Abstract
Cointroduction of plasmids into mammalian cells is commonly used to investigate transcription factor regulation of reporter genes or to normalize transfection efficiency. We report here that cotransfected DNA molecules commonly transfer enhancer elements from one plasmid to another. Using separate Renilla or Firefly luciferase reporters, we found that an estrogen response element (ERE) originally linked to one of the reporters stimulated expression of the non-ERE-containing reporter. Similar enhancer transfer was seen with the cytomegalovirus enhancer. This enhancer transfer effect was not seen when cells were transfected separately with the reporters and the extracts were then combined before luciferase assays. The degree of enhancer transfer increased with transfected plasmid concentration and was greater when linearized rather than circular plasmid DNA was used. We hypothesized that double-strand breaks and heteroligation of cointroduced DNA molecules mediated the transfer of regulatory elements from one molecule to another. PCR of transfected plasmid DNA confirmed nonhomologous end-joining (NHEJ) ligation of DNA fragments originally present in separate plasmids. The NHEJ reaction was enhanced by UV light treatment to introduce double-strand breaks, and it was greater after liposome-mediated transfection than after calcium-phosphate-mediated transfection. NHEJ also occurred after adenoviral transfer of DNA into cells. We conclude that NHEJ mediates the transfer of regulatory DNA elements among cointroduced DNA molecules. These findings indicate the need for caution when interpreting results of transfection experiments containing more than one plasmid and suggest a mechanism whereby viruses or other exogenous DNA might recombine to activate unrelated genes.
DNA regulatory elements generally function in cis and act by binding transcription factors. Thus, a transcription-regulating element on one DNA molecule does not affect transcription on another in trans, except in unusual situations in which an enhancer on one molecule has been artificially (6, 13, 14, 20) or spontaneously (3) placed in close proximity to another molecule. This concept of independent plasmid function is important in cotransfection experiments, which are commonly used to test the effects of one or more transcription factors on the regulation of a promoter linked to a reporter gene. Similarly, a separate reporter gene is often used as an internal control to correct for variations in transfection efficiency.
In cotransfection experiments, however, there is reason to doubt the assumption that transfected molecules always remain separate from one another. For example, it is known that stable transfectants often carry multiple, concatenated linearized plasmids (8, 16), suggestive of ligation between transfected circular plasmids before integration. The ability of a selectable marker to cointegrate with another gene is used to select stably transfected cells, probably because there is frequent cointegration of the two plasmids into the same site in the genome (2). These findings imply that a substantial portion of transfected DNA molecules can be ligated to one another before integration.
One possible mechanism of ligation is homologous recombination, which could occur through a homologous sequence, such as ori. However, in view of frequent integration of concatenated plasmids, homologous recombination is unlikely to account for this phenomenon. An alternative mechanism is nonhomologous end joining (NHEJ). DNA molecules introduced into cells are prone to double-strand breaks (DSBs), which allow cells to ligate them regardless of whether the two DNA ends are compatible, blunt, or mismatched (19). NHEJ involves many cellular factors, including DNA-dependent protein kinase, XRCC4, DNA ligase IV, and other components (18). NHEJ is relatively efficient and has been shown to recircularize transfected linearized plasmids (11, 17).
We observed that cotransfected reporter genes exhibit enhancer responses that were initially present on only one of the two reporter plasmids. We hypothesized that heteroligation of cointroduced DNA molecules might account for this effect by transferring an enhancer from one molecule to the other.
MATERIALS AND METHODS
Cell culture.
Estrogen receptor (ER)-positive MCF-7 cells (a gift from V. Craig Jordan) were maintained in Dulbecco's modified Eagle's medium-Ham's nutrient mixture F12 1:1 (DMEM-F12) with 8% (vol/vol) heat-inactivated fetal bovine serum (FBS) and antibiotics. During the experiments where 17β-estradiol (E2) was used, MCF-7 cells were cultured in phenol red-free DMEM-F12 containing 8% dextran/charcoal-stripped FBS (HyClone, Logan, Utah).
Vectors.
The pGL3 promoter vector SV-FL carries the firefly luciferase (FL) gene driven by the simian virus 40 (SV40) promoter. The phRL-TK vector TK-RL expresses the synthetic Renilla luciferase (RL) gene under the control of the herpes simplex virus thymidine kinase (HSV-TK) promoter. The phRL-CMV vector CMV-RL contains the RL gene driven by the cytomegalovirus (CMV) enhancer and promoter. These three plasmids were purchased from Promega (Madison, Wis.). The bovine growth hormone poly(A) sequence was obtained from pVAX1 (Invitrogen, Carlsbad, Calif.).
