Abstract
DNA single-strand break repair (SSBR) is important for maintaining genome stability and homeostasis. The current SSBR model derived from an in vitro-reconstituted reaction suggests that the SSBR complex mediated by X-ray repair cross-complementing protein 1 (XRCC1) is assembled sequentially at the site of damage. In this study, we provide biochemical data to demonstrate that two preformed XRCC1 protein complexes exist in cycling HeLa cells. One complex contains known enzymes that are important for SSBR, including DNA ligase 3 (DNL3), polynucleotide kinase 3′-phosphatase, and polymerase β; the other is a new complex that contains DNL3 and the ataxia with oculomotor apraxia type 1 (AOA) gene product aprataxin. We report the characterization of the new XRCC1 complex. XRCC1 is phosphorylated in vivo and in vitro by CK2, and CK2 phosphorylation of XRCC1 on S518, T519, and T523 largely determines aprataxin binding to XRCC1 though its FHA domain. An acute loss of aprataxin by small interfering RNA renders HeLa cells sensitive to methyl methanesulfonate treatment by a mechanism of shortened half-life of XRCC1. Thus, aprataxin plays a role to maintain the steady-state protein level of XRCC1. Collectively, these data provide insights into the SSBR molecular machinery in the cell and point to the involvement of aprataxin in SSBR, thus linking SSBR to the neurological disease AOA.
Several human syndromes whose gene products function in DNA damage response and repair characteristically exhibit defects with the development or maintenance of the nervous system (25). This defect is typified by ataxia-telangiectasia (A-T), which demonstrates an early-onset progressive cerebellar degeneration (17). ATM, the gene product mutated in this syndrome, is a central checkpoint kinase that is required for coordinating cellular responses to DNA damage, in particular to DNA double-strand breaks (DSB) (27). Mutations in the human Mre11 (hMre11), an important component in DSB repair, give rise to the A-T-like disorder that is nearly indistinguishable from A-T (29). Mre11 is found tightly associated with two other proteins, Nbs1 and Rad50 (33). This complex functions in the same pathway with ATM to activate the DNA damage response to DSB (23). Interestingly, Nijmegen breakage syndrome is also associated with mild neurological defects (6). Other syndromes that exhibit primary neurological symptoms and whose gene products are involved in DNA repair include xeroderma pigmentosum, Cockayne syndrome (24), and trichothiodystrophy (30). The gene products of these human syndromes are important components of the nucleotide excision repair pathway (14). These observations strongly suggest a link between DNA repair and neurological homeostasis.
Ataxia with oculomotor apraxia type 1 (AOA1) is another human syndrome that presents neurological features similar to those of A-T but does not exhibit the characteristic hypersensitivity to ionizing radiation (22). The AOA gene (APTX) was identified recently, and its gene product was named aprataxin (8, 21). Early-onset ataxia with hypoalbuminemia and AOA have been considered the same clinical entity because of the recent identification of a common mutation in the APTX gene. A new clinical entity named early-onset AOA and hypoalbuminemia has been proposed to explain these two diseases (28). Aprataxin is a modular protein composed of three domains, a forkhead-associated (FHA) domain, a histidine triad (HIT) domain, and a zinc finger domain. The FHA domain is a protein interaction module that binds phosphopeptides (9, 11), which may allow for regulated protein-protein interaction through phosphorylation of the binding partner. The N terminus of aprataxin that contains the FHA domain also shares distant homology with the N-terminal domain of the polynucleotide kinase 3′-phosphatase (PNK) (35). PNK is the rate-limiting enzyme in DNA single-strand break (SSB) repair (SSBR) (35). Based on the domain structure, aprataxin has been proposed to play a role in SSBR (3), although this has not been substantiated experimentally.
DNA SSBs are the most abundant lesions in cellular DNA (32). SSBs arise spontaneously during normal metabolism from direct attack by reactive oxygen species and as DNA repair intermediates during base excision repair (16). Cellular SSBs are repaired by the SSBR system that is mediated by the X-ray repair cross-complementing protein 1 (XRCC1) (4). XRCC1 was cloned more than 10 years ago by complementation of a mutant Chinese hamster ovary (CHO) cell line, EM9, which is hypersensitive to alkylating agents, including methyl methanesulfonate (MMS) and ethyl methanesulfonate (31). The entire SSBR reaction has been reconstituted in vitro and minimally requires five proteins: poly(ADP-ribose) polymerase (PARP1), XRCC1, PNK, DNA polymerase β (Polβ), and DNA ligase 3 (DNL3). A model for SSBR derived from in vitro experiments suggests that XRCC1 functions as a scaffold protein to coordinate the formation of an SSBR complex in a sequential fashion by which the preceding enzyme prepares the substrates for the subsequent enzyme (3). In this model, PARP1 functions as an SSB sensor to recruit the XRCC1/DNL3 heterodimer, followed by the temporal recruitment of PNK and Polβ to process the SSB so that DNL3 can ligate the nick at the last step. It is not known, however, whether the XRCC1-containing SSBR complex is preformed in the cell or assembled at the site of SSB as the model predicts.
It is also not clear how SSBR is regulated in the cell. Early work found that CK2 can phosphorylate XRCC1 in vitro (16). However, as CK2 can phosphorylate more than 300 proteins in vitro (20), the functional significance of CK2 phosphorylation of XRCC1 is not known. In fact, it is not known whether CK2 phosphorylates XRCC1 in vivo.
We report here that aprataxin interacts with XRCC1. We demonstrate that two preformed XRCC1-containing complexes exist in the cell. One contains PNK and the other contains aprataxin. We report the characterization of the interaction between XRCC1 and aprataxin. We found that XRCC1 is phosphorylated on seven sites in vivo. Three of these sites phosphorylated by CK2 are sufficient to regulate binding to the FHA domain of aprataxin. In addition, we present data to show that the acute loss of aprataxin by small interfering RNA (siRNA) renders HeLa cells sensitive to MMS through a mechanism that destabilizes XRCC1. These data provide insights into the SSBR molecular machinery in the cell, identify a role for aprataxin in SSBR, and suggest a potential link between SSBR with the neuronal disease AOA.
MATERIALS AND METHODS
Cloning, site-directed mutagenesis, generation of stable cell lines, and recombinant proteins.
Full-length cDNAs encoding APTX and PNK were amplified from the first-strand cDNA generated by RT-PCR using RNA isolated from HeLa cells and cloned into the Gateway PENTR TOPO vector (Invitrogen). The sequences were verified by DNA sequencing. Different expression vectors were generated by recombination reactions according to manufacturer's suggestions. Mammalian Gene Storm expression vector containing XRCC1-V5 (GS-XRCC1-V5) was purchased from Invitrogen. pGex-N-APTX encoding amino acids (aa) 1 to 174 of aprataxin was generated by subcloning APTX cDNA into the pGex-4T-1 vector, and pRSET-His-XRCC1 and pRSET-His-PNK were generated by subcloning cDNA into the pRSET vector (Invitrogen). Recombinant glutathione S-transferase (GST)-N-aprataxin, His-XRCC1, and His-PNK were expressed and purified according to standard procedures.
A QuikChange site-directed mutagenesis kit (Stratagene) was used to generate the S518/T519/T523/3A mutant by using the GS-XRCC1-V5 vector as a template. The FHA domain mutant I27A/R29A and AOA1 mutants V263G and P206L of aprataxin were generated the same way. The mutants were verified by sequencing. Stable cell lines of EM9-GS, EM9-XRCC1-WT, and EM9-3A were generated by transfection of EM9 cells with the respective plasmids and selection of single colonies that are resistant to zeocin.
