Abstract
V(D)J recombination requires binding and synapsis of a complementary (12/23) pair of recombination signal sequences (RSSs) by the RAG1 and RAG2 proteins, aided by a high-mobility group protein, HMG1 or HMG2. Double-strand DNA cleavage within this synaptic, or paired, complex is thought to involve DNA distortion or melting near the site of cleavage. Although V(D)J recombination normally occurs between RSSs located on the same DNA molecule (in cis), all previous studies that directly assessed RSS synapsis were performed with the two DNA substrates in trans. To overcome this limitation, we have developed a facilitated circularization assay using DNA substrates of reduced length to assess synapsis of RSSs in cis. We show that a 12/23 pair of RSSs is the preferred substrate for synapsis of cis RSSs and that the efficiency of pairing is dependent upon RAG1-RAG2 stoichiometry. Synapsis in cis occurs rapidly and is kinetically favored over synapsis of RSSs located in trans. This experimental system also allowed the generation of underwound DNA substrates containing pairs of RSSs in cis. Importantly, we found that the RAG proteins cleave such substrates substantially more efficiently than relaxed substrates and that underwinding may enhance RSS synapsis as well as RAG1/2-mediated catalysis. The energy stored in such underwound substrates may be used in the generation of DNA distortion and/or protein conformational changes needed for synapsis and cleavage. We propose that this unwinding is uniquely sensed during synapsis of an appropriate 12/23 pair of RSSs.
The process of immunoglobulin and T-cell receptor gene rearrangement, known as V(D)J recombination, occurs during lymphocyte differentiation and plays a major role in diversifying these antigen receptors in jawed vertebrates (55). Two steps precede DNA double-strand cleavage by the RAG1/RAG2 recombinase machinery. First, RAG1-RAG2 specifically bind to short DNA elements termed recombination signal sequences (RSSs) flanking the V, D, and J gene segments. RSSs are composed of two well-conserved sequences, a palindromic heptamer (consensus 5′-CACAGTG) and an A/T-rich nonamer (consensus 5′-ACAAAAACC) separated by a less-well-conserved spacer region of 12 or 23 bp in length (41). The second step, the synapsis of a pair of RSSs, determines both the identity and the type of gene segments to be adjoined. It is within this paired complex that DNA double-strand breaks are introduced between the heptamer and flanking coding region (referred to as hairpin formation, because cleavage generates a covalently sealed hairpin structure at the end of each coding segment).
In vivo, recombination occurs almost exclusively between gene segments flanked by a 12- and a 23-RSS (complementary RSSs) according to the 12/23 rule. The mechanisms enforcing this important restriction remain poorly understood at a molecular level. Biochemical experiments have revealed a more stringent adherence to the 12/23 rule in coupled cleavage assays (11, 56, 58) than in electrophoretic mobility shift assays that assess paired complex formation (22, 35, 52), leading to the notion that the 12/23 rule is imposed at both the synapsis and cleavage steps of the reaction. Coupled cleavage at the two RSSs in accordance with the 12/23 rule is critically dependent on HMG1/2 proteins and Mg2+ (11, 25, 46, 52, 56, 58).
HMG1 and -2 are ubiquitous, abundantly expressed nuclear proteins that bind nonspecifically in the minor groove of DNA via their two HMG box domains. They bind preferentially to distorted DNA, including cruciform four-way junction DNA structures (4, 13, 30, 40), and play a role in the repair of 1,2-cis-platinum DNA intrastrand adducts (39). HMG1 and -2 also bind to and preserve negatively supercoiled DNA structures (48). Intercalation of HMG box amino acids into DNA can prevent relaxation of negatively supercoiled DNA treated with topoisomerase I (21, 27, 50), and HMG1/2 proteins are able to unwind DNA, as demonstrated by their ability to reduce the linking number of circular plasmids covalently resealed in their presence (23, 24). Unwinding by intercalation is supported by the finding that intercalators such as ethidium bromide antagonize HMG1/2 binding to DNA and release these proteins from their complexes in nuclear chromatin (20). In V(D)J recombination, HMG1/2 proteins strongly enhance binding of the 23-RSS by the RAG proteins, perhaps by binding to the spacer-nonamer border, and are essential for efficient paired complex formation (34, 36, 51, 54). HMG1/2 have been suggested to play an architectural role in synapsis, helping to position the 12- and 23-RSS in the appropriate register for catalysis. Given the ability of HMG1/2 to unwind DNA and the negatively supercoiled state of DNA in chromatin, it is of particular interest to determine whether the underwound topological state influences RSS pairing, gene segment discrimination, or catalysis.
One mechanism by which DNA underwinding could contribute to DNA cleavage by the RAG proteins is by facilitating base unpairing. DNA melting at the heptamer-coding flank border has been implicated in RSS binding by the RAG proteins (2, 37, 51, 54) as well as in efficient DNA cleavage (9, 42, 44, 45). Underwinding of the DNA substrate could also contribute to synapsis or cleavage by other mechanisms, such as serving as a source of energy for conformational changes required during assembly of higher-order protein-DNA complexes, as in the case of Tn10 transposase (5, 8). In contrast to DNA melting, a potential role for DNA underwinding in the early steps of V(D)J recombination has not been systematically investigated.
Here, we investigate the role played by HMG1/2 proteins, RAG1/2 stoichiometry, and DNA topology in directing cis RSS synapsis and cleavage. All previous studies of RSS pairs located in cis have relied on DNA cleavage or the complete recombination reaction to serve as indirect indicators of RSS synapsis. To avoid this limitation, we have developed a facilitated circularization assay that allows assessment of cis RSS synapsis independent of DNA cleavage. We used this assay and conventional cleavage assays to address three questions: (i) to what extent is the 12/23 rule adhered to for synapsis of RSSs located in cis? (ii) what are the kinetics of RAG protein-mediated synapsis of RSSs located in cis? and (iii) does an underwound DNA topology influence synaptic complex assembly or DNA cleavage by the RAG proteins? Our experiments reveal factors and conditions that influence paired complex assembly and the 12/23 rule using RSSs in cis, independent of DNA cleavage, and demonstrate that underwound DNA is a strongly preferred substrate for DNA cleavage by the RAG proteins.
MATERIALS AND METHODS
Plasmids and DNA constructs.
Constructs used in the ligation and cleavage assays were synthesized starting from pJH290 (31) using a two-step PCR approach (33) (primer sequences are shown in Table 1). All substrates are 263 bp in length with XbaI sites at both ends (underlined in Table 1) and were generated by two PCR amplification steps. The same XbaI sites were used for cloning. IS95 was generated by a two-step PCR approach. In the first step, two amplification reactions were performed using pJH290 DNA as template, primers 1M and 2M for the first amplification, and primers 3M and 4M for the second amplification. The two amplification products obtained were used as template together with primers 1M and 4M for the second step of amplification. The products of the second step were cloned into the XbaI site of a modified pUC19 plasmid with no site for HindIII. The same strategy was used for cloning all other constructs described in this study (details and construct sequences are available upon request).
TABLE 1.
Primer sequences
| Primer | Sequencea |
|---|---|
| 1M | AGCTGCTCTAGAACAGCTATGACCATGATT |
| 2M | ACTGCAGGCAAGCTTAGATCAGACTGACTGCAGGGTTTTTGTTCCA |
| 3M | AGTCTGATCTAAGCTTGCCTGCAGTAACTTGTCGCGCCAATCGAG |
| 4M | TAGTGCTCTAGATTAGCTTCCTTAGCTCCT |
| 12-RSS Neg | TGGGCTGCAGGTCGACTGGCCATCTACAGACTGGAGCGGCCGCACTGCAGTCAGTCTGA |
| M13 Rev | AGGAAACAGCTATGACCATG |
| TN1 | GTAGTACTCCACTGTCTGGCTGTAGCGGCCGCCGCCTCGG GATCCTCTCA |
| TN2 | ACAGCCAGACAGTGGAGTACTACATGGCCAGGATCCCCGGGGATC |
| 12/12-1 | ATCGTGATCTAGAGATCCTGGCCAACAGCTATGACCATGACCATGATTACG |
| 12/12-2 | GGATCCCACAGTGGTAGTACTCCACACAAAAACCCTCGGGATC |
| 12/12-3 | GTGGAGTACTACCACTGTGGGATCC |
| 1m 23/23 | AGCTGCTCTAGATATGACCATGATTACGCC |
| 2m 23/23 | AGTACCATGGTTCCAGTCTGTAGCACTGTG |
| 3m 23/23 | CTACAGACTGGAACCATGGTACTAAAAAACCCTGCAGTCAGTCT |
| 4m 23/23 | TAGTGCTCTAGATCCTTAGCTCCTGAAAATC |
XbaI sites are underlined.
For both the RAG-mediated coupled cleavage and ligation assays, the 263-bp DNA substrates were body labeled with [α-32P]dCTP and obtained by thermal amplification using the appropriate primers, followed by extensive XbaI digestion and purification. The IS95 topoisomers used in the cleavage assays described below in Fig. 10 were obtained by ligating the XbaI cohesive ends of the 263-bp IS95 radioactively labeled DNA with T4 DNA ligase in the presence of HMG2. After ligation, the products were resolved on 5% acrylamide gels, and DNA from the bands corresponding to each topoisomer was purified.
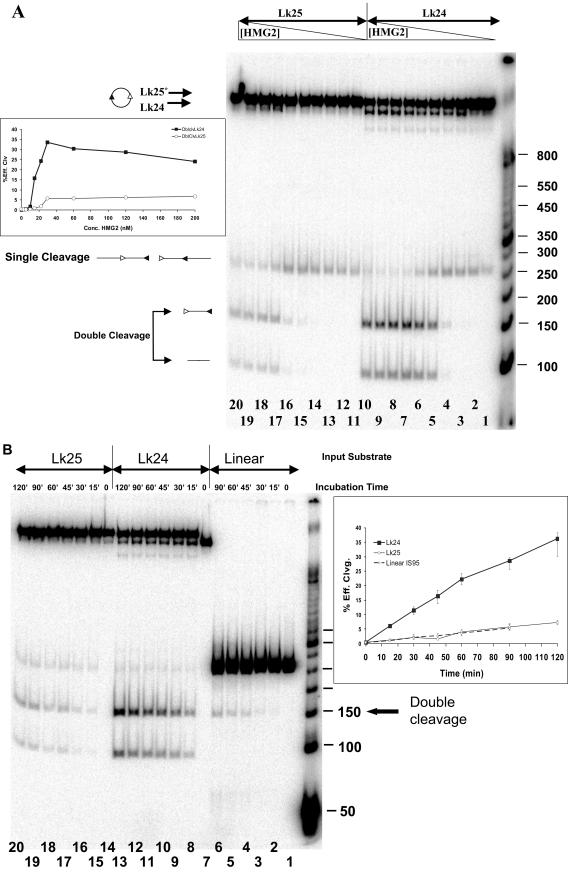
FIG.10.