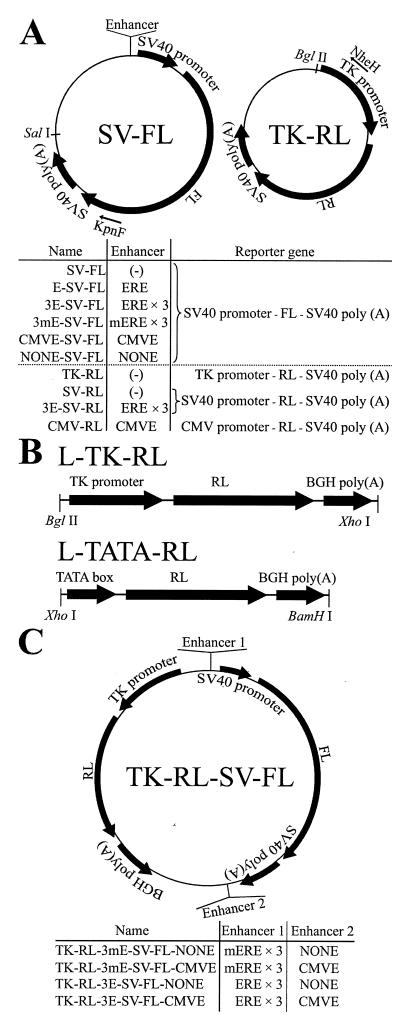
E-SV-FL, 3E-SV-FL (10), 3mE-SV-FL, CMVE-SV-FL, and NONE-SV-FL were constructed by inserting one estrogen response element (ERE) (GTCAGGTCACAGTGACCTGA), three EREs, three mutated EREs (GTCgtcgCACAGTGAtCaGA × 3), CMV enhancer (CMVE) (bases 3 to 657 of pCI-neo [Promega]), and its same-length negative control sequence NONE (bases 2517 to 3171 of pCI-neo) immediately before the SV40 promoter of SV-FL. SV-RL and 3E-SV-RL were constructed by switching the FL sequence of SV-FL and 3E-SV-FL with the RL gene. L-TK-RL and L-TATA-RL are linear reporters expressing RL under the control of the HSV-TK promoter and the minimal TATA box of the HSV-TK promoter starting from CATATTAAGG, respectively. They were excised from amplified plasmids and gel purified before transfection.
TK-RL-3(m)E-SV-FL-CMVE and TK-RL-3(m)E-SV-FL-NONE contain both the FL-expressing segment and the RL-expressing segment, which were inserted counterclockwise into the NotI site and bear (m)ERE × 3 and the CMVE or NONE, which was inserted into the BamHI site located immediately before the SalI site.
AdSV-FL is an adenoviral FL reporter constructed by using the sequence from SV-FL. AdCMV carries the CMVE and the CMV promoter, and AdAAT contains the human α1-antitrypsin gene promoter (−721 to + 44), which is active only in hepatocytes (4). In all adenoviral vectors, the E1a region (bases 393 to 1339) of the wild-type adenovirus type 5 309/356 was deleted. Recombinant adenoviruses were amplified and titrated as previously described (12).
Reporter gene introduction and luciferase assays.
For experiments using (m)ERE-bearing reporters, MCF-7 cells were seeded into 6-cm dishes (6 × 105 cells in 2.5 ml of DMEM-F12w/dcsFBS per dish) the day before transfection. Unless mentioned otherwise, 750 fmol of each reporter was transfected into one dish by incubating cells with either liposome-DNA complexes for 4 h (1) or calcium phosphate (CaP)-DNA precipitates for 6 h. Immediately after transfection, cells were trypsinized, mixed, and split into 24-well plates at a density of 1 × 105 cells per well, such that transfection efficiency would be equal among the wells. Twenty-four hours posttransfection, the medium was replaced with fresh medium containing either vehicle (0.1% [vol/vol] ethanol) or 1 nM E2 in triplicate. Cells were further cultured for 24 h, harvested in 1× Passive Lysis Buffer (Promega) and measured for FL activity as previously described (5). RL activity was measured using the Renilla luciferase assay kit (Promega), and protein concentration was determined by using the protein assay solution (Bio-Rad, Hercules, Calif.). FL and RL activities were normalized based on protein concentration. UV irradiation of plasmids was performed by placing plasmid droplets on a UV transilluminator (Foto/UV300; FOTODYNE, Hartland, Wis.) for 10 min.
In experiments using CMVE-SV-FL, DMEM-F12-containing regular FBS was used. MCF-7 cells (6 × 105) were transfected with the indicated amounts of plasmids in triplicate. Transfected cells were equally split into two dishes and cultured for 48 h. One dish was harvested for FL and RL assays, and the other was used for extraction of DNA, from which a genomic DNA segment (α1-antitrypsin gene promoter) (primers: 5′-GCATGCTTGGGAATGAAACT-3′ and 5′-ATTCACTGTCCCAGGTCA-3′ ) and a segment of TK-RL (primers: 5′-GTCCCAGGTCCACTTCGCATATTAAGG-3′ and 5′-GCTTAAGTTCGAGACTGTTGTGTCAGAA-3′) were simultaneously PCR amplified and electrophoresed in order to estimate the amount of transfected TK-RL relative to genomic DNA.
Adenoviral infection was performed with DMEM-F12 supplemented with regular FBS. Forty-eight hours after the first gene introduction (transfection or infection), FL activity was measured and normalized based on protein concentration.
When luciferase activities in vehicle- and E2-treated cells were compared, Student's t test was performed. In the FL assays after adenoviral infection, multivariate analyses were performed by analysis of variance followed by Fisher's protected least significant difference. In each graph, bars show means and standard deviations.
Analysis of NHEJ products in transfected cells.
DNA was extracted from transfected cells by using the DNeasy Tissue kit (QIAGEN, Valencia, Calif.). Isolated DNA was used as a template for PCR, and the NHEJ-mediated ligation was detected with primers KpnF and NheH. PCR products were electrophoresed, and DNA corresponding to approximately 150 to1,000 bp was purified, digested with NheI and KpnI, and ligated into the polylinker region of SV-FL. NHEJ products from 10 independent clones were sequenced, using RVprimer3 (Promega). KOD polymerase (Novagen, Hartland, Wis.) was used for all PCRs.
