Abstract
The expression status of the estrogen receptor alpha (ERα) and that of the epidermal growth factor receptor Her-2/neu frequently correlate inversely in breast cancers. While ERα-dependent cancers respond to antiestrogen therapy, Her-2/neu-overexpressing cancers typically display resistance to antiestrogens and poor prognosis. In this report we have explored the mechanism linking the loss of expression of ERα in breast cancer cells with overexpression of Her-2/neu, which signals constitutively via a phosphatidylinositol 3-kinase (PI3K)/Akt kinase pathway. We identify for the first time the Forkhead box protein FOXO3a (formerly termed FKHRL-1), which is inactivated by Akt, as a key regulator of ERα gene transcription. In breast cancer cell lines, expression of ERα was correlated with active FOXO3a levels. Ectopic FOXO3a expression induced ERα protein levels and promoter activity, while a dominant negative FOXO3a decreased ERα levels. By using transient transfection, mobility shift assays, and site-directed mutagenesis, two major functional Forkhead binding sites were identified in the human ERα promoter B. A chromatin immunoprecipitation assay confirmed FOXO3a binding at these two sites. Ectopic FOXO3a induced estrogen response element-driven reporter activity and expression of ERα target genes. The constitutively activated myristylated Akt reduced ERα expression, whereas agents that negatively affect the PI3K/Akt pathway, i.e., wortmannin, celecoxib, and the green tea polyphenol epigallocatechin-3 gallate, induced ERα. Thus, FOXO3a represents an important intracellular mediator of ERα expression, suggesting possible therapeutic intervention strategies for Her-2/neu-overexpressing refractory breast tumors.
Steroid hormones such as estrogen play important roles in breast cancer development. Most responses appear to be mediated through estrogen receptor alpha (ERα) and ERβ (10), although estrogen can elicit physiological events independent of ER (72, 77). More than 60% of human breast cancers are ERα positive (31). The presence of ERα is considered to be a good prognostic factor and correlates with a higher degree of differentiation of the tumor (41, 44) and increased disease-free survival (49). ERα positivity is also the major guideline for antiestrogen therapy and is a major target for selective ER modulators (51). In breast cancers, an inverse correlation has been noted between ERα status and the expression levels of epithelial growth factor receptor (EGFR) family members, including EGFR and Her-2/neu (1, 7, 36, 52, 69). Thus, breast cancers expressing high levels of EGFR and/or Her-2/neu usually express undetectable or very low levels of ERα (reviewed in reference 14). The cellular and molecular events that regulate ERα protein expression are poorly understood. Allelotyping studies indicate that physical loss of the ERα gene is not the main cause for the lack of expression in ERα-negative cells (57). ERα expression can also be regulated through epigenetic modification, e.g., methylation at the promoter, which has been reported to be responsible for the loss of ERα in a few but not most breast cancer cells (27, 76).
The EGFRs are a family of tyrosine kinases, which are generally activated by peptide ligand binding and subsequent receptor dimerization and tyrosine phosphorylation on the cytoplasmic tail (5). The family consists of the EGFR gene ERBB (HER1), ERBB2/HER2/neu, ERBB3/HER3, and ERBB4/HER4, with EGFR and Her-2/neu being overexpressed in a wide variety of tumors. Overexpression is sufficient to activate the ERBB2/HER2/neu receptor (13, 26), which has been seen in approximately 30% of breast cancers and is associated with poor prognosis and overall survival (24). In particular, it has been found associated with increased metastatic potential and resistance to chemotherapeutic agents (16). Her-2/neu receptor activation, via dimerization with other EGFR family members or itself, can lead to the activation of the phosphatidylinositol 3-kinase (PI3K) and thereby to the activation of the serine/threonine kinase Akt/protein kinase B (22, 66). Activated Akt is known to phosphorylate specific targets which promote survival (3, 34). Recent work has identified as targets of Akt the three members of the Forkhead box, group O subfamily Forkhead transcription factors FOXO1a, FOXO3a, and FOXO4, which were previously known as FKHR, FKHR-L1, and AFX, respectively (3). The activity of the mammalian Forkhead orthologues is controlled by this phosphorylation, i.e., Akt-phosphorylated Forkhead proteins bind to 14-3-3 protein and are transported to the cytoplasm (3). When hypophosphorylated, Forkhead proteins are released from 14-3-3 and translocate into the nucleus, where they transactivate target genes (43, 61, 68). Interestingly, treatment of ER-negative MDA-MB-231 breast cancer cells with the activating ligand EGF led to nuclear exclusion of Forkhead protein FKHR (28).
Given the inverse correlation between Her-2/neu and ERα status, the presence of a pathway connecting Her-2/neu and ERα expression seemed plausible, and several attempts have been made to delineate this pathway. It has been shown that overexpression of Her-2/neu converts estrogen-dependent tumor cells to an antiestrogen insensitive phenotype with decreased ERα expression (14, 38, 42, 55). Furthermore, treatment of ERα-positive MCF-7 cells with ErbB-activating ligands resulted in a substantial decrease in ERα mRNA and protein (59). The regulation of ERα expression by the PI3K/Akt pathway was also suggested by the correlation between PTEN, which suppresses PI3K activity, and ERα. Increased consumption of green tea has been closely associated with decreased numbers of axillary lymph node metastases among premenopausal patients with stage I or II breast cancer and with increased expression of ERα and progesterone receptor among postmenopausal women (45). Interestingly, our group has recently shown that green tea polyphenol epigallocatechin-3 gallate (EGCG) treatment of breast cancer cells inhibits Her-2/neu-mediated signaling via the PI3K/Akt pathway (54). Together these findings led us to hypothesize that the ERα gene is a downstream target of a Forkhead box transcription factor and thus subject to inhibition via a Her-2/neu-mediated PI3K to Akt kinase signaling pathway. Here we demonstrate that FOXO3a binding to two newly identified Forkhead sites upstream of ERα promoter B directly regulates promoter activity, leading to increased functional ERα levels. Treatment with agents that inhibit Her-2/neu to Akt kinase signaling, which enhances FOXO3a activity, elevate the level of ERα expression.
MATERIALS AND METHODS
Cell growth and treatment conditions.
The NF639 cell line (kindly provided by P. Leder, Harvard Medical School, Boston, Mass.) was derived from a mammary gland tumor in a mouse mammary tumor virus (MMTV)-Her-2/neu transgenic mouse and cultured as described previously (17). ERα-positive MCF-7 and ZR-75 cells and ER-negative Hs578T and MDA-MB-231 cells were purchased from the American Type Culture Collection (ATCC) and maintained in standard culturing medium as recommended by the ATCC. EGCG was purchased from LKT Laboratories Inc. Ponceau S and wortmannin were purchased from Sigma. Celecoxib was kindly provided by C.-S. Chen (Ohio State University, Columbus, Ohio).
Immunoblot analysis.
Whole-cell extracts (WCE) were isolated in radioimmunoprecipitation assay (RIPA) buffer supplemented with phosphatase inhibitors and sonicated for 3 s as described elsewhere (50, 54). For isolation of nuclei and cytoplasm, cells were incubated in hypotonic buffer and lysed and nuclei were collected by centrifugation, as described previously (30). The nuclear proteins were extracted with RIPA buffer for 15 min on ice. Protein samples (30 to 50 μg) were separated by electrophoresis in an 8 or 10% polyacrylamide-sodium dodecyl sulfate (SDS) gel and subjected to immunoblotting, as described elsewhere (53). Antibodies used included those for ERα (NeoMarker), anti-FOXO3a (Upstate Biotechnology), which recognizes both phosphorylated and hypophosphorylated FOXO3a, β-actin (Sigma), and RARα (Affinity BioReagents).
EMSA.
