SUMMARY
Cellular compartments that cannot be biochemically isolated are challenging to characterize. Here we demonstrate the proteomic characterization of the synaptic clefts that exist at both excitatory and inhibitory synapses. Normal brain function relies on the careful balance of these opposing neural connections, and understanding how this balance is achieved relies on knowledge of their protein compositions. Using a spatially restricted enzymatic tagging strategy, we mapped the proteomes of two of the most common excitatory and inhibitory synaptic clefts in living neurons. These proteomes reveal dozens of synaptic candidates, and assign numerous known synaptic proteins to a specific cleft type. The molecular differentiation of each cleft allowed us to identify Mdga2 as a potential specificity factor influencing Neuroligin-2's recruitment of presynaptic neurotransmitters at inhibitory synapses.
Graphical Abstract
INTRODUCTION
The mammalian brain is capable of complex cognition because individual nerve cells assemble into higher order circuits that receive, process, store, and transmit information. Central to this information flow are chemical synapses, specialized junctions between communicating neurons that mediate neurotransmitter release and recognition. Because synapse function, along with their formation, remodeling, and elimination, are so central to brain function, there is tremendous interest in dissecting the molecular architecture and functional properties of synapses.
Microscopy and mass spectrometry (MS)-based proteomics have been applied extensively to study the protein composition of synapses. Though powerful in its ability to provide spatial context, microscopy is limited when specific antibodies against target proteins are not available, or when recombinant tagging raises concerns about mislocalization or overexpression. Microscopy is also low-throughput, and more often applied to validate hypotheses than to perform an unbiased search for novel proteins.
MS-based proteomics, on the other hand, is ideally suited for high-throughput and unbiased detection of endogenous proteins. However, it sacrifices spatial information because analysis is performed after cell lysis. Fractionation schemes, such as to enrich entire synaptic terminals (i.e., synaptosomes) (Biesemann et al., 2014), synaptic vesicles (Takamori et al., 2006), the post-synaptic density (PSD) (Bayés et al., 2012), and the active zone (Boyken et al., 2013), recover some spatial information, but vary greatly in their degrees of purity. For example, mitochondrial, nuclear, and glial (Henn et al., 1976) contaminants are common in synaptosome and PSD preparations, and key proteins are frequently lost (Figure S1). In addition, fractionation usually blends across many synapse types. Synaptosome purifications, for example, do not distinguish between excitatory glutamate-releasing synapses and inhibitory GABA-releasing synapses, whose molecular compositions are quite different, due to their antagonistic functions. Finally, a major limitation of purification-based MS approaches is that many of the subdomains of the synapse are impossible to purify and therefore inaccessible to MS proteomic analysis. This includes the synaptic cleft and the inhibitory post-synaptic region, which lacks a detergent-insoluble “density” (in contrast to the excitatory PSD).
Recently, we (Rhee et al., 2013) and others (Roux et al., 2012) have reported approaches to proteomic mapping that bypass the need for organelle or subdomain purification, and instead target “promiscuous” tagging enzymes to the subcellular region of interest (APEX or BioID, respectively). In live cells, addition of a small molecule substrate triggers enzyme-catalyzed biotinylation of its neighboring endogenous proteins (Figure 1A-B). Subsequently, biotinylated proteins are enriched with streptavidin beads and identified by MS. The main advantages of this approach are that unpurifiable cellular regions, such as the synaptic cleft, can in principle be targeted for proteomic mapping, and the resulting data are potentially more accurate, because tagging is performed in living cells while membranes and protein complexes are still intact, and artifacts resulting from detergent lysis and serial centrifugation are avoided.
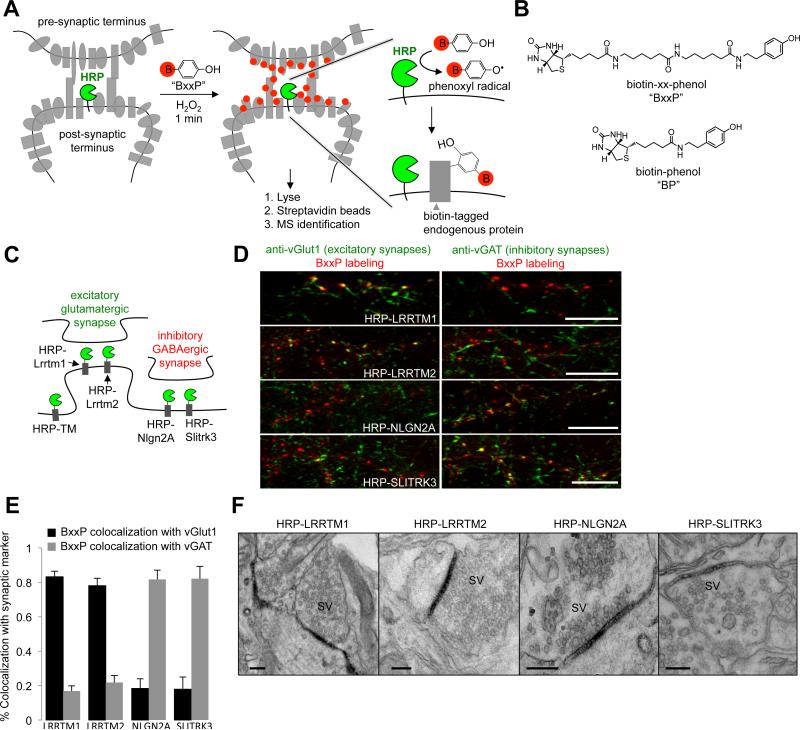
Figure 1. Design and characterization of peroxidase fusion constructs for proximity biotinylation.
(A) Scheme of peroxidase-mediated proteomic tagging in the synaptic cleft. Horseradish peroxidase (HRP) is genetically targeted to the cleft via fusion to a known cleft protein. The grey shapes are endogenous proteins residing inside and outside the synapse. To initiate labeling, the membrane impermeant biotin-phenol conjugate BxxP (red B = biotin; chemical structure in (B)) is added to the live neurons for 1 minute together with the oxidant H2O2. HRP converts BxxP into a phenoxyl radical, which covalently tags proximal endogenous proteins at electron rich side-chains such as Tyr (Rhee et al., 2013). Subsequently, neurons are lysed and biotinylated proteins are isolated using streptavidin beads for identification by mass spectrometry (MS). (B) Structure of BxxP and BP probes. (C) HRP fusion constructs employed in this study. HRP-TM is a general cell surface construct. (D) Fluorescence imaging of synaptic HRP fusion constructs with respect to excitatory and inhibitory synapse markers, vGlut1 and vGAT. For maximum detection sensitivity, the HRP constructs were visualized via BxxP labeling followed by neutravidin-AlexaFluor647 staining (red). Scale bars, 10 m. See also Figures S1-S4 for additional characterization of constructs and their expression levels. (E) Quantitation of colocalization extent for images in (D) plus 7 other fields of view containing >900 puncta per construct. Errors, ±1 s.d. (F) Electron microscopy (EM) of HRP fusion constructs. HRP catalyzes the polymerization and local deposition of diaminobenzidine, which recruits electron-dense osmium (Martell et al., 2012). SV, synaptic vesicles. Scale bars, 200 nm. See also Figure S2C for additional EM.
Though such technology could be highly enabling for the study of synapse molecular architecture, numerous hurdles must be surmounted. First, neither APEX nor BioID have been demonstrated in neurons. Second, we have only used APEX for proteomic mapping in membrane-enclosed compartments (the mitochondrial matrix (Rhee et al., 2013) and mitochondrial intermembrane space (Hung et al., 2014)), not “open” subcellular regions such as the synaptic cleft. It is unclear what spatial specificity is achievable in such an environment. BioID is expected to have a larger labeling radius than APEX, because the half-life of its reactive intermediate, biotin-AMP, is minutes in water, in contrast to <1 msec for the biotin-phenoxyl radical generated by APEX-catalyzed oxidation. Third, synapses constitute a tiny fraction of neurons by mass. It is unclear if existing protocols can enrich a small biotinylated proteome over the much larger nonbiotinylated proteome; previous APEX studies have targeted much more abundant structures, such as mitochondria. Fourth, it is challenging to localize APEX activity specifically to synaptic subdomains of interest. Though cleft-resident proteins are known, all of these also have pools elsewhere in the neuron, such as the secretory pathway. How can we achieve specific proteomic tagging only in the cleft and avoid capturing the endoplasmic reticulum (ER) or Golgi proteomes as well?
