Abstract
Background & Aims
Like all positive-sense RNA viruses, hepatitis C virus (HCV) induces host membrane alterations for its replication. In chronically infected cells, it is not known whether these viral replication organelles are being continually resupplied by newly synthesized viral proteins in situ, or whether they are generated de novo. Here we aimed to study temporal events in replication organelles formation and maturation.
Methods
Here we use pulse-chase labeling in combination with confocal microscopy, correlative-light electron microscopy and biochemical methods to identify temporally distinct populations of replication organelles in living cells and study the formation, morphogenesis as well as compositional and functional changes of replication organelles over time.
Results
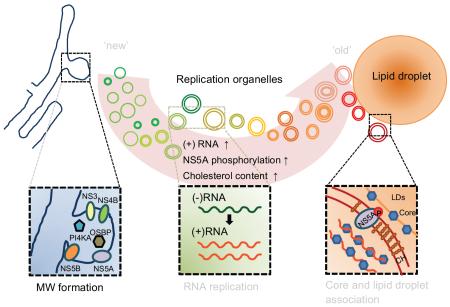
We found that HCV replication organelles are continuously generated de novo at spatially distinct sites from preformed ones. This process is accompanied by accumulated intracellular membrane alteration, increased cholesterol delivery, NS5A phosphorylation, and positive strand RNA content, and by eventual association with HCV core protein around LDs. Generation of spatially segregated foci requires viral NS5A and the host factors phosphatidylinositol 4-kinase PI4KA and oxysterol-binding protein, while association of foci with LDs requires cholesterol.
Conclusions
Thus, our results reveal that HCV replication organelles are not static structures, but instead are continuously generated and dynamically change in composition and possibly also in function.
Keywords: hepatitis C virus, viral replication, double membrane vesicles, SNAP, cholesterol
Graphical abstract
Introduction
Hepatitis C virus (HCV) is a prevalent and globally distributed human pathogen. More than 170 million people are chronically infected, of whom many will develop cirrhosis and/or hepatocellular carcinoma. The positive-sense RNA genome encodes a single polyprotein that is processed by cellular and viral proteases to generate 10 mature proteins [1], of which the NS3-5B region is sufficient to sustain viral RNA replication [2].
Like all positive-sense RNA viruses, HCV induces cytoplasmic membrane alterations in infected cells [3], which have been termed ‘replication organelles’ and are believed to be sites of HCV RNA synthesis. The membranes that form the HCV replication organelle are likely derived from the cellular endoplasmic reticulum (ER) and enriched in viral nonstructural proteins and HCV RNA [4, 5]. At the ultrastructural level, HCV replication organelles are characterized mostly by double-membrane vesicles (DMVs) and also by multi-membrane vesicles (MMVs) [6]. Recent studies have indicated that DMVs are the likely sites of HCV RNA replication and that NS5A is essential for their formation [6, 7]. This function of NS5A may be partially attributable to its ability to interact with various host factors that are essential for viral RNA replication. Among them are the lipid kinase phosphatidylinositol 4-kinase IIIα (PI4KA) [8-10] and its downstream effector, oxysterol-binding protein (OSBP) [11, 12]. These two proteins generate the unique lipid composition of the HCV replication organelle, which is enriched in phosphatidylinositol 4-phosphate (PI4P) [10, 13] and cholesterol [7, 14-16].
While progress has been made in defining early events of HCV replication organelle formation in acutely infected cells [6, 17], there remain many questions regarding the temporal regulation of its function and composition, as well as the turnover of HCV replication organelles in chronically infected cells. For instance, are replication organelles relatively static structures that are continually resupplied by viral proteins, or are new ones continually generated de novo? Furthermore, while HCV RNA synthesis occurs at the replication organelle, virion assembly is believed to occur at or near lipid droplets (LDs) [18]; how are HCV genomes destined for encapsidation physically transferred between these sites?
In this study, we used a pulse-chase fluorescent labeling approach that allows us to discriminate ‘old’ from ‘new’ NS5A-positive membranous structures (NS5A foci). ‘New’ NS5A foci form at sites distinct from ‘old’ NS5A foci even at short chase intervals, supporting a model of continuous de novo replication organelle formation instead of resupply of previously-formed ones. PI4KA, OSBP, and NS5A are required to initiate new NS5A foci. Cholesterol is preferentially trafficked to ‘old’ NS5A foci and is required for association of ‘old’ foci with HCV core protein and LDs. These findings collectively describe a process of continuous HCV replication organelle generation accompanied by alteration of lipid content and progressive association with sites of putative virion assembly that is dependent on cholesterol content.
Materials and Methods
Viruses and viral replicons
A sequence encoding the SNAPf tag [19] was inserted into domain III of NS5A in the context of the a full length HCV genome and a subgenomic replicon based on the genotype 2a JFH1 strain, termed here as FL-JFH1(NS5A/SNAP) and SGR-JFH1(NS5A/SNAP) respectively. Detailed information about these two constructs is described in the Supplementary Material.
Labeling of SNAP-tagged NS5A with fluorescent SNAP-tag substrates
Labeling of SNAP-tagged NS5A with SNAP-tag substrates (SNAP-Cell 505, SNAP-Cell TMR-Star, and SNAP Cell-Block; New England Biolabs, Ipswich, MA), was performed according to the manufacturer’s instructions. Briefly, cells were treated with complete medium containing 5μM SNAP-Cell 505, 3μM SNAP-Cell TMR-Star or 10μM SNAP Cell-Block for 15 min before they were washed out and replaced with fresh medium.
Correlative light-electron microscopy
SGR-JFH1(NS5A/SNAP) replicon cells seeded onto glass bottom dishes with gridded coverslips (MatTek, Ashland, MA) were labeled with STMR and S505 and then evaluated by confocal microscopy to identify cells of interest; their positions were recorded and DIC/confocal fluorescent images were acquired. Cells were immediately fixed and processed for EM sectioning. The sections were viewed on a JEOL JEM-1400 Plus transmission electron microscope at 80 kV. Further details are provided in Supplementary Material.
Quantitation of NS5A phosphorylation and negative: positive strand RNA ratios
SGR replicon cells were first labeled with Sblock and STMR to selectively label ‘old’ or ‘new’ NS5A before they were washed once with ice-cold PBS, lysed with 100 μM digitonin in PBS containing protease inhibitors, phosphatase inhibitors and RNase inhibitors, and centrifuged for 5 minutes at 12,000 × g. TMR-labeled NS5A-SNAP and associated RNA were then isolated by incubating the cell lysate with anti-TMR antibody for 1.5 hr at 4°C followed with Dynabeads Protein G (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. Immunoisolated material was then subjected to immunoblotting with chemiluminescence quantitation on an Odyssey imager (Li-Cor, Lincoln, NE) or to strand-specific qRT-PCR as previously described [20], and is described further in the Supplementary Material.
Statistics
The unpaired two-tailed Student’s t test was used to compare the means of control and experimental groups.
