Abstract
The Tor kinases are the targets of the immunosuppressive drug rapamycin and couple nutrient availability to cell growth. In the budding yeast Saccharomyces cerevisiae, the PP2A-related phosphatase Sit4 together with its regulatory subunit Tap42 mediates several Tor signaling events. Sit4 interacts with other potential regulatory proteins known as the Saps. Deletion of the SAP or SIT4 genes confers increased sensitivity to rapamycin and defects in expression of subsets of Tor-regulated genes. Sap155, Sap185, or Sap190 can restore these responses. Strains lacking Sap185 and Sap190 are hypersensitive to rapamycin, and this sensitivity is Gcn2 dependent and correlated with a defect in translation, constitutive eukaryotic initiation factor 2α hyperphosphorylation, induction of GCN4 translation, and hypersensitivity to amino acid starvation. We conclude that Tor signals via Sap-Sit4 complexes to control both transcriptional and translational programs that couple cell growth to amino acid availability.
The Tor kinases are key controllers of cell growth in eukaryotic cells. Rapamycin is a natural product that inhibits the activity of the Tor kinases in an evolutionarily conserved mechanism that leads to cell growth arrest in a manner similar to nutrient depletion. While a detailed understanding of the signals that regulate the activity of the Tor kinases is at this time unknown, recent evidence supports the notion that the Tor signaling program is dedicated to sensing amino acids. First, the tumor suppressor genes Tsc1 and Tsc2 were shown to act through the GTP binding protein Rheb to regulate Tor activity and, recently, homologs of Tsc1 and Tsc2 were found to control amino acid uptake in Schizosaccharomyces pombe (29, 47, 49). Second, a direct binding partner of Tor, GβL, was shown to regulate Tor kinase activity in response to leucine levels in the medium (26). The yeast homolog of GβL, Lst8, plays an important role in the regulation of amino acid levels as part of the Tor pathway and has been identified as a Tor interacting protein (8, 30, 51). The idea that Tor is involved in sensing an adequate supply of amino acids is also congruent with the results of genome-wide expression profiling that has identified genes whose expression is rapamycin sensitive. Hallmarks of the rapamycin-induced transcriptional program include the rapid repression of genes that promote growth, such as those involved in ribosome biogenesis, and concomitant induction of genes required for utilization of poor-quality nutrients and adaptation to environmental stress (5, 7, 17, 27, 37).
In Saccharomyces cerevisiae, the Tor kinases repress the transcription of genes subject to nitrogen catabolite repression, those involved in the retrograde response, the STRE genes, and subtelomeric genes encoding cell wall proteins. Simultaneously, Tor promotes the expression of genes involved in ribosome biogenesis, including the polymerase II-regulated ribosomal protein (RP) genes. Tor signaling controls these sets of genes either by affecting the nuclear localization of the transcription factors (5, 6, 12, 27) or the occupancy of factors that control histone acetylation at the target promoters (1, 39). While DNA-bound transcription factors that control the expression of the rapamycin-induced transcriptome have been identified in many cases, an understanding of the signaling events emanating from inactivation of the Tor kinases and sensed by these factors remains obscure.
In both yeast and mammals, many of the effects of Tor signaling are thought to be mediated by PP2A and PP2A-related phosphatases. In yeast a key phosphatase associated with Tor signaling is Sit4 (11). In some cases there has been a clear demonstration of a role for Sit4 in the regulation of genes subject to Tor control (5-7, 41); however, in other cases the involvement of Sit4 remains unclear (41). Sit4 is regulated, at least in part, by its interaction with Tap42 and its associated protein Tip41, as well as via a set of high-molecular-weight proteins known as the Sit4-associated proteins (Saps) (33). The exact role of these factors is as yet unclear. Tap42 is essential and is thought to either regulate the activity or the substrate specificity of Sit4. Mutant forms of Tap42 that are defective in binding to Sit4 confer rapamycin resistance and block many of the effects of Tor signaling (11, 12, 25, 50). Tap42 also plays a positive signaling role, and tap42 temperature-sensitive alleles impair gene induction at the nonpermissive temperature (7, 12). Tip41 is thought to promote the dissociation of Tap42 from Sit4 and, thus, favor the active form of the phosphatase (24).
The Sap proteins were identified biochemically as Sit4-associated proteins, which share significant homology and appear to compete with one another for binding to Sit4 (23, 33). In pairwise comparisons, Sap185 and Sap190 as well as Sap4 and Sap155 shared the highest similarity to each other and have been proposed to have overlapping functions (23, 33). Consistent with the idea that these genes have a redundant function, the two SAP gene pairs lie in syntenic regions of the yeast genome thought to have arisen from a genome endoduplication event (53). Importantly, cells lacking SAP155, SAP185, and SAP190 exhibit the same growth defect as cells with SIT4 deleted (33). These and other genetic data suggest that the Sap proteins act positively and in concert with Sit4, either by directing substrate specificity or by regulating phosphatase activity (23, 33).
In mammalian cells, flies, and nematodes, the Tor pathway is primarily associated with the regulation of translation in response to amino acid availability. In yeast cells, amino acid depletion results in the activation of the highly conserved Gcn2 kinase, which in turn phosphorylates the translation factor eukaryotic initiation factor 2α (eIF2α). This phosphorylation event slows the rate of GDP-GTP exchange on eIF2α and reduces the overall rate of translation initiation. In yeast, slowing of translation initiation selectively favors translation of the GCN4 mRNA. The leader region of the GCN4 mRNA features four short open reading frames that effect nutrient-regulated expression by a specialized reinitiation mechanism (18). Gcn4 is a transcriptional activator that directs expression of large, diverse sets of genes, most notably for those encoding amino acid biosynthetic enzymes (21, 34). This stringent mode of translational regulation is known as general amino acid control (GAAC). Recent work has identified cross talk between the Tor nutrient-sensing and the Gcn2 amino acid-sensing pathways (9, 28, 48). The Tor kinases were shown to promote the phosphorylation of Gcn2 at serine 577 (Ser577), as inhibition of Tor by rapamycin resulted in rapid dephosphorylation of this amino acid residue, Gcn2 activation, and phosphorylation of eIF2α.