Here we describe modifications to the APEX technique to enable successful proteomic mapping of the neuronal synaptic cleft. To begin to probe the vast diversity of synapses in the brain, we generate two independent proteomic lists: one representative of excitatory, glutamate-releasing synapses, and one representative of inhibitory, GABA-releasing synapses. We analyze these proteomes to show that specificity and depth-of-coverage are both higher than for previous proteomes obtained by biochemical fractionation. Follow-up imaging and western blotting provide synapse validation for ten proteomic hits. Finally, we perform functional studies in neuron culture on two post-synaptic membrane proteins identified in our excitatory and inhibitory proteomes, Mdga1 and Mdga2, and uncover differences in their regulatory effects on Nlgn2, suggesting potential roles in setting synapse specificity.
RESULTS
ESTABLISHING A PEROXIDASE-BASED PLATFORM FOR PROTEOMIC MAPPING OF THE SYNAPTIC CLEFT
Though APEX tagging has been performed in Drosophila larval muscle (Chen et al., 2015), reagent delivery to tissue was a concern, in addition to H2O2 toxicity. We therefore opted to use dissociated neuron culture, rather than intact brain tissue, to allow for rapid delivery and washout of biotin-phenol, H2O2, and subsequent peroxidase quenchers. At DIV19 (19 days in vitro), rat cortical neuron cultures displayed abundant staining of synapse markers (Bassoon, vGlut1, and vGAT), and synapses looked normal by electron microscopy (Figures 1F and S2C).
When designing the peroxidase fusion constructs to use for proteomic mapping in the synaptic cleft, our first concern was to maximize the activity of the peroxidase, because the proteomic region of interest is so small. APEX2 is the second-generation, more active variant of APEX (Lam et al., 2014). However, the commonly used horseradish peroxidase (HRP) enzyme is even more active than APEX2. For applications in the cytosol, nucleus, and mitochondria, HRP cannot be used, because its four structurally essential disulfide bonds do not form in reducing environments, leaving HRP inactive (Martell et al., 2012). However, HRP is active in the oxidizing secretory pathway and cell surface, and catalyzes the same labeling chemistry as APEX2 with biotin-phenol (Rees et al., 2015; Rhee et al., 2013). We therefore generated N-terminal, extracellular-facing fusions of HRP with three known glutamatergic excitatory synaptic cleft-resident proteins (Nlgn1, Lrrtm1, and Lrrtm2 (Linhoff et al., 2009; Song et al., 1999)) and two known GABAergic inhibitory synaptic cleft resident proteins (Slitrk3 and Nlgn2 (Chih et al., 2006; Takahashi et al., 2012; Varoqueaux et al., 2004)) (Figure 1C). Surprisingly, HRP-Nlgn1 exhibited poor specificity for excitatory over inhibitory synapses, by imaging and in a preliminary MS proteomic experiment (Figure S1) and was not characterized further. The remaining four constructs were highly enriched at synapses (Figure S2A-B), gave the expected preference for excitatory versus inhibitory synapses (Figure 1D-E), and showed specific and robust HRP activity in the cleft by electron microscopy (EM) (Figures 1F and S2C). Using lentiviral transduction, we titrated down expression levels to 40-70% that of the endogenous protein or mRNA counterpart (Figure S3A-C). Under these expression conditions, we did not observe changes in synapse size or density in transduced neurons (Figure S3E-F).
As expected, treatment with biotin-phenol (BP) and H2O2 gave labeling of both cell surface and intracellular proteins, likely in the secretory pathway proximal to ER and Golgi pools of the HRP fused proteins. We reasoned that we could improve specificity for the extracellular cleft proteome by rendering BP membrane impermeant. We synthesized a variant, called BxxP, with a long and polar polyamide linker (Figure 1B). Figure 2A shows that BxxP still gives robust biotinylation with HRP and H2O2, but no longer enters cells (no signal observed with intracellular constructs PSD95-APEX2 and APEX2-NES). When applied to the synaptic HRP fusion constructs, BxxP produced HRP- and H2O2-dependent biotinylation on the neuron surface that was punctate in appearance (Figures 1D, S4A-B). By contrast, HRP-TM, a control construct that targets HRP over the entire neuron surface, produced a diffuse rather than punctate BxxP labeling pattern.
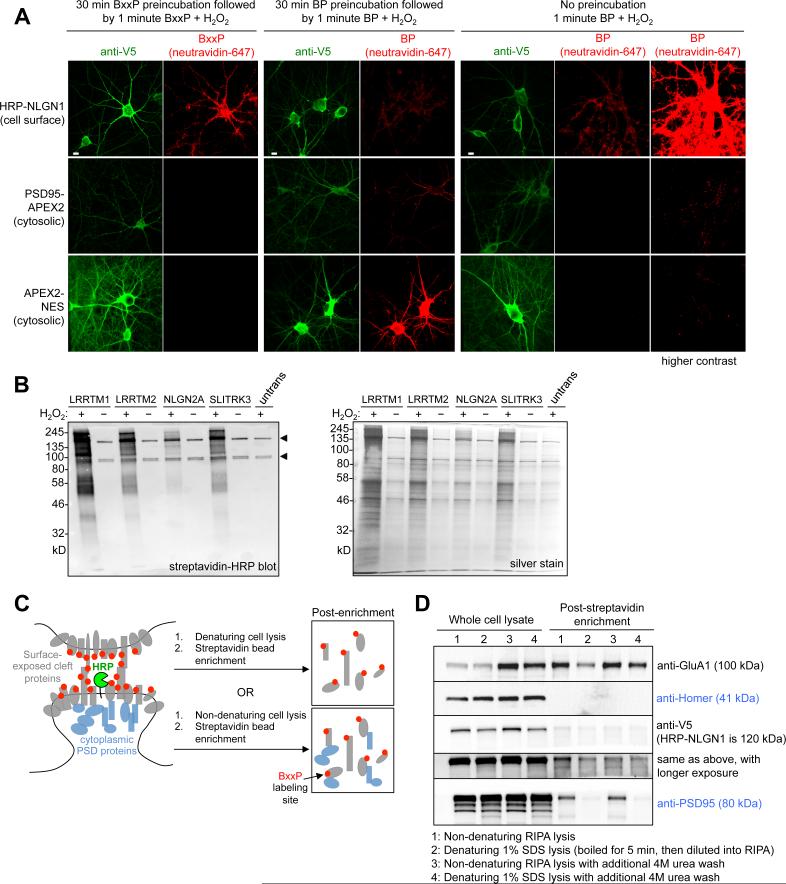
Figure 2. Development of BxxP probe for cell surface labeling and alternative streptavidin enrichment protocol.
(A) Characterization of BxxP membrane impermeability. Neurons expressing the indicated peroxidase fusion construct (left) were labeled live with BxxP or BP, then fixed and stained. HRP at the cell surface gives biotinylation with both BxxP and BP, whereas intracellular peroxidase fusion constructs show biotinylation with BP only. (B) Gel analysis of streptavidin-enriched lysates after live-neuron biotinylation with HRP fusion constructs. Untrans, untransfected neurons. Arrowheads point to endogenously biotinylated proteins (Chapman-Smith and Cronan, 1999). (C) Development of lysis conditions to separate biotinylated cleft proteins from cytosolic post-synaptic density (PSD) proteins. Our standard RIPA lysis (bottom) retrieved many intracellular PSD proteins (blue) along with biotinylated (red) cleft-exposed proteins (grey). (D) Four different lysis/wash conditions were tested to solubilize the PSD and retrieve proteins biotinylated by HRP-NLGN1 and BxxP. Blotting of streptavidin-enriched lysates for a cleft marker (GluA1) and intracellular markers (Homer and PSD95) showed that conditions 2 and 4 removed the latter while retaining the former. Condition 2 was used for large-scale proteomics. Condition 1 was used in previous studies (Hung et al., 2014; Rhee et al., 2013). The anti-V5 blot detects HRP-NLGN1. See also Figure S4.