Results
Characterization of a system for pulse-chase imaging of HCV NS5A as a marker of the HCV replication organelle
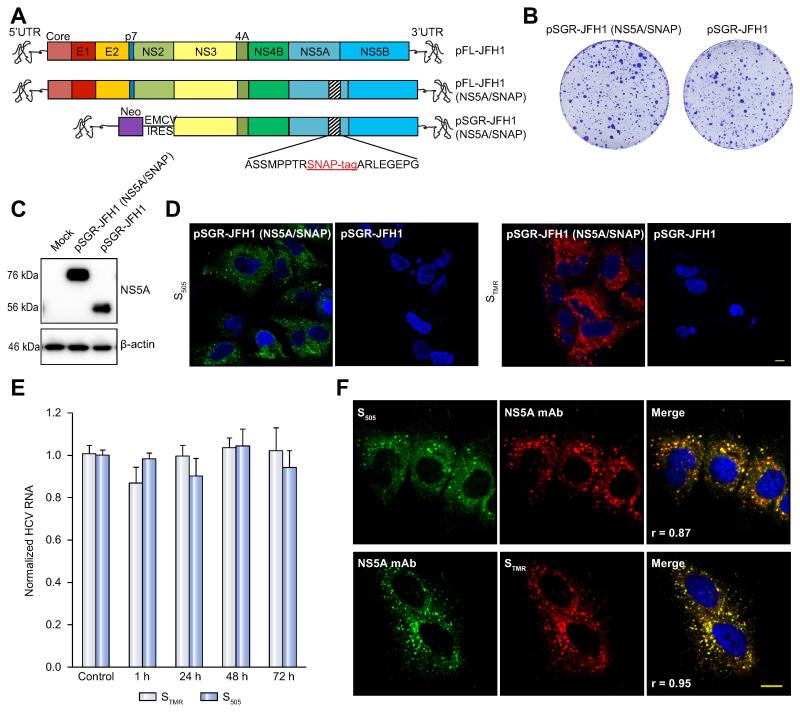
In order to study temporal aspects of HCV replication organelle formation and function, we genetically inserted a SNAP-tag into a known insertion-tolerant site within domain III of NS5A [21] as a marker of HCV replication organelles (Figure 1A). NS5A and tagged NS5A proteins have been widely used as an HCV replication organelle marker [6, 8-10, 21-23]. The SNAP-tag specifically, rapidly, and irreversibly forms a covalent bond with benzylguanine derivatives [24]. A colony formation assay showed that insertion of the SNAP tag into NS5A had minimal effects on replication compared to an untagged replicon (Figure 1B), and this was further confirmed by measuring the luciferase activity of replicons encoding a Renilla luciferase reporter or intracellular HCV RNA content in cells stably expressing replicons (Supplementary Figures 1A-C). Finally, NS5A-SNAP migrated as expected on SDS-PAGE (Figure 1C).
Figure 1. Development and characterization of SNAP tagged HCV genomes for live cell pulse-chase imaging.
(A). Schematic of viral constructs. A SNAP tag was inserted into a known tolerated insertion site within domain III of NS5A in the JFH1 strain to create plasmids encoding full-length FL-JFH1(NS5A/SNAP) and the subgenomic replicon SGR-JFH1(NS5A/SNAP).
(B). Colony formation assay of JFH-1 subgenomic replicons in Huh7.5.1 cells. Huh7.5.1 cells were transfected with in vitro transcribed RNA encoding the indicated constructs and G418-resistant colonies were visualized by Crystal Violet staining 21 days post-transfection.
(C). Immunoblot analysis of NS5A protein expression from cell lysates prepared from the indicated subgenomic replicon cells. β-actin is shown as a loading control.
(D). Living cells expressing the indicated subgenomic replicons with or without the SNAP tag were labeled with 5μM green fluorescent SNAP-Cell 505 (S505) or 3μM red fluorescent SNAP-Cell TMR-Star (STMR) for 15min. Nuclei were counterstained with DAPI. Scale bar, 10μm.
(E). SGR-JFH1(NS5A/SNAP) subgenomic replicon cells were either mock treated or stained with the indicated labeling reagent as in panel D and then incubated in fresh medium for the indicated times. HCV RNA was quantitated by qRT-PCR. Values are means ± SEM of three independent experiments and normalized to control.
(F). Specificity of SNAP labeling for NS5A protein. SGR-JFH1(NS5A/SNAP) subgenomic replicon cells were labeled with either STMR or S505 as above before fixing and immunostaining for NS5A. Scale bar, 10 μm.
Living NS5A-SNAP replicon cells were incubated with the green fluorescent cell-permeable SNAP-tag substrate SNAP-Cell 505 (S505) or the red fluorescent substrate SNAP-Cell TMR-star (STMR). Brightly labeled puncta were observed in cells stably expressing the NS5A-SNAP replicon but not in untagged replicon cells (Figure 1D). No decrease in HCV RNA content (Figure 1E) or reporter luciferase activity (Supplementary Figure 1D) was seen in cells labeled with either STMR or S505 and incubated for up to an additional 72 hr In addition, labeling with STMR did not change the half-life of NS5A-SNAP protein (Supplementary Figure 1E). There was near-complete overlap between NS5A immunostaining and NS5A-SNAP labeling by either S505 (Pearson’s correlation coefficient 0.87) or STMR (Pearson’s correlation coefficient 0.95) (Figure 1F).
Pulse-chase imaging of HCV replication organelle formation
We then tested sequential labeling of NS5A-SNAP, first with S505 to label pre-existing (‘old’) NS5A, followed by STMR to label newly synthesized (‘new’) NS5A after a chase period. Newly synthesized protein could be detected with STMR, but not when protein synthesis had been halted by cycloheximide treatment during the chase period (Supplementary Figure 1F), demonstrating specific labeling of newly synthesized protein. NS5A-SNAP labeling was efficiently blocked by the non-fluorescent compound SNAP-Cell Block (Sblock, Supplementary Figure 1G), and Sblock also did not inhibit HCV replication (Supplementary Figure 1H).