For nuclear extract preparation, nuclei were isolated and proteins were extracted using detergent lysis buffer as described previously (4). Oligonucleotides were prepared with BamHI restriction sites at the ends when used as probes or for cloning (15). The sequence of the FOXO3a binding site-containing oligonucleotide from the insulin-like growth factor binding protein IGFBP-1 gene was 5′-ATTGCTAGCAAGCAAAACAAACCGCTAGCTTA-3′ (termed insulin-responsive sequence [IRS]) (3), with the letters in bold indicating bases changed to G residues in the mutant form. The putative FOXO3a binding sites upstream of the ERα promoter were as follows: site 1 (−3160 bp), 5′-ACTGGATATAAATAAATATTGAAAAG-3′; site 4 (−2610 bp), 5′-ACTGCTTTCTGTAAACATGTGAAAAAT-3′, where the T and A residues in bold were mutated to C and T, respectively. Additional primers were site 2 (−3140 bp), 5′-CAGTATTGAAAATAAATACTGGATAT-3′, and site 3 (−3035 bp), 5′-TAGAAAGCCATAAAAATGTTAATGAT-3′. The Octomer-1 (Oct-1) sequence was 5′-TGTCGAATGCAAATCACTAGAA-3′. Oligonucleotides were labeled and used in an electrophoretic mobility shift assay (EMSA), as described previously (53). Where indicated, ∼500 ng of glutathione S-transferase (GST)-FOXO3a fusion protein or GST alone (kindly provided by S. Jeay, Boston University Medical School, Boston, Mass.) was added.
Plasmid constructs and site-directed mutagenesis.
The 3.9-kb human ERα promoter in pGL2Basic was kindly provided by Ronald Weigle (Stanford University School of Medicine, Stanford, Calif.) (12). Two shorter promoters, proA and proB, in pGL3Basic were kindly provided by Shin-Ichi Hayashi (Saitama Cancer Center Research Institute, Saitama, Japan) (71, 76). The expression vectors for wild-type (WT) FOXO3a, a constitutively active A3 FOXO3a mutant, and parental pECE vector were kindly provided by Michael Greenberg (Harvard Medical School) (3). In the A3 mutant, three sites of Akt phosphorylation of FOXO3a (T32, S253, and S315) were mutated to alanine residues. The AktM expression vector was generously provided by Z. Luo (Boston University School of Medicine). Mutagenesis was performed using either the QuikChange site-directed mutagenesis kit or the double priming method with DNA polymerase pfuTurbo (both from Stratagene) and confirmed by sequencing (Genetic Core Facility, Boston University Medical School). The primers used to make the mutations harbored the same mutations indicated above, with BamHI sequences omitted. For the construction of element-driven chloramphenicol acetyltransferase (CAT) reporter constructs, two copies of either WT or mutant forms of oligonucleotides corresponding to sites 1 and 4 were ligated into the BamHI site of the pBLCAT2 vector (no. 37527; ATCC), upstream of the thymidine kinase (TK) promoter.
Transfection and infection analyses.
NF639 cells were plated at 30% confluence 1 day prior to transfection into six-well or P100 tissue culture dishes for reporter assays or Western blot analysis, respectively. For reporter assays, cells were transfected in triplicate with the indicated amounts of DNA using FuGENE6 transfection reagent (Roche Diagnostics Corporation). After 30 to 48 h, cells were harvested and processed for reporter assays as described previously (33). Values are presented as the average ± the standard deviation (SD). For Western blot analysis, cells were harvested in RIPA buffer and processed as described above. MCF-7 and T47D cells were plated at 60% confluence, transfected using the GenePorter2 reagent (Gene Therapy Systems), and harvested with RIPA buffer as described above. For adenoviral infection, virus particles prepared as described previously (64) were added with an increasing multiplicity of infection to MCF-7 cells plated the previous day at a density of 5 × 105 cells/six-well dish, and cells were harvested after 40 h, as described above.
ChIP and semiquantitative PCR.
Cells (107) were fixed with 1% formaldehyde, pelleted, washed, and resuspended in 400 μl of SDS lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-Cl [pH 8.0], with protease inhibitors), as described elsewhere (40). After 10 min of incubation on ice, 600 μl of chromatin immunoprecipitation (ChIP) dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.0], and 16.7 mM NaCl, with protease inhibitors) was added, and genomic DNA was sheared by sonication to an average size of ∼500 bp. After removing 5% of the solution for evaluation of input complex, the lysates were precleared with 30 μl of salmon sperm DNA-protein A agarose beads (Upstate) for 1 h at 4°C, divided into two equal parts, and immunoprecipitated overnight in the cold using 4 μg of antibody against either FOXO3a or control rabbit immunoglobulin G (IgG). The immunocomplexes were collected using 30 μl of salmon sperm DNA-protein A agarose beads, washed sequentially with ChIP wash buffer 1 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 150 mM NaCl), ChIP wash buffer 2 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 500 mM NaCl), ChIP wash buffer 3 (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris; pH 8.0), and twice with TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA; pH 8.0). DNA-protein complexes were collected in ChIP elution buffer (1% SDS, 0.1 M NaHCO3) and disrupted by incubation at 65°C for 5 h in the presence of 312 mM NaCl and 0.06 μg of RNAse A/μl. Proteins were removed by overnight digestion with proteinase K at 37°C, and DNA fragments were purified by phenol-chloroform extraction and ethanol precipitation and analyzed by semiquantitative PCR. Primers used for the amplification of the S1/S4 FOXO3a site of ERα were (−2960 bp) 5′-CAGAGACCGGCCACTCCTG-3′ and (−2870 bp) 5′-GACACCCAATGGAGGCTTTGT-3′ (annealing at 70°C; 30 cycles). The control internal ERα primers were (−700 bp) 5′-GTCAGGCTGAGAGAATCTCAGA-3′ and (−610 bp) 5′-CTGAGGTCCTGGCAGGTTGC-3′ (annealing at 62°C; 32 cycles). The products were then resolved on a polyacrylamide gel and visualized using GelStar stain (Cambrex Inc.), as described in reference 39.
RESULTS
The abundance of hypophosphorylated nuclear FOXO3a correlates with ERα expression.
To assess whether activation of the Forkhead box protein correlates with the ERα status in breast cancer cells, we selected the human MCF-7 and ZR-75 cell lines (ERα positive) and Hs578T and MDA-MB-231 cells (ERα negative), as well as ERα-positive NF639 cells, which were derived from an MMTV-Her-2/neu transgenic mouse tumor (17, 53, 54). Western blot analysis confirmed ERα status. ERα protein levels were abundant in MCF-7 and ZR-75 cells, substantially lower in NF639 cells, and undetectable in Hs578T and MDA-MB-231 cells (Fig. 1A). Next, Western blot analysis was performed to assess FOXO1a, FOXO3a, and FOXO4 proteins. Using a FOXO3a antibody that recognizes both phosphorylated and hypophosphorylated protein, the ERα-positive MCF-7, ZR-75, and NF639 cells were all positive for nuclear FOXO3a, although the level in NF639 was again substantially lower. ERα-negative Hs578T and MDA-MB-231 cells were essentially negative for nuclear FOXO3a. Interestingly, an abundant level of faster-migrating, presumably active hypophosphorylated FOXO3a protein was seen in the WCE of the human MCF-7 and ZR-75 lines, while that of mouse NF639 cells displayed two bands, both of which appeared to migrate somewhat slower than the band seen in MCF-7 or ZR-75 cells (Fig. 1A). Treatment of the protein extract with protein phosphatase λ resulted in loss of the upper band and a shift to the faster-migrating band (data not shown). The existence of some hyperphosphorylated form of FOXO3a in NF639 cells is consistent with the activity of Her-2/neu/PI3K/Akt activity in these cells, as our group reported previously (53, 54). A similar correlation of FOXO3a and ERα expression was seen with ERα-positive T47D cells, whereas the expression of FOXO1a and FOXO4 did not show any specific correlation with ERα expression using the antibodies tested (data not shown). Thus, ERα status correlates with activation and nuclear localization of FOXO3a in breast cancer cell lines.
FIG. 1.
FOXO3a regulates ERα expression. (A) Samples (40 μg) of WCE or nuclear extracts (NE) from the indicated cell lines were analyzed by immunoblotting for FOXO3a (FOX), ERα, or β-actin (actin) as control for equal loading. (B) NF639 cells were transfected with 8 μg of pECE EV, WT FOXO3a, or A3 FOXO3a. Forty-eight hours after transfection, WCE were analyzed by immunoblotting for ERα, FOXO3a, and β-actin. (C) MCF-7 cells were infected with an increasing multiplicity of infection (MOI) of dnFOXO3a-expressing adenovirus. After 40 h, WCE were prepared and samples (40 μg) were analyzed by immunoblotting for FOXO3a and ERα. Equal loading was verified using β-actin immunoblotting and Ponceau S (Ponc.) staining.
