Abstract
Homeodomain containing transcription factors of the Hox family play critical roles in patterning the anteroposterior embryonic body axis, as well as in controlling several steps of organogenesis. Several Hox proteins have been shown to cooperate with members of the Pbx family for the recognition and activation of identified target enhancers. Hox proteins contact Pbx via a conserved hexapeptide motif. Previous biochemical studies provided evidence that critical amino acid substitutions in the hexapeptide sequence of Hoxa1 abolish its interaction with Pbx. As a result, these substitutions also abolish Hoxa1 activity on known target enhancers in cellular models, suggesting that Hoxa1 activity relies on its capacity to interact with Pbx. Here, we show that mice with mutations in the Hoxa1 hexapeptide display hindbrain, cranial nerve, and skeletal defects highly reminiscent of those reported for the Hoxa1 loss of function. Since similar hexapeptide mutations in the mouse Hoxb8 and the Drosophila AbdA proteins result in activity modulation and gain of function, our data demonstrate that the functional importance of the hexapeptide in vivo differs according to the Hox proteins.
Mammalian embryos are patterned along the anteroposterior axis of the body by the 39 homeotic (Hox) genes, which are organized in four chromosomal complexes (2, 10, 16). During development, the hindbrain is transiently divided into seven compartments, rhombomeres 1 to 7 (r1 to r7). The fate of the rhombomeres and their derivatives is controlled by the combined actions of Hox proteins (31, 38). Similarly, the identities of the different vertebrae along the vertebral column are also determined by the so-called homeotic code (2, 10). The numerous Hox gene inactivations performed to date further support the notion that some of these genes also contribute to patterning the proximodistal axis of the limbs, are involved in several steps of organogenesis, and may fulfill specific roles up to adulthood (7, 13, 41, 43).
Despite their apparently different functions in vivo, Hox proteins share very similar homeodomains and therefore bind to similar DNA sequences in vitro (22). Distinctive modulation of DNA binding and transcriptional activation by Hox proteins has been shown to arise from cooperative binding with partner proteins, like those of the Pbx family (4, 30, 40). The cooperative interaction between Hox and Pbx is mediated by a conserved hexapeptide sequence located N terminal to the Hox homeodomain. Point-mutational analysis has revealed that tryptophan and methionine residues of the hexapeptide are critical for this cooperative interaction (6, 17, 32). Very recent structural data and DNA binding analyses have provided evidence that the impact of Pbx on Hox DNA binding specificity and selectivity is higher for proteins encoded by the Hox genes lying at the 3′ side of the Hox complexes, like Hoxa1 and Hoxb1, than for those encoded by genes lying more 5′ (20). Therefore, it has been proposed that Hox-Pbx interaction may be important for Hox functions in vivo, in particular for the 3′ groups of paralogous genes.
Recently, the importance of the Hox-Pbx interaction for the achievement of the in vivo functions of Hox proteins has been directly investigated for the mouse Hoxb8 and the Drosophila AbdA proteins (27, 28). In both studies, the Hox protein hexapeptide sequences were mutated. The resulting phenotypes are indicative of activity changes relying on the modulation of DNA binding or the transregulatory potential of the proteins. Indeed, the Hoxb8 hexapeptide mutant behaves like a dominant negative, interfering with the activities of other Hox proteins (27). The equivalent mutation in AbdA did not affect either its ability to interact with Exd, the Drosophila homologue of Pbx, or its DNA binding capacity, but rather its transcriptional activation or repression activities (28). Thus, for both these proteins, the hexapeptide mutation resulted in some gain of function.
Hoxa1 is one of the first Hox genes to be expressed during embryonic development (29). In the mouse, its expression starts as early as 7.5 days postcoitum (dpc), is established in the neurectoderm and mesoderm at 8.0 dpc, and begins to retreat caudally by day 8.5 of gestation. This gene is thus very transiently expressed, but its functional inactivation results in perinatal lethality and in numerous malformations restricted to its early anterior domain of expression. Indeed, Hoxa1 null mice exhibit hindbrain segmentation and patterning defects that cause the abnormal development of cranial nerves, cranial ganglia, and branchial arch derivatives (3, 8, 24, 26).
Replacement of the WM amino acids by AA in the Hoxa1 hexapeptide prevents Hoxa1-Pbx interaction and cooperativity in binding DNA in vitro. However, this substitution does not affect the DNA binding ability of Hoxa1 monomers, which is of equally low affinity for both mutant and wild-type proteins (32, 33). In addition, it has been shown that the WM-to-AA Hoxa1 mutant protein Hoxa1WM-AA is inactive on distinct Hoxa1 target enhancers in transfected cells (35), suggesting that the mutation of the Hoxa1 hexapeptide results in a loss of function.
To directly assess the importance of the Hox-Pbx interaction for the Hoxa1 function in vivo, we generated recombinant mice to introduce a WM-to-AA substitution in the hexapeptide of the Hoxa1 protein. This substitution was the same as that generated for both biochemical and cellular approaches. The mutant mice present developmental defects of the hindbrain, cranial nerves, cranial ganglia, and branchial arch derivatives. These defects are highly similar to those reported for the Hoxa1 knockout, demonstrating that the in vivo function of Hoxa1 largely relies on the integrity of its hexapeptide. Since the WM-to-AA hexapeptide substitution in Hoxa1 resulted in a loss of function whereas the equivalent mutation in Hoxb8 or AbdA caused gain of activity, our data also demonstrate that the functional importance of the hexapeptide in vivo, and thus presumably that of the interaction with Pbx, clearly differs among the Hox proteins.
MATERIALS AND METHODS
Generating chimeric mice.
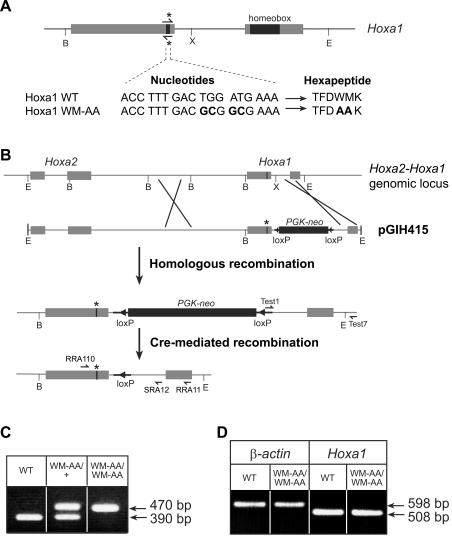
A 10.2-kb genomic fragment extending from an EcoRI site located 5′ of the Hoxa2 gene to an EcoRI site located 3′ of the Hoxa1 coding sequence was used for the construction of the targeting vector, pGIH415 (Fig. 1B). Nucleotide substitutions were introduced in the Hoxa1 hexapeptide by site-directed mutagenesis using a PCR approach (mutagenic primer, 5′-CGCAGACCTTTGACGCGCGAAAGTTAAAAGAAA-3′). A PGK-neomycin resistance gene cassette flanked by two loxP sites (15) was amplified and inserted into the unique XbaI site located in the intron of the Hoxa1 gene.
