Abstract
Myocardin is a transcriptional coactivator that regulates cardiac and smooth muscle gene expression by associating with serum response factor. We show that GATA transcription factors can either stimulate or suppress the transcriptional activity of myocardin, depending on the target gene. Modulation of myocardin activity by GATA4 is mediated by the physical interaction of myocardin with the DNA binding domain of GATA4 but does not require binding of GATA4 to DNA. Paradoxically, the transcription activation domain of GATA4 is dispensable for the stimulatory effect of GATA4 on myocardin activity but is required for repression of myocardin activity. The ability of GATA transcription factors to modulate myocardin activity provides a potential mechanism for fine tuning the expression of serum response factor target genes in a gene-specific manner.
Serum response factor (SRF) is a widely expressed transcription factor belonging to the MADS (MCM1, Agamous, Deficiens, SRF) box family of proteins (28, 34). SRF binds as a homodimer to a DNA consensus sequence known as a CArG box (CC[A/T]6GG), which is found in the control regions of numerous growth factor-regulated and muscle-specific genes (30). The spectrum of SRF target genes expressed by a cell is dependent on association of SRF with a wide variety of positive and negative cofactors, many of which are signal responsive and tissue restricted (35).
Myocardin is a powerful SRF coactivator expressed specifically in cardiac and smooth muscle cells (37, 38). Myocardin belongs to the SAP (scaffold-attachment factor A/B, Acinus, PIAS) domain family of nuclear proteins, which bind A/T-rich genomic regions known as scaffold or matrix attachment regions and have been implicated in chromatin remodeling (1). Although myocardin lacks sequence-specific DNA binding activity, it forms a stable DNA-protein complex with SRF, resulting in activation of SRF target genes in muscle cells (37). Myocardin is necessary (19) and sufficient (8, 39, 40) to activate smooth muscle gene expression in nonmuscle cells and uses SRF as an obligate partner in this process (38, 39). Expression of a dominant negative myocardin mutant in Xenopus embryos is also sufficient to extinguish cardiac gene expression (37), suggesting an essential role for myocardin or other members of the myocardin family in cardiogenesis. Two myocardin-related transcription factors (MRTFs), referred to as MRTF-A (21, 22, 32, 38) and MRTF-B (38), also interact with SRF and stimulate transcription through the CArG box, but these factors are not muscle restricted and are likely to modulate SRF activity in response to growth factor signaling (24).
SRF has also been shown to stimulate expression of smooth and cardiac muscle genes in association with a variety of homeodomain proteins (6, 9), LIM domain proteins (4), and GATA transcription factors (2, 26, 33). The six GATA factors share homology in two zinc finger domains that mediate DNA binding and cofactor interactions (5, 23). GATA4, -5, and 6 are expressed predominantly in cardiac and smooth muscle cell lineages, where they play diverse roles in differentiation, morphogenesis, and growth (15). GATA 4 is required for proper embryonic folding and heart tube formation (13, 25). GATA 4 and 5 have also been implicated in cardiac gene expression (14, 31), and GATA 6 is essential for mesoderm formation during gastrulation (12, 27).
Because myocardin and GATA factors both interact with SRF and participate in cardiac and smooth muscle gene expression, we investigated whether they might modulate each other's activities. Here, we show that GATA4 augments the activity of myocardin on some genes, such as the cardiac homeobox gene Nkx2.5, whereas it interferes with the activity of myocardin on other genes, such as the atrial natriuretic factor (ANF) gene. Modulation of myocardin activity by GATA4 is mediated by the direct physical interaction between the factors and is dependent on SRF DNA binding sites (CArG boxes) but not on GATA4 DNA binding. Modulation of myocardin activity by GATA4 provides a mechanism for fine tuning the expression of SRF target genes in a promoter-specific manner.
MATERIALS AND METHODS
Cell culture and transfection assays.
COS cell transfections and luciferase assays were performed as described (37). Unless otherwise indicated, 100 ng of reporter plasmid and 100 ng of each activator plasmid were used. The total amount of DNA per well was kept constant by adding the corresponding amount of expression vector without a cDNA insert.
The myocardin and SRF expression vectors have been described (6, 37, 38). Myocardin and GATA4 deletion mutants were generated through PCR-based mutagenesis with the QuikChange kit from Stratagene. GATA4 point mutants in the DNA binding domain were generously provided by Bruce Markham and have been described (7). All mutations were confirmed by DNA sequencing. The SM22-luciferase reporter contained the 1,343-bp promoter (17). The 4× SM22-CArG-luciferase reporter has been described (3). pCMV-lacZ was included as an internal control for variations in transfection efficiency.
The NK-tk-luciferase reporter was constructed by linking 5′ upstream sequences from −9432 to −8923 of the mouse Nkx2.5 gene (20) to a luciferase reporter with a thymidine kinase promoter. The NK-CArG-mutant-luciferase reporter has mutations in the CArG box from CCTTTTAAGG to AAGCTTAAGG. All the other reporters have been described previously (37).
GST protein binding assays.
The plasmid encoding a glutathione S-transferase (GST) fusion protein was transformed into Escherichia coli BL21-Codon Plus cells (Stratagene). The cells were grown at 37°C in 2XYT medium to an optical density of 1.0. Isopropylthiogalactopyranoside (IPTG) (50 μM) was then added to the culture to induce protein expression. After shaking at room temperature for 4 to 6 h, the cells were harvested and the GST protein was purified with glutathione beads according to Amersham's procedure.