Cell lines and antibodies.
EM9 and HeLa cells were purchased from the American Type Culture Collection. EM9 and its derivatives and HeLa cells were maintained in 10% fetal bovine serum-Dulbecco's modified Eagle's medium. Rabbit polyclonal antibodies against aprataxin (BL596), XRCC1 (A300-065A), CK2α (BL752, BL753, and BL754), CK2α′ (BL756), and V5 (A190-120A) were purchased from Bethyl Laboratories (Montgomery, Tex.). Mouse monoclonal antibodies against Polβ (clone 18S) and CK2β (6D5) were purchased from NeoMarkers and Santa Cruz Biotechnology, respectively. Mouse monoclonal FLAG antibody M2 was purchased from Sigma. Mouse monoclonal antibodies to PNK (BPCC1 and BPCC2) were made by immunizing mice with purified full-length recombinant PNK according to standard procedures.
Phospho-specific antibodies to singly phosphorylated S461 (BL606) or S475 (BL608), doubly phosphorylated S485 and T488 (BL610), and triply phosphorylated S518, T519, and T523 (BL603) were raised against the amino acid sequences of EETKAA(pS)PVLQED, DIEGVQ(pS)EGQDNG, NGAED(pS)GD(pT)EDELR, and DPYAG(pS)(pT)DEN(pT)DSEEHQ, respectively. All phospho-specific antibodies were affinity purified and recognized the phosphorylated and unphosphorylated peptides with a ratio greater than 99:1 by enzyme-linked immunosorbent assay (Bethyl Laboratories).
RNA interference, in vitro kinase assay, colony formation assay, immunoprecipitation (IP), and mass spectrometry.
The siRNA duplexes were synthesized by Dharmacon Research. The sequences targeting each gene were as follows: 5′-AACUCGACUCACUGUGCAGAA for XRCC1, 5′-AAUGUUCUCGACAGCAAGUAC for APTX, and 5′-AAAGCUGCGACUGAUAGAUUG for CK2α′. The siRNA of CK2α (siCK2α) and vimentin (siVimentin) were purchased from Dharmacon Research. HeLa cells were transfected with siRNA duplex by using Oligofectamine (Invitrogen) according to manufacturer's protocols and were usually treated with DNA-damaging agents 48 to 72 h after transfection (12).
An in vitro kinase assay using CK2 and XRCC1 and a colony formation assay were carried out as described previously (7), with minor modifications. Briefly, 3 days after siRNA transfection, HeLa cells were treated with different concentrations of MMS for 1 h and then cultured in fresh medium for at least 1 week before the colony was counted. For data presented in Fig. 6E, plasmids of GS-XRCC1 and GS vector were transfected again 2 days after APTX siRNA (siAPTX) transfection, and cells were treated with MMS 2 days later for colony formation assay. Procedures for IP, Western blotting, and identification of proteins with mass spectrometry were described previously (34).
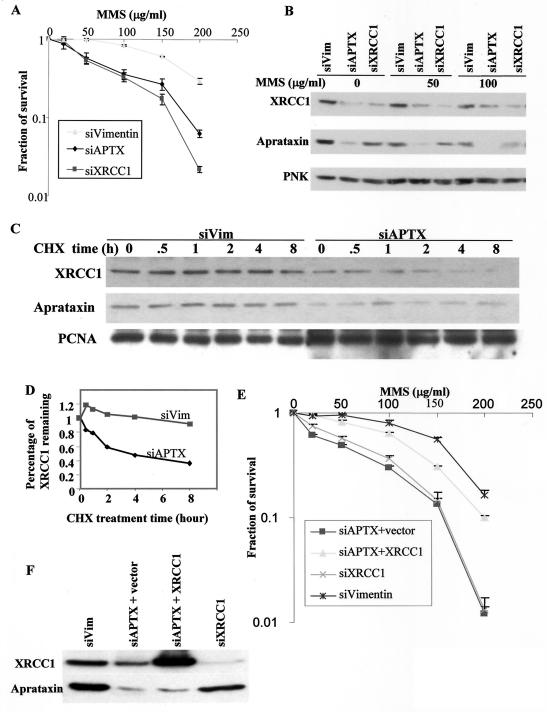
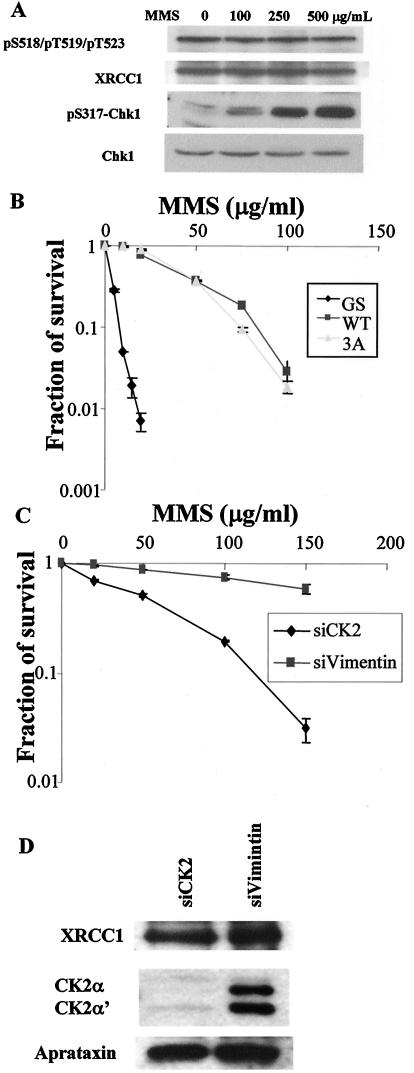
FIG. 6.
Possible role of aprataxin in SSBR. (A) Knockdown of aprataxin and XRCC1 sensitizes HeLa cells to MMS. Data from colony formation assays of HeLa cells transfected with the indicated siRNAs and treated with the indicated dose of MMS are shown. (B) Loss of aprataxin results in a decrease in the XRCC1 protein level. HeLa cells were transfected with the indicated siRNAs and treated with the indicated dose of MMS for 30 min, and whole-cell lysates were Western blotted for XRCC1, aprataxin, and PNK protein levels. (C and D) Half-life measurements of XRCC1 in siAPTX- and siVimentin-transfected HeLa cells. (E) Expression of XRCC1 in siAPTX-transfected cells rescues sensitivity to MMS. HeLa cells were transfected with the indicated siRNAs. For siAPTX-transfected HeLa cells, a vector control or XRCC1 cDNA was also transfected 2 days later and then evaluated for sensitivity to MMS by colony formation assay. (F) Western blot of HeLa cells that were used to evaluate MMS sensitivity described above (E).
Affinity measurement using fluorescence polarization.
Fluorescein isothiocyanate-labeled phosphopeptides based on XRCC1 with the sequence of YAGSTDENpTDSEEHQ, YAGSTDENpTDSAEHQ, or YAGpSpTDENpTDSEEHQ were first synthesized. For controls, an aliquot of the peptides was dephosphorylated with alkaline phosphatase. The phosphopeptide or dephosphorylated peptide was incubated with different concentrations of GST-aprataxin-FHA domain fusion proteins in a 96-well plate. Fluorescence polarization was measured on a Victor V plate reader (Perkin-Elmer).
Protein half-life measurement.