RESULTS
Evidence for ERE enhancer transfer between plasmids.
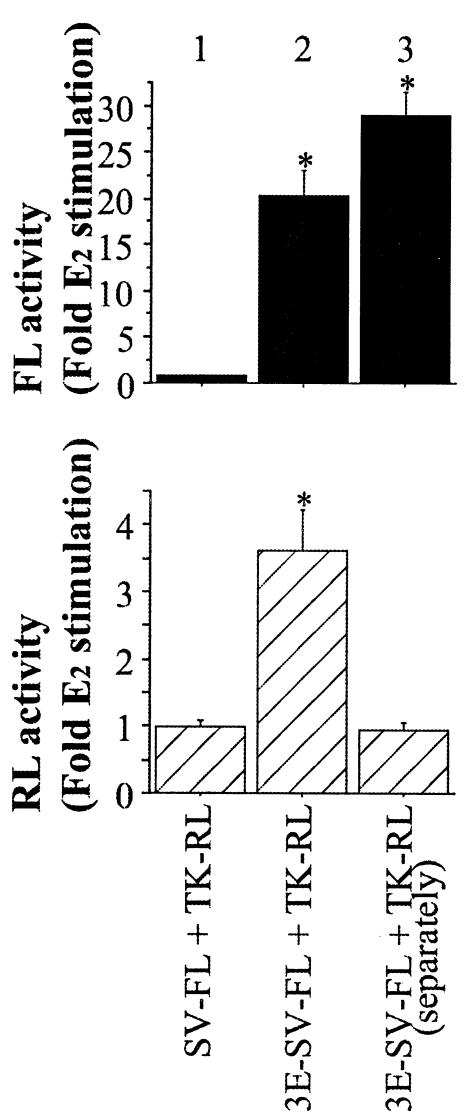
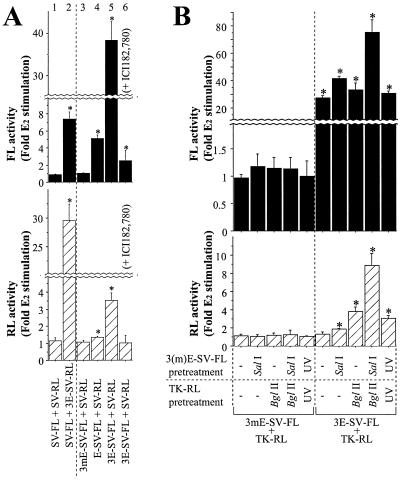
ER-positive MCF-7 cells were transfected with various combinations of plasmids expressing either FL or RL (Fig. 1A). Using this strategy, it was possible to detect potential interactions of regulatory elements of one plasmid with a cotransfected plasmid. When cells were liposome transfected with an ERE-containing promoter (3E-SV-FL) and a non-ERE-containing promoter (TK-RL), 1 nM E2 increased FL expression as anticipated because it contains EREs. Unexpectedly, RL activity also increased even though TK-RL contains no known EREs (Fig. 2, bar 2). This effect was not seen when the 3E-SV-FL and TK-RL plasmids were transfected separately into cells, even if the extracts were mixed and assayed as for the cotransfected cells (Fig. 2, bar 3). Thus, the ERE transfer effect is not caused by a latent ERE within the TK-RL plasmid or by some feature of the luciferase assay. Moreover, a similar ERE transfer phenomenon occurred with the reciprocal plasmid constructs. After cotransfection of an FL reporter without EREs (SV-FL) and an RL reporter with EREs (3E-SV-RL), E2 induced FL as well as RL activities (Fig. 3A, bar 2). E2 induction of RL and FL was dependent on the number of EREs in the cotransfected FL reporter (Fig. 3A, bars 3 to 5) and was inhibited by treatment with 100 nM ICI182,780, an ER antagonist (Fig. 3A, bar 6), confirming that ER-ERE-mediated transcription is responsible for activation of the cotransfected non-ERE-bearing reporter. Thus, the ERE in one plasmid can confer ERE responsiveness to a separate plasmid.
FIG. 1.
Structures of plasmid reporters. (A) SV-FL, TK-RL, and their related reporters. The nomenclature of plasmids containing various enhancers is indicated below each schematic. (B) Linearized RL reporters. (C) Plasmids expressing both FL and RL.
FIG. 2.
Transfer of ERE activity from one plasmid to another. Plasmids were liposome transfected into MCF-7 cells simultaneously (bars 1 and 2) or separately (bar 3). Asterisks denote E2-induced luciferase activities significantly greater (P < 0.05) than that of vehicle-treated cells, the mean of which was defined as 1.0.
FIG. 3.
The ERE enhancer transfer effect requires a functional ERE and ER. (A) Characterization of ERE enhancer transfer. MCF-7 cells liposome transfected with the indicated plasmids were treated with vehicle or E2. (B) Augmentation of the ERE enhancer transfer effect by linearization or by UV irradiation of plasmids. MCF-7 cells were CaP transfected with the plasmids. Asterisks indicate E2-induced luciferase activities significantly greater (P < 0.05) than that of vehicle-treated cells, the mean of which was defined as 1.0.
