Abstract
Peroxisome proliferator-activated receptors (PPAR) are ligand-activated transcription factors that form a subfamily of the nuclear receptor gene family. Since both flow and PPARγ have atheroprotective effects and extracellular signal-regulated kinase 5 (ERK5) kinase activity is significantly increased by flow, we investigated whether ERK5 kinase regulates PPARγ activity. We found that activation of ERK5 induced PPARγ1 activation in endothelial cells (ECs). However, we could not detect PPARγ phosphorylation by incubation with activated ERK5 in vitro, in contrast to ERK1/2 and JNK, suggesting a role for ERK5 as a scaffold. Endogenous PPARγ1 was coimmunoprecipitated with endogenous ERK5 in ECs. By mammalian two-hybrid analysis, we found that PPARγ1 associated with ERK5a at the hinge-helix 1 region of PPARγ1. Expressing a hinge-helix 1 region PPARγ1 fragment disrupted the ERK5a-PPARγ1 interaction, suggesting a critical role for hinge-helix 1 region of PPARγ in the ERK5-PPARγ interaction. Flow increased ERK5 and PPARγ1 activation, and the hinge-helix 1 region of the PPARγ1 fragment and dominant negative MEK5β significantly reduced flow-induced PPARγ activation. The dominant negative MEK5β also prevented flow-mediated inhibition of tumor necrosis factor alpha-mediated NF-κB activation and adhesion molecule expression, including vascular cellular adhesion molecule 1 and E-selectin, indicating a physiological role for ERK5 and PPARγ activation in flow-mediated antiinflammatory effects. We also found that ERK5 kinase activation was required, likely by inducing a conformational change in the NH2-terminal region of ERK5 that prevented association of ERK5 and PPARγ1. Furthermore, association of ERK5a and PPARγ1 disrupted the interaction of SMRT and PPARγ1, thereby inducing PPARγ activation. These data suggest that ERK5 mediates flow- and ligand-induced PPARγ activation via the interaction of ERK5 with the hinge-helix 1 region of PPARγ.
Peroxisome proliferator-activated receptors (PPAR) are ligand-activated transcription factors that form a subfamily of the nuclear receptor gene family. Among PPAR family members, the expression of PPARα and PPARγ has been reported in endothelial cells (ECs). Recently, Pasceri et al. reported that PPARγ activators inhibit expression of vascular cellular adhesion molecule 1 (VCAM-1) and intercellular adhesion molecule 1 (ICAM-1) in activated ECs and significantly reduce monocyte/macrophage homing to atherosclerotic plaques (23). Mitogen-activated protein (MAP) kinase signaling pathways have been shown to phosphorylate PPARγ and to decrease PPARγ transcriptional activity (7, 13). The NH2-terminal domain of PPARγ contains a consensus MAP kinase site in a region conserved between PPARγ1 and PPARγ2 isoforms (7, 13). Phosphorylation of PPARγ2 Ser112 (13) and PPARγ1 Ser82 (7) significantly inhibits both ligand-independent and ligand-dependent transcriptional activation by PPARγ. Phosphorylation-mediated transcriptional repression is due to a diminished ability of PPARγ to become transcriptionally activated by ligand rather than to a reduced capacity of the PPARγ-retinoid X receptor complex to heterodimerize its DNA binding site (7).
ERK5/BMK1 is a member of the MAP kinase family which is activated by redox and hyperosmotic stress, growth factors, and pathways involving certain G-protein-coupled receptors (12). Extracellular signal-regulated kinase 5 (ERK5) has a TEY sequence in its dual phosphorylation site, like ERK1/2, but it has unique carboxyl-terminal and loop-12 domains, suggesting that its regulation and function may be different from that of ERK1/2. The upstream kinase that phosphorylates ERK5 has been identified as MEK5 (17, 39). Like many MAP kinase family members, ERK5 plays a significant role in cell growth and differentiation, although emerging evidence suggests unique functional characteristics. Redox activation of ERK5 is documented to have an antiapoptotic effect (30), and ERK5 knockout mice have impaired cardiac and vascular development (28). It was reported that ERK5 regulates MEF2A, MEF2C, and MEF2D transcriptional activity (1, 16), but there are no reports on the regulation of nuclear receptors by ERK5.
Since both flow and PPARγ have atheroprotective effects and ERK5 kinase activity is significantly increased by flow, we investigated whether ERK5 kinase regulates PPARγ activity. In the present study, we show that activation of ERK5 induces PPARγ activation in ECs. PPARγ1 activation was induced by the association of activated ERK5a with the hinge-helix 1 region of PPARγ1 in a phosphorylation-independent manner, suggesting a role for ERK5 as a scaffold. ERK5 kinase activation was critical to reduce the inhibitory effect of the NH2-terminal region of ERK5 on the association of ERK5 and PPARγ (see Fig. 9, below). Thus, activation of ERK5 is a positive regulator for PPARγ1 activation via the interaction of the hinge-helix 1 domain of PPARγ1 and ERK5a.
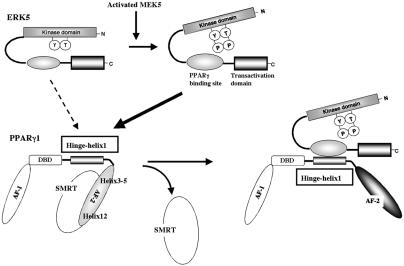
FIG. 9.
Model of the ERK5a-PPARγ1 interaction activating PPARγ1 activity. The position of H12 is regulated by a ligand. In the ligand binding receptor, H12 folds back to form part of the coactivator binding surface. By contrast, H12 inhibits corepressor binding to PPARγ and other nuclear receptors (29). The corepressor interaction surface requires H3, H4, and H5, thereby overlapping the coactivator interaction surface (14). In the present study we found a critical role for the hinge-helix 1 domain in regulating PPARγ1 transcriptional activity. The inactive NH2-terminal kinase domain of ERK5a partially inhibits the association of PPARγ1 on COOH-terminal ERK5 and also inhibits its transcriptional activity. Following activation, the inhibitory effect of NH2-terminal ERK5 decreases, and the middle region of ERK5a fully interacts with the hinge-helix 1 region of PPARγ1. The association of ERK5a with the hinge-helix 1 region of PPARγ1 releases corepressor SMRT and induces full activation of PPARγ1.
MATERIALS AND METHODS
Cell culture and transfection.
Human umbilical vein endothelial cells (HUVECs) were purchased from Cascade Biologics (Portland, Oreg.) and maintained in 2% low-serum growth supplement (Cascade Biologics) as described in the manufacturer's protocol. Cos7 and CHO cells were maintained in Dulbecco's modified Eagle's medium and Ham's F-12 medium (Invitrogen), respectively, and were supplemented with 10% fetal bovine serum as described previously. For transient-expression experiments, HUVECs and Cos7 cells were transfected with Lipofectamine Plus (Invitrogen) as described previously (37). It has been reported that flow decreases the expression of PPARγ in human-endothelial-cell-like (ECV304) cells (4). We used primarily cultured HUVECs in the present study. ECV304 is a cell line and has very different characteristics compared with our primary cell-cultured ECs, especially in response to flow. Very recently, another lab also reported that flow could induce PPARγ activation in HUVECs (Y. Liu, F. Rannou, K. Formentin, L. Zeng, T.-S. Lee, Y. Zhu, and J. Y. Shyy, abstr., Circulation 108:IV-133, 2003). Therefore, the difference between our data and previous data may be due to the nature of the different cell lines. BLMECs are bovine lung microvascular endothelial cells. These cells have an endothelial morphology similar to that of bovine aortic endothelial cells and share similar signaling transduction pathways, such as vascular endothelial growth factor, tumor necrosis factor (TNF), and platelet-derived growth factor. BLMECs at passage 3 to 8 were grown in MCDB-131 medium supplemented with 10% fetal bovine serum, heparin (15,300 U/liter; Sigma), hydrocortisone (2.76 μmol/liter; Sigma), bovine pituitary extract, epidermal growth factor (1.64 nmol/liter; Sigma), l-glutamine, and antibiotics (100 U of penicillin/ml and 68.6 mol of streptomycin/liter) in flasks precoated with 2% gelatin. For transient-expression experiments, cells were transfected with plasmids 1 day after plating.
For short-term flow experiments, HUVECs were plated on 60-mm dishes and cultured in Medium 200. The next day, the cells were transfected using the Lipofectamine Plus reagent method (Invitrogen). Transfection medium contained 2 μg of PPRE reporter plasmid, 1 μg of pSG5-PPARγ, and vector to provide equal amounts of transfected DNA with or without plasmid expressing the VP16-PPARγ1 amino acid 195 to 227 (aa 195-227) fragment. To control for variations in cell number and transfection efficiency, 20 ng of PRL-TK was cotransfected with a luciferase control reporter vector. After 24 h of transfection, the cells were stimulated with ciglitazone (5 μM). Three hours later, the cells were exposed for 20 min to flow (12 dynes/cm2) or no flow in flow buffer (Hank's balanced salt solution containing 5.5 mM glucose and 1.3 mM CaCl2) as described previously (34). After 16 h of ciglitazone stimulation, luciferase PPARγ transcriptional activity was assayed using a dual-luciferase reporter assay system (Promega), and luciferase luminescence was counted in a Luminometer (TD-20/20; Turner Design) and then normalized to cotransfected luciferase activity. For chronic flow experiments, a Large React angular parallel flow chamber (5 by 14 cm; Glyco Tech) was used for transfections and stimulated the cells by a constant flow. HUVECs or BLMECs were plated in the four wells (1.9 by 6.9 cm) of the chamber in full-serum medium. For the PPARγ1 reporter gene assay, the cells were transfected with 1 μg of (PPRE)3-tk-luc plasmid, 0.5 μg of pSG5-PPARγ1, 20 ng of PRL-TK with or without 1 μg of pACT-PPARγ1(aa 195-227) in OPTI-MEM, using Plus reagent and Lipofectamine. The total transfected DNA amount was normalized with no-insert vector plasmid DNA. For the Gal4-ERK5a reporter gene assay, the cells were transfected with 1 μg of pG5-luc plasmid and 0.5 μg of pBIND-ERK5a. After 24 h of transfection, flow was applied for the HUVECs at 5 dynes/cm2 using an Econo pump (model EP-1; Bio-Rad) for 6 to ∼9 h and then the cells were harvested for the reporter gene assay.