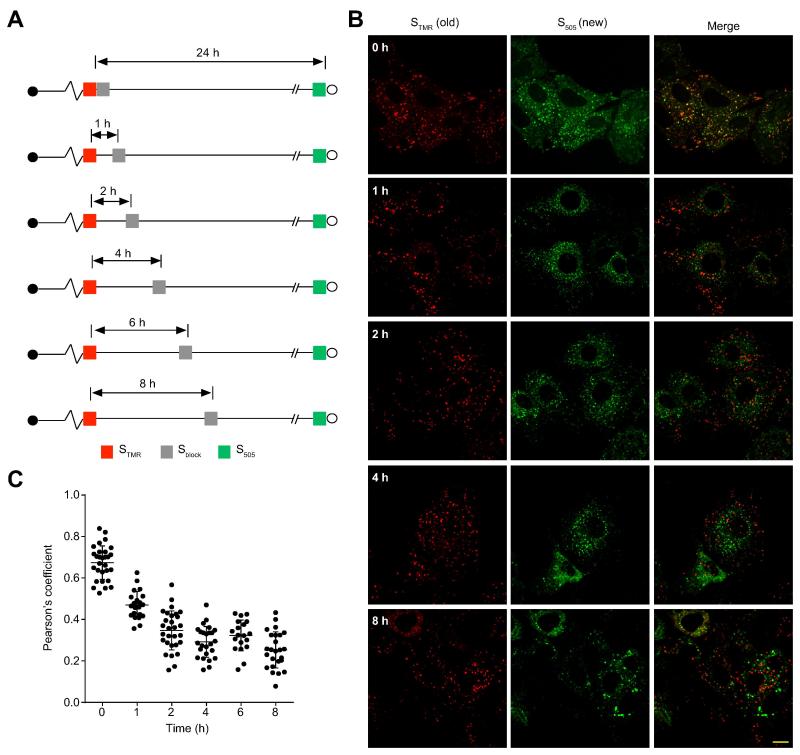
Having confirmed that this system is suitable for pulse-chase imaging in live cells, we tested whether replication organelles are stable structures that are resupplied by newly synthesized NS5A, or whether newly synthesized NS5A is directed to new replication organelles formed de novo. This can be determined by quantitating the colocalization between ‘old’ and ‘new’ NS5A molecules at varying chase times, as depicted in Figure 2A. Supplementary Figure 2 depicts the two models and their predictions regarding ‘old’ and ‘new’ NS5A colocalization with increasing chase times. ‘Old’ and ‘new’ NS5A signals displayed significant overlap only when Sblock was applied immediately after STMR labeling, while ‘new’ NS5A synthesized as little as 1 hr after ‘old’ NS5A labeling was found at foci distinct from ‘old’ NS5A foci (Figures 2B and C). The rapid loss of ‘old’ and ‘new’ NS5A colocalization was more consistent with newly synthesized NS5A being directed to de novo replication organelles formation rather than to resupplying previously formed ones. To exclude the possibility that the decrease in colocalization at later time points was due to variability in ‘new’ NS5A labeling duration, we fixed the ‘new’ NS5A labeling duration at 24 hr and obtained similar results (Supplementary Figure 3).
Figure 2. Pulse-chase imaging of SNAP-tagged NS5A.
(A). Schematic of experimental design. Parallel cultures of SGR-JFH1(NS5A/SNAP) subgenomic replicon cells were pulse labeled with STMR (red) and then treated with SNAP Cell-Block (Sblock) at the indicated times. Newly synthesized NS5A was labeled with S505 (green).
(B). Representative images of live cells labeled as described above. Scale bar, 10 μm.
(C). Quantitation of colocalization between STMR labeled NS5A and S505 labeled NS5A. Each point denotes the Pearson’s coefficient calculated from a single cell. Pearson’s coefficients greater than 0.5 are considered to be indicative of colocalization. Summary mean±SD values from 3 independent experiments are indicated for each timepoint.
Newly synthesized NS5A foci contain other components of the HCV replication complex
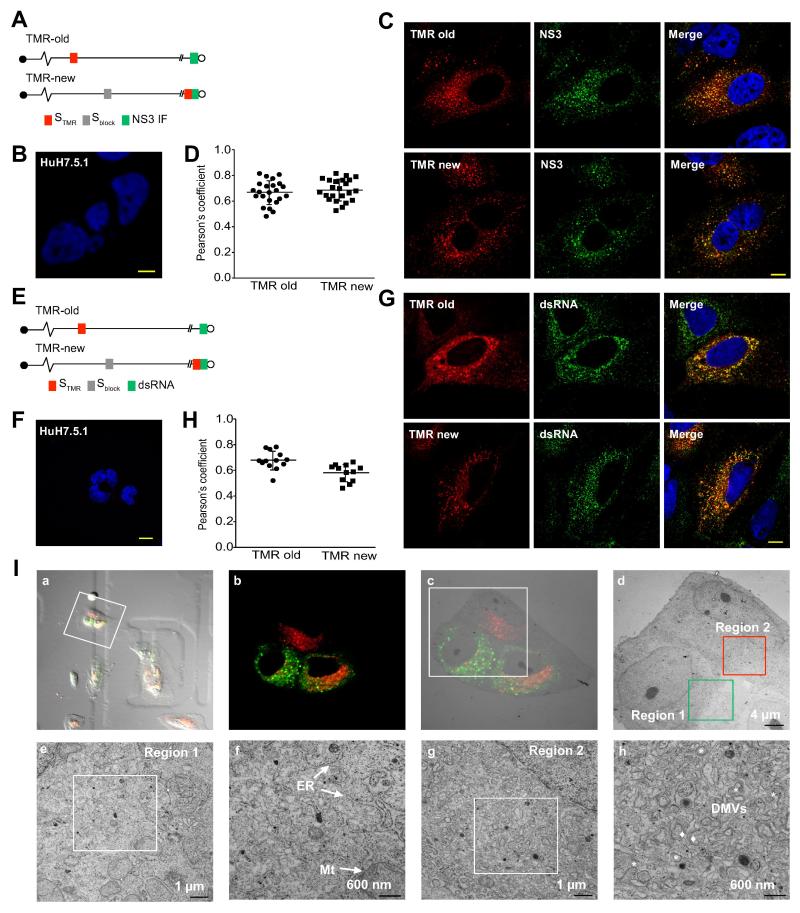
To determine whether NS5A-SNAP containing structures contain other components of the HCV replication complex, we selectively labeled cells for ‘old’ or ‘new’ NS5A followed by immunostaining for NS3 (Figure 3A). NS3 colocalized with both ‘old’ and ‘new’ NS5A foci (Figure 3C and D). In addition, HCV replication is believed to generate a double-stranded RNA (dsRNA) replicative intermediate, and anti-dsRNA antibodies have been used to visualize putative HCV replication complexes [25]. No dsRNA staining was detected in uninfected Huh 7.5.1 cells (Figure 3F), in contrast, dsRNA immunoreactivity colocalized with both ‘old’ NS5A and ‘new’ NS5A foci (Figures 3G and H), consistent with the presence of dsRNA replicative intermediates at both ‘new’ and ‘old’ NS5A foci. Unfortunately, we have not been able to reliably label newly synthesized HCV RNA using ribonucleoside analogs (not shown). We conclude that at least a subset of ‘new’ and ‘old’ NS5A foci contains HCV replicase components and a dsRNA intermediate, which suggests that they are indeed replication organelles.
Figure 3. Both ‘old’ and ‘new’ NS5A foci contain other components of the HCV replication complex.
(A). Schematic of experimental design. ‘Old’ NS5A in SGR-JFH1(NS5A/SNAP) subgenomic replicon cells was visualized with STMR labeling followed by a 24 hr chase (top), while ‘new’ NS5A was visualized by first blocking unlabeled NS5A by nonfluorescent Sblock followed by a 16 hr chase and labeling with STMR (bottom). Cells were then fixed and immunostained for NS3.
(B). Huh7.5.1 cells were fixed and immunostatined for NS3. Nuclei counterstained with DAPI. Scale bar, 10 μm.
(C). Representative images of cells labeled as described above. Scale bar, 10 μm.
(D). Quantitation of colocalization between STMR-labeled NS5A and NS3. Each point denotes the Pearson’s coefficient calculated from a single cell. Summary mean±SD values are indicated.
