, 0, 0.1, 0.5, and 1.0 μg of RMP1-508, respectively, plus 0.4 ng of Gal-VP16; bars 17 to 20, 0, 0.5, 1.0, and 2.0 μg of DMAP1, respectively, plus 0.4 ng of Gal-VP16; bars 21 to 24, 0, 0.5, 1.0, and 2.0 μg of DMAP1, respectively, plus 0.4 ng of Gal-VP16 and 0.5 μg of RMP1-508; bars 25 to 28, 0, 0.5, 1.0, and 2.0 μg of DMAP1, respectively, plus 0.4 ng of Gal-VP16 and 0.5 μg of RMPdCC. The error bars indicate standard deviations.
Next, we examined whether the corepressor activity of RMP was affected by DMAP1. The effect of RMP on Gal-VP16-driven transcriptional activation was examined in the presence or absence of DMAP1 (see Materials and Methods). RMP1-508 exhibited an inhibitory effect on Gal-VP16-driven transactivation in a dose-dependent manner (4) (Fig. 7C, bars 13 to 16) but did not have a detectable effect on the transcription of a reporter gene in the absence of Gal-VP16 (Fig. 7C, bars 5 to 8). DMAP1 exhibited no effect on the transcriptional activation of Gal-VP16 (Fig. 7C, bars 17 to 20) but augmented the corepressor activity of RMP1-508 in a dose-dependent manner (Fig. 7C, bars 21 to 24). In contrast, DMAP1 had no effect on the corepressor activity of RMPdCC (Fig. 7C, bars 25 to 28). Taken together, these results indicate that DMAP1 facilitates the nuclear localization and corepressor activities of RMP, probably through direct binding to the CC domain of RMP.
DISCUSSION
RPB5 is a common subunit of RNA polymerases I, II, and III. The crystal model of yeast RNA polymerase II shows that RPB5 is part of the lower jaw and that the N-terminal two-thirds of RPB5 is exposed and positioned close to DNA downstream of the initiation site (3). We identified human RPB5 to be a nuclear target of HBx, a multifunctional viral regulator of HBV which acts as a transcriptional coactivator in vivo and in vitro through direct interaction with the central part of RPB5 (10, 13, 14). The putative role of human RPB5 in activated transcription is consistent with the reported role of yeast RPB5 in the activated transcription of some promoters (12). The exposed domain of RPB5 seems to serve as a communicating surface, not only with HBx but also with factors involved in transcription, such as TFIIF subunit RAP30 and TFIIB (10, 27).
RMP was isolated during the search to find transcriptional modulators that interact with RPB5 (4) to understand the transcriptional modulation mechanism of HBx. Since RMP exhibits corepressor activity in the absence of HBx and antagonizes the coactivator activity of HBx by competitive binding to RPB5, we designated it RPB5-mediating protein (RMP) (4). However, its cDNA is reconstructed probably during cloning using a 5′ and 3′ rapid amplification of cDNA ends method due to template switching, since the 5′ sequence covering the first 25 aa is not from the sequence at the locus of the gene in human chromosome 19. RMP is distinct from the mediating protein families of the homoenzyme (15), and the molar ratio of RMP to RPB5 was <0.1 under normal growth conditions (unpublished data). The corepressor activity of RMP is rather weak, and cytoplasmic localization has been reported for a shorter isoform (aa 51 to 508). In this study, we report that RMP is mainly localized in the cytoplasm, accompanied by weak and diffuse nuclear localization. We describe the two elements, the NLS and the CC domain of RMP, that are important for subcellular localization. We isolated DMAP1, a putative interacting partner of RMP that facilitates the nuclear localization of RMP by interacting with the CC domain. DMAP1 is also a putative corepressor and associating factor of DNMT1, which is involved in gene silencing (20, 21, 22). DMAP1 augmented the corepressor activity of RMP, while DMAP1 had no effect on transcriptional activation driven by Gal-VP16 in the absence of RMP. The augmentation of the corepressor activity of RMP by DMAP1 might be due to efficient nuclear localization by DMAP1, but our preliminary results suggested that DMAP1 may play another role in transcriptional regulation, since it was recruited to the promoter by RMP (data not shown). The interaction of DMAP1 and RMP may facilitate the recruitment of DNMT1 in promoter regions that are important for the negative regulation of transcription by modifying chromatin structure. Such a possibility remains to be addressed. Homotypic interaction of RMP through the CC domain was also observed but did not affect the mostly cytoplasmic localization of RMP. Similar results were observed with RMP51-508 (or NNX3) (data not shown), the shorter isoform of RMP (26).
