Abstract
Background
Infection of plants by necrotizing pathogens can lead to the rapid and localized induction of a complex set of defense responses resulting in a restriction of pathogen growth and spread. Subsequently, an increase of plant resistance against a broad spectrum of pathogens is observed systemically. This plant immunity is known as Systemic Acquired Resistance. To identify components of the transduction pathway, we cloned and analysed the expression pattern of several mRNAs accumulating in cucumber plants after induction of Systemic Acquired Resistance.
Results
We tested on cucumber different compounds known to induce systemic acquired resistance. Among these, BTH (benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester) proved to be very effective. mRNA RT-PCR differential display was used to identify mRNA sequences induced 24 hours after the application of 10 μM BTH to cucumber plants. A cDNA library constructed from cucumber plants sprayed with 10 μM BTH was screened to get corresponding full length cDNAs. Among the identified cDNAs were those coding for a putative ras-related GTP-binding protein, a putative beta-1,4-N-Acetylglucosaminyltranferase III and a putative pathogenesis related protein. The time course of accumulation of the three corresponding mRNAs was analysed by northern blotting in plants treated by BTH or in plants infected by Colletotrichum lagenarium.
Conclusions
The mRNA RT-PCR differential display technique allowed the identification of three genes possibly involved in Systemic Acquired Resistance in cucumber. Pathogenesis-related proteins are known to be involved in plant defence against pathogens. GTP-binding protein and N-acetylglucosaminyltranferase III have been reported to be components of signal transduction pathways in mammals and plants.
Background
Infection of plants by necrotizing pathogens can lead to the induction of a complex set of defense responses resulting in a restriction of pathogen growth and spread. The infected leaves develop a hypersensitive response (HR), i.e., rapid, localized cell death occurring at the sites of pathogen entry [1]. Concomitant with the HR is the accumulation of salicylic acid and several classes of pathogenesis-related (PR) proteins, many of which exhibiting antimicrobial activity [2]. Subsequently, an enhancement of the plant defensive capacity against a broad spectrum of pathogens is observed. This resistance is expressed locally as well as in distal, uninfected tissues and can last for several weeks to months. It is known as Systemic Acquired Resistance or SAR [3].
Salicylic acid was identified as an important signal in the SAR transduction pathway. It was shown to accumulate during the onset of SAR in cucumber [4], tobacco [5], and Arabidopsis thaliana [6]. Exogenously supplied salicylic acid induces the same set of genes and resistance against the same spectrum of pathogens, as with pathogen-induced SAR [3]. Plants in which salicylic acid accumulation is prevented by over expression of a bacterial salicylate hydroxylase gene failed to develop SAR and/or exhibited increased susceptibility to pathogen infection [7,8].
Analysis of A. thaliana mutants has revealed that the NPR1 protein (also known as NIM1) is required for SAR induction and acts downstream of salicylic acid in the SAR pathway. Plants carrying mutations in this gene fail to express several PR genes and display enhanced susceptibility to infection [9,10]. The NPR1/NIM1 protein interacts with members of the TGA/OBF family of basic leucine zipper transcription factors. Some of these factors have been shown to bind to salicylic acid-responsive promoter elements of the PR-1 gene [11-13]. The salicylic acid induced interaction of NPR1 with TGA factors is localized in the nucleus [14], while cytoplasmic NPR1 appears to modulate crosstalk between salicylic acid- and jasmonate-dependent defense pathways [15]. Other components of the salicylic acid signaling pathway have been identified using genetic approaches in Arabidopsis thaliana [16]. They include PAD4 and EDS3 which activate EDS4, SID2 and EDS5 leading to salicylic acid accumulation [17].
Besides pathogens, SAR is also induced by exogenously applied chemicals. In addition to salicylic acid, well-known chemical inducers of PR genes include 2,6-dichloroisonicotinic acid (INA), benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester (BTH) and L-α-amino butyric acid (ABA) [18-23]. SAR induction appears to be governed by a complex signal transduction process [24] that involves a signal originating at the site of infection or treatment, and moving throughout the plant. Chemical inducers provide tools for dissection of the signal transduction pathway that regulates the defense responses (they enter the pathway at different points). On another hand, the cucumber plant provides a good model system for studying induced disease resistance [25]. Local and systemic increases in chitinase and/or peroxidase activity have been observed in response to inoculation with necrotizing pathogens or to treatment with SA, INA and BTH [26-28].
