Abstract
Aquaporins (AQPs) are channel-forming integral membrane proteins that facilitate the movement of water and many other small molecules. Compared to animals, plants contain a much higher number of AQPs in their genome. Homology-based identification of AQPs in sequenced species is feasible because of the high level of conservation of protein sequences across plant species. Genome-wide characterization of AQPs has highlighted several important aspects such as distribution, genetic organization, evolution and conserved features governing solute specificity. From a functional point of view, the understanding of AQP transport system has expanded rapidly with the help of transcriptomics and proteomics data. The efficient analysis of enormous amounts of data generated through omic scale studies has been facilitated through computational advancements. Prediction of protein tertiary structures, pore architecture, cavities, phosphorylation sites, heterodimerization, and co-expression networks has become more sophisticated and accurate with increasing computational tools and pipelines. However, the effectiveness of computational approaches is based on the understanding of physiological and biochemical properties, transport kinetics, solute specificity, molecular interactions, sequence variations, phylogeny and evolution of aquaporins. For this purpose, tools like Xenopus oocyte assays, yeast expression systems, artificial proteoliposomes, and lipid membranes have been efficiently exploited to study the many facets that influence solute transport by AQPs. In the present review, we discuss genome-wide identification of AQPs in plants in relation with recent advancements in analytical tools, and their availability and technological challenges as they apply to AQPs. An exhaustive review of omics resources available for AQP research is also provided in order to optimize their efficient utilization. Finally, a detailed catalog of computational tools and analytical pipelines is offered as a resource for AQP research.
Keywords: plant aquaporin, omic scale analysis, analytical approaches, yeast assay, Xenopus oocytes assay
Introduction
Aquaporins (AQPs) are channel-forming proteins that facilitate selective transport of water and many other small molecules like urea, silicon (Si) in the form of silicic acid, boron (B) in the form of boric acid, and CO2 across biological membranes. AQPs are present in almost all living organisms including eukaryotes and prokaryotes (Quigley et al., 2001; Tanghe et al., 2006; Benga and Huber, 2012; Benga, 2013). In animals, minor defects or changes in AQP configuration are known to cause many diseases such as hereditary nephrogenic diabetes insipidus, congenital cataracts and, more commonly, the inability to concentrate solutes in urine (Verkman, 2009; Benga and Huber, 2012). Similarly, plant AQPs have an important role in regulating the overall development of a plant, namely in the maintenance of hydraulic status under extreme conditions. As early as 1986, Benga et al. in a pioneer effort, reported the role of proteins in water transport (Benga et al., 1986a,b). Subsequently, Peter Agre's team proved through cRNA expression studies that those proteins, named aquaporin-1, were specific water channels (Preston et al., 1992), and Agre was awarded the Nobel Prize in chemistry in 2003 for his discovery (Agre, 2004). These findings have sparked a veritable explosion of work that has enhanced our understanding of the importance of AQPs in animals as well as in plants (Papadopoulos and Verkman, 2013; Deshmukh et al., 2015; Kitchen et al., 2015; Maurel et al., 2015; Kirscht et al., 2016; Srivastava et al., 2016).
AQPs from diverse origins have characteristic hourglass-like structures with six transmembranes (TM) alpha helices and two half TM alpha-helices with conserved NPA domains (asparagine–proline–alanine) (Jung et al., 1994; Murata et al., 2000). The two half alpha helices form a constraint in the center of the pore that regulates the selective transport of solutes through the pore. Another constraint known as the aromatic arginine (ar/R) selectivity filter (SF), formed mostly with four amino acid residues, plays also a major role in solute selectivity (Murata et al., 2000; Törnroth-Horsefield et al., 2006). Based on their phylogenetic distribution, plant AQPs are generally categorized into five major sub-families: plasma membrane intrinsic proteins (PIP), nodulin 26-like intrinsic proteins (NIPs), tonoplast intrinsic proteins (TIPs), small intrinsic proteins (SIPs), and uncharacterized intrinsic proteins (XIPs) (Quigley et al., 2001; Deshmukh et al., 2015). The phylogenic classification of each group is very well aligned with the functionality and characteristic features of AQPs (Grégoire et al., 2012; Deshmukh et al., 2015).
Availability of whole genome sequences for animal and plant species has facilitated genome-wide identification and classification of AQPs. For instance, compared to animals, plants have a larger number of AQPs ranging from 23 in Selaginella moellendorffii to 72 in soybean (Deshmukh et al., 2015). Apart from identifying novel genes, genome-wide studies have contributed to a better understanding of the molecular evolution of the AQP gene families (Gupta and Sankararamakrishnan, 2009; Deshmukh and Bélanger, 2016). Precise identification of conserved features along with their functional relevance has progressed rapidly with the availability of AQP sequences from many plant species. Similarly, transcriptome profiling of AQPs conducted in several plant species has helped to determine that AQPs have expression specific to tissue, growth stage, or environmental conditions (Gupta and Sankararamakrishnan, 2009).
Regulation of solute transport through AQPs is a very complex phenomenon that involves environmental stimuli, transcriptional changes, and post-translational modifications. For a better understanding of AQP-mediated transport systems, integration of information generated through different approaches such as genomics, transcriptomics, and proteomics is required. In addition, information about analytical tools and available resources is also important to properly characterize AQPs. In the present review, we discuss how different approaches and analytical tools exploiting the many available resources can contribute to the study of AQPs.
Genome-wide identification of AQPs
The initial genome-wide studies in Arabidopsis have paved the way to understand the distribution, characterization, and evolution of gene families in plants. The Arabidopsis genome has 35 AQPs that can be classified into four subfamilies based on phylogenetic distribution (Quigley et al., 2001). This classification was found to cluster fairly well with the functionality of AQPs. Subsequently, a second genome-wide study was performed in rice, which is considered as a model cereal crop and also represents a distinct monocot clade (Sakurai et al., 2005). Apart from the phylogenetic classification, solute-based classification of AQPs like aquaporins, aquaglyceroporins and S-aquaporins has also been used, particularly in animals (Benga, 2012). The information of AQPs in rice and Arabidopsis facilitated the monocot-dicot comparison that expanded the understanding of AQP gene families in plants. Later on, the genome sequencing of cucumber (Cucumis sativus) using next generation sequencing approaches started a new era characterized by a constant flow of reports of plant genome sequences and subsequent genome-wide AQP studies in plants (Huang et al., 2009; Deshmukh et al., 2013; Ariani and Gepts, 2015).
The genome sequences for moss (Physcomitrella patens) enabled the identification of 23 AQPs (Danielson and Johanson, 2008). In addition, the study added two new AQP subfamilies: Hybrid Intrinsic Proteins (HIP) and GlpF-like intrinsic proteins (GIPs) (Danielson and Johanson, 2008). Mosses, being primitive plants, are valuable for evolutionary studies, and the features observed in mosses are likely to be present in higher plants. In this regard, the seven AQP subfamilies found in mosses suggest that the diversion of AQPs was an early event and that higher plants lost two sub-families in the course of evolution. Subsequently, tissue-specific expression observed in vascular plants is argued to have evolved after the diversion of subfamilies. A recent study highlighting genome-wide comparison of AQPs in 25 plant species revealed several unique features about the subfamilies (Deshmukh et al., 2015). For instance, it is now clear that the XIP subfamily has been lost throughout the entire monocots, as well as within the Brassicaceae. In addition, Brassicaceae have also lost NIP2s from their genome (Deshmukh et al., 2015).
Most of the genome-wide studies have used AQP sequences reported in rice and Arabidopsis as a query to perform homology-based searches. However, it would be more accurate if a larger number of AQPs from different species could be included in the query sequences given that some subfamilies and groups are absent from Arabidopsis and rice. For example, NIP2s having characteristic G-S-G-R ar/R SF are missing from Arabidopsis, and, similarly, the entire XIP subfamily is absent in both Arabidopsis and rice (Table 1). In this paper, we have described over 1000 aquaporins from 26 plant species representing a wide range of families and clades (Supplementary Dataset 1). This exhaustive list will be useful as a query in genome-wide identification of AQPs in other plant species. The analytical steps required for genome-wide identification of AQPs are described in Figure S1.
Table 1.
Genome-wide identification and classification of aquaporins in 31 plant species.