Proteins translated in vitro were labeled with [35S]methionine with a TNT T7 reticulocyte lysate system (Promega). Glutathione beads conjugated with 1 μg of protein were incubated with 10 μl of TNT product at 4°C for 2 h in 500 μl GST binding buffer (20 mM Tris, pH 7.3, 150 mM NaCl, 0.5% NP-40, protease inhibitor cocktail from Roche, and 1 mM phenylmethylsulfonyl fluoride). The beads were washed three times with GST binding buffer. Fifty microliters of sodium dodecyl sulfate (SDS) loading buffer was then added to the beads. After boiling, 20 μl was loaded onto an SDS-polyacrylamide gel electrophoresis (PAGE) gel.
Reverse transcription-PCR.
Total RNA was isolated with Trizol reagent (Invitrogen). After treatment with DNase I, 1 μg of RNA was used as a template for reverse transcription with random hexamer primers that spanned introns in the genes. Sequences of primers are available upon request. Reverse transcription-PCRs were performed under conditions of linearity with respect to input RNA.
Gel mobility shift assays.
SRF and myocardin were translated in vitro with a TNT T7-coupled reticulocyte lysate system (Promega), and gel mobility shift assays were performed with double-stranded probes as described (3). The sequence of the top strand of the Nkx2.5 CArG box probe was GCCCCCCCAAGTTTAAATGCTCCTTTAAGGGCTTGAGTGTCTGCAGC (CArG box is in italics).
RESULTS
Synergistic activation of the Nkx2.5 enhancer by myocardin and GATA4.
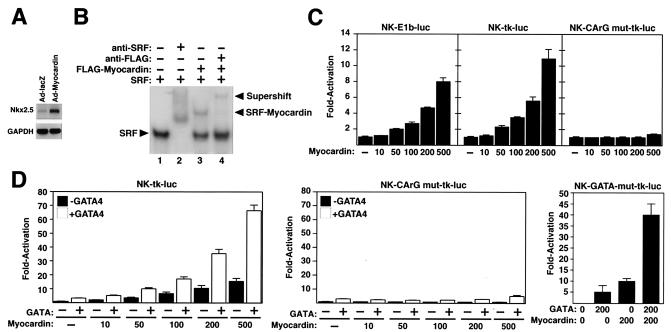
A dominant negative myocardin mutant suppresses Nkx2.5 expression in Xenopus embryos and in the P19 embryonal carcinoma cell line (36, 37). To test whether myocardin was able to stimulate the expression of Nkx2.5 in cardiac myocytes, we infected primary rat neonatal cardiac myocytes with an adenovirus encoding myocardin (Ad-myocardin). Indeed, the myocardin-expressing virus up-regulated Nkx2.5 expression by fivefold compared to a control virus expressing lacZ (Ad-lacZ) (Fig. 1A).
FIG. 1.
Myocardin and GATA4 synergistically activate the Nkx2.5 enhancer. (A) Primary neonatal cardiomyocytes were infected with adenoviruses encoding myocardin or lacZ (as a negative control). Four days later, expression of Nkx2.5 transcripts was measured by semiquantitative reverse transcription-PCR. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts were detected as a loading control. (B) Gel mobility shift assays were performed with a radiolabeled oligonucleotide corresponding to the CArG box in the Nkx2.5 enhancer and in vitro-translated SRF and myocardin proteins, as indicated. Myocardin formed a stable ternary complex with SRF on this sequence (lane 3). Anti-SRF antibody supershifted the SRF complex (lane 2), and anti-Flag antibody supershifted the ternary complex formed by SRF and Flag-myocardin (lane 4). (C) COS cells were transiently transfected with the luciferase reporter plasmids shown above each panel and the indicated amounts (in nanograms) of myocardin expression vector. Luciferase activity was assayed and is expressed as fold activation above the level of expression of the reporter gene alone. (D) COS cells were transiently transfected with the luciferase reporter plasmids shown above each panel and the indicated amounts (in nanograms) of myocardin expression vector with and without a GATA4 expression vector (100 ng), and luciferase activity was determined as in panel C.
Based on the above findings, we examined the early cardiac enhancer of the mouse Nkx2.5 gene for CArG boxes that might confer responsiveness to myocardin. The enhancer, which is located between bp −9432 and −8923 upstream of the Nkx2.5 gene, contains a single CArG box (20). In gel mobility shift assays, SRF bound avidly to this sequence and gave rise to a ternary complex in the presence of myocardin (Fig. 1B).
Transfection of COS cells with a myocardin expression vector and a luciferase reporter linked to the Nkx2.5 enhancer and the viral E1b promoter (NK-E1b-luc) showed that myocardin was able to transactivate the enhancer (Fig. 1C). Similar results were obtained when the enhancer was combined with the thymidine kinase (tk) basal promoter (NK-tk-luc), whereas an enhancer with a mutation in the CArG box (NK-CArGmut-tk-luc) was refractory to the activity of myocardin (Fig. 1C). Myocardin also activated the Nkx2.5 enhancer in transfected HeLa and 10T1/2 cells (data not shown).