Here, we dissect the role of the Sap proteins in Tor signaling. Deletion of either the four SAP genes or the SIT4 gene blocked expression of the rapamycin-induced NCR (nitrogen catabolite-repressed) genes as well as the Rtg1/3-regulated gene CIT2. We found that Sap155, Sap185, and Sap190, on their own, can mediate transcription of these Tor-regulated genes, whereas Sap4 is unable to suffice for this function. Remarkably, Sap185 and Sap190 are more effective that Sap155 and Sap4 at dampening the toxic effects of rapamycin. Genetic and biochemical data showed that Sap185, Sap190, and Sit4 are required for constitutive dephosphorylation of eIF2α. Accordingly, a sap185 sap190 mutant strain showed increased sensitivity to amino acid deprivation, constitutive phosphorylation of eIF2α, increased translation of GCN4 mRNA, and an exacerbated GAAC response upon Tor inhibition by rapamycin. Our results suggest that Tor signals via Sap-Sit4 phosphatase complexes to govern transcription and translation.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions.
Strains used in this study are listed in Table 1. Plasmids encoding the FLAG epitope-tagged Gcn2(pDH101) and FLAG epitope-tagged Gcn2-S577A(pCB149) as well as the GCN4-lacZ reporter in plasmid p180 were generously provided by Alan Hinnebusch and were described previously (9, 15, 19). Yeast was grown on yeast extract-peptone-dextrose (YEPD) or synthetic complete medium as described elsewhere (42). Rapamycin was added from concentrated stocks maintained in 5% Tween 20 and 95% ethanol. Unless otherwise noted, mutant yeast strains were constructed by PCR-mediated gene disruptions, replacing the entire open reading frame of the targeted gene with the kanMX, hygB, or NAT markers as described previously (16, 31). All gene deletions were confirmed by PCR. Yeast transformations were performed using the lithium acetate method (40). All strains used in this study were derived from the wild-type MLY41 (Σ1278b background) (32). Strains JRY40, JRY43, JRY44, JRY45, and JRY46 were created by crossing JRY29 to SCY115. This strain was sporulated, and the progeny were dissected and the genotypes were determined by PCR analysis. Strain JRY48 was generated by mating JRY29 to SCY51α. This strain was sporulated, and the progeny were verified by PCR analysis. Strain SCY94 was previously described (10).
TABLE 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| MLY41 | MATaura3-52 (Σ1278b background) | Lorenz et al. (32) |
| JRY32 | MATaura3-52 hisG-leu2::URA3-hisG | This study |
| SCY115 | MATα ura3-52 sap4::kanMX sap155::hygB | This study |
| SCY94 | MATaura3-52 sit4::kanMX | Cutler et al. (10) |
| SCY51 | MATα ura3-52 gcn2::kanMX | This study |
| JRY20 | MATaura3-52 sap185::hygB | This study |
| JRY21 | MATaura3-52 sap190::kanMX | This study |
| JRY29 | MATaura3-52 sap185::hygB sap190::kanMX | This study |
| JRY40 | MATaura3-52 sap4::kanMX sap155::hygB sap185::hygB sap190::kanMX | This study |
| JRY39 | MATaura3-52 tip41::NAT | This study |
| JRY43 | MATα ura3-52 sap4::kanMX sap155::hygB sap190::kanMX | This study |
| JRY44 | MATaura3-52 sap4::kanMX sap155::hygB sap185::hygB | This study |
| JRY45 | MATα ura3-52 sap155::hygB sap185::hygB sap190::kanMX | This study |
| JRY46 | MATα ura3-52 sap4::kanMX sap185::hygB sap190::kanMX | This study |
| JRY48 | MATα ura3-52 sap185::higB sap190::kanMX gcn2::kanMX | This study |
| JRY49 | MATα ura3-52 gln3::kanMX gcn4::kanMX | This study |
Northern and Western blotting.
RNA isolation and Northern blot analysis were performed as described previously (7), and specific signals were quantified with a Typhoon 9200 variable mode imager using the Image Quantifier 5.2 software (Molecular Dynamics). Whole-cell extracts were prepared from exponentially growing cultures treated with rapamycin as indicated. Cells were harvested and subjected to mechanical breakage with glass beads in lysis buffer containing 50 mM KHPO4 (pH 7.4), 50 mM KCl, 2 mM EDTA, 25 mM β-glycerophosphate, 25 mM NaF, 2 mM benzamidine, 0.5% Triton X-100, and 1 mM dithiothreitol, with the protease inhibitors leupeptin, aprotinin, and pepstatin added to 1 μg/ml and phenylmethylsulfonyl fluoride added to 0.5 mM. For GCN2 Ser577-P analysis, 3 mg of protein extract was immunoprecipitated with 30 μl of anti-Flag antibody immobilized in agarose beads (Sigma) for 1 to 2 h at 4°C. Immunoprecipitates were washed four times with lysis buffer, resolved in sodium dodecyl sulfate-Tris-glycine-4 to 12% polyacrylamide gels, transferred to polyvinylidene difluoride membranes, and probed with antibodies specific for Ser577-P (a kind gift of A. Hinnebusch). The blots were stripped and reprobed with anti-Flag antibody (Sigma). For analysis of eIF2α, 75 μg of protein was resolved on sodium dodecyl sulfate-Tris-glycine-4 to 20% polyacrylamide gels and subjected to Western blot analysis as above with antibodies specific to the phosphorylated Ser51 residue of eIF2α (Biosource International). For a loading control, immunoblots were probed with antibodies that recognized both the phosphorylated and unphosphorylated forms of eIF2α from yeast (a generous gift of A. Hinnebusch). Western blotting results were quantified by video densitometry with the NIH Image program, version 1.62.
Analysis of ribosome distribution on sucrose gradients.
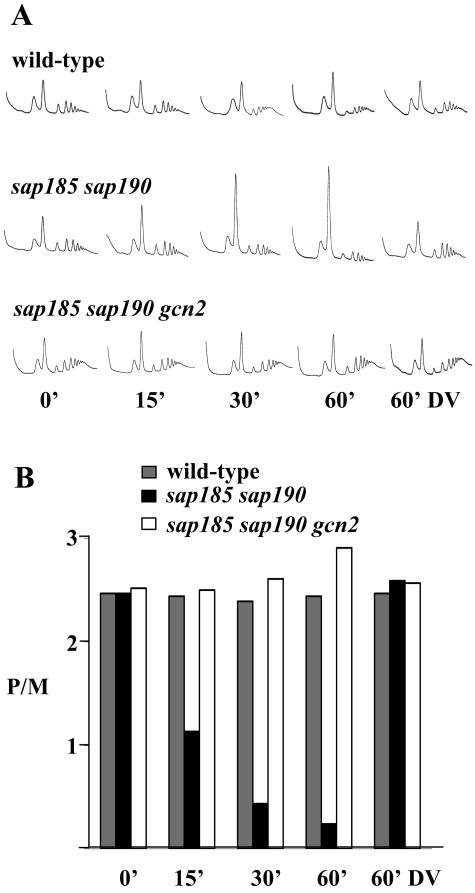
Yeast cultures were grown to an optical density at 600 nm of 0.7 and treated by addition of 200 nM rapamycin (an equal volume of drug vehicle alone was used as a control). Extracts were prepared in 100 μg of cycloheximide/ml, and these were layered onto 15-to-50% sucrose gradients. The gradients were sedimented via centrifugation at 285,000 × g for 2.5 h, and the A254 was measured continuously to yield the traces shown below in Fig. 6A (2).
