Abstract
Protein function elucidation often relies heavily on amino acid sequence analysis and other bioinformatics approaches. The reliance is extended to structure homology modeling for ligand docking and protein-protein interaction mapping. However, sequence analysis of RPA3313 exposes a large, unannotated class of hypothetical proteins mostly from the Rhizobiales order. In the absence of sequence and structure information, further functional elucidation of this class of proteins has been significantly hindered. A high quality NMR structure of RPA3313 reveals that the protein forms a novel split ββαβ fold with a conserved ligand binding pocket between the first β-strand and the N-terminus of the α-helix. Conserved residue analysis and protein-protein interaction prediction analyses reveal multiple protein binding sites and conserved functional residues. Results of a mass spectrometry proteomic analysis strongly point toward interaction with the ribosome and its subunits. The combined structural and proteomic analyses suggest that RPA3313 by itself or in a larger complex may assist in the transportation of substrates to or from the ribosome for further processing.
Keywords: NMR, protein structure, structural genomics, hypothetical proteins
Introduction
Rhodopseudomonas palustris is a unique organism known for its metabolic diversity and extensive distribution throughout the environment.1 It has the ability to grow under four distinct modes of metabolism (photoautotrophic, photoheterotrophic, chemoautotrophic, and chemoheterotrophic) on a wide assortment of carbon sources. R. palustris is typically found in soil and freshwater sources, but has also been discovered in swine waste and coastal sediments.2 As a purple non-sulfur photosynthetic bacterium, it belongs to the alphaproteobacterium order.3 Within this order exists many species which are similarly metabolically versatile, yet there are clear phylogenetic differences. In fact, based on 16S rRNA sequencing the inherent divergence in the order is not based on phototrophic ability but rather demonstrates a mixing of phototrophs and non-phototrophs.3 R. palustris is of particular biotechnological interest because it utilizes aromatic hydrocarbons as a carbon source under both aerobic and anaerobic growth conditions. Also, R. palustris fixes more nitrogen when grown on aromatic hydrocarbons relative to aliphatic substrates.4 Furthermore, growth of the organism can occur on aromatic substrates containing a range of functional moieties. The combination of all of these factors makes R. palustris a model organism for bioremediation, energy production, and other biotechnological applications.5–9
In 2004, the genome of R. palustris was sequenced and published along with a prediction of general gene functional classes.2 Approximately 15% of the genome is believed to be devoted solely to transport, which is surprising since prokaryotes usually commit only a third of this amount (5–10%) to transport.10 Nevertheless, this greater commitment of R. palustris to transport is consistent with its observed metabolic diversity. A larger assortment of transport proteins would be necessary for R. palustris to readily adapt to various carbon and energy sources; or to proliferate under changing respiration or environmental states. Conversely, 29% of the genome has been tentatively labeled as hypothetical or of unknown function. An additional 8% of the genome is only annotated with a general function. A follow-up LC-ES-MS/MS proteomics analysis of R. palustris included a more detailed functional annotation based on protein sequence analysis.11 But, the percentage of functionally uncharacterized or partially annotated proteins remained unchanged. Of particular note, the proteome of R. palustris was analyzed for each metabolic mode of growth. Thus, the relative expression rates of R. palustris proteins under each metabolic mode of growth are known.11 The large fraction of unannotated or partially annotated R. palustris genes presents a significant obstacle for the further development of biotechnological applications and hinders additional biochemical studies.
The R. palustris protein RPA3313 (7.45 kDa, 70 amino acids) is a hypothetical protein targeted for structural elucidation by the Structural Genomics Consortium at the University of Toronto (http://www.thesgc.org/). RPA3313 is currently classified by UniProtKB12 (Q6N4M4) as an uncharacterized protein. A BLAST search of the RPA3313 sequence reveals a group of 93 hypothetical yet conserved proteins (> 32% identity) from only the alphaproteobacterium order (Figure 1). Unfortunately, minimal structural or functional information was obtained from the sequence analysis since no structures of homologous proteins are present in the Protein Data Bank (http://www.rcsb.org/).13
Figure 1.
(A) A neighbor-join tree of the protein BLAST results of RPA3313 against non-redundant protein sequences. All of the sequence hits belong the alphaproteobacteria order and have identities >32%. The three colored groups are dominated by species belonging to the genera listed next to them. The tree highlights the fact that this protein is a member of an unannotated and a structurally uncharacterized class of proteins. (B) The BLAST sequence alignment of RPA3313 with the nine other Rhodopseudomonas proteins. The secondary structure is indicated above the sequence alignment and was generated with Polyview-2D.56 RPA3313 is indicated in the phylogenetic tree and sequence alignment with a dot.
RPA3313 was identified in the previously reported R. palustris proteomics study11 and was only observed to be expressed during photoautotrophic growth. Photoautotrophic organisms, such as R. palustris, sequester atmospheric CO2 and convert it to energy rich carbon sources. Ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) are found in photoautotrophic organisms and are responsible for most of the organic carbon in the environment. As the most abundant protein in nature, RubisCO is found in plants, bacteria, and archaea in at least four molecular forms.14 The genome of R. palustris contains multiple forms of RubisCO, which further contributes to its adaptability to diverse environmental conditions.15 The combined adaptability and RubisCO activity of R. palustris may be beneficial to biotechnological applications involving bulk removal of CO2 from the atmosphere. However, the photoautotrophic mode of metabolism in R. palustris and other alphaproteobacteria remains relatively unknown.16
Although the hypothetical protein RPA3313 has been experimentally verified as an expressed protein during photoautotrophic growth; the function and structure of RPA3313 still remains elusive. Structural approaches are a valuable alternative to obtaining a functional annotation when sequence similarity techniques fail and leaves a large class of functionally uncharacterized proteins.17,18 Thus, obtaining an NMR solution structure for R. palustris protein RPA3313 is expected to provide a better understanding of its general biological role and also provide a putative structure and function for the 93 homologous proteins (Figure 1). RPA3313 forms a novel split ββαβ fold with a conserved ligand binding pocket. The NMR structure combined with a bioinformatics analysis and mass spectrometry proteomics suggest RPA3313 may assist in the transportation of substrates to or from the ribosome for further processing.