| Plant species | PIP | TIP | NIP | SIP | XIP | AQP | References |
|---|---|---|---|---|---|---|---|
| Physcomitrella patens | 9 | 4 | 5 | 2 | 2 | 23 | Danielson and Johanson, 2008 |
| Selaginella moellendorffii | 3 | 3 | 8 | 1 | 3 | 19 | Anderberg et al., 2014; Deshmukh et al., 2015 |
| Picea abies | 18 | 6 | 13 | 2 | 0 | 39 | Deshmukh et al., 2015 |
| Musa acuminate | 21 | 18 | 9 | 3 | 0 | 51 | Deshmukh et al., 2015 |
| Oryza sativa | 11 | 10 | 11 | 2 | 0 | 34 | Sakurai et al., 2005 |
| Brachypodium distachyon | 11 | 10 | 9 | 2 | 0 | 32 | Deshmukh et al., 2015 |
| Sorghum bicolor | 14 | 13 | 10 | 3 | 0 | 40 | Deshmukh et al., 2015 |
| Zea mays | 10 | 13 | 13 | 7 | 0 | 43 | Deshmukh et al., 2015 |
| Setaria italic | 16 | 16 | 15 | 3 | 0 | 50 | Deshmukh et al., 2015 |
| Elaeis guineensis | 9 | 10 | 9 | 2 | 0 | 30 | Deshmukh et al., 2015 |
| Arabidopsis thaliana | 13 | 10 | 9 | 3 | 0 | 35 | Quigley et al., 2001 |
| Arabidopsis lyrata | 14 | 12 | 10 | 3 | 0 | 39 | Deshmukh et al., 2015 |
| Brassica rapa | 22 | 16 | 15 | 6 | 0 | 59 | Deshmukh et al., 2015 |
| Brassica oleracea | 25 | 19 | 17 | 6 | 0 | 67 | Deshmukh et al., 2015 |
| Carica papaya | 10 | 7 | 7 | 2 | 2 | 28 | Deshmukh et al., 2015 |
| Citrus sinensis | 11 | 9 | 8 | 3 | 3 | 34 | Deshmukh et al., 2015 |
| Citrus clementine | 14 | 10 | 9 | 3 | 1 | 37 | Deshmukh et al., 2015 |
| Vitis vinifera | 9 | 9 | 9 | 1 | 2 | 30 | Deshmukh et al., 2015 |
| Glycine max | 22 | 23 | 17 | 8 | 2 | 72 | Deshmukh et al., 2013 |
| Cajanus cajan | 12 | 13 | 10 | 4 | 1 | 40 | Deshmukh et al., 2015 |
| Fragaria vesca | 10 | 9 | 14 | 4 | 2 | 39 | Deshmukh et al., 2015 |
| Prunus persica | 7 | 8 | 9 | 3 | 2 | 29 | Deshmukh et al., 2015 |
| Ricinus communis | 10 | 9 | 8 | 4 | 5 | 36 | Deshmukh et al., 2015 |
| Populus trichocarpa | 15 | 18 | 11 | 7 | 7 | 58 | Gupta and Sankararamakrishnan, 2009 |
| Solanum tuberosum | 15 | 11 | 11 | 2 | 5 | 44 | Deshmukh et al., 2015 |
| Solanum lycopersicum | 14 | 10 | 11 | 3 | 6 | 44 | Deshmukh et al., 2015 |
| Phaseolus vulgaris | 12 | 13 | 10 | 4 | 2 | 41 | Ariani and Gepts, 2015 |
| Hevea brasiliensis Muell. Arg. | 15 | 17 | 9 | 4 | 6 | 51 | Zou et al., 2015 |
| Hordeum vulgare L | 11 | 7 | 4 | 2 | 0 | 22 | Hove et al., 2015 |
| Jatropha curcas | 9 | 9 | 8 | 4 | 2 | 32 | Zou et al., 2016 |
| Phyllostachys edulis | 10 | 6 | 8 | 2 | 0 | 26 | Sun et al., 2016 |
| Total 31 plant species | 402 | 348 | 316 | 105 | 53 | 1224 |
Transcriptomics studies for AQPs
Transcriptomics progressed initially with the technological improvement in chip-based expression profiling platforms (Schulze and Downward, 2001). Subsequently, the advancements in affordable sequencing technologies have greatly contributed to transcriptome sequencing (Burgess, 2016; Chen et al., 2016). As a result, transcriptomic resources have become widely available with RNA-seq studies performed on many plant species covering major crops, medicinal plants, model species and plant species important for evolutionary studies (www.ncbi.nlm.nih.gov/sra). Available transcriptomic resources are helpful to integrate information with genomics data for a better comprehension of gene functions (Movahedi et al., 2012; Patil et al., 2015; Sonah et al., 2016; Song et al., 2016). For instance, many of the recent studies highlighting genome-wide identification of AQPs have relied on transcriptomic resources to explain tissue-specific expression of those genes (Gupta and Sankararamakrishnan, 2009; Reuscher et al., 2013; Venkatesh et al., 2013; Deshmukh et al., 2015; Hu et al., 2015; Deokar and Tar'an, 2016; Deshmukh and Bélanger, 2016; Zou et al., 2016).
In a study of genome-wide identification of AQPs in soybean, we have used publically available RNA-seq and microarray data to elucidate the expression profile of AQPs across tissues (Deshmukh et al., 2013). Among particular observations, the study revealed a seed-specific expression for all members of the TIP3 subgroup. A similar type of seed-specific expression for TIP3s has been reported with rice and Arabidopsis, transcriptomic data (Deshmukh et al., 2013). These results suggest an important role of TIP3s in seed development, possibly in the desiccation process required for seed maturation. Similarly, Gupta and Sankararamakrishnan (2009) have used microarray data to perform genome-wide expression profiling of AQPs in poplar, and revealed higher expression of TIPs and PIPs in xylem tissues. Using a publicly available RNA-seq data for barley, Hove et al. (2015) observed a high level of HvNIP4;1 expression in inflorescences. Recently, the tissue-specific expression of NIP4s (AtNIP4; 1 and AtNIP4; 2) was found to be required for pollen development and pollination in Arabidopsis thaliana (Di Giorgio et al., 2016). Such information about expression profile is instrumental in defining substrate specificity and interdependency among AQPs.
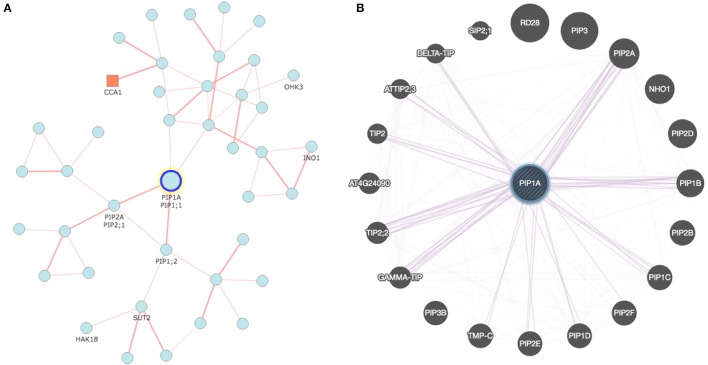
Interdependency of AQPs is a well-known phenomenon, particularly in the case of PIP1s and PIP2s (Yaneff et al., 2014). The AQP-mediated transport system is very complex and will rely on the conjugated action of distinct transporters to carry solutes from one tissue to another (Ma et al., 2007; Sakurai et al., 2015). In this context, recent developments in analytical tools offer great opportunities for construction of co-expression networks and several online tools are available for this purpose (Table 2). As an example, Figure 1 describes a co-expression network in rice using the online tool FREND that revealed the concerted role of PIP2-1, PIP1-1, and PIP1-2. Another tool, PlaNet, allows a comparative analysis of co-expression networks across plant species such as rice, soybean, Brachypodium, barley, Medicago, poplar and wheat. Furthermore, some tools exploit data from the literature, known protein domains, experimentally proven protein-protein interactions, genetic interactions based on QTL/GWAS studies, and information generated through proteomics interactions to infer specific roles of AQPs (Table 2). As part of an integrated omics approach, these tools will provide precise information about AQP-mediated molecular events in plants.
Table 2.
Tools available for the development of co-expression network using transcriptomic data from different plant species.
| S. no. | Database/Online tool | Plant species | Website | Other notes/Features |
|---|---|---|---|---|
| 1 | PlaNet | Arabidopsis, barley, Medicago truncatula, poplar, rice, soybean, Nicotiana tabacum, and wheat | http://aranet.mpimp-golm.mpg.de | Allows comparative analysis of co-expression networks across plant species |
| 2 | PLANEX | Arabidopsis thaliana, soybean, barley, rice, tomato, wheat, grape and maize | http://planex.plantbioinformatics.org | Gene Expression Omnibus (GEO) |
| 3 | ATTED-II | Arabidopsis, soybean, maize, rice, tomato, wheat, grape poplar, and muster | http://atted.jp | Uses known protein-protein interactions and functional annotations |
| 4 | CressExpress | Arabidopsis | http://cressexpress.org | Suitable for downstream data-mining, visualization, and analysis. |
| 5 | Genemania | Arabidopsis | http://genemania.org | GeneMANIA's database of 1800+ networks, containing over 500 million interactions across eight organisms |
| 6 | CORNET | Maize, Tool | https://bioinformatics.psb.ugent.be/cornet/ | Allows co-expression analysis using either predefined or user-defined groups of micro array experiments. |
| 7 | VTCdb | Grape | http://vtcdb.adelaide.edu.au/Home.aspx | Retrieves hierarchical optimized Gene Ontology enrichment and tissue/condition specificity genes within the module along with interactive network visualization and analysis via CytoscapeWeb. |
| 8 | CORE | Rice | https://core.ac.uk/display/8598313/tab/similar-list | Creates gene co-expression networks using both condition-dependent and condition-independent data |
| 9 | Xpressomics | Arabidopsis | https://xpressomics.com/search/ | Uses differential expression from expert-curated and analyzed raw data |
| 10 | CoP | Arabidopsis, soybean, barley, rice, poplar, wheat, grape, maize | http://webs2.kazusa.or.jp/kagiana/cop0911/ | Provides information about gene co-expression, specific gene expression, biological processes, and metabolic pathways that are mutually interconnected |
| 11 | RiceFREND | Rice | http://ricefrend.dna.affrc.go.jp/ | Based on a large collection of microarray data |
Figure 1.
Co-expression network developed for rice and Arabidopsis aquaporin genes: (A) Network analyzed with RiceFREND tool (http://ricefrend.dna.affrc.go.jp) showing interdependency of PIP1-1, PIP1-2, and PIP2-1 in rice; and (B) Network of Arabidopsis visualized with GENEMANIA tool (http://genemania.org) showing interaction of PIP1A with PIP2 and other genes. Network for all rice AQPs at the third hierarchical level is provided in Supplementary Data 2.
Proteomics contributions in AQP studies
Compared to genomics and transcriptomics, proteomic approaches have contributed limited efforts to the study of AQPs. This is mostly because of the costly and demanding methodological requirements of proteomic studies. Most of the large-scale proteomic analyses focusing on AQPs have been conducted with Arabidopsis, and rice (Table S1). In general, modern proteomic tools yield an enormous amount of valuable data that can be used to resolve complex molecular mechanisms, more specifically regarding post-translation modifications, protein expression, and protein-protein interactions (Deshmukh et al., 2014). However, relatively limited information is available for membrane proteins including AQPs because of constraints such as limited solubility, low expression and restricted use of restriction enzymes (Tan et al., 2008).
Solubility of AQPs is a very critical issue to perform efficient proteomic studies because of their higher hydrophobicity, difficult extraction, and presence in relatively small amounts. This issue prevents or limits reliance on standard approaches utilizing mass spectrometry (MS), matrix-assisted laser desorption ionization (MALDI), and electrospray ionization (ESI). Some of the pioneer studies, including those of Schindler et al. (1993) and Schey et al. (1992), developed procedures for improving solubilisation of membrane proteins by using organic solvents, acetonitrile, 2-propanol, hexafluoro-2-propanol and detergents. Better solubilisation also helps for a more efficient digestion. However, problems with dissolving membrane proteins remain even though the amount of AQPs obtained with fractionated plasma membrane was somewhat alleviated with the advancement in instrumentation that works at the nano-scale level (Table S1).