FIG. 6.
The sap185 sap190 mutant strain exhibits Gcn2-dependent rapamycin-hypersensitive translation. (A) Polysome traces from isogenic wild-type (MLY41) and mutant strains sap185 sap190 (JRY29) and sap185 sap190 gcn2 (JRY48). Strains were grown in YEPD medium, and either 200 nM rapamycin or drug vehicle (DV) was added for the indicated time periods. Polysomes were analyzed as described in Materials and Methods. The P/M ratio for each strain is plotted in panel B.
β-Galactosidase assays.
The GCN4-lacZ reporter in plasmid p180 contains the four upstream open reading frames required for proper regulation of GCN4 translation (19). Cultures of cells harboring p180 were grown overnight in synthetic complete medium lacking uracil and split and grown to early log phase before rapamycin was added, and cells were assayed for β-galactosidase activity using a permeabilized cell method (54). All results are an average of at least two independent determinations, with a 5 to 10% average deviation from the mean.
RESULTS
SAP genes have overlapping and specific roles in Tor signaling and growth.
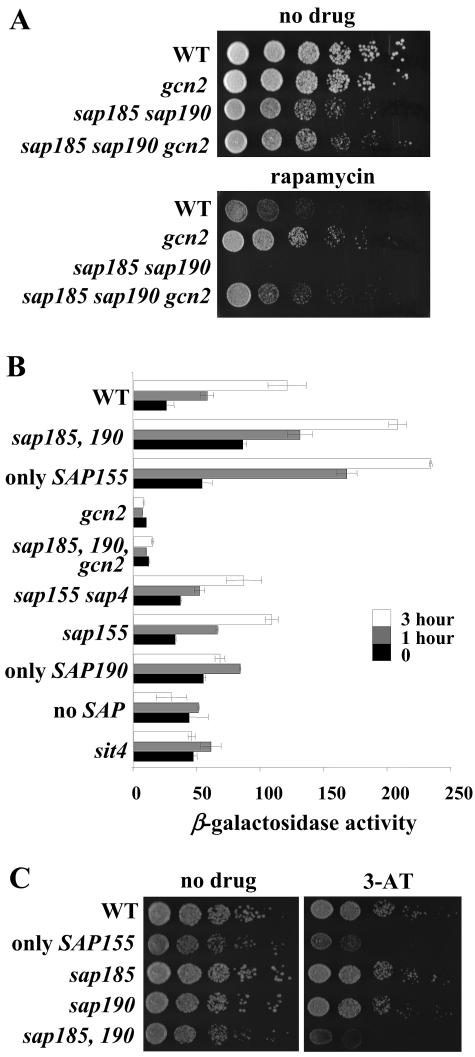
We sought to dissect Tor signaling downstream of the Sit4 phosphatase complex by examining the rapamycin-induced transcriptional profile in strains mutated for the different genes encoding individual Sit4-associated subunits. We constructed strains that had all four SAP genes deleted as well as strains that contained a pair or only one individual wild-type SAP gene. By this approach, we examined the contribution of each individual SAP gene to Tor signaling. In accord with earlier studies, we found that deletion of all four SAP genes resulted in a slow growth phenotype similar to that observed in a strain with sit4 deleted. A strain that expressed only Sap4 (sap155 sap190 sap185 mutant) also exhibited a slow growth phenotype, indicating that Sap4 is ineffective at providing Sap function (Fig. 1) (33). The quadruple sap4 sap155 sap185 sap190 mutant and the strain with sit4 deleted were both impaired for growth and, in addition, they were both extremely sensitive to rapamycin (Fig. 1). Cells expressing only Sap185 (sap4 sap155 sap190 mutant) or only Sap190 (sap4 sap155 sap185 mutant) grew at rates similar to wild-type cells and displayed a slightly higher tolerance to rapamycin, indicating that Sap190-Sit4 complexes are especially effective at dampening the toxic effects of rapamycin (Fig. 1 and data not shown). In fact, cells that contained only Sap185 or only Sap190 were as rapamycin resistant as a strain with tip41 deleted which is known to be rapamycin resistant (Fig. 1) (24). We found that sap185 or sap190 deletions alone did not significantly alter sensitivity to rapamycin while, in contrast, the sap185 sap190 double deletion resulted in a dramatic hypersensitivity to rapamycin even at concentrations as low as 20 nM (Fig. 1). Accordingly, cells that expressed only Sap155 were also hypersensitive to rapamycin (Fig. 1). We conclude that Sap185 and Sap190 share a redundant role in Tor signaling. One interpretation of our data is that Sap185-Sit4 and Sap190-Sit4 complexes may be responsible for dephosphorylation of targets that predispose cells to the toxic effects of rapamycin.
FIG. 1.
Individual Sap proteins play distinct roles in Tor signaling. Isogenic wild-type (WT; MLY41a), sit4 (SCY94), no-SAP (sap4 sap155 sap185 sap190; JRY40), only-SAP4 (sap155 sap185 sap190; JRY45), only-SAP185 (sap4 sap155 sap190; JRY43), only-SAP190 (sap4 sap155 sap185; JRY44), tip41 (JRY39), only-SAP155 (sap4 sap185 sap190; JRY46), sap185 (JRY20), sap190 (JRY21), and sap185 sap190 (JRY29) strains were grown overnight at 30°C in YEPD. Equivalent numbers of cells were serially diluted, and aliquots were spotted onto plates of YEPD or YEPD containing 0, 20, or 100 nM rapamycin (rapa). After 3 days of incubation at 30°C, the plates were photographed.
Sap proteins are required for the rapamycin-induced transcriptional profile of a subset of Tor-regulated genes.
