Abstract
Mitochondria play essential roles in cellular energy production via the oxidative phosphorylation system (OXPHOS) consisting of five multiprotein complexes and also in the initiation of apoptosis. NADH:ubiquinone oxidoreductase (complex I) is the largest complex that catalyzes the first step of electron transfer in the OXPHOS system. GRIM-19 was originally identified as a nuclear protein with apoptotic nature in interferon (IFN)- and all-trans-retinoic acid (RA)-induced tumor cells. To reveal its biological role, we generated mice deficient in GRIM-19 by gene targeting. Homologous deletion of GRIM-19 causes embryonic lethality at embryonic day 9.5. GRIM-19−/− blastocysts show retarded growth in vitro and, strikingly, display abnormal mitochondrial structure, morphology, and cellular distribution. We reexamined the cellular localization of GRIM-19 in various cell types and found its primary localization in the mitochondria. Furthermore, GRIM-19 is detected in the native form of mitochondrial complex I. Finally, we show that elimination of GRIM-19 destroys the assembly and electron transfer activity of complex I and also influences the other complexes in the mitochondrial respiratory chain. Our result demonstrates that GRIM-19, a gene product with a specific role in IFN-RA-induced cell death, is a functional component of mitochondrial complex I and is essential for early embryonic development.
Interferons (IFNs) regulate antiviral, antitumor, and immune responses in vertebrates by activating the JAK-STAT pathway and stimulating gene expression (29). All-trans-retinoic acid (RA), a metabolite of vitamin A, binds to specific nuclear receptors, induces expression of various genes, and inhibits growth of certain types of cancer (16). The combination of IFN and RA further suppresses cell growth and induces cell death in several tumor cell lines (15, 19). To understand the mechanism for this, an antisense knockout approach was utilized to identify genes that are involved in IFN-β-RA-inducible cell death in HeLa cells. These genes were identified and named GRIM (for genes associated with retinoid-interferon mortality) (11). GRIM-19 was one of such genes (2). GRIM-19 encodes a ∼16-kDa protein of 144 amino acids that was originally described primarily as a nuclear protein. It is expressed ubiquitously in various human tissues and at a high level in the heart and skeletal muscle and to a lesser extent in liver, kidney, and brain. While introduction of the antisense of GRIM-19 confers cell survival and growth, overexpression of GRIM-19 enhances the sensitivity of cells to IFN-RA-induced death. GRIM-19 was, therefore, suggested to be a death activator (2). Sequences with high identity to human GRIM-19 were also found in other vertebrates, including bovine, pig, mouse, rat, and chicken. In addition, homology sequences were also reported in lower species and plants such as Caenorhabditis elegans, Drosophila, and Arabidopsis thaliana (2, 8, 13), indicating that the GRIM-19 gene is evolutionarily conserved in most eukaryotes. However, the mechanism for its apoptotic role is largely unknown.
Mitochondria are the major source of energy in most eukaryotic cells which produce ATP through the oxidative phosphorylation system and the citric acid cycle. Mitochondria also regulate calcium homeostasis and modulate apoptosis through the release of cell death-inducing molecules (21, 23). The oxidative phosphorylation system is located in the inner membrane of mitochondria and comprises five individual complexes (complexes I to V) and two additional electron carriers, coenzyme Q10 and cytochrome c. These complexes are arranged as supercomplexes (9, 24). Complex I represents the largest and the least understood multimeric enzyme complex that catalyzes the first step in the electron transfer chain by transferring two electrons from NADH to ubiquinone and is coupled to the translocation of four protons across the inner mitochondrial membrane for ATP synthesis (21, 27, 33). Complex I is composed of more than 40 structural subunits. Seven are mitochondrion-encoded gene products, whereas the rest are encoded in the nucleus and transported into the mitochondria following synthesis in the cytoplasm (4, 28, 31). Complex I from bovine heart has been extensively studied to define its complete subunits (4, 8). A previously unidentified protein that is tightly associated and copurified with mitochondrial complex I was recently reported. Surprisingly, this novel protein, named B16.6, shares 83% identity with human GRIM-19 and is a bovine homologue of human GRIM-19 (8). However, the function of GRIM-19 in complex I is unclear.
Signal transducer and activator of transcription 3 (Stat3) belongs to a family of latent cytoplasmic transcription factors that are the key molecules in various cytokine signaling pathways (1, 7, 12). Recently, we have identified GRIM-19 as an interacting protein of Stat3 by yeast two-hybrid screening (17). We have showed that GRIM-19 specifically interacts with Stat3 but not with the other Stat family members, such as Stat1 and Stat5, in vivo and inhibits the transcriptional activity of Stat3. Subsequently, similar results were also reported by others (36). Furthermore, a recent report indicates that GRIM-19 interacts with a protein named GW112 that is highly expressed in colon cancers and has an antiapoptotic function (37). Based on these data, GRIM-19 seems to play pleiotropic roles within a cell.
To reveal the biological function of GRIM-19 and the possible links among these diverse biological processes that it is involved in, we generated mice deficient in GRIM-19 by gene targeting. Homology deletion of GRIM-19 causes early embryonic lethality at embryonic day 9.5 (E9.5). GRIM-19−/− blastocysts show an impaired growth of inner cell mass (ICM) and trophoblasts (TB), indicating that GRIM-19 is essential for early embryonic development. Further examination of mutant blastocysts revealed abnormal mitochondrial structure, morphology, and cellular distribution. Biochemical and cellular characterization of GRIM-19 indicates its primary mitochondrial localization and a key role in the complex I assembly and electron transfer ability. Together, our results demonstrate that GRIM-19 is an indispensable component of mitochondrial complex I that plays an essential role in the mitochondrial function and integrity.
MATERIALS AND METHODS
Cloning of the GRIM-19 gene and construction of the targeting plasmid.
Genomic clones of GRIM-19 were isolated by screening a mouse 129SVJ genomic library (Stratagene) with a mouse GRIM-19 cDNA (17) and subcloned into pBluescript II KS+ (Stratagene). The targeting vector was also constructed in pBluescript II KS+ by insertion of a 4.3-kb NotI-XbaI fragment comprising the 5′ flanking region of the GRIM-19 gene, a neomycin resistance expression cassette, a 1.5-kb XhoI-ClaI fragment of 3′ flanking DNA, and a herpes simplex virus thymidine kinase marker at the 3′ end shown in Fig. 1A. The final construct was verified by restriction enzyme digestion and partial sequencing.