FIG. 1.
Targeted mutagenesis of the Hoxa1 hexapeptide. (A) Point mutations introduced in the sequence of the Hoxa1 hexapeptide and the resulting WM-to-AA amino acid substitutions. The mutagenic primers used are indicated by half-arrows with asterisks. (B) Genomic organization of the Hoxa2-Hoxa1 genomic locus and structure of the targeting construct pGIH415 (top). The PGK-neomycin resistance cassette flanked by loxP sites was introduced into the unique XbaI site located in the intron of Hoxa1. Details of the mutant allele after homologous recombination and after Cre-mediated recombination are shown below. Sequences corresponding to the primers Test1, Test7, RRA110, and SRA12 used for animal genotyping are represented by half-arrows. Sequences corresponding to primers RRA110 and RRA11 used for RT-PCR analysis are also represented. The asterisk indicates the mutated hexapeptide. Shaded boxes, Hoxa2 and Hoxa1 coding sequences; solid boxes, PGK-neomycin (neo) resistance cassette; solid triangles, loxP sites; thick solid lines, additional nonhomologous sequences. B, BamHI; E, EcoRI; X, XbaI. (C) PCR analysis of DNAs isolated from wild-type (WT) and heterozygous (WM-AA/+) and homozygous (WM-AA/WM-AA) mutant embryos. PCR amplification with primers RRA110 and SRA12 shown in panel B gives rise to a 390-bp product for the wild-type allele and a 470-bp product for the mutated allele, due to the presence of the loxP tag. (D) RT-PCR analysis of mRNAs extracted from wild-type and homozygous mutant 8-dpc embryos. Amplification products of the expected size (508 bp) corresponding to the correctly spliced Hoxa1 mRNA are obtained at similar levels from homozygous mutant and wild-type embryos.
The targeting construct (pGIH415) was introduced in 129SvEv ES cells (Eurogentec), and recombinant ES clones were analyzed by PCR, sequencing, and Southern blotting. Chimeric mice were generated by the aggregation of ES cells with morulae. Two independent chimeric mice were obtained that provided germ line transmission of the Hoxa1 knocked-in allele. Chimeric males were crossed with a ubiquitously Cre-expressing line (PGK-Cre) (19) in order to remove the selection cassette. Afterwards, the F1 offspring were interbred.
Genotyping of animals and embryos.
To detect germ line transmission of the mutant Hoxa1 locus, chimeric males were crossed with CD1 females, and the genotype of the offspring was determined by PCR using primers Test1 (5′-CTCTCCTGAGTAGGACAAGC-3′) and Test7 (5′-GGAGTTAACTTCCAACCAAGG-3′). Test1 is complementary to the extremity of the PGK-neomycin cassette, and Test7 hybridizes to Hoxa1 sequence 3′ to the EcoRI site used to generate the targeting construct. After the offspring were crossed with PGK-Cre mice, primers complementary to sequences located at the end of the first exon (RRA110, 5′-ACCACTCATATGGACAAGAGC-3′) and at the end of the intron of Hoxa1 (SRA12, 5′-GCACACTATCTAACTATAGAC-3′) were used to give rise to a 390-bp product for the wild-type allele and a 470-bp product for the mutated allele. Embryos obtained from crosses between heterozygous Hoxa1WM-AA/+ mice were genotyped by PCR on DNA extracted from the yolk sac. The temperature cycling was 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s.
RNA extraction and RT-PCR analysis.
Total RNA was isolated from either wild-type or mutant homozygous litters using the TriPure isolation reagent (Roche). Two micrograms of RNA was then treated with DNase I and subjected to reverse transcription (RT) using the avian myeloblastosis virus reverse transcriptase (U.S. Biochemicals), after being primed with random hexamers. PCR amplifications were performed in a final volume of 25 μl with 5 μl of cDNA, each primer at 100 nM, deoxynucleoside triphosphate at 250 nM, 1 U of Taq DNA polymerase (Takara), and the buffer supplied with the enzyme. Temperature cycling was 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. As a control for cDNA synthesis, the β-actin sequence was amplified (25 amplification cycles with primers 5′-GGCATCGTGATGGACTCCG-3′ and 5′-GCTGGAAGGTGGACAGCGA-3′). After 40 cycles of PCR amplification with the Hoxa1 primers (RRA110 [5′-ACCACTCATATGGACAAGAGC-3′] and RRA11 [5′-GTGTCTGAGGTAGACGATGC-3′]), located in the first and second exons, respectively, the amplified fragment was sequenced to confirm the correct splicing of the mutant gene.
Whole-mount in situ hybridization and immunostaining.
Whole-mount in situ hybridization was carried out on intact embryos using digoxigenin-labeled riboprobes, as previously described (14). Immunohistochemistry was performed on 10.5-dpc embryos as described previously (23), using the monoclonal 2H3 antibody at a final concentration of 1.9 mg/ml and peroxidase-conjugated horse anti-mouse immunoglobulin G (Vector) diluted 1:500 as a secondary antibody.
In situ hybridization on serial coronal sections.
In situ hybridization was performed on coronal sections of 9.5-dpc embryos as described previously (34). Paraffin sections (6 μm thick) were serially distributed onto six slides. The first slide was stained with toluidine blue, and the remaining slides were hybridized with distinct 35S-labeled probes.
Analysis of skeletons.
Newborns were sacrificed, skinned, eviscerated, and fixed for 24 h in 95% ethanol-1% acetic acid. They were stained for 72 h in 80% ethanol-20% acetic acid-0.015% Alcian blue and then washed in 95% ethanol for 10 h and cleared in 1% KOH for 24 h. They were then stained overnight in 1% KOH-0.015% Alizarin red. The skeletons were cleared in 1% KOH-20% glycerol for 2 to 3 days and then dehydrated and stored in 50% ethanol-50% glycerol.