Plasmid construction.
pCMV-DN-SMRT was a kind gift from M. L. Privalsky (36). Mouse ERK5a, ERK5b, and the constitutively active form of MEK5α (CA-MEK5α) were cloned as described previously (33). Gal4-SRC-1 was constructed by inserting an EcoRV-EcoRV fragment, generated by PCR, into the pBIND vector (Promega). Gal4-SMRT was constructed by inserting SalI (blunt)-MulI (blunt) fragments, generated by PCR, into the pBIND vector. CA-MEKK1 was purchased from Stratagene. Gal4-PPARγ1, various deletions of Gal4-PPARγ1, and Gal4-ERK5a were created by cloning PCR-amplified DNA fragments corresponding to the different mouse PPARγ1 or ERK5a regions into the SalI and NotI sites of the pBIND vector. VP16-PPARγ1 and various deletions of VP16-ERK5a were created by cloning PCR-amplified DNA fragments corresponding to the different PPARγ or ERK5a regions into the SalI and NotI sites of the pACT vector (Promega). Gal4-ERK5a and VP16-ERK5a were created by inserting the mouse ERK5a isolated from pcDNA3.1-ERK5a into BamH1 and Not1 sites of the pBIND and pACT vectors, respectively.
Glutathione S-transferase (GST)-PPARγ1-truncated mutations (GST-PPARγ1-activation function 1 [AF-1] [aa 1-110], GST-PPARγ1-DNA binding domain [DBD] [aa 109-175], GST-PPARγ1-ligand binding domain [LBD] [aa 163-475]) were created by cloning PCR-amplified DNA fragments corresponding to the different PPARγ1 regions into the EcoRI and XhoI sites of the pGEX-KG vector (Amersham). The single or double mutations of PPARγ1 and ERK5a were created with the QuikChange site-directed mutagenesis kit (Stratagene). All constructs were verified by DNA sequencing.
In vitro phosphorylation of PPARγ1 by activated ERK5.
GST-PPARγ1-truncated mutant proteins were expressed in Escherichia coli and purified using glutathione-Sepharose 4B as described by the manufacturer (Pharmacia Biotech Inc.).
ERK5 activity was measured as previously described (2, 16). To determine whether PPARγ1 can be phosphorylated by activated ERK5, we performed an ERK5 in vitro kinase assay with GST-PPARγ1-AF-1, GST-PPARγ1-DBD, and GST-PPARγ1-LBD as the substrates.
Relative quantitative RT-PCR.
Total RNA isolation, first-strand cDNA synthesis, and relative quantitative reverse transcription-PCR (RT-PCR) using Ambion's Competimer technology were performed as described previously (3). Ambion's Competimer technology allowed us to modulate the amplification of 18S rRNA in the same linear range as the RNAs under study when amplified under the same conditions. The following primers were used for PCR analysis: VCAM-1, 5′-GAGCCTCAGATGTACTTTGGATGG-3′ (sense) and 5′-TAGAGAAAGAGTAGATCTCC-ACTCGG-3′ (antisense); E-selectin, 5′-TCTCACTTTTGTGCTTCTCC-3′ (sense) and 5′-TGGAGCCCAGTTTGTGGCT-3′ (antisense).
Mammalian one- or two-hybrid analysis.
HUVECs and Cos7 cells were plated in 12-well dishes at 2 × 105 cells/well and 24 h later transfected in Opti-MEN (Invitrogen) with the pG5-luc vector and various pBIND and pACT plasmids (Promega). The pG5-luc vector contains five Gal4 binding sites upstream of a minimal TATA box which, in turn, is upstream of the firefly luciferase gene. pBIND and pACT contain Gal4 and VP16, respectively, and were fused with PPARγ1, ERK5, silencing mediator of retinoid and thyroid hormone action (SMRT), or SRC-1 as indicated. Since pBIND also contains the Renilla luciferase gene, the expression and transfection efficiencies were normalized with the Renilla luciferase activity. Cells were collected 40 h after transfection except as indicated, and the luciferase activity was determined. Luciferase activity was assayed with a luciferase kit (Promega). Transfections were performed in triplicate, and each experiment was repeated at least two times.
Immunoprecipitation and Western blot analysis.
The cells were washed with phosphate-buffered saline and harvested in 0.5 ml of lysis buffer as described previously (37). Immunoprecipitation was performed as described previously with anti-ERK5 antibody (1) or anti-PPARγ antibody (Santa Cruz). Western blot analysis was performed as previously described (37). In brief, the blots were incubated for 4 h at room temperature with the anti-ERK5 (1), SMRT (Santa Cruz), VCAM-1 (Chemicon), or Xpress (Invitrogen) antibody, followed by incubation with horseradish peroxidase-conjugated secondary antibody (Amersham). Immunoreactive bands were visualized using enhanced chemiluminescence (Amersham).
Materials.
Ciglitazone (GR-205) and 15-deoxy-Δ12,14-prostaglandin J2 (PG-050) were from BIOMOL.
Statistical analysis.
Data are reported as means ± standard deviations (SD). Statistical analysis was performed with the StatView 4.0 package (ABACUS Concepts, Berkeley, Calif.). Differences were analyzed with a one-way or a two-way repeated-measures analysis of variance as appropriate, followed by Schéffe's correction.
RESULTS
MEK5-ERK5 enhanced PPARγ1 transcriptional activity in ECs.
To test the hypothesis that ERK5 regulates PPARγ1 transcriptional activity, we examined the effect of CA-MEK5α and ERK5 on PPARγ1 transcriptional activity. We coexpressed PPARγ1, CA-MEK5α, and ERK5 in HUVECs and examined PPARγ1-mediated transcriptional activity, as assayed by a luciferase reporter gene driven by three copies of a PPAR response element (PPRE) linked to a thymidine kinase (tk) promoter.
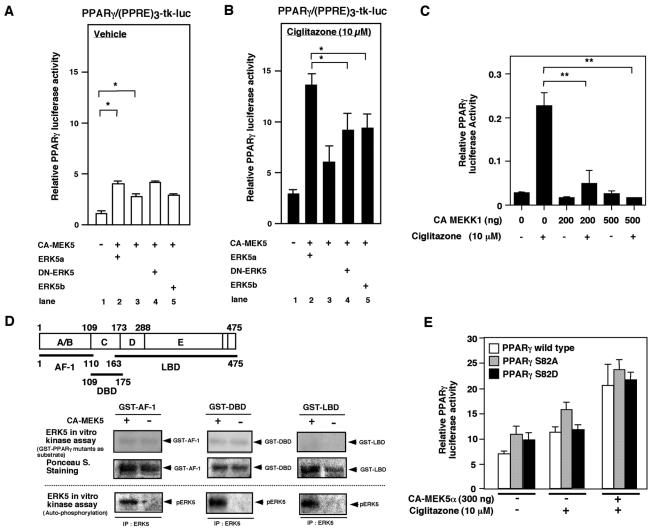
As shown in Fig. 1A, addition of CA-MEK5α significantly increased full-length PPARγ1 transcriptional activity (1.24 ± 0.34 versus 2.86 ± 0.31; P < 0.05) (lane 1 versus lane 3). Interestingly, cotransfection of CA-MEK5α and wild-type ERK5 (ERK5a) significantly enhanced PPARγ transcriptional activity to a greater extent than CA-MEK5α transfection alone (2.86 ± 0.31 versus 4.04 ± 0.34; P < 0.05) (lane 3 versus lane 2). We also found that CA-MEK5α expression enhanced transcriptional activity of a PPARγ1 ligand binding domain-truncated mutant (aa 162 to 475) in a ligand-dependent manner, suggesting that the NH2 terminal of the PPARγ1 region is likely not involved in the MEK5-ERK5-mediated effect on PPARγ1 activity (data not shown). PPARγ expression levels were not significantly different among the samples based on Western blot analyses (data not shown).
FIG. 1.
MEK5-ERK5 activation increases PPARγ1-mediated transactivation of the (PPRE)3-tk-luciferase reporter construct in HUVECs, which is independent of PPARγ1 S82 phosphorylation. (A and B) MEK5-ERK5 activation induced PPARγ1 transcriptional activity, but DN-ERK5 did not inhibit CA-MEK5α-mediated PPARγ activity. PPARγ1 transcriptional activity was measured by transfection of full-length PPARγ1 and the (PPRE)3-tk-luciferase reporter construct in HUVECs. PPARγ-mediated transactivation was determined with the transfection of wild-type ERK5 (ERK5a, lane2), empty vector (lane 3), or ERK5 mutants (DN-ERK5 [lane 4] or ERK5b [lane5]) with vehicle (A) or 10 μM ciglitazone (B). Results are the mean ± SD of three to six independent experiments. Luciferase activity of the (PPRE)3-tk-luc construct with CA-MEK5α and ERK5a in the absence of transfected PPARγ at ciglitazone concentrations of 0 and 10 μM were 0.8 ± 0.2 and 1.1 ± 0.2 (relative PPARγ luciferase activity), respectively. Luciferase activity of the TK promoter alone with CA-MEK5α and ERK5a was below 0.1 relative PPARγ luciferase activity. (C) Activation of MEKK1 inhibited PPARγ activation. CA-MEKK1, as indicated, was transfected in Cos7 cells, and pcDNA3.1 vector was used to provide equal amounts of transfected DNA. Results are the mean ± SD of three independent experiments. (D) ERK5 did not phosphorylate PPARγ1 in an in vitro kinase assay. CHO cells were transfected with vector or CA-MEK5α, and ERK5 was immunoprecipitated with ERK5 antibody. An immune complex kinase assay was then performed with GST or GST-PPARγ1 mutants (GST-PPARγ1-AF-1, GST-PPARγ1-DBD, and GST-PPARγ1-LBD). (E) CA-MEK5α- and/or ciglitazone-induced full-length PPARγ1 wild type or mutants (PPARγ1S82A or PPARγ1S82D) mediated transactivation of the (PPRE)3-tk-luciferase reporter construct in HUVECs. Results are the mean ± SD of three independent experiments.
