Abstract
Methylglyoxal (MG) is synthesized during glycolysis, although it inhibits cell growth in all types of organisms. Hence, it has long been asked why such a toxic metabolite is synthesized in vivo. Glyoxalase I is a major enzyme detoxifying MG. Here we show that the Yap1 transcription factor, which is critical for the oxidative-stress response in Saccharomyces cerevisiae, is constitutively concentrated in the nucleus and activates the expression of its target genes in a glyoxalase I-deficient mutant. Yap1 contains six cysteine residues in two cysteine-rich domains (CRDs), i.e., three cysteine residues clustering near the N terminus (n-CRD) and the remaining three cysteine residues near the C terminus (c-CRD). We reveal that any of the three cysteine residues in the c-CRD is sufficient for MG to allow Yap1 to translocate into the nucleus and to activate the expression of its target gene. A Yap1 mutant possessing only one cysteine residue in the c-CRD but no cysteine in the n-CRD and deletion of the basic leucine zipper domain can concentrate in the nucleus with MG treatment. However, substitution of all the cysteine residues in Yap1 abolishes the ability of this transcription factor to concentrate in the nucleus following MG treatment. The redox status of Yap1 is substantially unchanged, and protein(s) interaction with Yap1 through disulfide bond is hardly detected in cells treated with MG. Collectively, neither intermolecular nor intramolecular disulfide bond formation seems to be involved in Yap1 activation by MG. Moreover, we show that nucleocytoplasmic localization of Yap1 closely correlates with growth phase and intracellular MG level. We propose a novel regulatory pathway underlying Yap1 activation by a natural metabolite in the cell.
Oxygen respiration allows all types of aerobic organisms to produce ATP efficiently, although it inevitably generates harmful reactive oxygen species which consequently cause oxidative stress. Cells possess diverse defense systems against oxidative stress (for a review, see reference 6). Several antioxidant systems of the budding yeast Saccharomyces cerevisiae are regulated at the transcriptional level (31). The Yap1 transcription factor is a functional homologue of mammalian AP-1 in S. cerevisiae (20, 43), and the expression of several antioxidant genes is up-regulated by Yap1 under oxidative-stress-inducing conditions (42). Yap1 activity is considered to be regulated mainly through its nuclear localization; i.e., Yap1 is distributed in both the cytoplasm and nucleus, preferentially in the cytoplasm, under nonstress conditions, although it is concentrated in the nucleus on the exposure of cells to oxidative stress (35). Yap1 has a nuclear export signal (NES) sequence in the C terminus, and Crm1 (exportin) exports Yap1 from the nucleus to the cytoplasm under normal conditions. Under oxidative-stress-inducing conditions, the interaction between Yap1 and Crm1 is inhibited and Yap1 is consequently enriched in the nucleus, leading to elevated expression of its target genes (36, 68).
Three cysteine residues, which partially overlap the NES, exist in the C terminus of Yap1. These cysteine residues (C-terminal cysteine-rich domain [c-CRD]) are considered to constitute a redox sensor, and modification of these cysteine residues may block the interaction with Crm1 under oxidative stress-inducing conditions (68). We have demonstrated that the disulfide bond between Cys598 and Cys620 within the c-CRD in Yap1 is preferentially formed on H2O2 treatment (34). In addition, Delaunay et al. (11) reported that a disulfide bond between Cys303, which is in the n-CRD (N-terminal cysteine-rich domain), and Cys598 is formed. It has been reported that Gpx3, one of the glutathione peroxidase homologues in yeast (25), functions as a redox transducer for Yap1 (12).
Regarding deactivation of Yap1, we previously found that a thioredoxin deficiency (trx1Δ trx2Δ) caused the constitutive activation of Yap1: i.e., Yap1 was routinely concentrated in the nucleus and the basal expression levels of Yap1 target genes were increased (30). In this case, intracellular oxidation levels were twofold higher than those in the wild type. This elevated intracellular redox potential is considered to activate Yap1, since the constitutive activation of Yap1 was suppressed if cells were cultured anaerobically. Although a direct interaction of thioredoxin and Yap1 could not be detected by a two-hybrid assay (30), thioredoxin can be formally considered to be a negative regulator for Yap1 activity. We also clarified that the oxidized Yap1 is reduced directly by thioredoxin (34).
Several genes responsible for glutathione metabolism, such as glutathione synthesis (GSH1 and GSH2) and glutathione-dependent antioxidant systems (GPX2 and GLR1), are up-regulated by Yap1 (19, 25, 55, 58, 67). In addition to its antioxidant function, glutathione is important in detoxifying many harmful xenobiotics. In addition, glutathione functions as a detoxicant for endogenous toxic compounds such as methylglyoxal (MG: CH3COCHO) (27). MG is a typical 2-oxoaldehyde, and the major source of this aldehyde in eukaryotes is a β-elimination reaction carried out by triosephosphate isomerase, a glycolytic enzyme (29, 49, 50). MG was therefore once thought to be an intermediate of glycolysis. However, MG was found to inhibit the growth of various types of cells from microorganisms to mammals (14, 15, 56). MG exerts its toxicity through modification of macromolecules to diminish their biological activities. For example, MG reacts with guanine residues in both DNA and RNA to form an adduct (53). An interaction with proteins also contributes to MG toxicity (1). Detoxification of MG is performed by the glyoxalase system, which consists of the glyoxalase I and glyoxalase II enzymes. Glyoxalase I catalyzes the conversion of MG to S-d-lactoylglutathione in the presence of glutathione, while the glutathione thiol ester is hydrolyzed by glyoxalase II. Therefore, a deficiency of the structural gene for glyoxalase I (GLO1) and/or glyoxalase II (GLO2 and GLO4) in S. cerevisiae increases susceptibility to MG (3, 28). Although glyoxalase I is involved in a glutathione-requiring reaction, expression of the GLO1 gene is not up-regulated by Yap1 (26).
To gain insight into the cellular function of MG, we compared the gene expression profile between wild-type and glo1Δ cells by DNA microarray analysis. Since glyoxalase I is the major MG-metabolizing enzyme (28), the steady-state level of MG in the glo1Δ mutant was higher than that in the wild-type cells throughout the growth phase. We found that Yap1 is constitutively concentrated in the nucleus in the glo1Δ mutant. In contrast to the case involving thioredoxin deficiency, the constitutive activation of Yap1 in glo1Δ cells was observed under both aerobic and anaerobic conditions. Cysteine residues within the c-CRD in Yap1 are necessary for activation by MG, although no disulfide bond formation between such cysteine residues is involved in this event, because any one of the cysteine residues in the c-CRD of Yap1 is sufficient to activate this transcription factor with MG. We present evidence indicating that Yap1 activity is reversibly modulated by MG, a natural metabolite from glycolysis, without redox regulation.
MATERIALS AND METHODS
Strains, media, and growth conditions.
S. cerevisiae YPH250 (MATa trp1-Δ1 his3-Δ200 leu2-Δ1 lys2-801 ade2-101 ura3-52) and its isogenic diploid strain YPH274 (MATa/MATα trp1-Δ1/trp1-Δ1 his3-Δ200/his3-Δ200 leu2-Δ1/leu2-Δ1 lys2-801/lys2-801 ade2-101/ade2-101 ura3-52/ura3-52) were obtained from the Yeast Genetic Stock Center, University of California, Berkeley. SD minimal medium (2% glucose, 0.67% yeast nitrogen base without amino acids [pH 5.5]) supplemented with appropriate amino acids and bases was used for the culture of yeast cells. Unless otherwise stated, cells were cultured at 28°C with reciprocal shaking. For anaerobic culture, the medium was flushed with nitrogen gas for 2 min after subculturing of the cells, the vessel was sealed, and the cells were incubated at 28°C without shaking.
Gene disruption.
The GLO2 gene was cloned by PCR using the primers GLO2Sneo (5′-TTGGCGATGGCATTAACGAGGATCCCGCTT-3′) and GLO2Rneo (5′-GGTGCTAATGTCGACAATGTGTTCGGCCAA-3′). The GLO2Sneo and GLO2Rneo primers were designed to contain a BamHI site and SalI site (underlined), respectively. The PCR product containing the GLO2 gene was digested with BamHI and SalI and cloned into the BamHI and SalI sites of pUC19 to yield pUGLO2. The pUGLO2 plasmid was then digested with EcoRV and Eco47III and treated with Klenow, and the LEU2 gene was inserted to yield pUglo2ΔLEU2. The resultant plasmid was digested with PvuII and NcoI, and the glo2Δ::LEU2 cassette was used to disrupt the GLO2 gene.
The GLO4 gene was amplified by the primers GLO4S (5′-CCATGCAGGATCCATTATAACTATCAAAGG-3′) and GLO4R (5′-GGAAAAGAGAGAGAGTCGACGTCTGTGATG-3′). The GLO4S and GLO4R primers were designed to contain a BamHI site and SalI site (underlined), respectively. The PCR product containing the GLO4 gene was digested with BamHI and SalI and then cloned between the BamHI and SalI sites of pUC19 to construct pUGLO4. The pUGLO4 plasmid was digested with Ecol47III and treated with Klenow, and the HIS3 gene was inserted to give pUglo4ΔHIS3. The resultant plasmid was digested with SspI, and the glo4::HIS3 cassette was used to disrupt the GLO4 gene.
