Abstract
The Nrf2 transcription factor promotes survival following cellular insults that trigger oxidative damage. Nrf2 activity is opposed by the BTB/POZ domain protein Keap1. Keap1 is proposed to regulate Nrf2 activity strictly through its capacity to inhibit Nrf2 nuclear import. Recent work suggests that inhibition of Nrf2 may also depend upon ubiquitin-mediated proteolysis. To address the contribution of Keap1-dependent sequestration versus Nrf2 proteolysis, we identified the E3 ligase that regulates Nrf2 ubiquitination. We demonstrate that Keap1 is not solely a cytosolic anchor; rather, Keap1 is an adaptor that bridges Nrf2 to Cul3. We demonstrate that Cul3-Keap1 complexes regulate Nrf2 polyubiquitination both in vitro and in vivo. Inhibition of either Keap1 or Cul3 increases Nrf2 nuclear accumulation, leading to promiscuous activation of Nrf2-dependent gene expression. Our data demonstrate that Keap1 restrains Nrf2 activity via its capacity to target Nrf2 to a cytoplasmic Cul3-based E3 ligase and suggest a model in which Keap1 coordinately regulates both Nrf2 accumulation and access to target genes.
The Nrf2 transcription factor regulates the expression of antioxidant genes following cellular insults that induce oxidative stress (2, 3, 4, 13). Under homeostatic conditions, Nrf2 remains in an inactive cytoplasmic form through association with the bricabrac, tramtrack, and broad complex (BTB) domain-containing protein Keap1 (17). In response to endoplasmic reticulum stress or oxidative stress, Nrf2-Keap1 dissociation is triggered and Nrf2 accumulates in the nucleus, where it forms an active heterodimeric transcription factor, inducing the transcription of target genes involved in redox homeostasis (5, 6, 14, 15, 17).
Although it is clear that Keap1 can maintain Nrf2 in the cytoplasm, accumulation of many transcriptional regulators is also suppressed through the action of the 26S proteasome. Although it was initially thought that Nrf2 activation was strictly regulated through inhibition of nuclear import, increasing evidence indicates that Nrf2 protein levels are maintained at low levels through proteasome-mediated degradation (18, 20, 21, 24, 28). The fact that Keap1 has been implicated in both Nrf2 cytoplasmic sequestration and proteolysis suggests a model in which the regulation of Nrf2 activity is tightly regulated by proteolysis in the cytoplasmic compartment. Similar modes of regulation have been documented for other critical cellular regulators, such as p53 (9, 22) and cyclin D1 (8).
In general, proteins are targeted to the 26S proteasome through the covalent attachment of polyubiquitin chains. Ubiquitin conjugation is mediated by the sequential activities of an E1 enzyme, which mediates the ATP-dependent activation of ubiquitin, an E2 ubiquitin-conjugating enzyme (Ubc), and an E3 ubiquitin ligase; E2 and E3 function to coordinate the transfer of ubiquitin to the substrate protein. In addition to functioning in ubiquitin transfer, E3 generally drives substrate specificity and has thus been of intense interest.
The SCF ligases are among the best characterized of the known E3 ligases (7). The SCF complex is composed of Skp1, Cullin 1 (Cul1), an F-box protein that serves as a substrate specific adaptor protein, and the ring finger protein Rbx1/Roc1/Hrt1 (7). While SCF ligases containing Cul1 are known to regulate proteolytic degradation of a variety of cellular proteins, relatively few substrates have been identified for the related Cul3 protein or Cul3-containing complexes. Recent work from several groups revealed that Cul3 is targeted to ubiquitination substrates via adaptor proteins containing the BTB domain (10, 11, 23, 27). These BTB domain-containing proteins direct Cul3 binding, via the BTB domain, and substrate specificity through an independent protein-protein interaction domain; domains implicated in mediating substrate specific interactions include kelch repeat domains, ankyrin repeat domains, and MATH domains (10, 11, 27). The only documented substrate for the Cul3-BTB ligase thus far is the Caenorhabditis elegans MEI-1 protein, a regulator of meiotic progression (10, 23, 27).
The recent finding that BTB proteins can function as substrate-specific adaptors for Cul3-based E3 ligases suggests that Keap1 might bridge Nrf2 to Cul3. As such, Keap1 would participate directly in the regulation of Nrf2 polyubiquitination and subsequent 26S proteasome-mediated degradation. Here we demonstrate that in addition to maintaining Nrf2 in the cytoplasm, Keap1-Cul3 complexes act as Nrf2-specific E3 ubiquitin ligases that direct Nrf2 polyubiquitination and destruction via the 26S proteasome. We further demonstrate that both Keap1-dependent cytoplasmic sequestration and Cul3-dependent ubiquitination are required to prevent premature Nrf2 activation. Cellular stresses such endoplasmic reticulum stress and oxidative stress trigger release of Nrf2 from Keap1-Cul3 complexes, resulting in the accumulation of Nrf2 and increased expression of Nrf2 target genes. Our data reveal Nrf2 as a substrate for a Cul3-BTB (Keap1)-based E3 ligase, the first to be identified in mammalian cells.
MATERIALS AND METHODS
Tissue culture conditions, baculoviruses, and plasmids.
Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, antibiotics, and glutamine (Mediatech, Inc.). 293T cells were a gift from C. Sherr. PERK−/− mouse embryo fibroblasts (MEFs) were a gift from D. Cavener (29). Transfections were performed with Lipofectamine Plus reagent (Life Technologies). HA-Nrf2, GST-Nrf2, Myc-Keap1, His-Keap1, and the 4xARE reporter construct (containing four tandem repeats of the antioxidant response element (ARE) from glutathione S-transferase-Ya) were described previously (6).
The N-terminal 418 amino acids of human Cul3 were subcloned into pCDNA3 to result in Cul3N418. The His-Keap1 baculovirus expression vector was constructed by inserting His-Keap1 (6) into pVL1392 with XbaI and BamHI. The Cul3 baculoviral expression vector was generated by inserting Cul3 cDNA harboring an N-terminal Flag tag into pVL1393. The Cul3 and Keap1 baculoviruses were generated with the Baculogold transfection kit (Pharmingen). To knock down endogenous human Cul3, hairpins were synthesized and cloned into pSuperretro (Oligoengine) with target sequences 1 (GTACTAAGTCAGGTGTAAC) and 2 (GGTGCGAGAAGATGTACTA). Knockdown of human Keap1 was achieved by synthesizing hairpins that were cloned into the shuttle vector pSHAG and then into pMSCV target sequences 1 (GGGAGCAGGGCATGGAGGTGGTGTCCATTG) and 2 (ACCTGGAGCGAGGTGACCCGAATGACCAG). Vectors encoding short hairpin RNA against firefly luciferase were a gift from P. Klein. To achieve knockdown, 293T cells were transfected with 2 μg of the short hairpin RNA vectors and harvested 36 h posttransfection. Keap1 mutants (28) were gifts from M. Hannink, Ubc5 constructs (19) were gifts from A. M. Weissman, Flag-Cul3 was a gift from J. Singer, and His-ubiquitin was a gift from S. Fuchs. MG132, cycloheximide, tunicamycin, and tert-butyl-hydroquinone were purchased from Sigma.