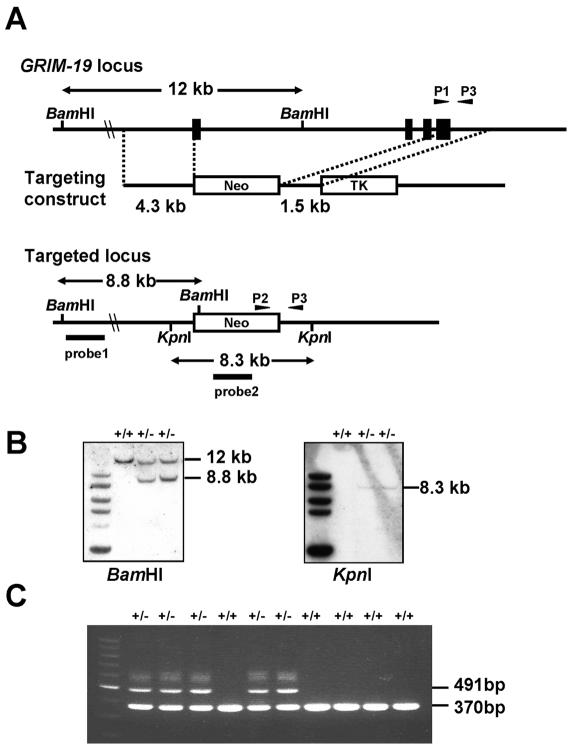
FIG. 1.
Disruption of the murine GRIM-19 gene. (A) Schematic presentation of wild-type and targeted GRIM-19 alleles and the gene targeting construct. Four exons are indicated with closed boxes. Arrowhead P1 indicates the primer specific for the wild-type allele, P2 indicates the primer for the GRIM-19− allele, and P3 indicates the primer for both the wild-type and GRIM-19− alleles. Probes 1 and 2 are used for Southern blot analysis. Neo, neomycin resistance cassette; TK, thymidine kinase marker. (B) Southern blot analysis of targeted ES cells. Genomic DNA from individual ES clones were digested with BamHI or KpnI and analyzed by Southern blotting with probe 1 (left panel) or probe 2 (right panel). The targeted clones are shown with the wild-type (12-kb) and mutant (8.8-kb) fragments as marked. (C) PCR genotyping of offspring from intercrossing two GRIM-19+/− mice. A 370-bp wild-type band and a 491-bp mutant band were amplified by primer pairs P1-P3 and P2-P3, respectively, as indicated in the legend of panel A.
Generation of GRIM-19 knockout mice.
The mouse protocol was approved by the animal research committee in the Institute of Molecular and Cell Biology. The targeting vector was linearized and electroporated into mouse embryonic stem (ES) cells derived from the 129SVJ strain. Clones resistant to G418 and ganciclovir were picked and screened for the targeted GRIM-19 allele by Southern blotting and PCR analysis. Two of the selected clones were injected separately into blastocysts derived from C57BL/6 mice and implanted into C57BL/6 pseudopregnant females. From one clone, two male mice carrying the targeted GRIM-19 allele in the germ line were obtained and either backcrossed to C57BL/SW or crossed to 129SVJ females. Offspring was genotyped by Southern blot analysis and PCR, and the heterozygous (GRIM-19+/−) animals were crossed to generate GRIM-19−/− embryos. The phenotypes of GRIM-19−/− embryos derived from both strains of mice were essentially the same.
Genotyping by Southern blotting and PCR analyses.
Southern blotting and PCR analyses were performed to genotype the targeted ES clones and weaned pups. For Southern blot analysis, 10 μg of genomic DNA was digested with BamHI or KpnI, separated on a 0.8% agarose gel, transferred onto a nylon membrane, and probed with probe 1 or 2 as shown in Fig. 1A. For PCR analysis, genomic DNA was used as the template in a reaction where the wild type and the mutant alleles were detected simultaneously. Primer 1 (5′-CTTAGGGCCTGAGCCAACGCAC-3′) and primer 3 (5′-GACTCACCTGCCTCCTGCCTGCC-3′) amplify a 370-bp fragment for the wild type, whereas primer 2 (5′-CGCCAATGACAAGACGCTGGG-3′) and primer 3 amplify a 491-bp fragment specific for the mutant allele.
Histological analysis.
Embryos at E5.5, 6.5, 7.5, and 8.5 were isolated together with uteruses of the mothers. The embryos and deciduas were fixed in 10% formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. The samples were examined by microscope (Leica DM4000 B) and photographed with a Leica DFC300 FX camera, and the image was analyzed with Leica FW4000 software. For genotyping by PCR analysis, embryo tissue was manually microdissected from the paraffin section and digested in 20 μl of buffer 2 of an Expand High Fidelity PCR system (Roche) containing 1.5 mM MgCl2, 1.6 mg of proteinase K per ml, and 0.5% Tween 20 at 56°C overnight. Proteinase K was then heat inactivated at 98°C for 16 min. An aliquot of 5 μl was used for PCR analysis as described above.
Isolation and culture of blastocysts in vitro.
GRIM-19 heterozygote males and females were intercrossed, and embryos at E3.5 were collected by flushing the uteruses of plugged females with M2 medium (Sigma). Blastocysts were then individually transferred to ES medium in culture slides (BD Biosciences) and cultured for 4 to 7 days. The blastocysts were genotyped by PCR after photography.
Transmission electron microscopy.
The blastocysts were fixed in 2.5% glutaraldehyde, rinsed in 0.1 M phosphate-buffered saline (PBS), and postfixed in 1% osmium tetroxide for 1 h. The sample was rinsed in phosphate buffer, dehydrated in ethanol, and embedded in resin. Semithin sections (0.5 μm) and ultrathin sections (90 to 95 nm) were cut with a diamond knife. Semithin sections were stained with toluidine blue and examined with a Zeiss microscope. Ultrathin sections were stained with uranyl acetate and lead citrate and examined with an EM208 transmission electron microscope (Philips Electron Optics).
BrdU incorporation and terminal deoxynucleotidyltransferase-mediated dUTP-fluorescein nick end labeling (TUNEL) assays.
Blastocyst outgrowth was labeled as described (18) by using a 5-bromo-2′-deoxyuridine (BrdU) Labeling and Detection Kit II (Roche). Briefly, BrdU was added to culture media to a final concentration of 10 μM and incubated for 12 h. Cells were then fixed with ethanol and incubated with anti-BrdU monoclonal antibody. A fluorescein-conjugated goat anti-mouse secondary antibody was used. Cleavage of genomic DNA during apoptosis was investigated with an In Situ Cell Death Detection Kit (Roche) according to the manufacturer's instruction. Positive control was digested with 100 U of DNase I per ml to induce DNA strand breaks prior to the labeling procedure. Cells were observed with a Bio-Rad Radiance 2000 laser scanning confocal microscope and analyzed by LaserSharp2000 software (Bio-Rad Laboratories). The figures were processed with Adobe Photoshop software.
Generation of mouse antibody against human GRIM-19.
Human GRIM-19 was cloned and expressed in bacteria as previously described (17). The mice were immunized by intraperitoneal injection with 100 μg of bacterially produced GRIM-19 protein and boosted with 50 μg of protein at the third, fourth, and fifth week. The blood was then routinely collected with a 1-week interval of boosting. The sera after nine boostings were directly used for immunofluorescence without further purification.
Immunofluorescence.