(E). Schematic of experimental design. Cells infected with JFH1(NS5A-SNAP) full length virus was labeled for “old” (top) and “new” (bottom) NS5A as in panels A. Cells were then fixed and immunostained for dsRNA.
(F). Huh7.5.1 cells were fixed and immunostatined for dsRNA. Nuclei counterstained with DAPI. Scale bar, 10 μm.
(G). Representative images of cells labeled as described above. Scale bar, 10 μm.
(H). Quantitation of colocalization between STMR-labeled NS5A and dsRNA. Each point denotes the Pearson’s coefficient calculated from a single cell. Summary mean±SD values are indicated.
(I). Correlative light-electron microscopy of SGR-JFH1(NS5A/SNAP) subgenomic replicon cells. Live replicon cells were stained with STMR (for old foci) and S505 (for new foci) before acquisition of fluorescence and DIC images followed by immediate processing for EM. (a) Merged DIC/epifluorescence image of live cells on gridded coverslips. (b) Confocal fluorescent image of cells of interest. (c) Merged EM and fluorescence microscopy images. (d) Low magnification electron micrograph depicting regions of interest containing predominantly ‘new’ NS5A foci (region 1) or ‘old’ NS5A foci (region 2). (e-f) higher magnification images of region 1 in panel d. (g-h) higher magnification images of region 2 in panel d. ER, endoplasmic reticulum; Mt, mitochondria; DMVs, double membrane vesicles. Asterisks indicate DMVs; diamonds indicate multimembrane vesicles (MMVs).
‘Old’ NS5A foci are associated with more prominent membrane rearrangements than ‘new’ NS5A foci
The time course of intracellular membrane rearrangements following acute HCV infection has been described [6]. We carried out correlative light-electron microscopy (CLEM) to characterize the ultrastructure of ‘old’ and ‘new’ NS5A foci in cells stably replicating HCV. Live cells were first labeled for ‘old’ and ‘new’ NS5A foci (Figure 3I, panel a). Cells were imaged by fluorescence microscopy (Figure 3I, panel b) and then immediately processed for EM with light/EM image correlation (Figure 3I, panel c). Prominent membrane rearrangements including DMVs and MMVs were readily detected in a region containing many ‘old’ NS5A foci (Figure 3I, panels d, g and h; Supplementary Figure 4). In contrast, in a region containing many ‘new’ NS5A foci, subjectively far fewer DMVs and no MMVs could be identified (Figure 3I, panels d, e and f; Supplementary Figure 4). Quantitative analysis could not be performed on the low numbers of cells that could be processed for CLEM. These results are consistent with the model that HCV replication organelle morphogenesis is a dynamic process in chronic HCV replication.
OSBP, PI4KA, and NS5A are required for new NS5A focus formation
The cellular proteins PI4KA and OSBP are essential for the integrity of HCV replication organelles; inhibiting either PI4KA or OSBP leads to the formation of large clusters of small DMVs [8, 10, 12]. Two questions that can be addressed by pulse-chase imaging are whether these proteins are required for the formation of new NS5A foci, and whether they are required to maintain the normal morphology of pre-existing foci. To this end, we examined the morphology and distribution of ‘old’ and ‘new’ NS5A foci upon treatment with inhibitors of PI4KA [26] and OSBP [27], as depicted in Figure 4A. In vehicle-treated cells, newly synthesized NS5A was found in small foci separated from old NS5A foci, as expected (Figure 4B, upper panels and Figure 4C). Inhibition of either PI4KA or OSBP during the chase time led to the appearance of both ‘old’ and ‘new’ NS5A in clusters (Figure 4B), suggesting that PI4KA and OSBP are required to maintain the integrity of preformed replication organelles. In addition, PI4KA or OSBP inhibition blocked de novo NS5A focus formation, indicating a requirement for these proteins in this process, and was also associated with ongoing NS5A synthesis at ‘old’ NS5A foci (Figures 4B and C), suggesting that PI4KA and OSBP are somehow required for termination of NS5A synthesis at ‘old’ NS5A foci.
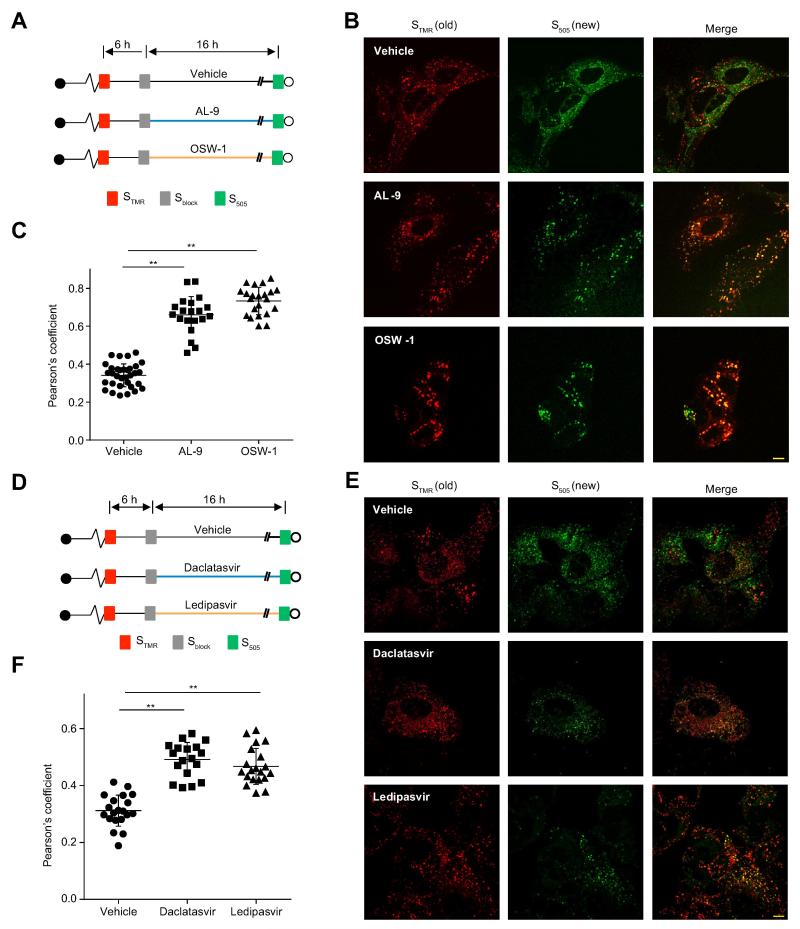
Figure 4. PI4KA, OSBP and NS5A are required to initiate new NS5A foci.
(A). Schematic of experimental design. SGR-JFH1 (NS5A/SNAP) subgenomic replicon cells were pulse labeled with STMR and treated with Sblock 6 hr later. Cells were then incubated with vehicle, AL-9 (4 μM), or OSW-1 (30 nM) for another 16 hr before newly synthesized NS5A was labeled with S505.
(B). Representative live-cell images of cells labeled as described above. Scale bar, 10μm.
(C). Quantitation of colocalization between STMR-labeled (old) and S505-labeled (new) NS5A. Each point denotes the Pearson’s coefficient calculated from a single cell. Summary mean±SD values representative of 3 independent experiments are indicated. ** p<0.001 versus vehicle.