Because transfection of ERK5a increased PPARγ1 activity, we examined the role of ERK5 kinase activity in PPARγ1 transcriptional activation. We cotransfected a dominant negative form of ERK5a (DN-ERK5 [dual phosphorylation site mutant T219A/Y221P]) or ERK5b (ATP binding site deleted, alternative splicing form [aa 78 to 806 of ERK5a]) (33) with CA-MEK5α and measured ciglitazone activation of PPARγ (Fig. 1A and B). Compared with cotransfection of CA-MEK5α and ERK5a, cotransfection of DN-ERK5 or ERK5b with CA-MEK5α significantly reduced ligand-mediated PPARγ1 activity (Fig. 1B, lane 2 versus lanes 4 and 5). These data suggested that ERK5 kinase activity is required for full stimulation of PPARγ1 transcriptional activity. However, DN-ERK5 or ERK5b did not inhibit CA-MEK5α-induced PPARγ1 activity (Fig. 1A and B, lane 3 versus lanes 4 and 5), suggesting that an ERK5 function besides endogenous kinase activity may be important. The scaffold function of ERK5 has previously been reported for MEF2, because the association of ERK5 with MEF2, but not MEF2 phosphorylation by ERK5, was regulated by MEF2 transcriptional activity (15). Therefore, association of ERK5 with PPARγ in addition to ERK5 kinase activity may regulate PPARγ1 transcriptional activity.
In contrast to ERK5, it has been reported that ERK1/2 and JNK inhibit PPARγ transcriptional activity through phosphorylation of Ser82 on PPARγ1 (6). Therefore, we investigated whether this inhibition by ERK1/2 and JNK could be observed in our cell system. For this purpose, we cotransfected full-length PPARγ1 and the luciferase reporter gene containing PPRE with or without CA-MEKK1 (as an upstream activator of ERK1/2 and JNK). We found that ciglitazone-induced PPARγ1 transcriptional activity was significantly inhibited by CA-MEKK1 transfection (Fig. 1C), demonstrating that ERK1/2 and JNK behaved as anticipated. We confirmed that CA-MEKK1 induced ERK1/2 and the JNK signaling pathway (8) (data not shown). These data support our finding that ERK5 differs from ERK1/2 and JNK with respect to PPARγ transcriptional activity.
ERK5 kinase did not phosphorylate PPARγ1 in vitro.
Since activation of ERK5 regulated PPARγ activity, as shown in Fig. 1A, we asked whether ERK5 could phosphorylate PPARγ in vitro. We cotransfected CA-MEK5α and Xpress-tagged ERK5a in Cos7 cells to activate ERK5a constitutively. Activated ERK5a was immunoprecipitated with an anti-ERK5 antibody, and an in vitro kinase assay was performed with GST, GST-PPARγ-AF-1 (aa 1 to 110, including Ser82, which is the ERK1/2 and JNK phosphorylation site), GST-PPARγ-DBD(aa 109-175), and GST-PPARγ-LBD(aa 163-475) as substrates. We did not use GST-PPARγ1 wild type, containing the complete sequence, because it was difficult to dissolve in lysis buffer and was easily degraded. As shown in Fig. 1D, transfection of CA-MEK5α activated ERK5 kinase, as shown by ERK5 autophosphorylation (Fig. 1D, bottom). However, ERK5 did not phosphorylate any PPARγ substrate (Fig. 1D, top).
Since ERK5, ERK1/2, and JNK phosphorylate similar proline-targeted consensus sequences, we mutated Ser82, which is the ERK1/2 and JNK phosphorylation site in PPARγ1, to alanine (S82A) or aspartate (S82D) and determined the effect of CA-MEK5α on PPARγ1 activity. As shown in Fig. 1E, we did not find any significant differences in ciglitazone-stimulated and/or CA-MEK5α-stimulated PPARγ1 activity between the wild type and PPARγ1 mutants. These data further suggest that the regulation of PPARγ1 activity by ERK5 is different from that of ERK1/2 and JNK.
Endogenous ERK5 associates with endogenous PPARγ in ECs.
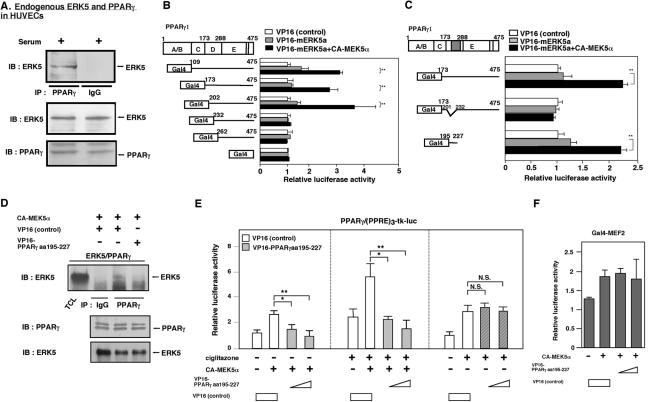
To investigate the potential interaction between ERK5 and PPARγ1, we analyzed their interaction by using coimmunoprecipitation. Since PPARγ is a nuclear receptor and ERK5 needs to be activated for its nuclear translocation, as our investigators previously described (33), we stimulated the cells with 10% serum for 30 min and immunoprecipitated with an anti-PPARγ antibody or rabbit immunoglobulin G (IgG) as a control. We found that endogenous PPARγ coimmunoprecipitated with endogenous ERK5 in ECs, but control rabbit IgG did not (Fig. 2A).
FIG. 2.
Endogenous ERK5 associates with endogenous PPARγ at the hinge-helix 1 region of PPARγ1, and the hinge-helix 1 region of the PPARγ1 fragment inhibited the ERK5-PPARγ interaction and CA-MEK5α-mediated PPARγ transcriptional activity. (A) HUVECs were stimulated with 10% serum for 30 min, whole-cell extract was immunoprecipitated with anti-PPARγ antibody or an equal amount of rabbit IgG, and Western blot analysis was performed with anti-ERK5 antibody (top). No difference in the amount of ERK5 (middle) or PPARγ (bottom) was observed in samples by Western blot analysis with anti-ERK5 (middle) or anti-PPARγ (bottom) antibody. (B and C) Association of activated ERK5a with PPARγ1 hinge-helix 1 was tested in a mammalian two-hybrid assay. The activation domain VP16 was fused to wild-type ERK5a and the PPARγ1 deletion mutants. Luciferase activity was normalized relative to the mean luciferase activity of the empty VP16 transfection (white bar; set as 1-fold). Constructs fused to the Gal4 binding domain were cotransfected with the Gal4-responsive luciferase reporter pG5-luc with or without cotransfection of CA-MEK5α in Cos7 cells for 40 h. The total transfected DNA amount was normalized with empty VP16 vector. Results are the mean ± SD of three independent experiments (B). (C) Association of activated ERK5a with PPARγ1 hinge-helix 1 was tested with PPARγ1 hinge-helix 1 truncated deletion mutant (PPARγ1 Δaa202-231) or small fragment of PPARγ1 (aa195-227) with or without CA-MEK5α or VP16-ERK5a, in a mammalian two-hybrid assay. The total transfected DNA amount was normalized with empty VP16 vector. (D) The VP16-PPARγ1(aa 195-227) fragment inhibited coimmunoprecipitation of ERK5 with PPARγ (top). No difference in the amount of PPARγ (middle) or ERK5 (bottom) was observed in samples by Western blot analysis with anti-PPARγ (middle) or anti-ERK5 (bottom) antibody. (E) Cells were transfected with plasmids expressing VP16 or the VP16-PPARγ1(aa 195-227) fragment and 1 μg of (PPRE)3-tk-luciferase, 0.5 μg of pSG5-PPARγ, and vector to provide equal amounts of transfected DNA, as described for Fig. 1. After 16 h of stimulation with or without ciglitazone, luciferase PPARγ1 transcriptional activity was assayed as described in the legend for Fig. 1. The total transfected DNA amount was normalized with empty VP16 vector. Results are the mean ± SD of three to six independent experiments. (F) Cotransfection of plasmid expressing the VP16-PPARγ1(aa 195-227) fragment did not inhibit CA-MEK5α-induced PPARγ1 activation in HUVECs. The total transfected DNA amount was normalized with empty VP16 vector.
To investigate the binding site of PPARγ1 with ERK5, we utilized a mammalian two-hybrid assay. A plasmid expressing the GAL4-DBD and the PPARγ (full-length or deletion mutants) was constructed by inserting PPARγ (including mutants) isolated from pSG5-PPARγ1 in frame into the pBIND vector. The plasmid expressing VP16-ERK5 (including mutants) was constructed by inserting the fragment of ERK5 into the VP16 activation domain containing plasmid pACT vector. As shown in Fig. 2B, hinge-helix 1 (aa 202 to 231) was required for the ERK5a-PPARγ1 interaction. To confirm the role of hinge-helix 1 region in the ERK5a-PPARγ1 interaction, we generated a truncated mutant form of PPARγ1 (Δaa202-231) and the PPARγ1(aa 195-227) fragment. As shown in Fig. 2C, deletion of hinge-helix 1 region completely inhibited the ERK5a-PPARγ1 interaction, but PPARγ1(aa 195-227) could associate with ERK5a, suggesting the critical role of hinge-helix 1 regions for the ERK5-PPARγ1 interaction.
Disruption of the ERK5-PPARγ1 interaction induced by the hinge-helix 1 fragment inhibited CA-MEK5α-induced PPARγ1 activity.