The aim of the present work was to identify and clone mRNAs accumulating after induction of SAR in cucumber plants. These sequences could correspond to proteins potentially involved in the signal transduction pathway leading to SAR or to proteins responsible for the state of resistance. In a first set of experiments, potential SAR inducing chemicals were tested in order to set up a reproducible system of activation of this resistance in cucumber. The mRNA RT-PCR differential display method [29] was then used to identify genes activated during SAR induction. Several differentially expressed cDNA were identified and some of them were cloned and sequenced. Their expression was analyzed by Northern blotting.
Results
Chemically induced resistance in cucumber
The SAR inducing activity of different chemicals has already been documented in one or several plant species [18-23]. To evaluate their activity on cucumber, cotyledons of two-week-old plants were infiltrated with 0.5 mM salicylic acid, 50 mM L-α-amino butyric acid, 0.78 mM 2-thiouracil, 1 mM thiamine or 20 mM BaCl2. Control plants were infiltrated with water. In a parallel experiment, four-week-old cucumber plants (with two true leaves) were sprayed with 5 mM salicylic acid, 50 mM L-α-amino butyric acid, 0.78 mM 2-thiouracil, 1 mM thiamine, 20 mM BaCl2 or 1 mM BTH. Control plants were sprayed with water or left untreated. Total RNAs were extracted from treated leaves 6 h, 24 h or 48 h after treatment. Induction of PR-8 gene was assayed by Northern blotting followed by hybridization with a cucumber PR-8 cDNA probe. PR-8 cucumber gene encodes an acidic chitinase which accumulates after tobacco necrosis virus (TNV) infection or salicylic acid induction and is considered as a SAR molecular marker [30].
In infiltration experiments (results not shown), control plants showed positive hybridization signals. To check whether this activation of the PR-8 gene could be due to wounding during infiltration, the effect of wounding alone was evaluated by making a cut from the midvein to the outer edge of the cotyledons. A strong signal was obtained after wounding which could be explained by induction of the acidic chitinase itself or of an isoform. Therefore infiltration did not prove to be an appropriate method for applying SAR inducers in cucumber, because it probably induced a wound response.
When applied by spraying, BTH, L-α-amino butyric acid and 2-thiouracil strongly induced PR-8 mRNA accumulation, while SA treatment led to a lower induction level (Fig. 1). Thiamine and BaCl2 induced slightly PR-8 expression (results not shown). RNA from control plants did not show any hybridization signal indicating that spraying itself did not induce PR-8 expression. BTH, which is an analogue of salicylic acid, showed the strongest PR-8 induction. But cucumber plants treated with 1 mM BTH showed a reduced growth (smaller leaves area and shorter internodes). As BTH was shown to be a very efficient activator of disease resistance in different plant species [20-23], we tried to induce SAR in cucumber plants with lower BTH concentrations (10 μM, 30 μM and 100 μM). PR-8 transcript accumulation was quite similar for the three concentrations (see Additional file 1) and no adverse effect on plant growth was observed.
Figure 1.
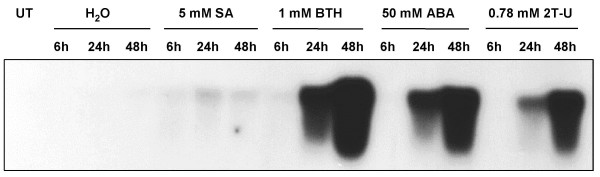
Time course of PR-8 mRNA accumulation in cucumber leaves in response to chemical treatments. Total RNA was extracted from cucumber leaves at various time after spraying with water, 5 mM salicylic acid, 1 mM BTH, 50 mM L-α-amino butyric acid (ABA), 0.78 mM 2-thiouracil (2T-U) or from untreated leaves (UT). RNA (10 μg) was fractionated by electrophoresis on agarose gel. Northern blot was probed with α-32P labeled PR-8 cDNA (loading of equal amounts of RNA was confirmed by ethidium bromide staining).