RESULTS
Knockin of the mouse Hoxa1 gene for a WM-to-AA substitution in the hexapeptide.
Point mutations were introduced in the Hoxa1 gene to replace the WM residues of the hexapeptide by AA (Fig. 1A). These amino acids substitutions did not alter the expression level and the stability of the mutated protein, as revealed by Western blots performed on extracts of cells transfected with Hoxa1 or Hoxa1WM-AA expression vectors (data not shown). The targeting construct (pGIH415) designed to introduce this WM-to-AA substitution in Hoxa1 is shown in Fig. 1B. To select the homologous recombination events in ES cells, a PGK-neomycin cassette flanked by loxP sites was inserted into the Hoxa1 intron. Recombinant ES cell clones were generated, and two independent chimeric mice were obtained that provided germ line transmission of the Hoxa1 knocked-in allele. Since the PGK-neomycin cassette could interfere with proper Hoxa1WM-AA gene expression (36, 37), the chimeric males were crossed with a ubiquitously Cre-expressing line (19). The resulting Cre-mediated recombination event left a short 82-bp insert within the intron of the Hoxa1WM-AA allele, useful for animal genotyping (Fig. 1C). The mutations inserted in the Hoxa1 gene did not prevent the splicing of the mRNA, as revealed by RT-PCR on total RNA extracted from either wild-type or homozygous mutant 8.0-dpc embryos (Fig. 1D). PCR amplification with primers flanking the splice junction (RRA110 and RRA11) gave rise to an expected 508-bp fragment in both wild-type and mutant mice. The sequencing of the RT-PCR product confirmed that the mRNA was correctly spliced.
The two knockin lines were derived independently, and no significant difference in either the penetrance or expressivity of the resulting phenotype was observed. The analysis of the Hoxa1WM-AA/+ mice revealed no abnormalities, as was the case for the Hoxa1+/− mice (8, 24).
Intercrossing the Hoxa1WM-AA/+ mice showed a Mendelian transmission of the knocked-in allele, indicating that homozygous mutant fetuses were not preferentially aborted. Among the homozygous mutant newborns, ∼25% died at birth or within 24 h and an additional 15% died within 2 weeks. The rate of viability (60%) observed for the Hoxa1WM-AA/WM-AA mice thus contrasts with the 100% perinatal lethality reported for the full inactivation of Hoxa1 (8, 24).
Hoxa1WM-AA/WM-AA hindbrain-patterning defects parallel Hoxa1 loss of function.
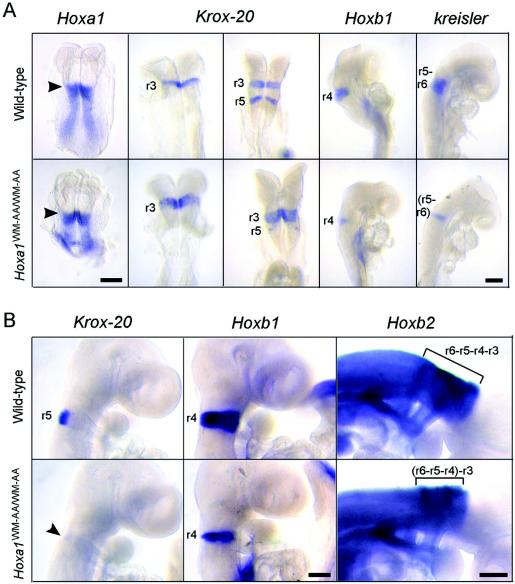
Since Hoxa1 is involved in hindbrain segmentation and patterning (3, 26), we used Krox-20, kreisler, and several Hox genes as rhombomeric markers to characterize hindbrain development in the Hoxa1WM-AA/WM-AA homozygotes. Whole-mount in situ hybridizations revealed that between 7.5 and 9.0 dpc, the Hoxa1 expression domains were not modified in the Hoxa1WM-AA/WM-AA homozygotes compared with wild-type embryos (Fig. 2A and data not shown). In control embryos, Krox-20 was first expressed in r3, then in r3 and r5 (Fig. 2A), and then only in r5 (Fig. 2B) at 8.0, 8.5, and 9.0 dpc, respectively. In contrast, the Krox-20 r3 domain was enlarged (5 of 5) (Fig. 2A) and the r5 domain was strongly reduced (12 of 12) (Fig. 2A and B) in mutant homozygotes. In addition, hybridizations with a Hoxb1 probe, an r4-specific marker (29), revealed only a thin strip of Hoxb1-expressing cells at the level of r4 (11 of 11) compared to the wild type (Fig. 2). The expression of kreisler, confined to r5 and r6 (9), was reduced in mutant homozygotes (five of five), showing only one rhombomere length (Fig. 2A), consistent with the drastic reduction of r5. Finally, in Hoxa1WM-AA/WM-AA mutant embryos, the length of the highly stained portion of the Hoxb2 territory (r3 to r6) (44) was shortened (seven of seven) (Fig. 2B), in agreement with the observed reduction of r4 and r5.
FIG. 2.
Analysis of hindbrain patterning by whole-mount in situ hybridization. (A) Wild-type (top) and mutant (bottom) embryos between 7.5 and 8.75 dpc hybridized with a probe for Hoxa1, Krox-20, Hoxb1, or kreisler. The arrowheads indicate the anterior limits of expression of Hoxa1, corresponding to the presumptive r3-r4 boundary. In the mutant, the r3 expression domain of Krox-20 is enlarged, while its r5 expression domain is drastically reduced. Moreover, the r4 expression domain of Hoxb1 is reduced, and the kreisler r5-r6 domain of expression is only one rhombomere long. (B) Wild-type (top) and mutant (bottom) 9.5-dpc embryos hybridized with a probe for Krox-20, Hoxb1, or Hoxb2. In the mutant, the r5 expression domain of Krox-20 is almost absent (arrowhead), the r4 expression domain of Hoxb1 is clearly reduced, and the highly stained portion of the Hoxb2 expression domain, corresponding to r3-r6, is shortened (bracket). Scale bars, 200 μm.