It is possible that the deletion mutant of the hinge-helix 1 region of PPARγ1 may change the tertiary structure of PPARγ1. To demonstrate the critical role of the hinge-helix 1 region for the ERK5-PPARγ1 interaction without mutating and destroying PPARγ1 structure, we determined whether the hinge-helix 1 fragment, which is the binding site of ERK5a, could disrupt the association of wild-type ERK5a and wild-type PPARγ. For this purpose, we generated six different hinge-helix 1 fragments. Our experimental approach was to fuse these peptide fragments with the VP16 active domain (which contains 46 aa). Since the VP16 active domain has a nuclear localization signal, fragments are able to translocate to the nucleus efficiently with this domain, and the fusion with the VP16 active domain will prevent degradation of these small peptide fragments in the cells. In addition, evaluation of the expression of small peptide fragments by Western blotting analysis is difficult, but we could easily detect the fused proteins by immunostaining with anti-VP16 antibody. We cotransfected cells with pcDNA-CA-MEK5α, pcDNA-ERK5a, or pSG5-PPARγ1 with empty VP16 construct. Cell lysates were tested for the effects of six different hinge-helix 1 fragments on ERK5 coprecipitation, and we immunoprecipitated with rabbit IgG or anti-rabbit PPARγ antibody and immunoblotted with anti-ERK5 antibody. As shown in Fig. 2D, ERK5 was coimmunoprecipitated by the anti-PPARγ antibody, but not by IgG. We found that cotransfection of the VP16-PPARγ1(aa 195-227) fragment, but not VP16 alone, significantly inhibited the coimmunoprecipitation of ERK5 with PPARγ. Among the tested fragments, the VP16-PPARγ1(aa 195-227) fragment had the most significant disrupting effect on the ERK5a-PPARγ1 interaction (data not shown). The expression levels of ERK5 and PPARγ were equal among the samples (Fig. 2D, lower). Since the VP16-PPARγ1(aa 195-227) fragment is too small to be detected by Western blotting, we confirmed the expression of the VP16 and VP16-PPARγ1(aa 195-227) fragment by immunostaining with anti-VP16 antibody (data not shown). We did not see any inhibitory effect with this VP16-PPARγ1(aa 195-227) fragment on the MEK5-ERK5a interaction, also suggesting the specific inhibitory effect of this fragment on PPARγ-ERK5 association (data not shown). These data support our findings from the mammalian two-hybrid assay that the hinge-helix 1 region of PPARγ is critical for ERK5a-PPARγ interaction.
Next, to demonstrate the critical role of the ERK5-PPARγ1 interaction for PPARγ1 activity, we determined whether the VP16-PPARγ1(aa 195-227) fragment could inhibit CA-MEK5α-ciglitazone-induced PPARγ1 activation. As shown in Fig. 2E, we found that VP16-fused PPARγ1(aa 195-227), but not VP16 alone or PPARγ1(aa 195-227) alone (data not shown), inhibited ciglitazone and/or CA-MEK5α-induced PPARγ1 activation. Of note, we did not observe significant inhibition of ciglitazone (alone)-induced PPARγ1 activation with the VP16-fused PPARγ1(aa 195-227) fragment, suggesting the specific effect of this fragment on the ERK5a-PPARγ interaction. To determine whether the inhibitory effect of VP16-PPARγ1(aa 195-227) is specific for CA-MEK5α-induced PPARγ1 activation, we investigated the effect of the VP16-PPARγ1(aa 195-227) fragment on CA-MEK5α-induced MEF2 activation. CA-MEK5α significantly induced MEF2 activation. In contrast to PPARγ1 activity, the VP16-PPARγ1(aa 195-227) fragment did not inhibit CA-MEK5α-induced MEF2 activation (Fig. 2F), suggesting the specific effect of VP16-PPARγ1(aa 195-227) on CA-MEK5α-induced PPARγ1 activation.
Flow enhances ciglitazone-induced PPARγ transcriptional activity via association of ERK5a kinase with the hinge-helix 1 region of PPARγ1.
In conduit arteries, steady laminar flow and physiological shear stress are atheroprotective, whereas turbulent flow and low shear stress are atherogenic (32). Previously, our group found that 20 min of steady flow significantly increased ERK5 activation in ECs (34).
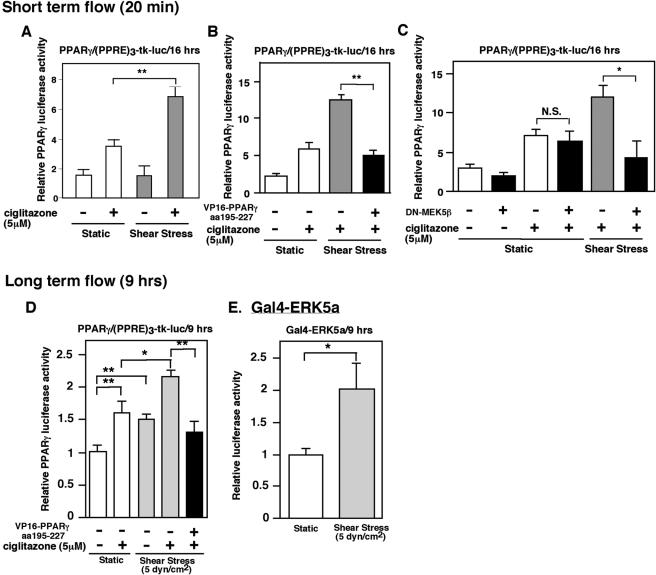
Since PPARγ activation has been reported to be atheroprotective (19, 20, 26), we studied the effect of flow on PPARγ transcriptional activity to determine the physiological relevance of the ERK5a-PPARγ1 interaction. We transfected (PPRE)3-tk-luciferase, pSG5-PPARγ, and vector to provide equal amounts of transfected DNA in HUVECs. As shown in Fig. 3A, flow (20 min) enhanced ciglitazone-induced PPARγ1 transcriptional activity (Fig. 3A), and cotransfection of the VP16-PPARγ1(aa 195-227) fragment significantly inhibited flow- and ciglitazone-induced PPARγ 1 transcriptional activity (Fig. 3B). To confirm the importance of ERK5, we utilized DN-MEK5β to inhibit ERK5 activation (5). Of note, DN-MEK5β inhibits ERK5 kinase activation but does not change ERK5 expression, which is different from that with DN-ERK5 or the ERK5b construct (5). As shown in Fig. 3C, DN-MEK5β could not inhibit ciglitazone-induced PPARγ activation but significantly inhibited flow-enhanced ciglitazone-mediated PPARγ activation. Furthermore, we found that 9 h of flow and the combination of flow and ciglitazone significantly increased PPARγ1 transcriptional activity in HUVECs. We did not find any significant change in PPARγ expression induced by flow (data not shown). Cotransfection of the VP16-PPARγ1(aa 195-227) fragment significantly inhibited ciglitazone- and flow-induced PPARγ1 transcriptional activity (Fig. 3D), similar to the effects of 20 min of flow described in Fig. 3B. In addition, 9 h of flow significantly increased ERK5 activation (2.0-fold [±0.4] increase; P < 0.01), as assayed with the Gal4-ERK5a construct in the one-hybrid mammalian assay (Fig. 3E). These data show a critical role for ERK5a association with PPARγ1 in flow-regulated PPARγ1 transcriptional activity and support the physiological significance of ERK5a and PPARγ1 interaction in ECs.
FIG. 3.
Flow-induced PPARγ1 transcriptional activation by the ERK5-PPARγ interaction and ERK5 activation. (A to C) Effect of short-term flow on PPARγ1 activity. At 24 h after transfection, growth-arrested HUVECs were stimulated by ciglitazone (5 μM), and then after 3 h of ciglitazone stimulation HUVECs were exposed for 20 min to flow (12 dynes/cm2) or no flow with or without plasmid expressing the VP16-PPARγ1(aa 195-227) fragment (B) or DN-MEK5β (C), as indicated. After 16 h of ciglitazone stimulation, luciferase PPARγ1 transcriptional activity was assayed as described in the legend for Fig. 1. (D and E) Effects of long-term flow on PPARγ1 and ERK5 activities. (D) Transfection medium contained 2 μg of PPRE reporter plasmid, 1 μg of pSG5-PPARγ, and vector to provide equal amounts of transfected DNA with or without plasmid expressing the VP16-PPARγ1(aa 195-227) fragment. At 48 h after transfection, growth-arrested HUVECs were exposed for 9 h to flow (5 dynes/cm2) or static condition with or without stimulation by ciglitazone (5 μM), as indicated. Luciferase PPARγ1 transcriptional activity was assayed as described in the legend for Fig. 1 after 9 h of ciglitazone or vehicle stimulation. (E) Gal4-ERK5a transcriptional activity was detected as described for Fig. 7a. Results are the mean ± SD of three independent experiments. *, P < 0.05; **, P < 0.01.
ERK5 activation is critical for the inhibitory effect of flow on TNF-α-mediated NF-κB activation.
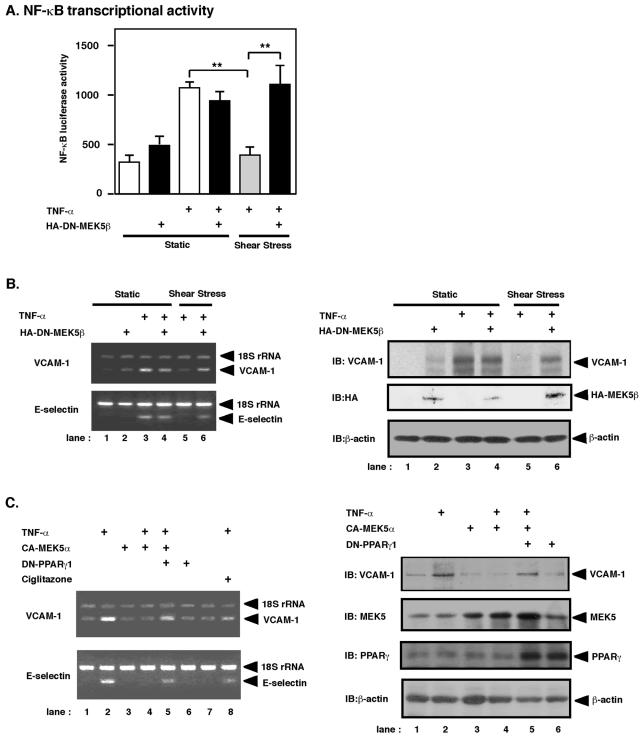
Previously, we and others found that both flow and PPARγ ligands inhibited TNF-α-mediated NF-κB activation (18, 32). To show a physiological role for ERK5/PPARγ activation by flow, we studied the role of ERK5 activation in the inhibitory effect of flow on TNF-α-mediated NF-κB activation. Since DN-MEK5β significantly inhibited flow (short-term flow) and PPARγ ligand-mediated PPARγ activation (Fig. 3C), we investigated whether DN-MEK5β could prevent the inhibitory effect of flow on TNF-α-induced NF-κB activation. As shown in Fig. 4A, DN-MEK5β significantly inhibited the ability of flow (6 h) to decrease TNF-α-mediated NF-κB activation. These data suggest that the inhibitory effect of flow on NF-κB activation is, at least partially, due to the activation of ERK5 and subsequent PPARγ activation.