We ran biotinylated neuron lysates on gel and analyzed them by streptavidin blotting. Figures S4C-D show that each HRP fusion construct biotinylates a wide range of endogenous proteins, in an H2O2-dependent manner. However, when we proceeded to enrich these biotinylated proteins using streptavidin-coated beads according to our previous protocol, we found that endogenous cytosolic proteins such as PSD95, which should not be tagged by HRP and BxxP, were also enriched (Figures 2C-D). We hypothesized that our previous enrichment protocol was not disassembling the detergent-insoluble and tightly crosslinked post-synaptic density (PSD), which was coming down with the biotinylated cleft proteome. We therefore modified the protocol by adding a 1% SDS lysis step with 10 min boiling to disrupt the PSD, and found that this removed undesired cytosolic proteins while preserving enrichment of desired synaptic surface proteins such as GluA1 (Figure 2D). Figure 2B shows streptavidin blot and silver stain visualization of our enriched biotinylated proteomes.
PROTEOMIC MAPPING OF EXCITATORY GLUTAMATERGIC AND INHIBITORY GABAERGIC SYNAPTIC CLEFTS
Previously, we found that a “ratiometric” APEX tagging strategy improved the spatial specificity of protein identifications in the mitochondrial intermembrane space (IMS), a compartment that is leaky to biotin phenoxyl radicals, due to porins in the outer mitochondrial membrane (Hung et al., 2016). In the ratiometric approach, for each detected protein, we compare its extent of biotinylation by targeted peroxidase (e.g., synaptic HRP fusion construct) versus non-targeted peroxidase (e.g., HRP-TM which targets HRP evenly over the entire neuron surface). If a protein is biotinylated more extensively by synaptic HRP than by HRP-TM, we retain it for our proteome. If it is biotinylated more extensively by HRP-TM than by synaptic HRP, we reject it; such proteins may be just outside of synapses, accessible to the biotin radical but not actually a synaptic protein.
Because the synaptic cleft is open and non-membrane-bounded, it represents an even greater challenge than the mitochondrial IMS in terms of spatial specificity. We reasoned that it would be essential to use the ratiometric tagging approach. We also wanted to capitalize on the availability of two validated HRP fusion constructs for each synaptic cleft type – Lrrtm1 and Lrrtm2 for the excitatory glutamatergic cleft, and Slitrk3 and Nlgn2 for the inhibitory GABAergic cleft. An endogenous protein enriched by two different excitatory HRP constructs is much more likely to be a true positive identification than a protein enriched by only one. With these considerations in mind, we designed the three proteomic experiments shown in Figure 3A. Each experiment combines four cellular samples: one biotinylated by an excitatory HRP fusion construct; one biotinylated by an inhibitory HRP fusion construct; one biotinylated by HRP-TM; and one non-biotinylated negative control. The four samples are separately lysed and enriched with streptavidin beads. After on-bead trypsin digestion to peptides, chemical iTRAQ tagging was performed to differentiate the samples by mass signature. The four samples were then combined and analyzed as a pooled mixture by liquid chromatography and tandem MS/MS.
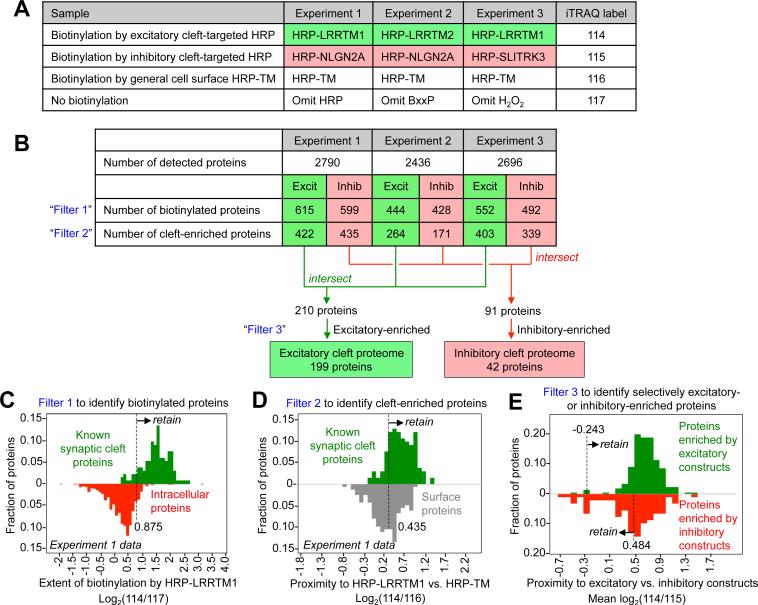
Figure 3. Design of proteomic study and cut-off analysis.
A) Design of three independent proteomic experiments. Each experiment consisted of four samples, which were separately enriched and tagged with unique iTRAQ labels (right). Mass spectrometry (MS) was performed on the mixture of four samples, resulting in four mass-shifted peaks of varying intensity per detected peptide. (B) Filtering of MS data to produce excitatory and inhibitory proteomic lists. The table shows the number of proteins remaining after each filtering step. In the first row, a protein was considered detected if two or more unique peptides were sequenced by MS. Filter 1 retains HRP-biotinylated proteins and removes non-specific bead binders, on the basis of 114/117 or 115/117 iTRAQ ratio. Filter 2 retains cleft-enriched proteins over general cell surface proteins, on the basis of 114/116 or 115/116 iTRAQ ratio. Filter 3 removes strongly inhibitory-enriched proteins (high 115/114 iTRAQ ratio) from the excitatory proteome, and vice versa for the inhibitory proteome. (C)-(E) Histograms that illustrate how Filters 1, 2, and 3 were applied. In (C) and (D), green shows the distribution of true positives. Red in (C) is the distribution of false positives. See also Figure S5 and Table S4.
As shown in Figure 3B, each experiment identified >2400 unique proteins. The vast majority of these were non-specific streptavidin bead binders, identifiable by their low 114/117 or 115/117 iTRAQ ratios. After removing these proteins (“Filter 1”, Figure 3C), we used the 114/116 and 115/116 iTRAQ ratios to identify proteins enriched by synaptic HRP constructs relative to the HRP-TM control construct (“Filter 2”, Figure 3D). Then, the three independent excitatory synaptic cleft datasets were intersected, as well as the three independent inhibitory synaptic cleft datasets. Figures S5B-C illustrate the importance of the three-way intersection to improve the quality of each proteomic list. The last filter (“Filter 3”, Figure 3E) was based on excitatory/inhibitory biotinylation ratio: we removed from the excitatory proteome the small number of proteins that were much more strongly biotinylated by inhibitory HRP constructs than excitatory HRP constructs, and vice versa. After these filtering steps, our final excitatory and inhibitory proteomic lists consisted of 199 (Table S1, Tab 1) and 42 (Table S1, Tab 2) unique proteins, respectively.
CHARACTERISTICS OF THE TWO PROTEOMIC LISTS
As expected, each proteomic list contains numerous ion channels, GPCRs, adhesion proteins, and transporters (Figure 4A). Both post-synaptic membrane proteins and pre-synaptic membrane proteins are represented, in addition to soluble secreted proteins such as Nptx1 (Figure 4B). We also observe some secreted proteins that may be of glial origin, such as Gpc6, which promotes excitatory synapse formation (Allen et al., 2012). Our excitatory synaptic cleft list contains all four AMPA receptor subunits and three NMDA receptor subunits, while seven GABAA receptor subunits appear in the inhibitory synaptic cleft list. The overlap between the two proteomic lists is 20 proteins (Table S1, Tab 3), which includes known dual-localized synaptic proteins such as Erbb4 (Krivosheya et al., 2008), Grik2 (Lerma, 2003), and Gabbr1 (Kulik et al., 2002).
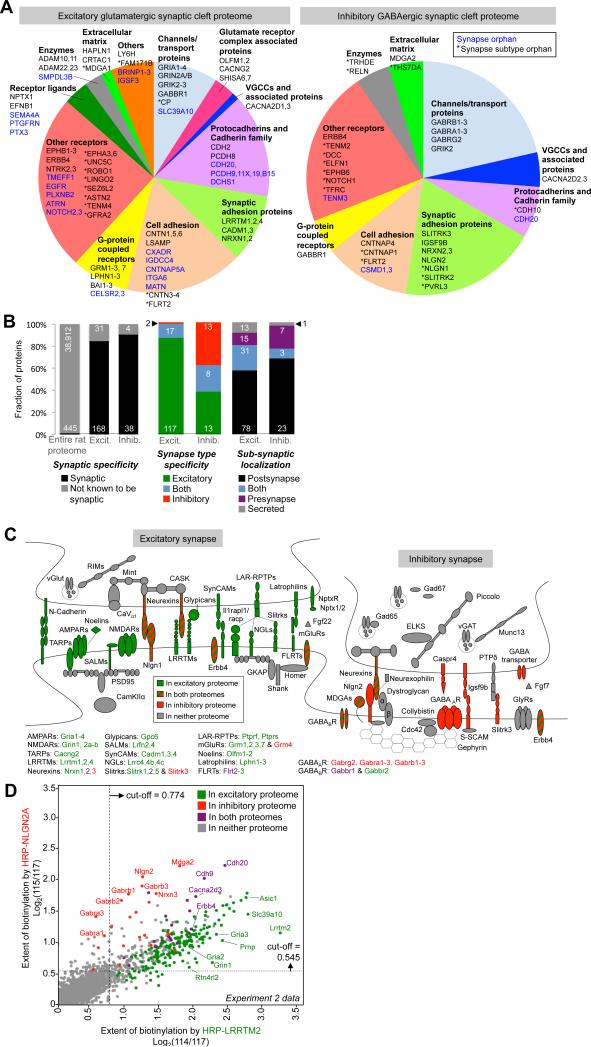
Figure 4. Specificity and coverage of excitatory and inhibitory synaptic cleft proteomes.