(D). Schematic of experimental design. SGR-JFH1 (NS5A/SNAP) subgenomic replicon cells were pulse labeled with STMR and treated with Sblock 6 hr later. Cells were then incubated with vehicle, daclatasvir (100pM), or ledipasvir (100 nM) for another 16 hr before newly synthesized NS5A was labeled with S505.
(E). Representative live-cell images of cells labeled as described above. Scale bar, 10 μm.
(F). Quantitation of colocalization between STMR-labeled (old) and S505-labeled (new) NS5A. Each point denotes the Pearson’s coefficient calculated from a single cell. Summary mean±SD values representative of 3 independent experiments are indicated. ** p<0.001 versus vehicle.
NS5A protein is essential for the formation of DMVs [6] and NS5A inhibitors block replication organelle formation [28]. However, although NS5A is essential for viral replication, NS5A inhibitors block HCV RNA replication with delayed kinetics compared to viral NS5B polymerase inhibitors [29], suggesting that NS5A inhibitors do not block the function of preexisting replication organelles. Here we assessed the effect of NS5A inhibitors on the formation of new replication organelles. New and old NS5A foci were labeled as described above and were either mock treated or treated with the FDA-approved NS5A inhibitors daclatasvir or ledipasvir (Figure 4D). Compared to vehicle-treated cells, NS5A inhibitors impeded de novo NS5A focus formation with a corresponding increase in colocalization between ‘old’ and ‘new’ NS5A proteins (Figure 4E and F).
To test whether reduced de novo HCV replication organelle formation caused by NS5A, PI4KA or OSBP inhibition was merely a nonspecific effect of inhibition of HCV replication and NS5A protein expression, we tested the effect of other HCV replication inhibitors on the formation of new NS5A foci. The NS5B inhibitor, 2'-C-methyladenosine (2′CMA), which used at a dose that inhibited HCV replication to a similar degree as NS5A, PI4KA or OSBP inhibitors (Supplementary Figure 5G), did not block ‘new’ NS5A foci from forming at distinct sites and did not increase the colocalization between ‘old’ and ‘new’ NS5A foci (Supplementary Figure 5A-C). Similar results were obtained when cells were treated with the microtubule inhibitors nocodazole or vinblastine (Supplementary Figure 5D-F), which inhibited HCV replication (Ref [30] and Supplementary Figure 5G) and disrupted microtubule formation as expected (Supplementary Figure 5H). These results indicate that the inhibition of new NS5A focus formation by NS5A, PI4KA, and OSBP inhibitors is specific and not simply an effect of inhibiting viral replication.
Cholesterol is preferentially trafficked to ‘old’ HCV replication organelles
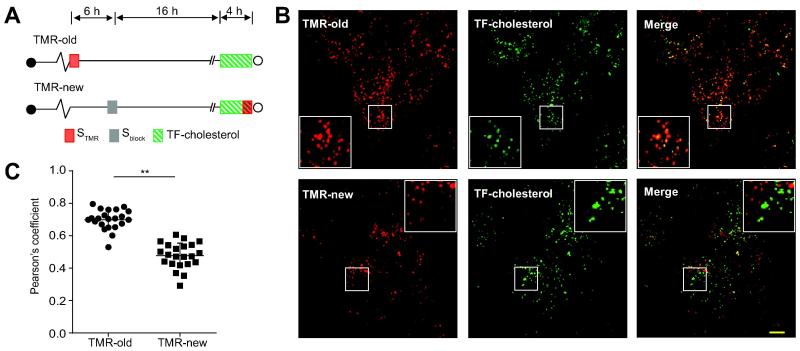
The continuous de novo formation of NS5A foci suggests the possibility that ‘new’ NS5A foci are compositionally and/or functionally distinct from foci containing ‘old’ NS5A. The HCV replication organelle is enriched in cholesterol [7, 14, 15], and PI4KA and OSBP are required for this enrichment [12]. We assessed the delivery of TopFluor-cholesterol (TF-cholesterol), to ‘new’ versus ‘old’ NS5A foci as depicted in Figure 5A. TF-cholesterol is a fluorescent cholesterol analog that closely mimics membrane partitioning and trafficking of native cholesterol [31] and has been used to monitor cholesterol trafficking to HCV replication organelles [12, 32]. While TF-cholesterol trafficked to both ‘old’ and ‘new’ NS5A foci, TF-cholesterol colocalized preferentially with ‘old’ NS5A foci (Figures 5B and C), suggesting that replication organelles become progressively enriched in cholesterol over time.
Figure 5. Preferential trafficking of a cholesterol analog to ‘old’ NS5A foci.
(A). Schematic of experimental design. SGR-JFH1(NS5A/SNAP) subgenomic replicon cells were either labeled with STMR (for old foci) or treated first with Sblock and 16 hr later with STMR (for new foci) immediately before imaging. 4μM fluorescent TopFluor-cholesterol (TF-cholesterol) was added 4 hr before imaging.
(B). Representative live-cell images. Scale bar, 10 μm.
(C). Quantitation of colocalization between STMR labeled NS5A and TF-cholesterol. Each point denotes the Pearson’s coefficient calculated from a single cell. Summary mean±SD values representative of 3 independent experiments are indicated. ** p<0.001.
NS5A focus aging is associated with increasing lipid droplet association
We do not understand how viral RNA at replication organelles reaches sites of virion assembly at or near LDs. In a static model of replication organelle resupply (Supplementary Figure 2), replication organelles containing newly synthesized NS5A are expected to localize near LDs. In contrast, in a de novo model where new replication organelles are continuously being initiated, newly synthesized NS5A foci might conceivably be distant from LDs, but with time should associate with LDs and HCV core protein.