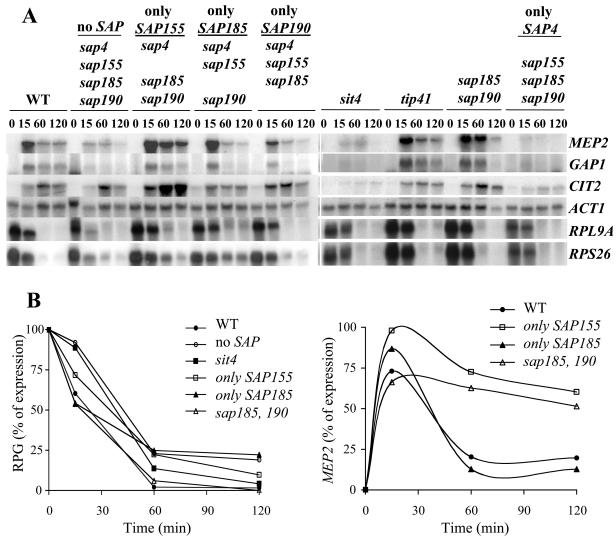
To gain insight into the mechanism(s) of the altered sensitivity to rapamycin caused by the SAP mutations, we tested the expression of genes subject to Tor control in the different sap and sit4 mutant strains. We observed that either deletion of all four SAP genes or deletion of SIT4 prevented the rapamycin-mediated induction of the NCR genes, such as MEP2 and GAP1. In addition, we found that induction of the Rtg-controlled gene CIT2 was normally expressed in the quadruple sap mutant but was impaired for induction in the sit4 mutant strain (Fig. 2A). In contrast, we observed only minor differences in the rapamycin-induced repression of the RP genes in these mutant strains, as quantified in Fig. 2B.
FIG. 2.
The Sap products are required for proper induction of a subset of Tor-regulated genes. (A) Exponentially growing cultures of isogenic wild-type (JRY32), no-SAP (sap4 sap155 sap185 sap190; JRY40), only-SAP155 (sap4 sap185 sap190; JRY46), only-SAP185 (sap4 sap155 sap190; JRY43), only-SAP190 (sap4 sap155 sap185; JRY44), sit4 (SCY94), tip41 (JRY39), sap185 sap190 (JRY29), and only-SAP4 (sap155 sap185 sap190; JRY45) strains were treated with 100 nM rapamycin for 0, 15, 60, or 120 min. RNA was prepared and analyzed by Northern blotting with radioactive probes that hybridize to the genes indicated at the right. (B) Northern blot signals for the RP genes (RPG) and MEP2 were quantified and normalized to the ACT1 loading control. Relative expression values for the two RP genes examined were averaged. The results shown in the graphs are the percentage of gene expression, with the time zero value as 100% (for the RP genes) or the strain with the maximal level of expression at 15 min as 100% (for MEP2).
Strains expressing only Sap185 (sap4 sap155 sap190 mutant) or only Sap190 (sap4 sap155 sap185 mutant) showed no defects in inducing the NCR genes or the retrograde response gene CIT2 upon treatment with rapamycin (Fig. 2A). Interestingly, cells expressing only Sap155 (sap4 sap185 sap190 mutant) and the sap185 sap190 double mutant showed an enhanced and sustained induction of CIT2 and MEP2 not observed in the wild-type strain (Fig. 2A and MEP2 quantification shown in B). We conclude that Sap155, Sap185, and Sap190 are on their own sufficient for induction of Tor-repressed genes. Furthermore, these results suggest that Sap155 is more efficient than the other Saps in directing expression of Tor-repressed genes. Alternatively, the absence of Sap185 and Sap190 function (in the sap185 sap190 and sap4 sap185 sap190 mutant strains) exacerbated the NCR response and the expression of CIT2 induced by rapamycin (see next section). Importantly, repression of the RP genes in response to rapamycin did not appear to be mediated via Sap-Sit4 complexes.
Tor control of Gcn2-dependent translation is mediated by Sap-Sit4 complexes.
With minor exceptions (i.e., enhanced and sustained expression of MEP2 and CIT2), the rapamycin-induced transcriptional profile of the sap185 sap190 mutant strain differed little from that observed in wild-type cells. While it is possible that these strains differ more substantially in the rapamycin-induced change in transcription of a subset of genes not examined in our studies, the dramatic hypersensitivity to rapamycin exhibited by this strain cannot readily be explained by differences in the transcription of known Tor target genes. Recent reports have suggested that the GAAC response can be activated by inhibition of Tor with rapamycin (9, 28, 48). Moreover, Tor is thought to negatively regulate this pathway by inhibiting the Gcn2 kinase (9, 28).
In response to amino acid depletion or inhibition of Tor by rapamycin, there is an overall decrease in translation. In part, this is achieved through the phosphorylation of eIF2α mediated by the activated Gcn2 kinase (9, 21). This response results in the translation of a subset of messages that include the mRNA for the transcription factor Gcn4. Gcn4 is required for the induction of a panoply of genes involved in amino acid biosynthesis, such as CIT2 and the transcription factor for the NCR genes Gln3 (21). Thus, we hypothesized that the sustained expression of MEP2 and CIT2 resulted from an exacerbated GAAC response elicited by the sap185 sap190 double mutation. To test this possibility, we compared the kinetics of induction of the Gcn4 target gene HIS7 and MEP2 in response to rapamycin (Fig. 3). Interestingly, expression of HIS7 in both the wild-type and the sap185 sap190 strains was delayed compared to that of MEP2. This result was consistent with the interpretation that Tor inhibition must first act to enable Gcn4 translation before activation of its target genes is achieved. In addition, the sap185 sap190 mutant strain exhibited a sustained MEP2 induction that was abolished by mutation of GCN2 (Fig. 3). These results suggest a role for Sap185 and Sap190 in the GAAC response regulated by Tor.
FIG. 3.
Rapamycin-induced activation of Gcn4 target genes requires Gcn2. Exponentially growing cultures of isogenic wild-type (MLY41), sap185 sap190 (JRY29), sap185 sap190 gcn2 (JRY48), and gcn4 gln3 (JRY49) mutant strains were treated with 100 nM rapamycin for 0, 15, or 60 min. RNA was prepared and analyzed by Northern blotting with radioactive probes that hybridize to the genes indicated at the left. The Northern blot specific signal for MEP2 was quantified as indicated in the Fig. 2 legend. The results are presented in the graph as the percentage of gene expression, assigning as 100% the expression level detected at 15 min.
We sought to examine the contribution of the Sap proteins to the GAAC response triggered by rapamycin. To this end, we tested if the altered sensitivity to rapamycin seen in the sap185 sap190 mutant strain was dependent upon Gcn2 activity. Deletion of GCN2 resulted in an increase in rapamycin resistance relative to that of the wild-type strain (Fig. 4A). Remarkably, the gcn2 mutation suppressed the rapamycin hypersensitivity of the sap185 sap190 mutant strain, and a gcn2 sap185 sap190 triple mutant strain was nearly as rapamycin resistant as the gcn2 mutant strain (Fig. 4A). These results suggest that the rapamycin hypersensitivity of the sap185 sap190 mutant strain is mediated by Gcn2. This result is reminiscent of earlier epistasis experiments with the activator of the NCR genes, Gln3, and its repressor, Ure2. While ure2 cells are rapamycin hypersensitive, ure2 gln3 double mutant cells are as rapamycin resistant as gln3 mutant cells (7).