Materials and Methods
Protein expression and purification
Uniformly 15N and 13C labeled RPA3313 samples were prepared for NMR structural studies as follows. The target sequence for RPA3313 (70 amino acids with a 21 amino acid histidine tag for purification, MGSSHHHHHHSSGRENLYFQG) was expressed from a pRI952 with glyT construct transformed into BL21(DE3) cells.19 Cells were grown in Luria-Bertani (LB) media at 37°C until an approximate optical density (OD600) of 0.6 and then spun down and transferred to M9 minimal media at 37°C containing 4% U-13C glucose and 1% U-15N NH4Cl. Expression of RPA3313 was induced after one hour of equilibration in the M9 media with isopropyl β-D-1-thiogalactopyranoside (IPTG). Cell lysates were collected 4 hours after induction with IPTG and purified with a Co2+ affinity column (HisPur Cobalt Resin, Thermo Scientific). Sample homogeneity was assessed by SDS-PAGE. Size exclusion chromatography and ESI-MS were used to confirm the monomeric solution state and exact mass of RPA3313 (Supplemental Figures S-2 and S-3, respectively). The protein sample was stored in an NMR sample tube in 18 mM 2-(4-morpholino)ethanesulfonic acid (MES) buffer with 0.01% sodium azide, 80 mM sodium chloride and 10% D2O at a pH of 5.6 (uncorrected).
NMR structure determination
All NMR experiments were collected with non-uniform sampling at 20% sparsity using a Poisson-gap schedule20 at 298K on a 700 MHz Bruker Avance III spectrometer equipped with a 5 mm QCI-P probe with cryogenically cooled carbon and proton channels. Backbone and side-chain assignments were completed using the standard triple resonance approach consisting of the following experiments: 1H-15N HSQC, 1H-13C HSQC, HNCO, HN(CA)CO, HNCA, HN(CO)CA, CCANH, CBCA(CO)NH, HNHA, HBHA(CO)NH, CC(CO)NH, HCC(CO)NH, HCCH-COSY, and HCCH-TOCSY.21,22 Identification of nuclear Overhauser effects (NOEs) was accomplished with 15N-edited NOESY-HSQC and 13C-edited NOESY-HSQC experiments using a mixing time of 150 ms. The resulting data was reconstructed using multidimensional decomposition (MDD) and processed in TopSpin 3.2 followed by evaluation in CCPNMR Analysis.23 Initial model generation according to backbone chemical shifts was undertaken using CS-ROSETTA24–26 on the open webserver at the BMRB (https://csrosetta.bmrb.wisc.edu/). The CS-ROSETTA software was only used for the creation of an initial model for RPA3313. CS-ROSETTA was not used to further refine the RPA3313 ensemble.
XPLOR-NIH version 2.37 was used to refine the initial model of target RPA3313.27,28 Briefly, the refinement involved 912 manually assigned NOE distance restraints, 66 hydrogen bond distance restraints, 30 3JNHα coupling constants, 128 13Cα/13Cβ chemical shifts, and 102 predicted dihedral angle restraints from TALOS+.29 1000 total structures were generated during the XPLOR-NIH structure refinement and the 20 lowest energy structures were subsequently subjected to water refinement according to the RECOORD conventions. The coordinate average structure for the water-refined models was further subjected to the same explicit water refinement method for energy minimization. The water-refined ensemble and average structure for target RPA3313 was analyzed with the PSVS software suite, which is comprised of commonly used structural validation packages.30–34 UCSF Chimera was used for the structural visualization and surface representation of RPA3313.35
Chemical crosslinking and in-gel digestion
Approximately 3 hours following the induction of RPA3313 in Lysogeny broth (LB) media, the E. coli culture was pelleted and resuspended in crosslinking buffer (1% paraformaldehyde, 1X PBS, pH 8, 37°C). Crosslinking was allowed to proceed for 15 minutes before quenching with 1.25 M glycine. The E. coli cells were lysed by sonication and RPA3313 with crosslinked binding partners was purified using the RPA3313 histidine tag and a Co2+ affinity column as described above in the protein purification section. The sample preparation procedure also efficiently removes the formaldehyde crosslinking. The purified proteins were then visualized by SDS-PAGE. Protein bands were excised before submission to the Nebraska Center for Mass Spectrometry for MS/MS analysis.
Rhodopseudomonas palustris (ATCC) was propagated in a filled flask of 500 mL of 112 medium at 30°C for several days until reaching stationary growth. Bacterial growth was red in color indicating that photoautotrophic had occurred. The culture was pelleted by centrifugation and resuspended in water prior to lysis by sonication. Extracted proteins were frozen and lyophilized overnight. Approximately 2 mg of pure RPA3313 was added to the R. palustris protein extract before the addition of crosslinking buffer. The crosslinking was performed similar to the E. coli crosslinking above. RPA3313 with crosslinked binding partners was purified using the RPA3313 histidine tag and a Co2+ affinity column as described above in the protein purification section.
MS proteomics
Protein bands separated with SDS–PAGE were digested in situ using a slightly modified version of a published method.36 Briefly, the samples were washed with 100 mM ammonium bicarbonate, reduced with 10 mM DTT, alkylated with 55 mM iodoacetamide, washed twice with 100 mM ammonium bicarbonate, and digested in situ with 10 ng/μL trypsin (Promega, Madison, WI, USA). Peptides were extracted with two 60 μL aliquots of 1:1 acetonitrile:water containing 1% formic acid. The extracts were dried down using a SpeedVac and then reconstituted into 15 uL of water + 0.1% formic acid. Four microliters of the extract solution was injected onto a trapping column (300 μm × 1 mm) in line with a 75 μm × 15 cm C18 reversed phase LC column (Waters, Milford, MA, USA). Peptides were eluted from the column using a water + 0.1% formic acid (A)/acetonitrile + 0.1% formic acid (B) gradient with a flow rate of 500 nL/min. The gradient was developed with the following time profile: 0 min, 5% B; 5 min, 5% B; 35 min, 35% B; 40 min, 45% B; 42 min, 60% B; 45 min, 90% B; 48 min, 90% B; and 50 min, 5% B.