FOXO3a induces ERα protein in NF639 cells.
To test directly whether FOXO3a can induce ERα expression, NF639 cells were transfected with vectors expressing either WT FOXO3a or A3 FOXO3a, a triple mutant form with the three sites of phosphorylation by Akt converted to alanine residues (T32A, S253A, and S315A) such that the protein can no longer be phosphorylated and inactivated by Akt (3), or the parental empty vector (EV) pECE as a control. Overexpression of WT or A3 FOXO3a increased the normalized level of ERα by 5.1-fold (±0.1) and 7.4-fold (±2.5), respectively, compared to cells transfected with pECE (Fig. 1B). The expression of the transfected FOXO3a protein was confirmed by immunoblotting (Fig. 1B). The slight retardation of the transfected WT and A3 FOXO3a proteins was likely due to the presence of the hemagglutinin tag. Overexpression of the A3 FOXO3a was more effective in inducing ERα expression, consistent with the constitutive activity of the A3 mutant (3). No significant changes in the ERα level were seen in the ERα-negative MDA-MB-231 breast cancer cell line in response to FOXO3a expression (data not shown). This is consistent with the reports that promoter hypermethylation is responsible for the loss of ERα in this cell line (75, 76).
We next assessed the effect of a dominant negative form of FOXO3a (dnFOXO3a), which contains the DNA binding domain but lacks the transactivation domain (64). ERα-positive MCF-7 cells were infected with an increasing multiplicity of dnFOXO3a-expressing adenovirus. WCE were prepared and subjected to immunoblotting for ERα, FOXO3a, and β-actin and stained with Ponceau S, which confirmed that loading was essentially equal. Increasing expression of dnFOXO3a resulted in a dose-dependent decrease in the ERα level (Fig. 1C). Taken together, the transfection analyses indicated that expression of FOXO3a is necessary and sufficient to maintain high ERα levels in ERα-positive NF639 and MCF-7 cells.
FOXO3a induces ERα promoter activity.
The long ∼4-kb ERα transcriptional unit (12, 70) has recently been characterized, and two proximal promoters (A and B) that are functional in breast cancer cells have been identified (71, 76) (Fig. 2A). NF639 cells were first cotransfected with ER3500-210 Luc, a 3.9-kb human ERα promoter construct (12) in the presence of WT or constitutively active A3 FOXO3a or the empty vector pECE, as control (Fig. 2B). WT and A3 FOXO3a induced ERα promoter activity by 4.8- and 5.7-fold, respectively, relative to that in pECE-cotransfected cells. Thus, FOXO3a induces ERα promoter activity.
FIG. 2.
FOXO3a induces ERα promoter activity. (A) Schematic representation of the ER3500-210Luc and ERα promoter A and B reporter constructs. (B) NF639 cells were cotransfected, in triplicate, with 0.5 μg of ER3500-210Luc in the presence of 2 μg of pECE parental vector, WT, or A3 FOXO3a expression vector. Equal amounts of protein were assessed for luciferase activity. Data are presented as fold induction (± SD) relative to that for the parental pECE vector, which was set as 1, and are representative of three independent experiments. (C) NF639 cells were transfected, in triplicate, with 0.5 μg of ERα promoter A or B luciferase reporter vectors and 0, 0.5, 1.0, or 1.5 μg of A3 FOXO3a expression vector and enough pECE to make up a total of 2 μg of DNA. After 48 h, cells were harvested and luciferase activities were measured. Data are presented as fold induction (± SD) relative to that with the pECE EV, which was set as 1, and are representative of three independent experiments. (Inset) WCE used for the luciferase assays were subjected to immunoblot analysis for FOXO3a protein.
To further locate the functional elements mediating this FOXO3a regulation, reporter constructs for proximal promoters A and B (promoter A construct, bp −1168 to +190; promoter B construct, bp −3284 to −1864) (27, 76) were similarly analyzed using an increasing dose of A3 FOXO3a expression vector. While promoter A showed a rather modest induction (1.9-fold) in response to FOXO3a, a large dose-dependent increase (8.6-fold) was seen with promoter B (Fig. 2C). Western blot analysis confirmed comparable expression of the transfected FOXO3a with both promoters (Fig. 2C, inset). Thus, promoter B, which is the one that is predominantly active in human breast cancers (71), responds potently to FOXO3a activity.
Identification of functional Forkhead binding sites in the ERα promoter.
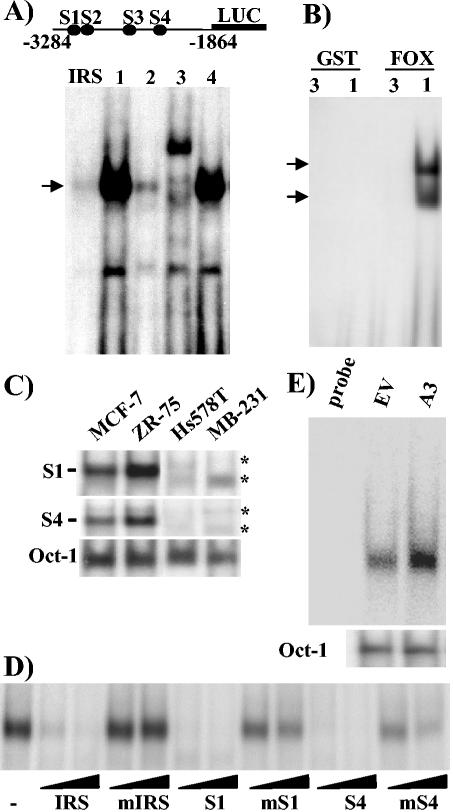
To locate putative cis-acting FOXO3a elements upstream of the human ERα promoter B, the sequence from bp −3284 to −1864 was scanned with MatInspector version 2.2 software (available at the website http://transfac.gbf.de/TRANSFAC/). Using 0.75 for the core similarity and 0.85 for the matrix similarity in the matrix group for vertebrates, the scanning retrieved four putative binding sites, designated S1 through S4 (Fig. 3A, upper panel). To begin to test the authenticity of these putative Forkhead binding sites, EMSA and cotransfection analyses were performed. Oligonucleotides of S1 through S4 were radiolabeled and used as probes with nuclear extracts from ZR-75 cells (Fig. 3A). Since a supershifting FOXO3a antibody is not currently available, as a control for FOXO3a binding the IRS Forkhead binding site from the IGFBP-1 gene was similarly subjected to EMSA to identify the position of the bona fide Forkhead band. A single band was seen with the IRS probe, as expected (3). The S1, S2, and S4 oligonucleotides all gave rise to a single major band which comigrated with the IRS complex, whereas the positions of the bands with S3 differed substantially. S1 and S4 oligonucleotides gave substantially higher band intensities than S2. (The IRS probe in this experiment had a somewhat lower specific activity and hence gave a weaker than typical signal.) Thus, three of the putative sites yielded a complex of the appropriate position. Since S1 and S4 showed much higher binding intensities, they were selected for further study.
FIG. 3.
FOXO3a binding sites are present upstream of ERα promoter B. (A) Schematic representation of the four putative FOXO3a binding sites in ERα promoter B. Each individual binding site, as well as the IRS, was end labeled and used in an EMSA with 15 μg of nuclear extract from ZR-75 cells. The arrow indicates the position of the major band observed. (B) Oligonucleotides containing sites S1 (1) and S3 (3) were incubated with GST or GST-FOXO3a (FOX) protein and subjected to EMSA. (C) Samples (15 μg) of nuclear extracts from MCF-7 and ZR-75 ERα-positive cells and Hs578T and MDA-MB-231 (MB-231) ERα-negative breast cancer cell lines were subjected to EMSA using as probe the oligonucleotides containing either the S1 and S4 binding sites or the Oct-1 sequence, as loading control. −, position of the protein complex unique to ERα-positive breast cancer cells; *, positions of protein complexes present in ERα-negative cell extracts. (D) Samples (15 μg) of nuclear extracts from ZR-75 cells were incubated with labeled IRS probe in the absence or presence of a 10- or 50-fold molar excess of WT or mutant (m) forms of the IRS, S1, or S4 oligonucleotides. (E) NF639 cells were transfected with pECE EV or A3 FOXO3a. After 40 h, cells were harvested and extracts were analyzed by EMSA. (Lower panel) Oct-1 was used as control probe and confirmed the presence of similar amounts of protein in each extract.