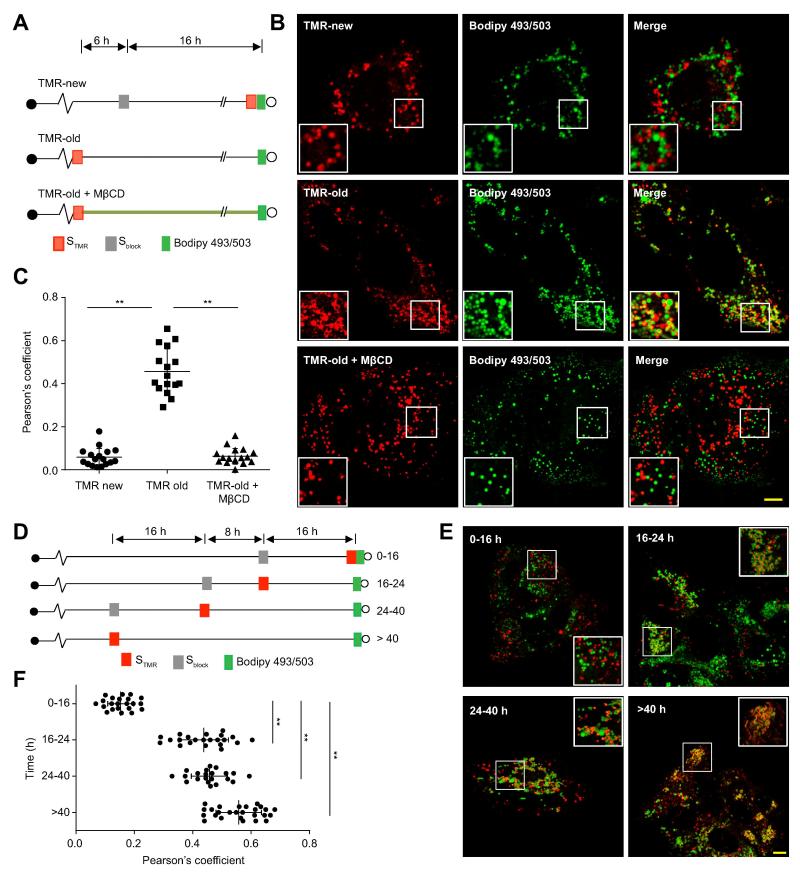
To test whether ‘old’ replication organelles are preferentially associated with LDs and core protein, cells were infected with a full-length infectious SNAP-tagged virus (Figure 1A), and ‘old’ or ‘new’ NS5A was selectively labeled while LDs were stained with the neutral lipid stain BODIPY 493/503 (Figure 6A). ‘Old’ NS5A was closely associated with LDs (Figures 6B, middle panels, and 6C) and core protein (Supplementary Figure 6A-C), often in a ring-like pattern encircling LDs, while ‘new’ NS5A was found in puncta that were not closely associated with LDs (Figures 6B, upper panels, and 6C) or core protein (Supplementary Figure 6A-C).
Figure 6. Cholesterol-dependent association of ‘old’ NS5A foci with lipid droplets.
(A). Schematic of experimental design to visualize ‘new’ and ‘old’ NS5A foci with cellular lipid droplets. Huh-7.5.1 cells infected with full-length JFH1(NS5A/SNAP) were either treated first with Sblock and 16 hr later with TMR (for new foci; top) or else pulse labeled with STMR (for old foci) and then either left untreated (middle) or treated with 1 mM methyl-β-cyclodextrin (MβCD; bottom) to deplete cellular cholesterol. Lipid droplets were labeled with BODIPY 493/503 before live cell imaging.
(B). Representative images of live cells labeled as described above. Scale bar, 10 μm.
(C). Quantitation of colocalization between STMR labeled NS5A and lipid droplets. Each point denotes the Pearson’s coefficient calculated from a single cell. Mean and errors (SD) are indicated. ** p<0.001.
(D). Schematic of experimental design to reveal the time-course of NS5A association with LDs. Huh-7.5.1 cells infected with full length JFH1(NS5A/SNAP) were treated with Sblock and STMR at the indicated time points to selectively label populations of NS5A molecules synthesized during the indicated intervals before imaging. Lipid droplets were visualized with BODIPY 493/503 before live cell imaging.
(E). Representative images of cells labeled as described. Scale bar, 10μm. Insets show enlargements of the boxed areas.
(F). Quantitation of colocalization between populations of NS5A molecules synthesized during varying intervals before imaging and lipid droplets. Each point denotes the Pearson’s coefficient calculated from a single cell. Mean and errors (SD) are indicated for each time point. ** p<0.001.
Since cholesterol is preferentially trafficked to ‘old’ replication organelles, we hypothesized that cholesterol enrichment might be necessary for replication organelles to associate with LDs. Indeed, acute depletion of cellular cholesterol with methyl-β-cyclodextrin (MβCD) prevented ‘old’ NS5A foci from colocalizing with LDs (Figures 6B bottom panel and 6C).
We next set out to characterize the kinetics of NS5A focus association with LDs and core protein by selectively labeling NS5A molecules synthesized during a specific time interval before LD labeling (Figure 6D). Little association was observed between LDs (Figures 6E and F) or core protein (Supplementary Figures 6D-F) and NS5A synthesized within the previous 16 hr. NS5A foci 16-24 hr old were more closely associated with LDs and core protein, and this association became even higher for foci containing NS5A synthesized at least 24 hr prior to LD or core protein labeling (Figures 6E and F, Supplementary Figures 6D-F).
To exclude the possibility that the decreased colocalization of ‘new’ NS5A or MβCD-treated ‘old’ NS5A foci with LDs was due to a decrease in the amount of labeled NS5A, we measured the amount of STMR labeled NS5A after anti-TMR immunoprecipitation (Supplementary Figure 7C). This anti-TMR immunoprecipitation is specific: NS5A and NS5A-associated HCV RNA were not immunoprecipitated by anti-TMR in cells expressing untagged NS5A or SNAP-tagged NS5A without STMR treatment as assessed by immunoblotting or qRT-PCR (Supplementary Figure 7A and B). As can be seen from Supplementary Figure 7D, no significant difference was observed in the amounts of NS5A immunoprecipitated. Similarly, if cells were selectively labeled during specific time intervals before immunoprecipitation (Supplementary Figure 7E), no differences were observed in the amount of immunoprecipitated NS5A by immunoblotting (Supplementary Figure 7F). These results suggest that the observed changes in HCV replication organelle association with LDs was not due to variations in the amount of labeled NS5A.
“Old” NS5A is more hyperphosphorylated and associated with a higher ratio of positive:negative strand genomes
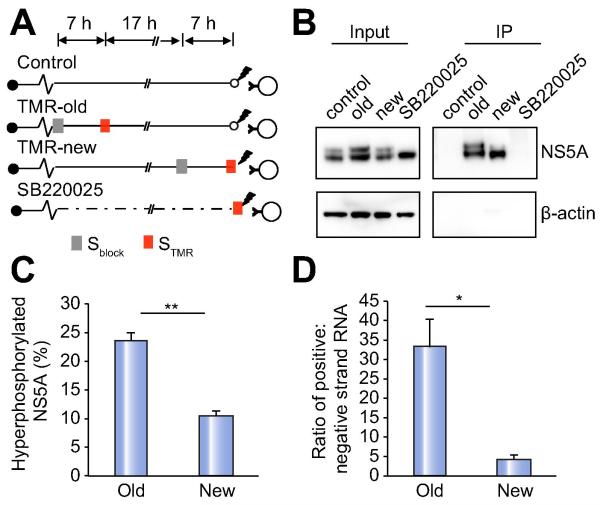
It has been suggested that the phosphorylation state of NS5A regulates switching of HCV RNA from replication to assembly [33, 34]. Given our observations that NS5A foci continually form de novo and become physically associated with LDs over time, we hypothesized that HCV RNA genomes at replication organelles might switch from viral RNA replication to virion assembly during this process, and that NS5A hyperphosphorylation should therefore be increased in ‘old’ replication organelles compared to ‘new’ replication organelles . Therefore, we assessed the phosphorylation state of ‘new’ and ‘old’ STMR-labeled NS5A molecules isolated by anti-TMR immunoprecipitation following detergent lysis (Figure 7A). We have not been able to affinity isolate NS5A in the absence of detergent using either this method or a different affinity tag [20]. A significantly higher fraction of ‘old’ NS5A was hyperphosphorylated compared to ‘new’ NS5A (24% vs 10%, Figures 7B and C), suggesting that a higher fraction of NS5A is indeed hyperphosphorylated in ‘old’ replication organelles. The small molecule kinase inhibitor SB 220025 inhibits NS5A hyperphosphorylation [35] and as expected, completely prevented the appearance of hyperphosphorylated NS5A (Figure 7B). However, no NS5A was immunoprecipitated after SB 220025 treatment because it inhibited STMR labeling of NS5A (Supplementary Figure 7G).