The eluting peptides were analyzed using a Synapt G2S Q-TOF tandem mass spectrometer (Waters, Milford, MA, USA) with electrospray ionization. Analyses were performed using data-dependent acquisition (DDA) with the following parameters: 0.7 sec. survey scan (380–200 Da) followed by up to four MS/MS acquisitions (50–2000 Da). The instrument was operated at a mass resolution of 18000. The instrument was calibrated using a solution of NaI in 1:1 water:acetonitrile. The MS/MS data were processed using Masslynx software (Micromass, Milford, MA, USA) to produce peak lists for database searching. Mascot (Matrix Science Ltd, London, UK) was used as the search engine. Data were searched against the National Centre for Biotechnology Information (NCBI) non-redundant database. The following search parameters were used: mass accuracy 20 ppm, enzyme specificity trypsin, fixed modification carboxyamidomethylcysteine (CAM), variable modification oxidized methionine. Protein identifications were based on random probability scores with a minimum value of 25.
Bioinformatics Analyses
The RPA3313 sequence (excluding the 21 residue histidine tag) and the NMR structure were submitted to the ConSurf webserver to identify evolutionary conserved residues.37–40 Structural comparison was done with PDBeFold41 and protein-protein interaction residues were predicted with cons-PPISP.42,43 Results from the ConSurf and cons-PPISP analyses were mapped onto the surface of the protein with UCSF Chimera.44 Surface hydrophobicity was also calculated within Chimera. The BLAST hits were visualized with the neighbor-join algorithm using Dendroscope.45
Results and Discussion
Solution Structure of R. palustris protein RPA3313
Backbone and side-chain resonance assignments for RPA3313 were made for the 68 assignable residues excluding the N-terminus histidine tag (Supplemental Figure S-1). The NMR assignments are nearly complete with 68 of 68 N, 68 of 68 HN, 68 of 68 Cα, 76 of 76 Hα, 60 of 60 Cβ, 93 of 97 Hβ, 33 of 55 Cγ, 57 of 60 Hγ, 13 of 28 Cδ, 32 of 39 Hδ, 3 of 12 Cε, 16 of 21 Hε, 0 of 9 Cζ, and 4 of 7 Hζ. The monomeric solution structure of RPA3313 was calculated using 912 distance restraints, 102 angle restraints, 30 3JNHα coupling constants, 128 13Cα/13Cβ chemical shifts, and an initial model generated using CS-ROSETTA.24–26 During structure generation, 1000 structures were initially created and the 20 lowest energy models were selected for further water refinement. A coordinate average of the 20 water-refined structures was subjected to water refinement for additional minimization. The water refined ensemble structures did not contain any distance violations >0.5 Å or dihedral angle violations >5°. Also, the NMR data agrees well with the calculated structures since the RMSD of the backbone secondary structure residues is 0.70±0.07 Å and the RMSD for heavy atoms is 1.2±0.12 Å. Complete structural statistics for the RPA3313 NMR structures are listed in Table 1. Chemical shift assignments have been submitted to the BMRB as entry 30070 and coordinate files have been uploaded to the PDB as entry 5JN6.
Table 1.
Structure Calculation Statisticsα
| rmsd for distance restraints (experimental) (Å) | <SA> |
|
|
|---|---|---|---|
| all (912) | 0.048±0.004 | 0.039 | |
| inter-residue sequential (|i−j| = 1) (269) | 0.019±0.009 | 0.002 | |
| inter-residue short-range (1 < |i−j| < 5) (238) | 0.075±0.006 | 0.067 | |
| inter-residue long-range (|i−j| ≥ 5) (83) | 0.070±0.019 | 0.054 | |
| intraresidue (256) | 0.007±0.003 | 0.005 | |
| H-bonds (66) | 0.022±0.007 | 0.024 | |
| rmsd for dihedral angle restraints (deg) (102) | 0.654±0.032 | 0.626 | |
| rmsd for 3JHNα restraints (Hz) (30) | 0.515±0.050 | 0.569 | |
| rmsd (covalent geometry) | |||
| bonds (Å) | 0.007±0.000 | 0.008 | |
| angles (deg) | 0.716±0.027 | 0.461 | |
| impropers (deg) | 1.075±0.090 | 0.977 | |
| energy (kcal/mol) | |||
| total | −1900.73±89.02 | −2161.44 | |
| bond | 29.05±3.24 | 30.99 | |
| angle | 87.92±8.88 | 96.49 | |
| dihedral | 3.58±1.76 | 2.43 | |
| impropers | 49.58±8.67 | 39.10 | |
| van der Waals | −184.53±10.91 | −191.49 | |
| NOE | 63.30±10.65 | 41.50 | |
| 3JHNα | 8.03±1.51 | 9.73 | |
| Cα and Cβ shifts | 77.89±10.00 | 65.75 | |
| RMSD from mean (residues 2-21, 27-55) (Å) | |||
| Backbone | 0.70±0.07 | ||
| Heavy Atoms | 1.20±0.12 |
<SA> represents the ensemble of the 20 water-refined simulated annealing structures.
represents the water refined average of the ensemble.
The overall quality of the RPA3313 NMR structure was assessed with the PSVS software suite (Table 2). All but one residue was located in the most favored region (98.3%) of the Ramachandran plot with the remaining residue in the allowed region (1.7%). PROCHECK further supported the dihedral angle quality of the RPA3313 NMR structure with Z-scores of 0.12 and −0.71 for ϕ, ψ angles and all angles, respectively. Overall model quality was further assessed with ProsaII that produced a good Z-score of −0.58. An excellent quality score of −0.45 was also obtained from a MolProbity analysis, which evaluates atom clashes in the 3D structure. The ProsaII, PROCHECK and MolProbity scores are consistent with other high-quality NMR structures deposited in the PDB. Conversely, the Verify3D structure assessment yielded only a modest score of −1.93, but the analysis is still within an acceptable range compared to other NMR structures. Verify3D measures agreement between the 3D structure and the primary sequence. The novel fold for the RPA3313 structure may be a factor in the relatively low Verify3D score.