In cucumber, SAR has been shown to be effective against anthracnose caused by Colletotrichum lagenarium and salicylic acid treatment was shown to induce resistance against this pathogen [31]. To confirm that BTH acts as a chemical inducer of SAR in cucumber, four-week-old plants were sprayed with water, 10 μM BTH, 50 μM BTH, or left untreated, 5 days prior to inoculation of the second true leaf with C. lagenarium. Two weeks after inoculation, the plants were scored for spreading lesions, i.e. lesions showing a larger size than the initial inoculated area. As shown in figure 2 and table 1, BTH treatment resulted in decreased symptoms. Control leaves inoculated with C. lagenarium showed large spreading lesions and thus extensive fungal growth. Treatment of plants with BTH prior to C. lagenarium inoculation resulted in a markedly decreased lesion formation (Fig. 2). In untreated plants and water treated plants, 67 and 64 % of inoculation sites showed spreading lesions respectively (Table 1). In BTH treated plants, only 23 % (10 μM BTH) or 9 % (50 μM BTH) of inoculated sites developed spreading lesions (Table 1).
Figure 2.

Local acquired resistance induced by 10 μM or 50 μM BTH. Four-week-old cucumber plants (with two true fully expanded leaves) were sprayed with H2O, 10 μM BTH, 50 μM BTH or left untreated. The second true leaf was infected with C. lagenarium 5 days later. Plants were photographed 2 weeks postinoculation.
Table 1.
Local acquired resistance induced by 10 μM or 50 μM BTH.
| untreated | H2O | 10 μM BTH | 50 μM BTH | |
| Exp.1 | 66 ± 24 % | 57 ± 25 % | 12 ± 9 % | 13 ± 11 % |
| Exp.2 | 68 ± 28 % | 71 ± 23 % | 34 ± 30 % | 5 ± 6 % |
| Mean value | 67 % | 64 % | 23 % | 9 % |
Four-week-old cucumber plants (with two true fully expanded leaves) were sprayed with H2O, 10 μM BTH, 50 μM BTH or left untreated. The second true leaf was infected with C. lagenarium 5 days later (ten infection sites per leaf). The percentages of spreading lesions (i.e. lesions showing a larger size than the initial inoculated area) observed 2 weeks after inoculation with C. lagenarium were calculated on the basis of 14 plants/treatment (experiment 1) and 16 plants/treatment (experiment 2) (means +/- standard deviations).
To check whether BTH is able to act systemically or is transported through cucumber plants, the third leaf of four-week-old plants was wrapped with a plastic film and the plants were sprayed with H2O, 10 μM BTH, 50 μM BTH or left untreated. This leaf was inoculated with C. lagenarium 7 days later. The percentage of spreading lesions on third leaves from pathogen-challenged untreated or H2O treated cucumber plants (infected controls) were respectively 86% and 83.5%. In BTH treated plants, 38 % (10 μM BTH) or 12 % (50 μM BTH) of inoculated sites developed spreading lesions (Table 2). These results suggest that BTH can activate resistance in untreated leaves.
Table 2.
Systemic acquired resistance induced by 10 μM or 50 μM BTH.
| untreated | H2O | 10 μM BTH | 50 μM BTH | |
| Exp.1 | 76 ± 20 % | 69 ± 17 % | 37 ± 19 % | 18 ± 16 % |
| Exp.2 | 96 ± 6 % | 98 ± 3 % | 39 ± 15 % | 7 ± 9 % |
| Mean value | 86 % | 83.5 % | 38 % | 12,5 % |
Four-week-old cucumber plants (with two true fully expanded leaves) were sprayed with H2O, 10 μM BTH, 50 μM BTH or left untreated. The third true leaf was infected with C. lagenarium 7 days later (ten infection sites per leaf). The percentages of spreading lesions (i.e. lesions showing a larger size than the initial inoculated area) observed 2 weeks after inoculation with C. lagenarium were calculated on the basis of 18 plants/treatment (experiment 1) and 17 plants/treatment (experiment 2) (means +/- standard deviations).