To test the possibility that the enhancer transfer effect might result from homologous recombination between cotransfected plasmids, we constructed linear reporters sharing no homologous sequences (Fig. 1B). When linearized L-TK-RL or L-TATA-RL was introduced into MCF-7 cells along with 3E-SV-FL, E2 still enhanced RL activity 2- to 2.5-fold (data not shown), demonstrating that homologous sequences are not required for the enhancer transfer effect. No enhancement was seen when a plasmid with mutated EREs (3mE-SV-FL) was used. Moreover, cotransfection of double-stranded ERE oligonucleotides (90 pmol), but not mutated ERE oligonucleotides, resulted in an approximately twofold induction of FL and RL expression in response to E2 in SV-FL-transfected cells and L-TK-RL-transfected cells, respectively (data not shown).
The enhancer transfer effect is dependent on the amount of ERE-containing plasmid in the liposome-mediated transfection mixture. When the amounts of 3E-SV-FL and TK-RL were reduced from 750 to 75 fmol, the E2-induced effect on RL activity decreased whereas the fold induction of FL by E2 was unchanged (data not shown). In contrast to liposome transfection, when CaP was used to cotransfect 3E-SV-FL and TK-RL, the transfer of ERE activity was negligible (Fig. 3B). This difference was not attributable to lower transfection efficiency, as there was minimal ERE transfer even when the amount of 3E-SV-RL plasmid was increased to a level that resulted in greater FL activity than in liposome-transfected cells (data not shown). These results suggest that the enhancer transfer phenomenon depends on the method of transfection as well as the amount of DNA.
Impact of plasmid structure on enhancer transfer.
When CaP was used to transfect linearized plasmids, the enhancer transfer effect was readily seen. As shown in Fig. 3B, SalI-digested 3E-SV-FL and BglII-digested TK-RL generated a stronger enhancer transfer effect than circular plasmids. Pretreatment of plasmids with UV light, which is known to generate DSBs, also enhanced the ERE transfer. After CaP transfection of UV-treated 3E-SV-FL and TK-RL, E2 increased RL activity about threefold (Fig. 3B), although the absolute levels of FL and RL were lower because of partial plasmid degradation. These results indicate that the presence of DSBs is associated with a greater enhancer transfer effect.
Evidence of nonhomologous end joining ligation between cotransfected plasmids.
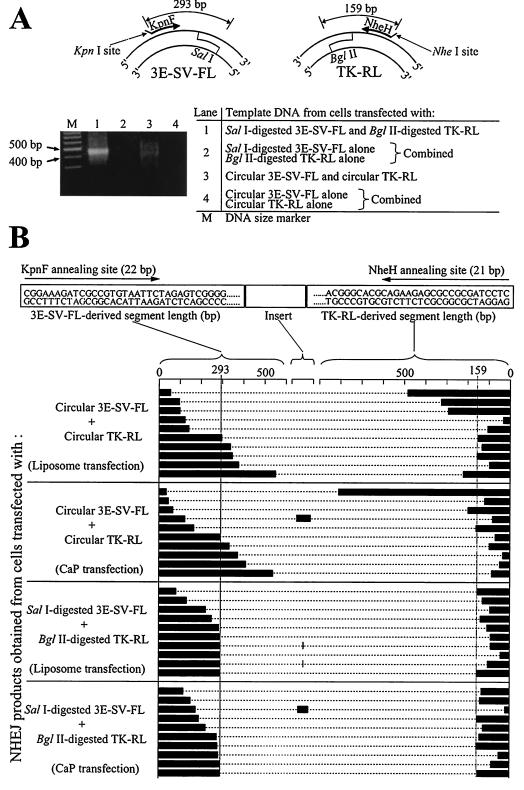
PCR using primers specific for each of two separate plasmids was used to assay directly for NHEJ (Fig. 4A). Using KpnF and NheH primers, PCR was performed on DNA extracted from cells after liposome transfection with circular 3E-SV-FL and TK-RL. This PCR resulted in DNA smears of variable length (Fig. 4A, lane 3). In contrast, PCR of DNA from cells liposome transfected with linearized SalI-digested 3E-SV-FL and BglII-digested TK-RL generated a more uniform product (Fig. 4A, lane 1), consistent with NHEJ between the SalI end of 3E-SV-FL and the BglII end of TK-RL. NHEJ products between two SalI ends of 3E-SV-FL or two BglII ends of TK-RL were not PCR amplified, perhaps because of predicted hairpin secondary structures of their sense and antisense strands. PCR of DNA isolated from cells CaP transfected with circular or linearized 3E-SV-FL and TK-RL produced similar, but less abundant, smears (data not shown). The DNA smears were not seen when cells were separately transfected with either 3E-SV-FL or TK-RL alone and DNA from these cells was combined and used as the PCR template (Fig. 4A, lanes 2 and 4).
FIG. 4.
Evidence of NHEJ-mediated plasmid ligation. (A) PCR amplification of NHEJ products obtained from MCF-7 cells liposome transfected with 3E-SV-FL and TK-RL, using primers KpnF and NheH. DNA used as the PCR template was extracted from cells 24 h posttransfection. (B) Composition of NHEJ products obtained from MCF-7 cells transfected with circular or linearized 3E-SV-FL and TK-RL. For each NHEJ product, the lengths of the 3E-SV-FL-derived segment, the TK-RL-derived segment, and the insert between them (if any) are shown.