Underwinding enhances cleavage by the RAG proteins. (A) Reactions were performed for 2 h at 37°C in the presence of 2.5 mM MgCl2, 3 nM DNA, 6 nM MBP-RAG1, 12.5 nM GST-RAG2, and increasing concentrations of HMG2. Lanes 1 to 10, Lk24 IS95 circular substrate; lanes 11 to 20, Lk25 IS95 relaxed circle. Reaction mixtures in lanes 1 to 10 and 11 to 20 contained the following concentrations of HMG2: 0, 2.5, 5, 10, 15, 22.5, 30, 60, 120, and 200 nM, respectively. (Inset) Quantitation of the amount of double cleavage, expressed as the percent efficiency of cleavage versus HMG2 concentration (see Materials and Methods). Dark boxes, Lk24 substrate; open diamonds, Lk25 substrate. Sizes of molecular size markers are indicated in base pairs on the right. (B) Kinetics of RAG-mediated coupled cleavage with linear, relaxed circular, and underwound circular substrates. Reactions were performed at 37°C in the presence of 2.5 mM MgCl2, 3 nM DNA, 6 nM MBP-RAG1, 12.5 nM GST-RAG2, and 120 nM HMG2 for the indicated length of time. Lanes 1 to 6, linear IS95 substrate; lanes 7 to 13, underwound Lk24 IS95 circular substrate; lanes 14 to 20, relaxed Lk25 IS95 circular substrate. Molecular size markers are the same as those used for panel A. (Inset) Quantitation of the amount of double cleavage, expressed as the percent efficiency of cleavage versus time. Data points represent average values obtained from three data sets, while error bars denote the extreme maximal and minimal values obtained from individual determinations.
Protein purification.
Throughout this work RAG1 denotes core RAG1 (amino acids [aa] 384 to 1008) fused at its N terminus to maltose binding protein (MBP-RAG1) and its catalytic site mutant MBP-RAG1-D708A. RAG2 denotes core RAG2 (aa 1 to 383) fused at its N terminus to glutathione S-transferase (GST-RAG2). These proteins were expressed and purified as previously described (15). Murine HMG2 (aa 1 to 185) lacking the C-terminal acidic domain was expressed and purified as described elsewhere (12).
Ligation assays.
All ligation and cleavage reactions were performed in the main buffer (MB), which contains 10 mM Tris-HCl (pH 7.4), 50 mM KCl, 2.5 mM MgCl2, and 1 mM ATP. Three types of ligation experiments were performed. In the first type, titration assays were performed with labeled substrate DNA (final concentration, 3 nM) equilibrated either with protein species (HMG2 or MBP-RAG1-D708A) or with ethidium bromide in increasing concentrations for 10 min at 25°C. Two hundred units of T4 DNA ligase (NEB) were then added with rapid mixing, and the reaction mixture was incubated an additional 2 min at the same temperature. Subsequently, each reaction was stopped by addition of EDTA to a final concentration of 16 mM. DNA was then deproteinized by proteinase K treatment and phenol-chloroform extraction and then ethanol precipitated. The purified DNA was resolved on 5% acrylamide-Tris-borate-EDTA native gels, which were dried and exposed for 24 h and imaged with a PhosphorImager (Molecular Dynamics). The scanned images were quantified using ImageQuant 5.2 (Molecular Dynamics).
The second type of assay (facilitated ligation) required the equilibration of labeled DNA substrate with various proteins in two steps. The labeled substrate DNA (final concentration, 3 nM) was equilibrated first with 22.5 nM HMG2 alone or mixed with 6 nM MBP-RAG1-D708A for 10 min at 25°C. In the second step, the protein species to be titrated (GST-RAG2) was added in increasing concentrations, with an additional 5-min incubation at 25°C. T4 DNA ligase was then added, and all subsequent steps were performed as described above.
In the third type of ligation experiment (time course ligation assays), 22.5 nM HMG2, 6 nM D708A-RAG1-MBP, and 200 U of T4 DNA ligase (NEB) with or without 12.5 nM GST-RAG2 were equilibrated for 10 min at 25°C, and the mixtures were then added with rapid mixing to labeled DNA substrate (final DNA concentration, 3 nM). Reactions were stopped and processed as described above. Data from ligation assays were plotted as efficiency of circle or multimer formation versus either the concentration of the species that was varied in the experiment or time. The efficiency of circle or multimer formation reflects the ratio between the quantified intensities of the bands corresponding to monomeric circle or linear multimer and the intensities of the bands corresponding to unligated substrate. In the case of the facilitated ligation assay, the data were expressed as the ratio of the efficiency of circle formation of the respective reaction with respect to the efficiency of circle formation in the control reaction lacking the titrated species. This ratio is denoted the fraction efficiency circularization.
Coupled cleavage assays.
Coupled cleavage assays were performed in MB by incubation of labeled DNA substrates (the same as those used in ligation assays) at a 3 nM final concentration with 22.5 nM HMG2 and 6 nM MBP-RAG1 for 10 min at 25°C. Subsequently, the species to be titrated in the reaction (GST-RAG2) was added in increasing concentrations to the mixtures equilibrated in the first step. For Fig. 10A, below, titration of HMG2 was performed by preincubating 3 nM substrate DNA with 6 nM MBP-RAG1 and 12.5 nM GST-RAG2 followed by addition of HMG2. The final mixtures were incubated either for 2 h at 37°C or as indicated and were then stopped by addition of EDTA to 16 mM followed by deproteinization and ethanol precipitation. The purified DNAs were separated on 5% acrylamide-TB nondenaturing gels. Gels were dried and visualized using a PhosphorImager. Data were plotted as percent efficiency of cleavage versus the concentration of the various species in the reaction mixtures. The percent efficiency of cleavage reflects the ratio between the band intensity corresponding to the cleaved DNA versus that of the total DNA in the reaction, expressed as a percentage.
Enzyme treatment of circular topoisomers.
The Lk24 and Lk25 circular topoisomers and the linear dimer obtained from ligation of IS95 in the presence of HMG2, isolated as previously described, were treated with exonuclease III (Exo III; NEB) or ClaI (NEB) to investigate their topology (circular versus linear) or with various topoisomerases to determine their state of underwinding. Each isolated topoisomer (3 nM) was incubated in one of the following 20-μl reaction mixtures: (i) 15 U of wheat germ topoisomerase I (Promega) for 1 h at 37°C in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol, and 2.5 mM Mg2+; (ii) 15 U of calf thymus topoisomerase I (Amersham-Pharmacia) for 1 h at 37°C in 35 mM Tris-HCl (pH 8), 72 mM KCl, 5 mM spermidine, 5 mM dithiothreitol, 5 mM Mg2+, and 0.01% bovine serum albumin; or (iii) 500 ng of Escherichia coli topoisomerase I (a generous gift of J. Wang) for 1 h at 30°C in 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 0.1 mM EDTA, and 2.5 mM Mg2+. The effect of DNA nicking on topoisomer mobility was assessed by incubating topoisomer (3 nM) with 2.5 ng of DNase I/ml for 3 min at 25°C in MB.
Statistical analysis of data.
For every set of experimental data (either cleavage or ligation assays), a minimum of three experiments were performed under identical conditions and the data points shown are average values. For each data set the extreme studentized deviate test (Microsoft Excel) was performed, resulting in exclusion of less than 5% of data points as outliers. For the facilitated ligation experiments of Fig. 7, below, an unpaired nonparametric t test (GraphPad Prism version 3.0) was used to assess statistical significance for pairs of data sets, as indicated in the legend to that figure.
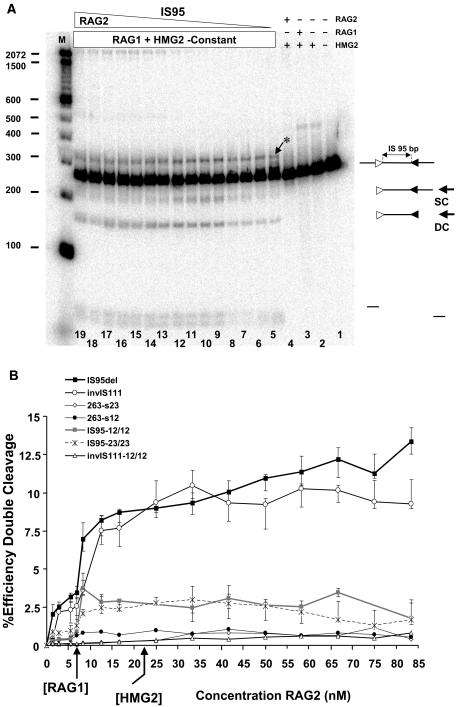
FIG. 7.