Differential expression of genes in BTH induced cucumber plants
Plants were sprayed with 10 μM BTH or water. Sampling was performed 24 h after treatment in order to point out early-induced differences in gene expression. RT-PCR products were separated on polyacrylamide gels. Bands that appeared on the display of RNA only from BTH treated plants, or only from water treated plants were likely to correspond to differentially expressed mRNAs. Twelve differentially expressed bands were identified (Table 3). Their sizes ranged between 250 and 450 bp. They corresponded to genes expressed in the BTH treated plants and not in the control ones or presenting a higher expression in the BTH treated plants compared to the control ones (Fig. 3). These bands were eluted from the gel, PCR amplified and cloned in the pGEM-T vector. In order to confirm the differential expression of these genes, reverse slot blot hybridizations were performed: DNA from four to ten clones obtained from each differentially displayed band was fixed on a membrane and hybridized with the corresponding reverse transcribed products from either BTH or H2O treated plants (Fig. 4). Six from the twelve fragments were confirmed to be differentially expressed: dd3, dd4, dd5, dd6, dd7 and dd11. Five bands (dd1, dd8, dd9, dd10 and dd12) were either expressed evenly between BTH and H2O treated plants (differential display false positives or signal masked by the expression of homologous genes). In the remaining case (dd2), no signal could be observed on the slot blots. This absence of hybridization signal could be due to a lack of sensitivity of the slot blot hybridization procedure.
Table 3.
Oligonucleotide primers used in RT-PCR that allowed to amplify the differentially displayed cDNA fragments.
| Clone name | Downstream primer used | Upstream primer used |
| dd1 | 5' T11AA | 5' CTGCTTGATG |
| dd2 | 5' T11CG | 5' GATCAATCGC |
| dd3 | 5' T11CG | 5' GATCAATCGC |
| dd4 | 5' T11CG | 5' TCGGTCATAG |
| dd5 | 5' T11GA | 5' AAACTCCGTC |
| dd6 | 5' T11GA | 5' TTTTGGCTCC |
| dd7 | 5' T11GA | 5' GATCTAACCG |
| dd8 | 5' T11GA | 5' GATCAATCGC |
| dd9 | 5' T11GA | 5' GATCAATCGC |
| dd10 | 5' T11GA | 5' TCGATACAGG |
| dd11 | 5' T11GA | 5' TACAACGAGG |
| dd12 | 5' T11GG | 5' TACAACGAGG |
Figure 3.
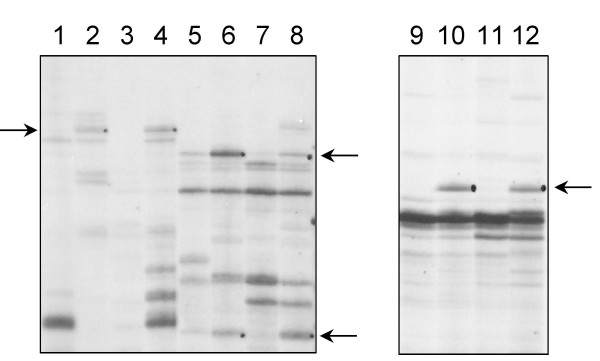
Zoom in on an auto-radiogram of differentially displayed RT-PCR products. The RT-PCR were performed with primers 5'T11GA and 5'GATCTAACCG (lane 1 to 4), primers 5'T11GA and 5'GATCAATCGC (lane 5 to 8), primers 5'T11GA and 5'TACAACGAGG (lanes 9 to 12). Uneven lanes: H2O treated plants. Even lanes: 10 μM BTH treated plants. The arrows show bands corresponding to transcripts accumulating only or presenting a higher expression in BTH treated plants.
Figure 4.
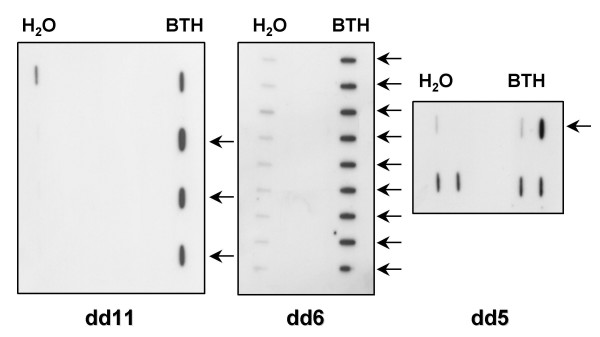
Slot blotting of PCR amplified fragments identified in differential display experiments. PCR amplified DNA from four dd11 clones, nine dd6 clones and four dd5 clones respectively were hybridized with the 32P labeled RT-PCR corresponding products from either H2O or BTH treated plants. The arrows show DNA from clones containing differentially displayed fragments.