HeLa cells were treated with 80 μM cycloheximide for the indicated time and harvested 3 days after transfection with siAPTX or siVimentin. The whole-cell lysates were separated by sodium dodecyl sulfate- polyacrylamide gel electrophoresis (SDS-PAGE), transferred to membrane, and Western blotted.
RESULTS
Two XRCC1-containing complexes in cycling HeLa cells.
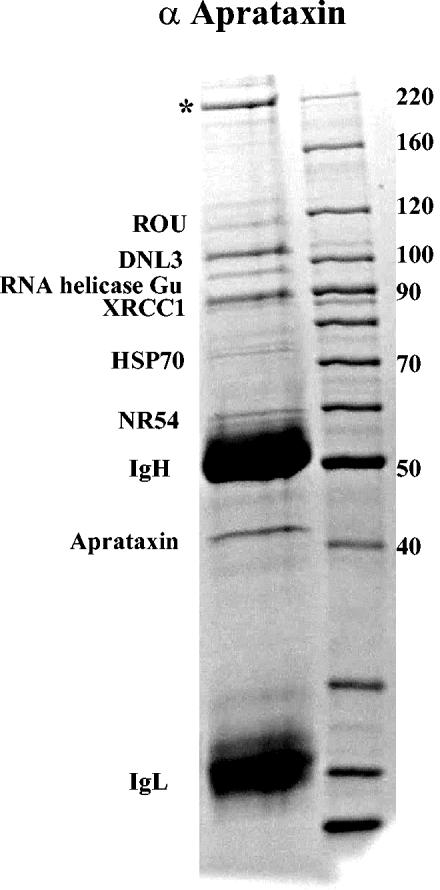
To identify proteins that were interacting with aprataxin, we first carried out IP of aprataxin from HeLa nuclear extracts (NE) made from cycling cells and identified the coimmunoprecipitated proteins by mass spectrometry (Fig. 1). Four major protein bands with similar staining intensities were identified as aprataxin, XRCC1, DNL3, and a protein of unknown function (labeled with an asterisk). Other minor bands were identified and labeled in Fig. 1. Since we often find these proteins in our proteomics analysis of protein complexes that are not related, we tentatively determine them to be nonspecific binding proteins. We then immunoprecipitated XRCC1 from HeLa NE and reciprocally identified aprataxin and DNL3, demonstrating that XRCC1 and aprataxin exist within a complex (data not shown). Notably, PNK is also identified in the XRCC1 IP but is absent in the aprataxin IP, suggesting that PNK may not be in a complex with aprataxin.
FIG. 1.
IP of the aprataxin complex. HeLa NE were immunoprecipitated with an affinity-purified antibody to aprataxin (α Aprataxin), resolved by SDS-4 to 20% PAGE, and stained with Coomassie blue. The major aprataxin-interacting proteins were identified by mass spectrometry. The band with an asterisk indicates a protein of unknown function.
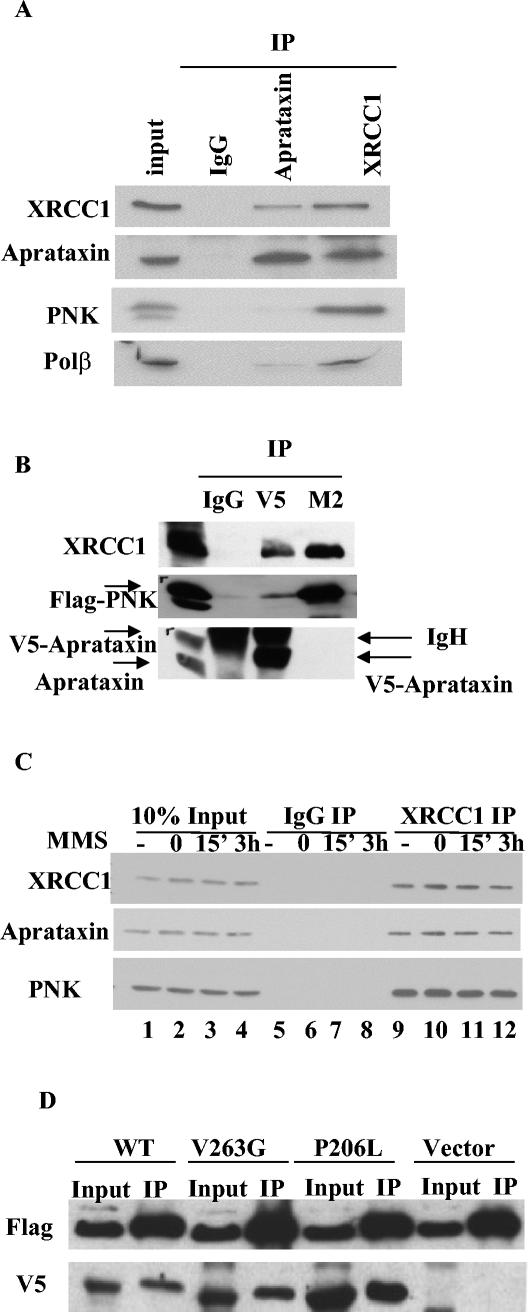
We carried out IP and Western experiments from HeLa NE to verify our results from mass spectrometric analysis. As shown in Fig. 2A, both aprataxin and XRCC1 coimmunoprecipitate with each other. While PNK and Polβ, two important enzymes in SSBR, coimmunoprecipitate with XRCC1, PNK is not detected and Polβ is detected just above the background level in the aprataxin IP. This finding is consistent with the large-scale IP result in which the level of Polβ is below the detection limit of Coomassie blue. To test whether the aprataxin antibody displaces binding of PNK to XRCC1, we transfected a V5-tagged aprataxin at the C terminus and a FLAG-tagged PNK at the N terminus individually in 293T cells and immunoprecipitated aprataxin and PNK with V5 and M2 antibodies, respectively. While both V5 and M2 antibodies coimmunoprecipitate the endogenous XRCC1, the M2 antibody does not coimmunoprecipitate aprataxin, and the V5 antibody coimmunoprecipitates only background level of PNK (Fig. 2B). This small amount of PNK that coimmunoprecipitates with the V5 antibody is likely the result of overexpression of FLAG-PNK. Protein-protein interaction data acquired from both endogenous and transfected proteins support a notion that there are two XRCC1-containing complexes in the cycling cells: one is the known XRCC1 complex that contains the essential SSBR enzymes PNK and Polβ and the other is a new complex that contains aprataxin.
FIG. 2.
Characterization of interactions of XRCC1 with aprataxin and PNK. (A) HeLa NE were immunoprecipitated with control antibody, antiaprataxin, and anti-XRCC1 antibodies and analyzed by Western blotting. (B) Transfection of 293T cells with V5-aprataxin and FLAG-PNK expression constructs. IP was carried out with control IgG, a rabbit polyclonal V5 antibody, and a mouse monoclonal FLAG antibody (M2). Western analysis was performed with antibodies to the indicated protein. (C) Interactions between XRCC1 with aprataxin and PNK are not induced by DNA damage caused by MMS. Whole-cell extracts made from untreated HeLa cells (lanes 1, 5, and 9) or HeLa cells treated with 300 μg of MMS/ml for 30 min and harvested immediately (lanes 2, 6, and 10) or allowed to recover for 15 min (lanes 3, 7, and 11) and 3 h (lanes 4, 8, and 12) are shown. IP was carried out by using control (lanes 5 to 8) and anti-XRCC1 (lanes 9 to 12) antibodies and Western blotted with the indicated antibodies. (D) Mutations in AOA1 do not disrupt interaction with XRCC1. Plasmids encoding FLAG-XRCC1 with the V5-aprataxin WT and the V263G and P206L mutants located in the central HIT domain were transfected in 293T cells. The FLAG-XRCC1 protein was immunoprecipitated with an M2 antibody, and coimmunoprecipitated aprataxin was detected with Western blotting for which a V5 antibody was used.