To confirm FOXO3a binding to the ERα sites, S1, which showed the strongest comigrating complex, and S3, which gave bands of alternative sizes, were used in EMSAs with GST-FOXO3a fusion protein or GST alone. S1 showed strong binding to GST-FOXO3a, but not to GST, while S3 did not bind to GST-FOXO3a or GST (Fig. 3B). Two bands were present with the S1 probe; the slower-migrating band likely represents full-length FOXO3a, and the faster-migrating band likely represents a degradation product, as two major bands could be visualized with Coomassie blue staining of the protein after purification (data not shown). Thus, the S1 site binds the FOXO3a protein.
To test whether the S1 and S4 Forkhead sites display the expected higher level of binding with nuclear extracts from ERα-positive versus -negative breast cancer cells, EMSA was performed with radiolabeled S1 and S4 oligonucleotide probes (Fig. 3C). Nuclear extracts from ERα-positive MCF-7 and ZR-75 cells bound avidly to S1 and S4, yielding a single complex, while those from ERα-negative Hs578T and MDA-MB-231 cells displayed only a low level of binding with distinct profiles. EMSA with the Oct-1 probe confirmed equal loading. Thus, higher levels of binding to S1 and S4 were seen with extracts from cells expressing elevated levels of FOXO3a.
To confirm the nature of the binding complexes for the S1 and S4 oligonucleotides, a competition EMSA was performed using WT IRS oligonucleotide as probe and WT and mutant competitor S1 and S4 oligonucleotides (Fig. 3D). As expected, WT but not mutant IRS competed for binding against itself. Importantly, WT S1 and S4 oligonucleotides efficiently competed for binding of ZR-75 nuclear extracts to the IRS oligonucleotide, whereas the mutant forms competed only poorly at the higher dose, suggesting the specificity of Forkhead binding.
Lastly, NF639 cells were transfected with A3 FOXO3a or EV DNA and extracts were made and subjected to EMSA as described above, with the S1 oligonucleotide as probe (Fig. 3E). A low level of binding was seen with NF639 cells, as expected based on the immunoblot in Fig. 1. Ectopic expression of A3 FOXO3a caused a specific increase in complex formation, compared to Oct-1 binding, consistent with an elevated level of FOXO3a protein (Fig. 1B and data not shown). Similar data were obtained with the S4 oligonucleotide as probe (data not shown). Taken together, these results indicate two strong FOXO3a binding sites are present upstream in the human ERα promoter (S1 and S4) and potentially one other more-weakly binding element (S2), while the S3 oligonucleotide does not appear to contain a Forkhead binding site.
S1 and S4 sites are functional Forkhead binding sites.
To test whether the S1 and S4 sites are functional, two copies of oligonucleotides containing either WT or mutant S1 and S4 binding sites were inserted upstream of the pBLCAT2 TK-CAT reporter vector, generating S1-CAT or MutS1-CAT and S4-CAT or MutS4-CAT vectors. Cotransfection of NF639 cells with S1-CAT or S4-CAT and an increasing amount of A3 FOXO3a showed a dose-dependent induction of CAT activity, with the highest at a dose of 2 μg, which was selected for further experiments (data not shown). The WT S1-CAT and S4-CAT reporter constructs were induced 3.0- and 3.9-fold, respectively, while the mutant constructs were essentially unaffected by 2 μg of A3 FOXO3a (i.e., only a ∼1.4- to 1.5-fold increase in activity was seen) (Fig. 4A). Taken together, the results demonstrate that the S1 and S4 sites represent functional Forkhead elements, responsive to FOXO3a.
FIG. 4.
The S1 and S4 sites contain functional Forkhead elements. (A) NF639 cells were cotransfected, in triplicate, with 0.5 μg of S1-CAT, MutS1-CAT, S4-CAT, or MutS4-CAT construct DNA with 2 μg of either pECE or A3 FOXO3a and processed as described above. Values for A3 FOXO3a are presented as the fold induction (± SD) relative to that of the pECE-cotransfected sample, which was set as 1. Data presented are representative of three independent experiments. (B) (Upper panel) Scheme of the proB-Luc construct. S1 and S4 Forkhead binding sites are indicated by the large black circles, and S2 is indicated by a smaller circle. (Bottom panel) WT, MutS4 (MS4), MutS1 (MS1), or MutS1/4 (MS1/4) proB-Luc constructs were cotransfected (1 μg), in triplicate, into NF639 cells with 1 μg of A3 FOXO3a or pECE vector. After 48 h, cells were harvested and processed as for Fig. 2. Data are presented as fold induction (± SD) relative to samples cotransfected with parental pECE vector, which was set as 1, and are representative of three independent experiments.
To test whether the S1 and S4 sites mediate signals leading to FOXO3a-dependent ERα promoter activity, site-directed mutagenesis was performed to introduce mutations at the S1 and S4 sites either individually or in combination in the promoter B construct, deriving MutS1-proB, MutS4-proB, or MutS1/4-proB, respectively (Fig. 4B). Mutation of either site individually had no substantial effect on responsiveness to FOXO3a; however, when both sites were mutated in combination, a 66.2% ± 6.0% loss in response to FOXO3a was observed. The responsiveness of the MutS1/4-proB construct was likely due either to the residual response of the mutant elements or from another weaker binding site(s) remaining in the promoter or to an indirect effect of FOXO3a. Overall, the data confirmed the importance of the S1 and S4 Forkhead sites in regulation of ERα B promoter activity by FOXO3a.
FOXO3a binds to S1/S4 sites on the endogenous ERα promoter.
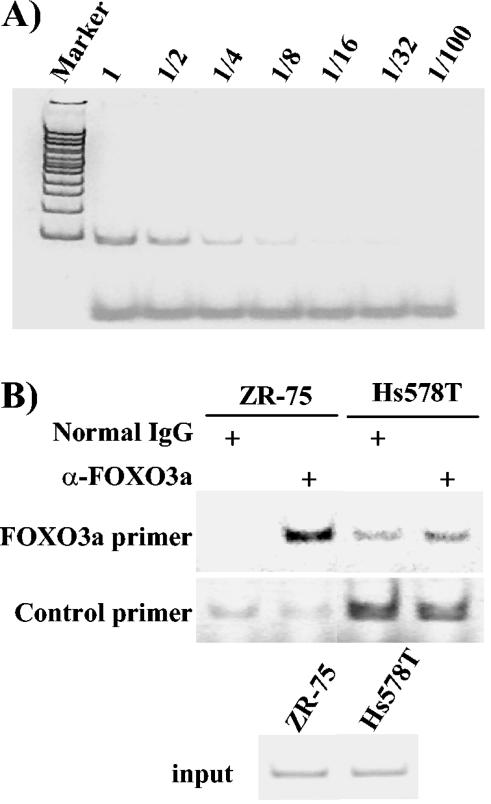
To confirm the binding of FOXO3a to the endogenous ERα promoter, ChIP assays were performed using a rabbit antibody specific for total FOXO3a versus normal IgG. Immunoprecipitated DNAs from ERα-positive ZR-75 cells and ERα-negative Hs578T cells were compared using semiquantitative PCR or real-time PCR. For the semiquantitative PCR, a condition of the PCR was selected within the linear, and thus quantitative, phase as shown in Fig. 5A. All subsequent PCRs were carried out within the linear range. The ERα-positive ZR-75 cells showed increased binding of FOXO3a to the S1 and S4 FOXO3a sites compared to that with normal rabbit IgG, consistent with the EMSA results shown above. In Hs578T cells, the FOXO3a antibody and control rabbit IgG pulled down comparable amounts of DNA fragments, which suggested a lack of specific binding. Both cell lines contained equal input material. To test for similar amounts of DNA in the FOXO3a versus IgG pairs, another pair of primers spanning an irrelevant region within the ERα gene was used. Equal amounts of DNA corresponding to this region were immunoprecipitated, indicating that the increased intensity of the amplified fragments was not due to random sample variation. In a separate experiment with real-time PCR, the ZR-75 cells gave a substantial signal, whereas the signal generated by the Hs578T sample was below the linear detection threshold (data not shown). These results indicate that FOXO3a is bound to the ERα promoter in ZR-75 cells.
FIG. 5.