The xpo1-1 allele was introduced to an xpo1Δ::LEU2 mutant in the YPH250 background as follows. The XPO1/CRM1 gene of YPH274 (MATa/MATα) was disrupted by replacement with LEU2 as described by Stade et al. (54). A plasmid carrying the xpo1-1 allele (pKW457) and pKW430 bearing NLSSV40-NESPKI-2 × GFP (54) were introduced into the resultant mutant (MATa/MATα XPO1/xpo1Δ::LEU2). Tetrad dissection was performed to isolate a strain in the same genetic background of YPH250, except for xpo1Δ::LEU2, with plasmid-borne xpo1-1 and the NLSSV40-NESPKI-2×GFP reporter gene.
The glo1::URA3, glo1Δ::HIS3, yap1Δ::HIS3, gpx3Δ::LEU2, tsa1Δ::TRP1, gsh1Δ::LEU2, trx1Δ::URA3, and trx2Δ::HIS3 mutants in the YPH250 background have been described previously (25, 26, 28, 30).
Reporter gene.
The Yap1-dependent reporter gene, which carried three AP-1 binding sites followed by an upstream activator site-less CYC1 gene and lacZ (37), was subcloned into YCp50. We designated this reporter gene AP-1-CYC1-lacZ. The DNA fragment containing AP-1-CYC1-lacZ was also cloned into a low-copy-number vector, pRS415. The FLR1-lacZ reporter gene (Ylp368/FLR1) was obtained from M. Raymond (45). The GLO1-lacZ gene (pRSGlac415) was described previously (26).
Enzyme assay.
β-Galactosidase activity was measured by the method of Miller (40). One unit of the activity was expressed as (A420 × 1,000) per hour (where A420 is the absorbance at 420 nm). Glyoxalase I activity was measured as described previously (24). One unit of glyoxalase I activity was defined as the amount of enzyme forming 1 μmol of S-d-lactoylglutathione per min at 25°C. The amount of protein was determined by the method of Bradford (4).
Measurement of intracellular MG content.
Cells were cultured in SD minimal medium with appropriate amino acids and bases. Cells were collected by centrifugation, washed once with 0.85% NaCl solution, and suspended in 1.0 ml of distilled water. They were boiled for 5 min and centrifuged at 1,500 × g for 5 min, and the resultant supernatants were used as an MG source. MG concentration was measured enzymatically (glyoxalase I; Sigma, St. Louis, Mo.). To measure cellular MG content in the MG-treated cells, cells were cultured in SD medium until the A610 was 0.5 and 8 mM MG was added. The cells were collected periodically, and cellular MG levels were measured as described above.
Western blotting.
To determine the effect of GLO1 deficiency on the steady-state levels of Yap1, cells of yap1Δ and yap1Δ glo1Δ mutants carrying pRS315-Yap1-9Myc, which was donated by M. Toledano, were cultured in SD medium until they reached an A610 of 1. They were disrupted with glass beads in 10 mM Tris-HCl buffer (pH 7.0) containing 1 mM β-mercaptoethanol. Homogenates were centrifuged at 12,000 × g for 15 min at 4°C. Cellular proteins (40 μg) in the supernatants were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were electrically transferred to a polyvinylidene difluoride membrane (Immobilon; Millipore). Yap1-9Myc protein was detected by an anti-c-Myc monoclonal antibody (9E 10; Roche Diagnostics GmbH, Mannheim, Germany). To determine the effect of exogenously added MG on the levels of Yap1, yap1Δ cells carrying pRS315-Yap1-9Myc were cultured in SD medium until an A610 of 1 was reached and 8 mM MG was added. Cells were collected periodically (0 to 3 h), and Yap1-9Myc protein levels were compared by Western blotting as described above.
To detect the biochemical changes in Yap1, the yap1Δ cells carrying pRS314 cp HA YAP1 (HA-tagged Yap1) (35) were treated with 8 mM MG for 60 min or 0.4 mM H2O2 for 15 min. Cellular proteins were extracted using trichloroacetic acid as described by Delaunay et al. (11), and the resultant proteins were subjected to nonreducing SDS-PAGE. The HA-tagged Yap1 was detected by anti-HA monoclonal antibody (Roche Diagnostics GmbH).
Fluorescence microscopy analysis.
Cells were cultured in SD minimal medium until an A610 of 0.5 was reached, and localization of green fluorescent protein (GFP)-tagged Yap1 was observed by fluorescence microscopy. To visualize DNA, the cells were stained with 1 μg of 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI) per ml.
Cloning and expression of gloA.
The structural gene for Escherichia coli glyoxalase I (gloA) (9) was amplified by PCR with the primers gloA-F (5′-AGCCATTTTGAGGATCCAAAGATGCGTCTT-3′) and gloA-R (5′-CGGGCGCGATGAGTCGACGCCCGGCAGGAG-3′). The gloA-F and gloA-R primers were designed to contain a BamHI site and SalI site, respectively (underlined). The PCR product was digested with BamHI and SalI, and the resultant fragment was cloned into the BamHI and SalI sites of the 2μm-based plasmid pG-1 (51), which contains the GAP promoter, PGK terminator, and TRP1 marker. The resultant plasmid (pG-1-gloA) was introduced into the glo1Δ mutant, and expression of gloA was confirmed by measurement of glyoxalase I activity.
Coimmunoprecipitation.
Cells of yap1Δ and yap1Δ glo1Δ mutants carrying the GFP-Yap1WT fusion protein were cultured to log phase, and cell extracts were prepared. Immunoprecipitation was carried out using a kit (Roche Molecular Biochemicals) with anti-glyoxalase I antibody (23) or anti-GFP antibody (Clontech) and protein A-agarose. Detailed procedures for immunoprecipitation were performed as specified by the manufacturer. After SDS-PAGE, Western blot analysis was conducted using anti-glyoxalase I antibody or anti-GFP antibody as the primary antibody and anti-rabbit immunoglobulin G antibody conjugated with horseradish peroxidase (New England Biolabs) as the secondary antibody. Immunoreactive protein was visualized with 4-chloro-1-naphthol and H2O2.
Cys-substituted mutants of Yap1.
GFP-Yap1 plasmids were based on the plasmid pRS cp-GFP-HA YAP1 (GFP-Yap1WT) (35). Detailed procedures for the construction of pRS cp-GFP-HA yap1 3Cys (GFP-Yap1p3Cys), where three cysteine residues were mutated to Thr (C303T, C310T, and C315T), were described previously (34). To construct pRS cp-GFP-HA yap1 6Cys (GFP-Yap1p6Cys), in which all six cysteine residues are substituted (C303T, C310, C315T, C598T, C620A, and C629T), the c-CRD region of pRS cp-GFP-HA yap1 cm46A5 (35) (yap1 gene with mutation of C598T, C620A, and C629T; GFP-Yap1pcm46A5) was isolated with BsmI and BstEII and replaced by the c-CRD region of pRS cp-GFP-HA yap1 3Cys. The construction of single-cysteine-substituted c-CRD mutants (GFP-Yap1C598T, GFP-Yap1C620A, and GFP-Yap1C629T) was described previously (35). GFP-Yap1 mutants that possess only one cysteine residue were constructed by a PCR-based method as described previously (35), using yap1 genes carrying a C598T, C620A, or C629T mutation as templates. The c-CRD regions of the resulting PCR fragments were digested with BsmI and SalI and cloned into pRS314 cp-GFP-HA yap1 3Cys to yield an expression vector for GFP-HA-fused Yap13Cys cm56, Yap13Cys cm46, and Yap13Cys cm45. To construct pRS cp-GFP Yap1dαH, the region of the yap1 gene corresponding to bZIP (amino acids 73 to 121) was replaced with a SmaI restriction site (CCCGGG encoding Pro-Gly) by the PCR-based method described above. To construct pRS cp-GFP yap1dαH 3Cys cm46, the AfeI-SalI restriction fragment of pRS cp-GFP Yap1dαH was replaced with the corresponding fragment of GFP-Yap13Cys cm56.
DNA microarray analysis.