FIG. 4.
Flow-inhibited TNF-α-mediated NF-κB activation and VCAM-1 expression by ERK5 and PPARγ activation. (A) HUVECs were transfected with pFR-Luc plasmid and pNF-kBLuc-plasmid. To control transfection efficiency, pRL-TK was transfected as a luciferase control reporter vector. After 24 h of transfection, HUVECs were treated with the following protocol: cells were maintained under static conditions for 20 min followed by vehicle (lanes 1 and 2) or TNF-α stimulation (20 ng/ml; lanes 3 and 4) under the same static conditions with (lanes 2 and 4) or without (lanes 1 and 3) DN-MEK5β transfection, or cells were subjected to flow (shear stress of 5 dynes/cm2) for 20 min followed by TNF-α stimulation (lanes 5 and 6) with (lane 6) or without (lane 5) DN-MEK5β transfection under continuous flow. After 6 h of TNF-α stimulation, luciferase NF-κB transcriptional activity was assayed using the dual-luciferase reporter assay system, and luciferase luminescence was counted in a Luminometer and then normalized to cotransfected luciferase activity as described in Materials and Methods. Results are the mean ± SD of three independent experiments. **, P < 0.01. (B) Effect of long-term flow on TNF-α-mediated VCAM-1 and E-selectin expression. After 24 h of transfection, BLMECs were treated in the following protocol: cells were maintained under static conditions for 60 min followed by vehicle (lanes 1 and 2) or TNF-α (20 ng/ml) stimulation (lanes 3 and 4) under the same static conditions with (lanes 2 and 4) or without (lanes 1 and 3)DN-MEK5β transfection, or cells were subjected to flow (shear stress of 5 dynes/cm2) for 60 min followed by TNF-α stimulation with (lane 6) or without (lane 5) DN-MEK5β transfection under continuous flow. (B, left) After 4 h of TNF-α stimulation, VCAM-1 (upper) and E-selectin (lower) mRNA levels were determined by relative quantitative RT-PCR. 18S rRNA was used as an internal control. (Right) After 6 h of TNF-α stimulation, VCAM-1, hemagglutinin-tagged MEK5β, and β-actin expression were determined by Western blot analysis. (C) Effect of ERK5 and PPARγ activation on TNF-α-mediated VCAM-1 expression. At 24 h after transfection, growth-arrested BLMECs were stimulated with TNF-α (20 ng/ml) with or without plasmid expressing CA-MEK5α or DN-PPARγ1, as indicated. (C, left) After 4 h of TNF-α stimulation, VCAM-1 (upper) and E-selectin (lower) mRNA levels were determined as described for panel B. (Right) VCAM-1, MEK5, PPARγ, and β-actin expression levels were determined by Western blot analysis.
ERK5 and PPARγ activation is critical for the inhibitory effect of flow on TNF-α-mediated VCAM-1 expression.
It is well documented that VCAM-1 and E-selectin expression is regulated by NF-κB activation (11). Since we determined the role of ERK5 for the inhibitory effect of flow on TNF-α-mediated NF-κB activation (Fig. 4A), first we investigated whether flow can inhibit TNF-α-mediated VCAM-1 and E-selectin mRNA expression via activation of ERK5. We used BLMECs in these particular experiments, because BLMECs have high transfection efficiency, as our investigators have previously described (24). As shown in Fig. 4B (left), TNF-α induced VCAM-1 and E-selectin mRNA expression after 4 h of stimulation (lane 3), and flow significantly inhibited VCAM-1 and E-selectin mRNA induction (lane 5), as previously described (9). We found that DN-MEK5β significantly blocked flow-mediated inhibition of TNF-α-mediated VCAM-1 and E-selectin mRNA expression (lane 6), suggesting a critical role for ERK5 activation in flow-mediated inhibition of VCAM-1 and E-selectin mRNA expression. To confirm that this mRNA regulation correlated with regulation at the protein level, we also determined whether flow could inhibit TNF-α-mediated VCAM-1 protein expression induction after 6 h of stimulation. As shown in Fig. 4B (right), consistent with the mRNA expression data (left), we found that DN-MEK5β significantly inhibited flow-mediated inhibition of VCAM-1 protein expression (Fig. 4B, right, lane 6).
Furthermore, to investigate the involvement of PPARγ activation in ERK5-mediated inhibition of VCAM-1 and E-selectin expression, we utilized a dominant negative form of PPARγ1 (DN-PPARγ1, L438A/E441A) and CA-MEK5α. As shown in Fig. 4C (left), TNF-α increased VCAM-1 and E-selectin mRNA expression (lane 2), and CA-MEK5α (lane 4) and ciglitazone (lane 8) significantly inhibited TNF-α-mediated VCAM-1 and E-selectin mRNA expression. Transfection of DN-PPARγ1 significantly decreased the inhibitory effect of ERK5 activation on VCAM-1 and E-selectin mRNA expression (lane 5), suggesting that the inhibitory effect of ERK5 activation is due to activation of PPARγ. As shown in Fig. 4C (right), consistent with the mRNA expression data (left), we also found that CA-MEK5α significantly inhibited TNF-α-mediated VCAM-1 protein expression (lane 4), and DN-PPARγ1 significantly recovered this inhibitory effect of ERK5 activation on VCAM-1 protein expression (Fig. 4C, right, lane 5).
Hinge-helix 1 region is critical for PPARγ1 transcriptional activity via its regulation of the association of SMRT with PPARγ1.
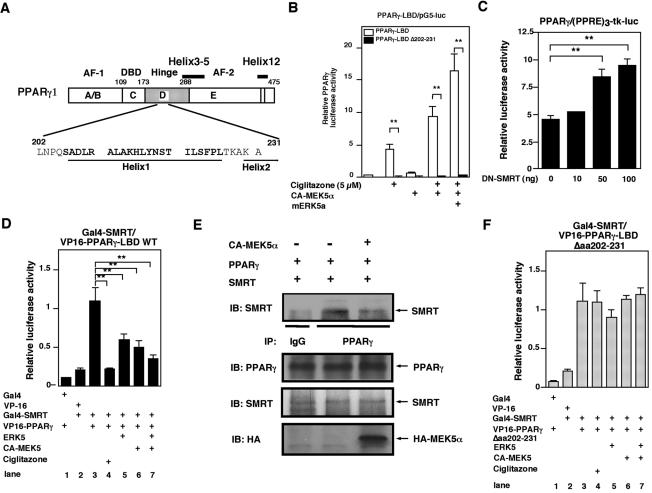
To determine the role of hinge-helix 1 in regulation of PPARγ1 transcriptional activity (Fig. 5A), we generated a hinge-helix 1 deletion mutant in PPARγ-LBD(aa 162-475). To measure PPARγ-LBD transcriptional activity, we used a mammalian one-hybrid assay (Fig. 5B). HUVECs were cotransfected with Gal4-PPARγ-LBD(aa 162-475) or Gal4-PPARγ-LBDΔ202-231 and pG5-luc with or without CA-MEK5α or ERK5a, and PPARγ-LBD transcriptional activities in response to ciglitazone and ERK5 were measured. As shown in Fig. 5B, CA-MEK5α/ERK5 significantly increased PPARγ1-LBD transcriptional activity, and deletion of hinge-helix 1 region from the LBD completely abolished PPARγ1-LBD transcriptional activity in ECs.
FIG. 5.
Activated ERK5 disrupts the association of corepressor SMRT with PPARγ1. (A) Scheme of the hinge-helix 1 region of PPARγ1. (B) The hinge-helix 1 region of PPARγ1 is critical for CA-MEK5α and ciglitazone-induced PPARγ1-mediated transactivation of the (PPRE)3-tk-luciferase reporter construct. (C) DN-SMRT increased PPARγ1-mediated transactivation of the (PPRE)3-tk-luciferase reporter construct. (D and F) Interaction of ERK5a with the hinge-helix 1 region of PPARγ1 disrupts SMRT/PPARγ1. Cos7 cells were transfected with plasmids expressing Gal4, VP16, Gal4-SMRT, ERK5a, or CA-MEK5α with wild-type VP16-PPARγ1-LBD (D) or VP16-PPARγ1-LBD Δaa202-231 (F), as indicated, in a mammalian two-hybrid assay. (E) ERK5 activation by cotransfection of CA-MEK5α and ERK5a inhibited coimmunoprecipitation of SMRT with PPARγ (top). No difference in the amount of PPARγ and SMRT was observed in samples by Western blot analysis with anti-PPARγ or anti-SMRT antibody (middle and bottom).
Previous studies have demonstrated that PPARγ interaction with the corepressor SMRT inhibits its activation. Helices 3 to 5 and 12 in the LBD are important for interaction with corepressors (14, 35). Since we found that the hinge-helix 1 domain is involved in the ERK5-PPARγ1 interaction, we investigated whether the interaction of activated ERK5 and PPARγ1 alters binding of SMRT to PPARγ1. We determined the expression of nuclear corepressor (N-CoR1) and SMRT mRNA in HUVECs by RT-PCR (data not shown). A dominant negative form of SMRT increased full-length PPARγ1 activity, suggesting a functional role for SMRT in HUVECs (Fig. 5C). As shown in Fig. 5D, two-hybrid analysis using Gal4-SMRT and VP16-wild-type PPARγ1-LBD(aa 173-475) indicated interaction of PPARγ1 and SMRT (lane 3), and ciglitazone inhibited this interaction (lane 4). Interestingly, cotransfection with ERK5a (lane 5), CA-MEK5α (lane 6), or ERK5a and CA-MEK5α (lane 7) significantly inhibited the interaction between corepressor SMRT and PPARγ1-LBD.