FOXO3a binds to the endogenous S1/S4 sites in the ERα promoter. (A) PCR conditions used to amplify the S1/S4 fragment. (B) ChIP was performed with either ERα-positive ZR-75 cells or ERα-negative Hs578T cells using FOXO3a antibody or normal rabbit IgG as control, as described in Materials and Methods. The FOXO3a primers are located in between the S1 and S4 sites, which are 550 bp from each other. The control primers, located at an irrelevant region, were used to show that a similar amount of DNA was present in each paired sample. (Lower panel) Input DNA before immunoprecipitation.
FOXO3a can induce functional ERα signaling.
To test whether the FOXO3a-induced ERα is capable of driving a functional receptor signaling cascade, a TK-ERE luciferase reporter vector was cotransfected into NF639 cells together with an increasing amount of either WT or A3 FOXO3a, or EV pECE as control. Expression of either WT or A3 FOXO3a dramatically induced estrogen response element (ERE) promoter activity, consistent with an induction at the ERα protein level. As expected, constitutively active A3 FOXO3a was somewhat more effective than the WT form (Fig. 6A).
FIG. 6.
FOXO3a induces ERα activity. (A) NF639 cells were plated in 12-well plates and transfected with pECE and 0.1, 0.2, or 0.4 μg of WT or A3 FOXO3a, together with 0.2 μg of TK-ERE luciferase vector. After 30 h, cells were harvested and analyzed for luciferase activity. Data are presented as fold induction (± SD) relative to samples cotransfected with parental pECE vector, which was set as 1, and are representative of three independent experiments. (B and C) NF639 cells were transfected with the indicated expression vectors, and after 30 h whole-cell or nuclear extracts were prepared. Samples of WCE (50 μg) were analyzed by immunoblotting for RARα (B), and nuclear extracts (15 μg) were analyzed for ERα (C). Analysis of β-actin levels confirmed equal loading.
RARα is a known target gene of ERα signaling (56). As a further test of whether the ERα protein induced by FOXO3a is functional, we next assessed the effects of transfection of NF639 cells with WT, A3 FOXO3a, or pECE with levels of RARα protein and nuclear levels of ERα, another indication of ERα activation. The transfection efficiency of the NF639 cells was in the range of 25% (data not shown). The level of expression of the endogenous target RARα gene (56) was induced by expression of either WT or A3 FOXO3a (Fig. 6B). Consistent with this increase in target gene level, expression of FOXO3a activity induced nuclear localization of ERα (Fig. 6C). Taken together, these results show that FOXO3a induces functional ERα signaling.
The Her-2/neu/PI3K/Akt signaling pathway regulates ERα expression.
Since FOXO3a is negatively regulated by Akt phosphorylation, the ability of the PI3K/Akt pathway to modulate ERα expression was next assessed. NF639 and MCF-7 ERα-positive cells were transfected with a vector expressing AktM, a constitutively active form of Akt (29, 53), and ERα protein levels were measured in WCE prepared after 48 h. AktM caused a decrease in ERα protein levels in both cell types. When normalized for loading, ERα levels were reduced to 50.4% ± 2.9% and 38.% ± 3.4% in NF639 and MCF-7 cells, respectively, upon expression of AktM (Fig. 7A). Thus, constitutive PI3K/Akt signaling leads to decreased ERα levels.
FIG. 7.
ERα expression is subjected to regulation by Akt kinase activity. (A) NF639 and MCF-7 cells were transfected with a vector expressing AktM, the constitutively active version of Akt. WCE were prepared after 48 h and subjected to immunoblot analysis for ERα and β-actin, as above. (B) NF639 cells were treated with 60 μg of EGCG/ml or the carrier solution, dimethyl sulfoxide, for 24 h, and WCE were analyzed by immunoblotting for ERα and β-actin (left panel) or cytoplasmic (CE) and nuclear (NE) proteins for FOXO3a and β-actin (right panel). (C) NF639 cells were treated with 25 or 50 μM celecoxib for 7 h, and WCE were analyzed by immunoblotting for ERα and β-actin. (D) NF639 cells were treated with 100 or 130 nM wortmannin for 7 h, and WCE were analyzed by immunoblotting for ERα, FOXO3a, and β-actin.
Since we have recently shown that the green tea polyphenol EGCG reduces the Her-2/neu/PI3K/Akt signaling pathway by inhibiting the constitutive tyrosine phosphorylation of the receptor (54), we next tested the effects of EGCG on ERα expression in NF639 cells. After 24 h of treatment with 60 μg of EGCG/ml, a 1.8-fold (±0.2) increase in normalized ERα protein level was observed (Fig. 7B, left panel). Consistent with the inhibition of Akt activity, EGCG treatment led to a reduction in the hyperphosphorylated, inactive form of FOXO3a in the cytoplasm and to nuclear accumulation of the hypophosphorylated, active FOXO3a (Fig. 7B, right panel). Celecoxib, which has been used as a COX-2 inhibitor (67), is now known to inhibit Akt activity (25). Treatment of NF639 cells with 25 or 50 μM celecoxib for 7 h resulted in a dose-dependent induction of ERα levels (Fig. 7C). Similar effects were seen upon treatment with the celecoxib derivative OSU-03013, which has retained the ability to inhibit Akt but is devoid of COX-2 inhibition (kindly provided by C.-S. Chen) (data not shown). Similarly, a 7-h treatment of NF639 cells with the selective PI3K inhibitor wortmannin at either 100 or 130 nM led to increased ERα levels (Fig. 7D), which was paralleled by the disappearance of the hyperphosphorylated form of FOXO3a, as expected. Taken together, these results demonstrate the PI3K/Akt/FOXO3a signaling pathway modulates ERα expression in breast cancer cells.
DISCUSSION
Here, we show that the Forkhead box protein FOXO3a, whose activity is repressed by the PI3K/Akt kinase signaling cascade, is an important transcriptional regulator of the gene encoding the steroid hormone receptor ERα. FOXO3a levels correlated with ERα expression in breast cancer cells. Ectopic expression of FOXO3a led to increased receptor expression and promoter activity in ERα-positive NF639 cells. Similarly, A3 FOXO3a increased receptor levels in ERα-positive human T47D breast cancer cells (data not shown). Conversely, expression of dnFOXO3a decreased ERα expression in MCF-7 cells. Two strongly binding Forkhead sites were identified in ERα promoter B which conferred responsiveness to FOXO3a when inserted upstream of the heterologous TK promoter. Mutation of these two sites in ERα promoter B abolished most of the response to FOXO3a. Furthermore, in vivo FOXO3a binding to these two upstream sites was substantially higher in ERα-positive ZR-75 cells than in ERα-negative Hs578T cells, as judged by a ChIP assay. Increased ERE-driven reporter activity and elevated levels of the endogenous ERα target gene RARα in NF639 cells indicated that the ERα protein induced by FOXO3a was functional. Similar experiments, performed using human T47D breast cancer cells, showed FOXO3a induced RNA levels of RARα and pS2, another ERα target gene, as well as of ERα (data not shown). Overexpression of Her-2/neu is known to correlate negatively with ERα levels and has also been implicated in the development of resistance to the antiestrogen tamoxifen (6, 32, 47, 62, 73). Our findings identify, for the first time, FOXO3a as a key intermediary in the mechanism controlling the inverse expression pattern between Her-2/neu and ERα levels. Given that simultaneous inhibition of EGFR and ERα signaling pathways leads to superior antitumor efficacy (8, 21), our findings suggest FOXO3a as a possible target in adjuvant therapeutic intervention strategies for Her-2/neu-overexpressing, antiestrogen-refractory breast tumors.
In addition to the direct regulation of ERα promoter activity reported here, it has been shown that Forkhead proteins can interact with ERα in a ligand-dependent manner; however, variable consequences were observed for this interaction. While Schuur and coworkers reported that the interaction of FKHR (FOX01a) and ERα enhances the transcriptional activity of ERα at the typical ERE site (63), Zhao et al. (78) reported that it inhibits this activity. It is possible that this variability is due to differences in the cell systems used, i.e., MCF-7 cells (63) versus HepG2 and COS cells (78). Our findings show that FOXO3a induces ERE activity and endogenous ERα target gene expression via the induction of ERα protein expression in breast cancer cells. Furthermore, signaling by a variety of growth factors results in tamoxifen resistance and decreased ERα expression, which is at least partially mediated through the mitogen-activated protein kinase pathway (48), although the protein factor(s) involved remains to be identified. Taken together, these studies indicate a complex mechanism of regulation of ERα activity by signaling cascades that remain to be fully elucidated.