FIG. 4.
The Sap proteins are required for regulation of GCN4 translation via Gcn2. (A) Isogenic wild-type (WT; MLY41a), gcn2 (SCY51), sap185 sap190 (JRY29), and sap185 sap190 gcn2 (JRY48) mutant strains were grown, serially diluted, and spotted onto plates of YEPD with or without 20 nM rapamycin as indicated in Fig. 1. After 3 days incubation at 30°C, the plates were photographed. (B) Cultures of isogenic wild-type (MLY41) or sap185 sap190 (JRY29), only-SAP155 (sap4 sap185 sap190; JRY46), gcn2 (SCY51), sap185 sap190 gcn2 (JRY48), sap155 sap4 (SCY115), sap155 (SCY108), only-SAP190 (sap4 sap155 sap185; JRY44), no-SAP (sap4 sap155 sap185 sap190; JRY40), and sit4 (SCY94) mutant strains harboring the GCN4-lacZ reporter plasmid p180 were grown to exponential phase in SD-Ura medium. Cultures were treated with 100 nM rapamycin for 0, 60, and 180 min and analyzed for β-galactosidase activity. (C) Isogenic wild-type (MLY41a), only-SAP155 (sap4 sap185 sap190; JRY46), sap185 (JRY20), sap190 (JRY21), and sap185 sap190 (JRY29) strains were spotted onto YEPD medium with or without 30 mM 3-AT. After 3 days of incubation at 30°C, the plates were photographed.
Next, we tested the effects of the sap185 sap190 double mutation on the activity of a GCN4-lacZ reporter containing the four upstream open reading frames required for proper regulation of GCN4 translation (19). The sap185 sap190 mutant strain exhibited a basal level of GCN4-lacZ reporter expression that was threefold higher than that in wild-type cells, and GCN4 expression was potently increased in response to rapamycin to an induced level roughly twice that of the wild-type strain (Fig. 4B). In accord with this result, the strain that had only Sap155 activated GCN4-lacZ to higher-than-wild-type levels, similar to that seen with the strain with sap185 and sap190 (Fig. 4B). Moreover, in agreement with previous findings (9, 28), both in the wild-type strain as well as in the sap185 sap190 mutant strain this activation was Gcn2 dependent (Fig. 4B). Interestingly, sap155 or sap4 sap155 mutant strains as well as the strain containing only Sap190 (sap4 sap155 sap185) activated the GCN4-lacZ reporter at levels nearly identical to those with the wild-type strain. These results indicated that Sap185 and Sap190 functions are required for the effective control of GCN4 expression downstream from Tor. Although strains lacking either the four SAP genes or the SIT4 gene showed a basal level of GCN4 expression higher than that with the wild-type strain, these mutant strains were severely impaired for induction of the GCN4-lacZ reporter in response to rapamycin (Fig. 4B). We reasoned that the lack of Sap185 and Sap190 functions might also alter the cells' sensitivity to the toxic histidine analog 3-amino triazole (3-AT). Consistent with our previous results, we observed that deletion of either sap185 or sap190 on their own resulted in no change in 3-AT sensitivity, while deletion of both sap185 and sap190 resulted in hypersensitivity to 3-AT (Fig. 4C). These results illustrated that Sap and Sit4 functions are required to direct Tor action to Gcn2-regulated translation. Moreover, Sap185 and Sap190 are required for a proper response to amino acid starvation.
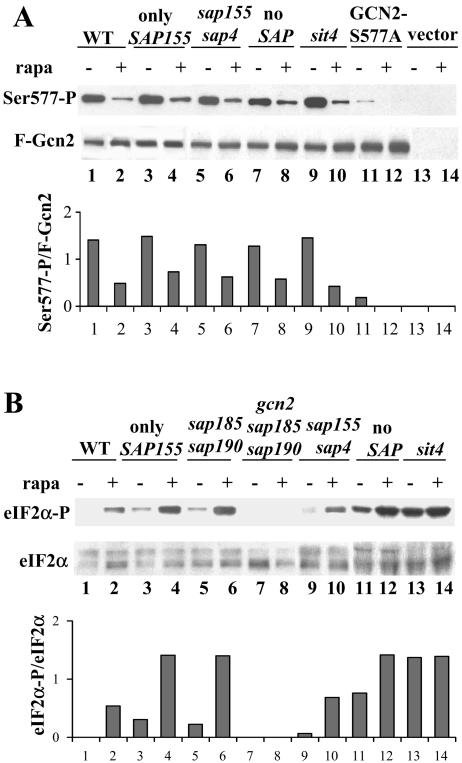
Rapamycin treatment promotes dephosphorylation of the Gcn2 kinase at Ser577, resulting in activation of the kinase towards eIF2α (9). We examined the roles of the various Sap and the Sit4 proteins on the rapamycin-induced dephosphorylation of Gcn2 at Ser577 by using antibodies that specifically recognize the phosphorylated form of the kinase. In agreement with earlier work, we observed that in wild-type cells rapamycin treatment resulted in a decrease in the amount of phosphorylated Ser 577 (Ser577-P) (9). We found that deletion of Sit4 did not block rapamycin-induced Gcn2 dephosphorylation at Ser577 (Fig. 5A). In addition, rapamycin treatment caused a decrease in Ser577-P in strains containing only Sap155 (sap4 sap185 sap190), lacking both SAP155 and SAP4, and in a strain lacking all four SAPs (sap4 sap155 sap185 sap190). We conclude that Sap-Sit4 complexes are not responsible for dephosphorylation of Gcn2 at Ser577.
FIG. 5.