To confirm that ERK5 activation disrupted the SMRT-PPARγ interaction, we transfected Cos7 cells with CA-MEK5α. As shown in Fig. 5E, we found that transfection of CA-MEK5α significantly inhibited the SMRT-PPARγ interaction, as measured by coimmunoprecipitation. When ERK5 is phosphorylated by CA-MEK5α, it can inhibit SMRT binding to PPARγ. Since the deletion mutant of hinge-helix 1 region in PPARγ1 had significantly reduced transcriptional activity, we examined whether the deletion of hinge-helix 1 interferes with the disruption of SMRT and PPARγ1 induced by ERK5 binding. As shown in Fig. 5F, the deletion mutant of hinge-helix 1 domain of PPARγ1 (VP16-PPARγΔaa202-231) associated with Gal4-SMRT (lane 3). Although ciglitazone disrupted the interaction of PPARγ1 wild type and SMRT as described previously (Fig. 5D), the deletion of hinge-helix 1 domain abolished ciglitazone-induced disruption of PPARγ1 and SMRT. In addition, activation of ERK5 was required for full interaction of ERK5a and PPARγ1, as shown in Fig. 2C, and we found that even this activated ERK5 could not interfere with the binding of SMRT and PPARγ1 (Fig. 5F, lanes 5 to 7).
Finally, to determine whether this SMRT-PPARγ disruption induced by activation of ERK5 is specific, we determined the effect of activated ERK5 on the interaction of PPARγ1 and coactivator SRC-1. As previously reported (21), the association of coactivator SRC-1 with PPARγ1 was induced by ciglitazone. In contrast to SMRT, we did not find any effect of CA-MEK5α/ERK5 on this interaction (data not shown), suggesting a specific effect of ERK5 on the PPARγ1 hinge-helix 1 region via binding of SMRT and PPARγ1.
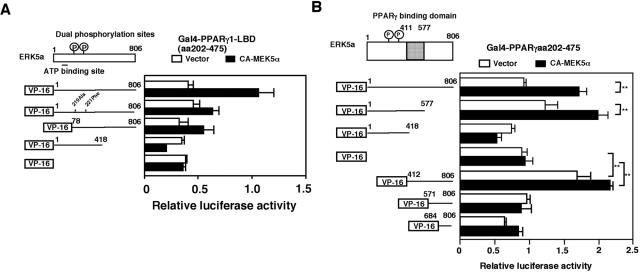
PPARγ1 binding site of ERK5.
To clarify the role of PPARγ1 association with ERK5, we determined the binding site of PPARγ1 on ERK5. A plasmid expressing VP16-ERK5a was constructed by inserting the fragment of ERK5a into the VP16 active domain plasmid pact as measured by pG5-luc activity (Fig. 6A). We found that cotransfection of CA-MEK5α induced association with PPARγ1-LBD(aa 202-475). Dominant negative forms of ERK5 (dual phosphorylation site mutants [ERK5a T219A/Y221F] and ERK5b) exhibited reduced PPARγ1 association as measured with pG5-luc. However, a truncated mutant of ERK5(aa 1-418) did not associate with PPARγ1, suggesting that the COOH-terminal region of ERK5a is required for association with PPARγ1 (Fig. 6A). Since dominant negative forms of ERK5 partially reduced the ERK5-PPARγ1 association and the ERK5 with the COOH terminal deleted did not associate with PPARγ1, we speculated that autophosphorylation of ERK5 was required for ERK5-PPARγ1 interaction. Therefore, we mutated several putative autophosphorylation sites in the COOH-terminal region of ERK5 and determined the ERK5-PPARγ1 interaction. However, every ERK5 putative autophosphorylation site mutant that we examined (S433A, S697A, T723A, and S793A) associated with PPARγ1 (data not shown). These data suggest that autophosphorylation of the ERK5 COOH-terminal region may not be required for PPARγ1-ERK5 interaction.
FIG. 6.
ERK5a binding site of PPARγ1. (A and B) Requirement of ERK5a kinase activity and the COOH-terminal region of ERK5 for the ERK5a-PPARγ1 interaction. (A) Cos7 cells were transfected with plasmids expressing wild-type Gal4-PPARγ1-LBD(aa 202-475), VP16, and CA-MEK5α with VP16-ERK5a, VP16-DN-ERK5, or VP16-ERK5(aa 1-418), as indicated, in a mammalian two-hybrid assay. (B) The middle region of ERK5 (aa 419 to 577) in COOH-terminal region is critical for ERK5a-PPARγ1 interaction. Cos7 cells were transfected with plasmids expressing Gal4-PPARγ1-LBD(aa 202-475), VP16, or CA-MEK5α with several COOH-terminal or NH2-terminal deletion mutants of VP16-ERK5a, as indicated, in a mammalian two-hybrid assay.
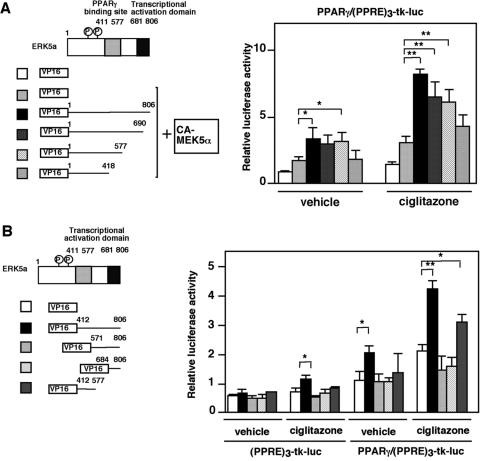
We next generated several deletion mutants of ERK5 to define the domains required for PPARγ association. ERK5 deletion mutants were cloned into the VP16 active domain plasmid pACT, and the interaction with Gal4-PPARγ1-LBD was determined in a two-hybrid mammalian assay. As shown in Fig. 6B, the deletion mutant ERK5a(aa 1-577), but not ERK5a(aa 1-418), associated with PPARγ1, suggesting that aa 419 to 577 contain the ERK5 binding domain for PPARγ1. To rule out the possibility of another binding site in COOH-terminal region, we also generated several small fragments of the COOH-terminal region of ERK5 and performed a two-hybrid mammalian assay. We found that ERK5 aa 412 to 806 associated with PPARγ1, but not ERK5 aa 571 to 806 and aa 684 to 806, suggesting that ERK5 aa 412 to 570 contains the only binding site of ERK5a for PPARγ1.
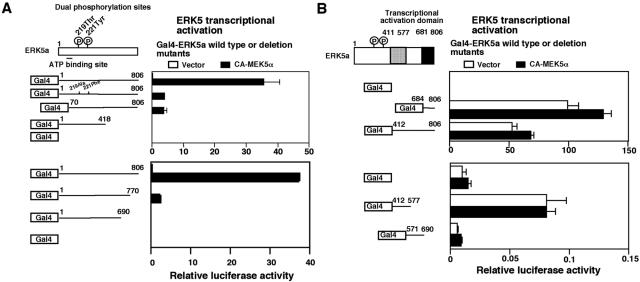
The COOH-terminal region of ERK5 has two transactivation domains.
Because ERK5 associates with PPARγ1 and enhances its activity, we investigated whether ERK5 itself exhibits any transcriptional activity, as reported previously (15). To evaluate ERK5 as an activator of transcription, we determined its effect on the transcriptional activation of a reporter gene in a one-hybrid mammalian assay. For this purpose, Cos7 cells were cotransfected with Gal4-ERK5a and its mutants with or without CA-MEK5α, and the effect on basal transcriptional activity was determined. As shown in Fig. 7A, ERK5a exhibited a very low transcriptional activity without CA-MEK5α transfection, but cotransfection of CA-MEK5α dramatically increased transcriptional activity. Dominant negative forms of ERK5 (DN-ERK5a and -ERK5b) significantly reduced transcriptional activity, suggesting that activated ERK5 is required for an active coactivator function. Analysis of COOH-terminal ERK5 deletion mutants showed that the coactivator function was associated with the COOH-terminal region of ERK5a (Fig. 7A, lower).
FIG. 7.
Transcriptional activation domains of ERK5a. (A) Cos7 cells were transfected with Gal4-dependent (Gal4-luc) reporter constructs with dominant negative forms of Gal4-DN-ERK5 or ERK5b (upper) and Gal4-ERK5 COOH-terminal deletion mutants (lower). Luciferase activity was measured in unstimulated cells. (B) Cos7 cells were transfected with Gal4-dependent (Gal4-luc) reporter constructs with several COOH-terminal fragments of Gal4-ERK5, as indicated. Luciferase activity was measured in unstimulated cells.
To identify the ERK5 transcriptional activation domain and the role of the NH2-terminal region, we also generated several COOH-terminal-truncated mutants of ERK5 and determined transcriptional activities in a mammalian one-hybrid assay. We found that the ERK5 COOH-terminal tail (aa 684 to 806) had very high transcriptional activity even without CA-MEK5α transfection (Fig. 7B, upper). The middle region of ERK5 (aa 412 to 577) also had a small but significant transcriptional activity (Fig. 7B, lower). These data suggest that the COOH-terminal ERK5 region has two transcriptional activator domains. Since full-length ERK5a required cotransfection of CA-MEK5α for full activation but the transcriptional activation domain fragments did not require CA-MEK5α cotransfection, our results suggest that NH2-terminal ERK5 may act as a negative regulator of these transactivation domains.
The COOH terminus of ERK5 transactivation domains is required for full activation of PPARγ1.
To determine the role of the two ERK5 transactivation domains on PPARγ1 activation, we generated several VP16-fused COOH-terminal ERK5 deletion mutants. As shown in Fig. 8A, CA-MEK5α induced PPARγ1 activity in HUVECs, and cotransfection of wild-type VP16-ERK5a enhanced its activity. Progressive deletion of COOH-terminal ERK5a gradually reduced ciglitazone- and CA-MEK5α-induced PPARγ1 activity. Complete deletion of the COOH-terminal region of ERK5 (ERK5[aa 1-418]) totally abolished the enhancing effect of ciglitazone and CA-MEK5α/ERK5a on PPARγ1 activation. Furthermore, as shown in Fig. 8B, we found that the entire COOH-terminal ERK5 (aa 412 to 806) and middle region of ERK5 (aa 412 to 577), which contains both the PPARγ1 binding site and transactivation domain, enhanced ciglitazone-induced PPARγ1 activation. However, the transactivation domain at the COOH-terminal tail of ERK5 (aa 684 to 806) alone could not induce PPARγ activity, suggesting a critical role for the middle region of ERK5 (aa 412 to 577) as a binding site of ERK5a with PPARγ1.