Table 2.
Structure Evaluation
| PSVS Z-score (residues 6-54) | |
| Verify3D | −1.93 |
| ProsaII (-ve) | −0.58 |
| Procheck (ϕ and ψ) | 0.12 |
| Procheck (all) | −0.71 |
| MolProbity | −0.45 |
| Ramachandran Space (all residues) | |
| most favored regions | 98.30% |
| allowed regions | 1.70% |
| disallowed regions | 0.00% |
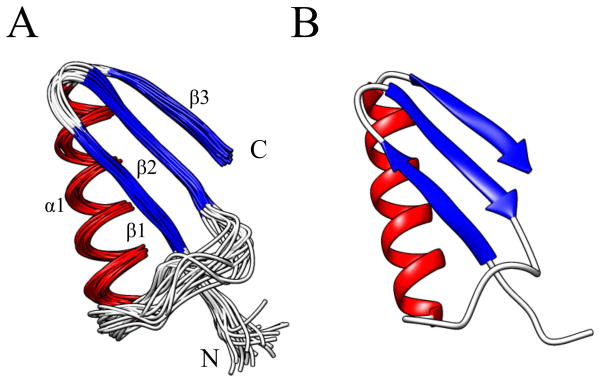
The structure of RPA3313 adopts a split ββαβ motif formed by 3 β-strands (β1-3) packed against an α-helix (Figure 2). There are no known structures of homologs to RPA3313 and a search against the PDB using PDBeFold did not yield any significant results. Although the ββαβ motif is ubiquitous, when the RPA3313 structure is compared to proteins with similar motifs there is either a different handedness, or the orientation of the β–sheet along the α-helix is askew. This is not uncommon for this type of fold, as the β–sheet typically curls or flexes to cover the hydrophobic core of the protein.46
Figure 2.
(A) An ensemble of the 20 best water-refined structures of RPA3313 for residues 6–54 and (B) the coordinate average structure of the ensemble after water refinement. The structures are colored according to secondary structure: red for α-helix, blue for β-strand, and white for loops and disordered regions. The structural elements and the N- and C-terminus are labeled. The disordered C-terminus has been excluded for clarity.
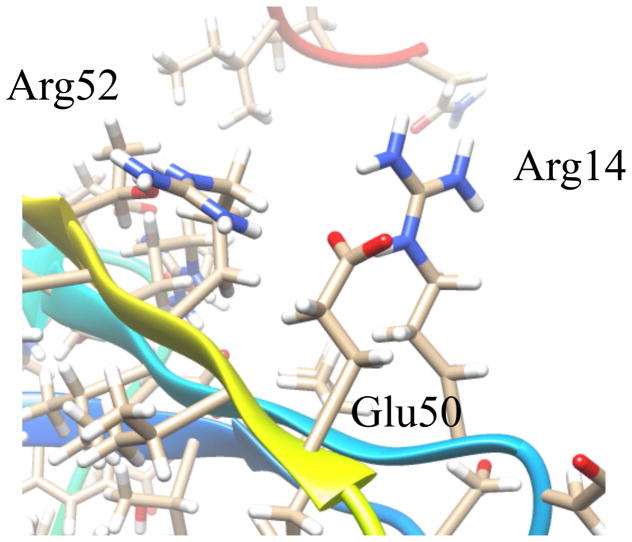
Starting at the N-terminus, the first 2 β-strands are formed antiparallel to one another and are connected by a β-hairpin turn. The initial residues at this terminus do not contribute to β1 and are disordered. At the beginning of β2, Trp15 creates significant bulk in the core of the protein near the β-hairpin. The indole side chain reaches from β2 toward the surface of the protein, which forces Gly10 to accommodate this structural perturbation. Both β1 and β2 have branched side chains forming the center of the protein. Connecting β2 to the α-helix is an extended loop region comprised mostly of negatively charged, polar residues. This loop outlines the top of a cavity formed with β1 and the N-terminus of the α-helix. An additional Tyr residue in the loop has its side chain in close proximity to Lys30, which marks the beginning of the α-helix. The length of the α-helix is approximately 17 residues and is terminated by Gly48. Alanine residues line the inside of the helix and polar residues, including one cysteine, create the solvent exposed surface. A γ-turn links the α-helix to β3, which runs parallel to β2. The side chain of Arg52 on β3 is angled toward the center of the β-sheet and creates a stacking interaction with Arg14 on β2 (Figure 3). This interaction is stabilized by Glu50, which may explain why the α-helix to β3 turn contains only 2 residues. The bottom of β3 is hydrophobic and consists of branched chain amino acids.
Figure 3.
Arginine stacking interaction on the surface of RPA3313. Arginines 14 and 52 interact across the top of the β-sheet and are stabilized by glutamate 50. Each of the residues is evolutionarily stable indicating the occurrence of a conserved interaction.
Following the last β-strand is the disordered C-terminus. At approximately 15 residues in length, this tail is mostly unstructured except for a small α-helical propensity centered on Val67. Seemingly uninteresting at first, the disordered C-terminus probably has a significant physiological function. Disordered termini are known to serve in a broad range of roles such as protein-protein interaction sites, chaperones, and signal processing.47 The proximity of the terminus to the large cavity on the surface of RPA3313 also suggests that it may potentially serve a role in activity (Figure 2b). Single-stranded DNA (ssDNA) binding proteins in prokaryotes maintain evolutionary conserved disordered C-termini that compete with the DNA binding site in order to exclude unwanted binders.48 Although it is not known if RPA3313 binds ssDNA, the mechanism of the competition between the disordered tail and ligand remains a possibility. Also, many photosynthetic organisms possess globular proteins that have extended termini and are involved in a wide array of functions.49 These extensions are highly variable and show little conservation between homologous species. However, they are necessary for host protein regulation and function. Since RPA3313 is expressed during photoautotrophic growth, it is possible that the disordered tail is involved in a light dependent mechanism.