Cells grown on glass coverslips in culture medium were first treated with 500 nM MitoTracker Red CMXRos (Molecular Probes) for 1 h before being fixed in 4% paraformaldehyde in PBS, permeabilized with 0.2% Triton X-100 in PBS, and blocked with 10% fetal bovine serum in PBS. GRIM-19 protein expression and localization in cells were then detected by using the GRIM-19 mouse primary antibody and a fluorescein isothiocyanate-coupled anti-mouse secondary antibody. The cells were then mounted in gel mount (Biomeda Corp.) and examined by using a Bio-Rad MRC 1024 laser scanning confocal microscope.
Mitochondrial isolation, proteinase K treatment, and cell fractionation.
Mitochondria were isolated following a protocol described previously (22). In brief, cells were harvested, washed with PBS, and resuspended in mitochondrial isolation buffer (MIB) containing 10 mM Tris-HCl (pH 7.4), 10 mM HEPES, 0.5 mM EDTA, 250 mM sucrose, 10 mg of bovine serum albumin per ml, 12% Percoll, and proteinase inhibitor cocktail (Roche). Cells in suspension underwent Dounce homogenization gently 10 times on ice with a loose pestle and 10 times with a tight pestle. A total of 3 ml of homogenate was layered onto a previously poured Percoll gradient (3.5 ml of 26% Percoll on 3.5 ml of 40% Percoll) and spun at 30,000 × g for 10 min. The second fraction containing mitochondria was removed and diluted 1:4 with cold MIB and centrifuged at 14,000 × g for 5 min. The pellet containing mitochondria was resuspended in MIB. Mitochondria were treated with proteinase K (100 ng/ml) in the absence or the presence of 1% Triton X-100 in MIB for various durations. The mitochondrial proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to Western blot analysis. The preparation of whole-cell lysates, separation of cytoplasmic and nuclear proteins, and Western blot analysis were performed as previously described (17).
Mitochondrial complex I assembly and oxidative phosphorylation assays.
Samples were prepared from mouse liver and blastocysts. Liver mitochondria were isolated as described above, and mitochondrial proteins were solubilized in 40 μl of solution containing 750 mM 6-aminocarproic acid, 50 mM bis-Tris (pH 7.0), and 5 μl of 10% dodecylmaltoside. The lysate was centrifuged, and the supernatant was used for further assays. For preparing samples from blastocysts, blastocysts were isolated and cultured in vitro. Twenty-seven normal or GRIM-19−/− blastocysts were combined and directly solubilized in 40 μl of the same solution described above. A total of 20 μg of solubilized proteins was used for further assays.
Blue native (BN)-PAGE (25, 26, 35) was used for separation of different mitochondrial complexes on a 5 to 18% polyacrylamide gradient. Respiratory complexes I to V were observed directly in gel (data not shown) and also by Western blot analysis by using mouse monoclonal antibodies against specific subunits of complexes I to V. The representative antibodies used were as follows: anti-OxPhos complex I 39-kDa, 30-kDa, and 17-kDa subunits; anti-OxPhos complex II 70-kDa subunit; anti-OxPhos complex III FeS subunit; anti-OxPhos complex IV subunit I; and anti-OxPhos complex V inhibitor protein (Molecular Probes). In-gel colorimetric reactions for oxidative phosphorylation of complex I and complex II were performed by BN-PAGE as described previously (35). Briefly, following BN-PAGE, the gels were rinsed with distilled water and then equilibrated in complex I reaction buffer (0.1 M Tris-HCl [pH 7.4]) for 10 min. The gels were subsequently incubated in fresh complex I reaction buffer plus 0.2 mM NADH as substrate and 0.2% nitroblue tetrazolium for 30 min. After this, the same BN gel was equilibrated with complex II reaction buffer (50 mM KH2PO4, 0.1 mM ATP, 4.5 mM EDTA [pH 7.4]) for 10 min, followed by incubation of the gel with fresh complex II reaction buffer with 10 mM succinate as substrate, 0.2% nitroblue tetrazolium, and 0.2 mM phenazine methosulfate for 30 min. The gels were destained overnight in 45% methanol and 10% acetic acid.
RESULTS
Disruption of the GRIM-19 gene.
To generate a deletion allele of the GRIM-19 gene, genomic clones containing the entire coding sequence of murine GRIM-19 were isolated from the mouse genomic library. The mouse GRIM-19 gene spans ∼8 kb containing four exons in the coding region. A targeting vector was constructed that consists of a neomycin resistance (neo) gene flanked by a 4.3-kb fragment derived from the 5′ portion and a 1.5-kb fragment from the 3′ portion of the GRIM-19 gene. In the targeted allele, 7 kb of the GRIM-19 gene including its four exons was replaced with the neo gene, thereby creating a null allele (Fig. 1A). ES cells were electroporated with the linearized targeting vector and selected in the presence of G418 and ganciclovir. Two ES clones bearing the targeted allele were isolated and confirmed by Southern blot (Fig. 1B) and PCR analyses (data not shown). These ES clones were microinjected into blastocysts derived from C57BL/6 mice to generate chimeric mice. From one clone, two male mice carrying the germ line transmission of the mutant allele were produced and either backcrossed to C57BL/SW females or crossed to 129SVJ females. The genotypes of the offspring were determined at the age of 20 days by PCR (Fig. 1C) and Southern blot analysis of DNA from the tail of the offspring (data not shown).
Loss of GRIM-19 leads to embryonic lethality.
Mice heterozygous for the targeted allele (GRIM-19+/−) were viable and fertile with no obvious phenotypic abnormalities. These mice were intercrossed to generate GRIM-19−/− mice and should have yielded Mendelian statistics of 25% wild-type, 50% heterozygous, and 25% homozygous mutants. However, of 421 mice examined, 33.7% were wild type and 66.3% were GRIM-19+/−. No GRIM-19−/− mice were obtained (Table 1). The results suggest that a homozygous deletion mutation of the GRIM-19 gene causes embryonic lethality.
TABLE 1.
Analysis of embryos from intercross of GRIM-19+/− mice
| Stage | No. of micea
|
|||
|---|---|---|---|---|
| +/+ (25%) | +/− (50%) | −/− (25%) | Total | |
| At weaning | 142 | 279b | 0 | 421 |
| E9.5 | 4 | 4 | 0 | 8 |
| E8.5 | 2 | 10 | 7 | 19 |
| E7.5 | 3 | 5 | 5 | 13 |
| E6.5 | 7 | 12 | 6 | 25 |
| E5.5 | 9 | 6 | 2 | 17 |
| E3.5 | 17 | 35 | 19 | 71 |
Genotyping by Southern blotting or PCR analysis.
Number of female mice, 127; number of male mice, 152.