Santoni et al. (2003) described the first comprehensive efforts for AQP research using proteomic approaches. They developed an inventory of Arabidopsis AQP isoforms expressed in root tissues that were characterized with MALDI and electro-ionization tandem MS. The study also provided key information about phosphorylation and other post-translational modifications of AQPs, particularly within the PIP subfamily. Nearly a decade after this pioneer work, a study by Mirzaei et al. (2012) demonstrated the effects of environmental factors like drought on AQP regulation in rice. The study described the AQP expression profile at precise physiological stages during the progression of drought over time. For this purpose, the authors used label-free quantitative shotgun proteomic approaches involving nano-LC-MS/MS to identify 1548 proteins including AQPs, and predicted the mechanisms involved in drought stress. Similarly, di Pietro et al. (2013) conducted extensive proteomic studies of Arabidopsis AQPs under different physiological conditions. They studied nine physiological treatments modulating root hydraulics over different time periods. They observed 55 AQP peptides undergoing significant changes with respect to different physiological conditions, including several post-translational modifications like methylation, acetylation, and phosphorylation.
Aquaporin tertiary structure
The precise definition of the molecular structure of a protein is very important to understand its function. It contributes to the elucidation of the specific activity of a protein and its interaction with other molecules including ligands and inhibitors. Since the discovery of the first AQP in human red blood cells and renal proximal tubules, several attempts have been made to resolve AQP structure (Preston et al., 1992). Initial work by Preston et al. (1994), using a-chymotrypsin digestion of intact oocytes and inside-out membrane vesicles, confirmed the cytoplasmic loops and orientations of the six transmembrane alpha helices predicted by the hydropathy analysis. In the same year, another study by Jung et al. (1994) predicted the hourglass model for AQPs based on the topological information and the positioning of the loops, more particularly loop B and loop E, which penetrate in the membrane from opposite sides to form a constrict harboring conserved NPA domains. This study was instrumental in predicting for the first time the role of NPA domains in the tight regulation of solute transport. Later, Murata et al. (2000) solved the atomic structure of AQP1 using electron crystallographic analysis that confirmed the earlier predictions. The AQP1 structure has 3.8 Å resolution describing highly conserved amino-acid residues that stabilize the fold, and form the hourglass structure. The structural model presented by Murata et al. (2000) showed the conserved hydrophobic residues lining the water channel at the center of the protein. The structure also showed constricts with a pore diameter of about 3 Å that provided a clue as to how the AQP was permeable to water but not the proton.
Up to now, about 51 AQP structures have been described using different approaches at varying levels of resolution (Table S2). Most of the solved structures belong to human and Escherichia coli AQPs representing eukaryotic and prokaryotic models. Apart from human, high-resolution AQP structures are also available from other animals such as rat, sheep, and cattle (Table S2). Compared to animals and prokaryotes, very few structures are available for plant AQPs. The first plant AQP structure, common bean TIP, was solved by Daniels et al. (1999) at low resolution (7.7 Å) using electron cryo-crystallography. In spite of the low resolution, the AQP structure showed resemblance with animal AQPs. Törnroth-Horsefield et al. (2006) described the first high resolution plant AQP structure (from spinach, SoPIP2;1) in its closed (2.1 Å resolution) and open conformation (3.9 Å resolution). The structure with closed and open conformation explained the gating mechanism in which loop D, through a displacement of up to 16 Å, widened the pore and acted as molecular gating. The mechanism of gating is found to be conserved across all plant species (Törnroth-Horsefield et al., 2006). Recently, the structure of Arabidopsis aquaporin AtTIP2;1 was determined at very high resolution (1.18 Å) (Kirscht et al., 2016). Most of the previously reported structures are from water-transporting AQPs (Table S2) but AtTIP2;1 is a model AQP for ammonia transport. Interestingly, Kirscht et al. (2016) discovered a fifth amino acid involved in the permeability of ammonia, expanding the complexity of solute specificity from the four amino acids ar/R SF. Owing to the increasing availability of high-resolution AQP structures, homology-based computational approaches used to predict 3D-structure have now become more efficient and sophisticated (Tables S2, S3).
Advances in computational methods during the last two decades have made it possible to predict structures more accurately. The success of computational methods can be attributed to the evolutionary conserved features of proteins, the relatively small number of unique protein fold in nature, and the ever increasing number of solved protein structures (www.rcsb.org; Koonin et al., 2002). Compared to proteins that have no similarity with known structures, prediction of structure for candidate AQPs is facilitated by the abundance of resolved AQP structures publicly available (Table S2). In addition, several online servers and tools available for the homology-based prediction of protein structures are helpful to pursue more advanced studies in AQPs (Table S3).
Molecular dynamics simulations are advanced computational approaches used for in silico reconstitution of protein structure in its native environment (Lindahl and Sansom, 2008). Molecular dynamics simulations became more advanced with the availability of very high-resolution 3-D structures, increased computing power, and improvement in the analytical algorithms. In addition, detailed information about the interaction between amino acid residues and the surrounding environment makes it possible to reconstitute protein structures in different environments (Lindahl and Sansom, 2008). More particularly, studies focusing on membrane proteins have provided information about the interaction between individual amino acids with the lipid molecules in the membrane environment. There are several methods and tools available for molecular dynamics simulations of membrane proteins like AQPs (Lindahl and Sansom, 2008). Recently, Sakurai et al. (2015) have performed molecular dynamics simulations to study silicic acid uptake through AQP (OsLsi1) coupled with another active transporter (OsLsi2) in rice roots. They developed a mathematical model using diffusion equation along with the effects of active transport by OsLsi2. The study provides s good example for the utilization of in vivo experimental data to calibrate the model.
Analytical and functional approaches
Xenopus oocyte assay
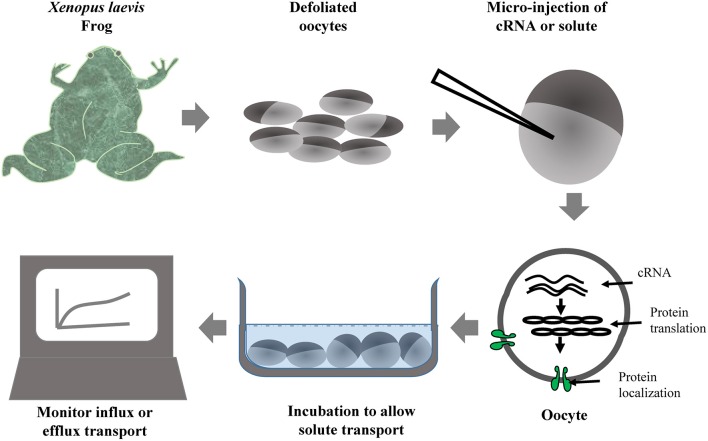
Oocytes of Xenopus laevis (African clawed frog) are commonly used for the evaluation of solute transport activity by AQPs and many other transporters. cRNA of foreign proteins can be easily injected and expressed in X. laevis oocytes. A cRNA volume of up to 50 nL can be injected in the oocyte, which allows production of large amounts of protein. The simplified steps involved in the X. laevis oocyte assay are provided in Figure 2. The transport of several different substrates by plant AQPs has been evaluated using X. laevis oocyte assay (Table 3). The oocyte assay conveniently allows the use of radiolabeled substrates to facilitate a better estimation of transport kinetics and also to increase the sensitivity of the assay (Ma et al., 2006). However, in the case of certain substrates such as silicic acid, many have used 68Ge as silicic acid surrogate, a method often criticized given that 68Ge was never shown to represent a perfectly interchangeable surrogate. In recent studies, silicic acid transport (influx and efflux) in oocytes was measured directly by atomic absorption spectrophotometry, a technical improvement that greatly facilitates the study of silicic acid movement in plants (Ma et al., 2006; Grégoire et al., 2012; Deshmukh et al., 2015; Carpentier et al., 2016; Vivancos et al., 2016). The measurement of change in volume of oocyte in response to osmolality of external solution is a simple and effective measure to study water transport by AQPs. The X. laevis oocyte system provides several advantages for the study of transporters. For instance, there is very low transport across the oocyte membrane through the endogenous transporters; therefore there is limited background effect and less ambiguity about solute transport. In addition, the relatively large size of X. laevis oocytes facilitates their manipulation and allows studying electrophoretic transporters using the two-electrode voltage clamp technique. Nevertheless, one has to keep in mind that the environment of a plant cell is drastically different from that of the X. laevis oocyte. Therefore, results obtained with the X. laevis oocyte assay need to be corroborated with the actual activity of the protein in plant cells.
Figure 2.
Simplified workflow of Xenopus oocyte assay used for the evaluation of solute transport by heterologously expressed foreign transporters including aquaporins.
Table 3.