(A) Proteins of each proteomic list, subdivided by functional class. Genes in blue have no prior connection to synapses (i.e., they are synapse orphans), while genes with asterisks(*) have no prior connection to that specific synapse type (but are known to be generally synaptic). (B) Graphs showing the synapse specificity (left) and synapse subtype specificity (middle) of the two proteomic lists. Excit. refers to the excitatory proteome of 199 proteins, and Inhib. refers to the inhibitory cleft proteome of 42 proteins. For synapse subtype analysis, only proteins with literature annotation as excitatory/inhibitory/both are included in the analysis; non-annotated synaptic proteins are excluded. On the far right, proteins are classified according to their sub-synaptic localization. Further details provided in Table S1, Tabs 1 and 2. (C) Cartoon depicting well-established excitatory (left) and inhibitory (right) synapse proteins. Proteins are colored according to whether they were detected in our excitatory proteome (green), inhibitory proteome (red), both (striped), or neither (grey). Proteins with multiple isoforms are listed below; purple font indicates detection in both proteomes. (D) Scatter plot showing the separation of proteins by E/I (excitatory/inhibitory) ratio. All proteins detected in Experiment 2 are plotted, by biotinylation extent in the inhibitory cleft (y-axis) versus excitatory cleft (x-axis). Each protein is colored according to whether it is present in either final proteomic list. Points corresponding to some well-established inhibitory, excitatory, and dual-localized synaptic proteins are labeled. Dashed lines indicate the cut-offs applied to the data (Filter 1). See also Figure S5D for additional E/I scatter plots.
To characterize the specificity of each proteomic list, we first checked for intracellular cytosolic proteins. Both lists lack any protein known to be intracellular (Table S1, Tabs 1-2, Column AN), attesting to the effectiveness of BxxP in restricting biotinylation to the cell surface. We then determined synapse specificity by calculating the fraction of each proteome with prior literature connection to synapses. 84% of the excitatory proteome, and 90% of the inhibitory proteome have previous synapse annotation (Figure 4B, left). The remaining 33 proteins (29 from the excitatory proteome, 2 from the inhibitory proteome, and 2 in both; see Table S2) that lack synapse annotation, which we call “synapse orphans”, could be false positives, or they could be newly discovered synaptic proteins. Below, we present imaging and western blotting data on ten of these synapse orphans, supporting the latter possibility.
To determine the excitatory versus inhibitory synapse specificity of each proteomic list, we analyzed the subset of proteins in each list with known excitatory or inhibitory synapse localization (Figure 4B, middle). The excitatory proteome is highly enriched for proteins known to reside at excitatory synapses or at both synapse types (98%). The inhibitory proteome is enriched for known inhibitory synapse proteins (62%), but also includes a significant number of proteins with excitatory annotation. Because the inhibitory synapse is poorly characterized, and the literature is biased towards assays of excitatory synapse localization, it is possible that many of these proteins are actually dual-localized to both excitatory and inhibitory synapses. Indeed, follow-up experiments described below and shown in Figure S6 suggest dual localization for four excitatory-annotated proteins that appear in our inhibitory synaptic cleft proteome.
Figure 4C provides an illustration of the synapse subtype specificity of both proteomic lists, showing for example that AMPA and NMDA receptor subunits are detected only in the excitatory proteome, while GABAA receptor components are detected exclusively in the inhibitory proteome. Interestingly, previous studies have suggested that some inhibitory synapse components “leak over” to excitatory synapses, and vice versa, perhaps to enable cross-talk or regulation between synapse types (Chen et al., 2012). Consistent with these studies, our data show that the inhibitory GABAB receptor subunit Gabbr1 resides at excitatory synapses as well (Kulik et al., 2002), and the excitatory kainate receptor Grik2 can also be found at inhibitory synapses (Lerma, 2003).
Due to our experimental design, every protein appearing in our lists is associated with an E/I (excitatory/inhibitory) ratio, based on the 114/115 iTRAQ ratio, that reflects its enrichment at excitatory versus inhibitory synapses. This can be visualized in the scatter plots shown in Figures 4D and S5D. Known excitatory synapse-specific proteins such as AMPA receptors (Gria2-3) and NMDA receptors (Grin1) appear below the diagonal, whereas known inhibitory synapse-specific proteins such as GABAA receptors (Gabra1,3, Gabrb1-3) lie above the diagonal. Dual localized proteins such as Erbb4 are close to the diagonal.
To characterize the sensitivity, or depth-of coverage, of each proteome, we generated separate lists of well-established excitatory cleft-resident or inhibitory cleft-resident proteins (Table S3). Of the 62 such excitatory proteins, our proteome contained 43 (69%). Of the 24 inhibitory proteins, we enriched 11 (46%). The proteins we failed to enrich may be sterically shielded from biotinylation through protein or membrane interactions in the live cell context. Alternatively, they could be dual-localized, with both a synaptic population and a non-synaptic population, and consequently removed by our Filter 2 step (Figures 3B and D), which considers the ratio of biotinylation by synapse-localized HRP versus general cell surface HRP-TM. Finally, some genes may not be expressed in the specific rat cortical neuron preparations that we used.
PROTEOMES REVEAL NEW SYNAPTIC PROTEINS
Within each proteome, ~85% of proteins have prior literature connection to synapses. The remaining ~15% are “synapse orphans”, with no previous literature assigning them to synapses. We found 29 such orphans in the excitatory cleft, 2 in the inhibitory cleft, and 2 in both clefts (some examples in blue font in Figure 4A; complete list of orphans in Table S2, Tab 1). Guided by the availability of commercial antibodies and transgenes for recombinant expression, we selected 14 excitatory synapse orphans for follow-up analysis (Figure 5A).
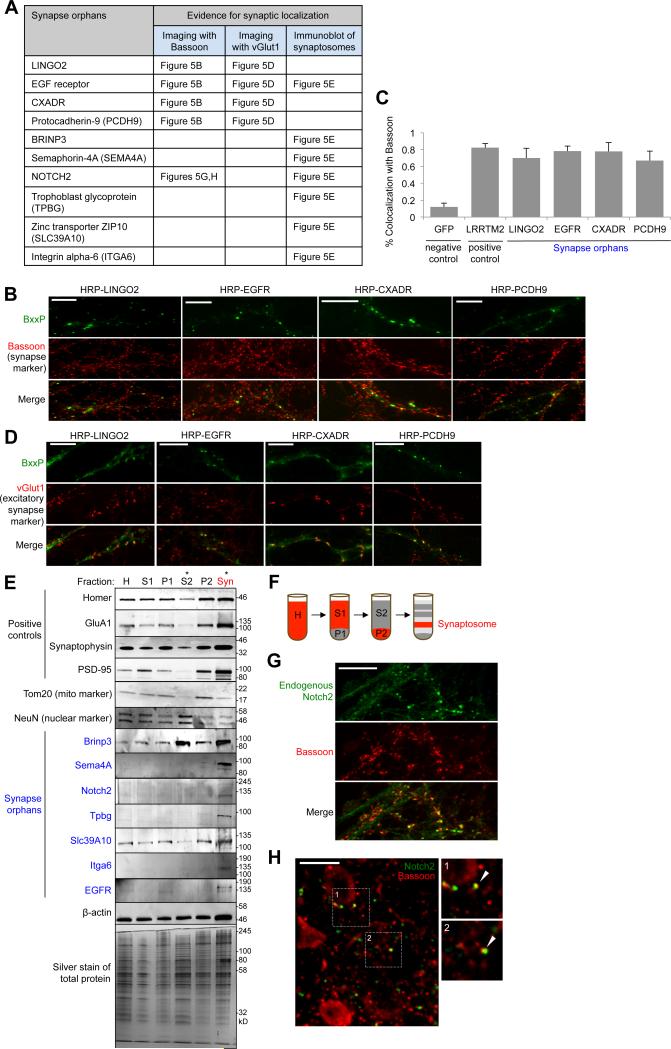
Figure 5. Imaging and synaptosome blotting of specific proteomic hits.