Sap185, Sap190, and Sit4 regulate the phosphorylation state of eIF2α. (A) Cultures of isogenic wild-type (WT; MLY41) and only-SAP155 (sap4 sap185 sap190; JRY46), sap155 sap4 (SCY115), no-SAP (sap4 sap155 sap185 sap190; JRY40), and sit4 (SCY94), harboring a centromeric plasmid with FLAG epitope-tagged Gcn2 (F-Gcn2) (pDH101), F-Gcn2-S577A (pCB149), or an appropriate vector control as indicated, were grown to early exponential phase in SD-Ura medium and treated with 100 nM rapamycin (rapa) for 0 or 20 min. Gcn2 was immunoprecipitated from whole-cell extracts by using the FLAG epitope, and Ser577-P levels were analyzed with antibodies that specifically recognize Ser577-P. The blot was stripped and reprobed to detect the F-Gcn2 protein as a loading control. (B) Exponentially growing cultures of isogenic wild-type (MLY41), only-SAP155 (sap4 sap185 sap190; JRY46), sap185 sap190 (JRY29), gcn2 sap185 sap190 (JRY48), sap155 sap4 (SCY115), no-SAP (sap4 sap155 sap185 sap190; JRY40), and sit4 (SCY94) mutant strains were grown in YEPD medium and treated with 100 nM rapamycin for 0 or 20 min. Whole-cell extracts were prepared and subjected to Western blotting with specific antibodies for eIF2α-P. As a loading control, this blot was stripped and reprobed with antisera that recognize both unphosphorylated and phosphorylated versions of yeast eIF2α. The results shown in panels A and B are representative of three independent experiments. We note that the levels of eIF2α-P were also determined in the extracts employed in the experiment shown in panel A, and the results observed were similar to the ones shown in panel B for the respective strains. Specific Western blotting signals were quantified, and the Gcn2-Ser577P/F-Gcn2 and eIF2α-P/eIF2α ratios were calculated and are shown in the graphs.
In response to amino acid depletion, a major role for the Gcn2 kinase is to phosphorylate eIF2α and to thereby inhibit translation initiation (reviewed in reference 20). Based on the marked increase in GCN4-lacZ induction exhibited by the sap185 sap190 mutant strain, we reasoned that this strain might display a more dramatic increase in eIF2α phosphorylation in response to rapamycin. We used antibodies that are specific for the phosphorylated form of eIF2α to examine eIF2α phosphorylation in protein extracts from cells treated with rapamycin. In agreement with previous reports, rapamycin treatment resulted in phosphorylation of eIF2α-P in the wild-type strain (Fig. 5B) (9, 28). A mutant with both sap155 and sap4 deleted showed increases in eIF2α-P similar to wild-type levels. Strikingly, mutation of both SAP185 and SAP190 in the SAP155-only (sap4 sap185 sap190) and sap185 sap190 mutant strains resulted in an increase in the basal level of eIF2α-P, and the level of eIF2α-P in these cells increased even further upon addition of rapamycin (Fig. 5B). Moreover, this phosphorylation was GCN2 dependent, as it was not observed in a strain lacking Gcn2. We observe a more pronounced increase in the basal level of eIF2α-P in strains lacking all Saps and Sit4 (Fig. 5B). These results correlate with the high basal level of GCN4 translation observed in the no-SAP and sit4 mutant strains (Fig. 4B). Taken together, our results demonstrate that Sap-Sit4 complexes are required for constitutively dephosphorylating a basal level of eIF2α-P, and they strongly suggest that Sap185-Sit4 or Sap190-Sit4 complexes are most efficient at decreasing a basal level of eIF2α-P produced by Gcn2.
If the dramatic hypersensitivity to rapamycin observed in a sap185 sap190 strain were due to hyperactivation of the GAAC pathway, we reasoned that there could be a difference in translation initiation in response to rapamycin. Earlier studies have shown that in wild-type cells of the W303 or the JK93d background rapamycin treatment results in a decreased polysome-to-monosome ratio (P/M) (4, 11). We consistently found that in wild-type cells of the Σ1278b background rapamycin treatment had no detectable effect on the P/M ratio (Fig. 6). This result may explain the increased resistance to rapamycin exhibited by the Σ1278b strain compared to either W303 or JK93d (data not shown). Remarkably, we observed that rapamycin treatment of the sap185 sap190 strain resulted in a dramatic increase in monosomes and a concomitant decrease in actively translating polysomes compared to the response in the wild-type strain (Fig. 6). Furthermore, this effect was Gcn2 dependent, as the sap185 sap190 gcn2 triple mutant showed a polysome profile that was as resistant to rapamycin as the wild-type strain.
DISCUSSION
Protein phosphorylation is a central regulatory mechanism by which cellular functions are controlled at the posttranslational level. A key question is how the activity and specificity of phosphatases are controlled when the number of protein kinases exceeds them by roughly fourfold (Saccharomyces Genome Database). S. cerevisiae has five catalytic subunits for PP2A, which are encoded by the SIT4, PPH21, PPH22, PPH3, and PPG1 genes. Their gene products share a high degree of sequence similarity and may have overlapping substrate specificities (44). How the specificity of these different catalytic subunits is achieved is not understood but likely involves interactions with regulatory and targeting subunits. Our present work provides evidence that the Sap proteins may be important for targeting Sit4 to specific substrates. We have shown that Sap185, Sap190, and Sit4 are required to dampen the toxic effects of rapamycin, and our studies strongly suggest that complexes comprised of Sap185-Sit4 and Sap190-Sit4 play a key role in Tor-regulated Gcn2-dependent translation.
Our present work defines the Sap proteins as effectors of Tor signaling and provides a platform to study further the complexities of how the Sit4 phosphatase controls events within the Tor signaling program. We provide evidence that specific Sap-Sit4 complexes act to control key steps within different branches of the Tor pathway. Consistent with previous reports, deletion of all four SAP genes had the same effect on cell growth as deletion of the SIT4 phosphatase gene, and the SAP genes could be divided into two functional groups: (i) Sap185 and Sap190 and (ii) Sap4 and Sap155. Sap185/Sap190-Sit4 complexes are capable of promoting growth and are required to confer wild-type levels of rapamycin resistance. One interpretation of these findings is that Tor signals via Sap185/Sap190-Sit4 complexes to constitutively dephosphorylate substrates that, in the phosphorylated form, would otherwise become toxic. Our results suggest that one such substrate is the translation initiation factor eIF2α. Deletion of both the SAP185 and SAP190 genes resulted in rapamycin hypersensitivity, constitutive eIF2α phosphorylation, increased GCN4 translation, and a marked increase in the M/P ratio in response to rapamycin. All of these effects were eliminated by deleting the gene encoding Gcn2, the kinase for eIF2α.
The growth defect in the strains lacking the four Saps or Sit4 did not appear to be caused by the constitutive phosphorylation of eIF2α observed in these strains, because the sap185 sap190 mutant strain, which also shows high levels of eIF2α, grew more efficiently than either the sit4 or the no-sap mutant strains. This indicated that there must be at least one more Sap-Sit4 target other than eIF2α which also regulates cell growth. This target may be another component of the translational machinery because, despite the high levels of eIF2α-P, the sit4 and the no-SAP mutant strains fail to induce GCN4 translation in response to rapamycin.