Reverse transcription-PCR.
For detection of Keap1 mRNA by reverse transcription-PCR, total RNA was extracted from 293T cells expressing short hairpin RNA against firefly luciferase or Keap1 with Trizol (Invitrogen) and digested with RQ1 DNase. Reverse transcription reactions were performed with Superscript II reverse transcriptase (Invitrogen) and oligo(dT) priming following the manufacturer's instructions. Keap1 was amplified with primers against a nonconserved region of Keap1 (5′-GGGAGGTGGCCAAGCAAGAGG-3′ and 5′-TCACCTGCGTGGGCTTGTGCAG-3′), and glyceraldehhyde-3-phosphate dehydrogenase was amplified as a control.
Immunofluorescence.
Cells proliferating on glass coverslips were cotransfected with plasmids encoding Myc-Keap1 and Flag-Cul3. Cells were fixed in 3% paraformaldehyde and permeabilized in 0.1% Triton in phosphate-buffered saline. The Myc epitope was detected with the Jac6 monoclonal antibody, and the Flag epitope was detected with the M2 monoclonal antibody (Sigma). Cells were stained with either fluorescein isothiocyanate-conjugated or biotinylated immunoglobulin G and Texas Red-streptavidin (Vector) secondary antibodies. DNA was detected with Hoechst dye 33258 (Sigma). Cells were visualized with a Nikon microscope fitted with appropriate filters.
Immunoprecipitation and immunoblotting.
To detect interactions between ectopically expressed proteins, 293T cells were transfected with the indicated expression vectors and treated as indicated. Cells were lysed in 50 mM Tris (pH 7.5)-1% NP-40-150 mM NaCl, and Flag-Cul3 was immunoprecipitated with the M2 monoclonal antibody (Sigma). Immunoprecipitated complexes and whole-cell lysates were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes (Osmonics) for immunoblot analysis. Myc-Keap1 and Myc-Keap1ΔBTB were detected with the 9E10 monoclonal antibody, hemagglutinin (HA)-Nrf2 was detected with either the 12CA5 monoclonal antibody or a polyclonal anti-Nrf2 antibody (Santa Cruz), and Flag-Cul3 and Cul3N418 were detected with a polyclonal anti-Cul3 antibody (Zymed or Santa Cruz). To detect endogenous protein levels and interactions, cells were lysed in EBC buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 0.5% NP-40, 1 mM EDTA). Endogenous Nrf2 was detected via immunoprecipitation with polyclonal anti-Nrf2 antibodies followed by immunoblot analysis with the same antibody. Endogenous Cul3 was detected via immunoblot analysis with anti-Cul3 polyclonal antibodies. In experiments utilizing proteasome inhibitors, buffers were supplemented with N-ethylmaleimide (Sigma). Transfection efficiency was routinely monitored via cotransfection with a vector encoding green fluorescent protein.
In vitro binding assays.
To assess Keap1 binding to Nrf2, glutathione S-transferase (GST)-Nrf2 immobilized on beads was mixed with Calicin, Keap1, or Keap1 mutants, in vitro transcribed and translated in the presence of [35S]methionine for 2 h at 4°C. Complexes were washed with NETN buffer (20 mM Tris [pH 8.0], 150 mM NaCl, 1 mM EDTA, 0.5% NP-40), resolved via SDS-PAGE, and visualized via autoradiography. To assess Nrf2 binding to Cul3, Nrf2, in vitro transcribed and translated in the presence of [35S]methionine was mixed with immunopurifed Cul3 or Keap1-Cul3 complexes from insect cells for 2 h at 4°C. Complexes were then washed with NETN lacking EDTA, resolved via SDS-PAGE, and visualized by autoradiography.
Reporter assays and cytotoxicity assays.
For reporter assays, 293T cells were transfected with the indicated plasmids and treated as indicated, and luciferase assays were carried out according to the manufacturer's instructions (dual luciferase reporter assay system; Promega) with a luminometer (PE Applied Biosystems). Firefly luciferase activity was normalized against Renilla luciferase activity from the same lysates. To assess cell death, wild-type and PERK−/− MEFs were transfected as indicated. Cells were treated with 2.5 μg of tunicamycin per ml for the indicated intervals and stained with propidium iodide. The percentage of propidium iodide-positive cells was determined via fluorescence and light microscopy.
Metabolic labeling.
293T cells were transfected as indicated and pooled 24 h posttransfection. Twelve hours later, the cells were cultured for 30 min in methionine- and cysteine-free Dulbecco's modified Eagle's medium supplemented with 10% dialyzed fetal calf serum and then pulsed with 150 μCi of [35S]methionine-cysteine (Amersham) per ml for 1 h. The cells were then changed to complete methionine- and cysteine-containing medium for the indicated intervals. Cells were lysed in NP-40 lysis buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid), and HA-Nrf2 was precipitated from cell lysates with anti-Nrf2 antibodies. Proteins were resolved by SDS-PAGE and visualized by autoradiography or by phosphorimaging.
In vivo ubiquitination assays.
293T cells transfected with the indicated plasmids were collected in phosphate-buffered saline, and 10% was used to make whole-cell lysates in EBC buffer. The remaining 90% were lysed in buffer 1 (6 M guanidine HCl, 0.1 M sodium phosphate, 0.01 M Tris [pH 8], 10 mM β-mercaptoethanol) supplemented with 5 mM imidazole. Cleared lysates were incubated with Talon affinity beads (BD Biosciences) overnight at 4°C. The beads were then sequentially washed with buffer 1, buffer 2 (8 M urea, 0.1 M sodium phosphate, 0.01 M Tris [pH 8], 10 mM β-mercaptoethanol), buffer 3 (8 M urea, 0.1 M sodium phosphate, 0.01 M Tris [pH 6.3], 10 mM β-mercaptoethanol, 0.2% Triton X-100), and buffer 4 (8 M urea, 0.1 M sodium phosphate, 0.01 M Tris [pH 6.3], 10 mM β-mercaptoethanol, 0.1% Triton X-100). All buffers were supplemented with N-ethylmaleimide. Bound proteins were eluted in sample buffer supplemented with 200 mM imidazole and resolved via SDS-PAGE. Following transfer to nitrocellulose membranes (Osmonics), the presence of Nrf2, Cul3, and Keap1 was detected via Western analysis.
In vitro ubiquitination assays.
Nrf2 ubiquitination was performed with in vitro [35S]methionine-labeled Nrf2 in the presence of Ubc5 (200 ng), ubiquitin (1 mg/ml), ubiquitin aldehyde (2 μM), in vitro-transcribed and -translated Keap1 or Keap1 mutants, ATP (4 mM), an ATP-regenerating system (20 mM creatine phosphate, 0.2 mg of creatine phosphokinase per ml) and protease and proteasome inhibitors at 30°C for 60 min. All chemicals were purchased from Sigma.
RESULTS
Formation of Nrf2-Cul3 complexes depends upon Keap1.