Myocardin is a relatively weak activator through single CArG boxes (37). We therefore tested whether its transcriptional potency might be enhanced in the presence of GATA4, which binds two essential GATA sites in the Nkx2.5 enhancer (20). As shown in Fig. 1D, GATA4 alone was a very weak activator of the Nkx2.5 enhancer, but it synergized with myocardin to activate the enhancer. The synergy between GATA4 and myocardin was abolished by mutation of the CArG box (NK-CArG mut-tk-luc; Fig. 1D). Surprisingly, however, the synergy was not affected when the two GATA sites were mutated (NK-GATA-mut-tk-luc; Fig. 1D). We conclude that myocardin transactivates the Nkx2.5 enhancer by associating with SRF on the CArG box in this enhancer and that GATA4 can potentiate the effect of myocardin on the enhancer through a mechanism independent of GATA4 DNA binding.
Repression of myocardin activity by GATA4.
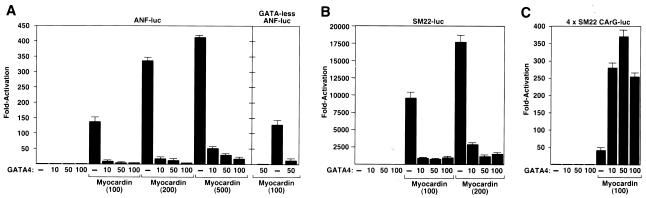
We next tested whether myocardin and GATA4 could cooperate to activate the ANF promoter, which contains a pair of binding sites for both factors (10, 16, 26). Myocardin activates this promoter much more effectively than the Nkx2.5 enhancer, at least in part because it contains two CArG boxes (Fig. 2A) (37). Unexpectedly, GATA4 potently repressed myocardin-dependent activation of the ANF promoter, such that as little as 10 ng of GATA4 expression plasmid resulted in a near-complete inhibition of ANF-luciferase expression (Fig. 2A).
FIG. 2.
Differential effects of GATA4 on myocardin-dependent transcription. COS cells were transiently transfected with luciferase reporter plasmids controlled by (A) the ANF promoter or a mutant promoter lacking the two GATA4 binding sites, (B) the SM22 promoter, or (C) four copies of the SM22 CArG box and the indicated amounts (in nanograms) of myocardin and GATA4 expression vectors. Luciferase activity was assayed and is expressed as fold activation above the level of expression of the reporter gene alone.
To determine whether the GATA sites in the promoter were required for repression, we examined the effects of myocardin and GATA4 on a mutant ANF promoter in which the two GATA sites were mutated. The mutant promoter (GATA-less ANF-luc) was activated by myocardin and repressed by GATA4 as effectively as the wild-type promoter (Fig. 2A). These results suggested that the repressive effect of GATA4 did not require direct binding of GATA4 to DNA.
To further investigate the potential modulation of myocardin activity by GATA4, we tested the effect of GATA4 on the SM22 promoter, which is extremely sensitive to myocardin and contains two essential CArG boxes but no GATA sites (8, 11). GATA4 potently suppressed the ability of myocardin to activate the SM22 promoter (Fig. 2B), supporting the notion that DNA binding by GATA4 was not involved in the repressive mechanism.
GATA4 enhances myocardin activity on a multimerized CArG box.
To determine whether the CArG box was sufficient to confer responsiveness of myocardin to GATA4, we tested a reporter gene containing the tk promoter linked to four tandem copies of the promoter-proximal SM22 CArG box (4xSM22 CArG-luc). As shown in Fig. 2C, this reporter was activated by myocardin but, in contrast to the behavior of the ANF and SM22 promoters, activation by myocardin was dramatically augmented by GATA4. In the absence of myocardin, GATA4 had no effect on the reporter, demonstrating that the stimulation of transcription by GATA4 required myocardin. Myocardin did not affect the expression of a reporter containing six tandem copies of the GATA binding site without a CArG box (data not shown), further suggesting that the positive and negative influences of GATA4 on myocardin were dependent on the presence of SRF binding sites and were independent of GATA4 DNA binding. Together, the above results demonstrated that GATA4 was able to discriminate between myocardin target genes and modulate myocardin activity, either positively or negatively, in a promoter context-dependent manner.
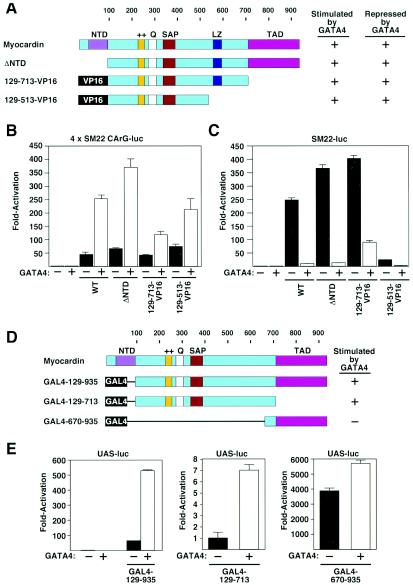
Mapping the GATA4-responsive region of myocardin.
To determine whether stimulation and suppression of myocardin activity by GATA4 were dependent on the same domain of the myocardin protein, we examined the effects of GATA4 on a series of myocardin deletion mutants (Fig. 3A to C). The transcriptional activity of a myocardin deletion mutant lacking the conserved N-terminal domain (ΔNTD) was stimulated by about an order of magnitude in the presence of GATA4 when assayed with the reporter controlled by the multimerized SM22 CArG box (4xSM22-CArG-luc; Fig. 3B). Deletion mutants in which residues from amino acid 513 to the C terminus, including the transcription activation domain (TAD), were deleted and replaced with VP16 (mutants 129-513-VP16 and 129-713-VP16) were also stimulated by GATA4 (Fig. 3A and B and data not shown). Deletion mutants that were stimulated by GATA4 on the multimerized SM22 CArG box reporter retained the ability to be repressed by GATA4 on the SM22 and ANF promoters (Fig. 3A to C and data not shown). These results suggested that the positive and negative effects of GATA4 were dependent on the same region of myocardin.