The dd3, dd4, dd5, dd6, dd7 and dd11 cDNA fragments were sequenced. Blast analysis did not lead to identification of any homology with known sequences. This is not surprising as mRNA differential display generates fragments corresponding to 3'untranslated regions for which homologies are difficult to find. Two of the six fragments were proved to be part of the same gene (dd5 and dd7). A cDNA library was constructed from cucumber plants sprayed with 10 μM BTH (leaves were sampled 24 h after treatment). The library was screened with the five differentially expressed fragments. Hits were obtained for four of these probes (corresponding to dd3, dd4, dd6 and dd11). Full length cDNAs corresponding to dd4, dd6 and dd11 were obtained. Homology between cloned cDNA and sequences in databases is presented in table 4. The cDNA dd3 matched to a putative RAS-related GTP-binding protein from A. thaliana. It will be referred to as CRG (cucumber RAS-related GTP-binding protein). The cDNA dd4 matched to a translation releasing factor 2. It will be referred to as CTR (cucumber translation releasing factor 2). The cDNA dd6 matched to a putative β-1,4-N-Acetylglucosaminyltranferase III from A. thaliana. This protein was named CGT (cucumber acetylglucosaminyltransferase III). The cDNA dd11 matched to a putative pathogenesis related protein from A. thaliana and, with slightly less homology, to a pathogenesis related protein from barley induced by fungal infection [32]. It will be referred to as CPR (cucumber pathogenesis-related protein).
Table 4.
Characteristics of isolated cDNA clones.
| Clone name | Clone length (bp) | Accession N° | Matching sequence from data base | Origin of matching sequence and accession number | Amino acid sequence % identity (*) |
| dd3 | 774 | AJ634908 | Ras-related GTP-binding protein | A. thaliana NP 177505 | 65 % (1e-73) |
| dd4 | 1533 | AJ635428 | Translation releasing factor 2 | A. thaliana NP 851097 | 70 % (7e-170) |
| dd6 | 2192 | AJ629867 | β-1,4-N-acetylglucosaminyltranferase | A. thaliana NP 172759 | 77 % (0.0) |
| dd11 | 1588 | AJ629866 | Pathogenesis-related protein | A. thaliana NP 565189 | 51 % (1e-51) |
* expected value of best match
PR-8, CPR, CRG and CGT expression analysis
To investigate the expression of PR-8, CPR, CRG and CGT genes in cucumber, the two first leaves of four-week old plants were sprayed with 10 μM BTH. In another batch of plants, the first leaf was inoculated with C. lagenarium. Total RNA was extracted from treated leaves or upper untreated leaves at various times after treatment. Accumulation of the PR-8, CPR, CRG and CGT mRNAs was assayed by Northern blotting followed by hybridization with the corresponding cDNA probes (fig. 5).
Figure 5.
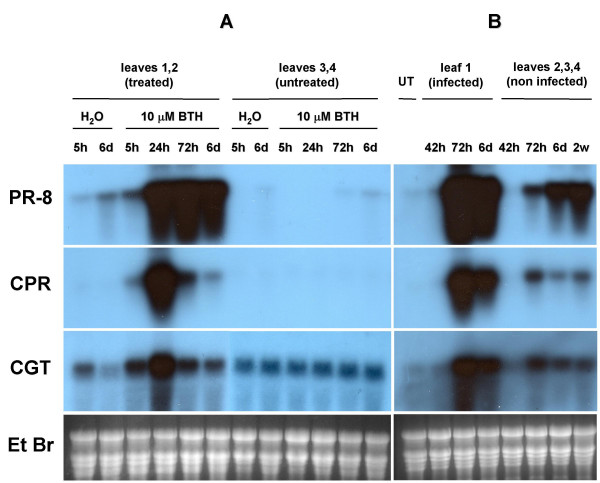
Time course of mRNA accumulation in cucumber leaves after BTH treatment (A) or C. lagenarium inoculation (B). A: Total RNA was extracted from cucumber leaves at various time after application of 10 μM BTH or water (mock treatment). RNA (15 μg) from leaves 1 and 2 (treated) and leaves 3 and 4 (untreated) was fractionated by electrophoresis on agarose gels. Identical Northern blots were probed with 32P-labeled PR-8, CPR and CGT cDNAs. B: Total RNA was extracted from cucumber leaves at various time after C. lagenarium inoculation. RNA (15 μg) from untreated leaves (UT), leaves 1 (infected) and leaves 2, 3 and 4 (non infected) was fractionated by electrophoresis on agarose gels. Identical Northern blots were probed with 32P-labeled PR-8, CPR and CGT cDNAs. Equal loading of RNA was confirmed by ethidium bromide (Et Br) staining. A representative gel is shown.