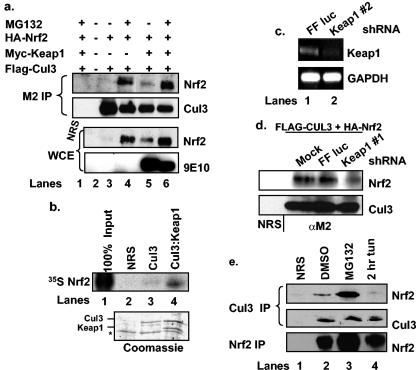
To begin to address the role of Cul3 in Nrf2 protein accumulation, we determined whether Nrf2 was present in Cul3 complexes in vivo. Lysates prepared from 293T cells transfected with vectors encoding Flag-Cul3 and HA-Nrf2 in the absence or presence of Myc-Keap1 were precipitated with the M2 monoclonal antibody. In the absence of ectopically expressed Keap1, Nrf2-Cul3 association was evident only when cells had been treated with the proteasome inhibitor MG132 (Fig. 1a, compare lanes 3 and 4). In cells expressing Cul3, Nrf2, and Keap1, a weak interaction was detected between Cul3 and Nrf2 (Fig. 1, lane 5). Both Nrf2 accumulation and association with Cul3 were restored by treatment of cells with MG132 (Fig. 1, lane 6). Coprecipitation of Myc-Keap1 was also detected in these experiments (data not shown). These data indicate that Nrf2 associates with Cul3 in cells and suggested that coexpression of Cul3 and Keap1 might specifically trigger Nrf2 proteolysis (see Fig. 3 to 7). We also tested the ability of Nrf2 to bind to a Keap1-Cul3 complex in vitro. Nrf2, in vitro transcribed and translated in the presence of [35S]methionine, was mixed with a purified Keap1-Cul3 complex or with purified Cul3 alone. While Nrf2 was able to efficiently bind the Keap1-Cul3 complex, binding was not observed between Nrf2 and Cul3 in the absence of Keap1 (Fig. 1b).
FIG. 1.
Nrf2 associates with a Cul3 complex in a Keap1-dependent manner. (a) 293T cells were transfected with plasmids encoding HA-Nrf2, Flag-Cul3, and Myc-Keap1 and treated as indicated. Flag-Cul3 was precipitated from whole-cell extracts (WCE) with the M2 monoclonal antibody. Cul3-associated Nrf2 was detected via immunoblot analysis. The presence of ectopic proteins was confirmed via immunoblot analysis. Lane 1 shows a control precipitation with a nonspecific antiserum (NRS). (b) Nrf2, in vitro transcribed and translated in the presence of [35S]methionine, was mixed with purified Flag-Cul3 (lane 3) or His-Keap1-Flag-Cul3 complexes (lane 4) from insect Sf9 cells. Lane 1 shows 100% input, and lane 2 shows a control precipitation with a nonspecific antiserum. Nrf2 was visualized by autoradiography. The bottom panel shows Coomassie staining. The asterisk indicates a nonspecific coprecipitating protein. (c) Following transfection of 293T cells with the indicated short hairpin RNA vectors, Keap1 mRNA levels were assessed by reverse transcription-PCR. (d) 293T cells were transfected with Flag-Cul3 and HA-Nrf2 in combination with short hairpin RNA vectors against firefly luciferase (lane 3) or Keap1 (lane 4). Following MG132 treatment, Flag-Cul3 was precipitated from cell lysates with the M2 antibody; associated Nrf2 was assessed via immunoblot analysis. Lane 1 shows a control precipitation with a nonspecific antiserum. (e) 293T cells were treated with 10 μM MG132 for 4 h (lane 3), 5 μg of tunicamycin per ml for 2 h (lane 4), or vehicle alone (lane 2). Cul3 was precipitated from whole-cell lysates with a Cul3-specific antibody, and Cul3-associated Nrf2 was assessed via immunoblot analysis. Total Nrf2 and Cul3 levels were determined via immunoblot analysis. Lane 1 shows a control precipitation with a nonspecific antiserum.
FIG. 3.
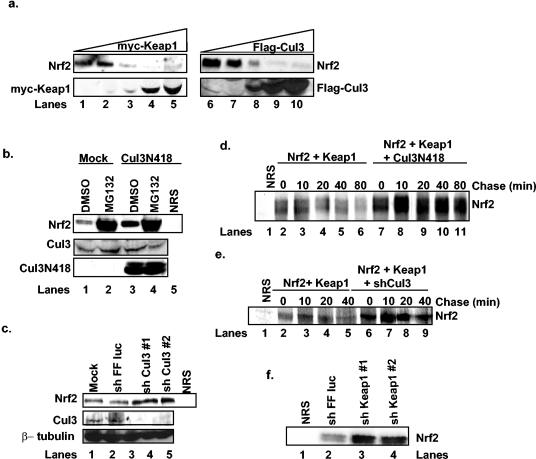
Inactivation of Cul3 stabilizes Nrf2. (a) 293T cells were transfected with increasing concentrations of plasmids encoding Myc-Keap1 (lanes 1 to 5) or Flag-Cul3 (lanes 6 to 10). The expression of Nrf2, Myc-Keap1, and Flag-Cul3 was assessed via immunoblot. (b) 293T cells were mock transfected (lanes 1 and 2) or transfected with a plasmid encoding the N-terminal 418 residues of Cul3 (lanes 3 to 5) and treated with dimethyl sulfoxide (DMSO) or 10 μM MG132 for 4 h. Total Nrf2 levels were detected via immunoprecipitation followed by immunoblot analysis, and total Cul3 levels were determined by immunoblot analysis. Lane 5 shows a control precipitation with a nonspecific antiserum. (c) 293T cells were mock transfected (lane 1) or transfected with plasmids expressing short hairpin RNAs against firefly luciferase (lane 2) or Cul3 (lanes 3 and 4). Total Nrf2 levels were detected by immunoprecipitation followed by immunoblot analysis. Total Cul3 and β-tubulin levels were determined via immunoblot analysis. Lane 5 shows a control precipitation with a nonspecific antiserum. (d) 293T cells transfected with plasmids encoding HA-Nrf2 and Myc-Keap1 in the absence (lanes 2 to 6) or presence of a plasmid encoding the N-terminal 418 residues of Cul3 (lanes 7 to 11) were pulsed with [35S]methionine followed by the addition of complete methionine-containing medium for the indicated intervals. Nrf2 was immunoprecipitated from whole-cell lysates, and proteins were resolved via SDS-PAGE and visualized by autoradiography. Lane 1 shows a control precipitation with a nonspecific antiserum. (e) 293T cells transfected with plasmids encoding HA-Nrf2 and Myc-Keap1 in the absence (lanes 2 to 5) or presence of a plasmid encoding short hairpin RNA against Cul3 (lanes 6 to 9) were pulsed with [35S]methionine followed by the addition of complete, methionine-containing medium for the indicated intervals. Nrf2 was precipitated from whole-cell lysates, and proteins were resolved via SDS-PAGE and visualized via phosphorimaging. Lane 1 shows a control precipitation with a nonspecific antiserum. (f) 293T cells were transfected with plasmids encoding short hairpin RNAs against firefly luciferase (lane 2) or Keap1 (lanes 3 and 4). Total Nrf2 levels were determined via immunoprecipitation followed by immunoblot analysis.