Figure 7. ‘Old’ NS5A species are more hyperphosphorylated and are associated with a higher ratio of positive:negative strand genomes.
(A). Schematic of experimental design to isolate ‘new’ and ‘old’ NS5A. Subgenomic replicon cells expressing NS5A-SNAP were either untreated or treated with 5μM SB 220025 and Sblock or STMR was applied at the indicated time points before detergent lysis and immunoprecipitation of TMR-labeled NS5A with anti-TMR antibody.
(B). Cell lysates (left panel) and immunoprecipitated TMR-NS5A (right panel) obtained as described above were subjected to immunoblotting for NS5A and β-actin.
(C). Quantitation of hyperphosphorylated ‘new’ versus ‘old’ NS5A, presented as a percentage of total NS5A; values are means±SEM of 5 independent experiments. ** p<0.001.
(D). Positive and negative strand RNA associated with ‘new’ and ‘old’ TMR-labeled NS5A was quantitated by strand-specific quantitative RT-PCR. Values shown are ratios of positive:negative strand RNA and are means±SEM of 4 independent experiments. * p<0.01.
In addition to increasing NS5A hyperphosphorylation, a model of de novo replication organelle generation and maturation predicts that ‘old’ replication organelles have more positive-strand genomes than ‘new’ replication organelles. This is in contrast to a model of replication organelle resupply, in which the amount of positive-strand genomes associated with ‘new’ and ‘old’ NS5A should remain relatively constant in cells stably infected with HCV. We conducted strand-specific quantitative RT-PCR to quantitate the amount of positive and negative strand HCV RNA associated with ‘old’ and ‘new’ NS5A immunoisolated by anti-TMR pulldown. The specificity of this strand-specific quantitation is shown in Supplementary Figure 7H. We detected negative strand HCV RNA bound to both immunoisolated ‘new’ and ‘old’ NS5A, consistent with our anti-dsRNA immunostaining results. The ratio of positive:negative strand HCV genomes was significantly higher in ‘old’ NS5A foci (33.1±7.3 vs 4.3±1.1 associated with ‘old’ versus ‘new’ NS5A, respectively; Figure 7D and Supplementary datasheet), consistent with a model of replication organelle maturation over time.
Discussion
Much remains to be learned about how HCV translation, replication and assembly events are regulated within the infected cell. For example, in the context of chronic HCV infection, it has not been known whether replication organelles are stable structures that are continually resupplied by newly synthesized viral proteins, or whether new replication organelles are continually being formed de novo. Our results are consistent with de novo formation, as newly-synthesized NS5A appears at sites spatially separated from preformed NS5A after relatively brief periods of as short as 1-2 hours. Eyre et al. have previously shown, also using SNAP-tagged NS5A, that NS5A synthesized between 0-24 hr is found at sites distinct from NS5A synthesized between 24-72 hr prior to imaging [23]. Our study agrees with but also extends this observation in several important ways. First, we examined multiple timepoints with varying chase periods to demonstrate that newly synthesized NS5A appears at distinct foci within 1-2 hours of chasing. Using multiple timepoints, as opposed to a single comparison, enables us to distinguish between ‘resupply’ versus ‘de novo’ models of replication organelle formation (Supplementary Figure 2), which cannot be determined with only two timepoints. In addition, we also characterized differences in composition and subcellular localization between ‘new’ and ‘old’ NS5A foci, and we demonstrated that several factors (PI4KA, OSBP, cholesterol, and NS5A) are necessary for the generation of new NS5A foci and/or association with LDs.
An important problem faced by a model of resupply is how a ‘static’ replication organelle is able to switch its associated positive-strand genomes among the mutually exclusive functions of translation, transcription of the negative strand replication intermediate, and assembly into virions. In contrast, a model of continuous de novo replication organelle formation could enable the switching of positive strand genomes from one function to another over time by regulation of protein and lipid composition, post-translational modifications of replication organelle components, and by physical association with other intracellular compartments. Several observations in this study are consistent with this model. First, we found compositional differences between ‘new’ and ‘old’ NS5A foci and a functional difference with regards to cholesterol trafficking. Specifically, a fluorescent cholesterol analog preferentially traffics to older NS5A foci and acute cholesterol depletion prevents association of ‘old’ replication organelles with LDs, suggesting that cholesterol enrichment of replication organelles is required for association with LDs . Second, ‘old’ NS5A foci are more likely to be associated with LDs and HCV core protein than ‘new’ foci; it takes roughly 16 hr for new NS5A foci to colocalize with LDs and core protein, which is consistent with the well-known 18-24 hr delay between HCV RNA transfection and the appearance of viral infectivity in the supernatant[36, 37]. Finally, we observed an increase in the density of DMVs and MMVs at ‘old’ NS5A foci compared to ‘new’ NS5A foci, suggesting possible differences in ultrastructural morphology that require confirmation in future studies.
PI4KA and its effector OSBP are required for HCV replication and maintenance of HCV replication organelle integrity [8-10, 12, 13, 26, 38], and NS5A is necessary and sufficient for DMV formation, albeit less efficiently when expressed alone [6]. In this study, we found that all three of these proteins are required for the de novo formation of NS5A foci. That other inhibitors of HCV replication did not block new NS5A focus formation indicates that this effect is not due simply to a general block in viral replication. In addition, inhibitors of these proteins unexpectedly led to the continued synthesis of NS5A at ‘old’ NS5A foci, suggesting that termination of viral protein translation at aged replication organelles is regulated somehow by these factors.
In summary, our results demonstrate spatial segregation and continuous turnover of temporally distinct sets of HCV replication organelles. Temporal changes in replication organelle composition may be associated with changes in replication organelle function and morphology, and we propose that this might be a mechanism that can facilitate separation of mutually exclusive processes in the viral infection cycle.
Supplementary Material
Lay summary.
Hepatitis C Virus (HCV) replication membrane structures are continuously generated at spatially distinct sites. New replication organelles are different in composition, and possibly also in function, compared to old replication organelles.
Acknowledgements
The authors thank Francis Chisari (Scripps Institute, La Jolla, CA), Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan), Charles Rice (Rockefeller University, New York, NY), Matthew Shair (Harvard University, Cambridge, MA), Raffaele De Francesco (Istituto Nazionale di Genetica Molecolare, Milano, Italy), Harry Greenberg (Stanford University, Palo Alto, CA), and Philippe Gallay (The Scripps Research Institute, La Jolla, CA) for their generous gifts of Huh7.5.1 cells, the infectious JFH-1 clone, anti-NS5A antibody (clone 9E10), OSW-1 and AL-9, anti-core antibody, and HCV NS5A inhibitors, respectively.
