Abstract
Gene duplication and loss are major driving forces in evolution. While many important genomic resources provide information on gene presence, there is a lack of tools giving equal importance to presence and absence information as well as web platforms enabling easy visual comparison of multiple domain-based protein occurences at once. Here, we present Aquerium, a platform for visualizing genomic presence and absence of biomolecules with a focus on protein domain architectures. The web server offers advanced domain organization querying against the database of pre-computed domains for ~26000 organisms and it can be utilized for identification of evolutionary events, such as fusion, disassociation, duplication and shuffling of protein domains. The tool also allows alternative inputs of custom entries or BLASTP results for visualization. Aquerium will be a useful tool for biologists who perform comparative genomic and evolutionary analyses. The web server is freely accessible at http://aquerium.utk.edu.
Keywords: protein, domain architecture, genomic occurrence, taxonomy tree, phylogenetic profile, genomic visualization
Introduction
Phylogenetic profiling is a method to detect functionally or physically interacting proteins by inferring their co-presence/absence in hierarchically clustered species1. If genes are gained or lost together, it is likely that their products participate in the same biological pathway, meaning that they interact functionally. The method was first described by Pellegrini et al. who investigated the coevolution patterns of Escherichia coli genes2. They demonstrated that gene groups that have similar occurrence profiles tend to be involved in the same pathways. Consequently, in addition to discovering protein-protein interactions, phylogenetic profiling can be used for protein function prediction. There has been a number of successful applications of this context-based method complemented by homology-based and experimental approaches3.
Homology is inferred through sequence-based similarity searches. Domain organization comparisons can also be used to infer homology and to identify protein families. Domains, defined as minimal structural and functional building blocks of proteins, are capable of folding autonomously and evolving independently. Single domain proteins (SDPs) were likely dominant in the early stage of life, whereas multi-domain proteins (MDPs) are enriched with the complexity of organisms4. In SDPs the domain itself functions alone while in MDPs domains work in collaboration to perform the protein function. Domains can exist in various arrangements in a protein and this flexibility enriches the diversity of protein families. The complexity of MDPs can be attributed to the evolutionary dynamic nature of domains. The evolutionary events, such as domain innovation, loss, duplication, fusion, disassociation and shuffling enable proteins and eventually organisms to adapt to their environment5. Particularly, domain shuffling, rather than de novo inventions from disordered sequences is the major evolutionary event to generate novel proteins4. It was suggested that the total number of unique protein domains decreased in the course of eukaryotic evolution. For instance, the last eukaryotic common ancestor had a larger unique domain pool than any of the current species6. Also, in mammals, a drop in the domain pool has been observed compared to the ancestral repository6. These observations suggest that reusing protein domains in various modifications and rearrangements drives protein evolution.
More complex organisms have relatively more complex MDPs7,8. This correlation may explain why gene number does not increase with organismal complexity9. Therefore, in order to understand complex networks, it is important to investigate the function of domains and how they collectively work together. Inferring the evolutionary relationships between domains is critically important in order to identify their functions and interactions.
Domains in protein sequences can be identified computationally, e.g. using. HMM (Hidden Markov Model) profiles. Pfam (Protein Families) database is a large collection of HMMs and underlying tools, which is one of the most popular resources for identifying protein domains10. Pfam-A, a manually curated subset of the database, currently (version 30.0) contains 16306 domain models. Another HMM utilizing resource, TIGRFAM, contains models for many full-length proteins so that it provides an easy detection for protein families11.
Biological networks diverge from their ancestor by protein or domain gain/loss and domain shuffling. Such diversity patterns can be detected by comparative analysis of domain architectures. For this reason, retrieving the domain organization of interest and visualizing its taxonomic distribution are the crucial steps in understanding the functional relationships within networks. In addition to Pfam, several other tools, such as SMART12, CDART13, DAhunter14 and PfamAlyzer15, provide domain architecture querying. These tools specialize in searching for protein homologies through similar domain architectures. However, none of the current resources allow advanced domain architecture querying while visualizing the domain presence and absence in the phylogenomic context. Furthermore, although there are softwares enabling visualization of the domain/protein of interest on the phylogenetic trees16–19, there is a lack of a webserver offering precomputed genomic occurences of domains to visualize genomic distributions of multiple queries at once.