FIG. 7.
In vitro reconstitution of Nrf2 ubiquitination. (a) In vitro-transcribed and -translated Nrf2 was mixed with ATP, ubiquitin, Ubc5, and in vitro-transcribed and -translated Keap1 in in vitro ubiquitination assays for 1 h at 30oC. In lanes 7 to 10, the reactions were carried out for 15, 30, 60, and 90 min, respectively. (b) In vitro-transcribed and -translated Keap1 (lane 1), Keap1ΔBTB (lane 4), Keap1ΔKelch (lane 3), or Calicin (lane 2) was mixed with in vitro-transcribed and -translated Nrf2, ATP, ubiquitin, and Ubc5 in in vitro ubiquitination assays, as in a, for 1 h at 30°C. (c) In vitro-transcribed and -translated Keap1 (lanes 1 to 3), Keap1ΔBTB (lanes 4 to 6), Keap1ΔKelch (lanes 7 to 9), and Calicin (lanes 10 to 12) were mixed with GST-Nrf2 (lanes 3, 6, 9, and 12) or the negative control, GST (lanes 2, 5, 8, and 11) and washed, and proteins were resolved via SDS-PAGE. Lanes 1, 4, 7, and 10 show 10% input. Proteins were visualized via autoradiography.
The in vitro binding data are consistent with Keap1's functioning as a bridge between Nrf2 and Cul3. However, the in vivo association of Nrf2 with Cul3 in the absence of ectopic Keap1 suggested either that Nrf2 can bind directly to Cul3 in cells or that association was mediated by endogenous Keap1. To address this issue, we knocked down endogenous Keap1 levels through the use of short hairpin RNAs. As suitable antibodies for detection of endogenous Keap1 are not available, we confirmed Keap1 knockdown by reverse transcription-PCR (Fig. 1c). In cells transfected with short hairpin RNA directed towards Keap1, we observed a marked decrease in the Nrf2-Cul3 association (Fig. 1d, compare lanes 2 to 4) that was comparable to the degree to which Keap1 was knocked down. Together, these results indicate that the interaction between Cul3 and Nrf2 is dependent upon the Keap1 protein and suggest that, following stress-dependent liberation of Nrf2 from Keap1, Nrf2 does not bind Cul3.
As the above interactions were assessed with ectopic protein, we were compelled to determine whether endogenous Nrf2 was associated with Cul3 complexes. Cul3 was precipitated from 293T cells with a Cul3-specific antiserum, and the presence of associated Nrf2 was assessed via immunoblot analysis with an Nrf2-specific antiserum. Nrf2 was detected in Cul3 precipitates in asynchronously proliferating cells (Fig. 1e, lane 2); increased association was apparent in cells treated with MG132 (lane 3).
We and others have previously shown that Nrf2 dissociates from Keap1 following endoplasmic reticulum stress or oxidative stress (6, 17). If Nrf2-Cul3 association depends upon Keap1 as an adaptor, then endoplasmic reticulum stress or oxidative stress should reduce the abundance of the Nrf2-Cul3 complex. We therefore determined whether endogenous Nrf2-Cul3 binding is regulated by cellular stress. Cul3 was precipitated from 293T cells treated with tunicamycin, a drug that elicits the unfolded-protein response and promotes Nrf2 nuclear localization (6), with a Cul3-specific antiserum, and the presence of associated Nrf2 was assessed via immunoblot analysis. In contrast to vehicle-treated cells, Cul3-Nrf2 association was decreased in cells treated with tunicamycin (5 μg/ml) (Fig. 1e, compare lanes 2 and 4).
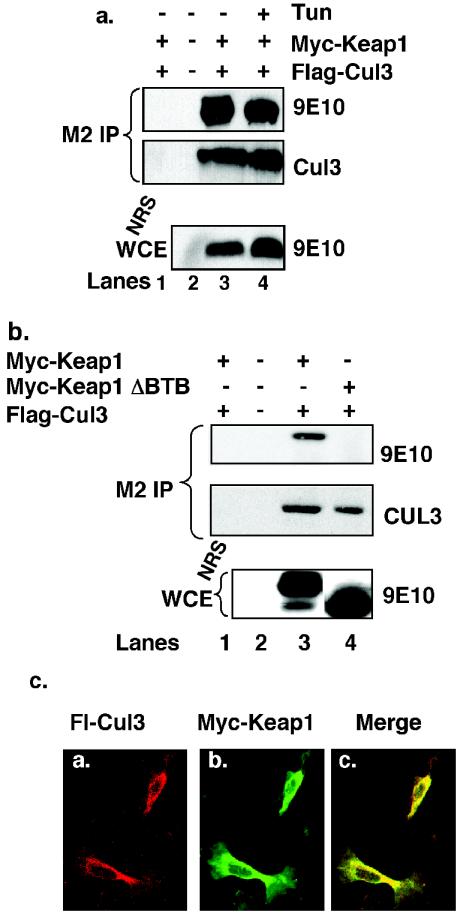
The data provided thus far demonstrate that Nrf2 associates with Cul3 in a Keap1-dependent manner and that endoplasmic reticulum stress induces the dissociation of Nrf2 from the Cul3 complex. Based on previous work, we reasoned that this dissociation reflects the disruption of Nrf2-Keap1 complexes rather than loss of Keap1-Cul3 interaction. Consistent with this notion, Cul3 remained bound to Keap1 throughout a course of tunicamycin treatment (Fig. 2a, compare lane 3 to 4). Thus, while endoplasmic reticulum stress triggers Nrf2 release from Keap1, it does not abolish Cul3-Keap1 association, suggesting that Keap1 is constitutively associated with Cul3.
FIG. 2.
Cul3 interacts with Keap1 through its BTB domain. (a) 293T cells transfected with plasmids encoding Myc-Keap1 and Flag-Cul3 were either left untreated (lanes 1 to 3) or treated with 5 μg/ of tunicamycin per ml for 1 h (lane 4). Flag-Cul3 was precipitated from whole-cell extracts with the M2 monoclonal antibody; Flag-Cul3 (middle panel) and Cul3-associated Keap1 were detected by immunoblot analysis with the 9E10 antibody (top panel). The presence of ectopically expressed Myc-Keap1 was confirmed by immunoblot analysis with the 9E10 antibody (bottom panel). Lane 1 is a control precipitation with an irrelevant antibody. (b) 293T cells were transfected with plasmids encoding Myc-Keap1, Myc-Keap1ΔBTB, or Flag-Cul3. Flag-Cul3 was precipitated from whole-cell lysates with the M2 monoclonal antibody, and Cul3-associated Keap1 was detected by immunoblot with the 9E10 antibody. Expression of ectopically expressed proteins was confirmed via immunoblot analysis. Lane 1 is a control precipitation with a nonspecific antiserum. (c) NIH 3T3 cells proliferating on glass coverslips were transfected with plasmids expressing Myc-Keap1 and Flag-Cul3. Cells were fixed and examined by indirect immunofluorescence for the presence of the Myc and Flag epitopes.