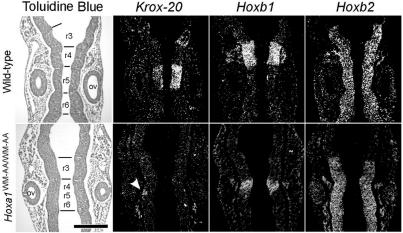
The modifications of Krox-20, Hoxb1, and Hoxb2 expression domains were confirmed by in situ hybridizations on coronal sections of 9.5-dpc mutant embryos (Fig. 3). In control embryos, the r5 expression of Krox-20 faced the anterior half of the otocysts. In contrast, in mutant embryos (five of five), only a very weak and narrow Krox-20 expression was detected at the level of r5, and it was restricted to the dorsal part of the neural tube. In controls, the r4 territory was well defined by Hoxb1-expressing cells, with a posterior boundary lying at the level of the anterior margin of the otocyst. In the homozygous mutants, the level of Hoxb1 expression in r4 was reduced (eight of eight). Furthermore, this territory appeared smaller, showed loosely defined borders, and was posteriorized so that its caudal boundary lay behind the anterior one-third of the otocyst. Computer-assisted superposition of the Hoxb1 and Krox-20 expression domains clearly showed that they were adjacent (data not shown) and thus represented the remnants of r4 and r5, respectively. The Hoxb2 expression domain in control and Hoxa1WM-AA/WM-AA embryos extended up to the r2-r3 boundary. However, only two rhombomeric bulges were discernible in Hoxa1WM-AA/WW-AA mutants (six of six), in place of four in the controls. These coronal sections thus also revealed that the rhombomeric segmentation was less pronounced in mutant than in wild-type embryos (Fig. 3, toluidine blue staining). Finally, the otocysts of mutant embryos appeared to be smaller and displaced rostrally and laterally (compare the positions of otic vesicles relative to r3 in Fig. 3).
FIG. 3.
Analysis of hindbrain patterning by in situ hybridization on serial coronal sections. Expression of Krox-20, Hoxb1, and Hoxb2 on serial coronal sections of 9.5-dpc wild-type (top) and mutant (bottom) embryos. Bright-field views after toluidine blue staining are presented on the left, and dark-field views after in situ hybridization are shown on the right. The arrowhead points to the Krox-20 r5 expression domain, drastically reduced in the mutant. ov, otic vesicle. Scale bar, 200 μm.
Taken together, these data show that the WM-to-AA substitution in the hexapeptide of Hoxa1 affects the hindbrain patterning from r3 to r6. r3 was enlarged, and r4, r5, and r6 were fused together in a unique bulge with its anterior half showing an r4 fate, followed by a tiny population of cells expressing r5 markers and a posterior domain with an r6 identity. These defects were observed with full penetrance and are very similar to those observed after knocking out the Hoxa1 gene (3, 26).
Modifications in gene expression in neural crests of Hoxa1WM-AA/WW-AA embryos.
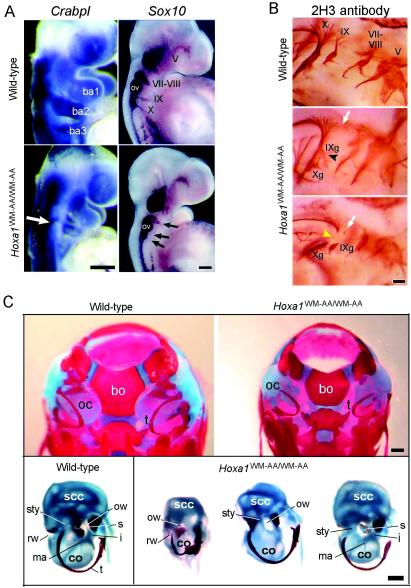
Since r3 to r6 were affected in the Hoxa1WM-AA/WW-AA embryos, we performed whole-mount in situ hybridizations with probes for genes expressed in the neural crest cells (ncc). The CrabpI gene encodes a retinoic acid binding protein strongly expressed in the neural tube and migrating cranial ncc (25). At the level of the hindbrain and pharyngeal arches, CrabpI is normally expressed in three distinct streams of ncc migrating into the first, second, and third pharyngeal arches (Fig. 4A). In about two-thirds of the Hoxa1WM-AA/WW-AA embryos, the CrabpI expression level in the second pharyngeal arch was significantly lower than in the wild type (three of five) (Fig. 4A).
FIG. 4.
Analysis of neural crest derivatives and skeletons. (A) Whole-mount in situ hybridization on wild-type (top) and homozygous mutant (bottom) embryos at 9.25 and 9.5 dpc. The embryos were hybridized with a CrabpI or a Sox10 probe. The open arrow indicates the reduction of the ncc population expressing CrabpI and migrating into the second branchial arch (ba2). The solid arrows indicate the reduction of neurogenic ncc at the level of nerve VII-VIII and the absence of Sox10 expression at the level of nerves IX and X. ba, branchial arch; ov, otic vesicle. (B) Immunostaining of cranial nerves of a 10.5-dpc wild-type embryo (top) and two Hoxa1WM-AA/WW-AA mutant embryos (bottom) (caudal is on the left). In the mutants, the open arrows indicate the absence of nerve IX dorsal roots, the solid arrowhead indicates the fusion of the inferior ganglion of nerve IX (IXg) with the inferior ganglion of nerve X (Xg), and the yellow arrowhead points to the lack of the interganglionic portion of nerve X. (C) (Top) Dorsal view of the skull of wild-type (left) and Hoxa1WM-AA/WW-AA mutant (right) newborns. In the mutant, the shape of the basioccipital bone (bo) is altered, the otic capsules (oc) are reduced, and the tympanic ring (t) is displaced. (Bottom) Dissected otic capsules and tympanic rings from a wild-type skull (left) and three Hoxa1WM-AA/WW-AA mutant skulls (right). co, cochlea; i, incus; ma, malleus; ow, oval window; rw, round window; s, stapes; scc, semicircular canals; sty, styloid process; t, tympanic ring. Scale bars, 200 (A and B) and 500 (C) μm.
The neurogenic ncc that contribute to the peripheral nervous system express Sox10, an HMG box transcription factor (18). In 9.5-dpc wild-type embryos, Sox10 was expressed in the superior ganglia of the cranial nerves (Fig. 4A). Ganglia V and VII-VIII were easily discernible, and migrating ncc were detected in the region of the superior ganglia of nerves IX and X. In Hoxa1WM-AA/WW-AA mutants, expression of Sox10 was strongly reduced in ganglia VII and VIII of 60% of the embryos (three of five) (Fig. 4A). In addition, the ncc taking part in nerve IX were either strongly reduced or absent, while they were always absent for nerve X (Fig. 4A).