To further assess the time of death, embryos from the heterozygous intercross were isolated with uterus, dissected, and genotyped at different embryonic days from E5.5 to E9.5 (Table 1). From E5.5 to E8.5, an average of 27% of total embryos was determined to be GRIM-19−/−. However, at E9.5, no GRIM-19−/− fetus was identified from deciduas containing normal fetuses, and ∼33% of deciduas were abnormal and contained no embryos, suggesting that GRIM-19−/− null embryos die before E9.5.
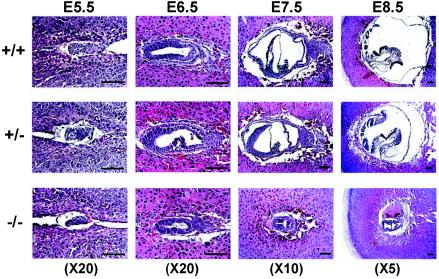
Histological analysis was performed to further characterize the GRIM-19−/− embryos at different gastrulation periods from E5.5 to E8.5 (Fig. 2). The genotype was determined by PCR amplification of DNA of microdissected embryonic tissue. The GRIM-19 null embryos were shown to be smaller than the wild-type and heterozygous embryos and disorganized at E5.5. The diminished GRIM-19−/− embryo persisted at E6.5. Furthermore, in contrast to the wild-type or GRIM-19+/− embryos which displayed normal gastrulation and early organogenesis at E7.5 and E8.5, respectively, GRIM-19 null embryos were much smaller and failed to undergo gastrulation at E7.5 and early organogenesis at E8.5. Although partial gastrulation was observed in the tiny embryo of the GRIM-19 null mutant at E8.5, this did not develop further as the embryos died at E9.5. These data indicate that the defects of GRIM-19−/− embryos appear at an early stage of embryonic development.
FIG. 2.
Histological analysis of embryos of wild-type, heterozygous, and homozygous mutants. Wild-type (+/+), GRIM-19+/−, and GRIM-19−/− embryos were dissected at 5 μm at E5.5, E6.5, E7.5, and E8.5 as indicated. The sections were stained with hematoxylin and eosin. The objective magnifications are indicated. Scale bar = 100 μm.
Retarded growth of GRIM-19−/− blastocysts in vitro.
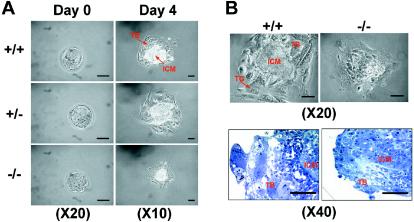
To further analyze the cause of the defect, we tried to establish ES cell lines with homologous deletions of GRIM-19 by several methods, but none of them was successful. We therefore studied the preimplantation stage embryos (blastocysts) isolated by uterine flushing at E3.5 and found that GRIM-19−/− embryos accounted for 27% of a total of 71 embryos (Table 1). Newly isolated GRIM-19−/− blastocysts (day 0) were viable and largely indistinguishable from those of the wild type. These blastocysts were cultured for various numbers of days in vitro to allow outgrowth and were eventually harvested for genotyping. The wild-type (GRIM-19+/+) and heterozygous (GRIM-19+/−) blastocysts for the targeted allele exhibited normal outgrowth of TB with giant cells (TG), which are necessary for inducing the decidual reaction during implantation, and of the ICM, which forms the future embryonic tissues in vivo. In contrast, in GRIM-19−/− blastocysts, the outgrowth of ICM was retarded. The sizes of the ICM and the TB were smaller, and no obvious TG cells were observed. Furthermore, a clear boundary between ICM and TB observed in the wild-type blastocysts was not seen in the mutant blastocysts (Fig. 3B, top panels). These observations were confirmed and displayed more clearly in the semithin sections stained with toluidine blue (bottom panels).
FIG. 3.
In vitro outgrowth of blastocysts. Intercrossed embryos at E3.5 were collected (day 0) and cultured for 4 days. The blastocysts were photographed at different objective magnifications as indicated. The bottom row of panel B shows semithin sections of the blastocysts stained with toluidine blue. TB, TG, and ICM are indicated by arrows. Scale bar = 50 μm.
Effect of GRIM-19 on proliferation and apoptosis in the blastocysts.
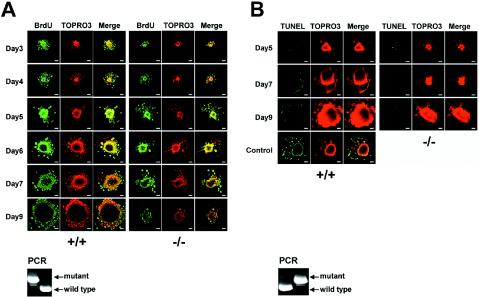
To further delineate the growth defect of the GRIM-19−/− blastocysts, we carried out BrdU incorporation assays during the blastocyst outgrowth. The DNA synthesis in the ICM and TB was detected in both wild-type and mutant blastocysts up to 5 days in culture even though the mutant blastocyst displayed smaller ICMs. The rate of DNA synthesis was decreased at days 6 and 7 and almost ceased at day 9 in the mutant blastocysts, as shown by the smaller ICMs and lower numbers of the labeled cells, indicating that DNA synthesis was diminished (Fig. 4A). To address whether the mutant blastocysts underwent spontaneous apoptosis, a TUNEL assay was performed in these blastocysts. No apparent apoptosis was observed in either wild-type or mutant blastocysts up to 9 days in culture (Fig. 4B). These results suggest that the growth retardation in the GRIM-19−/− blastocysts could be due to partially impaired DNA replication and cell proliferation in the ICM but not to cell death. The genotypes of the blastocysts were analyzed by PCR and are shown in Fig. 4.
FIG. 4.
Proliferation and apoptosis of the blastocysts. (A) BrdU incorporation during blastocyst outgrowth. E3.5 blastocysts from GRIM-19+/− intercrossed mice were isolated and cultured for various days. The cells were labeled with BrdU as described in Materials and Methods. TOPRO3 indicates nuclear staining. (B) TUNEL assay was performed as described in Materials and Methods. Control represents the normal blastocysts digested with DNase I prior to the TUNEL assay. The objective magnification is ×10. After assays, the genotypes of the blastocysts were determined by PCR, and representative results from one set of experiments are shown at the bottom of each panel. Scale bar = 50 μm.
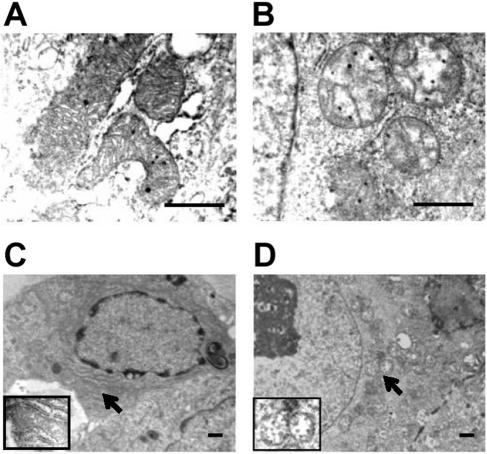
Abnormal structure, morphology, and distribution of mitochondria in GRIM-19−/− blastocysts.