Cells of the wild-type strain and the glo1Δ::HIS3 mutant were cultured in SD minimal medium until they reached an A610 of 1. Total RNA was extracted by the hot-phenol method (52), and poly(A)+ mRNA was isolated from total RNA by using a kit (Oligotex-dT30 <Super> mRNA purification kit; Takara Shuzo, Kyoto, Japan). cDNAs labeled with Cy3-dCTP or Cy5-dCTP were generated by using reverse transcriptase and the Cyscribe first-strand cDNA labeling kit (Amersham Pharmacia, Piscataway, N.J.). Fluorescent probes were purified using Qiaquick PCR purification columns (Qiagen GmbH, Hilden, Germany) and concentrated by a Microcon-30 filter (Amicon, Charlotte, N.C.). Hybridization was carried out at 37°C for 14 h in DIG Easy Hybe solution (Roche Diagnostics GmbH, Mannheim, Germany) containing 100 μg of salmon sperm DNA per ml. After hybridization, microarray slides (Eurogentec, Seraing, Belgium) were washed with 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% SDS and rinsed with 0.2× SSC at room temperature. Slides were scanned using an Affymetrix 417 Arrayer (Affymetrix, Santa Clara, Calif.). Images were processed using the microarray spot finding and quantification software ImaGene (BioDiscovery, Marina del Rey, Calif.). Fluorescence intensity values were normalized so that the medians of all Cy3 and Cy5 intensities were equal.
RESULTS
Yap1 is constitutively activated in a glo1Δ mutant.
MG is a natural metabolite derived from glycolysis, although it inhibits cell growth and in some cases it induces apoptosis (33). In addition, MG is likely to be involved in some pathological processes such as cancer and diabetes (32, 57). However, the intrinsic function of MG is not yet fully understood. To gain insight into the physiological function of MG in the cell, we performed DNA microarray analysis to compare the gene expression pattern between wild-type and glo1Δ cells. Because glyoxalase I is the major enzyme for the detoxification of MG, the steady-state level of MG in the glo1Δ mutant (5.25 ± 1.17 nmol/109 cells) was higher than that in the wild type (2.61 ± 0.39 nmol/109 cells). As a result of analysis; an increase (more than twofold) in the expression of approximately 10% of the entire set of yeast genes was observed in glo1Δ cells, with the genes for energy metabolism and stress response being among the 10%. Details of the microarray analysis will be discussed elsewhere.
We noted that many genes whose expression is regulated by Yap1 transcription factor were up-regulated in the glo1Δ mutant (Table 1). Yap1 is distributed predominantly in the cytoplasm under nonstress conditions, whereas it is concentrated in the nucleus under oxidative-stress-inducing conditions (35). Interestingly, Yap1 was constitutively concentrated in the nucleus in the glo1Δ mutant. We confirmed that GFP-Yap1 was observed in the nuclei of more than 87.6% of glo1Δ cells (n = 1,419). Consequently, the basal expression levels of the Yap1-dependent artificial reporter gene (AP-1-CYC1-lacZ) were increased (Fig.1A and B), although this was not the case for mutants defective in cytosolic glyoxalase II (glo2Δ), mitochondrial glyoxalase II (glo4Δ), and both of these enzymes (glo2Δ glo4Δ) (data not shown).
TABLE 1.
Effect of glyoxalase I deficiency on the expression of Yap1 target genes
| Genea | Product | Fold up-regulation |
|---|---|---|
| GSH1 | γ-Glutamylcysteine synthetase | 2.6 |
| GPX2 | Glutathione peroxidase | 6.4 |
| GTT2 | Glutathione S-transferase | 3.7 |
| TRX2 | Thioredoxin | 3.5 |
| TRR1 | Thioredoxin reductase | 3.4 |
| CYS3 | Cystathionine γ-lyase | 4.7 |
| SSA1 | Heat shock protein 70 | 2.9 |
| HSP78 | Mitochondrial heat shock protein | 2.4 |
| FLR1 | ABC transporter, fluconazole resistance | 3.2 |
| YCF1 | ABC transporter, cadmium resistance | 2.6 |
| GRE2 | Methylglyoxal reductase | 13.0 |
| TAL1 | Transaldolase | 3.0 |
| PGM2 | Phosphoglucomutase | 3.6 |
| PDI1 | Protein disulfide isomerase | 3.1 |
| YNL134c | Similarity to alcohol dehydrogenase | 2.1 |
The basis of some Yap1 targets is from the report by Lee et al. (38).
FIG. 1.
Effect of MG on activation of Yap1. (A) Cells of yap1Δ and yap1Δ glo1Δ carrying the GFP-Yap1 plasmid were cultured in SD medium to an A610 of 0.5, and localization of GFP-Yap1 was observed. (B) (Left panel) Cells of the wild type (WT) carrying the AP-1-CYC1-lacZ reporter plasmid were cultured in SD medium to an A610 of 0.5, and 8 mM MG was added. After a 3-h incubation, β-galactosidase activity was measured. (Right panel) For anaerobic culture, after cells of the wild type and the glo1Δ mutant with reporter gene were inoculated into SD medium, nitrogen gas was flushed through the flask, which was then sealed, and the cells were cultured without shaking at 28°C. (C) GFP-tagged Yap1 was expressed in each mutant. All mutants have a yap1Δ background. (D) GFP-Yap1 was expressed in the yap1Δ and yap1Δ gpx3Δ mutants. Cells were cultured in SD medium to an A610 of 0.5, and 8 mM MG or 0.4 mM H2O2 was added. Localization of GFP-Yap1 was monitored for 60 min (for MG) or 15 min (for H2O2) after the addition of each chemical. (E) Cells were cultured in YPD medium to an A610 of 0.5, diluted with 0.85% NaCl, and spotted onto YPD agar plates with or without 15 mM MG. The cells were cultured at 28°C for 2 days.
Previously, we observed a similar phenotype in terms of the nucleocytoplasmic localization of Yap1 in a cytosolic thioredoxin-deficient (trx1Δ trx2Δ) mutant (30). In this case, the constitutive activation of Yap1 was observed only when the mutant was cultured aerobically. In contrast, however, Yap1 activation in the glo1Δ mutant was also seen when cells were cultured anaerobically (Fig. 1B).
Recently, Delaunay et al. (12) reported that Gpx3, one of the glutathione peroxidase homologues in S. cerevisiae (25), is a redox transducer for Yap1. They proposed a model whereby Gpx3 is first oxidized by H2O2 and the oxidized Gpx3 (Gpx3-Cys-SOH) forms a mixed disulfide with Cys598 in Yap1, which is then reduced by Cys303 in Yap1 to form an intramolecular disulfide bond (Cys303-Cys598). Thus, Yap1 activation by H2O2 does not occur in a gpx3Δ mutant. However, the constitutive nuclear accumulation of Yap1 in the glo1Δ mutant was seen even in the gpx3Δ background (Fig. 1C). In addition, exogenously added MG allowed Yap1 to accumulate in the nucleus of the gpx3Δ cell (Fig. 1D). Furthermore, the gpx3Δ mutant did not exhibit hypersensitivity to MG (Fig. 1E).
On the other hand, Veal et al. (61) reported that Ybp1 functions in the Gpx3 pathway. They demonstrated that Ybp1 binds to Yap1 in vivo and stimulates the nuclear localization of Yap1 on exposure to H2O2 stress. Interestingly, W303-1A, a laboratory yeast strain, possesses a ybp1-1 mutation that abolishes the function of Ybp1 (61). We confirmed that the constitutive activation of Yap1 occurs in a glo1Δ mutant in a W303-1A background (data not shown).
Delaunay et al. (12) also reported that Gpx3 functions as a thioredoxin peroxidase. If an oxidized Gpx3 (Gpx3-Cys-SOH) is condensed with another cysteine residue in Gpx3 intramolecularly, an intramolecular disulfide bond is formed and thioredoxin reduces it, thereby accomplishing a thioredoxin-dependent H2O2 reducing reaction. S. cerevisiae possesses TSA1 coding for cytosolic thioredoxin peroxidase (7, 44). Recently, involvement of Tsa1 in H2O2-induced activation of Yap1 has been suggested (46). We determined whether Tsa1 is involved in the activation of Yap1 by MG, although Yap1 was constitutively concentrated in the nucleus of tsa1Δ cells in the glo1Δ background (Fig. 1C). Yap1 was found to be concentrated in the nucleus of tsa1Δ cells following treatment with MG (data not shown), and the tsa1Δ mutant did not exhibit susceptibility to MG (Fig. 1E). These results suggest that the underlying mechanism of Yap1 activation by MG is different from that by H2O2.
The intracellular MG level correlates with activation of Yap1.
The subcellular localization of Yap1 is regulated at the stage of nuclear export; i. e., Crm1 exports Yap1 from the nucleus to the cytoplasm under nonstress conditions, but interaction between Yap1 and Crm1 is inhibited under oxidative-stress conditions, thereby allowing Yap1 to remain in the nucleus (36, 68). We examined the function of Crm1 in the glo1Δ mutant by using the GFP-tagged Crm1-recognizable NES derived from the human protein kinase inhibitor α (PKIα) (64) with the nuclear localization signal (NLS) of the simian virus 40 (SV40) large T antigen (54). The resultant reporter protein (NLSSV40-NESPKI-2× GFP) is translocated to the nucleus but is immediately exported from the nucleus to the cytoplasm by Crm1, thereby allowing the cytoplasmic localization. If the function of Crm1 is impaired in glo1Δ cells, this reporter protein will remain in the nucleus.