Because Cul3 binds directly to BTB domains, we hypothesized that Keap1 should bind to Cul3 via its N-terminal BTB domain. To address this notion, 293T cells were transfected with plasmids encoding Flag-Cul3 and Myc-Keap1 or a Keap1 mutant which lacks the BTB domain. Lysates were subjected to precipitation with a Flag-specific antibody, and coprecipitating proteins were visualized by immunoblot with epitope-specific antibodies. As anticipated, Keap1 was detected in the Cul3 precipitates (Fig. 2b, lane 3). In contrast, Keap1ΔBTB did not coprecipitate with Cul3 (Fig. 2b, lane 4). These data demonstrate that Cul3-Keap1 association is dependent upon the Keap1 BTB domain. Consistent with the binding data, immunofluorescent staining revealed colocalization of Keap1 and Cul3 in the cytoplasm of asynchronously proliferating cells (Fig. 2c). These data suggest that Keap1 and Cul3 form cytoplasmic complexes.
Cul3 regulates Nrf2 degradation.
Our data provide evidence for protein complexes, which are minimally composed of Nrf2-Keap1-Cul3, consistent with the possibility that Cul3-Keap1 could direct Nrf2 ubiquitin-dependent proteolysis. We reasoned that if Cul3 and Keap1 regulated Nrf2 proteolysis, overexpression of Cul3 or Keap1 should result in reduced Nrf2 protein levels. We transfected 293T cells with increasing concentrations of plasmids encoding either Myc-Keap1 or Flag-Cul3 and assessed their effect on endogenous Nrf2 protein accumulation. With increasing levels of either Myc-Keap1 or Flag-Cul3, we noted a concomitant decrease in Nrf2 levels, as determined by immunoblot (Fig. 3a).
As an independent test of this hypothesis, we determined the ability of a dominant negative Cul3 mutant (Cul3N418), which is defective in Rbx1 binding but retains the capacity to associate with BTB domains (10), to increase steady-state Nrf2 levels. 293T cells were transfected with a plasmid encoding Cul3N418, and endogenous Nrf2 protein levels were assessed by immunoblot. Cells expressing Cul3N418 contained elevated Nrf2 levels compared to mock-transfected cells (Fig. 3b, compare lanes 1 and 3). We also used vectors encoding short hairpin RNAs to reduce endogenous Cul3 levels. 293T cells expressing the Cul3 short hairpin RNA contained significantly reduced Cul3 levels (Fig. 3c, compare lanes 1 to 4). As a consequence of Cul3 knockdown, Nrf2 levels were markedly higher in these cells than in either mock-transfected cells or cells expressing short hairpin RNA against firefly luciferase (compare lanes 1 to 4). These results demonstrate that a reduction in Cul3 function contributes to increased Nrf2 protein accumulation.
We next measured the half-life of Nrf2 in cells expressing Cul3N418 and short hairpin RNA directed towards Cul3. Pulse-chase analysis revealed that Nrf2 is rapidly turned over in control transfected cells (Fig. 3d, lanes 2 to 6; 3e, lanes 2 to 5), in agreement with previous data (20, 21, 24, 28). However, expression of Cul3N418 dramatically stabilized Nrf2, as little turnover was evident during the course of the experiment (Fig. 3d, lanes 7 to 11). Likewise, the Nrf2 half-life was significantly extended in cells expressing short hairpin RNA directed towards Cul3 (Fig. 3e, lanes 6 to 9). These data suggest that Cul3 and Keap1 are required for rapid Nrf2 proteolysis.
If Keap1 functions as the adaptor that bridges Nrf2 and Cul3, then loss of Keap1 should promote Nrf2 accumulation. Cells were transfected with a vector encoding two independent Keap1-specific short hairpin RNAs. As predicted, in cells expressing short hairpin RNAs specific for Keap1, basal Nrf2 protein accumulated relative to the level in those expressing control short hairpin RNA (Fig. 3f, compare lanes 2 to 4).
Cul3-dependent proteolysis limits Nrf2 transcriptional activity.
If Cul3-dependent proteolysis limits the threshold of Nrf2 accumulation in the absence of stress, loss of Cul3 should result in promiscuous Nrf2 activation and increased basal expression of Nrf2 target genes. To address the relative contribution of Cul3-dependent degradation of Nrf2 versus Keap1-dependent cytoplasmic sequestration, we assessed Nrf2 function in cells in which either Keap1 or Cul3 was knocked down by assessing expression of a luciferase reporter plasmid containing an Nrf2-responsive element antioxidant response element (ARE) (6). As expected, Keap1 knockdown dramatically increased Nrf2-dependent reporter activity, over 20-fold above that of vector-transfected cells (Fig. 4a), indicating that, in accordance with published results (25), Keap1 is necessary for basal repression of Nrf2 activity. In addition, Cul3 knockdown increased expression of the Nrf2 reporter plasmid approximately fourfold (Fig. 4a). It is important to note that reporter gene expression in these assays is measured without treating cells with agents known to trigger either endoplasmic reticulum stress or oxidative stress. The difference in reporter gene expression observed between the Keap1 and Cul3 knockdowns likely reflects the capacity of Keap1 to maintain Nrf2 in the cytoplasm under conditions of Cul3 knockdown and therefore prevent nuclear access to a majority of the accumulating Nrf2.
FIG. 4.
Cul3 contributes to the regulation of Nrf2-dependent gene expression. (a) 293T cells were transfected with the 4×ARE firefly luciferase reporter and a plasmid encoding Renilla luciferase in the absence or presence of plasmids expressing short hairpin RNA against Keap1 or Cul3. Cells were collected, and luciferase activity was measured with a luminometer. Error bars represent the standard deviation for three independent experiments. (b) Same as panel a except that cells were either mock treated or treated with 100 μM tBHQ for 16 h. (c) Mock-transfected wild-type MEFs and PERK−/− MEFs transfected as indicated were treated with 2.5 μg of tunicamycin (Tun) per ml for 0, 2, or 4 h. Cells were stained with propidium iodide, and the percentage of propidium iodide-positive cells (y axis) was determined. Error bars represent the standard deviation for three independent experiments.
Increased expression of an Nrf2 target gene in cells in which Cul3 is knocked down suggests that, in the absence of continued proteolysis, Nrf2 levels exceed those of Keap1, thereby permitting the accumulation of Keap1-free Nrf2. Implicit to this model is that a significant proportion of Nrf2 remains bound to Keap1 in inactive complexes. If so, cellular stress that liberates Nrf2 from Keap1 in the Cul3 knockdown cells should result in a synergistic increase in Nrf2-dependent gene expression. To test this possibility, cells were mock transfected or transfected with a Cul3-specific short hairpin RNA vector along with an Nrf2-dependent luciferase reporter plasmid. These cells were treated with vehicle control or the oxidative stress-inducing agent tert-butylhydroquinone (tBHQ; 100 μM). As above, knockdown of Cul3 resulted in a reproducible fourfold increase in basal ARE reporter activity (Fig. 4b, compare panels 1 and 3). Addition of tBHQ resulted in a greater than 30-fold induction of reporter activity (Fig. 4b, panel 4). Consistent with our hypothesis, we noted that tBHQ-dependent ARE reporter induction in the presence of Cul3 knockdown exceeded that achieved by tBHQ treatment alone (Fig. 4b, compare panels 2 and 4).