Conserved Residue Analysis
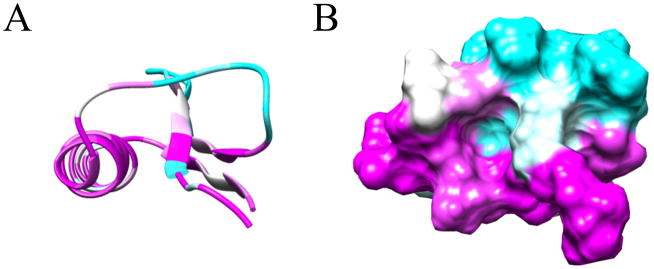
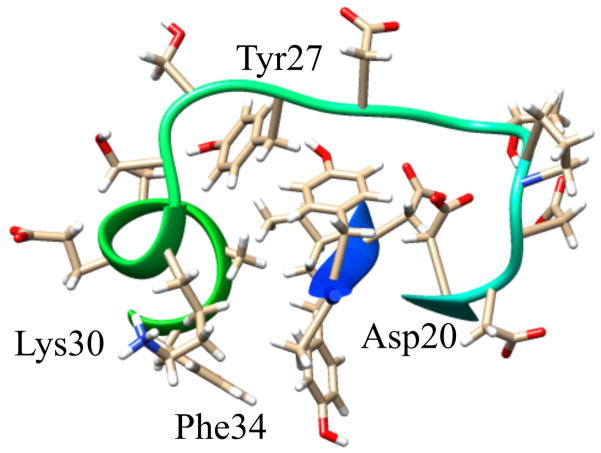
The structure of RPA3313 was submitted to the ConSurf server for conserved residue analysis. ConSurf identifies and scores residue conservation based on a BLAST search and a subsequent multiple sequence alignment. Plotted on a surface representation of the RPA3313 NMR structure are the ConSurf scores, which range from 0 (cyan) to 1 (magenta) with 1 signifying high conservation (Figure 4). Clearly visible is a conserved pocket between the extended loop and the top of β1. The deepest region of the cavity is defined by the peptide backbone of the α-helix and Tyr6. Conserved residues with side chains pointing into the pocket are Asp20, Tyr27, Lys30, and Phe34 (Figure 5). The Asp, Tyr, and Lys residues have the ability to form hydrogen bonds with a ligand. Additionally, Lys and Asp are possible metal coordinators and Tyr and Phe may be involved in π-π interactions with a ligand. Also conserved are small flexible residues Gly2, Ala4, and Gly25. Each residue is either at the top or the bottom of the pocket and likely contributes to important structure flexibility. These small residues would enable the protein to bend in order to accommodate a larger ligand, or to change the size of the entrance to the pocket based on other structural perturbations or modifications. Gly2 is doubly important as it follows the N-terminal start methionine. Small flexible residues trailing methionine enable truncation by an aminopeptidase and it is anticipated that the physiological form of RPA3313 lacks this initial methionine residue.50 Distal to the pocket, the γ-turn between α-helix and β3 is also highly conserved. It is possible that this turn also acts like a hinge between the β-sheet and α-helix to allow the protein to adjust to a possible change in the hydrophobic core resulting from binding a ligand or a protein-protein interaction. The aforementioned stacked arginine residues (14, 52) are also moderately conserved. The ConSurf score of approximately 0.5 indicate that this could be an evolutionary newer interaction or function. The stabilizing Glu50 residue has a higher conservation score (~0.7) than the arginines, but lower than the other residues in the γ-turn. This suggests that Glu50 may have a dual role in providing flexibility at the hinge in addition to stabilizing the arginine stacking interaction. Lastly, a highly conserved proline residue (56) exists at the end of β3. Proline is known to disrupt secondary structure formation and is most often found in disordered regions or turns. In this case, proline is acting as a terminator of a β-strand, which may assist in keeping the C-terminus residues in a disordered state. Furthermore, proline residues are associated with protein-protein interactions involving disordered protein regions.51 In these situations the disordered tail or region adopts an induced fit upon interaction or binding.
Figure 4.
ConSurf Analysis. (A) ConSurf per residue scores for the structure RPA3313. Conserved residues are magenta and non-conserved residues are cyan. (B) ConSurf scores mapped onto the molecular surface representation of RPA3313. A conserved pocket is formed between the N-terminus of the α-helix and the first β-strand. Residues found to be less conserved are mostly found in the loop regions and the disordered tail (C-terminus, not shown).
Figure 5.
Surface pocket and possible ligand binding site on RPA3313. Shown are the residues with side chains that point into the pocket of the protein. The evolutionarily conserved residues are labeled. The remaining residues may also participate in the function of the pocket even though they are not highly conserved.
Protein-Protein Interaction Site Prediction
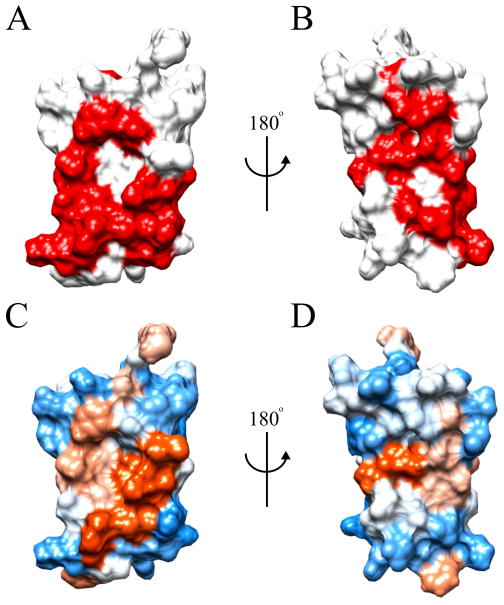
A further bioinformatics analysis of the structure of RPA3313 was carried out with cons-PPISP. The cons-PPISP server utilizes a neural network to predict position specific interaction sites on protein surfaces. Based on the output of cons-PPISP, it is possible to reliably identify clusters of residues that suggest a potential protein binding site. Two large sites were successfully identified that, when visualized on the surface of the RPA3313 NMR structure, lie opposite of one another (Figure 6). One potential protein binding site is between the β-sheet and α-helix on the bottom of RPA3313, while the other crosses the width of the β-sheet on the top of the protein. The bottom protein biding site consists of side chains from residues Tyr6, Trp15, Phe34, Cys38, Ser42, Ile45, Lys46, Glu50, Val51, Arg52, Ile53, and Thr54 (Figure 6a). Although mostly hydrophobic in composition (Figure 6c), these residues form a likely interaction hotspot due to their high abundance in other known protein-protein interactions.52 Furthermore, the surfaces of β-sheets are known to commonly participate in protein binding. A protein binding event at this bottom location on the RPA3313 surface could induce a significant reshuffling of the hydrophobic core as discussed earlier. The second predicted protein binding site runs perpendicular to the β-sheet and lies directly opposite the first predicted binding site (Figure 6b). Solvent exposed side chains from residues Val9, Tyr27, Ala32, Ala36, Ala39, and Asn43 populate the surface of the top protein binding site. This putative protein interaction site has both hydrophobic and hydrophilic regions (Figure 6d) indicating that multiple binding partners are possible.