First, to verify whether Crm1 in YPH250 background functions normally, NLSSV40-NESPKI-2×GFP was expressed in a temperature-sensitive mutant of Crm1 (xpo1-1) and the resultant transformant was shifted to the nonpermissive temperature. As a result, this reporter protein was distributed in the cytoplasm at the permissive temperature but was concentrated in the nucleus after a shift to the nonpermissive temperature; therefore, the function of Crm1 was confirmed to be normal in the strain used in this study (Fig. 2A). Next, we determined the localization of NLSSV40-NESPKI-2×GFP in the glo1Δ mutant. As shown in Fig. 2A, this reporter protein was distributed predominantly in the cytoplasm in glo1Δ cells. Similarly, exogenously added MG did not affect the localization of NLSSV40-NESPKI-2×GFP (Fig. 2A), indicating that Crm1 functions normally in the glo1Δ mutant.
FIG. 2.
The function of the Yap1 nuclear export machinery and the Yap1 protein level are normal in the glo1Δ mutant. (A) Temperature-sensitive xpo1-1 cells carrying pKW430 (NLSSV40-NESPKI-2×GFP) were cultured at 28°C (permissive temperature) to an A610 of 0.5 and shifted to 37°C (nonpermissive temperature). After 15 min, the localization of NLSSV40-NESPKI-2×GFP was monitored. Cells of the wild type (WT) and the glo1Δ mutant carrying pKW430 were cultured in SD medium to an A610 of 0.5, and localization of NLSSV40-NESPKI-2×GFP was observed. To determine the effect of exogenously added MG on the function of Crm1, wild-type cells carrying pKW430 were treated with 8 mM MG. No change in the localization of NLSSV40-NESPKI-2×GFP was seen at least up to 120 min after the addition of MG. The pictures shown in the figure were taken 60 min after the addition of MG. (B) To compare the steady-state level of Yap1 protein, cells of the yap1Δ (lane 1) and yap1Δ glo1Δ (lane 2) mutants carrying pRS315-Yap1-9Myc were cultured in SD medium to an A610 of 1.0, and cell extracts were prepared. To determine the effect of MG on the Yap1 protein level, yap1Δ cells carrying pRS315-Yap1-9Myc were cultured in SD medium to an A610 of 0.5. After the addition of 8 mM MG, the cells were incubated at 28°C and collected periodically. Lane 3, without MG for 1 h; lane 4, with 8 mM MG for 1 h. We confirmed that the Yap1 protein level did not change at least up to 3 h after MG treatment.
Thus, regarding the constitutive nuclear localization of Yap1 in the glo1Δ mutant, we considered three possible models. The first model is that glyoxalase I functions as a negative regulator for Yap1 through direct protein-protein interaction; i.e., glyoxalase I, which exists in the cytoplasm, may serve as an anchor protein for Yap1. A second model is that glyoxalase I deficiency influences the Yap1 protein level. A third model is that MG directly modulates the Yap1 activity.
To assess whether direct interaction occurs between Yap1 and glyoxalase I, we performed a coimmunoprecipitation assay. The GFP-Yap1WT fusion protein was expressed in the yap1Δ and yap1Δ glo1Δ mutants, and cell extracts were immunoprecipitaed with anti-glyoxalase I antibody and then Western blotted with anti-GFP antibody. However, no interaction between Yap1 and glyoxalase I was observed. Similarly, no direct interaction was observed when the anti-GFP antibody was used for the immunoprecipitation (data not shown).
Next, we compared the steady-state level of the Yap1 protein in wild-type and glo1Δ cells, because overexpression of YAP1 is reported to cause a similar phenotype in terms of Yap1 activation (63). The Yap1-9Myc protein was expressed by its own promoter in the yap1Δ and yap1Δ glo1Δ mutants. We confirmed that Myc-tagged Yap1 is functional in yap1Δ cells (data not shown). As shown in Fig. 2B, no difference was seen in the protein levels of Yap1 between the wild-type and glo1Δ cells. We confirmed that MG treatment did not affect the Yap1 protein levels (Fig. 2B).
Then we investigated whether MG is involved in the activation of Yap1. The steady-state level of MG in the glo1Δ mutant was higher than that in the wild type. Accumulation of MG was also observed in the cells cultured anaerobically (wild type, 3.33 ± 0.24 nmol/109 cells; glo1Δ, 6.68 ± 0.24 nmol/109 cells). Although we have confirmed that the yeast glyoxalase I per se does not function as a negative regulator for Yap1 through protein-protein interaction, we introduced the gloA gene coding for the glyoxalase I of E. coli (9) into the glo1Δ mutant to reduce the intracellular MG level. On expression of the gloA gene, the cellular MG content decreased to the normal level (vector control, 5.31 ± 0.86 nmol/109 cells; gloA expression, 2.14 ± 0.37 nmol/109 cells), and concomitantly the basal expression level of the Yap1-dependent reporter gene, AP-1-CYC1-lacZ, was reversed to the wild-type level (data not shown).
To determine the correlation between the intracellular MG level and Yap1 activity more directly, we added MG to the culture medium. As shown in Fig. 3, the intracellular MG level increased and subsequently Yap1 was concentrated in the nucleus, but the MG level reverted to normal when the cells were washed (Fig. 3). Consequently, Yap1 was immediately redistributed in the cytoplasm (Fig. 3). Since glyoxalase I is an efficient catalyst (kcat/Km = 1.89 × 106 M−1 s−1) (28), the turnover of intracellular MG is high. These results indicate that Yap1 activation directly correlates with the intracellular MG level. Importantly, these observations also suggest that activation of Yap1 by MG is reversible.
FIG. 3.
Yap1 activation by MG is reversible. GFP-Yap1 was expressed in the yap1Δ mutant and cultured in SD medium to an A610 of 0.5. At this point, the localization of GFP-Yap1 was monitored (time, 0 min), 8 mM MG was added to the culture medium, and the cells were incubated to monitor the localization of GFP-Yap1 periodically (time, 5 to 60 min). After 60 min, cells were collected by centrifugation, washed with 0.85% NaCl, and resuspended in fresh SD medium, and localization of GFP-Yap1 was monitored periodically (time, 1 to 5 min). The cellular MG content was measured when the A610 had reached 0.5, and 8 mM MG was added. The MG level was monitored after the addition of MG (30 and 60 min) and also after the removal of MG (5 min).
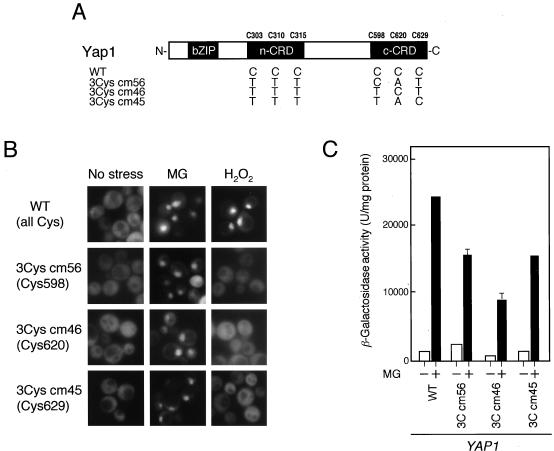
Importance of Cys residues in the c-CRD for Yap1 activation by MG.
Yap1 has six cysteine residues, three in the transactivation domain (Cys303, Cys310, and Cys315) (Coleman et al. [10] named it the N-terminal cysteine-rich domain [n-CRD]) and three at the C terminus (c-CRD; Cys598, Cys620, and Cys629) (Fig. 4A). Since the c-CRD overlaps the NES, deletion of the c-CRD causes the constitutive nuclear localization of Yap1 (35). Kuge et al. (35) reported that cysteine residues in the c-CRD are important for the regulation of the subcellular localization of Yap1. On the other hand, Coleman et al. (10) reported that the n-CRD contains trafficking information for Yap1. MG generates irreversible adducts with the arginine and lysine residues of protein (65). The c-CRD/NES of Yap1 contains Arg604, Lys610, Lys626, Lys628, and Arg632, and therefore it is conceivable that modification of these amino acid residues with MG might abolish the NES function and inhibit interaction with Crm1, leading to concentration of Yap1 in the nucleus of the glo1Δ mutant. However, as we have shown in Fig. 3, Yap1 activation by MG is reversible, and thus the possibility of modification of arginine and/or lysine residues scattered within the c-CRD/NES is lowered.
FIG. 4.
Importance of Cys residues in Yap1 for activation by MG. (A) Schematic of the Yap1 mutant protein. (B) GFP-tagged Yap1 of the wild type (WT) and each mutant was expressed in the yap1Δ and yap1Δ glo1Δ mutants. Cells were cultured in SD medium to an A610 of 0.5, and localization of GFP-Yap1 in yap1Δ glo1Δ cells was monitored. For yap1Δ cells, 8 mM MG or 0.4 mM H2O2 was added to the culture medium, the cells were incubated for a further 60 min (MG) or 15 min (H2O2), and localization of GFP-Yap1 was monitored. (C) Each GFP-tagged protein was also expressed in the yap1Δ mutant carrying the FLR1-lacZ reporter gene. After treatment with 8 mM MG for 3 h, the β-galactosidase activity was measured.