(A) Table summarizing results. 10 synapse orphans (proteins without prior literature connection to synapses) from our excitatory synaptic cleft proteome were analyzed. Figure panels showing relevant data are given. (B) Colocalization of four synapse orphans with pre-synaptic marker Bassoon. Genes were fused at their N-terminal ends to HRP, and visualized by BxxP labeling followed by neutravidin-AlexaFluor647 staining. (C) Quantitation of data in (B). >8 fields of view were analyzed for each construct. HRP-LRRTM2 was a positive control analyzed in parallel, and GFP was a non-synapse localized negative control. Errors, ±1 s.e.m. (D) Colocalization of orphans in (B) with excitatory synapse marker vGlut1. (E) Immunoblot detection of 7 synapse orphans (blue) in purified synaptosomes (Syn) derived from adult rat brain. Tom20 and NeuN are negative controls. Fractions defined in (F). Red tracks the synaptosomes after each fractionation step. (G, H) Confocal imaging of endogenous Notch2 in DIV19 cultured neurons (G) and adult rat brain tissue (H) along with synapse marker Bassoon. All scale bars, 10 μm. See also Figure S6.
To perform fluorescence imaging, we expressed 7 recombinant orphan proteins in DIV19 neuron cultures, via lentiviral transduction. We found that extremely low expression levels were essential to observe punctate rather than diffuse localization patterns, perhaps because mistargeting occurs upon overexpression. However, GFP and V5 epitope tags were not visible at such low expression levels; instead, we used HRP in combination with BxxP as a highly sensitive, amplifying imaging label (Figure S6 shows the superior sensitivity of HRP over Venus for fluorescence imaging). Figures 5B-D show that 4 orphan proteins from the excitatory proteome, tagged with HRP at their N-terminal ends, colocalize with endogenous Bassoon as well as vGlut1, a marker of glutamatergic synapses.
For eight synapse orphans, we obtained commercial antibodies, and used these to probe for the endogenous proteins in purified synaptosomes derived from adult rat brain. A concern when using neuron cultures is that synapses could form between neurons that do not normally contact each other in vivo. By contrast, our synaptosomes are derived from physiological synapses already present in the adult rat brain. For 7 orphans, we observed clear enrichment of the endogenous protein in synaptosome fractions compared to non-synaptosome fractions such as S2 (Figures 5E-F). The 8th antibody, against Notch3, failed to detect Notch3, even in whole neuron lysate (data not shown). Thus, for 7 synapse orphans, the combination of enrichment at synaptic clefts in live neuron culture (via HRP tagging) and enrichment in synaptosomes derived from adult brain tissue, provides strong and orthogonal evidence that these proteins are bona fide synaptic proteins.
One of these antibodies, against Notch2, also worked for visualization of the endogenous protein, both in neuron culture and in adult rat brain tissue. Figures 5G-H show colocalization of Notch2 and the synapse marker Bassoon, providing a third line of evidence that Notch2 is synaptically localized.
Altogether, of the 14 synapse orphans we analyzed by follow-up imaging and immunoblotting, positive identifications were made for 10 of them (Figure 5A). For the remaining 4, two were inconclusive (non-specific antibody for Notch3, and HRP tag disruption of surface trafficking for Matn2). Negative results were obtained for HRP-tagged Brinp2 and Smpdl3b, but we suspect that these soluble, secreted proteins may be especially sensitive to HRP tagging and could mislocalize. This is supported by the observation that HRP-tagged Brinp3 (homologous to Brinp2) also appeared non-synaptic by imaging, but due to availability of a specific antibody for this protein, we were able to assign endogenous Brinp3 to synapses via synaptosome immunoblotting (Figure 5E). Therefore, our statistics, albeit on a small sample size, suggest that our synaptic cleft proteomic lists have very low false positive rates (i.e., the 84-90% synapse specificity represents a lower bound) and may be a rich source of synapse protein candidates.
The proteomic datasets can also be mined for insights on the synapse sub-type specificity of known synaptic proteins. For example, for the 38 known synaptic proteins in our inhibitory synaptic cleft list, 17 were not previously known to reside at inhibitory GABAergic synapses specifically. Interestingly, the proteomic lists also highlight 11 proteins previously annotated as excitatory, that we detected at both excitatory and inhibitory synapses in our proteomic datasets. To follow up on 4 of these proteins, we performed fluorescence microscopy with HRP-tagged recombinant proteins introduced by lentiviral transduction. Figure S6 shows that all 4 of these proteins (Flrt2, EphB6, Dcc, and Elfn1) significantly colocalize with both vGlut1 and vGAT markers, suggesting that they reside at both synapse types, at least in neuron culture.
CD200 IS AN EXCITATORY SYNAPSE-LOCALIZED PROTEIN THAT REGULATES SYNAPSE NUMBERS IN THE VISUAL THALAMUS
For many of the proteins we enrich, the only prior evidence for synaptic localization was enrichment in a synaptosome MS study. As described above, the high false positive rates of such studies necessitate extensive follow up experimentation to distinguish true positives from false positives. By contrast, our lists are much more specific. Therefore, if a protein is detected in a synaptosome preparation and in our live cell proteomic map, it is much more likely to be a genuine synaptic protein. CD200 is one of the most highly enriched proteins in our excitatory synaptic cleft proteome. CD200 was previously identified in a synaptosome MS study (Biesemann et al., 2014) (which is why we did not classify it as a synapse orphan), but there has been no further characterization of this protein in neurons. We performed immunostaining of CD200 in brain tissue from wild-type mice and found that it was localized throughout the neuropil, as expected for an excitatory synaptic protein (Figure S6F). In the visual thalamus, CD200 expression was highest during early postnatal development (P10), which could indicate a role in synapse development or remodeling. We then used structured illumination microscopy (SIM) to assess whether CD200 colocalized with synaptic markers in vivo. Indeed, CD200 colocalized with both presynaptic marker vGlut2 and postsynaptic marker Homer in the P10 dorsal lateral geniculate nucleus of the thalamus (dLGN) (Figure S6G).
To probe the functional role of CD200 at synapses, we used high-resolution confocal microscopy to quantify the number of colocalized vGlut2 and Homer puncta in the dLGN of CD200 knockout (KO) mice at P10 (Figures S6H-I). We observed a significant reduction in synapse numbers compared to wild-type littermate controls, demonstrating that the function of CD200 is indeed relevant to the synapse. Future studies will be needed to determine whether CD200 plays a role in synapse formation or regulates an aspect of the synaptic refinement process.
MDGA1 AND MDGA2 HAVE DISTINCT LOCALIZATIONS AND REGULATORY FUNCTIONS
The availability of distinct proteomic datasets for the excitatory glutamatergic and inhibitory GABAergic synapses provides an opportunity to consider the complement of molecules that define and contribute to the specific formation of each synapse type. For example, specific synaptic adhesion proteins are known to play important roles in recruiting GABA vesicle-containing pre-synapses to GABA receptor-containing post-synaptic membranes, and glutamate vesicle-containing pre-synapses to glutamate receptor-containing post-synaptic membranes.
Our attention was drawn to two proteins, Mdga1 and Mdga2, that are not themselves synaptic adhesion proteins, but have been linked in previous studies to the well-studied adhesion protein Nlgn2. Lee et al., 2013 and Pettem et al., 2013 have shown that both Mdgas bind to Nlgn2, and that Mdga1 acts via Nlgn2 to downregulate inhibitory synapse formation. Due to high (~70%) sequence homology, Mdga2 is assumed to have the same localization and function as Mdga1, though Mdga2 has not previously been studied in neurons. Interestingly, we detected Mdga1 in our excitatory synaptic cleft proteome, and Mdga2 in our inhibitory proteome, which challenges this assumption and suggests that Mdga2 may have a different function at synapses than Mdga1.