List of important studies performed to evaluate different solutes transported by plant aquaporins using the Xenopus oocyte assay.
| Plant species | Gene | Solute | References |
|---|---|---|---|
| Arabidopsis | AtTIP2;1, AtTIP2;3 | Ammonium | Loqué et al., 2005; Liu et al., 2013 |
| Rice | Lsi1 (OsNIP2;1) | Arsenite | Zhao et al., 2010 |
| Arabidopsis | NIP5;1 | Boron | Takano et al., 2006 |
| Arabidopsis | NIP6;1 | Boron | Tanaka et al., 2008 |
| Nicotiana tabacum | NtAQP1 | CO2 | Uehlein et al., 2003 |
| Nicotiana tabacum | NtXIP1;1 | Glycerol, urea, boric acid | Bienert et al., 2011 |
| Soybean | Nodulin 26 | Glycerol, Water | Dean et al., 1999 |
| Nicotiana tabacum | NtAQP1 | Glycerol, Water, | Biela et al., 1999 |
| Poplar | PtNIP2-1 | Silicic acid | Deshmukh et al., 2015 |
| Tomato | SlNIP2-1 (Mutant) | Silicic acid | Deshmukh et al., 2015 |
| Barley | HvLsi1 | Silicic acid | Chiba et al., 2009 |
| Equisetum arvense | EaNIP3;1, EaNIP3;3 and EaNIP3;4 | Silicic acid | Grégoire et al., 2012 |
| Soybean | GmNIP2-1, GmNIP2-2 | Silicic acid | Deshmukh et al., 2013 |
| Rice | Lsi1 (OsNIP2;1) | Silicic acid (68Ge) | Ma et al., 2006 |
| Maize | ZmPIP1-5b | Urea, Water | Bousser et al., 2003 |
| Spinach | PM28A | Water | Johansson et al., 1998 |
| Maize | ZmPIP2a | Water | Chaumont et al., 2000 |
| Rice | OsPIP1;1 | Water | Liu et al., 2013 |
| Radish | VM23 | Water | Higuchi et al., 1998 |
| Olive | OePIP2.1, OeTIP1.1 | Water | Secchi et al., 2007 |
| Tomato | LeAqp2 | Water | Werner et al., 2001 |
| Walnut | JrPIP2,1 | Water | Sakr et al., 2003 |
| Soybean | GmTIP1-5, GmTIP2-5 | Water | Song et al., 2016 |
Translation of plant AQP transcripts in the oocyte may be altered by the differential codon preference between plants and Xenopus. For this reason, western blotting is often required to confirm protein expression/presence in the oocytes. However, codon optimization is rarely considered for plant AQPs when tested with oocytes, which raises the question, whether codon optimization is really a concern or not. Recently, Feng et al. (2013) reported better nitrate transport in oocytes with codon-optimized rice high affinity nitrate transporter. Similarly Bienert et al. (2014) also observed a significantly higher expression of ZmPIP1 and ZmPIP2 in yeast cells only after optimizing the codons. Nowadays, the use of synthesized DNA for gene-cloning related applications is becoming more common because of reduced costs and codon optimization can be routinely applied while synthesizing the gene for oocyte or yeast assays.
Evaluation of AQPs using yeast assays
Characteristic features of yeasts including ease of growth, short generation time, well established and easy transformation systems, and sequenced genomes, make them amenable as a heterologous system to study eukaryotic proteins. Several yeast species, including Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Pichia pastoris have been used as a tool to study foreign genes.
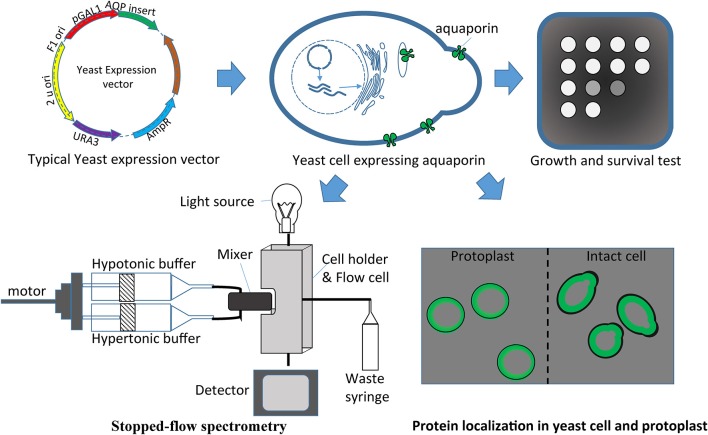
Numerous AQPs have been studied using a yeast expression system (Table 4). The study of water fluxes through AQPs in yeast assays is a particularly easy and affordable option. Water transport in yeast results in measurable cell volume changes (swelling or shrinking) in relatively short periods of time. The water transport through AQPs (as a hydrophilic passage) is much faster than the transport through the hydrophobic lipid bilayer membrane. This allows discrimination between water transport through the foreign AQP expressed in the yeast and the transport through the membrane. The volume change in yeast affects several physical parameters that can be used to make quantitative measurements essential to understand transport kinetics. Light absorption, light scattering or reflection with fluorescent dye are effective variables that are being used to monitor cell volume changes (Table 4, Figure 3). Given the small the size of the yeast that takes milliseconds to change volume in response to osmotic pressure, it requires a method known as stopped-flow spectrometry to take precise measurements. In stopped-flow spectrometry, protoplasts, vesicles or even intact cells are subjected simultaneously to hypotonic and hypertonic buffers. The rapidly mixing hypotonic and hypertonic solution stops transport in milliseconds, which allows taking measurements at the scale required to understand transport kinetics. Use of florescent dye in stopped-flow spectrometry increases precision of measurements. Recently, Sabir et al. (2014) evaluated grape AQPs for water conductivity using a stopped-flow fluorescence spectroscopy assay. They pre-loaded yeast cells with the non-fluorescent precursor 5-(and-6)-carboxyfluorescein diacetate (CFDA) that is permeable to membranes, and then intracellularly hydrolyzed CFDA to release the membrane impermeable fluorescent compound. Changes in cell volume in stopped-flow assay in response to osmotic changes resulted in changes in fluorescence intensity that can be measured to deduct transport kinetics.
Table 4.
List of plant aquaporins evaluated for transport of different solutes using yeast assays.
| Aquaporin | Plant | Yeast strain | Solute | Yeast assay | References |
|---|---|---|---|---|---|
| TIP1;1, TIP1;2 | Arabidopsis | Δtsa1,2, Δskn7, Δyap1, Δfps1, Δyfl054c | H2O2 | Spheroplast Swelling Assay, Survival test, Catalase Activity Assay, Fluorescence Assay, Bioimaging | Bienert et al., 2007 |
| AtNIP1;1, AtNIP2;1, AtNIP5;1,AtNIP6;1, AtNIP7;1 | Arabidopsis | W303-1A | As(III), As(V), antimonite | Yeast growth and survival test | Bienert et al., 2008 |
| AtPIP1;2, AtPIP2;3 | Arabidopsis | W303 | CO2 | Stopped-flow spectrometry | Heckwolf et al., 2011 |
| SIP1;1, SIP1;2, SIP2;1 | Arabidopsis | BJ5458 | H2O | Stopped-flow spectrometry | Ishikawa et al., 2005 |
| AtNIP7;1 | Arabidopsis | acr3Δ, fsp1Δ | As(III), As(V), | Yeast growth and survival test | Isayenkov and Maathuis, 2008 |
| HvNIP2;1 | Barley | INVSc2, SY1 | Boric acid, Ge, As | Yeast growth and survival test | Schnurbusch et al., 2010 |
| HvNIP1;1, HvNIP1;2, HvNIP2;1, HvNIP2;2 | Barley | ΔSKN7, ΔACR3 | H2O2, As(OH)3 | Yeast growth and survival test | Katsuhara et al., 2014 |
| VvTnPIP2;1, VvTnTIP1;1, VvTnTIP2;2, VvTnPIP1;4, VvTnPIP2;3, VvTnTIP4;1 | Grape | 10560-6B | H2O | Stopped-flow fluorescence spectroscopy | Sabir et al., 2014 |
| HaTIP1;1, HaPIP1;1, HaPIP1;2 | Helianthemum almeriense | SY1 | CO2, NH3, H2O | Stopped-flow spectrometry | Navarro-Ródenas et al., 2013 |
| VALT (TIP), PALT1 (PIP) | Hydrangea (Hydrangea macrophylla) | Δhsp150, BY4741 | Al | Yeast growth and survival test | Negishi et al., 2012 |
| LjNIP5;1, LjNIP6;1 | Lotus japonicus | W303-1A | As(III), As(V), antimonite | Yeast growth and survival test | Bienert et al., 2008 |
| LjNIP1 | Lotus japonicus | 31019b, YNVW1 | Amonia, Urea, H2O | Stopped-flow spectrometry, Yeast growth and survival test | Giovannetti et al., 2012 |
| ZmPIP1;2, ZmPIP2;5 | Maize | 31019b (Δmep1–3), BY4741 | H2O2 | Yeast growth and survival test, Codon optimization | Bienert et al., 2014 |
| PvTIP4;1 | Pteris vittata | Δfps1 and Δacr3 | As(III), As(V) | Yeast growth and survival test | He et al., 2016 |
| RsPIP1-2, RsPIP1-3, RsPIP2-1, RsPIP2-2 | Radish | BJ5458 | H20 | Stopped-flow spectrophotometry | Suga and Maeshima, 2004 |
| OsNIP1;1, OsNIP1;2, OsNIP2;1, OsNIP2;2, OsNIP3;1, OsNIP3;2, OsNIP3;3, OsNIP4;1 | Rice | ΔSKN7, ΔACR3 | H2O2, As(OH)3 | Yeast growth and survival test | Katsuhara et al., 2014 |
| OsNIP2;1, OsNIP2;2, OsNIP3;2 | Rice | W303-1A | As(III), As(V), antimonite | Yeast growth and survival test | Bienert et al., 2008 |
| TsTIP1;2 | Thellungiella salsuginea | aqy-null strain | H2O2 | Fluorescence assays, Yeast growth and survival test | Wang et al., 2014 |
| NtXIP1;1, StXIP1;1 | Tobacco | YNVW1 (Δdur3) | Urea | Yeast growth and survival test | Bienert et al., 2011 |
| TaTIP2;2 | Wheat | 31019b, BJ5458 | Ammonia | Stopped-flow spectrometry | Bertl and Kaldenhoff, 2007 |
Figure 3.
Yeast heterologous expression system used to evaluate aquaporins. Aquaporin cloned in the expression vector can be easily transferred in yeasts for different assays like growth and survival tests, protein localization, and/or stopped-flow spectrometry.
Apart from water, transport of many other solutes through AQPs is also studied with yeast systems. Commonly, yeast growth and survival are used to study solutes (Table 4). For instance, uptake of germanium (Ge), arsenate (As), boric acid and antimonite severely affect yeast growth, and this effect can be measured through heterologous expression of AQPs specific for such solutes. Similarly, mutant strains like YNVW1 carrying the deletion Δdur3 cannot grow on media with urea as sole nitrogen source, thus making such deletion strains useful to study urea transport by AQPs (Table 4).