To determine whether the cysteine residues that exist in the n-CRD and/or c-CRD are involved in the activation of Yap1 in the glo1Δ mutant, we constructed several cysteine-substituted mutants of Yap1 (Fig. 4A). In the Yap13Cys mutant, three cysteine residues in the n-CRD (Cys303, Cys310, and Cys315) were changed to threonine. Yap1cm46A5 contains the substitutions C598T, C620A, and C629T in the c-CRD. This mutant is insensitive to oxidative stress and is distributed in the cytoplasm even under oxidative-stress-inducing conditions (35). In Yap16Cys, the configurations of Yap13Cys and Yap1cm46A5 were combined. These cysteine-substituted yap1 mutant genes were fused to GFP gene, and the resultant GFP-Yap1 mutant proteins were expressed in the yap1Δ and yap1Δ glo1Δ mutants carrying the FLR1-lacZ reporter gene. The FLR1 gene is one of the Yap1 targets, and its promoter contains three Yap1 response elements (45).
As shown in Fig. 4B, the GFP-Yap1cm46A5 protein was present in the cytoplasm in the glo1Δ mutant. In addition, the GFP-Yap16Cys was not concentrated in the nucleus. These results indicate that (i) the c-CRD contains trafficking information for the constitutive nuclear localization of Yap1 in the glo1Δ mutant, (ii) cysteine residues in the c-CRD are critical, and (iii) arginine and/or lysine residues in the c-CRD are less feasible in the regulation of the nucleocytoplasmic localization of Yap1 in the glo1Δ mutant.
We confirmed the importance of the c-CRD for the constitutive nuclear localization of Yap1 in the glo1Δ mutant by using a fusion protein of the Gal4 DNA binding domain containing an NLS followed by GFP and the c-CRD (Gal4dbd-GFP-CRD) (Fig. 5A). This construct was proven to be exported from the nucleus by Crm1 and to be sensitive to oxidative stress (35). The Gal4dbd-GFP-CRD fusion protein was concentrated in the nucleus in the glo1Δ mutant under normal conditions (Fig. 5B). Additionally, this reporter protein accumulated in the nucleus following MG treatment (Fig. 5B). It does not contain the n-CRD, indicating that the c-CRD is sufficient for the constitutive nuclear localization of Yap1 in the glo1Δ mutant.
FIG. 5.
Importance of c-CRD for the nuclear localization of Yap1 following MG treatment. (A) Schematic of Yap1 and the Gal4-GFP-c-CRD reporter protein. (B) The Gal4-GFP-c-CRD reporter gene was expressed in wild-type (WT) and glo1Δ cells. Cells were cultured in SD medium to an A610 of 0.5, and localization of the reporter protein was monitored. Then 8 mM MG was added, and the cells were incubated for 60 min, at which time the localization of the reporter protein was again determined.
Next, we examined the effect of exogenously added MG on the localization of these Yap1 mutant proteins and evaluated their activity by using the FLR1-lacZ reporter gene. As shown in Fig. 4B, GFP-Yap13Cys was concentrated in the nucleus following MG treatment and the expression of FLR1-lacZ was induced. Neither GFP-Yap16Cys nor GFP-Yap1cm46A5 accumulated in the nucleus, and therefore no induction of the reporter gene was observed (Fig. 4C). In the case of H2O2 treatment, GFP-Yap13Cys was also concentrated in the nucleus (Fig. 4B); however, this mutant did not induce the reporter gene expression following H2O2 treatment for 1 h (data not shown). Previously, we demonstrated that a disulfide bond between Cys598 and Cys620 within the c-CRD of Yap1 is formed following H2O2 treatment (34). Although this GFP-Yap13Cys mutant protein was able to stay in the nucleus briefly (approximately 15 min), it redistributed to the cytoplasm rapidly compared with wild-type Yap1; i.e., GFP-Yap1WT continued to be present in the nucleus for 30 to 45 min under the same conditions. The disulfide bond between Cys303 in the n-CRD and Cys598 seems necessary for the stable nuclear localization of Yap1 (11, 34). Hence, one possible explanation for the absence of Yap1 target gene induction in cells expressing the GFP-Yap13Cys following H2O2 treatment is that this mutant Yap1 is not present in the nucleus long enough to induce target gene expression.
Any one of three Cys residues in c-CRD is sufficient for Yap1 activation by MG.
To determine whether disulfide bond formation is involved in the activation process of Yap1 by MG, we constructed a series of mutants that contains only one cysteine residue in the c-CRD, with all of the cysteine residues in the n-CRD being simultaneously substituted (GFP-Yap13Cys cm56, GFP-Yap13Cys cm46, and GFP-Yap13Cys cm45) (Fig. 6A). Interestingly, these Yap1 mutant proteins were able to concentrate in the nucleus following MG treatment (Fig. 6B), and, consequently, expression of the Yap1 target gene was induced (Fig. 6C). The nuclear localization of these single-cysteine mutants of Yap1 by MG was reversible; i.e., they were rapidly redistributed in the cytoplasm when MG was removed from the medium (data not shown). On the other hand, these mutants were no longer able to respond to H2O2 (Fig. 6B), because the proteins cannot form any intramolecular disulfide bonds. These results suggest that disulfide bond formation between the cysteine residues in Yap1 is not necessary for Yap1 activation by MG.
FIG. 6.
Any one of three Cys residues in the c-CRD is sufficient for Yap1 activation by MG. (A) Schematic of the Yap1 mutant protein. (B) GFP-tagged Yap1 of the wild type (WT) and each mutant was expressed in yap1Δ cells. When the A610 of the culture reached 0.5, 8 mM MG or 0.4 mM H2O2 was added to the culture medium, the cells were incubated for a further 60 min (MG) or 15 min (H2O2), and localization of GFP-Yap1 was then monitored. (C) Each GFP-tagged protein was also expressed in the yap1Δ mutant carrying the FLR1-lacZ reporter gene. After treatment with 8 mM MG for 3 h, the β-galactosidase activity was measured.
Finally, we deleted the bZIP domain of Yap1 (GFP-Yap1dαH) (Fig. 7A). Since the bZIP proteins usually interact physically with each other through the bZIP domain, such a bZIP domain-deleted Yap1 mutant is supposed to be difficult to approach closely, and therefore intermolecular disulfide bonds between cysteine residues, if any, seem less feasible. However, an intramolecular disulfide bond can be formed, and thus GFP-Yap1dαH was concentrated in the nucleus following H2O2 treatment (Fig. 7B). We then substituted the cysteine residues of this bZIP domain-deleted Yap1 mutant protein so that it contained only one cysteine in the c-CRD (GFP-Yap1dαH 3Cys cm46) (Fig. 7A). If the Yap1 NES was masked through an intermolecular disulfide bond formation between residual cysteine of GFP-Yap1dαH 3Cys cm46, this reporter protein would be expected to concentrate in the nucleus following H2O2 treatment. However, this mutant Yap1 protein was not concentrated in the nucleus after H2O2 treatment, and therefore, the intermolecular disulfide bond is thought to be formed only slightly even under conditions of oxidative stress. On the other hand, intriguingly, this mutant protein was able to concentrate in the nucleus following treatment with MG (Fig. 7B). Collectively, our data indicate that any one of three cysteine residues in the c-CRD is sufficient for Yap1 activation of MG.
FIG. 7.
No disulfide formation is involved in Yap1 activation by MG. (A) Schematic of the Yap1 mutant protein. (B) GFP-tagged Yap1 of the wild type (WT) and each mutant was expressed in yap1Δ cells. When the A610 of the culture reached 0.5, 8 mM MG or 0.4 mM H2O2 was added to the culture medium, cells were incubated for a further 60 min (MG) or 15 min (H2O2), and localization of GFP-Yap1 was then monitored.
The redox status of Yap1 is not changed by MG.
The data we have demonstrated thus far suggest that not only intramolecular but also intermolecular disulfide bond formation is less feasible in the regulatory mechanism of the nucleocytoplasmic localization of Yap1 by MG. Nevertheless, it remains possible that there are some MG sensor proteins in the cell, and that such proteins interact with cysteine residues in the c-CRD to mask the Yap1 NES and consequently block interaction with Crm1, leading to the nuclear localization of Yap1.
To address this possibility, we determined the biochemical changes in Yap1 following treatment with MG. Wild-type cells carrying HA-tagged Yap1 were treated with MG or H2O2, and cellular proteins were subjected to nonreducing SDS-PAGE. As shown in Fig. 8, the redox state of Yap1 was virtually unchanged by treatment with MG, whereas the oxidized form of Yap1 (disulfide bond between Cys303 in the n-CRD and Cys598 in the c-CRD) (11) emerged on H2O2 treatment. In addition, a band with low mobility on nonreducing SDS-PAGE appeared in the H2O2-treated sample, which is presumably the Yap1-Gpx3 complex (12). However, the low-mobility band was hardly detected in the MG-treated cells. Taken together, these results show that MG does not affect the redox state of Yap1. Furthermore, the possibility of the existence of an MG sensor protein(s), which masks the Yap1 NES to block the interaction with Crm1, can be excluded.