To address these problems, we developed Aquerium (architecture querying podium), a tool enabling biologists and bioinformaticians to understand the domain-based evolutionary history of proteins.
Materials and methods
Genomes from the NCBI genomes database (as of 12th of December 2014) which also had assembly records in the NCBI assembly database were selected. GenBank records for each genome were retrieved from the NCBI Entrez Genome database20. We created a proteome collection for each genome. In order to manage isoforms in eukaryotes, each protein was categorized under the gene identifier that it is coded by. If a gene has at least one protein isoform matching the query, the tool returns true (presence). If several isoforms match to query, only one of them is taken into account in order to eliminate redundancy in the matched gene number. The SeqDepot database21 was used locally to retrieve the pre-computed domain architectures from Pfam versions 27.0 and 28.0 and TIGRFAM versions 14 and 15. The local SeqDepot database was updated by running HMMER322 searches against domain databases for uncovered proteins. Domain hits are considered as “true” if their score is higher than the PFAM-determined domain-specific cutoff. Each domain model in PFAM database stands alone, meaning that the members of the same PFAM clan are considered as separate domains.
The NCBI taxonomy database23 was used to build the tree. In addition to eight major taxonomic ranks, we also included the five eukaryotic supergroups24. Protein identifier to taxonomic id mapping was performed using the NCBI Entrez tool. The resulting taxonomic tree can be visualized by using two sets of genomes: species-representative and full sets. Species-representative set (4934 genomes) was built by selecting only one representative for strains determined by their species-level taxonomic ids. The genomes with the largest number of genes among strains were selected as representative. The full set was composed of 26618 organisms. The tool also enables instant query generation with a given protein sequence by running HMMER-API to determine the domain architecture on the fly.
The data have been organized in a document based MongoDB database. Custom Python3 scripts were developed for searching the database. JavaScript was implemented in HTML5 to visualize the results. The final figures are drawn in Scalable Vector Graphics (SVG).
Results
Features
Advanced domain architecture querying
MDPs have various domain arrangements. In some proteins, the domain order is conserved, whereas other proteins are subjected to domain shuffling, duplication and loss. Diverged domain architectures might be indicators of modified or adapted function. For these reasons, it is important to enable extensive architecture querying.
Aquerium allows users to select the domain of interest, called “key domain”, to initialize the search. This field is mandatory and the algorithm will retrieve proteins that have at least one key domain. In the query page, a condition (“if” statement in Python syntax) can be specified to customize the query in terms of domain content and organization. This condition is used for enriched querying in which presence and absence of other domains can be examined. Moreover, the order of the domains, from N- to C-terminus can be specified and only proteins satisfying the given condition are retrieved. Specifying a condition is not necessary if the user is interested only in the presence and absence of the key domain. Domain search can be performed on species-representative and full sets and these sets can be filtered based on taxonomic units.
Visualization
Species are clustered based on their taxonomic ranks and represented as a sunburst tree on which each taxonomic class is drawn as an arc. The length of arcs scales to represent the number of species which are eventual descendants of the node. On the tree, there are nine taxonomic layers representing the major taxonomic ranks and supergroups for eukaryotes24. After taxonomic ranks, each outer ring represents the requested query. If there is any match in the corresponding genome, there will be a colored flag aligning with the organism on an outer circle.