DNA was isolated from each reaction mixture, and 10 clones were sequenced to characterize the PCR products. No NHEJ product was obtained from lanes 2 and 4 of Fig. 4A. In the products obtained from cells transfected with SalI-digested 3E-SV-FL and BglII-digested TK-RL, 3E-SV-FL-derived segments and TK-RL-derived segments were no longer than 293 bp (sequence from the KpnF annealing site to the 5′ overhang of the SalI end) and 159 bp (sequence from the NheH annealing site to the 5′ overhang of the BglII end), respectively (Fig. 4B), indicative of variable deletion from the DNA ends during the NHEJ process. On the other hand, the products of ligation between circular 3E-SV-FL and circular TK-RL contained 3E-SV-FL-derived sequences and TK-RL-derived segments which were frequently longer than 293 and 159 bp, respectively (Fig. 4B), indicating that NHEJ occurred between DSBs that were generated randomly in circular plasmids. NHEJ between 3E-SV-FL and TK-RL was also documented in cells CaP transfected with circular 3E-SV-FL and TK-RL even though there was no significant ERE transfer effect, indicating that ligation between the two plasmids occurred after CaP transfection but was not frequent enough to generate a detectable enhancer transfer. The long inserts occasionally found between the 3E-SV-FL-derived and TK-RL-derived segments were all from transfected plasmids and did not include MCF-7 cell genomic DNA. Similar NHEJ products were found from cells transfected with circular or linearized 3mE-SV-FL and TK-RL (data not shown).
Evidence for transfer of CMVE activity between plasmids.
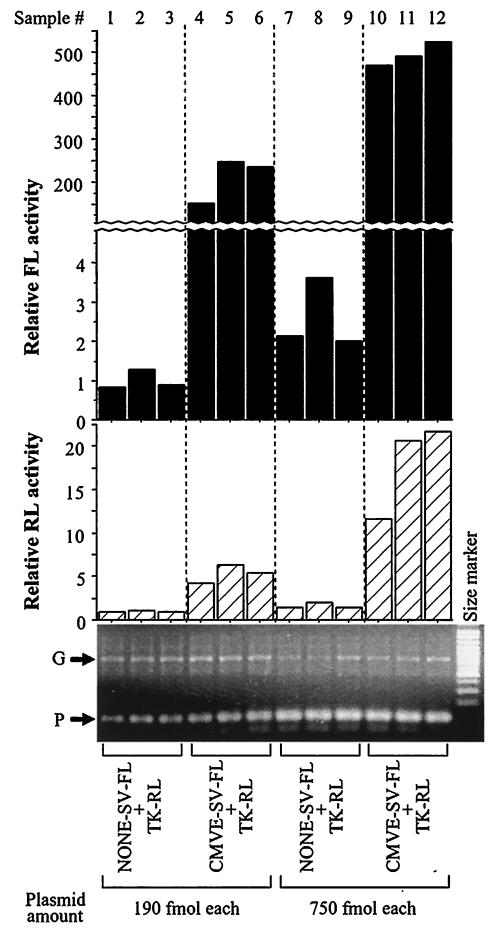
The enhancer transfer effect also occurred with the CMVE, which is commonly used in expression vectors. Both FL and RL activities were higher in MCF-7 cells liposome transfected with CMVE-SV-FL and TK-RL than in cells transfected with an equimolar amount of a non-CMVE-containing control (NONE-SV-FL) and TK-RL. PCR amplification of a genomic DNA sequence and a segment from TK-RL confirmed comparable transfection efficiency (Fig. 5). Similar results were obtained with CHO-K1, HeLa, BHK-21, and HEK293 cells, although the degree of the enhancer transfer effect varied among cell lines and by transfection technique (liposome > CaP) (data not shown). Thus, the enhancer transfer effect is seen with either an ERE or with the CMVE, indicating that this phenomenon occurs with different types of enhancers.
FIG. 5.
Transfer of CMVE activity between plasmids. MCF-7 cells liposome transfected in triplicate were split in two and harvested 48 h later for either luciferase assays or extraction of DNA. The DNA was used as a template for simultaneous PCR amplification of a genomic DNA sequence (G) or a TK-RL segment (P). NONE-SV-FL is a same-length negative control for CMVE-SV-FL. Results are given as relative luciferase activities, where the mean value for lanes 1 to 3 is equal to 1.0.
Enhancer transfer after adenoviral infection.
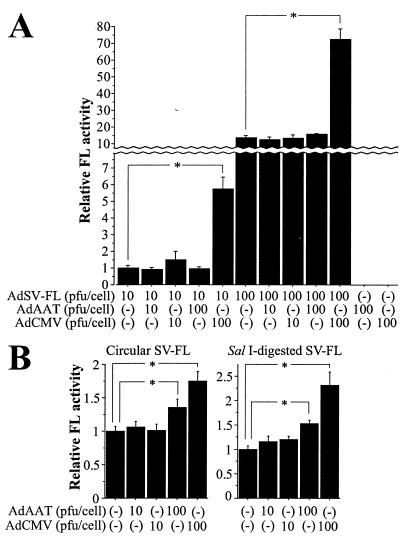
We considered the possibility that similar enhancer transfer effects might occur after infection with adenoviral vectors, whose genome consists of double-stranded linear DNA. Adenoviral vectors were constructed with (AdCMV) or without (AdAAT) the CMVE. In MCF-7 cells infected with AdSV-FL, coinfection of AdCMV, but not AdAAT, increased FL expression dose dependently, indicating transfer of CMVE activity (Fig. 6A). In cells liposome transfected with circular or linearized SV-FL, infection with either AdAAT or AdCMV stimulated FL activity, suggesting that endogenous adenoviral enhancers (7, 9) also exert enhancer transfer effects (Fig. 6B).