Figure 6.
cons-PPISP predictions and surface hydrophobicity. (A) Front and (B) back orientations of RPA3313 with predicted protein interaction sites colored red. (C) Front and (D) back surface hydrophobicity colored from blue (hydrophilic) to orange (hydrophobic).
Protein-Protein Crosslinking
A simple crosslinking experiment was carried out in order to determine possible protein binders to RPA3313. The crosslinking experiment was performed both in vivo in E. coli and in vitro with a proteome extract from R. palustris. The replicate crosslinking experiments were performed to reliably identify physiologically-relevant interaction partners to RPA3313. RPA3313 was overexpressed in E. coli and prior to the two distinct crosslinking experiments the cell culture was split into two separate samples. A small aliquot of the total cell culture was removed for the in vivo crosslinking experiment, and the remaining cell culture was then used to extract and purify the overexpressed RPA3313 protein.
Purified RPA3313 was spiked into a total protein extract from an R. palustris cell culture and the formaldehyde crosslinking was then performed in vitro. In contrast, the E. coli cell culture overexpressing RPA3313 was simply treated with formaldehyde for an in vivo crosslinking experiment. Formaldehyde was used to covalently link lysine side chains through amide bond formation and subsequently removed by heating the sample after purification. Following purification, the crosslinked proteins were resolved by SDS-PAGE and identified by MS/MS analysis. Proteins found to be crosslinked to RPA3313 belonged to ribosomal subunits in both E. coli and R. palustris (Table 3). Moreover, most of the protein component of the ribosome from both organisms was identified to bind RPA3313. Thus, RPA3313 appears likely to bind to the ribosome at one or multiple points. Since it is also possible that the ribosome remained intact during the crosslinking and purification process, the number of binding sites for RPA3313 on the ribosome remains undetermined. It is important to note that RPA3313 was only overexpressed in E. coli and not in R. palustris. This insures that the results are physiologically relevant and not a simple artifact of an overexpressed protein being crosslinked to equally abundant ribosomal proteins. In fact, an additional in vivo crosslinking experiment with a second overexpressed protein (human DJ-1) served as a negative control and verified that the RPA3313 results were not an artifact of a protein overexpression system. Despite identical experimental conditions and unlike RPA3313, human protein DJ-1 did not crosslink with any (as expected) E. coli proteins in vivo.
Table 3.
In vivo and in vitro Crosslinking Results
| Organism | Identified Ribosomal Proteins |
|---|---|
| E. coli | S2-11, S13, S15, S16, S18, S19, S21, L2-6, L9-11, L13-22, L24, L27, L28, L32 |
| R. palustris | S6-13, S15-17, S19-21, L1-3, L6, L7/12, L9, L10, L15, L17-19, L21, L23, L24, L30, L32, L33 |
| E. coli (DJ-1 control) | NA |
A previous study successfully sequenced and identified the ribosomal subunits from R. palustris.53 During the study, other uncharacterized proteins were purified with the ribosome, but none of them were identified as RPA3313. Like many ribosomal subunits from other organisms, some of the subunits from R. palustris contained disordered C-termini. The disordered C-terminus acts as an anchor and buries itself into the RNA core and also promotes proper assembly of the ribosome.54,55 Globular portions of the proteins are then exposed to the solvent to interact with other proteins. While RPA3313 is not part of the ribosome, its tertiary structure mimics a ribosomal subunit with the multiple protein-protein binding sites and a disordered C-terminus. RPA3313 may instead act as a chaperone or transporter for substrates traveling to or from the ribosome.
Conclusion
RPA3313 is a conserved protein from R. palustris and a member of functionally unannotated class of proteins in alphaproteobacteria. The purpose of this study was to structurally characterize this class of proteins and provide an initial functional characterization. An NMR solution structure reveals that RPA3313 adopts a novel globular split ββαβ motif followed by a disordered C-terminus tail. PSVS evaluation of the ensemble of the 20 lowest energy structures of RPA3313 produced generally good quality scores consistent with other high-quality NMR structures deposited in the PDB. Bioinformatics analyses led to the identification of several possible protein-protein interaction sites on the surface of RPA3313 and a large conserved pocket sandwiched between the β-sheet and α-helix. Crosslinking analysis revealed that RPA3313 interacts with the ribosome both in vivo and in vitro. Multiple ribosomal subunits were pulled down with RPA3313 in E. coli and in R. palustris, so the exact nature of the interaction between the two is unknown. In silico dockings, 15N NMR titrations, and ligand screenings were done in an attempt to determine the physiological role of RPA3313 (data not shown). However, a binder with a sub-millimolar binding constant was not found. It is possible that the tertiary structure of RPA3313 changes the shape of its binding pocket when in contact with another protein or in a much larger complex. Also, the C-terminus tail could be blocking or competing for the binding site and the lack of an N-terminus methionine truncation could also impede the binding site. The expression conditions of RPA3313 also remain unknown. It is possible that the protein is only expressed during certain metabolic modes of growth, as this protein is not found in evolutionary distant bacterial species with more limited metabolism. The combined structural and proteomic analyses in this study strongly suggests that RPA3313 by itself or in a larger complex may serve as a ribosomal transport protein.