FIG. 8.
Both transcriptional domains and the PPARγ1 binding site in the COOH-terminal region of ERK5 are critical to fully activate PPARγ1. (A and B) Transfection medium contained 1 μg of (PPRE)3-tk-luciferase, 0.5 μg of pSG5-PPARγ1, and vector to provide equal amounts of transfected DNA. pcDNA3.1-CA-MEK5α (A) and plasmids expressing deletion mutants of the COOH-terminal tail of ERK5a (A) or several VP16-fused truncated mutant fragments of the COOH-terminal region of ERK5a (B) were transfected in HUVECs as indicated, and the pcDNA3.1 vector was used to provide equal amounts of transfected DNA. After 24 h of transfection, growth-arrested HUVECs were stimulated with or without ciglitazone (5 μM). Luciferase PPARγ1 transcriptional activity was assayed as described in the legend for Fig. 1. Results are the mean ± SD of three independent experiments. Expression of full-length and truncated ERK5 was demonstrated by Western blotting with anti-ERK5 antibody (A, right panel). We also performed immunostaining with anti-VP16 antibody and confirmed expression of these constructs in HUVECs (data not shown).
DISCUSSION
In the present study, we propose a novel mechanism by which ERK5 regulates PPARγ1 transcriptional activity via association with the PPARγ1 hinge-helix 1 region (Fig. 9). Kasler et al. reported that the COOH-terminal region of ERK5 contained a MEF2-interacting domain and also a potent transcriptional activation domain. We found that the middle region of ERK5a, but not the COOH-terminal tail of ERK5a, associates with PPARγ1. The inactive NH2-terminal ERK5 kinase domain acts as a negative regulator of its COOH-terminal region, and the activation of ERK5 by CA-MEK5α disrupts this inhibitory effect of the NH2-terminal region of ERK5 on the COOH-terminal region, based on the following data: (i) dominant negative forms of ERK5 partially inhibited the activated ERK5-induced association between ERK5a and PPARγ1 (Fig. 6A); (ii) full-length ERK5a required cotransfection of CA-MEK5α for full activation, but COOH-terminal region fragments did not require CA-MEK5α cotransfection (Fig. 7A and B); and (iii) the COOH-terminal region of ERK5 could associate with PPARγ1 and increase PPARγ1 activation without CA-MEK5α transfection (Fig. 6 and 8B). We could not detect direct phosphorylation of PPARγ1 by ERK5a (Fig. 1D). Although dominant negative forms of ERK5 could partially associate with PPARγ1, ERK5a kinase activation was necessary for full association of ERK5a with PPARγ1 (Fig. 6). In addition, as shown in Fig. 1B, ERK5a kinase activation is important for full PPARγ1 activation. Our group and others have found that kinase activation of ERK5a initiates the nuclear translocation of ERK5a (16, 33). Therefore, both the disruption of the inhibitory effect of the NH2-terminal region of ERK5 (Fig. 9) and the nuclear translocation of ERK5a, which are induced by ERK5 activation, may be required to fully activate PPARγ1.
Flow increased ERK5 and PPARγ1 activation, and the hinge-helix 1 region of the PPARγ1 fragment significantly inhibited flow-induced PPARγ activation. To our knowledge, this is the first study to identify the important role of ERK5 and the hinge-helix 1 region (aa 202 to 231) of PPARγ1 in regulation of flow-induced PPARγ1 transcriptional activity. The ERK5 interaction with PPARγ was partially regulated by ERK5 kinase activation, which is similar to the ERK5 interaction with MEF2 (15). The likely mechanism for ERK5 to activate PPARγ1 is disruption of the SMRT and PPARγ1 interaction. These data suggest that ERK5 is a potent positive regulator of flow- and ligand-induced PPARγ activation via the interaction of ERK5 and the hinge-helix 1 region of PPARγ.
To determine the interaction of ERK5 and PPARγ, we utilized three different methods: (i) in vivo interaction between ERK5 and PPARγ by coimmunoprecipitation with endogenous ERK5 and endogenous PPARγ, (ii) a mammalian two-hybrid assay by generating VP16-ERK5a and Gal4-PPARγ1 construct and, most importantly, (iii) specific inhibition of coimmunoprecipitation of ERK5 and PPARγ by VP16-hinge-helix 1 fragment expression. These data strongly suggest that ERK5 and PPARγ associate in vivo. Furthermore, the inhibition of CA-MEK5α-induced, but not PPARγ ligand-induced, PPARγ transcriptional activation by VP16-hinge-helix 1 fragment expression suggests the physiological relevance of this ERK5-PPARγ interaction in regulating PPARγ transcriptional activity.
PPARγ1 contains two activation functions (AF) residing in the NH2-terminal A/B domain (AF-1) and the COOH-terminal end of the E domain (AF-2). The critical role of AF-2 activity for ligand-induced conformational change in the region of helix 12 (H12) of nuclear receptors has been well documented (22). The binding surface for the coactivator peptide is formed by helix loops H3, H4, part of H5, and H12. Pissios et al. have reported that helix 1 of the thyroid hormone receptor plays an important role in stabilizing the overall structure of the LBD upon binding hormone or the corepressor N-CoR1 (25). It has been reported that PGC-1 interacts with PPARγ in a ligand-independent fashion via the hinge region (PPARγ2 aa 181 to 227 and PPARγ1 aa 151 to 197), but not the helix 1 region, of the receptor. As shown in Fig. 1A and B and 2E, CA-MEK5α activated full-length PPARγ1 transcriptional activity in a ligand-independent manner. In contrast, the expression of PGC-1 alone does not induce PPARγ transcriptional activity without ligand (27). Therefore, although the binding sites of PGC-1 and ERK5a on PPARγ1 are near each other, the regulatory mechanisms of ERK5a action on PPARγ1 activity are quite different from that of PGC-1. Moreover, the complete inhibition of PPARγ1 transcriptional activity by deletion of the hinge-helix 1 region (PPARγ1 aa 202 to 231) supports the critical role of the hinge-helix 1 region in PPARγ1 transcriptional activity.
Flow stimulation of PPARγ1 activity, via activation of ERK5a, may contribute to the antiinflammatory and atheroprotective effects of flow. Previously, our investigators found that flow potently activates ERK5a (34). Flow also inhibits leukocyte binding, as well as ICAM-1 and VCAM-1 expression (31). PPARγ1 agonists similarly modulate the expression of proinflammatory cytokines (19), chemokines (10), and adhesion molecules (38) in ECs (20, 26). Activation of PPARγ itself has a potent antiinflammatory role, as shown by Wang et al., who reported that constitutive activation of PPARγ1 significantly inhibited expression of ICAM-1 and VCAM-1 in ECs (32). The finding that flow activates PPARγ1 transcriptional activity is unique, because growth factors and cytokines have been reported to inhibit PPARγ activation via ERK1/2 and JNK activation (6, 13). We have shown that flow stimulation of PPARγ is functional, since DN-MEK5β significantly prevented flow-mediated inhibition of TNF-α-induced VCAM-1 and E-selectin expression and the expression of DN-PPARγ1 significantly decreased the inhibitory effect of ERK5 on VCAM-1 and E-selectin expression.
We found a significant increase in PPARγ activation by the long-term flow regimen (9 h), but not by short-term flow (20 min). We anticipate that this is due to the balance between ERK1/2 and ERK5 activation by flow. As we explained in the introduction, ERK1/2 inhibits but ERK5 activates PPARγ transcriptional activity. Of note, flow-induced ERK1/2 activation is relatively temporary (10 to 40 min), while ERK5 activation is sustained for up to 6 h after stimulation (Fig. 3E and data not shown) (34). Therefore, since the effect of ERK1/2 activation on PPARγ might be stronger in a short-term flow regimen (20 min) than in a long-term flow regimen (9 h), we observed activation of PPARγ to a much greater extent in long-term flow.
Acknowledgments
We are grateful to Jay Yang, Michael Massett, Thunder Jalili, Burns C. Blaxall, and Joseph Miano for critical reading of the manuscript. We thank Keiji Fujiwara and Tamlyn Thomas for technical support and advice for the flow apparatus.
This work was supported by grants from the National Institutes of Health to J.A. (HL-66919, HL-65262, and HL73096-01A1) and to B.C.B. (HL-44721 and HL-49192).