On coronal sections, the facial-acoustic ganglion complex of control embryos was labeled by the Hoxb2 probe and located anterior to the otocyst (Fig. 3). In the Hoxa1WM-AA/WW-AA mutant embryos (six of six), this large ganglion complex was disorganized and abnormally positioned (Fig. 3). Indeed, the Hoxb2-labeled cells formed a narrow strip adjacent and parallel to the neuroepithelium at the level of the otocyst.
Defects in CrabpI, Sox10, and Hoxb2 gene expression were also reported for the Hoxa1 knockout mice (11, 12, 26), but in Hoxa1WM-AA/WW-AA mice, the reduction of the ncc streams revealed by these genetic markers was not as drastic as for the Hoxa1 loss of function (11, 12).
Development of cranial nerves is altered in Hoxa1WM-AA/WW-AA mutant embryos.
In order to analyze the formation of the cranial nerves, we immunostained 10.5-dpc embryos with a 2H3 monoclonal antibody, which labels the 155,000-molecular-weight neurofilament protein. As in Hoxa1 null mutants (8, 11, 26), defects in the combined superior ganglia IX-X led to the fusion of the proximal parts of the two nerves and to the loss of the brainstem connection of nerve IX (Fig. 4B). Moreover, inferior ganglion IX was often connected or fused with inferior ganglion X (11 of 20 ganglia, i.e., the left and right sides of 10 embryos [Fig. 4B]). Finally, both nerves IX and X frequently lacked their interganglionic portions (14 of 20 [Fig. 4B]). This defect was also observed but was reported to be fully penetrant in the Hoxa1 loss-of-function mutant (8, 26). In addition, nerve X was developed normally in ∼20% of the knockin embryos, which was not the case in the knockout embryos. Ganglion V was always normal, as was the case in Hoxa1 null mutants. However, unlike the Hoxa1 null homozygotes, ganglia VII and VIII were never fused with ganglion V in Hoxa1WM-AA/WW-AA embryos (8, 11, 26).
The Hoxa1 hexapeptide mutation results in abnormal development of occipital bone and ear.
Comparison of Alcian blue- and Alizarin red-stained skeletons of wild-type and Hoxa1WM-AA/WW-AA newborns revealed an abnormally shaped basioccipital bone in the mutant, lacking distinct concave lateral borders (13 out of 20 skulls) (Fig. 4C, top). At the level of the middle ear (Fig. 4C, bottom), the tympanic ring, which derives from the first-arch neural crest, was present but sometimes displaced (4 of 40), and in 2 cases out of 40, its shape was slightly altered. The malleus and incus, derived from the neural crest of the first arch, were always present, although the malleus occasionally appeared to be reduced in size or malformed (3 of 40). The stapes, a second-arch derivative, was generally present (absent in 3 cases out of 40), although it was sometimes partially formed.
The cartilaginous otic capsule, containing the inner ear labyrinth, was the most clearly abnormal cartilaginous structure in the Hoxa1WM-AA/WW-AA fetuses. The inner ear derives almost exclusively from the ectodermal otic placode, which invaginates to form the otocyst. The upper part of the otocyst develops to form the vestibule, which comprises the semicircular canals, the utricle, and the saccule. The lower part of the otocyst gives rise to the cochlea. In the most affected Hoxa1WM-AA/WW-AAhomozygotes, the semicircular canals were considerably dilated and had undergone a rudimentary morphogenesis (19 of 40), and the cochlea was severely reduced (10 of 40). In some cases, the styloid process, a second-arch skeletal element, was reduced in size (12 of 40). In more posterior regions, the Hoxa1WM-AA/WW-AA mice displayed no difference from the wild type in the number, size, and spacing of the vertebrae and ribs. The limbs were also normally developed.
Strikingly, all the cartilaginous and skeletal malformations observed in the Hoxa1WM-AA/WW-AA mice are reminiscent of those previously reported for the Hoxa1 knockout mice (1, 11, 24), although the latter presented more severe and more penetrant phenotypic alterations. Indeed, at the level of the middle ear ossicles, the tympanic ring was systematically rostrally displaced in the knockout mice, the malleus was malformed at a higher frequency than in the knockin mice (50%) (11), and the stapes was reported to be either absent (1) or fused to the otic capsule (11). The semicircular canal and cochlea defects, although similar, were also more penetrant for the Hoxa1 loss-of-function mutant (11).
DISCUSSION
The Hox-Pbx partnership in Hoxa1 function in vivo.
Numerous studies have highlighted Pbx proteins as key cofactors of the Hox transcription factors. The Hox-Pbx interaction involves a short hexapeptide in the Hox sequence. Replacing the conserved WM residues with alanines in the hexapeptide of Hoxa1 appeared to abolish its interaction with Pbx, as well as the Hox-Pbx heterodimeric binding to DNA (32, 33). However, these amino acid replacements did not alter the ability of the protein to bind DNA as a monomer (33). It has been further shown that the Hoxa1WM-AA mutant protein was inactive on distinct target enhancers in different cellular models, suggesting that the function of Hoxa1 is conditioned by its partnership with Pbx (35). However, this assumption, based on in vitro and ex vivo observation, awaited in vivo evidence. Here, we replaced the Hoxa1 gene by an allele coding for this Hoxa1WM-AA mutant protein. As a result, we observed that the embryonic structures affected in the Hoxa1WM-AA/WW-AA knockin mice correspond to those reported to be affected in the Hoxa1 knockout mice, so the phenotype of the Hoxa1WM-AA/WW-AA mutant is highly similar to that caused by the inactivation of Hoxa1. This demonstrates that the activity of Hoxa1 relies critically on its hexapeptide and thus presumably on its interaction with Pbx. Recently, structural data has provided explanations for the observation that Hox proteins encoded by the more 3′ Hox genes, like Hoxa1 or Hoxb1, depend to a larger extent on Pbx for their target site discrimination than those corresponding to the more 5′ genes (20). This Pbx dependency would explain why the Hoxa1 WM-to-AA mutation led to a phenotype resembling that of the loss of function, while similar mutations in more posterior genes do not (27, 28).