FIG. 3.
Mapping the region of myocardin that responds to GATA. (A) Schematic diagrams of myocardin mutants. The potential of each protein to be stimulated by GATA4 on the 4xSM22-CArG promoter or repressed by GATA4 on the SM22 promoter is indicated. (B and C) COS cells were transiently transfected with the luciferase reporter plasmids shown above each panel and expression vectors encoding GATA4 (100 ng) and myocardin or myocardin mutants (100 ng) shown in panel A, as indicated. Luciferase activity was assayed and is expressed as fold activation above the level of expression of the reporter gene alone. (D) Schematic diagrams of GAL4-myocardin fusion proteins. The potential of each protein to be stimulated by GATA4 is indicated. (E) COS cells were transiently transfected with an upstream activation sequence-luciferase reporter plasmid and expression vectors encoding GAL4-myocardin fusion proteins (100 ng) shown in panel D with and without a GATA4 expression vector (100 ng), as indicated. Luciferase activity was assayed and is expressed as fold activation above the level of expression of the reporter gene alone.
To determine whether the effects of GATA4 were mediated by myocardin or SRF (or another protein), we tested whether GATA4 affected the transcriptional activity of myocardin fused to the GAL4 DNA binding domain (Fig. 3D and E). GATA4 stimulated activity of a GAL4-myocardin fusion protein extending from amino acid 129 to the C terminus (GAL4-129-935). The TAD of myocardin, located between amino acid 713 and the C terminus, shows a greater than 10-fold increase in activity when it is isolated from the remainder of the protein (37). The TAD fused to the GAL4 DNA binding domain (GAL4-670-935) showed only a marginal response to GATA4. In contrast, residues 129 to 713, which are inactive alone (GAL4-129-713), showed about a sevenfold increase in transcriptional activity in the presence of GATA4. The finding that GATA4 was able to stimulate transcription via a portion of myocardin lacking its own transcription activation domain suggested that myocardin acted as a bridge between GATA4 and the transcriptional machinery (see below).
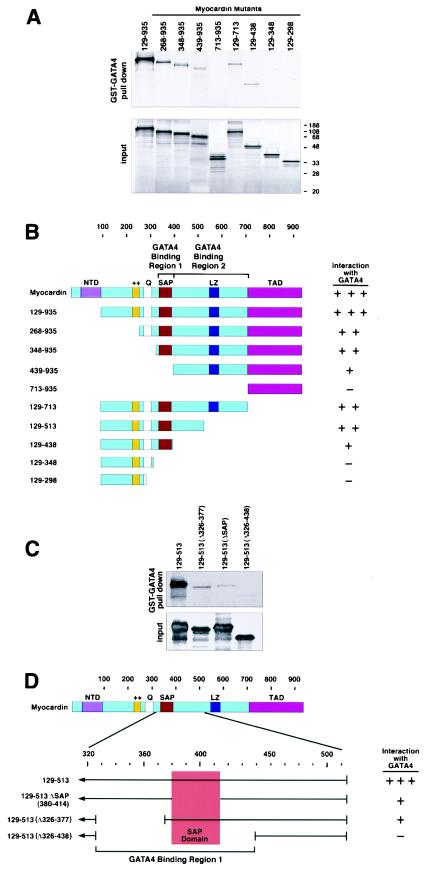
Mapping the GATA4 binding region of myocardin.
To determine whether the modulation of myocardin activity by GATA4 required direct physical interaction between the proteins, we performed GST pulldown assays with GST-GATA4 fusion protein and myocardin deletion mutants translated in vitro. As shown in Fig. 4, GATA4 interacted efficiently with myocardin. Amino-terminal deletion mutants up to residue 348 retained the ability to interact strongly with GATA4. A deletion mutant lacking residues 1 to 438 (mutant 439-935) was also able to interact with GATA4, albeit less strongly than mutants containing additional N-terminal sequences. The TAD of myocardin, contained within residues 713 to 935, did not interact with GATA4.
FIG.4.
Mapping the region of myocardin that interacts with GATA4. (A) A GST-GATA4 fusion protein encompassing amino acids 177 to 332 was incubated with [35S]methionine-labeled myocardin proteins translated in vitro. Myocardin proteins associated with GST-GATA4 are shown at the top, and 10% of the input proteins are shown at the bottom. (B) Summary of the GST-GATA4 pulldown and coimmunoprecipitation assays (data not shown). (C) GST pulldown assays were performed with the indicated myocardin mutants and GST-GATA4 as described for panel A. (D) Summary of the GST-GATA4 pulldown assays in panel C.
Deletions from the C terminus showed that residues 129 to 513 retained strong GATA4 binding activity. A smaller deletion mutant lacking additional C-terminal sequences (mutant 129-438) also bound GATA4, but its binding activity was reduced. Further deletions from the C-terminal end to residue 348 or 298 (mutants 129-348 and 129-298, respectively) abolished binding.