We further examined the morphology of cellular organelles in the blastocysts cultured for 4 days in vitro with transmission electron microscopy. In the normal blastocysts (eight were selected with normal size and morphology), the cytoplasmic organelles including the mitochondria appeared normal. The mitochondria were rod shaped with abundant and well-organized cristae in the matrix (Fig. 5A). Twenty-two blastocysts with smaller and impaired ICM outgrowth were also examined. The most drastic changes in these blastocysts were observed in the mitochondria. All the mitochondria were rounded and appeared distended (Fig. 5B). Furthermore, the internal structures of these mitochondria became interrupted with poorly developed cristae. In addition, after being cultured for 7 days, mitochondria of the mutant blastocysts tended to be clustered in the perinuclear region (Fig. 5D) in comparison to evenly distributed cytoplasmic mitochondria in the normal blastocysts (Fig. 5C). These results indicated that the GRIM-19 deficiency resulted in alterations in the mitochondrial structure, morphology, and cellular distribution, suggesting an essential role of GRIM-19 in mitochondrial formation. The mitochondrial abnormality can be detected as early as 3 days after culture in vitro, and no obvious apoptosis was detected in the mutant blastocysts (Fig. 4B). The changes in mitochondria, therefore, are unlikely to be a consequence of cell death.
FIG. 5.
Abnormal morphology and cellular distribution of mitochondria in the mutant blastocysts. Ultrathin sections of the blastocysts were examined with a transmission electron microscope. The mitochondria of the normal (A and C) and the mutant (B and D) cells are indicated by arrows. Scale bar = 1 μm.
Localization of GRIM-19 in mitochondria.
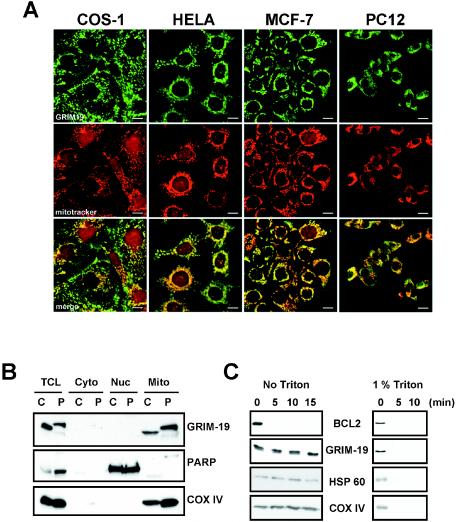
Human GRIM-19 was originally reported as a nuclear protein in HeLa cells (2). In our previous immunofluorescence experiments, transfected Myc-tagged GRIM-19 was detected with an anti-Myc antibody exclusively in the cytoplasm and colocalized with mitochondrial markers but not with endosome or lysosome markers. Furthermore, the endogenous GRIM-19 was detected primarily in the mitochondria with only trace amounts in the nucleus in human breast cancer cells when a rabbit polyclonal antibody against mouse GRIM-19 was used (17). However, a nuclear staining of GRIM-19 in HeLa cells was reported again recently (37). Since the GRIM-19−/− blastocysts showed mitochondrial abnormality, we decided to further examine the cellular localization of GRIM-19 in various cell types of different species, including HeLa (human cervical carcinoma endothelial cells), MCF-7 (human breast carcinoma endothelial cells), COS-1 (monkey kidney fibroblasts), PC12 (rat neuronal pheochromocytoma cells), and H9C2 (rat embryonic myoblasts) by immunofluorescence staining with a newly prepared mouse antibody against human GRIM-19 which displayed very low background. The results showed that GRIM-19 was exclusively localized in the cytoplasm with punctate staining that colocalized with MitoTracker Red in the cytoplasm in all cell lines examined (Fig. 6A and data not shown). Although MitoTracker showed a certain degree of nonspecific staining in the nucleus, no staining of GRIM-19 was detected in the nucleus. To further verify these results, COS-1 and PC12 cells were fractionated into cytoplasmic, mitochondrial, and nuclear portions, and GRIM-19 expression was determined. As shown in Fig. 6B, GRIM-19 was detected in the mitochondrial but not in the nuclear and the soluble cytoplasmic fractions in COS-1 and PC12 cells (top panel). Detection of the nuclear protein poly(ADP-ribose) polymerase (6) in the nuclear fraction and the mitochondrial protein cytochrome oxidase (complex IV) subunit IV (COX IV) in the mitochondrial fraction indicates an effective separation. These results further confirm that GRIM-19 is primarily localized in the mitochondria.
FIG. 6.
Cellular localization of GRIM-19. (A) Mitochondrial localization of GRIM-19 in various cell lines. Various cell types were grown in culture and treated with MitoTracker Red. After fixation, GRIM-19 protein expression and localization were detected by using a specific GRIM-19 mouse primary antibody and a fluorescein isothiocyanate-coupled anti-mouse secondary antibody (top frames). The cells were mounted and examined by using a Bio-Rad MRC 1024 laser scanning confocal microscope. Mitochondria were detected by MitoTracker Red (middle frames). Merged images are shown in the bottom frames. Scale bar = 10 μm. (B) COS-1 (C) and PC12 (P) cells were lysed and fractionated to cytoplasmic (Cyto), mitochondrial (Mito), and nuclear (Nuc) portions as described in Materials and Methods. The expression of GRIM-19 in total cell lysates and different fractions (containing 50 μg of proteins in each) were examined with Western blotting with antibody against GRIM-19. The blots were also probed with anti-poly(ADP-ribose) polymerase as a nuclear marker and anti-COX IV as a mitochondrial marker. (C) Submitochondrial localization of GRIM-19. Mitochondria were isolated from 293T cells and treated with 100 ng of proteinase K per ml for various times in the absence or the presence of 1% Triton X-100. The mitochondrial proteins were separated by SDS-PAGE and subjected to Western blot analysis with antibodies against BCL2, GRIM-19, heat shock protein 60 (HSP 60), and COX IV.
The submitochondrial localization of GRIM-19 was further investigated. Mitochondria were isolated and treated with proteinase K in the absence of Triton X-100. As shown in Fig. 6C, the mitochondrial outer membrane protein, BCL2, can be digested by proteinase K after a 5-min treatment. In contrast, GRIM-19 cannot be digested by proteinase K at various treatment durations under the same conditions. Similar results were observed for COX IV localized in the inner membrane and heat shock protein 60 (Hsp60) in the mitochondrial matrix. As a control, all the proteins could be digested when mitochondria were disrupted in the presence of Triton X-100. This result suggests that GRIM-19 is localized either in the inner membrane or in the matrix but not in the outer membrane of mitochondria.
GRIM-19 is a component of mitochondrial complex I.