To further investigate Mdga1 and Mdga2, we prepared recombinant fusions to HRP, since specific antibodies are not available, and imaged the proteins in DIV19 neurons. In agreement with the proteomic data, recombinant Mdga2 overlapped with the inhibitory marker vGAT exclusively (Figure 6A). Recombinant Mdga1, on the other hand, overlapped with both excitatory and inhibitory markers (Figure S7A). The discrepancy between this observation and our proteomic data could result from the incomplete coverage of the inhibitory proteome, which recalled only 46% of expected proteins. Previous imaging of GFP-Mdga1 also showed overlap with both excitatory and inhibitory terminals (Pettem et al., 2013). Thus, Mdga1 is most likely localized to both synapse types, while Mdga2 is specifically localized to GABAergic synapses only.
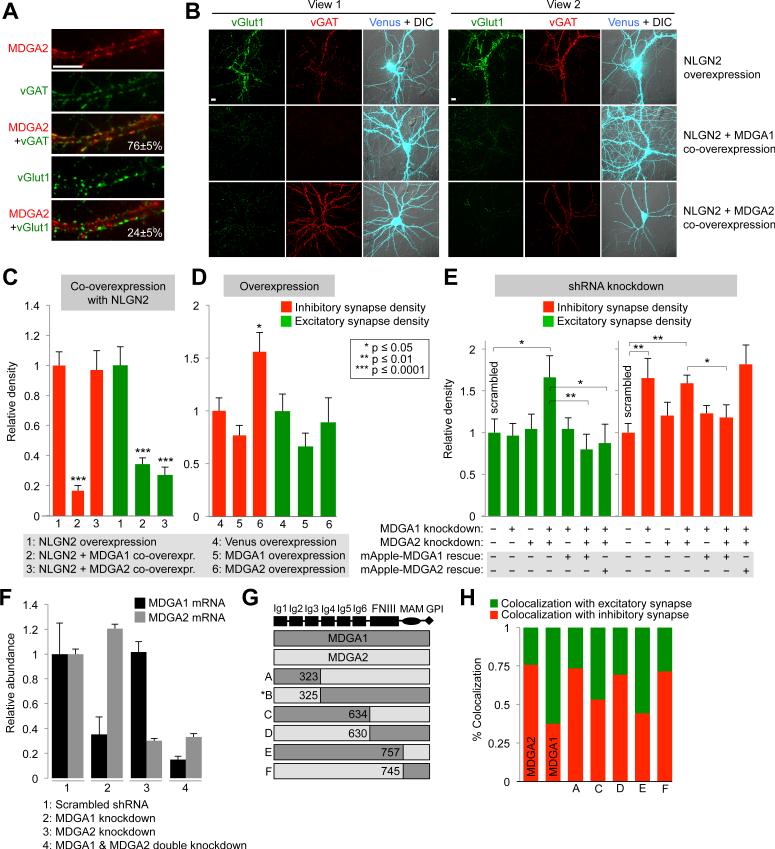
Figure 6. Mdga1 and Mdga2 have distinct synaptic localizations and functions.
(A) Fluorescence imaging of HRP-Mdga2, via BxxP labeling and neutravidin-AlexaFlour647 staining. Values give colocalization extent with inhibitory (vGAT) and excitatory (vGlut1) synapse markers analyzed from 8 fields of view each; errors, ± 1 s.d.; scale bar, 10 m. (B) Synaptogenesis assay based on overexpression of Nlgn2 to probe specificity of presynaptic vesicle recruitment. Nlgn2 was overexpressed along with Venus fluorescent protein (top), Venus-Mdga1 (middle), or Venus-Mdga2 (bottom). Two fields of view are shown per condition. Enhanced recruitment of excitatory or inhibitory synaptic vesicles to transfected neurons was assessed by staining with anti-vGlut1 and anti-vGAT antibodies, respectively. Images are representative of >20 transfected neurons per condition. Controls show that co-overexpression of Mdga1 or Mdga2 does not alter surface levels of V5-Nlgn2 (data not shown). (C) Quantitation of data in (B) along with 7 additional fields of view per condition. Synapse density is defined as total anti-vGlut1 or anti-vGAT intensity divided by area of transfected neuron. Errors, ± s.e.m. (D) Effect of Mdga1 or Mdga2 overexpression (without Nlgn2 co-overexpression) on excitatory and inhibitory vesicle densities, quantified as in (C). 12 fields of view analyzed per condition. Errors, ± s.e.m. (E) Effect of single or double knockdown of Mdga1 and/or Mdga2 on excitatory and inhibitory vesicle densities, quantified as in (C). Knockdowns (from 3 technical replicates per condition) verified by qPCR in (F) (errors, ± s.e.m.). Representative images in Figure S7D. 15 fields of view analyzed per condition. Errors, ± s.e.m. (G) Chimeras of MDGA1 and MDGA2 tested in (H). The parent genes each have 6 immunoglobulin (Ig) domains, a fibronectin type III (FNIII) domain, a memprin/A5 protein/receptor tyrosine phosphatase mu (MAM) domain, and a C-terminal GPI anchor. Chimera B exhibited poor surface trafficking and was not evaluated further. Numbers refer to amino acid residues at cross-over points. (H) Relative localization of chimeras to excitatory versus inhibitory synapses, assessed by imaging with anti-vGlut1 and anti-vGAT staining (images and error values shown in Figure S7E). See also Figure S7.
We sought to investigate the hypothesis that Mdga2 has a different function at synapses than Mdga1. Our assay capitalizes on the synaptogenic activity of Nlgn2 (Graf et al., 2004), which when overexpressed on the post-synaptic membrane, over-recruits both vGlut1- and vGAT-positive pre-synaptic terminals (Takahashi et al., 2012) (Figure S7B). When Mdga1 was cooverexpressed with Nlgn2, the enhanced recruitment of both vesicle types was suppressed (Figures 6B-C). Co-overexpression of Mdga2, however, suppressed selectively the recruitment of excitatory but not inhibitory vesicles.
Based on these observations, we hypothesized that Mdga1 and 2 both downregulate the trans-synaptic vesicle recruiting activity of Nlgn2 (which occurs via unknown presynaptic binding partner(s)) (Figure S7C). However, Mdga1 binds Nlgn2 in such a way that it blocks recruitment of both inhibitory and excitatory vesicles. MDGA2 binds Nlgn2 differently, blocking recruitment of only excitatory vesicles while allowing recruitment of inhibitory vesicles.
To further test this hypothesis, we performed shRNA knockdown of Mdga1, Mdga2, or both together (Figures 6E-F and S7D). The interpretation of the data requires the assumption that each Nlgn2 molecule binds to Mdga1 or 2, but not to both at once. In the case of Mdga1 knockdown, more Nlgn2 is freed to interact with Mdga2 instead. According to our model, Mdga2 promotes inhibitory vesicle recruitment but not excitatory. Correspondingly, we observe that Mdga1 knockdown causes an increase in inhibitory vesicle signal, but not excitatory signal. This is also consistent with previous observations (Lee et al., 2013; Pettem et al., 2013).
By itself, Mdga2 knockdown had no significant effect, but when combined with Mdga1 knockdown caused both inhibitory and excitatory vesicle signals to increase, consistent with full derepression of Nlgn2 activity. Because this phenotype is distinct from that caused by knockdown of Mdga1 only (increase in inhibitory vesicle density only), it suggests that Mdga2 plays a role in down-regulation of excitatory vesicle recruitment.
We also performed a gain-of-function assay by overexpressing only Mdga1 or Mdga2 (without Nlgn2 co-overexpression; Figure 6D). More Mdga2 in neurons might shift the equilibrium for Nlgn2, causing more of it to bind to Mdga2 than Mdga1. Accordingly we would expect to see increased inhibitory vesicle recruitment, with no effect on excitatory vesicle recruitment. Our data in Figure 6D shows this expected trend.
Fluorescence imaging of a panel of Mdga1/2 chimeras (Figure 6G) showed that their extracellular juxtamembrane Ig4-6 regions are responsible for their unique synaptic localizations (Figures 6H and S7E). Chimeras with the Ig4-6 region from Mdga1 exhibited “Mdga1-like” localization to both excitatory and inhibitory synapses, while chimeras with the Ig4-6 domain from Mdga2 exhibited “Mdga2-like” localization to inhibitory synapses only. Previous studies have shown that a different region of the Mdgas, the Ig1-3 domain, mediates cis-interactions with Nlgn2 (Lee et al., 2013; Pettem et al., 2013). Therefore, we postulate that the Mdgas target to excitatory and/or inhibitory synapses, governed by their Ig4-6 regions, and independent of interactions with Nlgn2. At inhibitory synapses, each Mdga then interacts with Nlgn2 to differentially regulate its activity. At excitatory synapses, perhaps the role of Mdga1 is to help prevent the invasion of inhibitory synaptic elements (such as pools of Nlgn2 itself) into excitatory terminals.