Recently, To et al. (2015) demonstrated rapid screening of an AQP mutant library to evaluate the effects of amino acid changes on solute transport. The authors developed a novel method that looks promising to study water transport ability for hundreds of AQPs simultaneously. The assay can be used to identify inhibitors as well as co-transporting molecules. The method exploits the property of yeast cells that show increased freezing tolerance with expression of functional AQPs. The rapid transport of water through AQP allows removal of water from freezing yeast cells that avoid formation of ice crystals thus preventing cell damage. With this method, a library of yeasts (preferably AQP mutant strains) transformed with different AQPs can grow in 96-well microplates that are exposed to freeze-thaw cycles. Only the yeasts transformed with functional AQPs will survive following freeze-thaw cycles, a quick way to assess AQP properties. Such high-throughput procedures will certainly expand the analytical power required to integrate omics scale research.
Mesophyll protoplast assay
Mesophyll protoplasts can be easily obtained for several plant species (Shen et al., 2014). More particularly, the procedure for the evaluation of AQPs using Arabidopsis, tobacco and maize mesophyll protoplasts is well established (Yoo et al., 2007; Besserer et al., 2012; Ma et al., 2015). There are several methods for the delivery of macromolecules into protoplasts including, electroporation, microinjection, and PEG–calcium fusion method (Sade et al., 2009; Shen et al., 2014). Transient expression of AQPs in plant mesophyll protoplast allows efficient study of solute transport and also the subcellular localization of the protein. Recently, Wang et al. (2015) exploited Arabidopsis protoplasts for the transient expression of an AQP from maize (MzPIP2;1) tagged with a green fluorescent protein to confirm the plasma membrane specific localization. Similarly, Chevalier et al. (2014) used the mesophyll protoplast assay to study subcellular localization of several maize AQPs belonging to the ZmPIP1s and ZmPIP2s subfamilies. They observed efficient localization in plasma membrane only for ZmPIP2s when expressed alone in the mesophyll protoplasts. They further swapped transmembrane domain-3 along with the ER export diacidic motif to demonstrate its requirement in the localization. Another study conducted using tobacco protoplasts showed the role of phosphoinositides in modulating aquaporin activity (Ma et al., 2015). AQP gene ZmPIP2;4 from maize was transiently expressed in tobacco protoplasts to evaluate phosphoinositides effect on AQP expression and water flux (Ma et al., 2015). In spite of the rather routine application of the mesophyll protoplast assay, a high-throughput method for AQP evaluation using this assay has not been developed yet. Such an assay would allow large-scale exploitation of native rather than surrogate membranes. However, the major limitation with this technique is the requirement of highly skilled expertise to handle protoplasts, and the relatively low rate of success when experiments are replicated across different labs.
Isolated vesicle assay: right-side out and inside-out
Vesicles isolated from different types of tissues and cell types are being used for the evaluation of AQPs. The vesicles can be easily obtained following ultracentrifugation-based fractioning. After obtaining vesicles separated from the other organelles and cytoplasm, AQPs can be studied with stopped-flow fluorescence spectroscopy that measures shrinking/swelling of the vesicle. Dordas et al. (2000) used plasma membrane vesicles obtained from squash (Cucurbita pepo) roots to study the role of AQPs in boric acid transport. They used mercuric chloride and phloretin, a well-known non-specific transporter inhibitor, to conclude that boric acid permeation occurred both through proteinaceous channels and diffusion through the membrane.
Isolated inverted vesicles are useful to measure uptake inside the vesicle by efflux transporter. The reversed membrane vesicle is known as inside-out where the apoplastic side is inside the vesicle. The inside out vesicles are generally used for the evaluation of efflux transporters and energy dependent transporters. Initially, Palmgren et al. (1990) developed the method to prepare inside-out and right-side-out (apoplastic side out) vesicles using sugar beet (Beta vulgaris L.) leaves. They used freezing and thawing to turn vesicles inside-out and subsequently separated the inside-out and right-side-out by repeating the phase partition step. ATPase assay is used to verify the proportion of inside-out vesicles, since the ATPase active site is situated on the cytoplasmic side of the membrane, and only sealed, inside-out vesicles efficiently perform ATP-dependent H+ pumping (Palmgren et al., 1990). Similarly, Sutka et al. (2005) used the stopped-flow technique to measure water transport activity of tonoplast vesicles. They were able to conclude that most of the AQPs located in the tonoplast membrane are sensitive to HgCl2 and few of them are inhibited by pH. Their study expanded the use of isolated vesicle assays to the tonoplast-specific AQPs. However, the major drawback of the system resides in its labor intensive procedure required to isolate plasma membrane vesicle fractions.
Transgenic approaches used for aquaporin research
The heterologous expression of AQPs in plants represents a useful approach for functional evaluation of AQPs, even though it can sometimes lead to experimental artifacts. Indeed, transgenic approaches are often criticized over the use of constitutive or non-specific promoters that express AQPs in tissues where natural expression is not observed. In the case where a plant trait is governed by genotypic variations, complementation assays with a transgenic approach is considered more reliable since expression of the transgenes are evaluated under the same conditions. In addition to contributing to the functional annotation of novel AQPs, transgenic approaches are also being exploited to develop crop plants with agronomically important traits (Table S4). Currently, Arabidopsis and tobacco remain the preferred source for the heterologous expression of genes including AQPs. In addition, extensive resource of T-DNA insertion mutant libraries available for Arabidopsis makes it an obvious choice for the functional study of novel AQPs. As a matter of fact, mutants are available for most of the AQPs identified in the Arabidopsis genome, which facilitates evaluation of any native Arabidopsis AQP as well as their homologs in other plant species.
Most of the AQP transgenic studies have evaluated responses against abiotic stresses like high salinity, drought, and cold (Table S4). In a notable effort, Jang et al. (2007) analyzed the effect of expression of AtPIP1;4 and AtPIP2;5 in Arabidopsis and tobacco transgenic plants under various abiotic stress conditions. They also noticed a change in endogenous AQP genes with the over expression of AtPIP1;4 and AtPIP2;5 transgenes. Another study by Peng et al. (2007) reported that the overexpression of PgTIP1 from Panax ginseng in transgenic Arabidopsis plants led to enhanced tolerance against salt-stress and drought, but lowered cold acclimation ability. The contrasting effect of overexpression of an AQP on drought and cold is expected since altered water movement has a contrasting effect over these stresses. However, many contradictory results with the heterologous expression of AQPs in transgenic plants have been reported. For instance, Peng et al. (2008) observed a significantly lowered freezing tolerance with overexpression of Rhododendron catawbiense AQP (RcPIP2) in Arabidopsis. Similarly, with the overexpression of a PIP in transgenic tobacco, Aharon et al. (2003) achieved improvement in plant growth under favorable growth conditions but not under drought or salt stress. AQPs are also extensively studied for physiological parameters like CO2 conductance and photosynthesis efficiency (Table S4). For instance, Katsuhara and Hanba (2008) evaluated the effect of HvPIP2;1, cloned from barley for the transport of water and CO2 conductance in a transgenic rice. In a rare study with biotic stress, Vivancos et al. (2015) showed that expression of a wheat NIP2 in Arabidopsis conferred higher Si absorption and better protection against powdery mildew.
Conclusions
Enormous progress has been achieved to understand solute transport in plants over last two decades following the discovery of the first AQP. Currently, over 30 plant species have been analyzed for genome-wide identification of AQPs. These efforts have highlighted the distribution and evolution of AQPs in plants and have also defined the phylogeny of AQPs and subsequent categorization into sub-families and groups. The dataset of over 1000 well-characterized AQPs provided here will be helpful to maximize the use of query sequences in homology-based AQP identification and subsequent characterization. Access to AQPs originating from diverse species is important to insure the identification of the entire set of AQPs in a given genome as well as their proper classification. In addition, the ever-growing resources of transcriptomic data should be exploited to characterize AQPs and refine our understanding of their role in relation with their tissue-specific expression. Integration of omics approaches to complex molecular systems like AQP-mediated transport has been facilitated lately with the development of powerful computational tools. Computational predictions however must be supported and validated by functional studies. For this purpose, biological assays such as Xenopus oocytes and yeast systems are now well-established approaches used to study solute specificity and transport kinetics of AQPs. In addition, several novel AQPs identified with omics efforts have been functionally annotated through transgenic approaches that have highlighted their beneficial role. With the current concerns over water resources, it is clear that a better understanding of AQP-mediated transport system in plants can only lead to the development and management of plants better adapted to changing environmental conditions.
Author contributions
RD, HS, and RRB compiled the data, drew the conclusions and wrote the manuscript.
Funding
The project was funded by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC), the Agri-Innovation program Growing Forward 2, SaskCanola and Agriculture and Agri-Food Canada and the Canada Research Chairs Program to RRB.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01896/full#supplementary-material
Proposed flowchart procedure for genome-wide identification of AQPs in plant species.
Proteomic approaches applied to the study of plant aquaporins (AQPs).
Details of aquaporin tertiary structures revealed with different methods. The data were otained by search perfomed with Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) on August 1st 2016.
Online tools and servers available for the prediction of tertiary structures of transmembrane proteins including aquaporins.
List of selected studies describing the effect of heterologous expression of aquaporins in plants.