During the preparation of this study, Gstaiger et al. reported a cDNA encoding 534 aa, called URI, which overlaps aa 26 to 508 of RMP. Human URI comprises α-class prefoldin (PFD) in aa 1 to 154, the RPB5 interaction region, a long acidic sequence, and a short conserved C-terminal sequence, all of which are evolutionarily conserved among eukaryotes (reference 5 and its supplemental data). Their findings show that URI/RMP is a negative transcriptional regulator under the control of TOR kinase and that URI/RMP occurs as a complex with STAP1, which is a member of the α-class PFDs; two other low-molecular-weight β-class PFDs; and RPB5. Interestingly, immunoaffinity purification using anti-STAP1 IgG allowed the recovery of not only URI/RMP, but also RPB5. However, the other RNA polymerase subunits seem not to be present (Fig. 1 of reference 5). The result strongly suggests multiple roles of RPB5 assembled in RNA polymerases and in complex with STAP1, implying a mechanism for regulating the dynamic distribution of RPB5 in the two complexes. The subcellular localization of the complex including URI/RMP and RPB5 remains unresolved. Yeast URI (Bud27p) has been documented as a cytoplasmic protein in the Saccharomyces Genome Database (SGD) (http://www.yeastgenome.org/). We demonstrated here that the RPB5-binding region does not contribute to the nuclear localization of RMP (Fig. 2A). Our preliminary confocal result using GFP-RPB5 showed that RPB5 is localized in the nucleus and cytoplasm and that the overexpression of RPB5 could not induce the nuclear localization of RMP (data not shown).
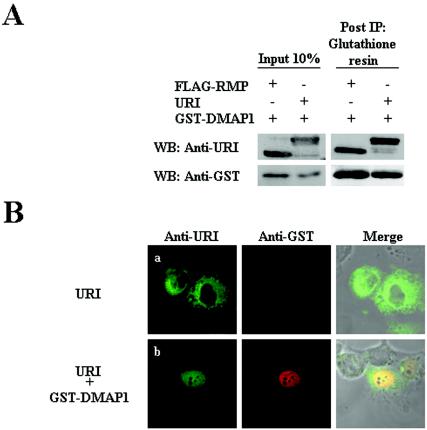
Since the RMP construct harbors a short stretch of the N-terminal CC, β-sheet, and C-terminal CC regions of PFD, there is a possibility that RMP, a truncated form of the full-length URI, but not URI may interact with DMAP1. To address this possibility, the subcellular localization of human URI (kindly provided by M. Gstaiger and W. Krek) was addressed in the presence and absence of DMAP1. URI was mainly localized in the cytoplasm in the absence of coexpression of DMAP1, and URI interacted with GST-DMAP1 in vivo, changing the subcellular localization of URI from the cytoplasm to the nucleus (Fig. 8). Therefore, the present observations with RMP reflect the properties of URI, although with the difference that diffuse signals of URI, but not RMP, seem to remain in the cytoplasm in the presence of DMAP1. Taken together, our results show that RMP/URI is a distinct protein in the PDF family, since it has dual roles in the cytoplasm and the nucleus and its subcellular localization is regulated by interacting partners. The previously reported roles of RPB5 in activating transcription and transcriptional modulation by HBx may need to be reevaluated, considering the new role of RPB5 that is not assembled in RNA polymerases (10, 12).
FIG. 8.