Supplementary Material
Acknowledgments
The authors would like to express their sincere gratitude to Cheryl Arrowsmith, Adelinda Yee and the Structural Genomics Consortium at the University of Toronto for their help in the early stages of the project and their gift of the RPA3313 clone. This work was supported, in part, by Award Number R21AI081154 from the National Institute of Allergy and Infectious Diseases Nebraska (R21AI081154), and the Tobacco Settlement Biomedical Research Development Fund. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases. Research was performed in facilities renovated with support from the NIH under Grant RR015468-01.
References
- 1.Evans K, Georgiou T, Hillon T, Fordham-Skelton A, Papiz M. Bacteriophytochromes control photosynthesis in Rhodopseudomonas palustris. Adv Photosynth Respir. 2009;28(Purple Phototrophic Bacteria):799–809. [Google Scholar]
- 2.Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JLTy, Peres C, Harrison FH, Gibson J, Harwood CS. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nature Biotechnology. 2004;22:55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
- 3.Kawasaki H, Hoshino Y, Yamasato K. Phylogenetic diversity of phototrophic purple non-sulfur bacteria in the Proteobacteria α group. FEMS Microbiology Letters. 1993;112:61–66. [Google Scholar]
- 4.Sasikala C, Ramana CV, Rao PR. Nitrogen fixation by Rhodopseudomonas palustris OU 11 with aromatic compounds as carbon source/electron donors. FEMS Microbiology Letters. 1994;122:75–78. [Google Scholar]
- 5.Akkerman I, Janssen M, Rocha J, Wijffels RH. Photobiological hydrogen production: photochemical efficiency and bioreactor design. Int J Hydrogen Energy. 2002;27(11–12):1195–1208. [Google Scholar]
- 6.Dagley S. Catabolism of aromatic compounds by microorganisms. Advan Microbial Physiol. 1971;6:1–46. doi: 10.1016/s0065-2911(08)60066-1. [DOI] [PubMed] [Google Scholar]
- 7.Romagnoli S, Tabita FR. Carbon dioxide metabolism and its regulation in nonsulfur purple photosynthetic bacteria. Adv Photosynth Respir. 2009;28(Purple Phototrophic Bacteria):563–576. [Google Scholar]
- 8.McKinlay JB. Systems Biology of Photobiological Hydrogen Production by Purple Non-sulfur Bacteria. Adv Photosynth Respir. 2014;38(Microbial BioEnergy: Hydrogen Production):155–176. [Google Scholar]
- 9.Tanawade SS, Bapat BA, Naikwade NS. Biofuels: use of Biotechnology to meet energy challenges. Int J Biomed Res. 2011;2(1):25–31. [Google Scholar]
- 10.Paulsen IT, Sliwinski MK, Saier MH. Microbial Genome Analyses: Global Comparisons of Transport Capabilities Based on Phylogenies, Bioenergetics and Substrate Specificities. Journal of Molecular Biology. 1998;277:573–592. doi: 10.1006/jmbi.1998.1609. [DOI] [PubMed] [Google Scholar]
- 11.VerBerkmoes NC, Shah MB, Lankford PK, Pelletier DA, Strader MB, Tabb DL, McDonald WH, Barton JW, Hurst GB, Hauser L, Davison BH, Beatty JT, Harwood CS, Tabita FR, Hettich RL, Larimer FW. Determination and comparison of the baseline proteomes of the versatile microbe Rhodopseudomonas palustris under its major metabolic states. Journal of Proteome Research. 2006;5:287–298. doi: 10.1021/pr0503230. [DOI] [PubMed] [Google Scholar]
- 12.Apweiler R, Bairoch A, Wu CH. Protein sequence databases. Curr Opin Chem Biol. 2004;8(1):76–80. doi: 10.1016/j.cbpa.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Research. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabita FR, Hanson TE, Satagopan S, Witte BH, Kreel NE. Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philos Trans R Soc, B. 2008;363(1504):2629–2640. doi: 10.1098/rstb.2008.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badger MR, Bek EJ. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exp Bot. 2008;59(7):1525–1541. doi: 10.1093/jxb/erm297. [DOI] [PubMed] [Google Scholar]
- 16.Bryant DA, Frigaard N-U. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 2006;14(11):488–496. doi: 10.1016/j.tim.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Skolnick J, Fetrow JS, Kolinski A. Structural genomics and its importance for gene function analysis. Nature Biotechnology. 2000;18:283–287. doi: 10.1038/73723. [DOI] [PubMed] [Google Scholar]
- 18.Baker D, Sali A. Protein Structure Prediction and Structural Genomics. Science. 2001:294. doi: 10.1126/science.1065659. [DOI] [PubMed] [Google Scholar]
- 19.Baca AM, Hol WGJ. Overcoming codon bias: A method for high-level overexpression of Plasmodium and other AT-rich parasite genes in Escherichia coli. Int J Parasitol. 2000;30(2):113–118. doi: 10.1016/s0020-7519(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 20.Hyberts SG, Takeuchi K, Wagner G. Poisson-Gap Sampling and FM Reconstruction for Enhancing Resolution and Sensitivity of Protein NMR Data. Journal of the American Chemical Society. 2010;132:2145–2147. doi: 10.1021/ja908004w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikura M, Kay LE, Bax A. A Novel Approach for Sequential Assignment of 1H, 13C, and 15N Spectra of Larger Proteins: Heteronuclear Triple-Resonance Three-Dimensional NMR Spectroscopy. Application to Calmodulint Biochemistry. 1990;29:4659–4667. doi: 10.1021/bi00471a022. [DOI] [PubMed] [Google Scholar]
- 22.Kay LE, Ikura M, Tschudin R, Bax A. Three-Dimensional Triple-Resonance NMR Spectroscopy of Isotopically Enriched Proteins. Journal of Magnetic Resonance. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN Data Model for NMR Spectroscopy: Development of a Software Pipeline. Proteins: Structure, Function and Bioinformatics. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 24.Shen Y, Bryan PN, He Y, Orban J, Baker D, Bax A. De novo structure generation using chemical shifts for proteins with high-sequence identity but different folds. Protein Science. 2010;19:349–356. doi: 10.1002/pro.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y, Lange O, Delaglio F, Rossi P, Aramini JM, Liu G, Eletsky A, Wu Y, Singarapu KK, Lemak A, Ignatchenko A, Arrowsmith CH, Szyperski T, Montelione GT, Baker D, Bax A. Consistent blind protein structure generation from NMR chemical shift data. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4685–4690. doi: 10.1073/pnas.0800256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. Journal of Biomolecular NMR. 2009;43:63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. Journal of Magnetic Resonance. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 28.Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor–NIH for NMR molecular structure determination. Progress in Nuclear Magnetic Resonance Spectroscopy. 2006;48:47–62. [Google Scholar]