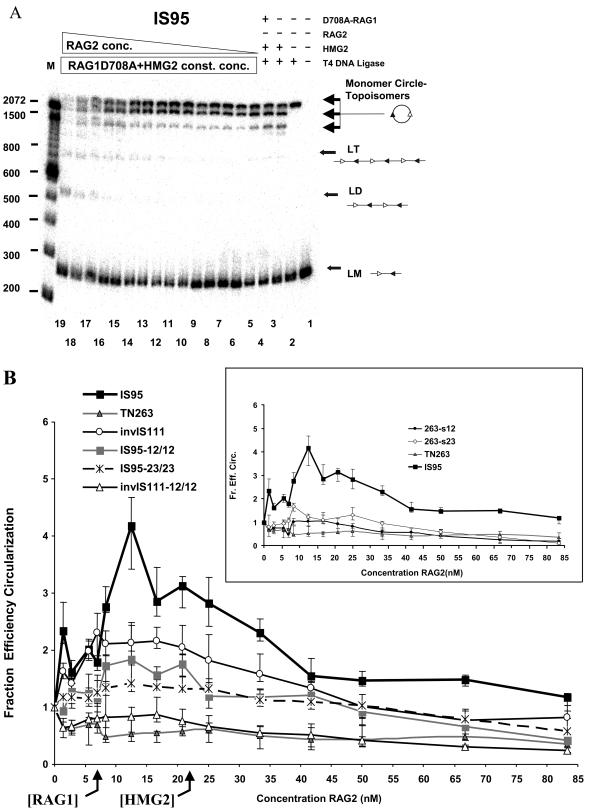
Synapsis of RSSs located in cis, as indicated, by facilitated ligation. (A) Effect of increasing GST-RAG2 concentration in a ligation mixture containing constant amounts of linear IS95 substrate (3 nM), HMG2 (22.5 nM), and MBP-RAG1-D708A (6 nM). Lanes 1 to 4, control reactions lacking various components, as indicated above the lanes; lanes 5 to 19, GST-RAG2 at 1.38, 2.77, 5.55, 6.94, 8.5, 10, 12.50, 16.6, 20.8, 25, 33, 41.65, 50, 66.67, or 83.3 nM, respectively. LT, linear trimer of IS95; other abbreviations are as for Fig. 3. Sizes of molecular mass markers are indicated in base pairs on the left. (B) Effect of RAG2 concentration on substrate circularization. Quantitated data from gels such as that inpanel A are represented as the fraction efficiency of circularization (see Materials and Methods) versus GST-RAG2 concentration. The main graph shows results for substrates containing two RSSs as well as for TN263, which lacks RSSs. The inset shows results for substrates containing a single RSS compared to IS95 and TN263. Data points represent average values obtained from at least three determinations. Data were analyzed statistically, and a small number of data points were excluded from calculations of average values as described in Materials and Methods. Error bars denote the extreme maximal and minimal values obtained from individual determinations, not the standard deviation. Differences in the values obtained were statistically significant for the following pairs of substrates in the range of RAG2 concentrations from 7 to 22 nM: IS95 versus TN263, P < 0.006; invIS111 versus TN263, P < 0.018; IS95-12/12 versus TN263, P < 0.044; IS95-23/23 versus TN263, P < 0.005; IS95 versus IS95-12/12, P < 0.044; IS95 versus IS95-23/23, P < 0.024; invIS111 versus invIS111-12/12, P < 0.049.
RESULTS
Experimental design.
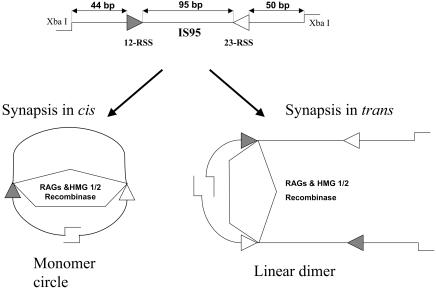
Previously, paired complex formation by the RAG proteins was studied using electrophoretic mobility shift assay and DNase I footprinting, using unlinked RSS oligonucleotides (in trans) (22, 35, 36, 52). To study this process on RSSs located in cis, we used a facilitated ligation method involving a DNA substrate in which the two RSSs to be paired were tethered and almost equally spaced from its two cohesive ends (Fig. 1). Ligation reactions were performed under buffer conditions that supported paired complex formation. The hypothesis underlying this approach was that formation of the RAG-RSS paired complex would bring the two cohesive ends into close proximity and facilitate ligation. The two ends might be located on the same DNA substrate (cis), in which case ligation would result in a monomer circle, or on distinct DNA substrates (trans), in which case ligation would result in linear multimer formation (Fig. 1). Experiments were performed with D708A mutant core RAG1 (aa 384 to 1008) and core RAG2 (aa 1 to 387) in the absence or presence of purified HMG2. The RAG1 active site mutant D708A was used because it is able to form all RAG1/RAG2/RSS complexes normally but cannot catalyze nicking or hairpin formation (15, 26, 29). The primary substrate used in our ligation assays, IS95, is 263 bp in length with 95 bp between the nonamers of the two RSSs and has XbaI cohesive ends (Fig. 1). The length of the substrate was chosen to be less than twice the value of the persistence length of double-stranded DNA (150 bp). A DNA circle of this length is mainly confined to lie in a single plane, and the writhe of such a ring can be neglected (57, 62, 63).
FIG. 1.
Schematic representation of the facilitated ligation assay and possible outcomes resulting from RAG-mediated paired complex formation on RSSs located in cis or in trans. At the top is shown the linear IS95 substrate (total length, 263 bp), which has cohesive ends generated by digestion with XbaI. Synapsis in cis and trans facilitates formation of monomer circles and linear dimers, respectively, upon ligation with T4 DNA ligase. In this and subsequent figures, filled and open triangles represent the 12- and 23-RSSs, respectively.
HMG2 and coupled cleavage of IS95.
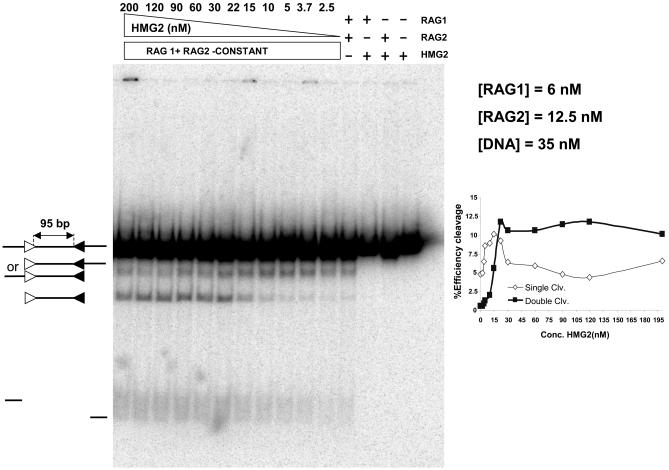
Because of its short intersignal distance, cleavage of IS95 by the RAG proteins would likely require HMG1/2 not only to facilitate RAG-RSS interactions but also to facilitate bending of the DNA substrate. To determine how cleavage of IS95 was influenced by HMG2 concentration, we performed coupled cleavage reactions in the presence of a range of concentrations of HMG2 and measured the levels of doubly and singly cleaved products. Reactions were performed using radioactively labeled IS95 substrate and constant amounts of the RAG proteins (Fig. 2). The amount of doubly cleaved product (Fig. 2, inset) increased dramatically as the concentration of HMG2 increased, reaching a plateau when the concentration of HMG2 approached that of the DNA. In contrast, single cleavage occurred at significant levels in the absence of HMG2 and first increased and then decreased as increasing amounts of HMG2 were added. The overall efficiency of cleavage increased in the range of a 5:1 to 1:1 molar ratio (DNA/HMG2). We conclude that coupled cleavage of IS95 is critically dependent on HMG2 and that a stoichiometric excess of HMG2 over DNA favors double over single cleavage.
FIG. 2.
HMG2 is essential for RAG-mediated coupled cleavage of IS95. Coupled cleavage reactions were performed in 2.5 mM Mg2+ with constant amounts of linear IS95 substrate, MBP-RAG1, and GST-RAG2, while HMG2 was added in increasing amounts in lanes 5 to 15. Lanes 1 to 4, control reactions lacking various components as indicated above the lanes; lanes 5 to 15, HMG2 at 2.5, 3.75, 5, 10, 15, 22.5, 30, 60, 90, 120, and 200 nM, respectively. (Inset) Quantitated data displayed as percent efficiency of cleavage versus HMG2 concentration (see Materials and Methods). Open symbols, cleavage at one RSS; closed symbols, cleavage at both RSSs.
HMG2 unwinds IS95.
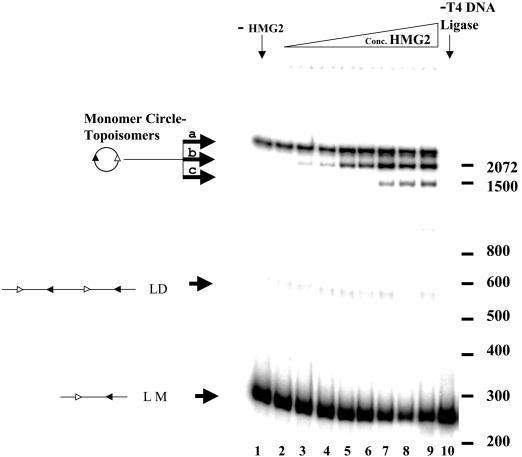
We next tested how ring closure reactions are affected by the two principal DNA binding proteins, HMG2 and RAG1, present in the reactions. While HMG proteins have previously been reported to facilitate ring closure reactions for DNA substrates smaller than the DNA persistence length, (38), these experiments had not been performed on substrates similar in size to IS95. Because HMG1/2 unwind DNA and modify the linking number of plasmids in recircularization assays (23, 24), we expected HMG2 to influence both the efficiency and the type of products of ligation of IS95. We performed ligation reactions with IS95 for 2 min in the presence of increasing concentrations of HMG2 (Fig. 3). Upon circularization, a 263-bp linear DNA molecule should generate a relaxed circle with a linking number (Lk) of 25, assuming a periodicity of 10.5 bp per turn (63). (Because these small circles do not accommodate writhe, Lk equals the twist number [Tw], and the Lk of the relaxed circle can be calculated as the integer obtained simply by dividing 263 by 10.5.) This species is seen in lane 1 (band a), where ligation was performed in the absence of HMG2. Addition of increasing concentrations of HMG2 first stimulated the formation of a higher-mobility circular topoisomer (band b, lanes 3 to 6) and then at higher concentrations the formation of a third topoisomer of even higher mobility (band c, lanes 7 to 9). Similar results were obtained when HMG1 was used (data not shown) and with TN263, a substrate identical in length to IS95 but lacking both RSSs (data not shown), demonstrating that the effect of HMG1/2 is not RSS dependent, as expected.
FIG. 3.
HMG2 induces formation of distinct topoisomeric circles in a circularization assay. The ring closure assay was performed with the IS95 substrate in the presence of increasing concentrations of HMG2. All ligation reactions were performed in the presence of 200 U of T4 DNA ligase (NEB) at 25°C for 2 min, with a DNA concentration of 3 nM and 2.5 mM Mg2+. Lane 1, no HMG2; lane 10, no T4 DNA ligase; lanes 2 to 9, HMG2 at 0.175, 0.35, 1.75, 3.5, 10.5, 21, 70, and 175 nM, respectively. LM, linear monomeric 263-bp IS95 substrate; LD, linear dimers of IS95; MC, monomeric circles. Sizes of molecular mass markers are indicated in base pairs on the right.