DISCUSSION
In this study, we develop and extend the APEX platform to achieve successful proteomic mapping of the synaptic cleft. We replace APEX2 with HRP, which is more active at the cell surface, and introduce the BxxP probe for restriction of peroxidase labeling to the neuronal cell surface. We apply an intersectional strategy, using two independent peroxidase fusion constructs targeting the same cellular locale, in order to dramatically improve the specificity of protein identifications. To adapt the technique to primary, non-dividing cells, we employ post-digestion iTRAQ chemical labeling for quantitation, rather than SILAC metabolic labeling, which requires protein turnover. Finally, to overcome background caused by the unique, detergent insoluble matrix underlying the post-synaptic membrane, we develop a denaturing lysis and streptavidin enrichment procedure that effectively separates intracellular proteins from cleft-exposed transmembrane proteins.
Our study opens the door for APEX to be applied to a greater diversity of cellular structures. This includes other unpurifiable subdomains of the synapse (e.g., the inhibitory post-synaptic region and synaptic ribbons), as well as smaller and more challenging domains across cell biology in general, for example, mitochondria-ER contact sites, RNA granules, the axon initial segment, and even macromolecular complexes. Because peroxidase-catalyzed proteomic tagging occurs in just 1 minute (in contrast to BioID which requires 18-24 hours of labeling), it should also be possible to map proteomes under different cell states, such as in response to drugs or LTP, or at different synapse maturities, or in models of brain disease.
The excitatory and inhibitory synaptic cleft proteomic lists generated by this study depart from existing synapse proteomes in several respects. First, they are much more specific. Due to contamination by mitochondrial, nuclear (Figures S1A-B), and glial proteins, synaptosome and PSD preparations typically have false discovery rates of 20-40% (Biesemann et al., 2014). This necessitates extensive follow-up experimentation to distinguish genuine synaptic proteins from false positives. By contrast, our datasets have false discovery rates of <10%, or likely much lower, based on our analysis of synapse orphans (Figure 5). Second, our lists have higher coverage (Figure S1C). By tagging in live cells and bypassing detergent lysis and serial centrifugation, we improve protein recovery, enabling us to identify dozens of proteins that were missed by previous synaptosome, PSD, and active zone preparations. These include soluble, secreted proteins such as Gpc6 and Reelin that may dissociate from organelles during centrifugation. Third, our lists chart an important subdomain of the synapse that has eluded previous MS studies because it cannot be purified. Fourth, our lists cleanly separate components of the glutamatergic synapse from components of the GABAergic synapse, in contrast to synaptosome preparations which blend across all synapse types, and PSD preparations which are applicable only to excitatory synapses because inhibitory synapses lack a PSD. Biesemann et al. 2014 have attempted to further purify synaptosomes by FACS, but their resulting vGlut1-enriched dataset is imperfect, containing inhibitory synapse components such as Gabrg2, Gabra1, Gabra5, and gephyrin. Here, using the power of genetic targeting, we achieve >98% specificity for excitatory components in our glutamatergic list, and >76% specificity for inhibitory components in our GABAergic list (value is corrected based on data shown in Figure S6).
Our inhibitory synapse proteome can also be compared to two previous studies that use immunoprecipitation-MS, rather than biochemical fractionation, to identify components of the GABAergic synapse (Heller et al., 2012; Kang et al., 2014). Though the Heller et al. study that uses GABAA receptor α1 immunoprecipitation is quite specific, both datasets miss the majority of known inhibitory synaptic cleft components (coverage <34%), probably because the baits used do not interact directly or stably with these proteins. By contrast, our mapping approach does not require direct interactions, because the biotinphenoxyl radical diffuses out of the peroxidase active site to tag endogenous proteins in the neighborhood of HRP/APEX.
Both synaptic cleft proteomic lists can be mined for biological insights or hypotheses. We have illustrated this by using our datasets to discover 10 synaptic proteins (validated by microscopy and/or synaptosome immunoblotting in Figure 5), and reveal a potential inhibitory synapse component for 4 known excitatory synaptic proteins (Figure S6). We also followed up on CD200, a protein previously linked to synapses only by crude synaptosome MS data. After observing strong enrichment in our excitatory proteome, we found that CD200 is highly expressed in the visual thalamus during periods of synaptic refinement, and its deletion perturbs the normal development of retinogeniculate synapses.
By also revealing different synaptic sub-type localizations for two known, homologous synaptic proteins (Mdga1 and Mdga2), our proteomic data inspired the hypothesis that these two proteins have different functional roles. Follow up experiments using gain of function and loss of function assays in neuron culture suggest that while Mdga1 may act to generally downregulate Nlgn2 activity, Mdga2 may function as a specificity factor at inhibitory synapses to downregulate Nlgn2's signaling with vGlut1 pre-synaptic terminals but not vGAT pre-synaptic terminals.
There are many more synapse orphans and synapse subtype orphans identified in our study, on which we did not perform follow up experimentation. Several of these are intriguing, and if validated, could open avenues for exploration. For example, Csmd1 is a synapse orphan identified in our inhibitory synaptic cleft proteome. Though no literature describes Csmd1 as a synaptic protein, the CSMD1 gene has been linked by GWAS studies to schizophrenia (Håvik et al., 2011), and the protein may be part of the complement pathway that facilitates synaptic pruning (Hong et al., 2016; Kraus et al., 2006). The detection of endogenous Csmd1 in the inhibitory cleft of live neurons suggests a possible link between inhibitory synapse elimination and schizophrenia that could be explored in future studies. We note that Csmd1 is a very difficult protein to study by conventional techniques, because there are no specific antibodies, and recombinant expression via standard techniques is not possible because the protein is enormous (~380 kDa). Proximity biotinylation and MS may be one of the only ways to investigate the subcellular localization of this protein. Interestingly, Csmd1 has been missed in all previous synaptosome, PSD, and active zone purifications.
In conclusion, our study departs from both classical synaptosome and PSD purifications, as well as previous APEX and BioID publications, and demonstrates that peroxidase-based proteomic mapping is a powerful technology for parsing the molecular properties of important nanoscale structures in biology. Mapped with few-nanometer spatial resolution and one minute temporal resolution, the excitatory and inhibitory synaptic cleft proteomes reported here can serve as rich resources for neuroscientists.
Supplementary Material
APEX proteomics extended to open cellular domains, cell surface, and primary cells
199 glutamatergic and 42 GABAergic synaptic cleft proteins identified
10 hits validated as synaptic proteins by imaging and/or synaptosome blotting
Mdga2 regulates specificity of presynaptic recruitment to inhibitory post-synapses
ACKNOWLEDGEMENTS
We thank N. Watson (Whitehead Institute Keck Microscopy Facility) for performing electron microscopy, J. Einstein for neuron cultures, A. Vignery (Yale) and J. Sedgwick (Schering-Plough) for CD200 knockout mice, and members of A. Ting's lab especially V. Hung, K. Cox, S. Lam, K. Pedram for assistance with data analysis and manuscript editing. K. Tye and G. Matthews assisted with brain slice imaging. J. Sanes, K. Shen and S. Slavoff provided experimental advice and feedback on the manuscript, and T. Hashimoto helped with data analysis. Funding was provided by the U.S. National Institutes of Health (R01-CA186568 to A.Y.T.; P41GM103412 and R01GM086197 to M.H.E.; R01-NS071008-01A1 and R01-NS092578 to B.S.) and the Howard Hughes Medical Institute Collaborative Initiative Award (A.Y.T. and S.A.C.) P.S.S. was supported by a Simons Center for the Social Brain Postdoctoral Fellowship and Feodor Lynen Research Fellowship from the Alexander von Humboldt Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
K.H.L. performed all experiments except those noted below. K.H.L. and P.S.S. performed shRNA knockdowns. K.H.L. and A.S.D. performed synaptosome purifications, qPCR, and Notch2 imaging in brain slice. N.D.U., T.S., and S.A.C. processed streptavidin-enriched proteomic material and performed mass spectrometry. P.S.S., T.J.D., and M.H.E. performed electron microscopy. E.K.L., D.K.W., and B.S. performed CD200-related experiments. K.H.L. and A.Y.T. conceived the study, designed experiments, and analyzed data. K.H.L. and A.Y.T. wrote and revised the manuscript.