FIG. 8.
Analysis of biochemical changes in Yap1. yap1Δ cells carrying HA-tagged Yap1 were cultured in SD medium to an A610 of 1.0 and then treated with 8 mM MG for 60 min or 0.4 mM H2O2 for 15 min. Cellular proteins were extracted as described in the text and subjected to nonreducing SDS-PAGE followed by Western blotting with anti-HA monoclonal antibody to detect HA-Yap1. The immunoreactive protein was detected using the ECL kit (Amersham Biosciences). The low-mobility band that appeared in H2O2-treated samples was assumed to be a Yap1-Gpx3 complex.
Physiological relevance of MG-dependent Yap1 activation.
Although MG is a ubiquitous natural metabolite, it causes disorders of cellular functions due to its high reactivity. Glyoxalase I is the major enzyme responsible for MG detoxification; nevertheless, GLO1 is not a target of Yap1 (26). However, as shown in Table 1, expression of GRE2 coding for methylglyoxal reductase (8), an alternative enzyme for MG detoxification (27), was strongly induced in glo1Δ cells. Hence, yeast cells seem armed with a Yap1-dependent MG-metabolizing enzyme (Gre2) as a backup system to prepare for glyoxalase I deficiency.
To gain further insights into the physiological correlation between Yap1 activation and MG metabolism, we monitored cellular MG level with respect to cell growth. As shown in Fig. 9, the cellular MG level increased with cell growth and reached its maximum during the transition period from log phase to stationary phase, which is referred to as the diauxic shift. We examined the intracellular localization of Yap1 periodically. As expected, the proportion of cells with the nuclear Yap1 increased synchronously in accordance with the cellular MG level, reaching maximum when the cellular MG level was also maximum. The steady-state level of MG in glo1Δ cells in the log phase was comparable to that of the maximal level of wild-type cells in the diauxic shift. The cellular MG level decreased gradually thereafter, which may be due to the increased expression of GLO1, although the induction mechanism of GLO1 expression in the diauxic shift has not yet been elucidated. Consequently, the proportion of cells with nuclear Yap1 decreased. Wiatrowski and Carlson (66) reported that carbon deprivation caused the nuclear localization of Yap1; however, although the glucose in the medium was almost exhausted after the diauxic shift, cells with the nuclear Yap1 were not observed, as far as we have examined. Thus, the changes in the proportion of cells with the nuclear Yap1 are probably linked to the cellular MG level. We also monitored the localization of GFP-tagged Yap13Cys cm46 and Yap16Cys mutants with respect to cell growth. Essentially, the result obtained with GFP-Yap13Cys cm46 was the same as that obtained with GFP-Yap1WT, whereas GFP-Yap16Cys was present in the cytoplasm throughout the growth phase (data not shown). Collectively, our data suggest that MG-dependent Yap1 activation occurs in yeast cells under normal growth conditions.
FIG. 9.
Correlation between cell growth, cellular MG level, and Yap1 localization. Cells were cultured in SD minimal medium, and cell growth (A610), glucose concentration in the medium, β-galactosidase activity driven by GLO1-lacZ, intracellular MG content, and subcellular localization of GFP-Yap1 were monitored periodically. The glucose concentration was determined using a kit (Glucose B-test Wako; Wako Pure Chemical Industries, Tokyo, Japan). The number of cells counted in each observation for localization of GFP-Yap1 was 500 to 1,000.
DISCUSSION
A sudden environmental change, such as in temperature, osmotic pressure, or redox conditions, induces the expression of a number of stress-responsive genes in both prokaryotes and eukaryotes, allowing the cells to become tolerant to these stress conditions. Each stress signal is sensed by a specific sensor molecule in the cells. In terms of oxidative-stress sensing, E. coli has two distinct transcription factors, OxyR and SoxR, that act as sensors for reactive oxygen species (31, 70). The former has two critical cysteine residues (Cys199 and Cys208), which form a disulfide bond when cells are exposed to oxidative stress elicited by H2O2 (69). Glutaredoxin reduces the disulfide bond of OxyR to down-regulate its activity (69). On the other hand, SoxR has a [2Fe-2S] cluster to sense superoxide anion radicals (13). In both cases, the transcription factor serves as a sensor and transducer of oxidative stress. Importantly, the oxidized form is the active form of the transcriptional regulator. In contrast, Ref-1 and thioredoxin are required to reduce an intermolecular disulfide bond in the mammalian AP-1 DNA binding domain to allow activation of the expression of its target genes under oxidative-stress-inducing conditions (22). This indicates that the reduced form of AP-1 is the active form. Recently, we found that cysteine residues in the c-CRD function as a redox sensor for Yap1 (34). A disulfide bond is formed between Cys598 and Cys620 when cells are challenged by H2O2 stress. Delaunay et al. (11) reported that a disulfide bond between Cys303 and Cys598 is also formed on exposure to H2O2. These disulfide bonds are reduced by thioredoxin in vitro. Intramolecular disulfide bond formation in the c-CRD may mask the NES, which subsequently blocks the interaction between Yap1 and Crm1 to allow Yap1 to concentrate in the nucleus (34). Yan et al. (68) reported that the interaction between Yap1 and Crm1 is strengthened by dithiothreitol in vitro. Presumably, thioredoxin reduces the disulfide bond(s) of Yap1 in vivo, and so thioredoxin deficiency causes constitutive nuclear localization of Yap1 (30). In addition, we found that a deficiency of thioredoxin reductase (trr1Δ) yielded the same phenotype in terms of the activation of Yap1 (5; S. Izawa and Y. Inoue, unpublished results). Taking into account the property of thioredoxin as a thiol-disulfide oxidoreductase, the oxidized form of Yap1 is the active form. Nevertheless, the present study has demonstrated that Yap1 activity seems to be regulated without redox modification in a glo1Δ mutant.
The intracellular MG content was increased in the glo1Δ mutant, although treatment of yeast cells with H2O2 did not increase the MG level (untreated, 2.14 ± 0.33 nmol/109 cells; treated, 2.17 ± 0.27 nmol/109 cells). Additionally, in a gsh1Δ mutant, intracellular oxidation levels were higher than those in the isogenic wild-type strain and basal expression levels of AP-1- CYC1-lacZ increased 3.5-fold; consequently, the majority of Yap1 was accumulated in the nucleus (data not shown), although the MG content was not increased (wild type, 2.60 ± 0.14 nmol/109 cells; gsh1Δ, 2.55 ± 0.06 nmol/109 cells), even though glutathione is required for the glyoxalase I reaction. Similarly, an increase in the MG content was not observed in a trx1Δ trx2Δ mutant (wild type, 2.60 ± 0.14 nmol/109 cells; trx1Δ trx2Δ, 2.70 ± 0.19 nmol/109 cells). These results, together with the fact that overexpression of TRX1 and TRX2 did not reverse the activation of Yap1 in the glo1Δ mutant (data not shown), suggest that there is no correlation between intracellular redox balance and the MG level in yeast cells.
We have demonstrated that (i) any one of the three cysteine residue(s) is sufficient for Yap1 to translocate into the nucleus and activate the expression of its target gene following MG treatment, (ii) the redox state of Yap1 per se is virtually unchanged with MG treatment, and (iii) the possibility of the existence of an MG sensor protein(s) that competes with Crm1 for binding to the Yap1 NES can be excluded. These observations lead us to propose that Yap1 may be a direct target of MG.
The occurrence of several adducts has been detected when MG and N-acetyl-l-arginine or N-acetyl-l-lysine are incubated in vitro (47). The c-CRD/NES of Yap1 contains two arginine residues (Arg604 and Arg632) and three lysine residues (Lys610, Lys626, and Lys628). Since we have demonstrated that cysteine-substituted mutant proteins GFP-Yap1cm46A5 and GFP-Yap16Cys were not able to locate to the nucleus and activate the expression of Yap1 target genes (Fig. 4), these arginine and/or lysine residues proximal to the cysteine residues in the c-CRD are not likely to be involved in the trafficking regulation of Yap1 by MG. We do not rule out the possibility of the modification of these amino acids by MG in vivo. Nevertheless, such modifications, if any, are less feasible as contributing at least to the MG-dependent localization of Yap1.
The most important aspect of Yap1 activation by MG is that it is a reversible reaction (Fig. 3). Lo et al. (39) reported that MG can form an adduct with N-acetyl-l-cysteine. However, Oya et al. (47) reported that 78% of the arginine and 27% of the lysine residues were lost if bovine serum albumin was incubated with high concentrations of MG for 24 h but that the levels of other amino acid residues were not changed. Therefore, arginine and lysine residues in protein may be preferentially modified by MG compared to cysteine residues, and modification of cysteine residues by MG may be labile and/or reversible. The chemical reaction between a thiol group and aldehyde at physiological pH yields hemithioacetal (62). For example, MG nonenzymatically condenses with glutathione to form hemithioacetal reversibly (Kd = 1 × 10−3 M) (60). One of the possible models that may account for the reversible activation of Yap1 by MG is that cysteine residues in the c-CRD of Yap1 may be modified by MG to form some adducts, presumably hemithioacetal, which may block the interaction between Yap1 and Crm1 which results in Yap1 concentrating in the nucleus. However, an in vivo adduct derived from MG and the cysteine residue of proteins has not yet been detected owing to its unstable and reversible nature (39).