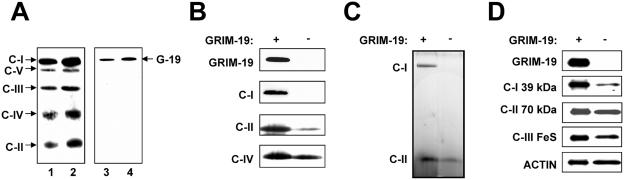
GRIM-19 was previously shown to be copurified with mitochondrial complex I in bovine heart (8). To obtain more direct evidence showing that GRIM-19 is a component of complex I, we carried out a BN-PAGE assay. BN-PAGE was developed as a method for analyzing membrane proteins and was shown to be particularly useful for characterizing the assembly of mitochondrial oxidative phosphorylation complexes and their enzymatic activities from tissues and cell lines (25, 26, 35). Mitochondria were isolated from mouse livers, and the enzyme complexes were resolved in BN-PAGE. Five major bands representing the oxidative phosphorylation complexes I to V were observed in gel (data not shown). The identity of the individual complexes was verified by Western blot analyses with antibodies against specific subunits of each complex (Fig. 7A, left panel). Complex I, the largest complex, was recognized by the antibody against the 39-kDa subunit (NDUFA9) (10). More importantly, the same complex was also recognized by anti-GRIM-19 antibody, whereas the other complexes were not (right panel). This result indicates that GRIM-19 is physically present in the native form of complex I.
FIG. 7.
Mitochondrial complex I assembly and activity assays. (A) Detection of GRIM-19 in complex I by BN-PAGE. Mitochondria from mouse liver were isolated, and 400 μg (lanes 1 and 3) and 600 μg (lanes 2 and 4) of mitochondrial proteins were solubilized and centrifuged. Aliquots of supernatants containing different mitochondrial respiratory complexes were separated by BN-PAGE as described in Materials and Methods and detected by Western blot analysis. Complexes I to V were detected by using mouse monoclonal antibodies against specific subunits of each complex that include the anti-OxPhos complex I 39-kDa subunit, anti-OxPhos complex II 70-kDa subunit, anti-OxPhos complex III FeS subunit, anti-OxPhos complex IV subunit I, and anti-OxPhos complex V inhibitor protein. GRIM-19 was detected in complex I with anti-GRIM-19 antibody (lanes 3 and 4). The positions of the complexes and GRIM-19 (G-19) are indicated by arrows. (B) GRIM-19 is essential for mitochondrial complex I assembly. Blastocysts were isolated and cultured in vitro for 7 days. Twenty-seven blastocysts containing the wild type and GRIM-19+/− and 27 blastocysts of GRIM-19−/− were combined and directly solubilized. A total of 20 μg of solubilized proteins was used to analyze the complex assembly as described in panel A. (C) GRIM-19 is essential for mitochondrial complex I activity. Samples from blastocysts were prepared as described in panel B. Solubilized protein (20 μg) was separated by BN-PAGE. In-gel colorimetric reactions for oxidative phosphorylation of complex I and complex II were performed as described in Materials and Methods. (D) Effect of GRIM-19 on the expression of subunits in the other complexes. Cell lysates from wild-type or GRIM-19−/− blastocysts were separated by SDS-PAGE and subjected to Western blot analysis with antibodies as indicated.
GRIM-19 is essential for mitochondrial complex I assembly and enzymatic activity.
We subsequently determined whether GRIM-19 is essential for the complex I function. As described above, BN-PAGE is a useful technique to analyze the assembly of intact multisubunit complexes of the mitochondrial respiratory chain and their enzymatic activity in oxidative phosphorylation. We adapted this technique to mouse blastocysts. Blastocysts were isolated from intercrossed mice and grown in vitro for 7 days. Twenty-seven blastocysts with normal outgrowth and ICM size (GRIM-19+/+ and GRIM-19+/−) and 27 blastocysts with retarded outgrowth of ICM (GRIM-19−/−) were combined. Proteins were extracted from these blastocysts and analyzed in two BN-PAGE assays in parallel, one for measuring the assembly of the complexes by Western blot analysis and another for enzymatic activity by an in-gel activity assay. As expected, GRIM-19 protein was detected in complex I of the wild-type blastocysts but was absent in the GRIM-19−/− blastocysts examined by Western blot analysis (Fig. 7B). Strikingly, complex I could not be detected by the antibody against the 39-kDa subunit of complex I (Fig. 7B) nor by two other antibodies against the 30- and 17-kDa subunits of complex I (data not shown) in the GRIM-19−/− blastocysts. In contrast, the wild-type blastocysts displayed a normal assembly of complex I. The assembly of complexes II and IV was also examined and shown to be at reduced levels in the GRIM-19−/− blastocysts (Fig. 7B). Furthermore, the electron transfer activity of complex I was examined and shown to be totally destroyed in the GRIM-19−/− blastocysts compared to that in the wild-type blastocysts. As a control, the enzymatic activity of complex II remained but was also reduced in the mutant blastocysts (Fig. 7C).
We also tested the protein level of subunits from different complexes. As expected, no GRIM-19 protein was detected in GRIM-19−/− blastocysts. The amount of the 39-kDa subunit of complex I was drastically decreased. In contrast, the amount of the 70-kDa subunit of complex II remained unchanged, although the level of subunit FeS of complex III was also reduced marginally (Fig. 7D). This suggests that without GRIM-19, other subunit proteins which cannot be correctly assembled into complexes will be degraded in the cell.
Together, our data suggest that GRIM-19 physically resides in complex I and plays an essential role in the assembly and the activity of complex I. Further, a defective complex I indirectly influences the assembly of other complexes to various degrees, indicating that there is tight communication between these complexes.
DISCUSSION
GRIM-19 is an essential component of mitochondrial complex I. Human GRIM-19 was originally identified as a cell death-related gene product in IFN-RA-treated breast cancer cell lines (2). Subsequently, its homolog was found in mitochondrial complex I purified from bovine heart (8). In agreement with the latter report, human GRIM-19 was also coimmunoprecipitated with an antibody against a complex I subunit, NDUFA6, from human heart mitochondria (20). These results raise a question about the role of GRIM-19 in cells: does it play a main role in the IFN-RA-induced pathway of cell death as a nuclear protein, or is it part of the mitochondrial complex I assembly? It is also possible that GRIM-19 participates in both fundamental biological processes. If so, how are the dual roles fulfilled? We attempted to address these questions by investigating the biological roles of GRIM-19 with genetic approaches. Our results show that a deficiency of GRIM-19 leads to the impaired assembly and consequent loss of enzymatic activity of complex I in mouse blastocysts. In addition, the assembly, enzymatic activity, and protein level of certain subunits in the other complexes of the mitochondrial respiratory chain were also altered to various degrees. Therefore, our data demonstrate that GRIM-19 is a component of complex I that is essential for the assembly of complex I and the integrity of the whole mitochondrial electron transfer chain.