To substantiate the idea of two XRCC1-containing complexes, we carried out column fractionation of HeLa NE. Aprataxin was found to dissociate from XRCC1 when proteins were eluted from P11 phosphocellulose or DEAE columns, preventing a biochemical separation of the two XRCC1 complexes (data not shown). The fractionation of HeLa NE on a Superose 6 gel filtration column was not sufficient to separate the aprataxin-XRCC1 and PNK-XRCC1 complexes (data shown). However, if the two XRCC1 complexes are of a comparable size, these complexes would not be separable by size exclusion. About one-half of PNK and aprataxin cofractionate with XRCC1 in the high-molecular-weight complexes, and the other half elutes in lower-molecular-weight complexes that are devoid of XRCC1. Thus, XRCC1 is limiting, and it appears that the abundances of XRCC1-PNK and XRCC1-aprataxin complexes are similar in HeLa NE.
To test whether interaction between XRCC1 and aprataxin or PNK is altered in the presence of DNA SSB, we treated cells with MMS at 300 μg/ml and examined the interaction of XRCC1 with aprataxin or PNK. No induced interaction was observed (Fig. 2C), suggesting that these XRCC1 complexes are preformed constitutively.
To investigate whether mutations in AOA1 affect XRCC1 and aprataxin interaction, we cotransfected plasmids encoding FLAG-XRCC1 with the wild-type (WT) V5-aprataxin and V263G and P206L mutations that are located in the central HIT domain in 293T cells, immunoprecipitated XRCC1 with a M2 antibody, and detected coimmunoprecipitated aprataxin with Western blotting by using a V5 antibody. As shown in Fig. 2D, mutations in the aprataxin HIT domain do not seem to abolish interaction with XRCC1. Therefore, a defect in the interaction of aprataxin with XRCC1 may not account for the mechanism of AOA disease.
XRCC1 is phosphorylated in vivo and in vitro by CK2.
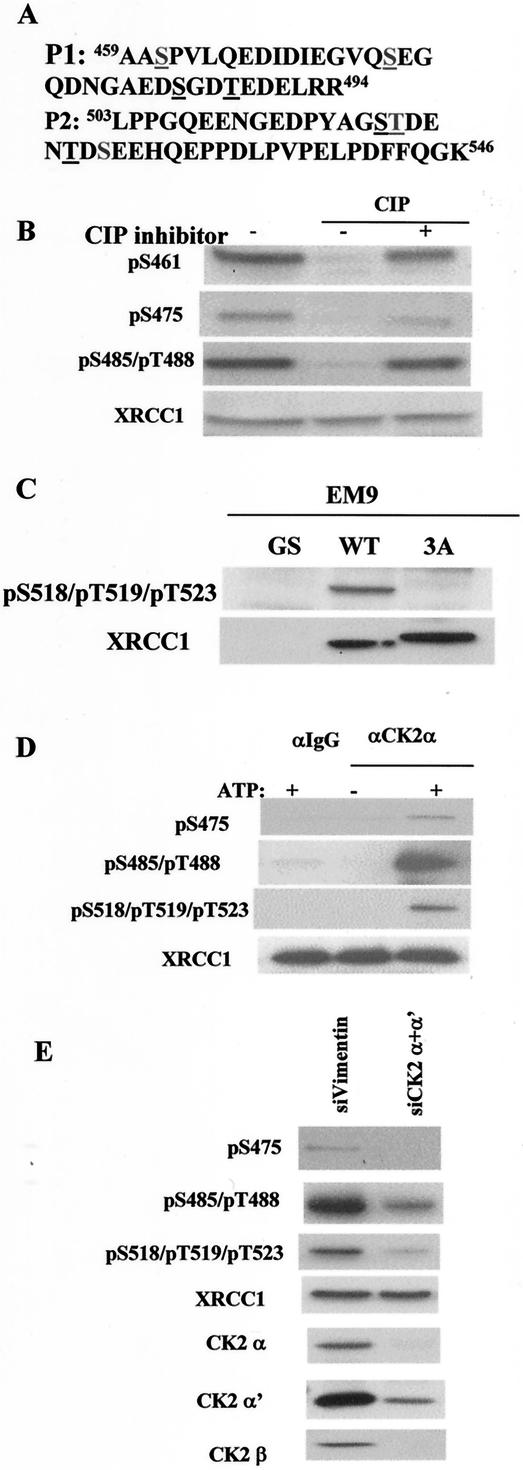
We also analyzed XRCC1 phosphorylation in vivo by using endogenous XRCC1 purified from cycling HeLa cells by IP and SDS-PAGE. Using mass spectrometry, we found two tryptic phosphopeptides (phosphopeptide 1 [P1] [aa 459 to 494] and P2 [aa 503 to 546]) (Fig. 3A) encompassing the linker region between the BRCT1 and BRCT2 domains. P1 was observed to be phosphorylated in vivo from one to four phosphates, and P2 is phosphorylated from three to four phosphates. The very large phosphopeptides prevented mass spectrometric sequencing to identify the exact phosphorylation sites (19). Inspection of the phosphopeptide sequences revealed that they contain CK2 phosphorylation consensus sites. To identify the exact phosphorylation sites in vivo, we made phospho-specific antibodies against individually phosphorylated pS461 and pS475, doubly phosphorylated pS485/pT488, and triply phosphorylated pS518/pT59/pT523. Among these sites, S461, S475, and S518 do not conform to CK2 consensus sites.
FIG. 3.
Characterization of XRCC1 phosphorylation. (A) Sequence of tryptic phospho-specific XRCC1 peptides identified by mass spectrometry analysis of XRCC1 immunoprecipitated from HeLa NE. The underlined amino acids are the sites of XRCC1 phosphorylation. P1 corresponds to aa 459 to 494 of XRCC1 and was found to contain from one to four phosphates. P2 corresponds to aa 503 to 546 of XRCC1 and contains three to four phosphates. (B) Confirmation of phosphorylation of XRCC1 at S461, S475, and S485/T488 in vivo with phospho-specific antibodies. HeLa whole-cell extracts were mock treated or treated with calf intestine phosphatase (CIP) in the presence (+) or absence (−) of a CIP inhibitor as indicated and Western blotted with the indicated antibodies. (C) Confirmation of XRCC1 phosphorylation at S518/T519/T523 in vivo. Whole-cell extracts made fromEM9-GS, EM9-XRCC1-WT, and EM9-XRCC1-3A cells were Western blotted with the pS518/pT519/pT523 and XRCC1 antibodies. (D) Phosphorylation of XRCC1 by CK2 IP-kinase assay. Control (IgG) or CK2α′ antibodies were used to immunoprecipitate from HeLa NE in the absence or presence of added ATP. The immunoprecipitates were used for IP-kinase assays with recombinant XRCC1. Phosphorylation of XRCC1 at different sites was examined by Western blotting. (E) Knockdown of CK2 subunits by siRNA and its effects on XRCC1 phosphorylation. HeLa cells were transfected with siVimentin or two siRNAs specific to the α and α′ subunits of CK2, and whole-cell extracts were Western blotted with the indicated antibodies.