References
- Agre P. (2004). Aquaporin water channels (Nobel lecture). Angew. Chem. Int. Ed. Engil. 43, 4278–4290. 10.1002/anie.200460804 [DOI] [PubMed] [Google Scholar]
- Aharon R., Shahak Y., Wininger S., Bendov R., Kapulnik Y., Galili G. (2003). Overexpression of a plasma membrane aquaporin in transgenic tobacco improves plant vigor under favorable growth conditions but not under drought or salt stress. Plant Cell 15, 439–447. 10.1105/tpc.009225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderberg H. I., Kjellbom P., Johanson U. (2014). Annotation of Selaginella moellendorffii major intrinsic proteins and the evolution of the protein family in terrestrial plants. Evolution of transporters in plants Front. Plant Sci. 3:33 10.3389/fpls.2012.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariani A., Gepts P. (2015). Genome-wide identification and characterization of aquaporin gene family in common bean (Phaseolus vulgaris L.). Mol. Genet. Genomics 290, 1771–1785. 10.1007/s00438-015-1038-2 [DOI] [PubMed] [Google Scholar]
- Benga G. (2012). On the definition, nomenclature and classification of water channel proteins (aquaporins and relatives). Mol. Aspects Med. 33, 514–517. 10.1016/j.mam.2012.04.003 [DOI] [PubMed] [Google Scholar]
- Benga G. (2013). Comparative studies of water permeability of red blood cells from humans and over 30 animal species: an overview of 20 years of collaboration with Philip Kuchel. Eur. Biophys. J. 42, 33–46. 10.1007/s00249-012-0868-7 [DOI] [PubMed] [Google Scholar]
- Benga G., Popescu O., Borza V., Pop V. I., Muresan A., Mocsy I., et al. (1986a). Water permeability in human erythrocytes: identification of membrane proteins involved in water transport. Eur. J. Cell Biol. 41, 252–262. [PubMed] [Google Scholar]
- Benga G., Popescu O., Pop V. I., Holmes R. P. (1986b). p-(Chloromercuri) benzenesulfonate binding by membrane proteins and the inhibition of water transport in human erythrocytes. Biochemistry 25, 1535–1538. 10.1021/bi00355a011 [DOI] [PubMed] [Google Scholar]
- Benga O., Huber V. J. (2012). Brain water channel proteins in health and disease. Mol. Aspects Med. 33, 562–578. 10.1016/j.mam.2012.03.008 [DOI] [PubMed] [Google Scholar]
- Bertl A., Kaldenhoff R. (2007). Function of a separate NH 3-pore in aquaporin TIP2; 2 from wheat. FEBS Lett. 581, 5413–5417. 10.1016/j.febslet.2007.10.034 [DOI] [PubMed] [Google Scholar]
- Besserer A., Burnotte E., Bienert G. P., Chevalier A. S., Errachid A., Grefen C., et al. (2012). Selective regulation of maize plasma membrane aquaporin trafficking and activity by the SNARE SYP121. Plant Cell 24, 3463–3481. 10.1105/tpc.112.101758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biela A., Grote K., Otto B., Hoth S., Hedrich R., Kaldenhoff R. (1999). The Nicotiana tabacum plasma membrane aquaporin NtAQP1 is mercury-insensitive and permeable for glycerol. Plant J. 18, 565–570. 10.1046/j.1365-313X.1999.00474.x [DOI] [PubMed] [Google Scholar]
- Bienert G. P., Bienert M. D., Jahn T. P., Boutry M., Chaumont F. (2011). Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J. 66, 306–317. 10.1111/j.1365-313X.2011.04496.x [DOI] [PubMed] [Google Scholar]
- Bienert G. P., Heinen R. B., Berny M. C., Chaumont F. (2014). Maize plasma membrane aquaporin ZmPIP2; 5, but not ZmPIP1; 2, facilitates transmembrane diffusion of hydrogen peroxide. Biochim. Biophys. Acta 1838, 216–222. 10.1016/j.bbamem.2013.08.011 [DOI] [PubMed] [Google Scholar]
- Bienert G. P., Møller A. L., Kristiansen K. A., Schulz A., Møller I. M., Schjoerring J. K., et al. (2007). Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 282, 1183–1192. 10.1074/jbc.M603761200 [DOI] [PubMed] [Google Scholar]
- Bienert G. P., Thorsen M., Schüssler M. D., Nilsson H. R., Wagner A., Tamás M. J., et al. (2008). A subgroup of plant aquaporins facilitate the bi-directional diffusion of As (OH) 3 and Sb (OH) 3 across membranes. BMC Biol. 6:26. 10.1186/1741-7007-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousser A., Sissoëff I., Roche O., Hoarau J., Mahé A. (2003). Cloning and characterization of ZmPIP1-5b, an aquaporin transporting water and urea. Plant Sci. 165, 21–31. 10.1016/S0168-9452(03)00117-1 [DOI] [Google Scholar]
- Burgess D. J. (2016). Gene expression: a space for transcriptomics. Nat. Rev. Genet. 17, 436–437. 10.1038/nrg.2016.94 [DOI] [PubMed] [Google Scholar]
- Carpentier G. A., Garneau A. P., Marcoux A.-A., Noël M., Frenette-Cotton R., Isenring P. (2016). Identification of key residues involved in Si transport by the aquaglyceroporins. J. Gen. Physiol. 148, 239–251. 10.1085/jgp.201611598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F., Barrieu F., Jung R., Chrispeels M. J. (2000). Plasma membrane intrinsic proteins from maize cluster in two sequence subgroups with differential aquaporin activity. Plant Physiol. 122, 1025–1034. 10.1104/pp.122.4.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Yao Q., Patil G. B., Agarwal G., Deshmukh R. K., Lin L., et al. (2016). Identification and comparative analysis of differential gene expression in soybean leaf tissue under drought and flooding stress revealed by RNA-Seq. Front. Plant Sci. 7:1044. 10.3389/fpls.2016.01044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier A. S., Bienert G. P., Chaumont F. (2014). A new LxxxA motif in the transmembrane Helix3 of maize aquaporins belonging to the plasma membrane intrinsic protein PIP2 group is required for their trafficking to the plasma membrane. Plant Physiol. 166, 125–138. 10.1104/pp.114.240945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y., Mitani N., Yamaji N., Ma J. F. (2009). HvLsi1 is a silicon influx transporter in barley. Plant J. 57, 810–818. 10.1111/j.1365-313X.2008.03728.x [DOI] [PubMed] [Google Scholar]
- Daniels M. J., Chrispeels M. J., Yeager M. (1999). Projection structure of a plant vacuole membrane aquaporin by electron cryo-crystallography. J. Mol. Biol. 294, 1337–1349. 10.1006/jmbi.1999.3293 [DOI] [PubMed] [Google Scholar]
- Danielson J. A., Johanson U. (2008). Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 8:45. 10.1186/1471-2229-8-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R. M., Rivers R. L., Zeidel M. L., Roberts D. M. (1999). Purification and functional reconstitution of soybean nodulin 26. An aquaporin with water and glycerol transport properties. Biochemistry 38, 347–353. 10.1021/bi982110c [DOI] [PubMed] [Google Scholar]
- Deokar A., Tar'an B. (2016). Genome-wide analysis of the aquaporin gene family in chickpea (Cicer arietinum L.). Front. Plant Sci. 7:1802. 10.3389/fpls.2016.01802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh R., Bélanger R. R. (2016). Molecular evolution of aquaporins and silicon influx in plants. Funct. Ecol. 30, 1277–1285. 10.1111/1365-2435.12570 [DOI] [Google Scholar]
- Deshmukh R., Sonah H., Patil G., Chen W., Prince S., Mutava R., et al. (2014). Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 5:244. 10.3389/fpls.2014.00244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh R. K., Vivancos J., Guérin V., Sonah H., Labbé C., Belzile F., et al. (2013). Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Mol. Biol. 83, 303–315. 10.1007/s11103-013-0087-3 [DOI] [PubMed] [Google Scholar]
- Deshmukh R. K., Vivancos J., Ramakrishnan G., Guérin V., Carpentier G., Sonah H., et al. (2015). A precise spacing between the NPA domains of aquaporins is essential for silicon permeability in plants. Plant J. 83, 489–500. 10.1111/tpj.12904 [DOI] [PubMed] [Google Scholar]
- Di Giorgio J. A., Bienert G. P., Ayub N. D., Yaneff A., Barberini M. L., Mecchia M. A., et al. (2016). Pollen-specific aquaporins NIP4; 1 and NIP4; 2 are required for pollen development and pollination in Arabidopsis thaliana. Plant Cell 28, 1053–1077. 10.1105/tpc.15.00776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Pietro M., Vialaret J., Li G.-W., Hem S., Prado K., Rossignol M., et al. (2013). Coordinated post-translational responses of aquaporins to abiotic and nutritional stimuli in Arabidopsis roots. Molecular & Cellular Proteomics 12, 3886–3897. 10.1074/mcp.M113.028241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dordas C., Chrispeels M. J., Brown P. H. (2000). Permeability and channel-mediated transport of boric acid across membrane vesicles isolated from squash roots. Plant physiology 124, 1349–1362. 10.1104/pp.124.3.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H., Xia X., Fan X., Xu G., Miller A. J. (2013). Optimizing plant transporter expression in Xenopus oocytes. Plant Methods 9:48. 10.1186/1746-4811-9-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannetti M., Balestrini R., Volpe V., Guether M., Straub D., Costa A., et al. (2012). Two putative-aquaporin genes are differentially expressed during arbuscular mycorrhizal symbiosis in Lotus japonicus. BMC Plant Biol. 12:186. 10.1186/1471-2229-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grégoire C., Rémus-Borel W., Vivancos J., Labbé C., Belzile F., Bélanger R. R. (2012). Discovery of a multigene family of aquaporin silicon transporters in the primitive plant Equisetum arvense. Plant J. 72, 320–330. 10.1111/j.1365-313X.2012.05082.x [DOI] [PubMed] [Google Scholar]
- Gupta A. B., Sankararamakrishnan R. (2009). Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol. 9:134. 10.1186/1471-2229-9-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Yan H., Chen Y., Shen H., Xu W., Zhang H., et al. (2016). An aquaporin PvTIP4; 1 from Pteris vittata may mediate arsenite uptake. New Phytol. 209, 746–761. 10.1111/nph.13637 [DOI] [PubMed] [Google Scholar]