The expression patterns of different ErbB members and ERα and ERβ during mammary gland development have been very well characterized (11, 60, 65). ERα expression is low during puberty and pregnancy and increases during lactation (60). Moreover, expression of ERα is associated with a higher degree of differentiation of tumors and lower speed of tumor cell proliferation (49), which is consistent with the fact that ERα-expressing cells are usually negative for proliferating cell nuclear antigen and Ki67 expression (9, 58, 60), two markers of proliferation. In fact, although ERα signaling can activate the cell cycle progression through either genomic or nongenomic pathways (74), estrogen-inducible genes can suppress tumor progression (19, 20). Since the Forkhead family of proteins has been shown to play a very important role in cell cycle blockade and apoptosis (3, 43, 46), our data suggest the activity of FOXO3a, which determines ERα status, is likely to be responsible, in part, for the observed inverse correlation between proliferation and ERα expression.
The Forkhead family of proteins have been found to be important regulators for differentiation in systems other than the breast, including myogenic differentiation, osteoblast maturation, thymocyte differentiation, and adipocyte differentiation (2, 18, 23, 37). In the mammary gland, expression of ErbB2 and ErbB3 in epithelial cells has been found to be high during puberty and pregnancy, when most of the ductal elongation and branching occurs, and is down-regulated throughout lactation, when the mammary epithelial cells are functionally differentiated (11). Tumor cells expressing high levels of Her-2/neu under the control of the MMTV-long terminal repeat promoter failed to express detectable levels of two milk proteins, whey acidic protein and β-casein, markers of differentiation (35). The role of FOXO3a in mammary differentiation is under investigation.
Acknowledgments
We thank B. Nikolajczyk, D. McDevit, and M. Liang for providing assistance with the ChIP assays. We thank P. Leder, M. Greenberg, R. Weigle, S. Hayashi, C.-S. Chen, Z. Luo, R. Spanjaard, K. Walsh, and S. Jeay for generously providing cell lines, cloned DNA, Akt inhibitor, RARα antibody, dnFOXO3a-expressing adenovirus, and the GST fusion protein.
This work was supported by National Institutes of Health grants PO1 ES11624 (G.E.S.) and RO1 CA36355 (G.E.S.).
REFERENCES
- 1.Biswas, D. K., A. P. Cruz, E. Gansberger, and A. B. Pardee. 2000. Epidermal growth factor-induced nuclear factor kappa B activation: a major pathway of cell-cycle progression in estrogen-receptor negative breast cancer cells. Proc. Natl. Acad. Sci. USA 97:8542-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bois, P. R., and G. C. Grosveld. 2003. FKHR (FOXO1a) is required for myotube fusion of primary mouse myoblasts. EMBO J. 22:1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 4.Brunet, A., G. Pages, and J. Pouyssegur. 1994. Constitutively active mutants of MAP kinase kinase (MEK1) induce growth factor-relaxation and oncogenicity when expressed in fibroblasts. Oncogene 9:3379-3387. [PubMed] [Google Scholar]
- 5.Burgess, A. W., H. S. Cho, C. Eigenbrot, K. M. Ferguson, T. P. Garrett, D. J. Leahy, M. A. Lemmon, M. X. Sliwkowski, C. W. Ward, and S. Yokoyama. 2003. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol. Cell 12:541-552. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, R. A., P. Bhat-Nakshatri, N. M. Patel, D. Constantinidou, S. Ali, and H. Nakshatri. 2001. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J. Biol. Chem. 276:9817-9824. [DOI] [PubMed] [Google Scholar]
- 7.Chrysogelos, S. A., and R. B. Dickson. 1994. EGF receptor expression, regulation, and function in breast cancer. Breast Cancer Res. Treat. 29:29-40. [DOI] [PubMed] [Google Scholar]
- 8.Clark, A. S., K. West, S. Streicher, and P. A. Dennis. 2002. Constitutive and inducible Akt activity promotes resistance to chemotherapy, trastuzumab, or tamoxifen in breast cancer cells. Mol. Cancer Ther. 1:707-717. [PubMed] [Google Scholar]
- 9.Clarke, R. B., A. Howell, C. S. Potten, and E. Anderson. 1997. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res. 57:4987-4991. [PubMed] [Google Scholar]
- 10.Couse, J. F., and K. S. Korach. 1999. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr. Rev. 20:358-417. [DOI] [PubMed] [Google Scholar]
- 11.Darcy, K. M., D. Zangani, A. L. Wohlhueter, R. Y. Huang, M. M. Vaughan, J. A. Russell, and M. M. Ip. 2000. Changes in ErbB2 (her-2/neu), ErbB3, and ErbB4 during growth, differentiation, and apoptosis of normal rat mammary epithelial cells. J. Histochem. Cytochem. 48:63-80. [DOI] [PubMed] [Google Scholar]
- 12.deConinck, E. C., L. A. McPherson, and R. J. Weigel. 1995. Transcriptional regulation of estrogen receptor in breast carcinomas. Mol. Cell. Biol. 15:2191-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Fiore, P. P., J. H. Pierce, M. H. Kraus, O. Segatto, C. R. King, and S. A. Aaronson. 1987. erbB-2 is a potent oncogene when overexpressed in NIH/3T3 cells. Science 237:178-182. [DOI] [PubMed] [Google Scholar]
- 14.Dowsett, M. 2001. Overexpression of HER-2 as a resistance mechanism to hormonal therapy for breast cancer. Endocr. Relat. Cancer 8:191-195. [DOI] [PubMed] [Google Scholar]
- 15.Duyao, M. P., A. J. Buckler, and G. E. Sonenshein. 1990. Interaction of an NF-kappa B-like factor with a site upstream of the c-myc promoter. Proc. Natl. Acad. Sci. USA 87:4727-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eccles, S. A. 2001. The role of c-erbB-2/HER2/neu in breast cancer progression and metastasis. J. Mammary Gland Biol. Neoplasia 6:393-406. [DOI] [PubMed] [Google Scholar]
- 17.Elson, A., and P. Leder. 1995. Protein-tyrosine phosphatase epsilon. An isoform specifically expressed in mouse mammary tumors initiated by v-Ha-ras or neu. J. Biol. Chem. 270:26116-26122. [DOI] [PubMed] [Google Scholar]
- 18.Farmer, S. R. 2003. The forkhead transcription factor Foxo1: a possible link between obesity and insulin resistance. Mol. Cell 11:6-8. [DOI] [PubMed] [Google Scholar]
- 19.Finlin, B. S., C. L. Gau, G. A. Murphy, H. Shao, T. Kimel, R. S. Seitz, Y. F. Chiu, D. Botstein, P. O. Brown, C. J. Der, F. Tamanoi, D. A. Andres, and C. M. Perou. 2001. RERG is a novel ras-related, estrogen-regulated and growth-inhibitory gene in breast cancer. J. Biol. Chem. 276:42259-42267. [DOI] [PubMed] [Google Scholar]
- 20.Fujita, N., D. L. Jaye, M. Kajita, C. Geigerman, C. S. Moreno, and P. A. Wade. 2003. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell 113:207-219. [DOI] [PubMed] [Google Scholar]
- 21.Gee, J. M., M. E. Harper, I. R. Hutcheson, T. A. Madden, D. Barrow, J. M. Knowlden, R. A. McClelland, N. Jordan, A. E. Wakeling, and R. I. Nicholson. 2003. The antiepidermal growth factor receptor agent gefitinib (ZD1839/Iressa) improves antihormone response and prevents development of resistance in breast cancer in vitro. Endocrinology 144:5105-5117. [DOI] [PubMed] [Google Scholar]
- 22.Grant, S., L. Qiao, and P. Dent. 2002. Roles of ERBB family receptor tyrosine kinases, and downstream signaling pathways, in the control of cell growth and survival. Front. Biosci. 7:d376-d389. [DOI] [PubMed] [Google Scholar]