The above deletions suggested that myocardin contained two GATA4-binding regions, one between residues 129 and 438 (region 1) and another between residues 439 and 713 (region 2) (Fig. 4B). Since the former region encompasses the SRF-binding domain and SAP domains, which are critical to the functions of myocardin, we created a series of smaller deletions within this region in order to further pinpoint this GATA4-binding domain. As shown in Fig. 4C and D, deletion of the SAP domain (amino acids 380 to 414) severely impaired but did not eliminate GATA4-binding activity (mutant 129-513ΔSAP). Similarly, deletion of residues 326 to 377 reduced GATA4 activity, and a deletion combining both regions (mutant Δ326 to 438) abolished GATA4 binding. We conclude that the GATA4-binding region 1 of myocardin maps to an extended sequence from 348 to 438 that encompasses the SAP domain. This GATA4-binding region of myocardin defined by GST pulldown assays correlated with the region of myocardin required for stimulation and suppression of myocardin activity by GATA4.
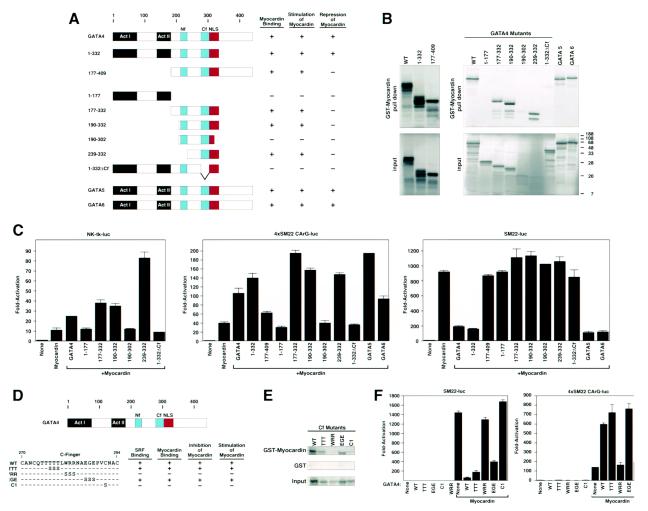
Mapping the myocardin-interacting region of GATA4.
The region of GATA4 that interacted with myocardin was mapped by coimmunoprecipitation and GST pulldown assays with a GST-myocardin fusion protein containing residues 129 to 510 and a series of GATA4 deletion mutants translated in vitro (Fig. 5A and B, and data not shown). Deletion of residues from amino acid 332 to the C terminus (mutant 1-332) did not affect binding of GATA4 to myocardin. Residues 1 to 177 of GATA4, which encompass the two TADs, did not interact with myocardin, whereas a strong interaction was observed with residues 177 to 332 and 190 to 332, which encompass the two zinc fingers and the nuclear localization sequence (NLS). Deletion of the N-terminal zinc finger (Nf) did not affect binding to myocardin (mutant 239 to 332), whereas deletion of the C-terminal zinc finger (Cf) or the NLS abolished myocardin binding (mutants 1-332ΔCf and 190-302, respectively). GATA5 and GATA6, which share extensive homology with GATA4, also interacted with myocardin (Fig. 5B).
FIG. 5.
Mapping the region of GATA4 that interacts with myocardin. (A) Schematic diagrams of GATA4 mutants and their activities. (B) A GST-myocardin fusion protein encompassing amino acids 129 to 510 was incubated with [35S]methionine-labeled GATA proteins translated in vitro. GATA4 proteins associated with GST-myocardin are shown at the top, and 10% of the input proteins are shown at the bottom. (C) COS cells were transiently transfected with the luciferase reporter plasmids shown above each panel and expression vectors encoding myocardin (100 ng) and GATA4 or GATA4 mutants (100 ng) shown in panel A, as indicated. Luciferase activity was assayed and is expressed as fold activation above the level of expression of the reporter alone. (D) Point mutants in Cf of GATA4 and their activities. (E) A GST-myocardin fusion protein encompassing amino acids 129 to 510 was incubated with [35S]methionine-labeled GATA proteins translated in vitro. GATA4 proteins associated with GST-myocardin or GST are shown at the top and middle, respectively, and 10% of the input proteins are shown at the bottom. (F) COS cells were transiently transfected with the luciferase reporter plasmids shown above each panel and expression vectors encoding myocardin (100 ng) and GATA4 or GATA4 mutants (100 ng) shown in panel D, as indicated. Luciferase activity was assayed and is expressed as fold activation above the level of expression of the reporter alone.
The ability of GATA4 deletion mutants to interact with myocardin correlated precisely with the ability to synergize with myocardin to activate the Nkx2.5 enhancer or the multimerized SM22 CArG box (Fig. 5A and C). Thus, only the Cf and NLS of GATA4 appeared to be required for myocardin binding and stimulation of myocardin activity. In contrast, binding to myocardin and repression of myocardin activity on the SM22-luciferase reporter could be uncoupled in certain deletion mutants. For example, repression required the amino-terminal TADs of GATA4 and the myocardin-binding region; deletion mutants containing only one domain or the other were unable to repress myocardin activity (e.g., mutants 1-177 and 177-332). In fact, the only GATA4 deletion mutant able to repress myocardin activity was mutant 1-332, which lacks the extreme C-terminal residues. GATA5 and GATA6 also activated and repressed myocardin activity as effectively as GATA4.
To further pinpoint the residues in the Cf of GATA4 that are critical for myocardin binding, we tested a series of point mutants for their abilities to bind the GST-myocardin fusion protein. Mutations that disrupted myocardin binding were also defective in stimulation of the multimerized SM22 CArG box and repression of the SM22 promoter with myocardin (Fig. 5D to F). The two mutants (WRR and C1) that were defective in myocardin binding also failed to interact with SRF (data not shown). Thus, we cannot distinguish whether GATA4 modulates myocardin activity on SRF-dependent promoters by associating with myocardin directly or with SRF or both. However, the ability of GATA4 to enhance the activity of a GAL4-myocardin fusion protein, which is SRF independent, suggests that GATA4 can act through a direct effect on myocardin.