The recent inactivation of the pbx2 and lzr/pbx4 genes in zebrafish has been shown to prevent hindbrain segmentation and to transform the r2-to-r6 territories into a default r1 identity (42). Since the hindbrain is patterned by the combinatorial action of Hox proteins of paralog groups 1 to 4, the phenotype conferred by pbx gene inactivation in zebra fish is consistent with a key partnership between 3′ Hox genes and Pbx in the control of hindbrain development. In the mouse, among the four Pbx genes, only Pbx1 has been experimentally inactivated (39). This inactivation resulted in skeletal malformations which do not phenocopy single or compound Hox gene knockouts, suggesting that not all Hox functions may be Pbx1 dependent. However, these results await the combined inactivation of multiple Pbx genes to allow conclusions about the reciprocal dependency of Hox and Pbx functions in mouse development.
The Hoxa1WM-AA/WW-AA phenotype compared to the Hoxa1 knockout phenotype.
Some aspects of the phenotype of Hoxa1WM-AA/WW-AA mice appear milder and less penetrant than in the Hoxa1 knockout mice. This holds for the inner and middle ear and for the neurogenic neural crest derivatives. This milder phenotype could account for the survival of roughly 60% of the Hoxa1WM-AA/WW-AA mice, while lethality was reported to be fully penetrant for the Hoxa1 null mice. The cause of the knockout lethality was attributed to anoxia and correlated with the lack of connection between the brainstem and nerve IX and X ganglia (24). In the case of the Hoxa1WM-AA/WW-AA mice, cranial nerves IX and X were also affected, but the defects were less penetrant and in some instances weaker.
The apparently lower expressivity and penetrance of the phenotype caused by the WM-to-AA substitution compared to the knockout may indicate that the hexapeptide mutation results in severe but not full loss of function of Hoxa1. It is possible that, contrary to the assumptions based on in vitro and ex vivo observations, Hoxa1 activity does not rely completely on its partnership with Pbx or that residual Hoxa1-Pbx interaction takes place in vivo despite the hexapeptide mutation. Alternatively, the milder phenotype displayed by the Hoxa1WM-AA/WW-AA mice may simply result from genetic background effects. The two Hoxa1 knockouts were generated in ES cells of 129Sv genotype, and chimeras were then crossed with C57BL/6 mice. Heterozygous offspring were either interbred or backcrossed in the C57BL/6 background. The Hoxa1WM-AA knockin was also generated in ES cells of 129Sv genotype. However, the chimeric males were crossed with hybrid PGK-Cre transgenic mice (BALB/c-C57BL/6) to remove the selection marker inserted in the Hoxa1 intron. The phenotype observed thus results from a hybrid background. It can be expected that stabilizing the Hoxa1WM-AA mutation in an inbred background will increase the penetrance and expressivity of the phenotype. Consistently, the phenotype of the Hoxb8 hexapeptide mutant appeared significantly more severe in an inbred (129SvEv) than in a hybrid (C57BL/6J-129SvEv) genetic background (27).
It is worth noting that two distinct Hoxa1 knockouts, generated by Lufkin et al. (24) and Chisaka et al. (8), respectively, did not show fully overlapping phenotypes. Notably, the latter survived up to 3.5 days and presented a more severe phenotype in ear morphogenesis, characterized by the lack of either one (1) or three ossicles (8) and by defects in the external ear. Moreover, the rhombencephalon of the knockout mice obtained by Chisaka et al. (8) did not exhibit the characteristic bulges defining the rhombomeres at 9.5 dpc, while these were discernible in the mutant described by Mark et al. (26). On the other hand, the Hoxa1 knockout mice reported by Mark et al. (26) presented a delayed closure of the neural tube at 9.5 dpc that was not apparent in the other Hoxa1 loss of function. It had been suggested that the observed differences between these two Hoxa1 null mice could result from differences in the mutant Hoxa1 alleles generated by the two groups (8, 26). Two transcripts are expressed from the Hoxa1 locus as a result of alternative splicing (21). One transcript encodes the entire Hoxa1 protein (331 amino acids), and the other gives rise to a shorter protein (133 amino acids) that does not contain the homeodomain. The Hoxa1 disruption generated by Lufkin et al. (24) is predicted to eliminate both transcripts, while that generated by Chisaka et al. (8) does not impair the expression of the short Hoxa1 protein. In our knockin mice, the expression of the shorter Hoxa1 protein is predicted not to be impaired, but the phenotype we observed was not more similar to one of the two reported knockouts. Indeed, similarly to Chisaka et al. (8), we observed a flat rhombencephalon and we did not observe a delay in the closure of the neural tube. However, the Hoxa1WM-AA/WW-AA mice did not exhibit the more severe phenotype in ear morphogenesis reported by these authors (8). Thus, although we cannot rule out the possibility that the 133-amino-acid protein may play a role in Hoxa1 function, this is not sufficient to account for the differences among the three phenotypes.
A final difference that can cause phenotype differences between the two Hoxa1 knockout alleles and the knockin allele resides in the persistence of the selection marker in the Hoxa1 locus in both knockout strategies, whereas it has been removed in the knockin mice. Importantly, in some knockouts, the presence of selection markers in recombinant loci has been associated with the misregulation of neighboring genes, which in turn resulted in phenotypic alterations not attributable to the gene inactivation per se (36, 37).
The Hoxa1 hexapeptide mutation compared to other Hox hexapeptide mutations.
While the Hoxa1 hexapeptide mutation resulted in a loss of function, similar mutations in other Hox proteins have been shown to lead to very different activity changes. For Labial, the Drosophila homologue of Hoxa1, it has been proposed that the hexapeptide inhibits Lab function by impairing DNA binding, as well as its transcriptional activation potential. An Exd/Pbx-induced conformational change would thus relieve these inhibitions. Indeed, Chan et al. (5) showed that while Lab does not bind efficiently to DNA in the absence of Exd, a hexapeptide mutation allows Lab to bind DNA as a monomer and renders it hyperactive. This therefore contrasts with the effect of the WM-to-AA substitution, which does not emancipate Hoxa1 from its partnership with Pbx. Hoxa1 and Hoxa1WM-AA have been shown to bind DNA as monomers with similarly low efficiencies (33), and the mutant protein is inactive on target enhancers in culture cells (35). If the hexapeptide of Hoxa1 displayed some inhibitory effect, as proposed for Lab, the hexapeptide mutation would be expected to result in some gain of function and would be neomorphic by comparison with the Hoxa1 knockout, which is not the case.