URI can interact with DMAP1 that affects subcellular localization of URI. (A) COS 1 cells were cotransfected with plasmids expressing FLAG-RMP and GST-DMAP1 or with plasmids expressing URI and GST-DMAP1. Total lysates of the cells were immunoprecipitated (IP) using glutathione-Sepharose 4B resin. The coprecipitated proteins were detected by Western blotting (WB) with a mouse monoclonal antibody. The presence or absence of each protein in cotransfected lysates is shown as + or −, respectively. (B) HLE cells were transfected with expression vectors encoding URI (a) or URI and GST-DMAP1 (b). URI proteins were detected with mouse monoclonal anti-URI antibodies and visualized by fluorescein isothiocyanate-conjugated goat anti-mouse antibodies. GST-DMAP1 was visualized with rabbit polyclonal anti-GST antibodies and Texas Red-conjugated goat anti-rabbit IgG secondary antibodies.
Acknowledgments
We are grateful to M. Gstaiger and W. Krek for their generous gift of full-length cDNA of the URI protein and mouse monoclonal anti-URI antibodies. We also thank Y. Hirose for providing the pFR-Luc plasmid, T. Ito for supplying yeast strain PJ69-4A, and members of the laboratory for their helpful comments and constructive discussions. We thank F. Momoshima, K. Kuwabara, and M. Yasukawa for their technical assistance.
This work was supported in part by Grant-in-aid for Scientific Research category B no. 111480200 and Grant-in-aid for Scientific Research on Priority Areas Cancer, Mechanism of Oncogenesis and Anti-oncogenesis no. 12213050 from the Ministry of Education, Sports, Culture and Technology of Japan.
REFERENCES
- 1.Cheong, J. H., M. Yi, Y. Lin, and S. Murakami. 1995. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 14:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung, W.-H., J. L. Craighead, W.-H. Chang, C. Ezeokonkwo, A. Bareket-Samish, R. D. Kornberg, and F. J. Asturias. 2003. RNA polymerase II/TFIIF structure and conserved organization of the initiation complex. Mol. Cell 12:1003-1013. [DOI] [PubMed] [Google Scholar]
- 3.Cramer, P. D., A. Bushnell, J. Fu, A. L. Gnatt, B. Maier-Davis, N. E. Thompson, R. R. Burgess, A. M. Edwards, P. R. David, and R. D. Kornberg. 2000. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288:640-649. [DOI] [PubMed] [Google Scholar]
- 4.Dorjsuren, D., Y. Lin, W. Wei, T. Yamashita, T. Nomura, N. Hayashi, and S. Murakami. 1998. RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Mol. Cell. Biol. 18:7546-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gstaiger, M., B. Luke, D. Hess, E. J. Oakeley, C. Wirbelauer, M. Blondel, M. Vigneron, M. Peter, and W. Krek. 2003. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302:1208-1212. [DOI] [PubMed] [Google Scholar]
- 6.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim, Y. J., S. Bjorklund, M. Li, H. Sayer, and R. D. Kornberg. 1994. A multiprotein complex mediator of transcriptional activation and its interaction with C-terminal domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 8.Koleske, A. J., and R. A. Young. 1994. An RNA polymerase II holoenzyme responsive to activators. Nature 368:466-469. [DOI] [PubMed] [Google Scholar]
- 9.Lewis, B. A., and D. Reinberg. 2003. The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 116:3667-3675. [DOI] [PubMed] [Google Scholar]
- 10.Lin, Y., H. Tang, T. Nomura, D. Dorjsuren, N. Hayashi, W. Wei, T. Ohta, R. Roeder, and S. Murakami. 1998. The hepatitis B virus X protein is a co-activator of activated transcription that modulates the transcription machinery and distal binding activators. J. Biol. Chem. 273:27097-27103. [DOI] [PubMed] [Google Scholar]
- 11.Lin, Y., T. Nomura, J. H. Cheong, D. Dorjsuren, K. Iida, and S. Murakami. 1997. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcriptional factor IIB and RNA polymerase II subunit 5. J. Biol. Chem. 272:7132-7139. [DOI] [PubMed] [Google Scholar]