FIG. 6.
Enhancer transfer by adenoviral DNA. (A) CMVE transfer between adenoviral vectors. MCF-7 cells were incubated with viruses for 4 h in 24-well plates in triplicate. Forty-eight hours later, cells were harvested and FL activity was measured; the activity was normalized based on protein concentration. Asterisks indicate a statistically significant (P < 0.05) difference. (B) Effects of adenoviral DNA on reporter plasmid activity. Immediately after liposome transfection of either circular or linearized SV-FL (5.6 pmol) into MCF-7 cells (4.5 × 106), the cells were equally split into 45 wells, followed by overnight adenovirus infection in triplicate. Normalized FL activity was measured 48 h posttransfection. Asterisks denote a significant (P < 0.05) difference.
Similar to transfected plasmid DNA, NHEJ products were generated between SV-FL and the end of AdCMV in MCF-7 cells transfected with circular or linearized SV-FL and infected with AdCMV (data not shown). PCR was performed using KpnF and a primer that annealed to both the left-hand and right-hand inverted terminal repeats of adenoviruses. When DNA extracted from cells infected with AdCMV alone was combined with DNA isolated from cells transfected only with SV-FL, no such NHEJ PCR products were obtained. NHEJ products between two adenoviral DNA ends were undetectable, probably due to inefficient PCR amplification of their hairpin-like secondary structures. These results indicate that adenoviral DNA can also become ligated to cointroduced DNA and exert enhancer transfer effects.
Evidence for functional interactions between transferred enhancers.
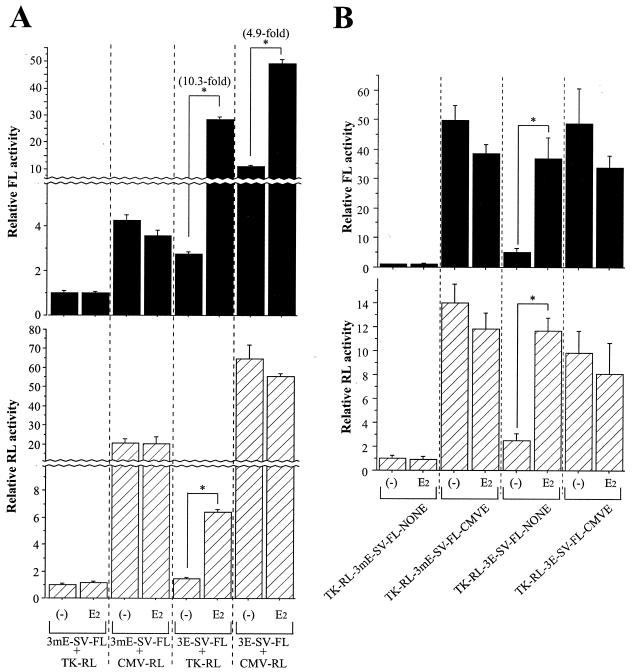
In the absence of E2, MCF-7 cells liposome transfected with 3E-SV-FL and CMV-RL exhibited higher FL and RL activities than cells transfected with 3E-SV-FL and TK-RL, compatible with the transfer of the CMVE to 3E-SV-FL by CMV-RL (Fig. 7A). However, the fold induction of FL activity by E2 was consistently lower (4.9-fold) in cells transfected with a plasmid combination containing the CMVE (3E-SV-FL and CMV-RL) compared to cells (10.3-fold) transfected with a plasmid combination lacking the CMVE (3E-SV-FL and TK-RL). These results suggest that an ERE-containing reporter might be less responsive to E2 after being ligated to the CMVE by the NHEJ mechanism. Therefore, we tested E2 responsiveness of reporter plasmids constructed to carry both the ERE (or mutant ERE) and the CMVE (or NONE) (Fig. 1C). After CaP transfection of TK-RL-3E-SV-FL-CMVE into MCF-7 cells, E2 induced neither FL nor RL activity despite the presence of the canonical EREs (Fig. 7B), indicating that when the CMVE and the ERE are present on the same DNA molecule, the strong enhancer properties of the CMVE blunts E2-ER-ERE-mediated activation of either the SV40 or the HSV-TK promoter. Because of this masking phenomenon, the presence of the CMVE on any of the cotransfected plasmids has the potential to dampen ERE responsiveness.
FIG. 7.
Transfer of the CMVE reduces E2-induced ERE activity. (A) ERE-mediated transcription by E2 is reduced by cotransfected CMVE-containing plasmids. MCF-7 cells were liposome transfected with 3E-SV-FL and CMV-RL and compared with those transfected with 3E-SV-FL and TK-RL. Asterisks indicate significantly greater luciferase activities (P < 0.05) in E2-treated cells than in vehicle-treated cells. The mean activity of reactions for the first bars is defined as 1.0. (B) E2-ER-ERE-mediated transcription is impaired by locating the CMVE on the same DNA molecule. MCF-7 cells were CaP transfected with each plasmid. Asterisks denote significantly greater luciferase activities (P < 0.05) in E2-treated cells than in vehicle-treated cells. The mean activity of reactions for the first bars is defined as 1.0.