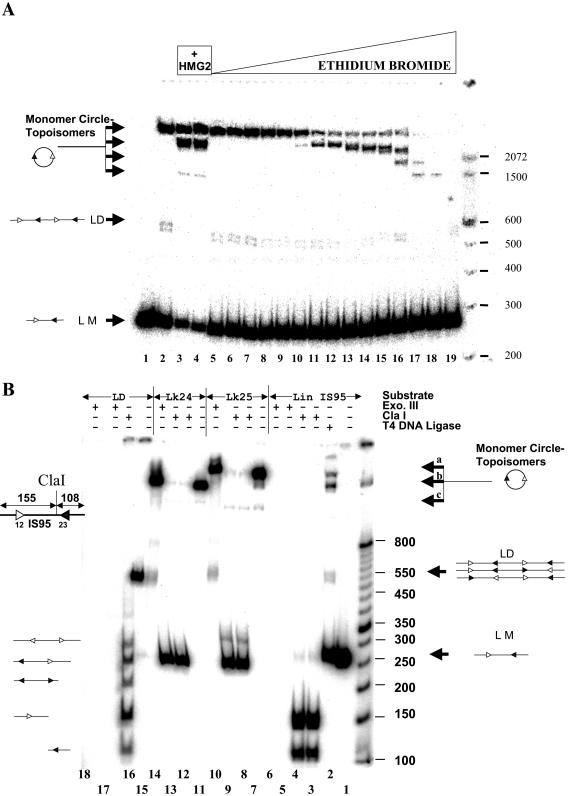
Three types of experiments were performed to determine the nature of the high-mobility topoisomers. First, we performed ligation reactions with IS95 in the presence of ethidium bromide (Fig. 4A). Ethidium bromide unwinds DNA by intercalation, and as its concentration is increased higher-mobility topoisomers abruptly appear. These topoisomers had the same mobility as those induced by HMG2, suggesting that the higher-mobility forms generated in the presence of HMG2 were underwound.
FIG. 4.
Characterization of ligation products. (A) Ligation products generated in the presence of ethidium bromide have mobilities similar to those induced by HMG2. A ring closure assay was performed using 3 nM IS95 substrate in the presence of increasing concentrations of ethidium bromide. Lane 1, no T4 DNA ligase; lane 2, T4 DNA ligase alone; lanes 3 and 4, ligation in the presence of 21 and 35 nM HMG2; lanes 5 to 19, ethidium bromide at 6.67, 13.34, 20, 26.7, 40, 66.7, 133.4, 267, 400, 533, and 677 ng/ml and 1.34, 2.67, 3.34, and 4 μg/ml, respectively. LM, linear monomeric 263-bp IS95 substrate; LD, linear dimers of IS95; MC, monomeric circles. Sizes of molecular mass markers are indicated in base pairs on the right. (B) Analysis of products generated by ligation of IS95 in the presence of HMG2. Substrates (as indicated at top) were treated withvarious enzymes as indicated above the lanes. Lane 1, linear IS95; lane 2, ligation of IS95 (3 nM) in the presence of 22.5 nM HMG2; lanes 3 and 4, 3 nM IS95 linear substrate digested with ClaI (0.5 U for 30 min [lane 3] or 1 U for 2 h [lane 4]); lanes 5 and 6, 3 nM IS95 linear substrate digested with Exo III (0.5 U for 15 min [lane 5] or 1 U for 2 h [lane 6]); lane 7, purified Lk25 topoisomer (band a); lanes 8 and 9, 3 nM Lk25 digested with ClaI as in lanes 3 and 4; lane 10, Lk25 digested with Exo III (1 U for 2 h); lane 11, purified Lk24 topoisomer (band b); lanes 12 and 13, 3 nM Lk24 digested with ClaI as in lanes 3 and 4; lane 14, Lk24 digested with Exo III (1 U for 2 h); lane 15, purified LD of IS95; lane 16, 3 nM LD digested with ClaI (1 U for 2 h); lanes 17 and 18, 3 nM LD digested with Exo III as in lanes 5 and 6.
Second, we tested the structure of the major ligation products by enzymatic digestion. We gel purified the high-mobility (band b) and low-mobility (band a) topoisomer products and the presumptive linear dimer species (LD) and subjected them to Exo III and ClaI digestion (Fig. 4B). Exo III should degrade only linear species, while digestion with ClaI (which has one asymmetric cleavage site in IS95) reveals concatamer formation. Bands a and b were Exo III resistant and linearized by ClaI, while the linear dimer was Exo III sensitive and gave the expected pattern of bands with ClaI (Fig. 4B). These results support our assignment of bands a and b as circular monomeric topoisomers that differ in mobility based upon their linking number difference.
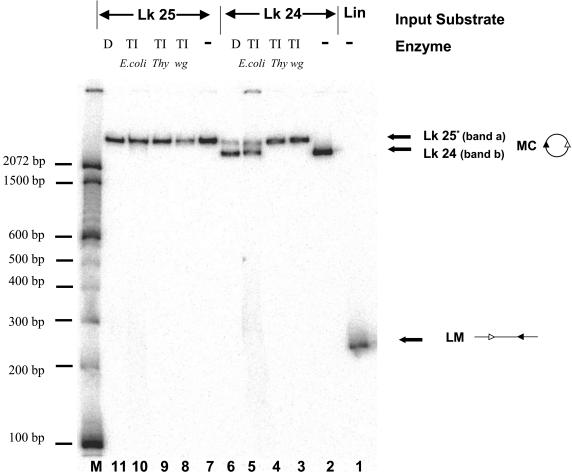
Third, we treated the purified topoisomers with topoisomerase I from different sources (Fig. 5). Treatment of the low-mobility topoisomer (band a) with either topoisomerase I or DNase I (to nick the DNA) did not change its mobility, strongly arguing that this species corresponds to the relaxed circle with linking number (Lk) of 25. Treatment with E. coli topoisomerase I, which relaxes only negatively supercoiled DNA, or with DNase I partially converted the high-mobility topoisomer (band b) into a species with the same mobility as band a. We conclude that the high-mobility topoisomer is underwound with respect to the relaxed circle, likely by one full helical turn, and has an Lk of 24. These data also demonstrate that the nicked circle and the intact relaxed circle have similar mobilities.
FIG. 5.
The fast-migrating topoisomer generated by ligation of IS95 in the presence of HMG2 is unwound by one helical turn with respect to the relaxed 263-bp circle with an Lk of 25. Lane 1, linear monomer 263-bp IS95; lanes 2 to 5, purified band b (Lk24) untreated or treated with wheat germ topoisomerase I (TI wg), calf thymus topoisomerase I (TI thy), E. coli topoisomerase I (TI E. coli), or DNase I (D), as indicated above the lanes; lanes 7 to 11, purified band a (Lk25) untreated or treated as for Lk24, as indicated above the lanes. Sizes of molecular mass markers are indicated in base pairs on the left.
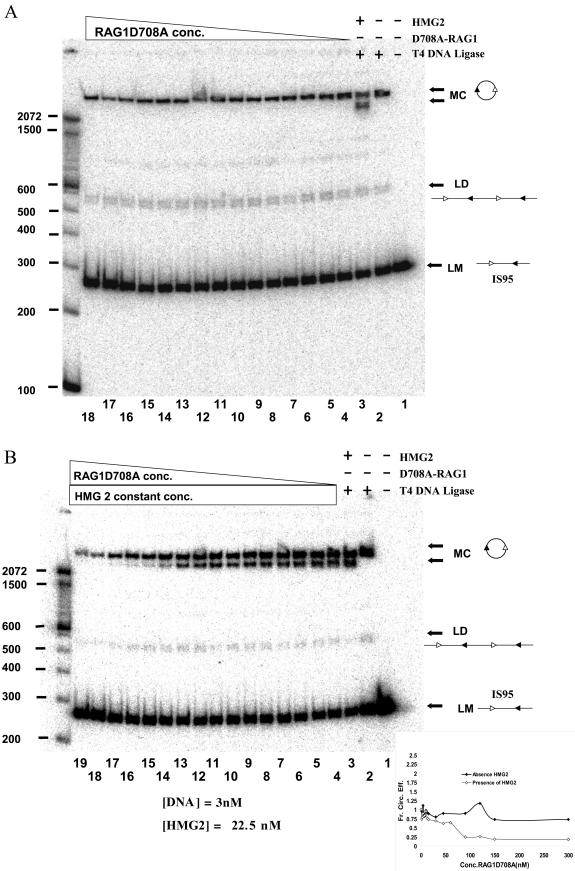
RAG1 alters the DNA binding properties of HMG2.
RAG1 binds to RSS sites either alone or in complex with RAG2 (7, 10, 43, 49, 53). It was therefore important to determine whether RAG1 (in the absence of RAG2) influenced DNA topology and the efficiency of ring closure in the absence or presence of HMG2. When increasing amounts of RAG1-D708A were added to ligation reactions performed with IS95 (Fig. 6A) or TN263 (data not shown) in the absence of HMG2, no change in the efficiency of circle formation or in circle topology was observed (a single topoisomer with an Lk of 25 was formed). When a similar experiment was performed adding RAG1-D708A to a preincubated mixture of HMG2 and either IS95 (Fig. 6B) or TN263 (data not shown), a progressive loss of the Lk24 species was observed as the concentration of RAG1-D708A increased. At a 6:1 RAG1-D708A/HMG2 molar ratio, the ability of HMG2 to stimulate formation of the Lk24 circle was abolished, and only the Lk25 product was observed (Fig. 6B, lanes 17 to 19 and inset, and data not shown). Since this effect occurs with an excess of RAG1 versus HMG2 and is independent of the presence of RSSs in the reaction, we infer that it is most likely caused by RAG1 interacting with HMG2. This interaction might prevent HMG2 from binding to DNA (under these conditions) or might alter the mode of interaction so that unwinding is no longer introduced into the DNA. The former possibility seems most likely, because addition of excess RAG1 also eliminates the enhancement of ligation induced by HMG2 (Fig. 6B) and because fluorescence polarization anisotropy experiments have indicated that excess RAG1 prevents interaction of HMG2 with DNA (unpublished data). A direct interaction between RAG1 and HMG1/2 has been demonstrated previously in pull-down experiments (1).
FIG.6.