A recent report concluded that Tor negatively regulates Gcn2 activity, in part by dephosphorylation of Ser577 (9). We also observed a decrease in Ser577-P in response to rapamycin treatment; however, in contrast to findings presented in this previous report with regard to Sit4 (9), in our studies this effect does not appear to be mediated by Sit4 or any of the Sap proteins. In accord with Cherkasova and Hinnebusch, we observed no significant differences in rapamycin-induced Gcn2 Ser577 dephosphorylation when comparing the wild-type strain with strains lacking the PP2A isoforms (the pph21 pph22 double mutant or the pph3 single mutant strains [data not shown]). The possibility that Sap185/190-Sit4 complexes are required for the dephosphorylation of eIF2α would allow for the levels of eIF2α-P to be controlled independently of Gcn2 Ser577 and may explain the effects observed thus far. Our studies do not exclude models in which Sap-Sit4 complexes regulate Gcn2 activity in a Ser577-independent manner. Furthermore, we also favor the previous suggestion that Gcn2-Ser577 may be targeted by the Tor kinase or a Tor-regulated kinase (9). A former study identified eIF2α-Ser51 as a substrate for the type 1 protein phosphatase Glc7 (52). Together with our study, these results indicate that phosphorylation of eIF2α is controlled by multiple phosphatases. Regulation of a single phosphorylated residue by multiple phosphatases is not an uncommon mechanism, and one such example is the Ser845 residue in the AMPA receptor GluR1 subunit, which appears to be dually regulated by both PP1 and calcineurin (43, 46).
Earlier work established that individual Sit4-Sap complexes vary in response to growth signals, such as carbon source and amino acid availability, and are cell cycle regulated (33, 45). It is also notable that the Saps have been shown to be phospho-proteins and are dephosphorylated in a Sit4-dependent fashion (33). Thus, it is possible that Tor signaling regulates the phosphorylation state of the Sap proteins and thereby their interaction with Sit4. One obvious candidate for regulation of the Sit4-Sap complexes is Tap42, especially in light of the fact that Tap42 plays both positive and negative roles in Sit4-dependent processes (12). An attractive model is that in response to Tor signaling, Tap42 promotes the formation of specific Sap-Sit4 complexes. Given the multitude of PP2A-associated factors, this process is likely to be complex. However, given that mammals express homologs of both Tap42 and Tip41, as well as the Saps, further insights from yeast are likely to apply to our understanding of Tor signaling mechanics in mammalian cells that can be targeted for therapy.
The mechanisms by which the Tor kinases are inhibited by rapamycin have been conserved from yeast to humans throughout a billion years of evolution. Work in multicellular eukaryotes has established Tor as a pathway dedicated to promoting protein synthesis and cell growth in response to amino acid availability. Our findings, and several related recent reports, extend this paradigm in yeast to a greater extent than previously appreciated (8, 9, 28). The rapamycin-induced gene programs of the retrograde response genes as well as the Gln3-controlled genes subject to NCR repression are instrumental in the synthesis of amino acids. The finding that Gcn4, the key transcription factor for amino acid biosynthesis genes, is induced by a similar pathway further supports this model. Gcn4 and Gln3 share a number of target genes, and it is striking that Tor controls these two transcription factors by different molecular strategies: in the case of Gln3 it controls by regulating its interaction with a negative regulator, Ure2, which restrains it to the cytoplasm under noninducing conditions, and in the case of Gcn4 it controls by regulating its translation. Furthermore, studies analyzing the transcriptome regulated by the GAAC response identified the GLN3 gene as a target for Gcn4 (34). Our studies support a model in which an increased translation of GCN4 leads to increased expression of Gln3 target genes, explaining the enhanced expression of the NCR genes in response to rapamycin observed in the sap185 sap190 and the sap4 sap185 sap190 mutant strains.
Earlier, it was demonstrated that Tor signaling is required for the maintenance of the histone acetylase Esa1 at the promoters of rapamycin-sensitive RP genes (38). It is therefore intriguing that recent proteomic studies have reported an Esa1-Sap185 interaction (22). We have examined this interaction in detail, but several lines of evidence suggest that the Esa1-Sap185 interaction does not play a significant role in the regulation of RP genes by the Tor pathway. First, this interaction is largely unaffected by rapamycin treatment. Second, we have been unsuccessful at localizing Sap185 to the promoters of RP genes by chromatin immunoprecipitation experiments. Third, and most importantly, we find that rapamycin-induced repression of the RP genes is independent of any of the Sap proteins as well as Sit4. These findings are congruent with a recent study which found that the RP genes are regulated by Tor in a Tap42-independent fashion (12).
Rapamycin is currently enjoying clinical success, being used as a drug to help patients cope with transplant rejection, complications following cardiac surgery, and most recently as a chemotherapeutic agent. Rapamycin displays remarkable cytotoxic activity against certain types of tumors, most notably those containing mutations in the tumor suppressor gene PTEN (35, 36). These studies have generated considerable excitement among oncologists and have helped lay the groundwork for “synthetic lethal” strategies to combat cancer. In this approach, certain mutations in tumors that favor their growth may predispose them to molecules that inhibit dysregulated signaling pathways (13, 14). Given our findings on the role of Tor in translational control in yeast, it may be of considerable practical importance that PTEN has been implicated in the control of translation in mammalian cells (3). By analogy with our findings that yeast sap185 sap190 mutant strains are exquisitely hypersensitive to rapamycin, yet exhibit normal growth, tumor cells with similar mutations would be amenable to eradication with rapamycin.
Acknowledgments
We are in debt to Alan Hinnebusch and Vera Cherkasova for generous gifts of plasmids and antisera and for criticism. We thank Joseph Heitman, Daniel Lew, and Raphael Valdivia for critical and constructive reading of the manuscript.
This work was supported by K22 award CA94925-01 from the NCI (to M.E.C.) and by Wellcome Trust project grant 067328/Z/02/Z (to M.P.A).