- 23.Hatta, M., H. Daitoku, H. Matsuzaki, Y. Deyama, Y. Yoshimura, K. Suzuki, A. Matsumoto, and A. Fukamizu. 2002. Regulation of alkaline phosphatase promoter activity by forkhead transcription factor FKHR. Int. J. Mol. Med. 9:147-152. [PubMed] [Google Scholar]
- 24.Hortobagyi, G. N., M. C. Hung, and A. U. Buzdar. 1999. Recent developments in breast cancer therapy. Semin. Oncol. 26:11-20. [PubMed] [Google Scholar]
- 25.Hsu, A. L., T. T. Ching, D. S. Wang, X. Song, V. M. Rangnekar, and C. S. Chen. 2000. The cyclooxygenase-2 inhibitor celecoxib induces apoptosis by blocking Akt activation in human prostate cancer cells independently of Bcl-2. J. Biol. Chem. 275:11397-11403. [DOI] [PubMed] [Google Scholar]
- 26.Hudziak, R. M., J. Schlessinger, and A. Ullrich. 1987. Increased expression of the putative growth factor receptor p185HER2 causes transformation and tumorigenesis of NIH 3T3 cells. Proc. Natl. Acad. Sci. USA 84:7159-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwase, H., Y. Omoto, H. Iwata, T. Toyama, Y. Hara, Y. Ando, Y. Ito, Y. Fujii, and S. Kobayashi. 1999. DNA methylation analysis at distal and proximal promoter regions of the oestrogen receptor gene in breast cancers. Br. J. Cancer 80:1982-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson, J. G., J. I. Kreisberg, A. P. Koterba, D. Yee, and M. G. Brattain. 2000. Phosphorylation and nuclear exclusion of the forkhead transcription factor FKHR after epidermal growth factor treatment in human breast cancer cells. Oncogene 19:4574-4581. [DOI] [PubMed] [Google Scholar]
- 29.Jeay, S., S. Pianetti, H. M. Kagan, and G. E. Sonenshein. 2003. Lysyl oxidase inhibits ras-mediated transformation by preventing activation of NF-kappa B. Mol. Cell. Biol. 23:2251-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang, H. Y., C. Petrovas, and G. E. Sonenshein. 2002. RelB-p50 NF-kappa B complexes are selectively induced by cytomegalovirus immediate-early protein 1: differential regulation of Bcl-xL promoter activity by NF-kappa B family members. J. Virol. 76:5737-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keen, J. C., and N. E. Davidson. 2003. The biology of breast carcinoma. Cancer 97:825-833. [DOI] [PubMed] [Google Scholar]
- 32.Kurokawa, H., and C. L. Arteaga. 2003. ErbB (HER) receptors can abrogate antiestrogen action in human breast cancer by multiple signaling mechanisms. Clin. Cancer Res. 9:511S-515S. [PubMed] [Google Scholar]
- 33.La Rosa, F. A., J. W. Pierce, and G. E. Sonenshein. 1994. Differential regulation of the c-myc oncogene promoter by the NF-kappa B rel family of transcription factors. Mol. Cell. Biol. 14:1039-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawlor, M. A., and D. R. Alessi. 2001. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J. Cell Sci. 114:2903-2910. [DOI] [PubMed] [Google Scholar]
- 35.Lazar, H., A. Baltzer, C. Gimmi, A. Marti, and R. Jaggi. 2000. Over-expression of erbB-2/neu is paralleled by inhibition of mouse- mammary-epithelial-cell differentiation and developmental apoptosis. Int. J. Cancer 85:578-583. [PubMed] [Google Scholar]
- 36.Lee, C. S., A. deFazio, C. J. Ormandy, and R. L. Sutherland. 1996. Inverse regulation of oestrogen receptor and epidermal growth factor receptor gene expression in MCF-7 breast cancer cells treated with phorbol ester. J. Steroid Biochem. Mol. Biol. 58:267-275. [DOI] [PubMed] [Google Scholar]
- 37.Leenders, H., S. Whiffield, C. Benoist, and D. Mathis. 2000. Role of the forkhead transcription family member, FKHR, in thymocyte differentiation. Eur. J. Immunol. 30:2980-2990. [DOI] [PubMed] [Google Scholar]
- 38.Liu, Y., D. el-Ashry, D. Chen, I. Y. Ding, and F. G. Kern. 1995. MCF-7 breast cancer cells overexpressing transfected c-erbB-2 have an in vitro growth advantage in estrogen-depleted conditions and reduced estrogen-dependence and tamoxifen-sensitivity in vivo. Breast Cancer Res. Treat. 34:97-117. [DOI] [PubMed] [Google Scholar]
- 39.Lu, J., M. J. Pazin, and K. Ravid. 2004. Properties of ets-1 binding to chromatin and its effect on platelet factor 4 gene expression. Mol. Cell. Biol. 24:428-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarthy, K. M., D. McDevit, A. Andreucci, R. Reeves, and B. S. Nikolajczyk. 2003. HMGA1 co-activates transcription in B cells through indirect association with DNA. J. Biol. Chem. 278:42106-42114. [DOI] [PubMed] [Google Scholar]
- 41.McCarty, K. S., Jr., T. K. Barton, B. F. Fetter, B. H. Woodard, J. A. Mossler, W. Reeves, J. Daly, W. E. Wilkinson, and K. S. McCarty, Sr. 1980. Correlation of estrogen and progesterone receptors with histologic differentiation in mammary carcinoma. Cancer 46:2851-2858. [DOI] [PubMed] [Google Scholar]
- 42.McClelland, R. A., D. Barrow, T. A. Madden, C. M. Dutkowski, J. Pamment, J. M. Knowlden, J. M. Gee, and R. I. Nicholson. 2001. Enhanced epidermal growth factor receptor signaling in MCF7 breast cancer cells after long-term culture in the presence of the pure antiestrogen ICI 182,780 (Faslodex). Endocrinology 142:2776-2788. [DOI] [PubMed] [Google Scholar]
- 43.Medema, R. H., G. J. Kops, J. L. Bos, and B. M. Burgering. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404:782-787. [DOI] [PubMed] [Google Scholar]
- 44.Mossler, J. A., K. S. McCarty, Jr., and W. W. Johnston. 1981. The correlation of cytologic grade and steroid receptor content in effusions of metastatic breast carcinoma. Acta Cytol. 25:653-658. [PubMed] [Google Scholar]
- 45.Nakachi, K., K. Suemasu, K. Suga, T. Takeo, K. Imai, and Y. Higashi. 1998. Influence of drinking green tea on breast cancer malignancy among Japanese patients. Jpn. J. Cancer Res. 89:254-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura, N., S. Ramaswamy, F. Vazquez, S. Signoretti, M. Loda, and W. R. Sellers. 2000. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol. Cell. Biol. 20:8969-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholson, R. I., I. R. Hutcheson, M. E. Harper, J. M. Knowlden, D. Barrow, R. A. McClelland, H. E. Jones, A. E. Wakeling, and J. M. Gee. 2001. Modulation of epidermal growth factor receptor in endocrine-resistant, oestrogen receptor-positive breast cancer. Endocr. Relat. Cancer 8:175-182. [DOI] [PubMed] [Google Scholar]
- 48.Oh, A. S., L. A. Lorant, J. N. Holloway, D. L. Miller, F. G. Kern, and D. El-Ashry. 2001. Hyperactivation of MAPK induces loss of ERα expression in breast cancer cells. Mol. Endocrinol. 15:1344-1359. [DOI] [PubMed] [Google Scholar]
- 49.Osborne, C. K. 1998. Steroid hormone receptors in breast cancer management. Breast Cancer Res. Treat. 51:227-238. [DOI] [PubMed] [Google Scholar]
- 50.Pages, G., A. Brunet, G. L'Allemain, and J. Pouyssegur. 1994. Constitutive mutant and putative regulatory serine phosphorylation site of mammalian MAP kinase kinase (MEK1). EMBO J. 13:3003-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park, W. C., and V. C. Jordan. 2002. Selective estrogen receptor modulators (SERMs) and their roles in breast cancer prevention. Trends Mol. Med. 8:82-88. [DOI] [PubMed] [Google Scholar]