DISCUSSION
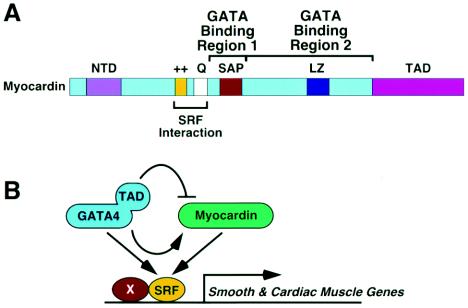
The major finding of this study is that GATA4 can stimulate or suppress myocardin activity in a target gene-specific manner. Modulation of myocardin activity by GATA4 is mediated, at least in part, by the direct interaction of GATA4 with myocardin and does not require binding of GATA4 to DNA. Paradoxically, stimulation of myocardin activity by GATA4 requires only the DNA binding domain, but not the TAD of GATA4, whereas repression of myocardin activity by GATA4 requires the DNA binding domain and the TAD (shown in Fig. 6). These findings reveal an unusual complexity to the mechanism(s) involved in myocardin- and GATA-dependent gene regulation with the potential to provide gene-specific transcriptional control and fine tuning of the expression of SRF target genes.
FIG. 6.
Schematic diagram of myocardin and a model. (A) The domains of myocardin are shown. (B) The regions of GATA4 that associate with myocardin and mediate activation and repression are shown.
Interaction of GATA4 with myocardin.
Our results demonstrate that GATA4 can interact with two regions of myocardin. GATA4-binding region 1 of myocardin maps to residues 326 to 438. This region encompasses the SAP domain (residues 380 to 414), but deletions to either side of this domain impair GATA4 binding, indicating that binding requires an extended sequence of myocardin such that deletions throughout this region perturb this protein-protein interaction. GATA4 can also interact with residues 439 to 713 independently of binding region 1. We have not further defined this region, which includes the TAD, since replacing it with VP16 does not diminish the responsiveness of myocardin to GATA4.
What mechanism(s) might account for the opposite effects of GATA factors on myocardin activity? Because GATA4 can stimulate myocardin activity on some regulatory regions (e.g., the Nkx2.5 enhancer) and inhibit activity on others (e.g., the ANF promoter), it is likely that the differential effects of GATA4 are dependent on other factors that bind myocardin target genes. Stimulation of myocardin activity by GATA4 was observed with an artificial reporter containing multimers of the SRF binding site, which suggests to us that this type of stimulatory activity may reflect a simpler mechanism and that repression may involve more complex interactions between GATA4 and other nuclear factors. The ANF and SM22 promoters were both repressed by GATA4 in the presence of myocardin, suggesting that they are regulated by the same mechanism. The ANF promoter contains two GATA binding sites, neither of which is required for repression by GATA4, and the SM22 promoter contains no GATA sites. Inspection of these promoters has not revealed any obvious binding sites for common factors that might mediate the repressive influence of GATA4.
Understanding the mechanisms involved in GATA-dependent modulation of myocardin activity is complicated by the fact that both myocardin and GATA4 interact with SRF (2, 26, 37). This raises the possibility that some of the observed effects of GATA4 may reflect competition between myocardin and GATA4 for interaction with SRF or recruitment of both factors by SRF, independent of their interaction with each other. The finding that GATA4 can stimulate the activity of a GAL4-myocardin fusion protein that activates transcription independently of SRF suggests that the stimulatory effects of GATA4 do not require its direct association with SRF and are likely to be mediated by direct physical association with myocardin. This conclusion is supported by the precise correlation between the ability of GATA4 mutants to interact with myocardin and to stimulate myocardin activity.
Although GATA4 interacts with myocardin and enhances myocardin activity, myocardin cannot stimulate transcription through GATA binding sites. The inability of myocardin to activate transcription by tethering to GATA factors on DNA may be explained by the fact that myocardin interacts with the same residues in the C-terminal zinc finger of GATA4 that mediate GATA4 DNA binding, which could preclude the formation of a stable GATA-myocardin ternary complex on DNA. In contrast, GATA4 interacts with domains of myocardin that are not required for association with SRF. Therefore, GATA4 can interact with myocardin without perturbing myocardin's ability to interact with SRF.
It is intriguing that stimulation of myocardin activity requires only the Cf and NLS of GATA4, whereas suppression of myocardin activity requires these domains in addition to the N-terminal transcription activation domain. The ability to separate the stimulatory and suppressive effects of GATA4 by deletion of the TAD suggests that these occur through distinct mechanisms. We can envision at least three mechanisms that might account for the ability of GATA4 to stimulate myocardin activity. GATA4 could induce a conformational change in myocardin that augments its transcriptional activity, possibly by unmasking the TAD or stabilizing its interaction with the transcriptional machinery. GATA4 could recruit a coactivator to the myocardin-SRF complex, or it could displace an inhibitory protein from the complex.
With respect to the mechanism for GATA4-mediated suppression of myocardin activity, the requirement for the TAD in this process suggests that the presence of GATA4 may cause “squelching” (29) such that this domain competes with an activation domain of another factor, which could be myocardin itself, SRF, or another factor required for activation of the set of genes that are suppressed by GATA4. Another mechanism may involve competition between myocardin and GATA4 for SRF interaction. It is not obvious, however, how the TAD of GATA4 would contribute to such a repressive mechanism.