Loss of the endoplasmic reticulum stress-inducible PERK kinase greatly sensitizes cells to the proapoptotic effects of the glycosylation inhibitor tunicamycin (12). We have previously shown that overexpression of Nrf2, a downstream target of PERK, restores cellular redox balance in PERK−/− murine embryonic fibroblasts (MEFs) challenged with tunicamycin, consistent with Nrf2's functioning as a mediator of PERK-dependent survival (5). As independent confirmation that loss of Cul3 function permits promiscuous Nrf2 activation, we assessed whether overexpression of the dominant negative Cul3N418 mutant would decrease the sensitivity of PERK-deficient MEFs to endoplasmic reticulum stress-induced cell death. PERK−/− MEFs were transfected with empty vector or vectors encoding Cul3N418 or, as a control, Nrf2. Cells were then treated with 2.5 μg of tunicamycin per ml, and cell death was assessed by propidium iodide exclusion (Fig. 4c). Tunicamycin treatment rapidly induced cell death in PERK−/− MEFs relative to that observed for wild-type MEFs. As predicted, ectopic expression of Nrf2 reduced the early onset of cell death noted for PERK−/− MEFs, as did expression of Cul3N418. Taken together, these data demonstrate that Cul3 and Keap1 mediate Nrf2 protein stability and activity and together oppose Nrf2-dependent, ARE-dependent gene expression.
Endoplasmic reticulum stress-dependent Nrf2-Keap1 dissociation reduces Nrf2 proteolysis.
Endoplasmic reticulum stress or oxidative stress is predicted to promote increased Nrf2 accumulation, given that it reduces Nrf2-Cul3 binding (Fig. 1e). We assessed Nrf2 levels in 293T cells that were either left untreated or treated with tunicamycin (5 μg/ml) or tBHQ (100 μM). Increased levels of Nrf2 were observed in cells that had been treated with tunicamycin or tBHQ (Fig. 5a, compare lanes 1 to 3). Longer exposures were required for detection of Nrf2 in untreated cells (data not shown). These results are in agreement with previous data that demonstrated PERK-dependent increases in Nrf2 levels in response to glucose deprivation (5) and oxidative stress (18, 20, 21, 24, 28).
FIG. 5.
Endoplasmic reticulum or oxidative stress stabilizes Nrf2. (a) 293T cells were mock treated (lane 1), treated with 100 μM tBHQ for 8 h (lane 2), or treated with 5 μg of tunicamycin (Tun) (lane 3) per ml for 1 h. Total Nrf2 levels were detected via immunoprecipitation followed by immunoblot analysis. Lane 4 shows a control precipitation with a nonspecific antiserum. (b) NIH 3T3 cells were treated for 30 min with dimethyl sulfoxide (DMSO) (lanes 2 to 5) or with 5 μg of tunicamycin per ml (lanes 6 to 9) before the addition of 10 μM cycloheximide (CHX) for the indicated intervals. Nrf2 was detected by immunoprecipitation followed by immunoblot analysis. Lane 1 shows a control precipitation with a nonspecific antiserum.
While PERK activity enhances Nrf2 protein accumulation, the previous experiment did not address whether Nrf2 is more stable following the cellular stress. To address this issue, NIH 3T3 cells were left untreated or treated with tunicamycin for 30 min. Following induction of the endoplasmic reticulum stress response, cells were exposed to cycloheximide for different intervals and Nrf2 loss was assessed by immunoblot with an Nrf2-specific antiserum. As expected, the half-life of Nrf2 was shorter in untreated cells than in tunicamycin-treated cells (Fig. 5b, compare lanes 2 to 5 and 6 to 9). Together, these results suggest that Nrf2 stability increases following endoplasmic reticulum or oxidative stress via the dissolution of Cul3-Keap1-Nrf2 complex formation.
Cul3 promotes Nrf2 ubiquitination.
Our data demonstrate that Cul3 mediates Nrf2 protein stability under homeostatic conditions. As Cul3 is known to regulate protein degradation via its capacity to direct polyubiquitination, we hypothesized that Cul3-Keap1 complexes likely direct Nrf2 polyubiquitination. In agreement with previous data, we found that treatment of cells with proteasome inhibitors led to Nrf2 accumulation as well as the accumulation of higher-molecular-weight forms of Nrf2, consistent with ubiquitin conjugation (Fig. 6a, compare lanes 2 and 3).
FIG. 6.
Cul3 promotes Nrf2 polyubiquitination. (a) Lysates were collected from 293T cells that were mock treated (lane 2) or treated with 10 μM MG132 (lane 3) for 2 h. Nrf2 was detected via immunoprecipitation followed by immunoblot analysis. Lane 1 shows a control precipitation. (b) 293T cells were transfected with plasmids encoding 6xHis-ubiquitin, Nrf2, Myc-Keap1, Flag-Cul3, and Cul3N418 in the indicated combinations. Cells were lysed under denaturing conditions, and ubiquitin-containing complexes were affinity purified and resolved via SDS-PAGE. The presence of Nrf2, Cul3, and Keap1 was determined via immunoblot analysis.
To more directly assess whether Nrf2 is subject to polyubiquitination and to assess the importance of Cul3 in this process, we performed in vivo ubiquitination assays. 293T cells were transfected with combinations of plasmids encoding 6× His-ubiquitin, Nrf2, Myc-Keap1, Flag-Cul3, and Cul3N418. Ubiquitin conjugates were purified from the lysates via Ni2+ affinity chromatography under denaturing conditions, and the presence of ubiquitin-conjugated Nrf2 was assessed via immunoblot analysis with Nrf2 antiserum (Fig. 6b, top panel). Expression of the indicated proteins was confirmed in whole-cell lysates (bottom panels). In the absence of ectopically expressed Cul3, little ubiquitinated Nrf2 was seen (Fig. 6b, top panel, lanes 1 to 4), although detectable levels of Nrf2 ubiquitination were observed in the presence of ectopic Keap1 (Fig. 6b, top panel, lane 4). Cul3 expression, in the absence or presence of ectopically expressed Keap1, greatly enhanced Nrf2 polyubiquitination (Fig. 6b, lanes 5 and 6). As expected, expression of Cul3N418, which is defective for Rbx1 association, did not promote Nrf2 ubiquitination (Fig. 6b, lane 7). We also noted decreased endogenous Nrf2 polyubiquitination in cells expressing short hairpin RNA against Cul3 (data not shown).