DISCUSSION
We have shown that an enhancer on one DNA molecule can affect the transcriptional activity of another molecule cointroduced into cells. Although this effect initially appears to reflect an action in trans, we found it is actually a cis action of enhancer transfer after NHEJ-mediated ligation. This phenomenon is clearly different from “squelching” (15), where cotransfected expression vectors compete for limited amounts of transcription factors or transcriptional coactivators.
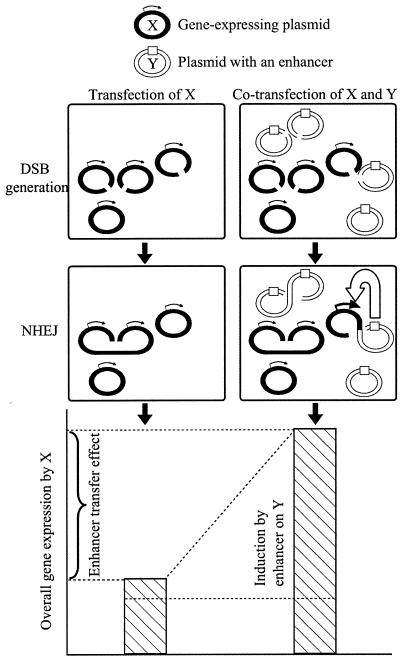
Our findings that enhancer activity can be transferred from one plasmid to another have important implications for the design and interpretation of transfection experiments that contain more than one plasmid. Since NHEJ has been documented in many cell lines, enhancer transfer effects are likely to occur, depending on the frequency of ligation and the strength of the enhancer (Fig. 8). Even if NHEJ does not complete covalent ligation, the formation of protein bridges between DNA molecules may be sufficient for an enhancer to act on another molecule (14).
FIG. 8.
Proposed mechanism for the enhancer transfer effect. A portion of plasmid X molecules become ligated to cointroduced Y molecules by NHEJ, leading to enhanced gene expression. The degree of the enhancer transfer effect depends on the frequency of heteroligation between X and Y and the strength of the enhancer on Y.
An internal control reporter is commonly used to correct for transfection efficiency in reporter gene assays. Our study demonstrates that introducing an additional plasmid creates at least two sources of error. First, enhancer sequences within the internal control reporter might be transferred to an experimental plasmid. Second, enhancer sequences within the experimental plasmid might be transferred to the internal control, altering its level of expression and leading to incorrect corrections of transfection efficiency. Based on our results, the degree of error introduced is likely to be affected by the method of transfection, plasmid concentration, extent of DSBs, and the strength of the enhancers. In view of these findings, the use of an internal control plasmid should be avoided whenever possible. Experimental replicates can be used to assess variation in transfection efficiency, particularly when comparisons are made within the same cell line. The protein concentration of the cell lysate can also be used to correct for variability in cell number or protein extraction. In principle, it is also possible to directly determine the amount of transfected DNA by using PCR (Fig. 5). Although this is not practical when analyzing large numbers of samples, it might be used to assess transfection efficiency in different cell lines or when using different transfection methods. Our results also offer strategies to minimize enhancer transfer. If a small amount of plasmid DNA is cotransfected, enhancer transfer can be reduced and may be negligible. For example, when 3E-SV-FL was liposome transfected into MCF-7 cells along with 1/20 the molar amount of CMV-RL, the CMVE transfer effect on basal FL activity was not detected and the E2-induced fold induction of FL activity was comparable to that seen in cells transfected with 3E-SV-FL alone (data not shown). Because DSBs are necessary for NHEJ, high-quality circular plasmids are less likely to undergo enhancer transfer than linear or partially degraded DNA.
Another common situation in which enhancer transfer may be problematic is when separate plasmids are used to express various transcription factors in combination with reporter genes. In this circumstance, a strong enhancer such as the CMVE may perturb the activity of the promoter driving the reporter gene. Importantly, an enhancer such as the CMVE can either stimulate or mask other DNA regulatory elements, depending on the context of the promoter it joins. When the CMVE is introduced in combination with a relatively weak minimal promoter (e.g., TK or SV40), it increases basal expression. On the other hand, the CMVE dampens E2 stimulation of the ERE. Thus, it is difficult to predict the consequences of enhancer transfer between plasmids, particularly when complex native promoter sequences are used. To some extent, the effects of cotransfecting plasmids with strong enhancers are recognized and have led to the common practice of using equivalent amounts of an empty expression vector or a vector expressing an irrelevant protein. In addition to the measures described above to minimize NHEJ, it is preferable to use a control plasmid with the same backbone, ideally expressing a mutant form of the transcription factor.
Apart from transfection experiments, our observations demonstrate that DNA introduced by adenoviruses can also undergo ligation by NHEJ. Thus, native adenoviral enhancers, or enhancers present in exogenous genes carried by the virus, can be transferred to other reporters. In addition, NHEJ might affect the structure of adenoviral DNA during viral propagation as well as after infection.