Effect of RAG1 on the products of ligation. (A) RAG1 by itself does not influence ligation. Ring closure assays were performed in the presence of 3 nM IS95 equilibrated with increasing amounts of MBP-RAG1-D708A prior to ligation. Lanes 1 to 3, control reactions lacking various components, as indicated above the lanes; lanes 4 to 18, MBP-RAG1-D708A at 1.25, 2.5, 3.75, 5, 7.5, 10, 12.5, 15, 30, 45, 60, 90, 120, 150, and 300 nM, respectively (all lanes [4 to 18] lack HMG2). Sizes of molecular mass markers are indicated in base pairs on the left. (B) RAG1 inhibits ligation in the presence of HMG2. Ring closure assays were performed in the presence of 3 nM IS95 equilibrated with 22.5 nM HMG2 and increasing amounts of MBP-RAG1-D708A prior to ligation. Lanes 1 to 3, control reactions lacking various components, as indicated above the lanes; lanes 4 to 19, MBP-RAG1-D708A at 1.25, 2.5, 3.75, 5, 7.5, 10, 12.5, 15, 30, 45, 60, 90, 120, 150, 300, and 900 nM, respectively (all lanes [4 to 19] also contained 22.5 nM HMG2). Sizes of molecular mass markers are indicated in base pairs on the left. LM, linear monomeric 263-bp IS95 substrate; LD, linear dimers of IS95; MC, monomeric circles. The inset shows the quantitation of the data from panels A and B, represented graphically as the fraction circularization efficiency (see Materials and Methods) versus the concentration of MBP-RAG1-D708A.
Paired complex assembly with RSSs located in cis.
We next used the facilitated ligation assay to characterize RAG1/2-mediated synapsis of RSSs located in cis, investigating the effect of varying the RAG1/RAG2 ratio. Ring closure reactions were performed by preincubating labeled DNA substrate with constant amounts of HMG2 and RAG1-D708A, then adding increasing concentrations of RAG2, and subsequently adding T4 DNA ligase for 2 min. The titration experiment was performed a minimum of three times for each substrate (typically four to six times), and band intensities were quantitated using a phosphorimager. A typical experiment using the IS95 substrate is shown in Fig. 7A. An increase in circle formation was reproducibly detected when all three proteins were present and was seen most clearly at intermediate concentrations of RAG2 (compare lanes 10 to 13 with the control lane 4; note the decrease in substrate and increase in both major topoisomers). A similar experiment was performed in which increasing concentrations of RAG2 were added to ligation mixtures in the absence of RAG1-D708A; no enhancement of ligation above that obtained in the HMG2 control reaction was observed (data not shown), demonstrating that both RAG proteins are required for facilitated ligation. We also created a substrate analogous to IS95 but with a longer distance (264 bp) between the 12- and 23-RSSs. This substrate, IS264, also exhibited a significant enhancement of circularization, specifically in the presence of both RAG proteins and HMG2 (data not shown), demonstrating that the facilitated ligation assay is applicable to longer substrates that more closely resemble the spacing between certain endogenous D and J gene segments.
The quantitated band intensities from multiple experiments were used to calculate an average fraction efficiency of circularization (as defined in Materials and Methods) at each RAG2 concentration for each substrate, which was plotted against RAG2 concentration (Fig. 7B). Because the fraction efficiency of circularization is a ratio, calculated as the circularization efficiency with all three proteins present divided by the circularization efficiency in the absence of RAG2, a value of 1 indicates that no stimulation has occurred—that is, the ligation efficiency is equal to that seen with only RAG1-D708A and HMG2. The error bars in Fig. 4B represent the range spanned by the minimum and maximum values obtained at each RAG2 concentration (rather than the standard deviation, which is typically much smaller).
The maximal enhancement of circle formation with IS95 (three- to fourfold increase versus the control reaction) occurred with close-to-equimolar amounts of RAG1-D708A and RAG2 (1:1 to 1:2 molar ratios) (Fig. 7B). No enhancement of ligation was seen at any concentration of RAG2 when using the TN263 substrate that lacks RSSs (Fig. 7B). A large molar excess of RAG2 over RAG1 (6:1 to 10:1) inhibited circle formation but enhanced generation of linear dimers and trimers (ligation in trans) (Fig. 7A, lanes 16 to 19). Enhanced trans-ligation may have been due to RAG2-GST dimerization (via the GST domain) at very high protein concentrations, since addition of free GST protein inhibits this multimerization effect while causing a less substantial reduction in the efficiency of circle formation (data not shown).
To ascertain whether facilitated ligation and DNA cleavage are optimal in the same range of RAG1 and RAG2 concentrations, we performed coupled cleavage reactions in which IS95 was preincubated with constant amounts of RAG1 and HMG2, with subsequent addition of increasing amounts of RAG2. Cleavage increased substantially to near-maximal values as the RAG1/RAG2 ratio increased from 1:1 to 1:2 (representative gel in Fig. 8A; quantitation of double cleavage in B), closely paralleling the results obtained in the facilitated ligation assay. However, higher levels of RAG2 were not inhibitory for double cleavage as they were for ligation. As expected, no cleavage products were detected when TN263 was incubated with RAG1/RAG2/HMG2 (data not shown). We conclude that the cleavage and ligation assays responded comparably at low and intermediate RAG2 concentrations, consistent with the idea that facilitated ligation, like cleavage, relies on formation of the paired complex.
FIG. 8.
Effect of RAG2 concentration on coupled cleavage of RSSs located in cis. (A) Effect of increasing GST-RAG2 concentration on cleavage of linear IS95 substrate (3 nM) in reaction mixtures containing HMG2 (22.5 nM) and MBP-RAG1-D708A (6 nM). Lanes 1 to 4, control reactions lacking various components, as indicated above the lanes; lanes 5 to 19 GST-RAG2 at 1.38, 2.77, 5.55, 6.94, 8.5, 12.50, 16.6, 25, 33, 41.7, 50, 58.3, 66.7, 75, and 83.3 nM, respectively. SC, product of single cleavage at either of the RSSs; DC, double cleavage product. The asterisk indicates a band created by nicking of the substrate at both RSSs. Sizes of molecular mass markers are indicated in base pairs on the left. (B) Effect of RAG2 concentration on coupled cleavage. Quantitated data from gels such as that of panel A are represented as the percent efficiency of double cleavage (see Materials and Methods) versus GST-RAG2 concentration. Data points represent average values obtained from at least three data sets, while error bars denote the extreme maximal and minimal values obtained from individual determinations.
Ring closure and coupled cleavage assays were also performed for an inversional substrate, invIS111. invIS111 was created by inverting the 12-RSS in IS95, yielding a substrate with 111 bp between the last (heptamer-distal) nucleotides of the two nonamers. Results obtained with invIS111 broadly paralleled those with IS95: facilitated ligation (more than twofold enhancement over the control [Fig. 7B]) and coupled cleavage (Fig. 8B) were maximal, or nearly maximal, as the concentration of RAG2 equaled or slightly exceeded that of RAG1.
Facilitated ligation reactions were also performed by preincubating labeled substrates with HMG2 and RAG2 and adding increasing concentrations of RAG1-D708A. Although this resulted in enhanced circle formation over the same range of RAG1 and RAG2 concentrations as with titration of RAG2, the magnitude of the enhancement was about twofold lower than was seen in RAG2 titration experiments (data not shown). We consider that the lower level of facilitated ligation seen in the RAG1 titration experiments is a result of a different order of addition of the reagents and, for higher RAG1 concentrations, sequestration of HMG2 by the excess RAG1 (Fig. 6B).
In conclusion, facilitated circularization assays on both IS95 and invIS111 substrates were consistent with paired complex formation using 12/23 RSS pairs located in cis. The results suggest that RAG1/RAG2 ratios between 1:1 and 1:2 are optimal for paired complex formation.
Facilitated circularization occurs preferentially for 12/23 substrates.
Does synapsis in cis obey the 12/23 rule? To address this question, we determined whether facilitated circularization could be detected with substrates containing 12/12 or 23/23 RSS pairs (substrates IS95-12/12, IS95-23/23, and invIS111-12/12), as well as with substrates containing only a single RSS (substrates 263-s12 and 263-s23). All substrates were 263 bp in length, and assays were performed as described in the previous section, with the results presented in Fig. 7B. The only substrate that clearly exhibited enhanced circle formation (1.5- to 1.7-fold over the control) was IS95-12/12 (gray squares). Statistical analysis indicated that ligation of this substrate was significantly greater than that of TN263 but lower than that of IS95 (see Materials and Methods). IS95-23/23 exhibited a maximal value of 1.3, only marginally above the amount of ligation seen with this substrate in the absence of RAG2. While synapsis of the two 23-RSSs may be occurring in this substrate, our assay is not sufficiently sensitive to detect it unambiguously. None of the other control substrates tested in this assay (invIS111-12/12, 263-s12, and 263-s23) supported substantial facilitated circularization (Fig. 7B and inset). These important controls demonstrated that the presence of one RSS does not support facilitated ligation and that two identical (12/12 or 23/23) RSSs in cis yield significantly reduced levels of ligation relative to an appropriate 12/23 pair. The results strongly suggest that much of the facilitated ligation seen with the 12/23 substrates IS95 and invIS111 is due to paired complex formation rather than nonspecific interactions between RAG complexes bound to two RSSs (or one RSS and a non-RSs) on the DNA. When the concentration of RAG2 exceeded that of RAG1-D708A by sixfold or more, all of these alternate substrates, except TN263, showed an increase in linear multimer production (data not shown).
We also performed coupled cleavage assays with these control substrates (Fig. 8B). Double cleavage is three- to fourfold less efficient for IS95-12/12 and IS95-23/23 than for IS95. Curiously, double cleavage of invIS111-12/12 was below the level of detection, greatly reduced from that seen with invIS111 (Fig. 8B). Similarly, invIS111-12/12 gave no evidence of facilitated ligation, yielding results very similar to those with TN263 (Fig. 7B). Thus, in these short substrates a deletional orientation of RSSs supports detectable synapsis and double cleavage in violation of the 12/23 rule, while an inversional orientation does not (see Discussion). As expected, single RSS-bearing substrates (263-s12 and 263-s23) also did not yield detectable doubly cleaved product (Fig. 8B). These results demonstrated that, with our substrates, efficient RAG1/2-mediated coupled cleavage requires the presence of an appropriate RSS pair, and they are consistent with our conclusion that the deficit in facilitated circularization seen with substrates lacking a 12/23 RSS pair is best explained by their relative inability to support RAG-mediated synapsis.
Kinetics of RSS synapsis in cis.