As shown in Fig. 3B, phospho-specific antibodies against pS461, pS475, and doubly phosphorylated pS485/pT488 recognize XRCC1 in a phosphorylation-dependent manner. Thus, these sites can be phosphorylated in vivo in HeLa cells. Since P2 contains three phosphorylation sites (S518, T519, and T523) that are fully phosphorylated in vivo, and the rest of the sites are not fully phosphorylated, we concentrated our biochemical and functional analysis on the triple-phosphorylation sites.
To verify that the phospho-specific antibody against triply phosphorylated pS518/pT59/pT523 is sequence specific, we generated stable EM9-WT, EM9-3A, and EM9-GS cell lines that are complemented with XRCC1-WT, XRCC1-3A (where S518, T519, and T523 are mutated to Ala), and an empty vector, respectively. This triple-phospho-specific antibody is specific to the three phosphorylation sites since it recognizes only WT XRCC1 and not the 3A mutant (Fig. 3C); it also recognizes XRCC1 from cycling HeLa cells but does not recognize the dephosphorylated XRCC1, demonstrating that it is phosphorylation specific (data not shown). These results also show that S518, T519, and T523 are phosphorylated in vivo in both EM9 and HeLa cells.
To test whether CK2 phosphorylates XRCC1 in vitro, we immunoprecipitated the CK2 kinase from HeLa cells and carried out an immunocomplex kinase assay with a recombinant His-tagged XRCC1 protein purified from Escherichia coli. Western blotting with the phospho-specific antibodies showed that XRCC1 is phosphorylated at six sites by CK2 in an ATP-dependent manner in vitro (Fig. 3D), despite the fact that S475 and S518 do not strictly conform to CK2 consensus sites. Phosphorylation of S461, which does not conform to the CK2 site and is farther away from the CK2 consensus site cluster, is not phosphorylated by CK2.
We used siRNA to demonstrate that XRCC1 phosphorylation depends on CK2 in vivo. Since there are two independent kinase catalytic subunits (α and α′) in HeLa cells, we transfected HeLa cells with siRNA specific to these two subunits. Down-regulation of CK2 attenuates phosphorylation of XRCC1 at these sites significantly, including S475, which is not a CK2 consensus site (Fig. 3E). Residual CK2 activity after RNA interference most likely can account for the remaining phosphorylation. We conclude that CK2 phosphorylates XRCC1 both in vitro and in vivo.
To test whether MMS treatment modulates XRCC1 phosphorylation, we treated cells with various doses of MMS and examined phosphorylation by Western blotting. Phosphorylation at the triply phosphorylated sites S518, T519, and T523 as well as the other three in vivo CK2-dependent sites does not change in response to MMS (Fig. 4A and data not shown), in agreement with previously published observations that CK2 is constitutively active (18).
FIG. 4.
Phosphorylation of XRCC1 at S518/T519/T523 in response to DNA damage caused by MMS and its requirement for cellular survival. (A) Phosphorylation of XRCC1 at S518/T519/T523 is not induced by MMS treatment. Whole-cell extracts made from HeLa cells that were treated with the indicated dose of MMS for 30 min were Western blotted with the indicated antibodies. Phosphorylation of Chk1 at Ser317 was used to confirm that MMS activates the DNA damage response. (B) Phosphorylation of XRCC1 at S518/T519/T523 is not required for cellular survival of MMS treatment in EM9 cells. Data from colony formation assays of the EM-GS, EM9-XRCC1-WT, and EM9-XRCC1-3A cells in response to MMS are shown. (C) Requirement of CK2 in cellular survival of MMS treatment. Data from colony formation assays of HeLa cells transfected with siVimentin and siCK2 are shown. (D) Western blots to measure the protein levels of XRCC1, CK2, and aprataxin in siVimentin- and siCK2-transfected HeLa cells.
We used the colony formation assay to test MMS sensitivity of the stable EM9-WT, EM9-3A, and EM9-GS cell lines to evaluate the functional consequence of XRCC1 phosphorylation. The EM9-3A cell line exhibits MMS sensitivity similar to that of EM9-WT, while the EM9-GS cell line is hypersensitive (Fig. 4B). Therefore, the phosphorylation of XRCC1 at S518, T519, and T523 is not required for cellular survival in response to MMS. In agreement with this conclusion, the EM9-WT and EM9-3A cell lines display similar SSBR capacity when tested with the Comet assay (data not shown).
Because S518, T519, and T523 are phosphorylated by CK2, we investigated the requirement of CK2 for cellular survival in response to MMS. As shown in Fig. 4C, CK2 is required for survival. Down-regulation of CK2 does not significantly change XRCC1 and aprataxin protein levels (Fig. 4D), ruling out the possibility that the requirement of CK2 for survival is met through the regulation of XRCC1 and aprataxin protein levels.
The FHA domain of aprataxin binds the phosphorylated S518, T519, and T523 of XRCC1.
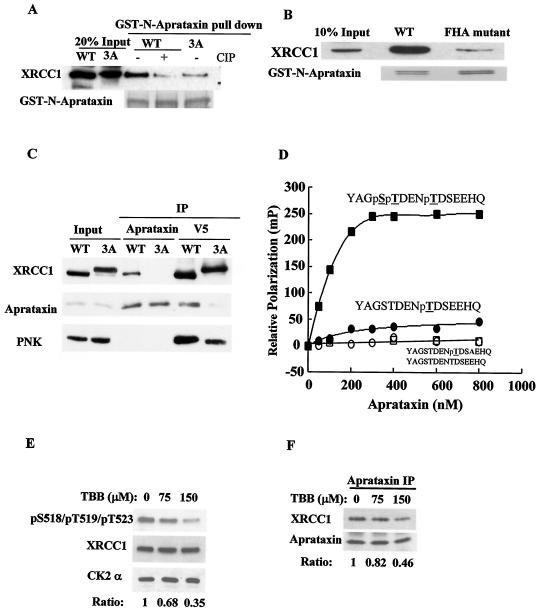
To test whether the FHA domain of aprataxin binds XRCC1 in a phosphorylation-dependent manner, we made a GST fusion of the N terminus of aprataxin containing the FHA domain and used it to pull down XRCC1 from whole-cell lysates made from EM9-WT and EM9-3A. As shown in Fig. 5A, only the WT XRCC1, not the XRCC1-3A mutant, can be pulled down by the GST-N-aprataxin, and dephosphorylation of XRCC1 diminishes the interaction to the same extent as the 3A mutant (Fig. 5A).
FIG. 5.