- 29.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. Journal of Biomolecular NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharya A, Tejero R, Montelione GT. Evaluating Protein Structures Determined by Structural Genomics Consortia Tools for Structure Quality Evaluation. Proteins: Structure, Function and Bioinformatics. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 31.Sippl MJ. Recognition of Errors in Three-Dimensional Structures of Proteins. Proteins: Structure, Function, and Genetics. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 32.Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 33.Lovell SC, Davis IW, Arendall WB, III, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure Validation by Cα Geometry: φ, ψ and Cβ, Deviation. Proteins: Structure, Function, and Genetics. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera — A Visualization System for Exploratory Research and Analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 37.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Research. 2005;33:299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Celniker G, Nimrod G, Ashkenazy H, Glaser F, Martz E, Mayrose I, Pupko T, Ben-Tal N. ConSurf: Using Evolutionary Data to Raise Testable Hypotheses about Protein Function. Israel Journal of Chemistry. 2013;53:199–206. [Google Scholar]
- 39.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: Identification of Functional Regions in Proteins by Surface-Mapping of Phylogenetic Information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 40.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Research. 2010;38:529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallographica Section D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, Zhou HX. Prediction of interface residues in protein-protein complexes by a consensus neural network method: Test against NMR data. Proteins: Structure, Function and Bioinformatics. 2005;61:21–35. doi: 10.1002/prot.20514. [DOI] [PubMed] [Google Scholar]
- 43.Zhou H-x, Shan Y. Prediction of Protein Interaction Sites From Sequence Profile and Residue Neighbor List. Proteins: Structure, Function, and Genetics. 2001;44:336–343. doi: 10.1002/prot.1099. [DOI] [PubMed] [Google Scholar]
- 44.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Huson DH, Scornavacca C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Systematic Biology. 2012;0:1–7. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 46.Orengo CA, Thornton JM. Alpha plus beta folds revisited: some favoured motifs. Current Biology. 1993;1:105–120. doi: 10.1016/0969-2126(93)90026-d. [DOI] [PubMed] [Google Scholar]
- 47.Uversky VN. The most important thing is the tail: Multitudinous functionalities of intrinsically disordered protein termini. FEBS Letters. 2013;587:1891–1901. doi: 10.1016/j.febslet.2013.04.042. [DOI] [PubMed] [Google Scholar]
- 48.Marintcheva B, Marintchev A, Wagner G, Richardson CC. Acidic C-terminal tail of the ssDNA-binding protein of bacteriophage T7 and ssDNA compete for the same binding surface. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1855–1860. doi: 10.1073/pnas.0711919105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thieulin-pardo G, Avilan L, Kojadinovic M, Gontero B. Fairy “tails”: flexibility and function of intrinsically disordered extensions in the photosynthetic world. Frontiers in Molecular Biosciences. 2015;2:1–18. doi: 10.3389/fmolb.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-bassat A, Bauer K, Chang S-y, Myambo KEN, Boosman A. Processing of the Initiation Methionine from Proteins: Properties of the Escherichia coli Methionine Aminopeptidase and Its Gene Structure. Journal of Bacteriology. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Theillet F-x, Kalmar L, Tompa P, Han K-h, Dunker AK, Daughdrill GW, Uversky VN, Theillet F-x, Kalmar L, Tompa P, Han K-h, Selenko P, Dunker AK. The alphabet of intrinsic disorder I. Act like a Pro: On the abundance and roles of proline residues in intrinsically disordered proteins. Intrinsically Disordered Proteins. 2013:1. doi: 10.4161/idp.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma B, Elkayam T, Wolfson H, Nussinov R. Protein-protein interactions: Structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strader MB, VerBerkmoes NC, Tabb DL, Connelly HM, Barton JW, Bruce BD, Pelletier DA, Davison BH, Hettich RL, Larimer FW, Hurst GB. Characterization of the 70S Ribosome from Rhodopseudomonas palustris Using and Integrated “Top-Down” and “Bottom-Up” Mass Spectrometric Approach. Journal of Proteome Research. 2008;3:965–978. doi: 10.1021/pr049940z. [DOI] [PubMed] [Google Scholar]
- 54.Peng Z, Oldfield CJ, Xue B, Mizianty MJ, Dunker AK, Kurgan L, Uversky VN. A creature with a hundred waggly tails: intrinsically disordered proteins in the ribosome. Cellular and Molecular Life Sciences. 2014;71:1477–1504. doi: 10.1007/s00018-013-1446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brodersen DE, WMC, Carter AP, Wimberly BT, Ramakrishnan V. Crystal Structure of the 30 S Ribosomal Subunit from Thermus thermophilus: Structure of the Proteins and their Interactions with 16 S RNA. Journal of Molecular Biology. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 56.Porollo AA, Adamczak R, Meller J. POLYVIEW: a flexible visualization tool for structural and functional annotations of proteins. Bioinformatics. 2004;20(15):2460–2462. doi: 10.1093/bioinformatics/bth248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.