In the “zoom” mode, each taxonomic node, represented by an arc, is zoomable on click. The sunburst is redrawn and shows only the selected node and its children in a circular layout. Extensive coloring options are offered on the fly, allowing to produce publication-quality figures. The coloring of flags can also be performed as a heatmap depending on the quantity of each flag. Multiple layers can be visualized on the same tree. Users can visualize up to 10 outer layers in the same tree. In the “Arc” mode, clicking on a node will redirect the user to another web page where they can visualize the associated organisms and the domain architectures on a collapsible tree layout.
Data export
The sunburst tree can be exported in scalable vector graphics (SVG). The compiled data can be downloaded in semicolon separated file (CSV) format, which includes the taxonomic identifiers and the number of occurrences for each organism. A JSON-formatted file containing taxonomically classified organism information is also available to retrieve. Moreover, protein sequence (in FASTA format) download option for a desired taxonomic unit is available.
Custom input for visualization
In addition to protein domains, the sunburst tree can be produced with any other type of genomic data. Users can input a custom table containing NCBI taxonomic id followed by numeric or binary occurrence profiles in CSV format and visualize the results. Up to 10 flag layers can be visualized in a single request.
Aquerium web server also offers visualizing BLASTP hits on the tree. Users must download BLASTP25 results (in XML format) from NCBI and upload it to the Aquerium web server.
Illustrations
In order to exemplify Aquerium performance, we presented two independent test cases which show potential applications to similar problems.
Identification of a domain fusion event
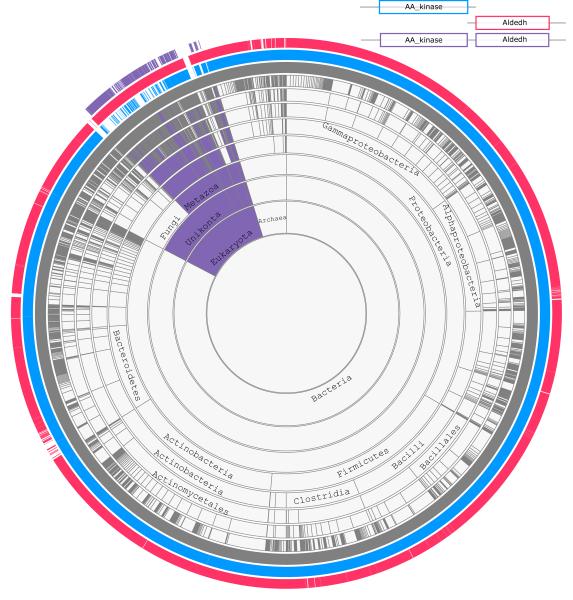
Amino acid kinase (AA_kinase) and Aldehyde dyhydrogenase (Aldedh) domain families are universal and seen in all domains of life with minor absences in few parasitic clades. These domains usually comprise a single domain protein, such as E. coli glutamate-5-kinase and γ-glutamyl phosphate reductase. Human δ-1-pyrroline-5-carboxylate synthetase has evolved as a fusion product of AA_kinase and Aldedh domains26. Figure 1 shows the presences and absences of these domains. The outmost layer shows the occurrence of these two domains together in a single protein. In all supergroups of eukaryotes, these two domains are fused. The observed pattern of inheritance suggests that the fusion of these domains has occurred in the common ancestor of eukaryotes, and the common ancestor of fungi lost it.
Figure 1.
Illustration of a domain fusion event. Fused proteins containing both Aldedh and AA_kinase domains are found in all represented eukaryotic supergroups, suggesting that the fusion occurred in the last eukaryotic common ancestor.