In eukaryotes, the subunit composition of complex I is mostly defined by direct analysis of the proteins in the purified complex. This approach provides important information about the structural subunits of the complexes. In general, however, it is difficult to establish whether any subunit is a bona fide subunit unless a key functional role has been identified. The genetic approach provides such functional assays. GRIM-19, to our best knowledge, is the first subunit of complex I that is knocked out in mice. Cytochrome c, the only water-soluble component of the electron transfer chain, has been knocked out in mice. A deficiency of cytochrome c also causes embryonic lethality at E10.5 in mouse (14). Similar to the function of cytochrome c, the function of GRIM-19 is also critical for early embryonic development in mouse.
Mitochondrial complex I is the largest and the least understood among the respiratory chain complexes. Assembly of the complexes of the mitochondrial respiratory chain is a complicated, poorly understood process that requires various assembly factors. Defects in assembly of these complexes contribute to several identified mitochondrial diseases and possibly to cancers and neuronal degenerative diseases (5, 32). A deficiency of complex I is also frequently the cause for oxidative phosphorylation disorder (30). Prokaryotic complex I is simpler in that it consists of 13 to 14 subunits (34). The corresponding 14 subunits in eukaryotic complex I, which includes 7 subunits encoded by mitochondrial DNA and the rest encoded by nuclear genes, are defined as core subunits for the redox and proton translocating activities, whereas the other subunits are considered peripheral subunits which may carry out additional functions (31, 34). Since GRIM-19 is not a core subunit, perhaps GRIM-19 functions mainly in the assembly of complex I. Furthermore, because GRIM-19 is found to be tightly associated with complex Iλ, a subcomplex of complex I consisting of 14 subunits and representing the arm protruding into the mitochondrial matrix (8), GRIM-19 is likely to be involved in complex Iλ assembly. The 39-kDa subunit has been shown to be involved in the initial step of complex I assembly in patients with complex I deficiency (3). In our BN-PAGE assay, no complex I assembly intermediates or subcomplexes were detected with antibody against the 39-kDa subunit in the GRIM-19−/− blastocysts (Fig. 7B and data not shown). Moreover, this subunit can be coimmunoprecipitated with GRIM-19 antibody in mouse brain lysates and interacts directly with GRIM-19 in vitro (H. Lu and X. Cao, unpublished data). Therefore, GRIM-19 may play critical roles in the early assembly of the subcomplex of complex I. Conditional mutants of GRIM-19 or other subunits at a later developmental stage of mice could be used as models for human mitochondrial diseases.
Cellular localization of GRIM-19.
The issue of whether GRIM-19 is a nuclear protein or a mitochondrial protein has been controversial. To address this, various cells from different tissues and species have been examined, and it has been demonstrated by immunofluorescence with both mouse anti-human GRIM-19 (Fig. 6A) and rabbit anti-mouse GRIM-19 (17) that the primary cellular localization of GRIM-19 is in the mitochondria. The nuclear GRIM-19, if any, only exists in a trace amount. The results were verified by cellular fractionations (Fig. 6B). In addition, our complex I assembly assays clearly indicate that the primary location of GRIM-19 is in complex I in mitochondria (Fig. 7A and B). The reason that GRIM-19 was detected strongly in the nucleus with immunofluorescence staining in some reports (2, 37) could be due to nonspecific reactions of the antibodies to the nuclear protein(s), since we found that the normal rabbit immunoglobulin G showed nonspecific nuclear staining under confocal microscopy when a high power level was utilized (data not shown). On the other hand, it is still possible that GRIM-19 is able to be released from mitochondria and translocated to another cellular compartment including the nucleus under certain circumstances. However, nuclear translocation was not observed by treatment of the cells with IFN-β and RA in MCF-7 cells as well as in other cell types (reference 17 and data not shown). Taken together, our data demonstrate that the primary cellular localization of GRIM-19 is in the mitochondria.
Other possible functions of GRIM-19.
In spite of its primary role in mitochondrial complex I, other functions of GRIM-19 are not excluded. As described above, GRIM-19 was originally identified in IFN-β- and RA-treated cancer cells. Such treatment enhances the mRNA and protein levels of GRIM-19. Since overexpressing its antisense RNA improves the survival of the cell, whereas overexpression of the gene enhances death in IFN-β- and RA-treated cells, GRIM-19 is postulated to be a cell death-promoting gene. Although the molecular basis for the gene's apoptotic nature is largely unknown, involvement of caspases has been suggested (2). Interestingly, the recent report identified a protein, GW112, that is localized in mitochondria and interacts with GRIM-19 (37). This protein antagonizes the apoptotic role of GRIM-19 in the IFN-β- and RA-treated cells. Cytochrome c plays dual roles in the mitochondrial respiratory chain and apoptosis. Based on the data obtained so far, GRIM-19 may represent another potentially important gene involved in the function of mitochondria in life and death within a cell. Furthermore, since it has been controversial whether mitochondrion-mediated cell death depends on the activity of oxidative phosphorylation (21), further studies of the mechanism of the biological role of GRIM-19 will provide insight to these fundamental questions.
Acknowledgments
This work was supported by the Agency for Science, Technology and Research of Singapore. X. Cao is an adjunct staff member of the Department of Biochemistry, National University of Singapore.
We are grateful to Christopher J. Leaver and Mohammed Sabar for protocols of the mitochondrial complex assays and for valuable suggestions. We thank Baohong Lin and Jie Li for technical assistance. We also thank the staffs in the In Vivo Model System Facility, Histology Unit, and DNA Sequencing and Analysis Facility in our institute and the Electron Microscopy Facility in the Department of Anatomy of the National University of Singapore. We are grateful to Wanjin Hong for valuable comments on the manuscript and Cheh Peng Lim for reading the manuscript critically.