Interactions between XRCC1 and aprataxin or PNK are mediated by phosphorylation of XRCC1 at S518/T519/T523. (A) The interaction between the N terminus of aprataxin and XRCC1 is phosphorylation dependent. The purified GST-N-aprataxin was mixed with whole-cell extracts from EM9-XRCC1-WT or EM9-XRCC1-3A cells. Where indicated, the extracts prepared from the EM9-XRCC1-WT cells were pretreated with CIP. GST beads were used in pull-down experiments and Western blotted for XRCC1. GST-N-aprataxin was detected with Coomassie blue. (B) The FHA domain of aprataxin mediates interaction with XRCC1. GST pull down of XRCC1 from whole-cell extracts of EM9-XRCC1-WT with the GST-N-aprataxin WT and the GST-N-aprataxin FHA mutant (I27A/R29A). (C) 293T cells were transfected with WT V5-XRCC1 or the V5-XRCC1-3A mutant and immunoprecipitated for aprataxin or V5 and Western blotted with the indicated antibody. (D) Binding of GST-N-aprataxin to phosphorylated XRCC1 peptides measured by fluorescence polarization. Fluorescein isothiocyanate-labeled phosphopeptides of YAGSTDENpTDSEEHQ, YAGSTDENpTDSAEHQ, or YAGpSpTDENpTDSEEHQ were synthesized (underlining indicates phosphorylated residues, and boldface indicates the Ala mutation at P+3), and an aliquot of the peptides were dephosphorylated with alkaline phosphatase for controls. (E) Western blots measuring XRCC1 phosphorylation when HeLa cells were treated with TBB for 12 h at the indicated concentration. The ratio of the signal intensity of the phospho-specific antibody to that of XRCC1, which is normalized to the untreated sample, is tabulated. (F) Inhibition of CK2 kinase activity leads to dissociation of aprataxin from XRCC1. Aprataxin was immunoprecipitated from HeLa cells treated with TBB, and coimmunoprecipitated XRCC1 was Western blotted. The ratio of the signal intensity of XRCC1 to that of aprataxin, which is normalized to the untreated sample, is tabulated.
To demonstrate that the FHA domain is responsible for binding, we mutated two conserved residues, I27 and R29, in the FHA domain of GST-N-aprataxin to Ala. This FHA mutant significantly diminishes interaction with XRCC1 (Fig. 5B). Thus, phosphorylation of XRCC1 at S518, T519, and T523 regulates binding to aprataxin through the FHA domain in vitro.
To examine whether phosphorylation at S518, T519, and T523 regulates binding to aprataxin in vivo, V5-XRCC1-WT or V5-XRCC1-3A was transfected into 293T cells, and IP with antibodies against V5 or aprataxin was carried out. Endogenous aprataxin can coimmunoprecipitate only with V5-XRCC1-WT but not V5-XRCC1-3A; in the reciprocal experiment, V5-XRCC1-WT, but not V5-XRCC1-3A, can coimmunoprecipitate aprataxin (Fig. 5C), demonstrating that phosphorylation of XRCC1 at S518, T519, and T523 regulates its binding to aprataxin in vivo. Surprisingly, XRCC1-3A also coimmunoprecipitates significantly less endogenous PNK than XRCC1-WT does, indicating that phosphorylation of XRCC1 at the triple sites may also strengthen the interaction with PNK. However, PNK seems able to bind XRCC1 with a higher affinity than that of aprataxin when the triple-phosphorylation sites are mutated.
We estimated binding affinity by using fluorescence polarization (Fig. 5D). A peptide where only T523 (pT1) is phosphorylated binds to the aprataxin FHA domain with an apparent Kd of approximately 210 nM. Because the FHA domain has been reported to prefer acidic amino acids at the P+3 position (three amino acid residues C terminal to the phosphorylation sites), we synthesized a mutant pT1 peptide where the amino acid residue E at the P+3 position was substituted to A (pT1-EA). This mutation completely abolishes the binding of pT1 to aprataxin, indicating that the amino acid E is important for aprataxin FHA domain recognition. Significantly, additional phosphorylation of the two upstream residues (S518 and T519) dramatically enhances the binding of the pT1 peptide to the FHA domain. This was reflected by both a twofold decrease in the apparent Kd value (∼90 nM) and a fivefold increase in maximum binding.
To further demonstrate that CK2 phosphorylation of XRCC1 is important for aprataxin binding, we treated HeLa cells with a CK2 inhibitor (26), 4,5,6,7-tetrabromo-2-azabenzamidazole (TBB), that results in the attenuation of XRCC1 phosphorylation in a dose-dependent manner (Fig. 5E). Reduction in XRCC1 phosphorylation leads to dissociation of XRCC1 from aprataxin (Fig. 5F). Thus, CK2 kinase activity is required for XRCC1 and aprataxin binding.
An acute loss of aprataxin results in a lower steady-state protein level of XRCC1.
Since the binding of aprataxin to XRCC1 does not seem to be required for cellular survival in response to MMS treatment (Fig. 4B), we tested whether aprataxin itself is required. We transfected HeLa cells with siAPTX and XRCC1 siRNA and tested their sensitivity to MMS by using the colony formation assay. An acute loss of APTX and XRCC1 renders HeLa cells sensitive to MMS (Fig. 6A), suggesting a functional link between aprataxin and XRCC1.
Western blotting of cell lysates from HeLa cells transfected with different siRNA revealed that an acute loss of aprataxin leads to a reduced protein level of XRCC1 (Fig. 6B). A loss of XRCC1, however, has a minimal effect on aprataxin; neither the loss of aprataxin nor the loss of XRCC1 has an effect on the protein level of PNK (Fig. 6B). We measured XRCC1 protein stability when cells were transfected with different siRNA. When protein synthesis is inhibited with 80 μM of cycloheximide, XRCC1 protein is degraded much faster in siAPTX-transfected cells than in siVimentin-transfected cells (Fig. 6C and D). Therefore, aprataxin is required to stabilize XRCC1 in HeLa cells. This finding suggests an indirect mechanism for the requirement of aprataxin for cellular survival in response to MMS treatment in which aprataxin is required to maintain the steady-state protein level of XRCC1, which is essential for SSBR.
To substantiate this idea, we overexpressed XRCC1 after transfection of siAPTX in HeLa cells and measured MMS sensitivity. Exogenously expressed XRCC1 rescues MMS sensitivity of HeLa cells transfected with siAPTX to a significant degree (Fig. 6E and F). This result supports the notion that MMS sensitivity of HeLa cells after an acute loss of aprataxin may be due to the reduced protein level of XRCC1.
DISCUSSION
In this study, we provided biochemical data to demonstrate that two preformed XRCC1 protein complexes exist in cycling HeLa cells. One complex contains known enzymes that are important for SSBR, including DNL3, PNK, and Polβ, and the other contains DNL3 and the AOA gene (APTX) product aprataxin. The latter complex is a newly identified XRCC1 complex. Using mass spectrometry and phospho-specific antibodies, we show that XRCC1 is phosphorylated in vivo on at least seven sites and that CK2 phosphorylates XRCC1 at six of these sites in vitro and is required to phosphorylate them in vivo. We also show that XRCC1 phosphorylated on S518, T519, and T523 by CK2 is the in vivo target of the FHA domain of aprataxin. Interestingly, phosphorylation of XRCC1 on S518, T519, and T523 also modulates PNK binding to XRCC1 but to a lesser extent than aprataxin. Functionally, the phosphorylation of XRCC1 that is important for aprataxin binding does not seem to be required for cellular survival in response to MMS or SSBR in EM9 cells, but an acute loss of aprataxin by siRNA renders HeLa cells sensitive to MMS. We provide data to show that the acute loss of APTX leads to the destabilization of XRCC1. Therefore, one function of aprataxin is to maintain the steady-state protein level of XRCC1. Collectively, these data reveal a new component of the SSBR molecular machinery in the cell and potentially link SSBR to the neurological disease AOA.
Two preformed XRCC1-containing protein complexes in cycling cells.