REFERENCES
- 1.Abe, J., M. Kusuhara, R. J. Ulevitch, B. C. Berk, and J. D. Lee. 1996. Big mitogen-activated protein kinase 1 (BMK1) is a redox-sensitive kinase. J. Biol. Chem. 271:16586-16590. [DOI] [PubMed] [Google Scholar]
- 2.Abe, J., M. Takahashi, M. Ishida, J. D. Lee, and B. C. Berk. 1997. c-Src is required for oxidative stress-mediated activation of big mitogen-activated protein kinase 1. J. Biol. Chem. 272:20389-20394. [DOI] [PubMed] [Google Scholar]
- 3.Aizawa, T., H. Wei, J. M. Miano, J. Abe, B. C. Berk, and C. Yan. 2003. Role of phosphodiesterase 3 in NO/cGMP-mediated antiinflammatory effects in vascular smooth muscle cells. Circ. Res. 93:406-413. [DOI] [PubMed] [Google Scholar]
- 4.Ameshima, S., H. Golpon, C. D. Cool, D. Chan, R. W. Vandivier, S. J. Gardai, M. Wick, R. A. Nemenoff, M. W. Geraci, and N. F. Voelkel. 2003. Peroxisome proliferator-activated receptor gamma (PPARγ) expression is decreased in pulmonary hypertension and affects endothelial cell growth. Circ. Res. 92:1162-1169. [DOI] [PubMed] [Google Scholar]
- 5.Cameron, S. J., J. Abe, S. Malik, W. Che, and J. Yang. 2004. Differential role of MEK5α and MEK5β in BMK1/ERK5 activation. J. Biol. Chem. 279:1506-1512. [DOI] [PubMed] [Google Scholar]
- 6.Camp, H. S., and S. R. Tafuri. 1997. Regulation of peroxisome proliferator-activated receptor gamma activity by mitogen-activated protein kinase. J. Biol. Chem. 272:10811-10816. [DOI] [PubMed] [Google Scholar]
- 7.Camp, H. S., S. R. Tafuri, and T. Leff. 1999. c-Jun N-terminal kinase phosphorylates peroxisome proliferator-activated receptor-gamma 1 and negatively regulates its transcriptional activity. Endocrinology 140:392-397. [DOI] [PubMed] [Google Scholar]
- 8.Che, W., N. Lerner-Marmarosh, Q. Huang, M. Osawa, S. Ohta, M. Yoshizumi, M. Glassman, J. D. Lee, C. Yan, B. C. Berk, and J. Abe. 2002. Insulin-like growth factor-1 enhances inflammatory responses in endothelial cells: role of Gab1 and MEKK3 in TNF-alpha-induced c-Jun and NF-κB activation and adhesion molecule expression. Circ. Res. 90:1222-1230. [DOI] [PubMed] [Google Scholar]
- 9.Chiu, J. J., P. L. Lee, C. N. Chen, C. I. Lee, S. F. Chang, L. J. Chen, S. C. Lien, Y. C. Ko, S. Usami, and S. Chien. 2004. Shear stress increases ICAM-1 and decreases VCAM-1 and E-selectin expressions induced by tumor necrosis factor-α in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 24:73-79. [DOI] [PubMed] [Google Scholar]
- 10.Collins, A. R., W. P. Meehan, U. Kintscher, S. Jackson, S. Wakino, G. Noh, W. Palinski, W. A. Hsueh, and R. E. Law. 2001. Troglitazone inhibits formation of early atherosclerotic lesions in diabetic and nondiabetic low density lipoprotein receptor-deficient mice. Arterioscler. Thromb. Vasc. Biol. 21:365-371. [DOI] [PubMed] [Google Scholar]
- 11.Collins, T., M. A. Read, A. S. Neish, M. Z. Whitley, D. Thanos, and T. Maniatis. 1995. Transcriptional regulation of endothelial cell adhesion molecules: NF-κB and cytokine-inducible enhancers. FASEB J. 9:899-909. [PubMed] [Google Scholar]
- 12.Gutkind, J. S. 2000. Regulation of mitogen-activated protein kinase signaling networks by G protein-coupled receptors. Sci. STKE 2000:RE1. [DOI] [PubMed] [Google Scholar]
- 13.Hu, E., J. B. Kim, P. Sarraf, and B. M. Spiegelman. 1996. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARγ. Science 274:2100-2103. [DOI] [PubMed] [Google Scholar]
- 14.Hu, X., and M. A. Lazar. 1999. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature 402:93-96. [DOI] [PubMed] [Google Scholar]
- 15.Kasler, H. G., J. Victoria, O. Duramad, and A. Winoto. 2000. ERK5 is a novel type of mitogen-activated protein kinase containing a transcriptional activation domain. Mol. Cell. Biol. 20:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato, Y., V. V. Kravchenko, R. I. Tapping, J. Han, R. J. Ulevitch, and J. D. Lee. 1997. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 16:7054-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, J. D., R. J. Ulevitch, and J. Han. 1995. Primary structure of BMK1: a new mammalian MAP kinase. Biochem. Biophys. Res. Commun. 213:715-724. [DOI] [PubMed] [Google Scholar]
- 18.Lerner-Marmarosh, N., M. Yoshizumi, W. Che, J. Surapisitchat, H. Kawakatsu, M. Akaike, B. Ding, Q. Huang, C. Yan, B. C. Berk, and J. I. Abe. 2003. Inhibition of tumor necrosis factor-α-induced SHP-2 phosphatase activity by shear stress: a mechanism to reduce endothelial inflammation. Arterioscler. Thromb. Vasc. Biol. 23:1775-1781. [DOI] [PubMed] [Google Scholar]
- 19.Li, A. C., K. K. Brown, M. J. Silvestre, T. M. Willson, W. Palinski, and C. K. Glass. 2000. Peroxisome proliferator-activated receptor gamma ligands inhibit development of atherosclerosis in LDL receptor-deficient mice. J. Clin. Investig. 106:523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marx, N., T. Bourcier, G. K. Sukhova, P. Libby, and J. Plutzky. 1999. PPARγ activation in human endothelial cells increases plasminogen activator inhibitor type-1 expression: PPARγ as a potential mediator in vascular disease. Arterioscler. Thromb. Vasc. Biol. 19:546-551. [DOI] [PubMed] [Google Scholar]
- 21.McInerney, E. M., D. W. Rose, S. E. Flynn, S. Westin, T. M. Mullen, A. Krones, J. Inostroza, J. Torchia, R. T. Nolte, N. Assa-Munt, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1998. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 12:3357-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moras, D., and H. Gronemeyer. 1998. The nuclear receptor ligand-binding domain: structure and function. Curr. Opin. Cell Biol. 10:384-391. [DOI] [PubMed] [Google Scholar]
- 23.Pasceri, V., H. D. Wu, J. T. Willerson, and E. T. Yeh. 2000. Modulation of vascular inflammation in vitro and in vivo by peroxisome proliferator-activated receptor-gamma activators. Circulation 101:235-238. [DOI] [PubMed] [Google Scholar]
- 24.Pi, X., C. Yan, and B. C. Berk. 2004. Big mitogen-activated protein kinase (BMK1)/ERK5 protects endothelial cells from apoptosis. Circ. Res. 94:362-369. [DOI] [PubMed] [Google Scholar]
- 25.Pissios, P., I. Tzameli, P. Kushner, and D. D. Moore. 2000. Dynamic stabilization of nuclear receptor ligand binding domains by hormone or corepressor binding. Mol. Cell 6:245-253. [DOI] [PubMed] [Google Scholar]
- 26.Plutzky, J. 2001. Inflammatory pathways in atherosclerosis and acute coronary syndromes. Am. J. Cardiol. 88:10K-15K. [DOI] [PubMed] [Google Scholar]
- 27.Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright, and B. M. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829-839. [DOI] [PubMed] [Google Scholar]
- 28.Regan, C. P., W. Li, D. M. Boucher, S. Spatz, M. S. Su, and K. Kuida. 2002. Erk5 null mice display multiple extraembryonic vascular and embryonic cardiovascular defects. Proc. Natl. Acad. Sci. USA 99:9248-9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulman, I. G., H. Juguilon, and R. M. Evans. 1996. Activation and repression by nuclear hormone receptors: hormone modulates an equilibrium between active and repressive states. Mol. Cell. Biol. 16:3807-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzaki, Y., M. Yoshizumi, S. Kagami, A. H. Koyama, Y. Taketani, H. Houchi, K. Tsuchiya, E. Takeda, and T. Tamaki. 2002. Hydrogen peroxide stimulates c-Src-mediated big mitogen-activated protein kinase 1 (BMK1) and the MEF2C signaling pathway in PC12 cells: potential role in cell survival following oxidative insults. J. Biol. Chem. 277:9614-9621. [DOI] [PubMed] [Google Scholar]
- 31.Traub, O., and B. C. Berk. 1998. Laminar shear stress: mechanisms by which endothelial cells transduce an atheroprotective force. Arterioscler. Thromb. Vasc. Biol. 18:677-685. [DOI] [PubMed] [Google Scholar]
- 32.Wang, N., L. Verna, N. G. Chen, J. Chen, H. Li, B. M. Forman, and M. B. Stemerman. 2002. Constitutive activation of peroxisome proliferator-activated receptor-gamma suppresses pro-inflammatory adhesion molecules in human vascular endothelial cells. J. Biol. Chem. 277:34176-34181. [DOI] [PubMed] [Google Scholar]
- 33.Yan, C., H. Luo, J. D. Lee, J. Abe, and B. C. Berk. 2001. Molecular cloning of mouse ERK5/BMK1 splice variants and characterization of ERK5 functional domains. J. Biol. Chem. 276:10870-10878. [DOI] [PubMed] [Google Scholar]
- 34.Yan, C., M. Takahashi, M. Okuda, J. D. Lee, and B. C. Berk. 1999. Fluid shear stress stimulates big mitogen-activated protein kinase 1 (BMK1) activity in endothelial cells: dependence on tyrosine kinases and intracellular calcium. J. Biol. Chem. 274:143-150. [DOI] [PubMed] [Google Scholar]
- 35.Yan, Z., and A. M. Jetten. 2000. Characterization of the repressor function of the nuclear orphan receptor retinoid receptor-related testis-associated receptor/germ cell nuclear factor. J. Biol. Chem. 275:35077-35085. [DOI] [PubMed] [Google Scholar]
- 36.Yoh, S. M., V. K. Chatterjee, and M. L. Privalsky. 1997. Thyroid hormone resistance syndrome manifests as an aberrant interaction between mutant T3 receptors and transcriptional corepressors. Mol. Endocrinol. 11:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshizumi, M., J. Abe, J. Haendeler, Q. Huang, and B. C. Berk. 2000. Src and cas mediate JNK activation but not ERK1/2 and p38 kinases by reactive oxygen species. J. Biol. Chem. 275:11706-11712. [DOI] [PubMed] [Google Scholar]
- 38.Yue, T. L., J. Chen, W. Bao, P. K. Narayanan, A. Bril, W. Jiang, P. G. Lysko, J. L. Gu, R. Boyce, D. M. Zimmerman, T. K. Hart, R. E. Buckingham, and E. H. Ohlstein. 2001. In vivo myocardial protection from ischemia/reperfusion injury by the peroxisome proliferator-activated receptor-gamma agonist rosiglitazone. Circulation 104:2588-2594. [DOI] [PubMed] [Google Scholar]
- 39.Zhou, G., Z. Q. Bao, and J. E. Dixon. 1995. Components of a new human protein kinase signal transduction pathway. J. Biol. Chem. 270:12665-12669. [DOI] [PubMed] [Google Scholar]