Our finding that GATA4 can suppress the activity of myocardin on SRF target genes is somewhat surprising in light of numerous previous studies demonstrating that GATA4 can stimulate SRF-dependent transcription (2, 26, 33). The stimulatory effect of GATA4 on SRF is mediated by the direct interaction of the SRF MADS box with the Cf of GATA4 (2, 26, 33), the same region that we showed to mediate interaction of GATA4 with myocardin. This seeming discrepancy from previous studies may be explained, at least in part, by the fact that SRF and GATA4 are relatively weak activators that stimulate transcription by about an order of magnitude under optimal conditions. In contrast, myocardin stimulates expression of target genes with two or more SRF binding sites by several orders of magnitude. Thus, a modest stimulatory effect of GATA4 on SRF activity would be overcome by the dramatic repressive effect of GATA4 on myocardin activity.
Modulation of myocardin activity through the stoichiometry of its partners.
We have previously shown that the transcriptional activity of myocardin is exquisitely sensitive to the level of SRF, such that relatively minor increases in SRF expression above an optimal level result in pronounced suppression of myocardin activity analogous to the effects of GATA4 seen in this study (37). These findings illustrate the importance of precise control over the stoichiometry of these transcriptional activators. Given the importance of myocardin and GATA factors in smooth and cardiac muscle development, it will be especially interesting to explore whether perturbations in the relative levels of expression of the genes encoding these factors in vivo influences development of the cardiovascular system. The importance of GATA factors and the myocardin-related transcription factor MRTF-A in hematopoietic development also suggests that the types of regulatory interactions described in this study will be of importance in that system as well.
Acknowledgments
We thank Bruce Markham and Robert Schwartz for reagents and Yibin Wang for assistance with adenoviruses. We are also grateful to Alisha Tizenor for assistance with graphics and Jennifer Page for editorial assistance.
This work was supported by grants from the National Institutes of Health, the Donald W. Reynolds Clinical Cardiovascular Research Center, and the Robert A. Welch Foundation to E.N.O. D.Z.W. was supported by the Muscular Dystrophy Association and a start-up fund from the University of North Carolina.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 2000. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25:112-114. [DOI] [PubMed] [Google Scholar]
- 2.Belaguli, N. S., J. L. Sepulveda, V. Nigam, F. Charron, M. Nemer, and R. J. Schwartz. 2000. Cardiac tissue enriched factors serum response factor and GATA-4 are mutual coregulators. Mol. Cell. Biol. 20:7550-7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, P. S., L. Li, J. McAnally, and E. N. Olson. 2001. Muscle specificity encoded by specific serum response factor-binding sites. J. Biol. Chem. 276:17206-17212. [DOI] [PubMed] [Google Scholar]
- 4.Chang, D. F., N. S. Belaguli, D. Iyer, W. B. Roberts, S. P. Wu, X. R. Dong, J. G. Marx, M. S. Moore, M. C. Beckerle, M. W. Majesky, and R. J. Schwartz. 2003. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev. Cell. 4:107-118. [DOI] [PubMed] [Google Scholar]
- 5.Charron, F., and M. Nemer. 1999. GATA transcription factors and cardiac development. Semin. Cell Dev. Biol. 10:85-91. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C. Y., and R. J. Schwartz. 1996. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Mol. Cell. Biol. 16:6372-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai, Y. S., and B. E. Markham. 2001. p300 Functions as a coactivator of transcription factor GATA-4. J. Biol. Chem. 276:37178-37185. [DOI] [PubMed] [Google Scholar]
- 8.Du, K. L., H. S. Ip, J. Li, M. Chen, F. Dandre, W. Yu, M. M. Lu, G. K. Owens, and M. S. Parmacek. 2003. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol. Cell. Biol. 23:2425-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grueneberg, D. A., S. Natesan, C. Alexandre, and M. Z. Gilman. 1992. Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor. Science 257:1089-1095. [DOI] [PubMed] [Google Scholar]
- 10.Hines, W. A., J. Thorburn, and A. Thorburn. 1999. A low-affinity serum response element allows other transcription factors to activate inducible gene expression in cardiac myocytes. Mol. Cell. Biol. 19:1841-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, S., H. S. Ip, M. M. Lu, C. Clendenin, and M. S. Parmacek. 1997. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol. Cell. Biol. 17:2266-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koutsourakis, M., A. Langeveld, R. Patient, R. Beddington, and F. Grosveld. 1999. The transcription factor GATA6 is essential for early extraembryonic development. Development 126:723-732. [PubMed] [Google Scholar]
- 13.Kuo, C. T., E. E. Morrisey, R. Anandappa, K. Sigrist, M. M. Lu, M. S. Parmacek, C. Soudais, and J. M. Leiden. 1997. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 11:1048-1060. [DOI] [PubMed] [Google Scholar]
- 14.Latinkic, B. V., S. Kotecha, and T. J. Mohun. 2003. Induction of cardiomyocytes by GATA4 in Xenopus ectodermal explants. Development 130:3865-3876. [DOI] [PubMed] [Google Scholar]