The reconstituted in vitro SSBR assay has provided much insight into the molecular mechanism of SSBR. The current recruitment model based on this assay describes the SSBR reaction as an ordered sequential handoff reaction at the site of SSB organized by the scaffold protein XRCC1 (4). Our isolated XRCC1 complexes suggest that there is a preformed steady-state XRCC1 complex containing DNL3, PNK, and Polβ in the cell. This complex may be recruited by the SSB recognition factor PARP1 (13). We did observe PARP1 in the XRCC1 IP, but the fact that PARP1 is also found in control immunoglobulin G (IgG) prevented us from concluding that PARP1 is also a component of the steady-state XRCC1 complex. We feel that PARP1 may be loosely associated with the XRCC1 complex. Since this known XRCC1 complex contains all necessary components for SSBR, this is a presumed SSBR complex. The isolation of an XRCC1 complex containing most of the essential components for SSBR suggests that either SSBR is constitutively active in cycling cells or the basic machinery for SSBR is preassembled and is ready to be recruited to the site of SSB for repair in response to DNA damage.
The surprising finding here is that aprataxin and XRCC1-DNL3 form another steady-state complex independent of PNK and possibly Polβ. The relationship between these two XRCC1-containing complexes and how their abundance is regulated are not yet known. Our analysis of XRCC1 phosphorylation provides a clue. We found that phosphorylation of XRCC1 at three sites is sufficient to regulate binding of XRCC1 to the FHA domain of aprataxin. Interestingly, phosphorylation of XRCC1 also regulates binding to the FHA domain of PNK, although the precise phosphorylation sites that regulate such binding are not mapped (19). One can envision a scenario in which the three phosphorylation sites at S518, T519, and T523 determine a conformation of XRCC1 that allows aprataxin and PNK to bind to the same region competitively, thus producing a mutually exclusive PNK-XRCC1 complex or aprataxin-XRCC1 complex. The extent of XRCC1 phosphorylation on different sites can vary greatly in the cell. The region spanning aa 503 to 546 that contains S518, T519, and T523, the major determinant for aprataxin binding, is fully phosphorylated with three phosphates, but the region spanning aa 459 to 494 that contains S461, S475, S485, and T488 is phosphorylated in various degrees from one to four phosphates (19). Thus, phosphorylation of S518, T519, and T523 is much preferred to that of other sites to allow binding of aprataxin constitutively. In addition, we found that a mutation at S518, T519, and T523 leads to reduced phosphorylation in the region of aa 459 to 494 including S475, S485, and T488 (Y. Wang and J. Qin, unpublished data); but under such conditions, binding of PNK is only weakened, while binding of aprataxin is abolished. Therefore, the relative ratio of these two complexes can be determined by the relative abundance of PNK and aprataxin in the cell as well as their relative affinity to phosphorylated and unphosphorylated XRCC1. Since we did not observe a free form of XRCC1 but did observe free forms of aprataxin and PNK, we concluded that all cellular XRCC1 is in protein complexes.
The mode of binding of CK2-phosphorylated XRCC1 to aprataxin is intriguing. Phosphorylations of S518 and T519 in addition to T523 contribute to aprataxin binding significantly, by which the triply phosphorylated XRCC1 binds much more strongly than does the singly phosphorylated XRCC1 at T523, which itself confers to the FHA domain binding motif pTXX(D/E) (10). Thus, the extent of XRCC1 phosphorylation by CK2 determines binding affinity of aprataxin with XRCC1, which in turn may determine the relative ratios of the aprataxin-XRCC1 and PNK-XRCC1 complexes. Since PNK seems to interact with underphosphorylated XRCC1 with a higher affinity than aprataxin does, the extent of XRCC1 phosphorylation should play an important role in modulating interaction with PNK. Thus, it is likely that CK2 can modulate the relative abundance of the PNK-XRCC1 and aprataxin-XRCC1 complexes by fine tuning XRCC1 phosphorylation. CK2 was reported to change activity and abundance or cellular localization, depending on the cellular condition that promotes proliferation or differentiation (18). CK2 is also expressed and functions in neuronal cells (1). It is possible that CK2 can function as a rheostat in cells including neurons to modify the SSBR process.
Is there a role for aprataxin in regulating SSBR?
We have shown that aprataxin is required for survival in response to MMS in HeLa cells. This can be explained to some degree by our observation that a loss of APTX results in a reduced XRCC1 protein level. It is a common phenomenon that the loss of a protein leads to destabilization of its binding partner. For example, the loss of XRCC1 leads to a decreased protein level of DNL3 (5); a decreased level of DSB repair protein Mre11 destabilizes its binding partners Rad50 and Nbs1 (29), and the loss of the checkpoint kinase ATR leads to the destabilization of its binding partner ATRIP (7). The reduction of XRCC1 in siAPTX-transfected cells is not as severe as the examples cited above, which may be explained by the observation that the PNK level does not change significantly, so it is possible that the remaining XRCC1 can be stabilized by PNK. This reduced level of XRCC1 is not sufficient to support cellular survival in response to MMS. Such an idea is supported by our finding that overexpression of XRCC1 in an APTX knockdown background can rescue MMS sensitivity to a significant degree. Thus, aprataxin plays a role in maintaining the steady-state level of the important SSBR protein XRCC1.
Although we have firmly established that aprataxin interacts with XRCC1, such interaction does not seem to be important for SSBR when determined by the MMS sensitivity or Comet assay of the EM9-WT and EM9-3A cells. Since the 3A mutant can still allow PNK to bind, although with lower affinity, this is apparently sufficient for cellular survival in response to MMS. This finding is consistent with the finding that EM9 cells complemented with XRCC1-CKM, where all consensus CK2 sites are mutated, are not sensitive to H2O2 but are defective in SSBR when assayed by the Comet assay (19). CK2, however, is required for cellular survival in response to MMS, suggesting that CK2 may regulate proteins in both the DNA damage response and repair pathways. These observations suggest that CK2 may be a kinase that coordinates SSBR and cell death.
Most of the disease-causing mutations of AOA are localized to the central HIT domain that is predicted to have adenosine 5′-monophosphoramide hydrolase activity (2). These mutations (at least for P206L and V263G that we have tested) do not impair binding to XRCC1. It is possible that aprataxin has an enzymatic role in SSBR in addition to that of stabilizing XRCC1, but the enzymatic role is too subtle to be revealed by our rather gross colony formation and Comet assays. The identification of a separation-of-function mutant that retains XRCC1 binding but is defective in HIT domain function will be required to establish such a role. It is interesting that a fibroblast cell line derived from a patient with AOA does not seem to exhibit lower XRCC1 protein levels (K. W. Caldecott, personal communication). As almost all AOA patients have mutations in the middle portion of the protein (8, 21), it is possible that this patient cell line retains the N terminus of aprataxin, where the FHA domain resides. Thus, it is possible that the remaining N-terminal aprataxin fragment can stabilize the XRCC1 protein through its FHA domain. As a result, the XRCC1 protein level is normal and the patient cell line is not grossly sensitive to MMS. In fact, it has been shown that the patient cell line is slightly sensitive to H2O2 but is proficient in SSBR assayed by the Comet assay (15). Thus, the HIT domain and the C terminus of aprataxin may be more important for the underlining molecular mechanism of the cause of AOA. Even though we have firmly established that aprataxin is important for cellular survival of MMS treatment and that the FHA domain is important for binding to XRCC1, we have not yet revealed the molecular mechanism for AOA disease. It is an attractive model to test whether the HIT domain and the C terminus of aprataxin are important for some aspects of SSBR such as the fidelity of SSBR, for example. Such fidelity is important for neuronal cell homeostasis, but it is too subtle to be revealed in proliferating cells by the colony formation and Comet assays. We do not know whether data presented here apply to neuronal cells, where the action of these proteins may be more relevant to the link between SSBR and neurodegeneration. SSBR occurs at least in terminally differentiated cells, and results obtained from proliferating cells should provide a road map for future work with postmitotic neuronal cells.