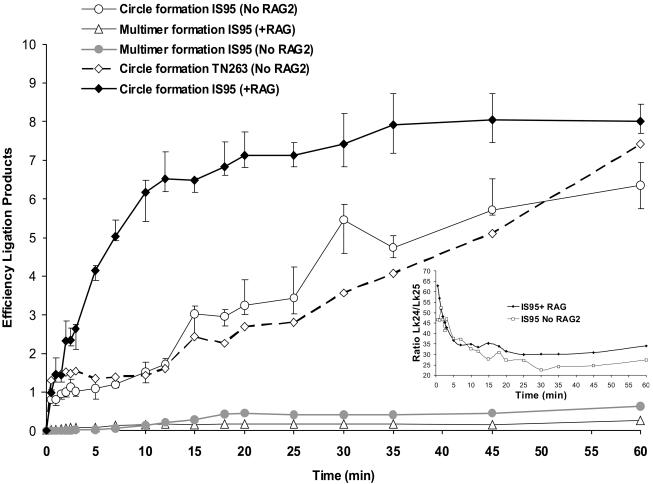
We next examined the time course of RAG1/2-facilitated circularization. IS95 was incubated with a mixture of RAG1, RAG2, HMG2, and T4 DNA ligase for times ranging from 30 s to 1 h, without prior equilibration of proteins with DNA (see Materials and Methods). For comparison, control reactions were performed in which RAG2 was omitted from the reaction mixture. The results are presented in Fig. 9. Note that in this figure, values on the y axis provide a direct measure of the amount of ligation product (ratio of intensity of the product band to that of the substrate band) rather than a relative efficiency of ligation and that data obtained from complete and control reactions are shown. For IS95, the complete reaction exhibited enhanced circularization at very early time points (as early as 1.5 min) relative to the control reaction lacking RAG2 (Fig. 9). Enhancement was maximal in the first 12 to 15 min, at which point circularization in complete reaction began to plateau, while it steadily increased in the control reaction, as expected for a reaction containing limiting amounts of the DNA substrate (3 nM). To rule out nonspecific DNA binding and synapsis by oligomers of RAG1, we measured the kinetics of circularization of TN263 in reactions containing RAG1-D708A and HMG2 but lacking RAG2 (Fig. 9). No significant differences were seen between IS95 and TN263 in reactions lacking RAG2. Multimeric linear species were only detectable at later time points and accumulated to higher levels in the control reaction lacking RAG2 than in the complete reaction (Fig. 9). While both the complete and control reactions showed a strong preference for formation of circles versus linear multimers, this preference was considerably greater for the complete reaction (maximal preference, 30- to 45-fold) than for control reactions (≈10-fold). We conclude that synaptic complex assembly, as assessed by the RAG-mediated ligation assay, occurs rapidly (within 1 min) and has a preference for RSSs located in cis at the substrate concentrations used in our experiments.
FIG. 9.
Kinetics of RAG-mediated synapsis of cis RSSs. Complete ligation reactions were performed by adding a mixture containing 200 U of T4 DNA ligase, 12.5 nM GST-RAG2, and 6 nM MBP-RAG1-D708A to a solution containing 3 nM IS95 DNA in the presence of 2.5 mM MgCl2 at 25°C, followed by quenching with 16 mM EDTA at the appropriate time intervals. For control reactions, GST-RAG2 was omitted from the reaction mixture. The main figure represents quantitation of monomer circle and linear multimer formation with time. In the case of circle formation, the efficiency of ligation expresses the ratio between the amount of circle formed by ligation versus the amount of unligated monomer, while in the case of multimers the efficiency expresses the ratio between the amount of linear dimer and trimer generated versus the amount of unligated monomer. (Inset) Ratio of underwound Lk24 to relaxed Lk25 product plotted versus time. Data points represent average values obtained from at least three data sets (except for circle formation with TN263, which represents a single data set), while error bars denote the extreme maximal and minimal values obtained from individual determinations.
The ratio between the two circular monomeric topoisomers (Lk24 and Lk25) decreased with incubation time (Fig. 9, inset). Interestingly, this ratio was higher in the presence of both RAG proteins than in control reactions, both at very early time points (1 to 2 min) and at later time points (>15 min). Hence, the presence of both RAG proteins (and the potential for paired complex formation) favors generation of the underwound circular topoisomer. This observation raises the possibility that underwinding creates a preferred substrate for RAG-mediated RSS synapsis.
DNA underwinding facilitates RAG1/2-mediated catalysis.
To determine whether DNA underwinding influences DNA cleavage by the RAG proteins, the two circular topoisomers Lk24 and Lk25 (generated by ligating IS95 in presence of HMG2) were purified and subjected to cleavage by the RAG proteins in the presence of increasing amounts of HMG2 (Fig. 10A; concentration of DNA substrate, 3 nM). The singly cleaved product was the only species generated at the lowest levels of HMG2 for both substrates (see Fig. 2 for cleavage of linear IS95; DNA concentration, 35 nM). As the concentration of HMG2 was increased, double cleavage for both circular substrates increased substantially, although to much higher levels with the underwound Lk24 substrate (Fig. 10A and inset). Cleavage of both substrates saturated at an HMG concentration of 30 nM and, importantly, even at the highest concentrations of HMG2 (a 40- to 66-fold molar excess over DNA substrate), the underwound circle was cleaved substantially more efficiently than the relaxed circle. This rules out the possibility that Lk24 cleaves more efficiently merely because it binds HMG2 better (HMG proteins are known to bind preferentially to underwound substrates [6, 48]).
We then determined the kinetics of cleavage for the two circular topoisomers and linear IS95 in the presence of a saturating concentration of HMG2 (120 nM, a 40-fold molar excess over DNA) (Fig. 10B). Double cleavage of the underwound Lk24 topoisomer was approximately fivefold greater than for either of the other two substrates at all time points examined (Fig. 10B, inset). In addition, similar results were obtained with the longer IS264 substrate (total length, 430 bp): one of its underwound circular topoisomers underwent double cleavage substantially more efficiently than the relaxed circle or the linear substrate (data not shown). Hence, the effect of underwinding was not restricted to our small 263-bp substrates. We conclude that underwinding of the DNA substrate strongly stimulates coupled cleavage by the RAG proteins. While it is possible that HMG1/2 contribute to substrate underwinding during V(D)J recombination, it is clear that DNA underwinding does not eliminate the requirement for HMG1/2 in the cleavage reaction and, hence, HMG1/2 must play other roles in facilitating coupled cleavage.
It is interesting that within the first 15 min of the reaction, well over half of the Lk24 substrate was converted to a slower-mobility form that comigrated with the relaxed circle Lk25 (Fig. 10B, lane 8). This was the result of RAG-mediated RSS nicking, as evidenced by several observations: the change in mobility was not seen with the TN263 substrate (lacking RSSs), it did not occur when catalytically inactive RAG1-D708A was used instead of RAG1 (data not shown), and the mobility change was reproduced by limited DNase I treatment of Lk24 (Fig. 5). Since nicking occurs rapidly but the underwound substrate remains a strongly preferred target for RAG-mediated double cleavage for at least another 105 min, we infer that nicking by the RAG proteins does not simply dissipate the energy of the underwound state (discussed below). Rather, the RAG proteins may remain tightly bound to the nicked RSS, as previously reported (19), thus preventing swiveling of the DNA and relaxation of the supercoiled state. Since nicking occurs more rapidly than coupled cleavage (60, 61) and can occur prior to synapsis (59), this tight binding of the RAG proteins to a nicked RSS could help maintain the negative supercoiling associated with DNA in chromatin during V(D)J recombination. This may have important implications for RSS synapsis and cleavage in vivo by the RAG proteins.
DISCUSSION
Using the facilitated ligation assay, we have been able to address several novel aspects of V(D)J recombination. First, we have been able to examine paired complex formation involving RSSs located in cis in the absence of cleavage. Second, we have been able to explore the extent to which 12/23 selectivity is imposed at the level of synapsis for cis RSSs. Third, we have addressed the role played by DNA unwinding in RAG-mediated catalysis.
Paired complex formation on complementary RSSs located in cis.
The facilitated ligation assay described here provides the first methodology with which to directly study RAG-mediated synapsis of RSSs located in cis. Using DNA cleavage as a measure of paired complex formation is not desirable, since mechanistic and kinetic considerations will almost certainly be quite different for synapsis and catalysis. Physiological V(D)J recombination occurs between RSSs located in cis, making it important to examine synapsis with this configuration of RSSs.
Several observations argue that this assay measures paired complex formation: enhanced circle formation requires the presence of two RSSs on the substrate and is significantly greater when these RSSs are an appropriate 12/23 pair; enhanced ligation is seen only in the presence of both RAG proteins and HMG2; and the optimal ratio of RAG1 to RAG2 (1:1 to 1:2) is the same for enhanced ligation and coupled cleavage.
Use of this assay has provided several insights concerning the early steps of V(D)J recombination. First, paired complex formation with cis RSSs occurs rapidly, with enhanced ligation evident within 1 to 2 min of adding the RAG1/2 and HMG2 proteins to the substrate. This agrees with a recent analysis of the kinetics of the cleavage phase of V(D)J recombination using oligonucleotide RSS substrates in trans (60, 61). One study, which measured the kinetics of DNA nicking and hairpin formation and used computer modeling to calculate rate constants, found that significant synapsis occurs on a time scale of minutes and that nicking and synapsis occur much more rapidly than hairpin formation (61). Our study, which made use of the active-site mutant D708A RAG1 protein for ligation assays, revealed the additional point that nicking is not required for rapid synapsis (in cis). Second, we have demonstrated that, for RSSs in cis, the 12/23 rule is imposed in part at the step of synapsis. And third, we have demonstrated that DNA underwinding stimulates coupled cleavage at cis RSSs. These latter two points are discussed below.
The primary limitation of the facilitated ligation assay is that of low sensitivity. While the assay clearly detects paired complex formation involving two consensus RSSs, the enhancement of ligation is relatively subtle (Fig. 7A) and detection of low-level or inefficient synapsis would likely be difficult (as may be the case for the IS95-23/23 substrate). Low sensitivity can be explained by the fact that circularization occurs robustly in the presence of HMG2 alone (Fig. 3) and, hence, if RAG-mediated synapsis of the RSSs is to be detectable as enhanced circularization, it must make an already rapid and efficient process even more efficient. The assay is therefore limited by the potential of paired complex formation to increase the effective concentration of the substrate ends relative to one another when DNA bending by HMG2 has already dramatically increased this parameter.