- 52.Perou, C. M., T. Sorlie, M. B. Eisen, M. van de Rijn, S. S. Jeffrey, C. A. Rees, J. R. Pollack, D. T. Ross, H. Johnsen, L. A. Akslen, O. Fluge, A. Pergamenschikov, C. Williams, S. X. Zhu, P. E. Lonning, A. L. Borresen-Dale, P. O. Brown, and D. Botstein. 2000. Molecular portraits of human breast tumours. Nature 406:747-752. [DOI] [PubMed] [Google Scholar]
- 53.Pianetti, S., M. Arsura, R. Romieu-Mourez, R. J. Coffey, and G. E. Sonenshein. 2001. Her-2/neu overexpression induces NF-κB via a PI3-kinase/Akt pathway involving calpain-mediated degradation of IκB-α that can be inhibited by the tumor suppressor PTEN. Oncogene 20:1287-1299. [DOI] [PubMed] [Google Scholar]
- 54.Pianetti, S., S. Guo, K. T. Kavanagh, and G. E. Sonenshein. 2002. Green tea polyphenol epigallocatechin-3 gallate inhibits Her-2/neu signaling, proliferation, and transformed phenotype of breast cancer cells. Cancer Res. 62:652-655. [PubMed] [Google Scholar]
- 55.Pietras, R. J., J. Arboleda, D. M. Reese, N. Wongvipat, M. D. Pegram, L. Ramos, C. M. Gorman, M. G. Parker, M. X. Sliwkowski, and D. J. Slamon. 1995. HER-2 tyrosine kinase pathway targets estrogen receptor and promotes hormone-independent growth in human breast cancer cells. Oncogene 10:2435-2446. [PubMed] [Google Scholar]
- 56.Rishi, A. K., Z. M. Shao, R. G. Baumann, X. S. Li, M. S. Sheikh, S. Kimura, N. Bashirelahi, and J. A. Fontana. 1995. Estradiol regulation of the human retinoic acid receptor alpha gene in human breast carcinoma cells is mediated via an imperfect half-palindromic estrogen response element and Sp1 motifs. Cancer Res. 55:4999-5006. [PubMed] [Google Scholar]
- 57.Rodriguez, C., A. Causse, E. Ursule, and C. Theillet. 2000. At least five regions of imbalance on 6q in breast tumors, combining losses and gains. Genes Chromosomes Cancer 27:76-84. [PubMed] [Google Scholar]
- 58.Russo, J., X. Ao, C. Grill, and I. H. Russo. 1999. Pattern of distribution of cells positive for estrogen receptor alpha and progesterone receptor in relation to proliferating cells in the mammary gland. Breast Cancer Res. Treat. 53:217-227. [DOI] [PubMed] [Google Scholar]
- 59.Saceda, M., T. W. Grunt, R. Colomer, M. E. Lippman, R. Lupu, and M. B. Martin. 1996. Regulation of estrogen receptor concentration and activity by an ErbB/HER ligand in breast carcinoma cell lines. Endocrinology 137:4322-4330. [DOI] [PubMed] [Google Scholar]
- 60.Saji, S., E. V. Jensen, S. Nilsson, T. Rylander, M. Warner, and J. A. Gustafsson. 2000. Estrogen receptors alpha and beta in the rodent mammary gland. Proc. Natl. Acad. Sci. USA 97:337-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samatar, A. A., L. Wang, A. Mirza, S. Koseoglu, S. Liu, and C. C. Kumar. 2002. Transforming growth factor-beta 2 is a transcriptional target for Akt/protein kinase B via forkhead transcription factor. J. Biol. Chem. 277:28118-28126. [DOI] [PubMed] [Google Scholar]
- 62.Schiff, R., S. Massarweh, J. Shou, and C. K. Osborne. 2003. Breast cancer endocrine resistance: how growth factor signaling and estrogen receptor coregulators modulate response. Clin. Cancer Res. 9:447S-454S. [PubMed] [Google Scholar]
- 63.Schuur, E. R., A. V. Loktev, M. Sharma, Z. Sun, R. A. Roth, and R. J. Weigel. 2001. Ligand-dependent interaction of estrogen receptor-alpha with members of the forkhead transcription factor family. J. Biol. Chem. 276:33554-33560. [DOI] [PubMed] [Google Scholar]
- 64.Skurk, C., H. Maatz, H. S. Kim, J. Yang, M. R. Abid, W. C. Aird, and K. Walsh. 2004. The Akt-regulated forkhead transcription factor FOXO3a controls endothelial cell viability through modulation of the caspase-8 inhibitor FLIP. J. Biol. Chem. 279:1513-1525. [DOI] [PubMed] [Google Scholar]
- 65.Stern, D. F. 2003. ErbBs in mammary development. Exp. Cell Res. 284:89-98. [DOI] [PubMed] [Google Scholar]
- 66.Stern, D. F. 2000. Tyrosine kinase signalling in breast cancer: ErbB family receptor tyrosine kinases. Breast Cancer Res. 2:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subbaramaiah, K., and A. J. Dannenberg. 2003. Cyclooxygenase 2: a molecular target for cancer prevention and treatment. Trends Pharmacol. Sci. 24:96-102. [DOI] [PubMed] [Google Scholar]
- 68.Suhara, T., H. S. Kim, L. A. Kirshenbaum, and K. Walsh. 2002. Suppression of Akt signaling induces Fas ligand expression: involvement of caspase and Jun kinase activation in Akt-mediated Fas ligand regulation. Mol. Cell. Biol. 22:680-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tagliabue, E., S. Menard, J. F. Robertson, and L. Harris. 1999. c-erbB-2 expression in primary breast cancer. Int. J. Biol. Markers 14:16-26. [DOI] [PubMed] [Google Scholar]
- 70.Tang, Z., I. Treilleux, and M. Brown. 1997. A transcriptional enhancer required for the differential expression of the human estrogen receptor in breast cancers. Mol. Cell. Biol. 17:1274-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanimoto, K., H. Eguchi, T. Yoshida, K. Hajiro-Nakanishi, and S. Hayashi. 1999. Regulation of estrogen receptor alpha gene mediated by promoter B responsible for its enhanced expression in human breast cancer. Nucleic Acids Res. 27:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsai, E. M., S. C. Wang, J. N. Lee, and M. C. Hung. 2001. Akt activation by estrogen in estrogen receptor-negative breast cancer cells. Cancer Res. 61:8390-8392. [PubMed] [Google Scholar]
- 73.Witters, L. M., R. Kumar, V. M. Chinchilli, and A. Lipton. 1997. Enhanced anti-proliferative activity of the combination of tamoxifen plus HER-2-neu antibody. Breast Cancer Res. Treat. 42:1-5. [DOI] [PubMed] [Google Scholar]
- 74.Wong, C. W., C. McNally, E. Nickbarg, B. S. Komm, and B. J. Cheskis. 2002. Estrogen receptor-interacting protein that modulates its nongenomic activity-crosstalk with Src/Erk phosphorylation cascade. Proc. Natl. Acad. Sci. USA 99:14783-14788. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Yang, X., D. L. Phillips, A. T. Ferguson, W. G. Nelson, J. G. Herman, and N. E. Davidson. 2001. Synergistic activation of functional estrogen receptor (ER)-alpha by DNA methyltransferase and histone deacetylase inhibition in human ER-alpha-negative breast cancer cells. Cancer Res. 61:7025-7029. [PubMed] [Google Scholar]
- 76.Yoshida, T., H. Eguchi, K. Nakachi, K. Tanimoto, Y. Higashi, K. Suemasu, Y. Iino, Y. Morishita, and S. Hayashi. 2000. Distinct mechanisms of loss of estrogen receptor alpha gene expression in human breast cancer: methylation of the gene and alteration of trans-acting factors. Carcinogenesis 21:2193-2201. [DOI] [PubMed] [Google Scholar]
- 77.Yu, X., R. V. Rajala, J. F. McGinnis, F. Li, R. E. Anderson, X. Yan, S. Li, R. V. Elias, R. R. Knapp, and W. Cao. 2004. Involvement of insulin/phosphoinositide 3-kinase/Akt signal pathway in 17β-estradiol-mediated neuroprotection. J. Biol. Chem. 279:13086-13094. [DOI] [PubMed] [Google Scholar]
- 78.Zhao, H. H., R. E. Herrera, E. Coronado-Heinsohn, M. C. Yang, J. H. Ludes-Meyers, K. J. Seybold-Tilson, Z. Nawaz, D. Yee, F. G. Barr, S. G. Diab, P. H. Brown, S. A. Fuqua, and C. K. Osborne. 2001. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J. Biol. Chem. 276:27907-27912. [DOI] [PubMed] [Google Scholar]