Amino acid substitutions in the hexapeptides of Hoxb8 and AbdA in vivo led to phenotypes corresponding to activity changes due to the modification of DNA binding or transregulatory potential (27, 28). Indeed, the phenotype caused by the Hoxb8 mutation corresponded to homeotic transformations similar to those observed for the loss of function of other Hox genes, such as Hoxa7, Hoxb7, or Hoxb9. These, together with genetic tests, provided evidence that the hexapeptide mutation did not result in Hoxb8 loss of function but rather in a dominant negative interfering with the activity of homologous Hox proteins (27). Similarly, amino acid substitutions in the hexapeptide of AbdA, which is among the closest homologues of Hoxb8 in Drosophila, did not lead to a loss of function. Rather, these substitutions affected the transregulatory function of AbdA in a way that depends on the target transcription enhancers (28). Therefore, for both of these proteins, the hexapeptide mutation resulted in gain of function.
In conclusion, the phenotype of recombinant mice with mutations in the hexapeptide of Hoxa1 demonstrates that the activity of this protein critically, if not fully, relies on the integrity of its hexapeptide, which strongly suggests that the function of Hoxa1 is conditioned by its partnership with Pbx cofactors. In addition, since the Hoxa1WM-AA protein behaves as a loss of function while similar mutations in other Hox proteins provided gains of function, our study also demonstrates that the hexapeptide assumes a different functional importance, and maybe distinct roles, among Hox proteins in vivo.
Acknowledgments
This work was supported by the EC in the framework of the BIOTECH program (grant CT98 0227), the National Fund for Scientific Research (FNRS, Belgium), the Télévie, and the Fonds Spéciaux de Recherche of the Université Catholique de Louvain. S.R. is a Scientific Research Worker FNRS-Télévie. A.G. held a Wellcome Trust Career Development Grant.
We are grateful to F. M. Rijli and X. Lampe for helpful discussions and critical comments on the manuscript. Our gratitude goes to C. Matis for the RNA samples of embryonic origin. The 2H3 monoclonal antibody developed by Jessel and Dodd was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the Department of Biological Sciences, University of Iowa, Iowa City.
REFERENCES
- 1.Barrow, J. R., and M. R. Capecchi. 1999. Compensatory defects associated with mutations in Hoxa1 restore normal palatogenesis to Hoxa2 mutants. Development 126:5011-5026. [DOI] [PubMed] [Google Scholar]
- 2.Capecchi, M. R. 1997. Hox genes and mammalian development. Cold Spring Harbor Symp. Quant. Biol. 62:273-281. [PubMed] [Google Scholar]
- 3.Carpenter, E. M., G. M. Goddard, O. Chisaka, N. R. Manley, and M. R. Capecchi. 1993. Loss of Hoxa-1 (Hox-1.6) function results in the reorganization of the murine hindbrain. Development 118:1063-1075. [DOI] [PubMed] [Google Scholar]
- 4.Chan, S.-K., L. Jaffe, M. Capovilla, J. Botas, and R. Mann. 1994. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 78:603-615. [DOI] [PubMed] [Google Scholar]
- 5.Chan, S.-W., H. Pöpperl, R. Krumlauf, and R. S. Mann. 1996. An extradenticle-induced conformational change in a HOX protein overcomes an inhibitory function of the conserved hexapeptide motif. EMBO J. 15:2476-2487. [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, C.-P., W.-Y. Shen, S. Rozenfeld, H. F. Lawrence, C. Largman, and M. L. Cleary. 1995. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 9:663-674. [DOI] [PubMed] [Google Scholar]
- 7.Chen, F., and M. R. Capecchi. 1999. Paralogous mouse Hox genes, Hoxa9, Hoxb9 and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proc. Natl. Acad. Sci. USA 96:541-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chisaka, O., T. Musci, and M. R. Capecchi. 1992. Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene, Hox-1.6. Nature 355:516-520. [DOI] [PubMed] [Google Scholar]
- 9.Cordes, S. P., and G. S. Barsh. 1994. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell 79:1025-1034. [DOI] [PubMed] [Google Scholar]
- 10.Favier, B., and P. Dollé. 1997. Developmental functions of mammalian Hox genes. Mol. Hum. Reprod. 3:115-131. [DOI] [PubMed] [Google Scholar]
- 11.Gavalas, A., M. Studer, A. Lumsden, F. M. Rijli, R. Krumlauf, and P. Chambon. 1998. Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development 125:1123-1136. [DOI] [PubMed] [Google Scholar]
- 12.Gavalas, A., P. Trainor, L. Ariza-McNaughton, and R. Krumlauf. 2001. Synergy between Hoxa1 and Hoxb1: the relationship between arch patterning and the generation of cranial neural crest. Development 128:3017-3027. [DOI] [PubMed] [Google Scholar]
- 13.Godwin, A. R., and M. R. Capecchi. 1998. Hoxc13 mutant mice lack external hair. Genes Dev. 12:11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gofflot, F., M. Hall, and G. M. Morris-Kay. 1998. Genetic patterning of the posterior neuropore region of curly tail mouse embryos: deficiency of Wnt5a expression. Int. J. Dev. Biol. 42:637-644. [PubMed] [Google Scholar]
- 15.Kellendonk, C., F. Tronche, A.-P. Monaghan, P.-O. Angrand, F. Stewart, and G. Schütz. 1996. Regulation of Cre recombinase activity by the synthetic steroid RU 486. Nucleic Acids Res. 24:1404-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kmita, M., and D. Duboule. 2003. Organizing axes in time and space; 25 years of colinear tinkering. Science 301:331-333. [DOI] [PubMed] [Google Scholar]
- 17.Knoepfler, P. S., and M. P. Kamps. 1995. The pentapeptide motif of Hox proteins is required for cooperative DNA binding with Pbx1a, physically contacts Pbx1, and enhances DNA binding by Pbx1. Mol. Cell. Biol. 15:5811-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhlbrodt, K., B. Herbarth, E. Sock, I. Hermans-Borgmeyer, and M. Wegner. 1998. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 18:237-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lallemand, Y., V. Luria, R. Haffner-Krausz, and P. Lonai. 1998. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res. 7:105-112. [DOI] [PubMed] [Google Scholar]