- Heckwolf M., Pater D., Hanson D. T., Kaldenhoff R. (2011). The Arabidopsis thaliana aquaporin AtPIP1; 2 is a physiologically relevant CO2 transport facilitator. Plant J. 67, 795–804. 10.1111/j.1365-313X.2011.04634.x [DOI] [PubMed] [Google Scholar]
- Higuchi T., Suga S., Tsuchiya T., Hisada H., Morishima S., Okada Y., et al. (1998). Molecular cloning, water channel activity and tissue specific expression of two isoforms of radish vacuolar aquaporin1. Plant Cell Physiol. 39, 905–913. 10.1093/oxfordjournals.pcp.a029453 [DOI] [PubMed] [Google Scholar]
- Hove R. M., Ziemann M., Bhave M. (2015). Identification and expression analysis of the barley (Hordeum vulgare L.) aquaporin gene family. PLoS ONE 10:e0128025. 10.1371/journal.pone.0128025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Hou X., Huang C., Yan Y., Tie W., Ding Z., et al. (2015). Genome-wide identification and expression analyses of aquaporin gene family during development and abiotic stress in banana. Int. J. Mol. Sci. 16, 19728–19751. 10.3390/ijms160819728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Li R., Zhang Z., Li L., Gu X., Fan W., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41, 1275–1281. 10.1038/ng.475 [DOI] [PubMed] [Google Scholar]
- Isayenkov S. V., Maathuis F. J. (2008). The Arabidopsis thaliana aquaglyceroporin AtNIP7; 1 is a pathway for arsenite uptake. FEBS Lett. 582, 1625–1628. 10.1016/j.febslet.2008.04.022 [DOI] [PubMed] [Google Scholar]
- Ishikawa F., Suga S., Uemura T., Sato M. H., Maeshima M. (2005). Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Lett. 579, 5814–5820. 10.1016/j.febslet.2005.09.076 [DOI] [PubMed] [Google Scholar]
- Jang J. Y., Lee S. H., Rhee J. Y., Chung G. C., Ahn S. J., Kang H. (2007). Transgenic Arabidopsis and tobacco plants overexpressing an aquaporin respond differently to various abiotic stresses. Plant Mol. Biol. 64, 621–632. 10.1007/s11103-007-9181-8 [DOI] [PubMed] [Google Scholar]
- Johansson I., Karlsson M., Shukla V. K., Chrispeels M. J., Larsson C., Kjellbom P. (1998). Water transport activity of the plasma membrane aquaporin PM28A is regulated by phosphorylation. Plant Cell 10, 451–459. 10.1105/tpc.10.3.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. S., Preston G. M., Smith B. L., Guggino W. B., Agre P. (1994). Molecular structure of the water channel through aquaporin CHIP. The hourglass model. J. Biol. Chem. 269, 14648–14654. [PubMed] [Google Scholar]
- Katsuhara M., Hanba Y. T. (2008). Barley plasma membrane intrinsic proteins (PIP aquaporins) as water and CO2 transporters. Pflügers Arch. 456, 687–691. 10.1007/s00424-007-0434-9 [DOI] [PubMed] [Google Scholar]
- Katsuhara M., Sasano S., Horie T., Matsumoto T., Rhee J., Shibasaka M. (2014). Functional and molecular characteristics of rice and barley NIP aquaporins transporting water, hydrogen peroxide and arsenite. Plant Biotechnol. 31, 213–219. 10.5511/plantbiotechnology.14.0421a [DOI] [Google Scholar]
- Kirscht A., Kaptan S. S., Bienert G. P., Chaumont F., Nissen P., de Groot B. L., et al. (2016). Crystal structure of an ammonia-permeable aquaporin. PLoS Biol. 14:e1002411. 10.1371/journal.pbio.1002411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen P., Day R. E., Salman M. M., Conner M. T., Bill R. M., Conner A. C. (2015). Beyond water homeostasis: diverse functional roles of mammalian aquaporins. Biochim. Biophys. Acta 1850, 2410–2421. 10.1016/j.bbagen.2015.08.023 [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Wolf Y. I., Karev G. P. (2002). The structure of the protein universe and genome evolution. Nature 420, 218–223. 10.1038/nature01256 [DOI] [PubMed] [Google Scholar]
- Lindahl E., Sansom M. S. (2008). Membrane proteins: molecular dynamics simulations. Curr. Opin. Struct. Biol. 18, 425–431. 10.1016/j.sbi.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Liu C., Fukumoto T., Matsumoto T., Gena P., Frascaria D., Kaneko T., et al. (2013). Aquaporin OsPIP1; 1 promotes rice salt resistance and seed germination. Plant Physiol. Biochem. 63, 151–158. 10.1016/j.plaphy.2012.11.018 [DOI] [PubMed] [Google Scholar]
- Loqué D., Ludewig U., Yuan L., von Wirén N. (2005). Tonoplast intrinsic proteins AtTIP2; 1 and AtTIP2; 3 facilitate NH3 transport into the vacuole. Plant Physiol. 137, 671–680. 10.1104/pp.104.051268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. F., Tamai K., Yamaji N., Mitani N., Konishi S., Katsuhara M., et al. (2006). A silicon transporter in rice. Nature 440, 688–691. 10.1038/nature04590 [DOI] [PubMed] [Google Scholar]
- Ma J. F., Yamaji N., Mitani N., Tamai K., Konishi S., Fujiwara T., et al. (2007). An efflux transporter of silicon in rice. Nature 448, 209–212. 10.1038/nature05964 [DOI] [PubMed] [Google Scholar]
- Ma X., Shatil-Cohen A., Ben-Dor S., Wigoda N., Perera I. Y., Im Y. J., et al. (2015). Do phosphoinositides regulate membrane water permeability of tobacco protoplasts by enhancing the aquaporin pathway? Planta 241, 741–755. 10.1007/s00425-014-2216-x [DOI] [PubMed] [Google Scholar]
- Maurel C., Boursiac Y., Luu D.-T., Santoni V., Shahzad Z., Verdoucq L. (2015). Aquaporins in plants. Physiol. Rev. 95, 1321–1358. 10.1152/physrev.00008.2015 [DOI] [PubMed] [Google Scholar]
- Mirzaei M., Pascovici D., Atwell B. J., Haynes P. A. (2012). Differential regulation of aquaporins, small GTPases and V-ATPases proteins in rice leaves subjected to drought stress and recovery. Proteomics 12, 864–877. 10.1002/pmic.201100389 [DOI] [PubMed] [Google Scholar]
- Movahedi S., Van Bel M., Heyndrickx K. S., Vandepoele K. (2012). Comparative co-expression analysis in plant biology. Plant Cell Environ. 35, 1787–1798. 10.1111/j.1365-3040.2012.02517.x [DOI] [PubMed] [Google Scholar]
- Murata K., Mitsuoka K., Hirai T., Walz T., Agre P., Heymann J. B., et al. (2000). Structural determinants of water permeation through aquaporin-1. Nature 407, 599–605. 10.1038/35036519 [DOI] [PubMed] [Google Scholar]
- Navarro-Ródenas A., Bárzana G., Nicolás E., Carra A., Schubert A., Morte A. (2013). Expression analysis of aquaporins from desert truffle mycorrhizal symbiosis reveals a fine-tuned regulation under drought. Mol. Plant-Microbe Interact. 26, 1068–1078. 10.1094/MPMI-07-12-0178-R [DOI] [PubMed] [Google Scholar]
- Negishi T., Oshima K., Hattori M., Kanai M., Mano S., Nishimura M., et al. (2012). Tonoplast-and plasma membrane-localized aquaporin-family transporters in blue hydrangea sepals of aluminum hyperaccumulating plant. PLoS ONE 7:e43189. 10.1371/journal.pone.0043189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren M. G., Askerlund P., Fredrikson K., Widell S., Sommarin M., Larsson C. (1990). Sealed inside-out and right-side-out plasma membrane vesicles optimal conditions for formation and separation. Plant Physiol. 92, 871–880. 10.1104/pp.92.4.871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos M. C., Verkman A. S. (2013). Aquaporin water channels in the nervous system. Nat. Rev. Neurosci. 14, 265–277. 10.1038/nrn3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil G., Valliyodan B., Deshmukh R., Prince S., Nicander B., Zhao M., et al. (2015). Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics 16:520. 10.1186/s12864-015-1730-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Arora R., Li G., Wang X., Fessehaie A. (2008). Rhododendron catawbiense plasma membrane intrinsic proteins are aquaporins, and their over-expression compromises constitutive freezing tolerance and cold acclimation ability of transgenic Arabidopsis plants. Plant Cell Environ. 31, 1275–1289. 10.1111/j.1365-3040.2008.01840.x [DOI] [PubMed] [Google Scholar]
- Peng Y., Lin W., Cai W., Arora R. (2007). Overexpression of a Panax ginseng tonoplast aquaporin alters salt tolerance, drought tolerance and cold acclimation ability in transgenic Arabidopsis plants. Planta 226, 729–740. 10.1007/s00425-007-0520-4 [DOI] [PubMed] [Google Scholar]
- Preston G. M., Carroll T. P., Guggino W. B., Agre P. (1992). Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science 256:385. 10.1126/science.256.5055.385 [DOI] [PubMed] [Google Scholar]
- Preston G. M., Jung J. S., Guggino W. B., Agre P. (1994). Membrane topology of aquaporin CHIP. Analysis of functional epitope-scanning mutants by vectorial proteolysis. J. Biol. Chem. 269, 1668–1673. [PubMed] [Google Scholar]
- Quigley F., Rosenberg J. M., Shachar-Hill Y., Bohnert H. J. (2001). From genome to function: the Arabidopsis aquaporins. Genome Biol. 3:research0001.0001–0001.0017. 10.1186/gb-2001-3-1-research0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuscher S., Akiyama M., Mori C., Aoki K., Shibata D., Shiratake K. (2013). Genome-wide identification and expression analysis of aquaporins in tomato. PLoS ONE 8:e79052. 10.1371/journal.pone.0079052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir F., Leandro M. J., Martins A. P., Loureiro-Dias M. C., Moura T. F., Soveral G., et al. (2014). Exploring three PIPs and three TIPs of grapevine for transport of water and atypical substrates through heterologous expression in aqy-null yeast. PLoS ONE 9:e102087. 10.1371/journal.pone.0102087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade N., Vinocur B. J., Diber A., Shatil A., Ronen G., Nissan H., et al. (2009). Improving plant stress tolerance and yield production: is the tonoplast aquaporin SlTIP2; 2 a key to isohydric to anisohydric conversion? New Phytol. 181, 651–661. 10.1111/j.1469-8137.2008.02689.x [DOI] [PubMed] [Google Scholar]