Competing interests statement
The Massachusetts Institute of Technology has filed a patent covering the peroxidase-based proteomic mapping technology. The authors will deposit the genetic constructs used in this work with Addgene (www.addgene.org/).
REFERENCES
- Allen NJ, Bennett ML, Foo LC, Wang GX, Chakraborty C, Smith SJ, Barres BA. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 2012;486:410–414. doi: 10.1038/nature11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayés A, Collins MO, Croning MDR, van de Lagemaat LN, Choudhary JS, Grant SGN. Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins. PLoS One. 2012;7:e46683. doi: 10.1371/journal.pone.0046683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesemann C, Grønborg M, Luquet E, Wichert SP, Bernard V, Bungers SR, Cooper B, Varoqueaux F, Li L, Byrne JA, et al. Proteomic screening of glutamatergic mouse brain synaptosomes isolated by fluorescence activated sorting. EMBO J. 2014;33:157–170. doi: 10.1002/embj.201386120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyken J, Grønborg M, Riedel D, Urlaub H, Jahn R, Chua JJE. Molecular profiling of synaptic vesicle docking sites reveals novel proteins but few differences between glutamatergic and GABAergic synapses. Neuron. 2013;78:285–297. doi: 10.1016/j.neuron.2013.02.027. [DOI] [PubMed] [Google Scholar]
- Chapman-Smith A, Cronan JE. In vivo enzymatic protein biotinylation. Biomol. Eng. 1999;16:119–125. doi: 10.1016/s1050-3862(99)00046-7. [DOI] [PubMed] [Google Scholar]
- Chen C-L, Hu Y, Udeshi ND, Lau TY, Wirtz-Peitz F, He L, Ting AY, Carr SA, Perrimon N. Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc. Natl. Acad. Sci. U. S. A. 2015 doi: 10.1073/pnas.1515623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Villa KL, Cha JW, So PTC, Kubota Y, Nedivi E. Clustered dynamics of inhibitory synapses and dendritic spines in the adult neocortex. Neuron. 2012;74:361–373. doi: 10.1016/j.neuron.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin S-X, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håvik B, Le Hellard S, Rietschel M, Lybæk H, Djurovic S, Mattheisen M, Mühleisen TW, Degenhardt F, Priebe L, Maier W, et al. The complement control-related genes CSMD1 and CSMD2 associate to schizophrenia. Biol. Psychiatry. 2011;70:35–42. doi: 10.1016/j.biopsych.2011.01.030. [DOI] [PubMed] [Google Scholar]
- Heller EA, Zhang W, Selimi F, Earnheart JC, Ślimak MA, Santos-Torres J, Ibañez-Tallon I, Aoki C, Chait BT, Heintz N. The biochemical anatomy of cortical inhibitory synapses. PLoS One. 2012;7:e39572. doi: 10.1371/journal.pone.0039572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn FA, Anderson DJ, Rustad DG. Glial contamination of synaptosomal fractions. Brain Res. 1976;101:341–344. doi: 10.1016/0006-8993(76)90274-2. [DOI] [PubMed] [Google Scholar]
- Hoek RM, Ruuls SR, Murphy CA, Wright GJ, Goddard R, Zurawski SM, Blom B, Homola ME, Streit WJ, Brown MH, et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200). Science. 2000;290:1768–1771. doi: 10.1126/science.290.5497.1768. [DOI] [PubMed] [Google Scholar]
- Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V, Zou P, Rhee H-W, Udeshi ND, Cracan V, Svinkina T, Carr SA, Mootha VK, Ting AY. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell. 2014;55:332–341. doi: 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA, Ting AY. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc. 2016;11:456–475. doi: 10.1038/nprot.2016.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Ge Y, Cassidy RM, Lam V, Luo L, Moon K-M, Lewis R, Molday RS, Wong ROL, Foster LJ, et al. A combined transgenic proteomic analysis and regulated trafficking of neuroligin-2. J. Biol. Chem. 2014;289:29350–29364. doi: 10.1074/jbc.M114.549279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus DM, Elliott GS, Chute H, Horan T, Pfenninger KH, Sanford SD, Foster S, Scully S, Welcher AA, Holers VM. CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelial tissues. J. Immunol. 2006;176:4419–4430. doi: 10.4049/jimmunol.176.7.4419. [DOI] [PubMed] [Google Scholar]
- Krivosheya D, Tapia L, Levinson JN, Huang K, Kang Y, Hines R, Ting AK, Craig AM, Mei L, Bamji SX, et al. ErbB4-neuregulin signaling modulates synapse development and dendritic arborization through distinct mechanisms. J. Biol. Chem. 2008;283:32944–32956. doi: 10.1074/jbc.M800073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulik A, Nakadate K, Nyíri G, Notomi T, Malitschek B, Bettler B, Shigemoto R. Distinct localization of GABA(B) receptors relative to synaptic sites in the rat cerebellum and ventrobasal thalamus. Eur. J. Neurosci. 2002;15:291–307. doi: 10.1046/j.0953-816x.2001.01855.x. [DOI] [PubMed] [Google Scholar]
- Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods. 2014;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Kim Y, Lee S-J, Qiang Y, Lee D, Lee HW, Kim H, Je HS, Südhof TC, Ko J. MDGAs interact selectively with neuroligin-2 but not other neuroligins to regulate inhibitory synapse development. Proc. Natl. Acad. Sci. U. S. A. 2013;110:336–341. doi: 10.1073/pnas.1219987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerma J. Roles and rules of kainate receptors in synaptic transmission. Nat. Rev. Neurosci. 2003;4:481–495. doi: 10.1038/nrn1118. [DOI] [PubMed] [Google Scholar]
- Linhoff MW, Laurén J, Cassidy RM, Dobie FA, Takahashi H, Nygaard HB, Airaksinen MS, Strittmatter SM, Craig AM. An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron. 2009;61:734–749. doi: 10.1016/j.neuron.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong S-E, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettem KL, Yokomaku D, Takahashi H, Ge Y, Craig AM. Interaction between autism-linked MDGAs and neuroligins suppresses inhibitory synapse development. J. Cell Biol. 2013;200:321–336. doi: 10.1083/jcb.201206028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia M, Wang T, Avramopoulos D, Valle D, Thomas G, Huganir RL, Goes FS, Potash JB, Zandi PP. SynaptomeDB: an ontology-based knowledgebase for synaptic genes. Bioinformatics. 2012;28:897–899. doi: 10.1093/bioinformatics/bts040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Rees JS, Li X-W, Perrett S, Lilley KS, Jackson AP. Selective Proteomic Proximity Labeling Assay Using Tyramide (SPPLAT): A Quantitative Method for the Proteomic Analysis of Localized Membrane-Bound Protein Clusters. Curr. Protoc. Protein Sci. 2015;80:19, 27, 1–18. doi: 10.1002/0471140864.ps1927s80. [DOI] [PubMed] [Google Scholar]
- Rhee H-W, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JY, Ichtchenko K, Südhof TC, Brose N. Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1100–1105. doi: 10.1073/pnas.96.3.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svinkina T, Gu H, Silva JC, Mertins P, Qiao J, Fereshetian S, Jaffe JD, Kuhn E, Udeshi ND, Carr SA. Deep, quantitative coverage of the lysine acetylome using novel anti-acetyl-lysine antibodies and an optimized proteomic workflow. Mol. Cell. Proteomics. 2015;14:2429–2440. doi: 10.1074/mcp.O114.047555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Katayama K-I, Sohya K, Miyamoto H, Prasad T, Matsumoto Y, Ota M, Yasuda H, Tsumoto T, Aruga J, et al. Selective control of inhibitory synapse development by Slitrk3-PTPδ trans-synaptic interaction. Nat. Neurosci. 2012;15:389–398. doi: 10.1038/nn.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamori S, Holt M, Stenius K, Lemke EA, Grønborg M, Riedel D, Urlaub H, Schenck S, Brügger B, Ringler P, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Jamain S, Brose N. Neuroligin 2 is exclusively localized to inhibitory synapses. Eur. J. Cell Biol. 2004;83:449–456. doi: 10.1078/0171-9335-00410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.