REFERENCES
- 1.Ai, W., P. G. Bertram, C. K. Tsang, T. F. Chan, and X. F. Zheng. 2002. Regulation of subtelomeric silencing during stress response. Mol. Cell 10:1295-1305. [DOI] [PubMed] [Google Scholar]
- 2.Ashe, M. P., S. K. De Long, and A. B. Sachs. 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11:833-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backman, S., V. Stambolic, and T. Mak. 2002. PTEN function in mammalian cell size regulation. Curr. Opin. Neurobiol. 12:516-522. [DOI] [PubMed] [Google Scholar]
- 4.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 6.Bertram, P. G., J. H. Choi, J. Carvalho, W. Ai, C. Zeng, T. F. Chan, and X. F. Zheng. 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275:35727-35733. [DOI] [PubMed] [Google Scholar]
- 7.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, E. J., and C. A. Kaiser. 2003. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J. Cell Biol. 161:333-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherkasova, V. A., and A. G. Hinnebusch. 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev. 17:859-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cutler, N. S., X. Pan, J. Heitman, and M. E. Cardenas. 2001. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell 12:4103-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Como, C. J., and K. T. Arndt. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10:1904-1916. [DOI] [PubMed] [Google Scholar]
- 12.Duvel, K., A. Santhanam, S. Garrett, L. Schneper, and J. R. Broach. 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11:1467-1478. [DOI] [PubMed] [Google Scholar]
- 13.Garber, K. 2001. Rapamycin's resurrection: a new way to target the cancer cell cycle. J. Natl. Cancer Inst. 93:1517-1519. [DOI] [PubMed] [Google Scholar]
- 14.Garber, K. 2002. Synthetic lethality: killing cancer with cancer. J. Natl. Cancer Inst. 94:1666-1668. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Barrio, M., J. Dong, V. A. Cherkasova, X. Zhang, F. Zhang, S. Ufano, R. Lai, J. Qin, and A. G. Hinnebusch. 2002. Serine 577 is phosphorylated and negatively affects the tRNA binding and eIF2α kinase activities of GCN2. J. Biol. Chem. 277:30675-30683. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 17.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinnebusch, A. 2000. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes, p. 185-243. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Hinnebusch, A. G. 1985. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 5:2349-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinnebusch, A. G. 1990. Involvement of an initiation factor and protein phosphorylation in translational control of GCN4 mRNA. Trends Biochem. Sci. 15:148-152. [DOI] [PubMed] [Google Scholar]
- 21.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 23.Jablonowski, D., A. R. Butler, L. Fichtner, D. Gardiner, R. Schaffrath, and M. J. Stark. 2001. Sit4p protein phosphatase is required for sensitivity of Saccharomyces cerevisiae to Kluyveromyces lactis zymocin. Genetics 159:1479-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacinto, E., B. Guo, K. T. Arndt, T. Schmelzle, and M. N. Hall. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8:1017-1026. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, Y., and J. R. Broach. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18:2782-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, D. H., D. Sarbassov dos, S. M. Ali, R. R. Latek, K. V. Guntur, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2003. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11:895-904. [DOI] [PubMed] [Google Scholar]
- 27.Komeili, A., K. P. Wedaman, E. K. O'Shea, and T. Powers. 2000. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151:863-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubota, H., T. Obata, K. Ota, T. Sasaki, and T. Ito. 2003. Rapamycin-induced translational derepression of GCN4 mRNA involves a novel mechanism for activation of the eIF2α kinase GCN2. J. Biol. Chem. 278:20457-20460. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y., M. N. Corradetti, K. Inoki, and K. L. Guan. 2004. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem. Sci. 29:32-38. [DOI] [PubMed] [Google Scholar]
- 30.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457-468. [DOI] [PubMed] [Google Scholar]
- 31.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz, M. C., and J. Heitman. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 16:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luke, M. M., F. Della Seta, C. J. Di Como, H. Sugimoto, R. Kobayashi, and K. T. Arndt. 1996. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 16:2744-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neshat, M. S., I. K. Mellinghoff, C. Tran, B. Stiles, G. Thomas, R. Petersen, P. Frost, J. J. Gibbons, H. Wu, and C. L. Sawyers. 2001. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc. Natl. Acad. Sci. USA 98:10314-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Podsypanina, K., R. T. Lee, C. Politis, I. Hennessy, A. Crane, J. Puc, M. Neshat, H. Wang, L. Yang, J. Gibbons, P. Frost, V. Dreisbach, J. Blenis, Z. Gaciong, P. Fisher, C. Sawyers, L. Hedrick-Ellenson, and R. Parsons. 2001. An inhibitor of mTOR reduces neoplasia and normalizes p70/S6 kinase activity in Pten+/− mice. Proc. Natl. Acad. Sci. USA 98:10320-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10:987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rohde, J., J. Heitman, and M. E. Cardenas. 2001. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 276:9583-9586. [DOI] [PubMed] [Google Scholar]
- 39.Rohde, J. R., and M. E. Cardenas. 2003. The Tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 23:629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 41.Shamji, A. F., F. G. Kuruvilla, and S. L. Schreiber. 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10:1574-1581. [DOI] [PubMed] [Google Scholar]
- 42.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 43.Snyder, G. L., S. Galdi, A. A. Fienberg, P. Allen, A. C. Nairn, and P. Greengard. 2003. Regulation of AMPA receptor dephosphorylation by glutamate receptor agonists. Neuropharmacology 45:703-713. [DOI] [PubMed] [Google Scholar]
- 44.Stark, M. J. 1996. Yeast protein serine/threonine phosphatases: multiple roles and diverse regulation. Yeast 12:1647-1675. [DOI] [PubMed] [Google Scholar]
- 45.Sutton, A., F. Lin, and K. T. Arndt. 1991. The SIT4 protein phosphatase is required in late G1 for progression into S phase. Cold Spring Harbor Symp. Quant. Biol. 56:75-81. [DOI] [PubMed] [Google Scholar]
- 46.Tavalin, S. J., M. Colledge, J. W. Hell, L. K. Langeberg, R. L. Huganir, and J. D. Scott. 2002. Regulation of GluR1 by the A-kinase anchoring protein 79 (AKAP79) signaling complex shares properties with long-term depression. J. Neurosci. 22:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tee, A. R., B. D. Manning, P. P. Roux, L. C. Cantley, and J. Blenis. 2003. Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13:1259-1268. [DOI] [PubMed] [Google Scholar]
- 48.Valenzuela, L., C. Aranda, and A. Gonzalez. 2001. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J. Bacteriol. 183:2331-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Slegtenhorst, M., E. Carr, R. Stoyanova, W. Kruger, and E. P. Henske. 2004. tsc1+ and tsc2+ regulate arginine uptake and metabolism in Schizosaccharomyces pombe. J. Biol. Chem. 279:12706-12713. [DOI] [PubMed] [Google Scholar]
- 50.Wang, H., and Y. Jiang. 2003. The Tap42-protein phosphatase type 2A catalytic subunit complex is required for cell cycle-dependent distribution of actin in yeast. Mol. Cell. Biol. 23:3116-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wedaman, K. P., A. Reinke, S. Anderson, J. Yates III, J. M. McCaffery, and T. Powers. 2003. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14:1204-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wek, R. C., J. F. Cannon, T. E. Dever, and A. G. Hinnebusch. 1992. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2 alpha kinase GCN2. Mol. Cell. Biol. 12:5700-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708-713. [DOI] [PubMed] [Google Scholar]
- 54.Yocum, R. R., S. Hanley, R. West, Jr., and M. Ptashne. 1984. Use of lacZ fusions to delimit regulatory elements of the inducible divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1985-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]