- Sakr S., Alves G., Morillon R., Maurel K., Decourteix M., Guilliot A., et al. (2003). Plasma membrane aquaporins are involved in winter embolism recovery in walnut tree. Plant Physiol. 133, 630–641. 10.1104/pp.103.027797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai G., Satake A., Yamaji N., Mitani-Ueno N., Yokozawa M., Feugier F. G., et al. (2015). In silico simulation modeling reveals the importance of the Casparian strip for efficient silicon uptake in rice roots. Plant Cell Physiol. 56, 631–639. 10.1093/pcp/pcv017 [DOI] [PubMed] [Google Scholar]
- Sakurai J., Ishikawa F., Yamaguchi T., Uemura M., Maeshima M. (2005). Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 46, 1568–1577. 10.1093/pcp/pci172 [DOI] [PubMed] [Google Scholar]
- Santoni V., Vinh J., Pflieger D., Sommerer N., Maurel C. (2003). A proteomic study reveals novel insights into the diversity of aquaporin forms expressed in the plasma membrane of plant roots. Biochem. J. 373, 289–296. 10.1042/bj20030159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schey K. L., Papac D. I., Knapp D. R., Crouch R. K. (1992). Matrix-assisted laser desorption mass spectrometry of rhodopsin and bacteriorhodopsin. Biophys. J. 63:1240. 10.1016/s0006-3495(92)81699-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler P. A., Van Dorsselaer A., Falick A. M. (1993). Analysis of hydrophobic proteins and peptides by electrospray ionization mass spectrometry. Anal. Biochem. 213, 256–263. 10.1006/abio.1993.1418 [DOI] [PubMed] [Google Scholar]
- Schnurbusch T., Hayes J., Hrmova M., Baumann U., Ramesh S. A., Tyerman S. D., et al. (2010). Boron toxicity tolerance in barley through reduced expression of the multifunctional aquaporin HvNIP2; 1. Plant Physiol. 153, 1706–1715. 10.1104/pp.110.158832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A., Downward J. (2001). Navigating gene expression using microarrays—a technology review. Nat. Cell Biol. 3, E190–E195. 10.1038/35087138 [DOI] [PubMed] [Google Scholar]
- Secchi F., Lovisolo C., Uehlein N., Kaldenhoff R., Schubert A. (2007). Isolation and functional characterization of three aquaporins from olive (Olea europaea L.). Planta 225, 381–392. 10.1007/s00425-006-0365-2 [DOI] [PubMed] [Google Scholar]
- Shen J., Fu J., Ma J., Wang X., Gao C., Zhuang C., et al. (2014). Isolation, culture, and transient transformation of plant protoplasts. Curr. Protoc. Cell Biol. 63, 2.8.1-2.8.17 10.1002/0471143030.cb0208s63 [DOI] [PubMed] [Google Scholar]
- Sonah H., Chavan S., Katara J., Chaudhary J., Kadam S., Patil G., et al. (2016). Genome-wide identification and characterization of Xylanase Inhibitor Protein (XIP) genes in cereals. Indian J. Genet. Plant Breed. 76, 159–166. 10.5958/0975-6906.2016.00036.5 [DOI] [Google Scholar]
- Song L., Nguyen N., Deshmukh R. K., Patil G. B., Prince S. J., Valliyodan B., et al. (2016). Soybean TIP gene family analysis and characterization of GmTIP1; 5 and GmTIP2; 5 in water transport. Front. Plant Sci. 7:1564. 10.3389/fpls.2016.01564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. K., Penna S., Nguyen D. V., Tran L.-S. (2016). Multifaceted roles of aquaporins as molecular conduits in plant responses to abiotic stresses. Crit. Rev. Biotechnol. 36, 389–398. 10.3109/07388551.2014.973367 [DOI] [PubMed] [Google Scholar]
- Suga S., Maeshima M. (2004). Water channel activity of radish plasma membrane aquaporins heterologously expressed in yeast and their modification by site-directed mutagenesis. Plant Cell Physiol. 45, 823–830. 10.1093/pcp/pch120 [DOI] [PubMed] [Google Scholar]
- Sun H., Li L., Lou Y., Zhao H., Gao Z. (2016). Genome-wide identification and characterization of aquaporin gene family in moso bamboo (Phyllostachys edulis). Mol. Biol. Rep. 43, 437–450. 10.1007/s11033-016-3973-3 [DOI] [PubMed] [Google Scholar]
- Sutka M., Alleva K., Parisi M., Amodeo G. (2005). Tonoplast vesicles of Beta vulgaris storage root show functional aquaporins regulated by protons. Biol. Cell 97, 837–846. 10.1042/BC20040121 [DOI] [PubMed] [Google Scholar]
- Takano J., Wada M., Ludewig U., Schaaf G., von Wirén N., Fujiwara T. (2006). The Arabidopsis major intrinsic protein NIP5; 1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18, 1498–1509. 10.1105/tpc.106.041640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S., Tan H. T., Chung M. (2008). Membrane proteins and membrane proteomics. Proteomics 8, 3924–3932. 10.1002/pmic.200800597 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Wallace I. S., Takano J., Roberts D. M., Fujiwara T. (2008). NIP6; 1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 20, 2860–2875. 10.1105/tpc.108.058628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanghe A., Van Dijck P., Thevelein J. M. (2006). Why do microorganisms have aquaporins? Trends Microbiol. 14, 78–85. 10.1016/j.tim.2005.12.001 [DOI] [PubMed] [Google Scholar]
- To J., Yeo C. Y., Soon C. H., Torres J. (2015). A generic high-throughput assay to detect aquaporin functional mutants: potential application to discovery of aquaporin inhibitors. Biochim. Biophys. Acta 1850, 1869–1876. 10.1016/j.bbagen.2015.05.019 [DOI] [PubMed] [Google Scholar]
- Törnroth-Horsefield S., Wang Y., Hedfalk K., Johanson U., Karlsson M., Tajkhorshid E., et al. (2006). Structural mechanism of plant aquaporin gating. Nature 439, 688–694. 10.1038/nature04316 [DOI] [PubMed] [Google Scholar]
- Uehlein N., Lovisolo C., Siefritz F., Kaldenhoff R. (2003). The tobacco aquaporin NtAQP1 is a membrane CO2 pore with physiological functions. Nature 425, 734–737. 10.1038/nature02027 [DOI] [PubMed] [Google Scholar]
- Venkatesh J., Yu J.-W., Park S. W. (2013). Genome-wide analysis and expression profiling of the Solanum tuberosum aquaporins. Plant Physiol. Biochem. 73, 392–404. 10.1016/j.plaphy.2013.10.025 [DOI] [PubMed] [Google Scholar]
- Verkman A. (2009). Aquaporins: translating bench research to human disease. J. Exp. Biol. 212, 1707–1715. 10.1242/jeb.024125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivancos J., Deshmukh R., Grégoire C., Rémus-Borel W., Belzile F., Bélanger R. R. (2016). Identification and characterization of silicon efflux transporters in horsetail (Equisetum arvense). J. Plant Physiol. 200, 82–89. 10.1016/j.jplph.2016.06.011 [DOI] [PubMed] [Google Scholar]
- Vivancos J., Labbé C., Menzies J. G., Bélanger R. R. (2015). Silicon-mediated resistance of Arabidopsis against powdery mildew involves mechanisms other than the salicylic acid (SA)-dependent defence pathway. Mol. Plant Pathol. 16, 572–582. 10.1111/mpp.12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-L., Chen A.-P., Zhong N.-Q., Liu N., Wu X.-M., Wang F., et al. (2014). The Thellungiella salsuginea tonoplast aquaporin TsTIP1; 2 functions in protection against multiple abiotic stresses. Plant Cell Physiol. 55, 148–161. 10.1093/pcp/pct166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Li Q., Lei Q., Feng C., Gao Y., Zheng X., et al. (2015). MzPIP2; 1: an aquaporin involved in radial water movement in both water uptake and transportation, altered the drought and salt tolerance of transgenic Arabidopsis. PLoS ONE 10:e0142446. 10.1371/journal.pone.0142446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M., Uehlein N., Proksch P., Kaldenhoff R. (2001). Characterization of two tomato aquaporins and expression during the incompatible interaction of tomato with the plant parasite Cuscuta reflexa. Planta 213, 550–555. 10.1007/s004250100533 [DOI] [PubMed] [Google Scholar]
- Yaneff A., Sigaut L., Marquez M., Alleva K., Pietrasanta L. I., Amodeo G. (2014). Heteromerization of PIP aquaporins affects their intrinsic permeability. Proc. Natl. Acad. Sci. U.S.A. 111, 231–236. 10.1073/pnas.1316537111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- Zhao F. J., Ago Y., Mitani N., Li R. Y., Su Y. H., Yamaji N., et al. (2010). The role of the rice aquaporin Lsi1 in arsenite efflux from roots. New Phytol. 186, 392–399. 10.1111/j.1469-8137.2010.03192.x [DOI] [PubMed] [Google Scholar]
- Zou Z., Gong J., An F., Xie G., Wang J., Mo Y., et al. (2015). Genome-wide identification of rubber tree (Hevea brasiliensis Muell. Arg.) aquaporin genes and their response to ethephon stimulation in the laticifer, a rubber-producing tissue. BMC Genomics 16:1001. 10.1186/s12864-015-2152-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Yang L., Gong J., Mo Y., Wang J., Cao J., et al. (2016). Genome-wide identification of Jatropha curcas aquaporin genes and the comparative analysis provides insights into the gene family expansion and evolution in Hevea brasiliensis. Front. Plant Sci. 7:395. 10.3389/fpls.2016.00395 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proposed flowchart procedure for genome-wide identification of AQPs in plant species.
Proteomic approaches applied to the study of plant aquaporins (AQPs).
Details of aquaporin tertiary structures revealed with different methods. The data were otained by search perfomed with Protein Data Bank (http://www.rcsb.org/pdb/home/home.do) on August 1st 2016.
Online tools and servers available for the prediction of tertiary structures of transmembrane proteins including aquaporins.
List of selected studies describing the effect of heterologous expression of aquaporins in plants.