Coexisting proteins and abundance correlation
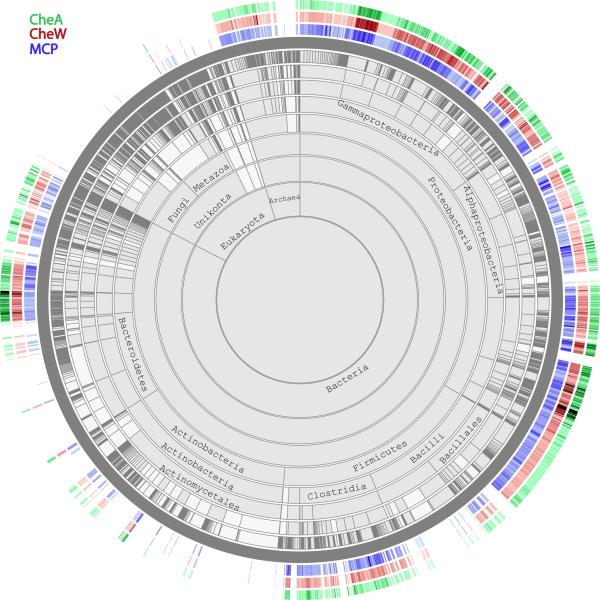
The signaling complex in bacterial chemotaxis, which has been conserved since the common bacterial ancestor27, consists of MCPs (chemoreceptors), CheA (a kinase) and CheW (an adaptor). These three proteins are found together in 98% of genomes that encode chemotaxis genes27. Figure 2 shows the phlyetic distributions of these three proteins. Satisfactorily, in the vast majority of cases, all three proteins are either present or absent in genomes indicating the presence or absence of chemotaxis as a cellular function. This test case serves as a control for true negatives. The relative abundances of these proteins in genomes (some genomes have several different types of the signaling complex encoded by different sets of genes) also correlate. This is visualized using the heatmap option revealing the number of hits for each organism. Increased abundance is shown by a change of color intensity from light to dark.
Figure 2.
Interacting proteins coevolve. Chemotaxis proteins MCP, CheA and CheW are known to be interacting with each other. They show similar patterns of not only occurrence, but also relative abundance.
Discussion
There are several software packages enabling phylogenetic profiling for genes/proteins. DoMosaics18 is the software performing the closest task to Aquerium. This software requires users to perform the computations locally. Despite being flexible in terms of data input/output, it comes with the requirement of being familiar with the current bioinformatics tools. ETE317 is a significantly useful python package offering various ways to represent data on the phylogenetic tree given as an input. Both of these tools are designed to be used by computational biologists. PhyloGene server focuses on the coevolution of eukaryotic proteins with a set of species19. This tools has a drawback of containing no prokaryotic data and domain-based remote homologs cannot be found using the server. We think that Aquerium fills a gap in the field by offering an easy-querying of domain architectures for both computational and experimental scientists with no requirement of coding experience.
The presence of genetic material in a genome is almost never questioned except for the possibility of contamination. On the other hand, the absence is always questioned and negative information should be treated cautiously. Being confident about the absence of particular genes/proteins/domains in genomes is challenging for two main reasons: (i) genomes may be incomplete, erroneous or contaminated and (ii) genes may not be identified due to computational limitations. However, the absences of two or more genes/proteins/domains that are consistently observed in independent samplings strongly suggests that the absence is true (Figure 2). Independent co-evolution can be identified by large-scale analyzes; as the number of samples increases, the likelihood of finding independent cases also increases.
Conclusions
Aquerium enables exploring a variety of phenomena in a genomic context, ranging from evolution of individual domains to inferring potential protein-protein interactions, by placing a nearly equal weight on the presence and the absence of genomic entities, such as genes, proteins and their domains. Thus, we expect this tool to be useful to many biologists working within the genomic landscape.
Acknowledgments
We thank Luke Ulrich and Aaron Fleetwood for helpful comments on the manuscript.
Funding This work was supported in part by the National Institutes of Health grants GM072285 and DE024463 (to I.B.Z).
Footnotes
Competing interests The authors declare that they have no competing interests.
Authors' contributions O.A. conceived the problem, designed and built the algorithm. I.B.Z. provided substantial advice and guidance during all phases of project. O.A. and I.B.Z. wrote the manuscript.