Recently, Azevedo et al. reported that cysteine residues in the c-CRD can be modified by a cysteine-modifying chemical, N-ethylmaleimide, that allows Yap1 to accumulate in the nucleus (2). However, this is not surprising when the nature of the chemical (cysteine modifier) is considered. Furthermore, such a chemical is xenobiotic and not a natural metabolite. In addition to the reversibility of Yap1 activation by MG, attention should be focused on the fact that MG is a natural metabolite synthesized by the ubiquitous energy-generating system, glycolysis.
To explore the physiological relevance of MG-dependent Yap1 activation under normal yeast growth conditions, we monitored MG level with respect to cell growth. The cellular MG level reached maximum during the diauxic shift, and the proportion of cells with the nuclear Yap1 rose concomitantly with the increase in the cellular MG level (Fig. 9). The steady-state level of MG in the log phase for the glo1Δ mutant, in which cell Yap1 is constitutively concentrated in the nucleus, was almost comparable to that for wild-type cells during the diauxic shift (Fig. 9). Energy metabolism markedly changes before and after the diauxic shift; i.e., ATP production depends largely on glycolysis before the shift (fermentative growth), whereas it depends on oxidative phosphorylation in mitochondria during a postdiauxic shift (respiratory growth) (17). Additionally, the production of several stress proteins increases after the diauxic shift, and thus cells in the stationary phase usually exhibit higher tolerance to several environmental stresses (21). Yap1 is one of the transcription factors responsible for the stress response in yeast.
The alterations in protein function due to the posttranslational modification can be provoked by phosphorylation, redox regulation (disulfide bond formation), glutathionylation, ADP ribosylation, acetylation, and so on (16, 18, 41, 48, 59). To our knowledge, this is the first report presenting the possibility of the modulation of a transcription factor function due to potential modification by MG. This study provides new insights into the processes underlying the activation of Yap1. Importantly, intracellular MG levels vary depending on the growth phase, and therefore, the metabolic competence of cells seems to be a factor in this regulatory system. Considering that MG is derived from glycolysis, which is the root of metabolism, we would hypothesize that MG may function as a metabolic signaling molecule.
Acknowledgments
We thank W. S. Moye-Rowley for pSM27, M. Raymond for Ylp368/FLR1, K. Weis for pKW430 and the xpo1-1 mutant (KWY121), and M. Toledano for pRS315-Yap1-9Myc.
This study was partially supported by grants from BRAIN.
REFERENCES
- 1.Abdulnur, S. F. 1976. The interactions of glyoxals with proteins and DNA in relation to cancer. Int. J. Quant. Chem. Quant. Biol. Symp. 3:59-64. [Google Scholar]
- 2.Azevedo, D., F. Tacnet, A. Delaunay, C. Rodrigues-Pousada, and M. B. Toledano. 2003. Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Radic. Biol. Med. 35:889-900. [DOI] [PubMed] [Google Scholar]
- 3.Bito, A., M. Haider, I. Hadler, and M. Breitenbach. 1997. Identification and phenotypic analysis of two glyoxalase II encoding genes from Saccharomyces cerevisiae, GLO2 and GLO4, and intracellular localization of the corresponding proteins. J. Biol. Chem. 272:21509-21519. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Carmel-Harel, O., R. Stearman, A. P. Gasch, D. Botstein, P. O. Brown, and G. Storz. 2001. Role of thioredoxin reductase in the Yap1-dependent response to oxidative stress in Saccharomyces cerevisiae. Mol. Microbiol. 39:595-605. [DOI] [PubMed] [Google Scholar]
- 6.Carmel-Harel, O., and G. Storz. 2000. Roles of the glutathione- and thioredoxin-dependent reduction systems in Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Annu. Rev. Microbiol. 54:439-461. [DOI] [PubMed] [Google Scholar]
- 7.Chae, H. Z., I. H. Kim, K. Kim, and S. G. Rhee. 1993. Cloning, sequencing, and mutation of thiol-specific antioxidant gene of Saccharomyces cerevisiae. J. Biol. Chem. 268:16815-16821. [PubMed] [Google Scholar]
- 8.Chen, C. N., L. Porubleva, G. Shearer, M. Svrakic, L. G. Holden, J. L. Dover, M. Johnston, P. R. Chitnis, and D. H. Kohl. 2003. Associating protein activities with their genes: rapid identification of a gene encoding a methylglyoxal reductase in the yeast Saccharomyces cerevisiae. Yeast 20:545-554. [DOI] [PubMed] [Google Scholar]
- 9.Clugston, S. L., J. F. Barnard, R. Kinach, D. Miedema, R. Ruman, E. Daub, and J. F. Honek. 1998. Overproduction and characterization of a dimeric non-zinc glyoxalase I from Escherichia coli: evidence for optimal activation by nickel ions. Biochemistry 37:8754-8763. [DOI] [PubMed] [Google Scholar]
- 10.Coleman, S. T., E. A. Epping, S. M. Steggerda, and W. S. Moye-Rowley. 1999. Yap1 activates gene transcripiton in an oxidant-specific fashion. Mol. Cell. Biol. 19:8302-8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delaunay, A., A.-D. Isnard, and M. B. Toledano. 2000. H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19:5157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delaunay, A., D. Pflieger, M. B. Barrault, J. Vinh, and M. B. Toledano. 2002. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111:471-481. [DOI] [PubMed] [Google Scholar]
- 13.Ding, H., and B. Demple. 1998. Thiol-mediated disassembly and reassembly of [2Fe-2S] clusters in the redox-regulated transcription factor SoxR. Biochemistry 37:17280-17286. [DOI] [PubMed] [Google Scholar]
- 14.Egyud, L. G., and A. Szent-Gyorgyi. 1966. On the regulation of cell division. Proc. Natl. Acad. Sci. USA 56:203-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fodor, G., J.-P. Sachetto, A. Szent-Gyorgyi, and L. B. Egyud. 1967. Ketone aldehydes in animal tissues. Proc. Natl. Acad. Sci. USA 57:1644-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frand, A. R., J. W. Cuozzo., and C. A. Kaiser. 2000. Pathways for protein disulphide bond formation. Trends Cell Biol. 10:203-210. [DOI] [PubMed] [Google Scholar]
- 17.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghezzi, P., and V. Bonetto. 2003. Redox proteomics: identification of oxidatively modified proteins. Proteomics 3:1145-1153. [DOI] [PubMed] [Google Scholar]
- 19.Grant, C. M., L. P. Collinson, J. H. Roe, and I. W. Dawes. 1996. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol. Microbiol. 21:171-179. [DOI] [PubMed] [Google Scholar]
- 20.Harshman, K. D., W. S. Moye-Rowley, and C. S. Parker. 1988. Transcriptional activation by the SV40 AP-1 recognition element in yeast is mediated by a factor similar to AP-1 that is distinct from GCN4. Cell 53:321-330. [DOI] [PubMed] [Google Scholar]
- 21.Herman, P. K. 2002. Stationary phase in yeast. Curr. Opin. Microbiol. 5:602-607. [DOI] [PubMed] [Google Scholar]
- 22.Hirota, K., M. Matsui, S. Iwata, A. Nishiyama, K. Mori, and J. Yodoi. 1997. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc. Natl. Acad. Sci. USA 94:3633-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue, Y., B.-Y. Choi, K. Murata, and A. Kimura. 1990. Sexual response of Saccharomyces cerevisiae: phosphorylation of yeast glyoxalase I by a cell extract of mating factor-treated cells. J. Biochem. (Tokyo) 108:4-6. [DOI] [PubMed] [Google Scholar]
- 24.Inoue, Y., H. Rhee, K. Watanabe, K. Murata, and A. Kimura. 1987. Metabolism of 2-ketoaldehydes in mold: purification and characterization of glyoxalase I from Aspergillus niger. J. Biochem. (Tokyo) 102:583-589. [DOI] [PubMed] [Google Scholar]
- 25.Inoue, Y., T. Matsuda, K. Sugiyama, S. Izawa, and A. Kimura. 1999. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J. Biol. Chem. 274:27002-27009. [DOI] [PubMed] [Google Scholar]
- 26.Inoue, Y., Y. Tsujimoto, and A. Kimura. 1998. Expression of the glyoxalase I gene of Saccharomyces cerevisiae is regulated by high osmolarity glycerol mitogen-activated protein kinase pathway in osmotic stress response. J. Biol. Chem. 273:2977-2983. [DOI] [PubMed] [Google Scholar]