- 20.LaRonde-LeBlanc, N. A., and C. Wolberger. 2003. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 17:2060-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.La Rosa, G. J., and L. J. Gudas. 1998. Early retinoic acid-induced F9 teratocarcinoma stem cell gene ERA-1: alternate splicing creates transcripts for a homeobox-containing protein and one lacking the homeobox. Mol. Cell. Biol. 8:3906-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laughon, A. 1991. DNA-binding specificity of homeodomains. Biochemistry 30:11357-11367. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, P. M., M. P. Dunn, J. A. McMahon, M. Logan, J. F. Martin, B. St-Jacques, and A. P. McMahon. 2001. Cholesterol modification of Sonic Hedgehog is required for long-range signalling activity and effective modulation of signalling by Ptc1. Cell 105:599-612. [DOI] [PubMed] [Google Scholar]
- 24.Lufkin, T., A. Dierich, M. LeMeur, M. Mark, and P. Chambon. 1991. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell 66:1105-1119. [DOI] [PubMed] [Google Scholar]
- 25.Maden, M., C. Horton, A. Graham, L. Leonard, J. Pizzey, G. Siegenthaler, A. Lumsden, and U. Eriksson. 1992. Domains of cellular retinoic acid-binding protein I (CRABPI) expression in the hindbrain and neural crest of the mouse embryo. Mech. Dev. 37:13-23. [DOI] [PubMed] [Google Scholar]
- 26.Mark, M., T. Lufkin, J.-L. Vonesch, E. Ruberte, J.-C. Olivo, P. Dollé, P. Gorry, A. Lumsden, and P. Chambon. 1993. Two rhombomeres are altered in Hoxa-1 mutant mice. Development 119:319-338. [DOI] [PubMed] [Google Scholar]
- 27.Medina-Martínez, O., and R. Ramírez-Solis. 2003. In vivo mutagenesis of the Hoxb8 hexapeptide domain leads to dominant homeotic transformations that mimic the loss-of-function mutations in genes of the Hoxb cluster. Dev. Biol. 264:77-90. [DOI] [PubMed] [Google Scholar]
- 28.Merabet, S., Z. Kambris, M. Capovilla, H. Bérenger, J. Pradel, and Y. Graba. 2003. The hexapeptide and linker regions of the AbdA Hox protein regulate its activating and repressive functions. Dev. Cell. 4:761-768. [DOI] [PubMed] [Google Scholar]
- 29.Murphy, P., and R. E. Hill. 1991. Expression of the mouse labial-like homeobox-containing genes, Hox-2.9 and Hox-1.6, during segmentation of the hindbrain. Development 111:61-74. [DOI] [PubMed] [Google Scholar]
- 30.Neuteboom, S. T., and C. Murre. 1997. Pbx raises the DNA binding specificity but not the selectivity of Antennapedia Hox proteins. Mol. Cell. Biol. 17:4696-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nonchev, S., A. Maconochie, A. Gould, A. Morrison, and R. Krumlauf. 1997. Cross-regulatory interactions between Hox genes and the control of segmental expression in the vertebrate central nervous system. Cold Spring Harbor Symp. Quant. Biol. 62:313-323. [PubMed] [Google Scholar]
- 32.Phelan, M. L., I. Rambaldi, and M. S. Featherstone. 1995. Cooperative interactions between HOX and PBX proteins mediated by a conserved peptide motif. Mol. Cell. Biol. 15:3989-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phelan, M. L., and M. S. Featherstone. 1997. Distinct Hox N-terminal arm residues are responsible for specificity of DNA recognition by HOX monomers and HOX-PBX heterodimers. J. Biol. Chem. 272:8635-8643. [DOI] [PubMed] [Google Scholar]
- 34.Picard, J. J., F. Clotman, G. van Maele-Fabry, E. Menegola, A. Bastin, and E. Giavini. 1997. Alterations in expression domains of developmental genes induced in mouse embryos exposed to valproate, p. 161-167. In S. Klug and R. Thiel (ed.), Methods in developmental toxicology and biology. Blackwell Science, Berlin, Germany.
- 35.Remacle, S., C. Shaw-Jackson, C. Matis, X. Lampe, J. Picard, and R. Rezsohazy. 2002. Changing homeodomain residues 2 and 3 of Hoxa1 alters its activity in a cell-type and enhancer dependent manner. Nucleic Acids Res. 30:2663-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren, S.-H., P.-O. Angrand, and F. M. Rijli. 2002. Targeted insertion results in a rhombomere 2-specific Hoxa2 knockdown and ectopic activation of Hoxa1 expression. Dev. Dyn. 225:305-315. [DOI] [PubMed] [Google Scholar]
- 37.Rijli, F. M., P. Dollé, V. Fraulob, M. LeMeur, and P. Chambon. 1994. Insertion of a targeting construct in a Hoxd-10 allele can influence the control of Hoxd-9 expression. Dev. Dyn. 201:366-377. [DOI] [PubMed] [Google Scholar]
- 38.Rijli, F. M., A. Gavalas, and P. Chambon. 1998. Segmentation and specification in the branchial region of the head: the role of the Hox selector genes. Int. J. Dev. Biol. 42:393-401. [PubMed] [Google Scholar]
- 39.Selleri, L., M. J. Depew, Y. Jacobs, S. K. Chanda, K. Y. Tsang, K. S. E. Cheah, J. L. R. Rubenstein, S. O'Gorman, and M. L. Cleary. 2001. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 128:3543-3557. [DOI] [PubMed] [Google Scholar]
- 40.Van Dijk, M. A., and C. Murre. 1994. Extradenticle raises the DNA-binding specificity of homeotic selector gene products. Cell 78:617-624. [DOI] [PubMed] [Google Scholar]
- 41.van Oostveen, J., J. Bijl, F. Raaphorst, J. Walboomers, and C. Meijer. 1999. The role of homeobox genes in normal hematopoiesis and hematological malignancies. Leukemia 13:1675-1690. [DOI] [PubMed] [Google Scholar]
- 42.Waskiewicz, A. J., H. A. Rikhof, and C. B. Moens. 2002. Eliminating zebrafish Pbx proteins reveals a hindbrain ground state. Dev. Cell 3:723-733. [DOI] [PubMed] [Google Scholar]
- 43.Wellik, D. M., and M. R. Capecchi. 2003. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science 307:363-367. [DOI] [PubMed] [Google Scholar]
- 44.Wilkinson, D. G., S. Bhatt, M. Cook, E. Boncinelli, and R. Krumlauf. 1989. Segmental expression of Hox-2 homeobox-containing genes in the developing mouse hindbrain. Nature 341:405-409. [DOI] [PubMed] [Google Scholar]