- 12.Miyao, T., and N. A. Woychik. 1998. RNA polymerase subunit RPB5 plays a role in transcriptional activation. Proc. Natl. Acad. Sci. USA 95:15281-15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakami, S. 2001. Hepatitis B virus X protein: multifunctional viral regulator. J. Gastroenterol. 36:651-660. [DOI] [PubMed] [Google Scholar]
- 14.Murakami, S. 1999. Hepatitis B virus X protein: structure, function and biology. Intervirology 42:81-99. [DOI] [PubMed] [Google Scholar]
- 15.Naar, A. M., B. D. Lemon, and R. Tjian. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70:475-501. [DOI] [PubMed] [Google Scholar]
- 16.Naar, A. M., D. J. Taatjes, W. Zhai, E. Nogales, and R. Tjian. 2002. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 16:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 18.Petermann, R., B. M. Mossier, D. N. Aryee, V. Khazak, E. A. Golemis, and H. Kovar. 1998. Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene 17:603-610. [DOI] [PubMed] [Google Scholar]
- 19.Pillai, B., V. Sampath, N. Sharma, and P. Sadhale. 2001. Rpb4, a non-essential subunit of core RNA polymerase II of Saccharomyces cerevisiae is important for activated transcription of a subset of genes. J. Biol. Chem. 276:30641-30647. [DOI] [PubMed] [Google Scholar]
- 20.Robert, M. F., S. Morin, N. Beaulieu, F. Gauthier, I. C. Chute, A. Barsalou, and A. R. Macleod. 2003. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 33:61-65. [DOI] [PubMed] [Google Scholar]
- 21.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 22.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 23.Schlegel, B. P., V. J. Green, J. A. Ladias, and J. D. Parvin. 2000. BRCA1 interaction with RNA polymerase II reveals a role for hRPB2 and hRPB10α in activated transcription. Proc. Natl. Acad. Sci. USA 97:3148-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sentenac, A. 1985. Eukaryotic RNA polymerases. Crit. Rev. Biochem. 18:31-91. [DOI] [PubMed] [Google Scholar]
- 25.Tan, Q., K. L. Linask, R. H. Ebright, and N. A. Woychik. 2000. Activation mutants in yeast RNA polymerase II subunit RPB3 provide evidence for a structurally conserved surface required for activation in eukaryotes and bacteria. Genes Dev. 14:339-348. [PMC free article] [PubMed] [Google Scholar]
- 26.Van Leuven., F., S. Torrekens, D. Moechars, C. Hilliker, M. Buellens, M. Bollen, and J. Delabie. 1998. Molecular cloning of a gene on chromosome 19q12 coding for a novel intracellular protein: analysis of expression in human and mouse tissues and in human tumor cells, particularly Reed-Sternberg cells in Hodgkin disease. Genomics 54:511-520. [DOI] [PubMed] [Google Scholar]
- 27.Wei, W., D. Dorjsuren, Y. Lin, W. Qin, T. Nomura, N. Hayashi, and S. Murakami. 2001. Direct interaction between the subunit RAP30 of transcription factor IIF (TFIIF) and RNA polymerase subunit 5, which contributes to the association between TFIIF and RNA polymerase II. J. Biol. Chem. 276:12266-12273. [DOI] [PubMed] [Google Scholar]
- 28.Woychik, N. A., and M. Hampsey. 2002. The RNA polymerase II machinery: structure illuminates function. Cell 108:453-463. [DOI] [PubMed] [Google Scholar]
- 29.Wu, S. Y., T. Zhou, and C.-M. Chiang. 2003. Human mediator enhances activator-facilitated recruitment of RNA polymerase II and promoter recognition by TATA-binding protein (TBP) independently of TBP-associated factors. Mol. Cell. Biol. 23:6229-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokoyama, N., N. Hayashi, T. Seki, N. Pante, T. Ohba, K. Nishii, K. Kuma, T. Hayashida, T. Miyata, U. Aebi, M. Fukui, and T. Nishimoto. 1995. A giant nucleopore protein that binds Ran/TC4. Nature 376:184-188. [DOI] [PubMed] [Google Scholar]