Deletional and inversional orientations of the RSSs.
The RAG proteins are able to perform recombination between two complementary RSSs located either in a deletional or inversional orientation. Previous studies have demonstrated that short intersignal distances inhibit both coupled cleavage (11) and V(D)J recombination (47). These studies also indicated that for inversional substrates this inhibition is manifested at longer intersignal distances than for deletional substrates, suggesting that for synapsis of closely spaced RSSs, an inversional orientation places greater strain on the intersignal DNA than does a deletional orientation. We have examined this issue using the ring closure assay with substrates that have a longer intersignal distance than those for which inhibition was observed in the studies mentioned above. In our assays, both deletional (IS95) and inversional (invIS111) orientations supported facilitated ligation (Fig. 7B) and coupled cleavage (Fig. 8B). Although the deletional substrate allowed slightly better circle formation, there was no major difference in the RAG-mediated coupled cleavage efficiencies observed for the two substrates (Fig. 8B). Therefore, with our substrates, inversional and deletional orientations support synapsis and coupled cleavage of complementary RSSs almost equally well. A difference between the two orientations is apparent, however, for substrates containing an inappropriate 12/12 pair of RSSs: the deletional 12/12 substrate supports detectable enhanced ligation and cleavage, while the comparable inversional substrate does not. If, as suggested by the discussion above, RSS synapsis of the inversional substrate engenders a greater energetic cost than for the deletional substrate, this finding may indicate a reduced stability of the paired complex formed with a 12/12 RSS pair compared with a 12/23 RSS pair. We propose that the paired complex formed with an inappropriate RSS pair is less able to accommodate the strain created by the inversional orientation than is that formed by the appropriate 12/23 pair.
The 12/23 rule is imposed in part at the level of paired complex formation.
Three factors, the RAG proteins, an appropriate divalent metal ion, and HMG1/2, have been shown to play an important role in favoring synapsis of an appropriate 12/23 pair of RSSs (11, 22, 35, 52, 56, 58). Previously, direct assessment of paired complex assembly and the 12/23 rule used oligonucleotide substrates (RSSs in trans) (22, 36, 52), while other studies indirectly assessed synapsis by measuring the double cleavage efficiency on substrates with RSSs located in cis (9, 10, 46, 58). These latter studies revealed strong adherence to the 12/23 rule and in one case were interpreted as suggesting that the 12/23 rule is imposed primarily at the step of catalysis of hairpin formation (58). In contrast, studies with trans RSSs indicated that significant enforcement of the 12/23 rule occurred at the step of synapsis (22, 25, 52). We present here evidence that RAG-facilitated circularization occurs preferentially for substrates containing a 12- and a 23-RSS (Fig. 7B, IS95 and invIS111), with less, but detectable, facilitated ligation observed for a substrate containing two 12-RSSs. Our data argue that the 12/23 rule is imposed to some extent at the step of synapsis and is fully consistent with an additional 12/23 checkpoint at the step of hairpin formation.
DNA unwinding stimulates coupled cleavage by the RAG proteins.
Ligation of a 263-bp DNA template in the presence of HMG2 leads to the formation of two predominant circular topoisomers, one with an Lk of 25 (the relaxed circle) and one underwound by one full helical turn, with an Lk of 24. When we examined the kinetics of ring closure, we found that the presence of both RAG proteins favored the formation of the Lk24 circle in the first 1 to 2 min of the reaction and also at later time points (Fig. 9, inset). Ligation of a 263-bp linear DNA at 25°C in the absence of HMG2 resulted in only the Lk25 relaxed circle (Fig. 3), indicating, as expected, that generation of the underwound Lk24 topoisomer is thermodynamically unfavorable. The finding that the Lk24/Lk25 ratio was higher for ligation in the presence of RAG1/RAG2 than for ligation in absence of RAG2 raised the possibility that the RAG proteins preferentially synapse underwound RSSs, thereby increasing the amount of Lk24 circle generated.
What might be the advantage of preferential synapsis of underwound RSSs? One attractive possibility is that this occurs in order to facilitate the catalytic step that follows. If this were the case, then coupled cleavage should occur more efficiently with an underwound substrate than with a relaxed substrate. Our data demonstrate that this is the case: underwinding enhances double cleavage at a wide range of HMG2 concentrations (Fig. 10A) and dramatically stimulates double cleavage at all time points tested in the presence of saturating HMG2 concentrations (Fig. 10B).
Substrate underwinding could be envisioned to stimulate cleavage by the RAG proteins by a variety of mechanisms. Given that nicking at one RSS is required for hairpin formation at the other, one possibility is that DNA unwinding contributes to hairpin formation by increasing the rate of nicking. This is not likely to be a major effect, because nicking occurs rapidly with relaxed (oligonucleotide) substrates and does not appear to be rate limiting for hairpin formation (60, 61). A second possibility is that underwinding facilitates the formation of a specific configuration of protein and DNA in the synaptic complex. Given the considerable evidence suggesting DNA melting near the site of cleavage (14, 16), it is reasonable to think that this configuration would include base unpairing near the hairpin scissile phosphate, as well as a (perhaps tightly coupled) conformational change in the proteins. We therefore imagine that the energy stored in the underwound substrate is used to facilitate a conformational change required for hairpin formation.
One attractive possibility, suggested by studies with the Tn10 transposase, is that DNA bending by HMG1/2 provides the needed energy in the absence of underwinding. The DNA bending protein integration host factor plays several critical roles in Tn10 transposition, acting both as an architectural component of transposase-DNA complexes and as a mediator of a DNA loop whose unfolding helps orchestrate structural changes at the site of cleavage and communication between catalytic events at the two ends of the transposon (17). Chalmers and colleagues have proposed that DNA bending by the integration host factor and by the HMG1/2 play parallel roles in Tn10 and RAG synaptic complexes, respectively, allowing the nucleoprotein complex to sense when one catalytic step (e.g., nicking) has been completed and to drive conformational changes needed for the next (e.g., hairpin formation) (8). While DNA underwinding might assist HMG1/2 in performing such a function, it does not substitute for HMG1/2. This is to be expected if HMG1/2 have other functions (e.g., architectural) in formation of the paired complex.
Our data demonstrate that Lk24 is nicked rapidly but maintains its advantage in double cleavage for a long time period thereafter (Fig. 10B), strongly suggesting that the advantage conferred by the underwound state is maintained after nicking. This could happen in at least two ways. First, because the RAG proteins remain tightly bound to nicked RSSs (19), they might prevent swiveling of the DNA and relaxation of the supercoiling (note that nicking by the RAG proteins can occur prior to synapsis). In this case, the energy of the underwound state would remain available to drive a conformational change after synapsis. Alternatively, the energy of underwinding might exert its effect (e.g., facilitating a conformational change) at the time of nicking, even if nicking occurs prior to synapsis.
It is useful to consider our findings in the context of the DNA unwinding and torsional stress that RSSs likely experience within negatively supercoiled, chromatinized DNA in lymphocytes undergoing V(D)J recombination. RAG1/2 cannot bind nucleosomal RSSs in vitro (3, 18, 28, 32), although HMG1/2 group proteins may facilitate such interactions (28). HMG1/2 group proteins have been implicated in preventing relaxation of negatively supercoiled DNA by topoisomerase I (48) and are thought to facilitate nucleosome assembly on postreplication DNA both by preserving negative DNA supercoiling and by DNA bending (20). In addition, it is known that the toroidal superhelicity associated with nucleosomal DNA can be preserved as a moderate degree of “local” negative supercoiling when short stretches of DNA are released from the nucleosomal structure (57). We speculate that the superhelical tension under which accessible RSSs are bound by the RAG proteins in partially uncondensed chromatin resembles that in the unwound Lk24 circular substrate and that this facilitates RSS synapsis and catalysis of hairpin formation by the RAG complex. It is plausible that some aspect of the architecture of the paired complex ensures that a complementary 12/23 RSS pair is best able to take advantage of such unwinding both for synapsis and subsequent hairpin formation.
In conclusion, we have developed an in vitro, facilitated ligation system to assess RAG-mediated synapsis of RSSs located in cis. Using this system, we have characterized the contributions of the RAG1, RAG2, and HMG2 proteins to circularization, revealing that paired complex formation with cis RSSs is dependent upon the presence of HMG2, requires close-to-equimolar ratios of RAG1 and RAG2, and exhibits an RSS preference in keeping with the 12/23 rule. Finally, we find that DNA unwinding facilitates RAG-mediated coupled cleavage. It will now be of interest to determine the mechanism by which substrate underwinding influences synapsis and cleavage.
Acknowledgments
We thank James C. Wang (Harvard University) for his generous gift of purified E. coli topoisomerase I. We also thank Yuk-Ching Tse-Dinh (New York Medical College) for providing us with AS 17 E. coli for expressing E. coli topoisomerase I. Discussions with Octavian Henegariu, Nigel Grindley, Sebastian Fugmann, and Chia-Lun Tsai were particularly helpful during the course of this work.