References
- 1.Skunca N, Dessimoz C. Phylogenetic profiling: how much input data is enough? PLoS One. 2015;10(2):e0114701. doi: 10.1371/journal.pone.0114701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci U S A. 1999;96(8):4285–8. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kensche PR, van Noort V, Dutilh BE, Huynen MA. Practical and theoretical advances in predicting the function of a protein by its phylogenetic distribution. J R Soc Interface. 2008;5(19):151–70. doi: 10.1098/rsif.2007.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Roberto RB, Peisajovich SG. The role of domain shuffling in the evolution of signaling networks. J Exp Zool B Mol Dev Evol. 2014;322(2):65–72. doi: 10.1002/jez.b.22551. [DOI] [PubMed] [Google Scholar]
- 5.Kummerfeld SK, Teichmann SA. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 2005;21(1):25–30. doi: 10.1016/j.tig.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Zmasek CM, Godzik A. Strong functional patterns in the evolution of eukaryotic genomes revealed by the reconstruction of ancestral protein domain repertoires. Genome Biol. 2011;12(1):R4. doi: 10.1186/gb-2011-12-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das S, Yu L, Gaitatzes C, Rogers R, Freeman J, Bienkowska J, Adams RM, Smith TF, Lindelien J. Biology's new Rosetta stone. Nature. 1997;385(6611):29–30. doi: 10.1038/385029a0. [DOI] [PubMed] [Google Scholar]
- 8.Wolf YI, Brenner SE, Bash PA, Koonin EV. Distribution of protein folds in the three superkingdoms of life. Genome Res. 1999;9(1):17–26. [PubMed] [Google Scholar]
- 9.Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420(6912):218–23. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 10.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. TIGRFAMs and Genome Properties in 2013. Nucleic Acids Res. 2013;41(Database issue):D387–95. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43(Database issue):D257–60. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Res. 2002;12(10):1619–23. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee B, Lee D. DAhunter: a web-based server that identifies homologous proteins by comparing domain architecture. Nucleic Acids Res. 2008;36(Web Server issue):W60–4. doi: 10.1093/nar/gkn172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollich V, Sonnhammer EL. PfamAlyzer: domain-centric homology search. Bioinformatics. 2007;23(24):3382–3. doi: 10.1093/bioinformatics/btm521. [DOI] [PubMed] [Google Scholar]
- 16.Csuros M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26(15):1910–2. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- 17.Huerta-Cepas J, Serra F, Bork P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol Biol Evol. 2016;33(6):1635–8. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore AD, Held A, Terrapon N, Weiner J., 3rd Bornberg-Bauer E. DoMosaics: software for domain arrangement visualization and domain-centric analysis of proteins. Bioinformatics. 2014;30(2):282–3. doi: 10.1093/bioinformatics/btt640. [DOI] [PubMed] [Google Scholar]
- 19.Sadreyev IR, Ji F, Cohen E, Ruvkun G, Tabach Y. PhyloGene server for identification and visualization of co-evolving proteins using normalized phylogenetic profiles. Nucleic Acids Res. 2015;43(W1):W154–9. doi: 10.1093/nar/gkv452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibney G, Baxevanis AD. Searching NCBI Databases Using Entrez. Curr Protoc Hum Genet. 2011;Chapter 6(Unit6 10) doi: 10.1002/0471142905.hg0610s71. [DOI] [PubMed] [Google Scholar]
- 21.Ulrich LE, Zhulin IB. SeqDepot: streamlined database of biological sequences and precomputed features. Bioinformatics. 2014;30(2):295–7. doi: 10.1093/bioinformatics/btt658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013;41(12):e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012;40(Database issue):D136–43. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koonin EV. Preview. The incredible expanding ancestor of eukaryotes. Cell. 2010;140(5):606–8. doi: 10.1016/j.cell.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, et al. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41(Web Server issue):W29–33. doi: 10.1093/nar/gkt282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcotte EM, Pellegrini M, Ng H-L, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285(5428):751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 27.Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3(128):ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]