- 27.Inoue, Y., and A. Kimura. 1995. MG and regulation of its metabolism in microorganisms. Adv. Microb. Physiol. 37:177-227. [DOI] [PubMed] [Google Scholar]
- 28.Inoue, Y., and A. Kimura. 1996. Identification of the structural gene for glyoxalase I from Saccharomyces cerevisiae. J. Biol. Chem. 271:25958-25965. [PubMed] [Google Scholar]
- 29.Iyengar, R., and I. A. Rose. 1981. Concentration of activated intermediates of the fructose-1,6-bisphosphate aldolase and triosephosphate isomerase reactions. Biochemistry 20:1223-1229. [DOI] [PubMed] [Google Scholar]
- 30.Izawa, S., K. Maeda, K. Sugiyama, J. Mano, Y. Inoue, and A. Kimura. 1999. Thioredoxin deficiency causes the constitutive activation of Yap1, an AP-1-like transcription factor in Saccharomyces cerevisiae. J. Biol. Chem. 274:28459-28465. [DOI] [PubMed] [Google Scholar]
- 31.Jamieson, D. J., and G. Storz. 1997. Transcriptional regulators of oxidative stress responses, p. 91-115. In J. G. Scandalios (ed.), Oxidative stress and the molecular biology of antioxidant defenses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Kalapos, M. P. 1999. Methylglyoxal in living organisms: chemistry, biochemistry, toxicology and biological implications. Toxicol. Lett. 110:145-175. [DOI] [PubMed] [Google Scholar]
- 33.Kang, Y., L. G. Edwards, and P. J. Thornalley. 1996. Effect of methylglyoxal on human leukaemia 60 cell growth: modification of DNA G1 growth arrest and induction of apoptosis. Leuk. Res. 20:397-405. [DOI] [PubMed] [Google Scholar]
- 34.Kuge, S., M. Arita, A. Murayama, K. Maeta, S. Izawa, Y. Inoue, and A. Nomoto. 2001. Regulation of the yeast Yap1 nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol. Cell. Biol. 21:6139-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuge, S., N. Jones, and A. Nomoto. 1997. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 16:1710-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuge, S., T. Toda, N. lizuka, and A. Nomoto. 1998. Crm1 (Xpol) dependent nuclear export of the budding yeast transcription factor yAP-1 is sensitive to oxidative stress. Genes Cells 3:521-532. [DOI] [PubMed] [Google Scholar]
- 37.Kuge, S., and N. Jones. 1994. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274:16040-16046. [DOI] [PubMed] [Google Scholar]
- 39.Lo, T. W. C., M. E. Westwood, A. C. McLellan, T. Selwood, and P. J. Thornalley. 1994. Binding and modification of proteins by MG under physiological conditions. A kinetic and mechanistic study with Nα- acetylarginine, Nα-acetylcysteine, and Nα-acetyllysine, and bovine serum albumin. J. Biol. Chem. 269:32299-32305. [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Moore, B. R., and S. J. Free. 1985. Protein modification and its biological role. Int. J. Biochem. 17:283-289. [DOI] [PubMed] [Google Scholar]
- 42.Moye-Rowley, W. S. 2003. Regulation of the transcriptional response to oxidative stress in fungi: similarities and differences. Eukaryot. Cell 2:381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moye-Rowley, W. S., K. D. Harshman, and C. S. Parker. 1989. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 3:283-292. [DOI] [PubMed] [Google Scholar]
- 44.Netto, L. E. S., H. Z. Chae, S. W. Kang, S. G. Rhee, and E. R. Stadtman. 1996. Removal of hydrogen peroxide by thiol-specific antioxidant enzyme (TSA) is involved with its antioxidant properties. TSA possesses thiol peroxidase activity. J. Biol. Chem. 271:15315-15321. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen, D. T., A. M. Alarco, and M. Raymond. 2001. Multiple Yap1- binding sites mediate induction of the yeast major facilitator FLR1 gene in response to drugs, oxidants, and alkylating agents. J. Biol. Chem. 276:1138-1145. [DOI] [PubMed] [Google Scholar]
- 46.Okazaki, S., A. Naganuma, and S. Kuge. Peroxiredoxin-mediated redox regulation of the nuclear localization of Yap1, a transcription factor in budding yeast. Antioxidant Redox Signaling, in press. [DOI] [PubMed]
- 47.Oya, T., N. Hattori, Y. Mizuno, S. Miyata, S. Maeda, T. Osawa, and K. Uchida. 1999. MG modification of protein. Chemical and immunochemical characterization of MG-arginine adducts. J. Biol. Chem. 274:18492-18502. [DOI] [PubMed] [Google Scholar]
- 48.Polevoda, B., and F. Sherman. 2002. The diversity of acetylated proteins. Genome Biol. 3:0006.1-0006.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richard, J. P. 1991. Kinetic parameters for the elimination reaction catalyzed by triosephosphate isomerase and an estimation of the reaction's physiological significance. Biochemistry 30:4581-4585. [DOI] [PubMed] [Google Scholar]
- 50.Richard, J. P. 1993. Mechanism for the formation of MG from triosephosphates. Biochem. Soc. Trans. 21:549-553. [DOI] [PubMed] [Google Scholar]
- 51.Schena, M., D. Picard, and K. R. Yamamoto. 1991. Vectors for constitutive and inducible gene expression in yeast. Methods Enzymol. 194:389-398. [DOI] [PubMed] [Google Scholar]
- 52.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shapiro, R., B. I. Cohen, S. J. Shiuey, and H. Maurer. 1969. On the reaction of guanine with glyoxal, pyruvaldehyde, and kethoxal, and the structure of the acylguanines. A new synthesis of N2-alkylguanines. Biochemistry 8:238-245. [DOI] [PubMed] [Google Scholar]
- 54.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 55.Sugiyama, K., S. Izawa, and Y. Inoue. 2000. The Yap1-dependent induction of glutathione synthesis in heat shock response of Saccharomyces cerevisiae. J. Biol. Chem. 275:15535-15540. [DOI] [PubMed] [Google Scholar]
- 56.Szent-Gyorgyi, A., L. G. Egyud, and J. A. McLaughlin. 1967. Keto- aldehydes and cell division. Science 155:539-541. [DOI] [PubMed] [Google Scholar]
- 57.Thornalley, P. J. 1993. Modification of the glyoxalase system in disease processes and prospects for therapeutic strategies. Biochem. Soc. Trans. 21:531-534. [DOI] [PubMed] [Google Scholar]
- 58.Tsuzi, D., K. Maeta, Y. Takatsume, S. Izawa, and Y. Inoue. 2004. Regulation of the yeast phospholipid hydroperoxide glutathione peroxidase GPX2 by oxidative stress is mediated by Yap1 and Skn7. FEBS Lett. 565:148-154. [DOI] [PubMed] [Google Scholar]
- 59.Ueda, K., and O. Hayaishi. 1985. ADP-ribosylation. Annu. Rev. Biochem. 54:73-100. [DOI] [PubMed] [Google Scholar]
- 60.Vander Jagt, D. L. 1993. Glyoxalase II: molecular characteristics, kinetics and mechanism. Biochem. Soc. Trans. 21:522-527. [DOI] [PubMed] [Google Scholar]
- 61.Veal, E. A., S. J. Ross, P. Malakasi, E. Peacock, and B. A. Morgan. 2003. Ybp1 is required for the hydrogen peroxide-induced oxidation of the Yap1 transcription factor. J. Biol. Chem. 278:30896-30904. [DOI] [PubMed] [Google Scholar]
- 62.Weber, A. L. 1984. Prebiotic formation of ′energy-rich' thioesters from glyceraldehyde and N-acetylcysteine. Origins Life Evol. Biosph. 15:17-27. [DOI] [PubMed] [Google Scholar]
- 63.Wemmie, J. A., M. S. Szczypka, D. J. Thiele, and W. S. Moye-Rowley. 1994. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J. Biol. Chem. 269:32592-32597. [PubMed] [Google Scholar]
- 64.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 65.Westwood, M. E., and P. J. Thornalley. 1995. Molecular characteristics of methylglyoxal-modified bovine and human serum albumins. Comparison with glucose-derived advanced glycation endproduct-modified serum albumins. J. Protein Chem. 14:359-372. [DOI] [PubMed] [Google Scholar]
- 66.Wiatrowski, H. A., and M. Carlson. 2003. Yap1 accumulates in the nucleus in response to carbon stress in Saccharomyces cerevisiae. Eukaryot. Cell 2:19-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu, A. L., and W. S. Moye-Rowley. 1994. GSH1, which encodes γ- glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol. Cell. Biol. 14:5832-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan, C., L. H. Lee, and L. I. Davis. 1998. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 17:7416-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng, M., F. Aslund, and G. Storz. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718-1721. [DOI] [PubMed] [Google Scholar]
- 70.Zheng, M., and G. Storz. 2000. Redox sensing by prokaryotic transcription factors. Biochem. Pharmacol. 59:1-6. [DOI] [PubMed] [Google Scholar]