REFERENCES
- 1.Akira, S., Y. Nishio, M. Inoue, X. J. Wang, S. Wei, T. Matsusaka, K. Yoshida, T. Sudo, M. Naruto, and T. Kishimoto. 1994. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 77:63-71. [DOI] [PubMed] [Google Scholar]
- 2.Angell, J. E., D. J. Lindner, P. S. Shapiro, E. R. Hofmann, and D. V. Kalvakolanu. 2000. Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J. Biol. Chem. 275:33416-33426. [DOI] [PubMed] [Google Scholar]
- 3.Antonicka, H., I. Ogilvie, T. Taivassalo, R. P. Anitori, R. G. Haller, J. Vissing, N. G. Kennaway, and E. A. Shoubridge. 2003. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J. Biol. Chem. 278:43081-43088. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, J., I. M. Fearnley, R. J. Shannon, J. Hirst, and J. E. Walker. 2003. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol. Cell. Proteomics 2:117-126. [DOI] [PubMed] [Google Scholar]
- 5.Dahl, H. H., and D. R. Thorburn. 2001. Mitochondrial diseases: beyond the magic circle. Am. J. Med. Genet. 106:1-3. [DOI] [PubMed] [Google Scholar]
- 6.D'Amours, D., S. Desnoyers, I. D'Silva, and G. G. Poirier. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342:249-268. [PMC free article] [PubMed] [Google Scholar]
- 7.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 8.Fearnley, I. M., J. Carroll, R. J. Shannon, M. J. Runswick, J. E. Walker, J. Hirst. 2001. GRIM-19, a cell death regulatory gene product, is a subunit of bovine mitochondrial NADH:ubiquinone oxidoreductase (complex I). J. Biol. Chem. 276:38345-38348. [DOI] [PubMed] [Google Scholar]
- 9.Hatefi, Y. 1985. The mitochondrial electron transport and oxidative phosphorylation system. Annu. Rev. Biochem. 54:1015-1069. [DOI] [PubMed] [Google Scholar]
- 10.Hirst, J., J. Carroll, I. M. Fearnley, R. J. Shannon, and J. E. Walker. 2003. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta 1604:135-150. [DOI] [PubMed] [Google Scholar]
- 11.Hofman, E. R., M. Boyanapalli, D. J. Lindner, X. Weihua, B. A. Hassel, R. Jagus, P. L. Gutierrez, and D. V. Kalvakolanu. 1998. Thioredoxin reductase mediates cell death effects of the combination of beta interferon and retinoic acid. Mol. Cell. Biol. 18:6493-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ihle, J. N. 2001. The Stat family in cytokine signaling. Curr. Opin. Cell Biol. 13:211-217. [DOI] [PubMed] [Google Scholar]
- 13.Lai, C. H., C. Y. Chou, L. Y. Ch'ang, C. S. Liu, and W. Lin. 2000. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 10:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, K., Y. Li, J. M. Shelton, J. A. Richardson, E. Spencer, Z. J Chen, X. Wang, and R. S. Williams. 2000. Cytochrome c deficiency causes embryonic lethality and attenuates stress-induced apoptosis. Cell 101:389-399. [DOI] [PubMed] [Google Scholar]
- 15.Lindner, D. J., E. C. Borden, and D. V. Kalvakolanu. 1997. Synergistic antitumor effects of a combination of interferons and retinoic acid on human tumor cells in vitro and in vivo. Clin. Cancer Res. 3:931-937. [PubMed] [Google Scholar]
- 16.Love, J. M., and L. J. Gudas. 1994. Vitamin A, differentiation and cancer. Curr. Opin. Cell Biol. 6:825-831. [DOI] [PubMed] [Google Scholar]
- 17.Lufei, C., J. Ma, G. Huang, T. Zhang, V. Novotny-Diermayr, C. T. Ong, and X. Cao. 2003. GRIM-19, a death-regulatory gene product, suppresses Stat3 activity via functional interaction. EMBO J. 22:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lykke-Andersen, K., L. Schaefer, S. Menon, X. W. Deng, J. B. Miller, and N. Wei. 2003. Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol. Cell. Biol. 23:6790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore, D. M., D. V. Kalvakolanu, S. M. Lippman, J. J. Kavanagh, W. K. Hong, E. C. Borden, M. Paredes-Espinoza, and I. H. Krakoff. 1994. Retinoic acid and interferon in human cancer: mechanistic and clinical studies. Semin. Hematol. 31:31-37. [PubMed] [Google Scholar]
- 20.Murray, J., B, Zhang, S. W. Taylor, D. Oglesbee, E. Fahy, M. F. Marusich, S. S. Ghosh, and R. A. Capaldi. 2003. The subunit composition of the human NADH dehydrogenase obtained by rapid one-step immunopurification. J. Biol. Chem. 278:13619-13622. [DOI] [PubMed] [Google Scholar]
- 21.Newmeyer, D. D., and S. Ferguson-Miller. 2003. Mitochondria: releasing power for life and unleashing the machineries of death. Cell 112:481-490. [DOI] [PubMed] [Google Scholar]
- 22.Rajapakse, N., K. Shimizu, M. Payne, and D. Busija. 2001. Isolation and characterization of intact mitochondria from neonatal rat brain. Brain Res. Protoc. 8:176-183. [DOI] [PubMed] [Google Scholar]
- 23.Saraste, M. 1999. Oxidative phosphorylation at the fin de siècle. Science 283:1488-1493. [DOI] [PubMed] [Google Scholar]
- 24.Schägger, H., and K. Pfeiffer. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 19:1777-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schägger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 26.Schägger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 27.Schultz, B. E., and S. I. Chan. 2001. Structures and proton-pumping strategies of mitochondrial respiratory enzymes. Annu. Rev. Biophys. Biomol. Struct. 30:23-65. [DOI] [PubMed] [Google Scholar]
- 28.Skehel, J. M., I. M. Fearnley, and J. E. Walker. 1998. NADH:ubiquinone oxidoreductase from bovine heart mitochondria: sequence of a novel 17.2-kDa subunit. FEBS Lett. 438:301-305. [DOI] [PubMed] [Google Scholar]
- 29.Stark, G. R., I. M. Kerr, B. R. Williams, R H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 30.Triepels, R. H., L. P. Van Den Heuvel, J. M. Trijbels, and J. A. Smeitink. 2001. Respiratory chain complex I deficiency. Am. J. Med. Genet. 106:37-45. [DOI] [PubMed] [Google Scholar]
- 31.Walker, J. E. 1992. The NADH:ubiquinone oxidoreductase (complex I) of respiratory chains. Q. Rev. Biophys. 25:253-324. [DOI] [PubMed] [Google Scholar]
- 32.Wallace, D. C. 1999. Mitochondrial diseases in man and mouse. Science 283:1482-1488. [DOI] [PubMed] [Google Scholar]
- 33.Weiss, H., T. Friedrich, G. Hofhaus, and D. Preis. 1991. The respiratory-chain NADH dehydrogenase (complex I) of mitochondria. Eur. J. Biochem. 197:563-576. [DOI] [PubMed] [Google Scholar]
- 34.Yagi, T., T. Yano, S. Di Bernardo, and A. Matsuno-Yagi. 1998. Procaryotic complex I (NDH-1), an overview. Biochim. Biophys. Acta 1364:125-133. [DOI] [PubMed] [Google Scholar]
- 35.Zerbetto, E., L. Vergani, and F. Dabbeni-Sala. 1997. Quantification of muscle mitochondrial oxidative phosphorylation enzymes via histochemical staining of blue native polyacrylamide gels. Electrophoresis 18:2059-2064. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, J., J. Yang, S. K. Roy, S. Tininini, J. Hu, J. F. Bromberg, V. Poli, G. R. Stark, and D. V. Kalvakolanu. 2003. The cell death regulator GRIM-19 is an inhibitor of signal transducer and activator of transcription 3. Proc. Natl. Acad. Sci. USA 100:9342-9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, X., Q. Huang, Z. Yang, Y. Li, and C. Y. Li. 2004. GW112, a novel antiapoptotic protein that promotes tumor growth. Cancer Res. 64:2474-2481. [DOI] [PubMed] [Google Scholar]