In summary, we have demonstrated that cointroduced DNA molecules can be ligated to one another by NHEJ, thereby allowing an enhancer on one molecule to act on another. These findings have important implications for the interpretation of cotransfection experiments, as enhancers on expression vectors or within viral DNA can alter the expression of reporter genes or other expression vectors.
Acknowledgments
We thank Alan Wakeling for providing ICI182,780, Wen-Xia Gu and Tom Kotlar for determining nucleotide sequences, Susan Park and Christine Glidewell-Kenney for critically reading the manuscript, and Tetsuya Igarashi for information on the ERE transfer effect.
This work was supported in part by NIH grants P50 CA 89018 and PO1 HD 21921.
REFERENCES
- 1.Campbell, M. J. 1995. Lipofection reagents prepared by a simple ethanol injection technique. BioTechniques 18:1027-1032. [PubMed] [Google Scholar]
- 2.Chen, C., and L. A. Chasin. 1998. Cointegration of DNA molecules introduced into mammalian cells by electroporation. Somat. Cell Mol. Genet. 24:249-256. [DOI] [PubMed] [Google Scholar]
- 3.Chen, J. L., K. L. Huisinga, M. M. Viering, S. A. Ou, C. T. Wu, and P. K. Geyer. 2002. Enhancer action in trans is permitted throughout the Drosophila genome. Proc. Natl. Acad. Sci. USA 99:3723-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciliberto, G., L. Dente, and R. Cortese. 1985. Cell-specific expression of a transfected human α1-antitrypsin gene. Cell 41:531-540. [DOI] [PubMed] [Google Scholar]
- 5.Duan, W. R., J. L. Shin, and J. L. Jameson. 1999. Estradiol suppresses phosphorylation of cyclic adenosine 3′,5′-monophosphate response element binding protein (CREB) in the pituitary: evidence for indirect action via gonadotropin-releasing hormone. Mol. Endocrinol. 13:1338-1352. [DOI] [PubMed] [Google Scholar]
- 6.Dunaway, M., and P. Dröge. 1989. Transactivation of the Xenopus rRNA gene promoter by its enhancer. Nature 341:657-659. [DOI] [PubMed] [Google Scholar]
- 7.Hearing, P., and T. Shenk. 1983. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell 33:695-703. [DOI] [PubMed] [Google Scholar]
- 8.Hoglund, M., T. Siden, and D. Rohme. 1992. Different pathways for chromosomal integration of transfected circular pSVneo plasmids in normal and established rodent cells. Gene 116:215-222. [DOI] [PubMed] [Google Scholar]
- 9.Imperiale, M. J., R. P. Hart, and J. R. Nevins. 1985. An enhancer-like element in the adenovirus E2 promoter contains sequences essential for uninduced and E1A-induced transcription. Proc. Natl. Acad. Sci. USA 82:381-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa, T., K. Takano, J. Yasufuku-Takano, T. Fujita, T. Igarashi, M. Miura, and K. Hata. 2001. Estrogenic impurities in labware. Nat. Biotechnol. 19:812. [DOI] [PubMed] [Google Scholar]
- 11.Kabotyanski, E. B., L. Gomelsky, J.-O. Han, T. D. Stamato, and D. B. Roth. 1998. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 26:5333-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, E. J., L. M. Anderson, B. Thimmapaya, and J. L. Jameson. 1999. Targeted expression of toxic genes directed by pituitary hormone promoters: a potential strategy for adenovirus-mediated gene therapy of pituitary tumors. Endocrinology 84:786-794. [DOI] [PubMed] [Google Scholar]
- 13.Mahmoudi, T., K. R. Katsani, and C. P. Verrijzer. 2002. GAGA can mediate enhancer function in trans by linking two separate DNA molecules. EMBO J. 21:1775-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller, H. P., J. M. Sogo, and W. Schaffner. 1989. An enhancer stimulates transcription in trans when attached to the promoter via a protein bridge. Cell 58:767-777. [DOI] [PubMed] [Google Scholar]
- 15.Natesan, S., V. M. Rivera, E. Molinari, and M. Gilman. 1997. Transcriptional squelching re-examined. Nature 390:349-350. [DOI] [PubMed] [Google Scholar]
- 16.Simone, P. D., A. D. Leonardo, G. Costanzo, R. Melfi, and G. Spinelli. 2001. The sea urchin sns insulator blocks CMV enhancer following integration in human cells. Biochem. Biophys. Res. Commun. 284:987-992. [DOI] [PubMed] [Google Scholar]
- 17.Smith, J., E. Riballo, B. Kysela, C. Baldeyron, K. Manolis, C. Masson, M. R. Lieber, D. Papadopoulo, and P. Jeggo. 2003. Impact of DNA ligase IV on the fidelty of end joining in human cells. Nucleic Acids Res. 31:2157-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valerie, K., and L. F. Povirk. 2003. Regulation and mechanism of mammalian double-strand break repair. Oncogene 22:5792-5812. [DOI] [PubMed] [Google Scholar]
- 19.Wake, C. T., T. Gudewicz, T. Porter, A. White, and J. H. Wilson. 1984. How damaged is the biologically active subpopulation of transfected DNA? Mol. Cell. Biol. 4:387-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wedel, A., D. S. Weiss, D. Popham, P. Dröge, and S. Kustu. 1990. A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science 248:486-490. [DOI] [PubMed] [Google Scholar]