Financial support: This work was supported by grants DK097374 from the National Institutes of Health and the Greenview Foundation Hepatitis C Research Fund (to AWT). Microscopy was performed at the University of Michigan Microscopy & Image Analysis Laboratory with support from the University of Michigan Center for Gastrointestinal Research (NIDDK 5P30DK034933).
Abbreviations
- HCV
hepatitis C virus
- LDs
lipid droplets
- ER
endoplasmic reticulum
- DMVs
double-membrane vesicles
- MMVs
multi-membrane vesicles
- PI4KA
phosphatidylinositol 4-kinase IIIα
- OSBP
oxysterol-binding protein
- PI4P
phosphatidylinositol 4-phosphate
- CLEM
correlative light-electron microscopy
- MβCD
methyl-β-cyclodextrin
Footnotes
Conflict of interest: The authors who have taken part in this study declared that they do not have anything to disclose regarding funding or conflict of interest with respect to this manuscript.
Authors’ contributions: Conceived and designed the experiments: AT and HW. Performed the experiments: HW. Analyzed the data: HW, AT. Wrote the paper: AT and HW.
References
- [1].Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- [2].Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- [3].den Boon JA, Diaz A, Ahlquist P. Cytoplasmic viral replication complexes. Cell Host Microbe. 2010;8:77–85. doi: 10.1016/j.chom.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Egger D, Wölk B, Gosert R, Bianchi L, Blum HE, Moradpour D, et al. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, et al. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J Virol. 2013 doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, et al. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Amako Y, Sarkeshik A, Hotta H, Yates J, 3rd, Siddiqui A. Role of oxysterol binding protein in hepatitis C virus infection. J Virol. 2009;83:9237–9246. doi: 10.1128/JVI.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang H, Perry JW, Lauring AS, Neddermann P, De Francesco R, Tai AW. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology. 2014;146:1373–1385. e1371–1311. doi: 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. Hepatitis C Virus Stimulates the Phosphatidylinositol 4-Kinase III Alpha-Dependent Phosphatidylinositol 4-Phosphate Production That Is Essential for Its Replication. J Virol. 2011;85:8870–8883. doi: 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology. 2004;324:451–461. doi: 10.1016/j.virol.2004.03.034. [DOI] [PubMed] [Google Scholar]
- [15].Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J Virol. 2003;77:4160–4168. doi: 10.1128/JVI.77.7.4160-4168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Paul D, Madan V, Bartenschlager R. Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe. 2014;16:569–579. doi: 10.1016/j.chom.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shulla A, Randall G. Spatiotemporal analysis of hepatitis C virus infection. PLoS Pathog. 2015;11:e1004758. doi: 10.1371/journal.ppat.1004758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- [19].Sun X, Zhang A, Baker B, Sun L, Howard A, Buswell J, et al. Development of SNAP-tag fluorogenic probes for wash-free fluorescence imaging. Chembiochem. 2011;12:2217–2226. doi: 10.1002/cbic.201100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Salloum S, Wang H, Ferguson C, Parton RG, Tai AW. Rab18 Binds to Hepatitis C Virus NS5A and Promotes Interaction between Sites of Viral Replication and Lipid Droplets. PLoS Pathog. 2013;9:e1003513. doi: 10.1371/journal.ppat.1003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, et al. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J Virol. 2004;78:7400–7409. doi: 10.1128/JVI.78.14.7400-7409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wölk B, Büchele B, Moradpour D, Rice CM. A dynamic view of hepatitis C virus replication complexes. J Virol. 2008;82:10519–10531. doi: 10.1128/JVI.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Eyre NS, Fiches GN, Aloia AL, Helbig KJ, McCartney EM, McErlean CS, et al. Dynamic imaging of the hepatitis C virus NS5A protein during a productive infection. J Virol. 2014 doi: 10.1128/JVI.02490-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- [25].Targett-Adams P, Boulant S, McLauchlan J. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J Virol. 2008;82:2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bianco A, Reghellin V, Donnici L, Fenu S, Alvarez R, Baruffa C, et al. Metabolism of Phosphatidylinositol 4-Kinase IIIalpha-Dependent PI4P Is Subverted by HCV and Is Targeted by a 4-Anilino Quinazoline with Antiviral Activity. PLoS Pathog. 2012;8:e1002576. doi: 10.1371/journal.ppat.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Burgett AW, Poulsen TB, Wangkanont K, Anderson DR, Kikuchi C, Shimada K, et al. Natural products reveal cancer cell dependence on oxysterol-binding proteins. Nat Chem Biol. 2011;7:639–647. doi: 10.1038/nchembio.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Berger C, Romero-Brey I, Radujkovic D, Terreux R, Zayas M, Paul D, et al. Daclatasvir-Like Inhibitors of NS5A Block Early Biogenesis of Hepatitis C Virus-Induced Membranous Replication Factories, Independent of RNA Replication. Gastroenterology. 2014 doi: 10.1053/j.gastro.2014.07.019. [DOI] [PubMed] [Google Scholar]
- [29].Targett-Adams P, Graham EJ, Middleton J, Palmer A, Shaw SM, Lavender H, et al. Small Molecules Targeting Hepatitis C Virus-Encoded NS5A Cause Subcellular Redistribution of their Target: Insights into Compound Mode of Action. J Virol. 2011 doi: 10.1128/JVI.00215-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gentzsch J, Hinkelmann B, Kaderali L, Irschik H, Jansen R, Sasse F, et al. Hepatitis C virus complete life cycle screen for identification of small molecules with pro- or antiviral activity. Antiviral Res. 2011;89:136–148. doi: 10.1016/j.antiviral.2010.12.005. [DOI] [PubMed] [Google Scholar]
- [31].Holtta-Vuori M, Uronen RL, Repakova J, Salonen E, Vattulainen I, Panula P, et al. BODIPY-cholesterol: a new tool to visualize sterol trafficking in living cells and organisms. Traffic. 2008;9:1839–1849. doi: 10.1111/j.1600-0854.2008.00801.x. [DOI] [PubMed] [Google Scholar]
- [32].Shanmugam S, Saravanabalaji D, Yi M. Detergent-resistant membrane association of NS2 and E2 during hepatitis C virus replication. J Virol. 2015;89:4562–4574. doi: 10.1128/JVI.00123-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Evans MJ, Rice CM, Goff SP. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci U S A. 2004;101:13038–13043. doi: 10.1073/pnas.0405152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Masaki T, Matsunaga S, Takahashi H, Nakashima K, Kimura Y, Ito M, et al. Involvement of hepatitis C virus NS5A hyperphosphorylation mediated by casein kinase I-alpha in infectious virus production. J Virol. 2014;88:7541–7555. doi: 10.1128/JVI.03170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Qiu D, Lemm JA, O'Boyle DR, 2nd, Sun JH, Nower PT, Nguyen V, et al. The effects of NS5A inhibitors on NS5A phosphorylation, polyprotein processing and localization. J Gen Virol. 2011;92:2502–2511. doi: 10.1099/vir.0.034801-0. [DOI] [PubMed] [Google Scholar]
- [36].Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- [37].Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, et al. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.