Acknowledgments
We thank Grant Steward for critical reading of the manuscript.
This work was supported by grant CA92584 from the NIH. J.Q. is a recipient of a career development award from the Department of Defense Breast Cancer Research Program (DAMD17-00-1-0146).
REFERENCES
- 1.Blanquet, P. R. 2000. Casein kinase 2 as a potentially important enzyme in the nervous system. Prog. Neurobiol. 60:211-246. [DOI] [PubMed] [Google Scholar]
- 2.Brenner, C. 2002. Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry 41:9003-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldecott, K. W. 2003. DNA single-strand break repair and spinocerebellar ataxia. Cell 112:7-10. [DOI] [PubMed] [Google Scholar]
- 4.Caldecott, K. W. 2003. XRCC1 and DNA strand break repair. DNA Repair 2:955-969. [DOI] [PubMed] [Google Scholar]
- 5.Caldecott, K. W., J. D. Tucker, L. H. Stanker, and L. H. Thompson. 1995. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 23:4836-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates III, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. [DOI] [PubMed] [Google Scholar]
- 7.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 8.Date, H., O. Onodera, H. Tanaka, K. Iwabuchi, K. Uekawa, S. Igarashi, R. Koike, T. Hiroi, T. Yuasa, Y. Awaya, T. Sakai, T. Takahashi, H. Nagatomo, Y. Sekijima, I. Kawachi, Y. Takiyama, M. Nishizawa, N. Fukuhara, K. Saito, S. Sugano, and S. Tsuji. 2001. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat. Genet. 29:184-188. [DOI] [PubMed] [Google Scholar]
- 9.Durocher, D., J. Henckel, A. R. Fersht, and S. P. Jackson. 1999. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell 4:387-394. [DOI] [PubMed] [Google Scholar]
- 10.Durocher, D., and S. P. Jackson. 2002. The FHA domain. FEBS Lett. 513:58-66. [DOI] [PubMed] [Google Scholar]
- 11.Durocher, D., I. A. Taylor, D. Sarbassova, L. F. Haire, S. L. Westcott, S. P. Jackson, S. J. Smerdon, and M. B. Yaffe. 2000. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6:1169-1182. [DOI] [PubMed] [Google Scholar]
- 12.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 13.El Khamisy, S. F., M. Masutani, H. Suzuki, and K. W. Caldecott. 2003. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 31:5526-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedberg, E. C. 2001. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer 1:22-33. [DOI] [PubMed] [Google Scholar]
- 15.Gueven, N., O. J. Becherel, A. W. Kijas, P. Chen, O. Howe, J. H. Rudolph, R. Gatti, H. Date, O. Onodera, G. Taucher-Scholz, and M. F. Lavin. 2004. Aprataxin, a novel protein that protects against genotoxic stress. Hum. Mol. Genet. 13:1081-1093. [DOI] [PubMed] [Google Scholar]
- 16.Kubota, Y., R. A. Nash, A. Klungland, P. Schar, D. E. Barnes, and T. Lindahl. 1996. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 15:6662-6670. [PMC free article] [PubMed] [Google Scholar]
- 17.Lavin, M. F., and Y. Shiloh. 1997. The genetic defect in ataxia-telangiectasia. Annu. Rev. Immunol. 15:177-202. [DOI] [PubMed] [Google Scholar]
- 18.Litchfield, D. W. 2003. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 369:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loizou, J. I., S. F. El Khamisy, A. Zlatanou, D. J. Moore, D. W. Chan, J. Qin, S. Sarno, F. Meggio, L. A. Pinna, and K. W. Caldecott. 2004. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell 117:17-28. [DOI] [PubMed] [Google Scholar]
- 20.Meggio, F., and L. A. Pinna. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17:349-368. [DOI] [PubMed] [Google Scholar]
- 21.Moreira, M. C., C. Barbot, N. Tachi, N. Kozuka, E. Uchida, T. Gibson, P. Mendonca, M. Costa, J. Barros, T. Yanagisawa, M. Watanabe, Y. Ikeda, M. Aoki, T. Nagata, P. Coutinho, J. Sequeiros, and M. Koenig. 2001. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat. Genet. 29:189-193. [DOI] [PubMed] [Google Scholar]
- 22.Nemeth, A. H., E. Bochukova, E. Dunne, S. M. Huson, J. Elston, M. A. Hannan, M. Jackson, C. J. Chapman, and A. M. Taylor. 2000. Autosomal recessive cerebellar ataxia with oculomotor apraxia (ataxia-telangiectasia-like syndrome) is linked to chromosome 9q34. Am. J. Hum. Genet. 67:1320-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrini, J. H., and T. H. Stracker. 2003. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 13:458-462. [DOI] [PubMed] [Google Scholar]
- 24.Rapin, I., Y. Lindenbaum, D. W. Dickson, K. H. Kraemer, and J. H. Robbins. 2000. Cockayne syndrome and xeroderma pigmentosum. Neurology 55:1442-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolig, R. L., and P. J. McKinnon. 2000. Linking DNA damage and neurodegeneration. Trends Neurosci. 23:417-424. [DOI] [PubMed] [Google Scholar]
- 26.Sarno, S., H. Reddy, F. Meggio, M. Ruzzene, S. P. Davies, A. Donella-Deana, D. Shugar, and L. A. Pinna. 2001. Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 (′casein kinase-2′). FEBS Lett. 496:44-48. [DOI] [PubMed] [Google Scholar]
- 27.Shiloh, Y. 2001. ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11:71-77. [DOI] [PubMed] [Google Scholar]
- 28.Shimazaki, H., Y. Takiyama, K. Sakoe, K. Ikeguchi, K. Niijima, J. Kaneko, M. Namekawa, T. Ogawa, H. Date, S. Tsuji, I. Nakano, and M. Nishizawa. 2002. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia: the aprataxin gene mutations. Neurology 59:590-595. [DOI] [PubMed] [Google Scholar]
- 29.Stewart, G. S., R. S. Maser, T. Stankovic, D. A. Bressan, M. I. Kaplan, N. G. Jaspers, A. Raams, P. J. Byrd, J. H. Petrini, and A. M. Taylor. 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99:577-587. [DOI] [PubMed] [Google Scholar]
- 30.Taylor, E. M., B. C. Broughton, E. Botta, M. Stefanini, A. Sarasin, N. G. Jaspers, H. Fawcett, S. A. Harcourt, C. F. Arlett, and A. R. Lehmann. 1997. Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene. Proc. Natl. Acad. Sci. USA 94:8658-8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson, L. H., K. W. Brookman, N. J. Jones, S. A. Allen, and A. V. Carrano. 1990. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol. Cell. Biol. 10:6160-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson, L. H., and M. G. West. 2000. XRCC1 keeps DNA from getting stranded. Mutat. Res. 459:1-18. [DOI] [PubMed] [Google Scholar]
- 33.Trujillo, K. M., S. S. Yuan, E. Y. Lee, and P. Sung. 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 273:21447-21450. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927-939. [PMC free article] [PubMed] [Google Scholar]
- 35.Whitehouse, C. J., R. M. Taylor, A. Thistlethwaite, H. Zhang, F. Karimi-Busheri, D. D. Lasko, M. Weinfeld, and K. W. Caldecott. 2001. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 104:107-117. [DOI] [PubMed] [Google Scholar]