This work was supported by grant AI32524 to D.G.S. from the National Institutes of Health. M.C. is a postdoctoral associate, and D.G.S. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Aidinis, V., T. Bonaldi, M. Beltrame, S. Santagata, M. E. Bianchi, and E. Spanopoulou. 1999. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol. Cell. Biol. 19:6532-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akamatsu, Y., and M. A. Oettinger. 1998. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol. Cell. Biol. 18:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann, M., A. Mamais, F. McBlane, H. Xiao, and J. Boyes. 2003. Regulation of V(D)J recombination by nucleosome positioning at recombination signal sequences. EMBO J. 22:5197-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi, M. E., L. Falciola, S. Ferrari, and D. M. Lilley. 1992. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 11:1055-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalmers, R., A. Guhathakurta, H. Benjamin, and N. Kleckner. 1998. IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell 93:897-908. [DOI] [PubMed] [Google Scholar]
- 6.Churchill, M. E., D. N. Jones, T. Glaser, H. Hefner, M. A. Searles, and A. A. Travers. 1995. HMG-D is an architecture-specific protein that preferentially binds to DNA containing the dinucleotide TG. EMBO J. 14:1264-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciubotaru, M., L. M. Ptaszek, G. A. Baker, S. N. Baker, F. V. Bright, and D. G. Schatz. 2003. RAG1-DNA binding in V(D)J recombination. Specificity and DNA-induced conformational changes revealed by fluorescence and CD spectroscopy. J. Biol. Chem. 278:5584-5596. [DOI] [PubMed] [Google Scholar]
- 8.Crellin, P., S. Sewitz, and R. Chalmers. 2004. DNA looping and catalysis: the IHF-folded arm of Tn10 promotes conformational changes and hairpin resolution. Mol. Cell 13:537-547. [DOI] [PubMed] [Google Scholar]
- 9.Cuomo, C. A., C. L. Mundy, and M. A. Oettinger. 1996. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol. 16:5683-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Difilippantonio, M. J., C. J. McMahan, Q. M. Eastman, E. Spanopoulou, and D. G. Schatz. 1996. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell 87:253-262. [DOI] [PubMed] [Google Scholar]
- 11.Eastman, Q. M., T. M. Leu, and D. G. Schatz. 1996. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature 380:85-88. [DOI] [PubMed] [Google Scholar]
- 12.Eastman, Q. M., I. J. Villey, and D. G. Schatz. 1999. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol. Cell. Biol. 19:3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falciola, L., A. I. Murchie, D. M. Lilley, and M. Bianchi. 1994. Mutational analysis of the DNA binding domain A of chromosomal protein HMG1. Nucleic Acids Res. 22:285-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 15.Fugmann, S. D., I. J. Villey, L. M. Ptaszek, and D. G. Schatz. 2000. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell 5:97-107. [DOI] [PubMed] [Google Scholar]
- 16.Gellert, M. 2002. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71:101-132. [DOI] [PubMed] [Google Scholar]
- 17.Gellert, M., J. E. Hesse, K. Hiom, M. Melek, M. Modesti, T. T. Paull, D. A. Ramsden, and D. C. van Gent. 1999. V(D)J recombination: links to transposition and double-strand break repair. Cold Spring Harbor Symp. Quant. Biol. 64:161-167. [DOI] [PubMed] [Google Scholar]
- 18.Golding, A., S. Chandler, E. Ballestar, A. P. Wolffe, and M. S. Schlissel. 1999. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. EMBO J. 18:3712-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grawunder, U., and M. R. Lieber. 1997. A complex of RAG-1 and RAG-2 proteins persists on DNA after single-strand cleavage at V(D)J recombination signal sequences. Nucleic Acids Res. 25:1375-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosschedl, R., K. Giese, and J. Pagel. 1994. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 10:94-100. [DOI] [PubMed] [Google Scholar]
- 21.He, Q., U. M. Ohndorf, and S. J. Lippard. 2000. Intercalating residues determine the mode of HMG1 domains A and B binding to cisplatin-modified DNA. Biochemistry 39:14426-14435. [DOI] [PubMed] [Google Scholar]
- 22.Hiom, K., and M. Gellert. 1998. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell 1:1011-1019. [DOI] [PubMed] [Google Scholar]
- 23.Javaherian, K., J. F. Liu, and J. C. Wang. 1978. Nonhistone proteins HMG1 and HMG2 change the DNA helical structure. Science 199:1345-1346. [DOI] [PubMed] [Google Scholar]
- 24.Javaherian, K., M. Sadeghi, and L. F. Liu. 1979. Nonhistone proteins HMG1 and HMG2 unwind DNA double helix. Nucleic Acids Res. 6:3569-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, J. M., and M. Gellert. 2002. Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J. 21:4162-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, D. R., Y. Dai, C. L. Mundy, W. Yang, and M. A. Oettinger. 1999. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 13:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klass, J., F. V. T. Murphy, S. Fouts, M. Serenil, A. Changela, J. Siple, and M. E. Churchill. 2003. The role of intercalating residues in chromosomal high-mobility-group protein DNA binding, bending and specificity. Nucleic Acids Res. 31:2852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon, J., A. N. Imbalzano, A. Matthews, and M. A. Oettinger. 1998. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol. Cell 2:829-839. [DOI] [PubMed] [Google Scholar]
- 29.Landree, M. A., J. A. Wibbenmeyer, and D. B. Roth. 1999. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 13:3059-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landsman, D., and M. Bustin. 1993. A signature for the HMG-1 box DNA-binding proteins. Bioessays 15:539-546. [DOI] [PubMed] [Google Scholar]
- 31.Lieber, M. R., J. E. Hesse, S. Lewis, G. C. Bosma, N. Rosenberg, K. Mizuuchi, M. J. Bosma, and M. Gellert. 1988. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell 55:7-16. [DOI] [PubMed] [Google Scholar]
- 32.McBlane, F., and J. Boyes. 2000. Stimulation of V(D)J recombination by histone acetylation. Curr. Biol. 10:483-486. [DOI] [PubMed] [Google Scholar]
- 33.Mikaelian, I., and A. Sergeant. 1992. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 20:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mo, X., T. Bailin, S. Noggle, and M. J. Sadofsky. 2000. A highly ordered structure in V(D)J recombination cleavage complexes is facilitated by HMG1. Nucleic Acids Res. 28:1228-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mundy, C. L., N. Patenge, A. G. Matthews, and M. A. Oettinger. 2002. Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol. 22:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagawa, F., M. Kodama, T. Nishihara, K. Ishiguro, and H. Sakano. 2002. Footprint analysis of recombination signal sequences in the 12/23 synaptic complex of V(D)J recombination. Mol. Cell. Biol. 22:7217-7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peak, M. M., J. L. Arbuckle, and K. K. Rodgers. 2003. The central domain of core RAG1 preferentially recognizes single-stranded recombination signal sequence heptamer. J. Biol. Chem. 278:18235-18240. [DOI] [PubMed] [Google Scholar]
- 38.Pil, P. M., C. S. Chow, and S. J. Lippard. 1993. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc. Natl. Acad. Sci. USA 90:9465-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pil, P. M., and S. J. Lippard. 1992. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science 256:234-237. [DOI] [PubMed] [Google Scholar]
- 40.Pohler, J. R. G., D. G. Norman, J. Bramham, M. E. Bianchi, and D. M. Lilley. 1998. HMG box proteins bind to four-way DNA junctions in their open conformation. EMBO J. 17:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramsden, D. A., K. Baetz, and G. E. Wu. 1994. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 22:1785-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramsden, D. A., J. F. McBlane, D. C. van Gent, and M. Gellert. 1996. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 15:3197-3206. [PMC free article] [PubMed] [Google Scholar]
- 43.Rodgers, K. K., I. J. Villey, L. Ptaszek, E. Corbett, D. G. Schatz, and J. E. Coleman. 1999. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 27:2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santagata, S., V. Aidinis, and E. Spanopoulou. 1998. The effect of Me2+ cofactors at the initial stages of V(D)J recombination. J. Biol. Chem. 273:16325-16331. [DOI] [PubMed] [Google Scholar]
- 45.Santagata, S., E. Besmer, A. Villa, F. Bozzi, J. S. Allingham, C. Sobacchi, D. B. Haniford, P. Vezzoni, M. C. Nussenzweig, Z. Q. Pan, and P. Cortes. 1999. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol. Cell 4:935-947. [DOI] [PubMed] [Google Scholar]
- 46.Sawchuk, D. J., F. Weis-Garcia, S. Malik, E. Besmer, M. Bustin, M. C. Nussenzweig, and P. Cortes. 1997. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J. Exp. Med. 185:2025-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheehan, K. M., and M. R. Lieber. 1993. V(D)J recombination: signal and coding joint resolution are uncoupled and depend on parallel synapsis of the sites. Mol. Cell. Biol. 13:1363-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheflin, L. G., and S. W. Spaulding. 1989. High mobility group protein 1 preferentially conserves torsion in negatively supercoiled DNA. Biochemistry 28:5658-5664. [DOI] [PubMed] [Google Scholar]
- 49.Spanopoulou, E., F. Zaitseva, F. H. Wang, S. Santagata, D. Baltimore, and G. Panayotou. 1996. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell 87:263-276. [DOI] [PubMed] [Google Scholar]
- 50.Stros, M., and E. Muselikova. 2000. A role of basic residues and the putative intercalating phenylalanine of the HMG-1 box B in DNA supercoiling and binding to four-way DNA junctions. J. Biol. Chem. 275:35699-35707. [DOI] [PubMed] [Google Scholar]
- 51.Swanson, P. C. 2002. Fine structure and activity of discrete RAG-HMG complexes on V(D)J recombination signals. Mol. Cell. Biol. 22:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swanson, P. C. 2002. A RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol. Cell. Biol. 22:7790-7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swanson, P. C., and S. Desiderio. 1999. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 19:3674-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swanson, P. C., and S. Desiderio. 1998. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity 9:115-125. [DOI] [PubMed] [Google Scholar]
- 55.Tonegawa, S. 1983. Somatic generation of antibody diversity. Nature 302:575-581. [DOI] [PubMed] [Google Scholar]
- 56.van Gent, D. C., D. A. Ramsden, and M. Gellert. 1996. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell 85:107-113. [DOI] [PubMed] [Google Scholar]
- 57.Vologodskii, A. 1992. Topology and physics of circular DNA. CRC Press, Boca Raton, Fla.
- 58.West, R. B., and M. R. Lieber. 1998. The RAG-HMG1 complex enforces the 12/23 rule of V(D)J recombination specifically at the double-hairpin formation step. Mol. Cell. Biol. 18:6408-6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu, K., and M. R. Lieber. 1999. Mechanistic basis for coding end sequence effects in the initiation of V(D)J recombination. Mol. Cell. Biol. 19:8094-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, K., and M. R. Lieber. 2000. The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol. Cell. Biol. 20:7914-7921.11027262 [Google Scholar]
- 61.Yu, K., A. Taghva, Y. Ma, and M. R. Lieber. 2004. Kinetic analysis of the nicking and hairpin formation steps in V(D)J recombination. DNA Repair 3:67-75. [DOI] [PubMed] [Google Scholar]
- 62.Zeman, S. M., and D. M. Crothers. 2001. Simultaneous measurement of binding constants and unwinding angles by gel electrophoresis. Methods Enzymol. 340:51-68. [DOI] [PubMed] [Google Scholar]
- 63.Zeman, S. M., K. M. Depew, S. J. Danishefsky, and D. M. Crothers. 1998. Simultaneous determination of helical unwinding angles and intrinsic association constants in ligand-DNA complexes: the interaction between DNA and calichearubicin B. Proc. Natl. Acad. Sci. USA 95:4327-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]