- 15.Laverriere, A. C., C. MacNeill, C. Mueller, R. E. Poelmann, J. B. Burch, and T. Evans. 1994. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J. Biol. Chem. 269:23177-23184. [PubMed] [Google Scholar]
- 16.Lee, Y., T. Shioi, H. Kasahara, S. M. Jobe, R. J. Wiese, B. E. Markham, and S. Izumo. 1998. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol. Cell. Biol. 18:3120-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, L., J. M. Miano, B. Mercer, and E. N. Olson. 1996. Expression of the SM22alpha promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J. Cell Biol. 132:849-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, L., Z. Liu, B. Mercer, P. Overbeek, and E. N. Olson. 1997. Evidence for serum response factor-mediated regulatory networks governing SM22alpha transcription in smooth, skeletal, and cardiac muscle cells. Dev. Biol. 187:311-321. [DOI] [PubMed] [Google Scholar]
- 19.Li, S., D. Wang, Z. Wang, J. Richardson, and E. N. Olson. 2003. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc. Natl. Acad. Sci. USA 100:9366-9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lien, C. L., C. Wu, B. Mercer, R. Webb, J. A. Richardson, and E. N. Olson. 1999. Control of early cardiac-specific transcription of an Nkx2-5 by a GATA-dependent enhancer. Development 126:75-84. [DOI] [PubMed] [Google Scholar]
- 21.Ma, Z., S. W. Morris, V. Valentine, M. Li, J. A. Herbrick, X. Cui, D. Bouman, Y. Li, P. K. Mehta, D. Nizetic, Y. Kaneko, G. C. Chan, L. C. Chan, J. Squire, S. W. Scherer, and J. K. Hitzler. 2001. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 28:220-221. [DOI] [PubMed] [Google Scholar]
- 22.Mercher, T., M. B. Coniat, R. Monni, M. Mauchauffe, F. N. Khac, L. Gressin, F. Mugneret, T. Leblanc, N. Dastugue, R. Berger, and O. A. Bernard. 2001. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc. Natl. Acad. Sci. USA 98:5776-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merika, M., and S. H. Orkin. 1993. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 13:3999-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miralles, F., G. Posern, A. I. Zaromytidou, and R. Treisman. 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113:329-342. [DOI] [PubMed] [Google Scholar]
- 25.Molkentin, J. D., Q. Lin, S. A. Duncan, and E. N. Olson. 1997. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 11:1061-1072. [DOI] [PubMed] [Google Scholar]
- 26.Morin, S., P. Paradis, A. Aries, and M. Nemer. 2001. Serum response factor-GATA ternary complex required for nuclear signaling by a G-protein-coupled receptor. Mol. Cell. Biol. 21:1036-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrisey, E. E., Z. Tang, K. Sigrist, M. M. Lu, F. Jiang, H. S. Ip, and M. S. Parmacek. 1998. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 12:3579-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norman, C., M. Runswick, R. Pollock, and R. Treisman. 1988. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55:989-1003. [DOI] [PubMed] [Google Scholar]
- 29.Ptashne, M. 1988. How eukaryotic transcriptional activators work. Nature 335:683-689. [DOI] [PubMed] [Google Scholar]
- 30.Reecy, J., N. S. Belaguli, and R. J. Schwartz. 1998. SRF/homeobox protein interactions, p. 273-290. In R. Harvey and N. Rosenthal (ed.), Heart development. Academic Press, San Diego, Calif.
- 31.Reiter, J. F., J. Alexander, A. Rodaway, D. Yelon, R. Patient, N. Holder, and D. Y. Stainier. 1999. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 13:2983-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasazuki, T., T. Sawada, S. Sakon, T. Kitamura, T. Kishi, T. Okazaki, M. Katano, M. Tanaka, M. Watanabe, H. Yagita, K. Okumura, and H. Nakano. 2002. Identification of a novel transcriptional activator, BSAC, by a functional cloning to inhibit tumor necrosis factor-induced cell death. J. Biol. Chem. 277:28853-28860. [DOI] [PubMed] [Google Scholar]
- 33.Sepulveda, J. L., S. Vlahopoulos, D. Iyer, N. Belaguli, and R. J. Schwartz. 2002. Combinatorial expression of GATA4, Nkx2-5, and serum response factor directs early cardiac gene activity. J. Biol. Chem. 277:25775-25782. [DOI] [PubMed] [Google Scholar]
- 34.Shore, P., and A. D. Sharrocks. 1995. The MADS-box family of transcription factors. Eur. J. Biochem. 229:1-13. [DOI] [PubMed] [Google Scholar]
- 35.Treisman, R. 1994. Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev. 4:96-101. [DOI] [PubMed] [Google Scholar]
- 36.Ueyama, T., H. Kasahara, T. Ishiwata, Q. Nie, and S. Izumo. 2003. Myocardin expression is regulated by Nkx2.5, and its function is required for cardiomyogenesis. Mol. Cell. Biol. 23:9222-9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, D., P. S. Chang, Z. Wang, L. Sutherland, J. A. Richardson, E. Small, P. A. Krieg, and E. N. Olson. 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105:851-862. [DOI] [PubMed] [Google Scholar]
- 38.Wang, D. Z., S. Li, D. Hockemeyer, L. Sutherland, Z. Wang, G. Schratt, J. A. Richardson, A. Nordheim, and E. N. Olson. 2002. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl. Acad. Sci. USA 99:14855-14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Z., D. Z. Wang, G. C. Pipes, and E. N. Olson. 2003. Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl. Acad. Sci. USA 100:7129-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida, T., S. Sinha, F. Dandre, B. R. Wamhoff, M. H. Hoofnagle, B. E. Kremer, D. Z. Wang, E. N. Olson, and G. K. Owens. 2003. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circ. Res. 92:856-864. [DOI] [PubMed] [Google Scholar]