We next asked whether Cul3-Keap1 complexes could promote Nrf2 ubiquitination in vitro. Rbx1-Cul3-Keap1 complexes were assembled by coupled transcription and translation of Keap1 in reticulocyte extracts. These complexes were then mixed with recombinant Ubc5, ubiquitin, ATP, and [35S]methionine-labeled Nrf2. In the presence of Ubc5, high-molecular-weight forms of Nrf2 were readily apparent (Fig. 7a, lanes 4 and 5). This alteration in Nrf2 mobility was strictly dependent upon Keap1-Nrf2 interactions, as incubation with empty reticulocyte lysates or with Keap1ΔKelch complexes, a mutant Keap1 that cannot bind Nrf2 (Fig. 7c), could not support Nrf2 ubiquitination (Fig. 7b, lane 3). Importantly, Calicin, a BTB and kelch domain-containing protein that binds to Cul3 (data not shown) but not Nrf2 (Fig. 7c), could not promote Nrf2 ubiquitination (Fig. 7b, lane 2). To assess the importance of Cul3 in Nrf2 ubiquitination, the reactions were carried out in the presence of lysates expressing Keap1ΔBTB, a mutant that can bind Nrf2 (Fig. 7c) but not Cul3 (Fig. 2b). Ubiquitination of Nrf2 was not supported in these reactions (Fig. 7b, lane 4), indicating that direct association of Cul3 with Keap1 is necessary for Keap1-dependent Nrf2 ubiquitination. Taken together, our in vivo and in vitro data strongly indicate that Keap1 serves as a specificity factor for Cul3-dependent Nrf2 ubiquitination and degradation.
DISCUSSION
Keap1 and Cul3 are the core subunits of an Nrf2-specific SCF3 ubiquitin ligase.
The Nrf2 transcription factor regulates inducible expression of antioxidant and drug metabolism genes (1, 6, 16, 26). Under normal growth conditions, Nrf2 activity is buffered by the BTB domain-containing protein Keap1 (17, 20, 25, 28, 30). While the Keap1-dependent regulation of Nrf2 was thought to reflect cytosolic sequestration of Nrf2, accumulating evidence suggests that Keap1 binding might also target Nrf2 for proteolysis (18, 20, 21, 24, 28). Whether Keap1 participates directly in Nrf2 proteolysis was not established in the previous studies. Recent work from several laboratories has implicated proteins containing the BTB domain as scaffolding proteins that target Cul3-based ubiquitin ligase complexes to specific protein substrates (10, 11, 23, 27). Here, we demonstrate that Keap1-Cul3 complexes function as critical subunits of an E3 ligase that targets Nrf2 for 26S proteasome-dependent destruction. We demonstrate that in cells cultured under homeostatic conditions, endogenous Nrf2 is present in Cul3 complexes. Initiation of either an endoplasmic reticulum stress response or oxidative stress, both of which are known to trigger dissociation of Keap1-Nrf2 complexes (6, 17), results in the accumulation of Keap1- and Cul3-free Nrf2, suggesting that Nrf2-Cul3 association is dependent upon Keap1. Consistent with this notion, knockdown of endogenous Keap1 with short hairpin RNA efficiently reduced Nrf2-Cul3 association.
While the interdependence of Cul3 and Nrf2 on Keap1 for association is suggestive, it does not demonstrate that Nrf2 is subject to Keap1-Cul3-mediated polyubiquitination and subsequent degradation. Several pieces of data support our conclusion that Keap1 and Cul3 constitute essential components of an Nrf2-specific E3 ligase. First, inhibition of Cul3 via overexpression of a dominant inhibitory allele or expression of Cul3-specific short hairpin RNA resulted in an increase in steady-state Nrf2 levels that correlated with decreased Nrf2 turnover, as assessed by pulse-chase analysis. Additionally, the increased Nrf2 accumulation correlated with increased Nrf2 activity, as assessed by analysis of Nrf2-dependent gene expression. This suggests that Cul3 provides an essential level of regulation in the maintenance of regulated Nrf2 activity. Second, agents that trigger Nrf2-Keap1 dissociation, such as tunicamycin, also promote Nrf2 accumulation and decreased Nrf2 proteolysis. Third, overexpression of Cul3 increased Nrf2 polyubiquitination and decreased Nrf2 protein levels, while knockdown of Cul3 (or expression of dominant inhibitory Cul3) decreased Nrf2 polyubiquitination. Finally, we demonstrated that Cul3-Keap1 complexes can support Nrf2 polyubiquitination in vitro.
Once again, ubiquitination was dependent upon the Cul3-Keap1 interaction, as a Keap1 mutant lacking the BTB domain did not support Nrf2 ubiquitination. The inability of the Keap1ΔKelch mutant to support Nrf2 ubiquitination demonstrates that direct Keap1-Nrf2 association is also required. In addition, it is important to note that ubiquitination of Nrf2 is presumably dependent upon Rbx1 or an Rbx1-like factor, given that Cul3 mutants deficient in Rbx1 binding both stabilized Nrf2 and inhibited Nrf2 polyubiquitination. Collectively, our data strongly suggest that Keap1 functions as the substrate-specific adaptor for an SCF3-type E3 ligase (27) that targets Nrf2 for Cul3-dependent ubiquitination. As such, Nrf2 is the first identified mammalian target of a BTB domain-containing ubiquitin ligase.
Cul3 as a negative regulator of a cellular stress response pathway.
Under homeostatic conditions, Nrf2 is maintained in the cytoplasm via association with Keap1 (17). Targeted deletion of Keap1 results in constitutive nuclear accumulation of Nrf2 and ensuing overexpression of Nrf2 target genes, culminating in postnatal lethality due to hyperkeratosis in the esophagus and forestomach (25). The compound deletion of Keap1 and Nrf2 rescues this phenotype, demonstrating that Nrf2 is the critical downstream target of Keap1 (25). Likewise, our data demonstrate that chronic knockdown of Cul3 also results in constitutive Nrf2 activation and increased basal expression of Nrf2 target genes, consistent with Cul3's functioning as a negative regulator of Nrf2. These results are consistent with other work suggesting that cytoplasmic proteolysis maintains continuously low Nrf2 protein levels (18, 20, 21, 24, 28).
Our data demonstrate that Keap1 opposes Nrf2 activation via two complementary mechanisms. The first is cytoplasmic sequestration (17). The second is by bridging Nrf2 to Cul3 and thereby targeting Nrf2 for proteolysis. This mode of regulation ensures that in response to cellular stresses such as endoplasmic reticulum stress, cells are capable of quickly activating a pool of Nrf2, leading to gene expression patterns that counteract the toxic effects brought on by the stress.
If Keap1 can prevent Nrf2 nuclear entry, is Cul3-dependent proteolysis an essential regulatory component? Loss of Cul3 did not provide the robust activation of Nrf2-dependent gene expression that is witnessed upon Keap1 loss. This likely reflects the capacity of Keap1, which is still present in Cul3 knockdown experiments, to maintain a majority of Nrf2 in the cytoplasm. The strong induction of Nrf2 activity upon Keap1 inactivation, in contrast to that observed in the Cul3 knockdown, reflects relief of both Nrf2 degradation and Keap1-dependent sequestration. However, as loss of Cul3 does permit activation of Nrf2 in the absence of stress and increased cell survival following endoplasmic reticulum stress, Cul3-dependent degradation is a critical regulatory mode for Nrf2. Our data suggest a model in which Keap1 is the central negative regulator of Nrf2 but is also likely to be the limiting component. The rapid degradation of Nrf2 therefore maintains a balance of Nrf2-Keap1 and thereby prevents promiscuous activation of Nrf2 in the absence of an appropriate signal.
Unlike the canonical SCF E3 ligase, for which substrate recognition relies upon both substrate phosphorylation and a substrate-specific F-box constituent, Nrf2-Keap1 binding is negatively regulated by Nrf2 phosphorylation. Cellular stresses such as those initiated by glucose/nutrient restriction (5, 6) and treatment of cells with agents that promote oxidative damage (15) trigger phosphorylation-dependent Nrf2-Keap1 dissociation, allowing the nuclear accumulation of Nrf2 and subsequent increase in the expression of Nrf2 target genes (5, 6, 15). Under conditions of endoplasmic reticulum stress, the PERK kinase triggers Nrf2 phosphorylation (6), while under conditions of oxidative stress, protein kinase C-dependent phosphorylation (15) triggers Nrf2-Keap1 dissociation. Our results place Cul3 as a critical negative regulator of stress-induced gene transcription. The identification of BTB domain-containing proteins, of which there are more than 200 in mammals, as regulators of Cul3-mediated proteolysis suggests that Cul3 is likely to regulate numerous cellular processes, similar to the many pathways known to be regulated by other Cullin family members.
Acknowledgments
We thank J. Singer for providing Flag-Cul3, S. Fuchs for helpful advice and providing His-ubiquitin, G. Hannon for providing the pSHAG and pMSCV vectors, C. J. Sherr for providing 293T cells, P. S. Klein for providing short hairpin RNA vectors against firefly luciferase, A. M. Weissman for providing Ubc5 constructs, Y. W. Kan and J. Chan for providing Nrf2 cDNA, D. Cavener for providing PERK−/− MEFs, M. Hannink for providing mutant Keap1 constructs, M. Olson for providing the Jac6 antibody, and R. Woolery for excellent technical assistance.
This work was supported by the Abramson Family Cancer Research Institute and National Institutes of Health (NIH) grant CA104838 (J.A.D.), NIH grant AG11085 (J.W.H.), and Department of Defense grant DAMD17-02-1-0284 (J.J.).
REFERENCES
- 1.Alam, J., D. Stewart, C. Touchard, S. Boinapally, A. M. Choi, and J. L. Cook. 1999. Nrf2, a cap'n'collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 274:26071-26078. [DOI] [PubMed] [Google Scholar]
- 2.Buetler, T. M., E. P. Gallagher, C. Wang, D. L. Stahl, J. D. Hayes, and D. L. Eaton. 1995. Induction of phase I and phase II drug-metabolizing enzyme mRNA, protein, and activity by BHA, ethoxyquin, and oltipraz. Toxicol. Appl. Pharmacol. 135:45-57. [DOI] [PubMed] [Google Scholar]
- 3.Chan, J. Y., and M. Kwong. 2000. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim. Biophys. Acta 1517:19-26. [DOI] [PubMed] [Google Scholar]
- 4.Chan, K., and Y. W. Kan. 1999. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl. Acad. Sci. USA 96:12731-12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullinan, S. B., and J. A. Diehl. 2004. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following ER stress. J. Biol. Chem. 279:20076-20087. [DOI] [PubMed] [Google Scholar]
- 6.Cullinan, S. B., D. Zhang, M. Hannink, E. Arvisais, R. J. Kaufman, and J. A. Diehl. 2003. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol. Cell. Biol. 23:7198-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 8.Diehl, J. A., M. Cheng, M. F. Roussel, and C. J. Sherr. 1998. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12:3499-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freedman, D. A., and A. J. Levine. 1998. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 18:7288-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa, M., Y. J. He, C. Borchers, and Y. Xiong. 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5:1001-1007. [DOI] [PubMed] [Google Scholar]
- 11.Geyer, R., S. Wee, S. Anderson, J. Yates, and D. A. Wolf. 2003. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell 12:783-790. [DOI] [PubMed] [Google Scholar]
- 12.Harding, H. P., Y. Zhang, A. Bertolotti, H. Zeng, and D. Ron. 2000. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5:897-904. [DOI] [PubMed] [Google Scholar]
- 13.Hayes, J. D., S. A. Chanas, C. J. Henderson, M. McMahon, C. Sun, G. J. Moffat, C. R. Wolf, and M. Yamamoto. 2000. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochem. Soc. Trans. 28:33-41. [DOI] [PubMed] [Google Scholar]
- 14.Hayes, J. D., and M. McMahon. 2001. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett. 174:103-113. [DOI] [PubMed] [Google Scholar]
- 15.Huang, H. C., T. Nguyen, and C. B. Pickett. 2000. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc. Natl. Acad. Sci. USA 97:12475-12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh, K., T. Chiba, S. Takahashi, T. Ishii, K. Igarashi, Y. Katoh, T. Oyake, N. Hayashi, K. Satoh, I. Hatayama, M. Yamamoto, and Y. Nabeshima. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236:313-322. [DOI] [PubMed] [Google Scholar]
- 17.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, T. O'Connor, and M. Yamamoto. 2003. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8:379-391. [DOI] [PubMed] [Google Scholar]
- 19.Jensen, J. P., P. W. Bates, M. Yang, R. D. Vierstra, and A. M. Weissman. 1995. Identification of a family of closely related human ubiquitin conjugating enzymes. J. Biol. Chem. 270:30408-30414. [DOI] [PubMed] [Google Scholar]
- 20.McMahon, M., K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 278:21592-21600. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen, T., P. J. Sherratt, H. C. Huang, C. S. Yang, and C. B. Pickett. 2003. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 278:4536-4541. [DOI] [PubMed] [Google Scholar]
- 22.O'Keefe, K., H. Li, and Y. Zhang. 2003. Nucleocytoplasmic shuttling of p53 is essential for MDM2-mediated cytoplasmic degradation but not ubiquitination. Mol. Cell. Biol. 23:6396-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pintard, L., J. H. Willis, A. Willems, J. L. Johnson, M. Srayko, T. Kurz, S. Glaser, P. E. Mains, M. Tyers, B. Bowerman, and M. Peter. 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425:311-316. [DOI] [PubMed] [Google Scholar]
- 24.Stewart, D., E. Killeen, R. Naquin, S. Alam, and J. Alam. 2003. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J. Biol. Chem. 278:2396-2402. [DOI] [PubMed] [Google Scholar]
- 25.Wakabayashi, N., K. Itoh, J. Wakabayashi, H. Motohashi, S. Noda, S. Takahashi, S. Imakado, T. Kotsuji, F. Otsuka, D. R. Roop, T. Harada, J. D. Engel, and M. Yamamoto. 2003. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 35:238-245. [DOI] [PubMed] [Google Scholar]
- 26.Wild, A. C., H. R. Moinova, and R. T. Mulcahy. 1999. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 274:33627-33636. [DOI] [PubMed] [Google Scholar]
- 27.Xu, L., Y. Wei, J. Reboul, P. Vaglio, T. H. Shin, M. Vidal, S. J. Elledge, and J. W. Harper. 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425:316-321. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, D. D., and M. Hannink. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23:8137-8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, P., B. McGrath, S. Li, A. Frank, F. Zambito, J. Reinert, M. Gannon, K. Ma, K. McNaughton, and D. R. Cavener. 2002. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell. Biol. 22:3864-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zipper, L. M., and R. T. Mulcahy. 2002. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J. Biol. Chem. 277:36544-36552. [DOI] [PubMed] [Google Scholar]