Expression of PR-8 gene was induced at a high level by both 10 μM BTH spraying and C. lagenarium infection in the leaves submitted to the treatment. PR-8 mRNA could be detected as soon as 5 h after BTH application and a maximum of expression could be observed after 72 h. PR-8 expression was observed as from 72 h after inoculation with C. lagenarium. No significant PR-8 mRNA accumulation could be detected in the untreated leaves from BTH sprayed plants, while in non infected leaves PR-8 expression was observed 72 h after infection by C. lagenarium; the level of expression was still increasing 2 weeks after C. lagenarium inoculation.
CPR gene expression was strongly induced in both 10 μM BTH treated and C. lagenarium inoculated leaves. CPR mRNA could be detected as soon as 5 h after BTH application and a maximum of expression could be observed after 24 h. CPR expression was observed 72 h after infection by C. lagenarium. No expression of CPR gene could be observed in untreated leaves from BTH sprayed plants, while CPR mRNA accumulation could be detected in uninfected leaves from C. lagenarium infected plants 72 h after inoculation.
CGT gene showed a significant basal level of expression as it could be observed in control plants (fig. 5). Nevertheless the expression of CGT was increased both by spraying with 10 μM BTH (with a maximum at 24 h) and by inoculation with C. lagenarium (with a maximum at 72 h) in the leaves submitted to the treatment. A systemic induction could also be observed in untreated leaves from C. lagenarium inoculated plants (as from 72 h after infection).
The expression level of the CRG gene was too low to allow us to show an induction of the expression (data not shown).
Expression of the same genes was analyzed in wounded cucumber plants. A cut was made from the midvein to the outer edge of the first leaf. Total RNA was extracted from the cut leaf or the upper leaf, 24 and 48 h after wounding. An induction of the expression of PR-8 and CPR genes was only observed in the cut leaf 24 h after wounding. No expression was observed later nor in the uncut upper leaf. The CGT and CRG genes were not induced by wounding (results not shown).
Expression of these genes was also analyzed in cucumber plants showing rhizobacteria-induced systemic resistance. Plants were grown in compost drenched with a Pseudomonas putida BTP1 suspension, a plant growth-promoting rhizobacteria known to promote induced systemic resistance in cucumber [33]. Total RNA was extracted from cotyledons collected three weeks after sowing. PR-8, CPR, CGT and CRG did not show any induction of expression in these conditions (results not shown).
Discussion
We tested here two different techniques and several compounds to induce SAR in cucumber. The expression of PR-8 gene, which encodes an acidic chitinase from cucumber, was used as an indicator of the activation of the defense response. The accumulation of this acidic chitinase after tobacco necrosis virus infection or salicylic acid induction correlates with induced resistance [30]. Among the different compounds tested, BTH applied by spraying proved to be the stronger inducer of PR-8 mRNA accumulation. BTH sprayed at a concentration of 10 μM was still able to induce PR-8 gene expression. The protective effects of BTH on cucumber were confirmed in challenge experiments with C. lagenarium. Spraying cucumber plants with BTH (10 μM or 50 μM) allowed effective local and systemic protection against C. lagenarium. BTH was recently identified as a safe, reliable and non phytotoxic plant protection agent by scientists at Novartis. Exogenous application of BTH to tobacco, A. thaliana, wheat and cucumber has been shown to activate a number of SAR associated genes leading to enhanced plant protection against various pathogens [20-22,28].
Identifying SAR induced genes could provide clues to elucidate the signal transduction pathway leading to plant defense responses. We used RT-PCR differential display in BTH-treated cucumber plants to detect SAR associated mRNAs. We have identified four BTH-inducible genes: a putative ras-related GTP binding protein (CRG), a putative translation releasing factor 2 (CTR), a putative β-1,4-N-acetylglucosaminyltranferase III (CGT) and a putative pathogenesis related protein (CPR). Expression of PR-8, CPR and CGT genes was shown to be induced by 10 μM BTH treatment as well as C. lagenarium infection. The response to BTH was observed earlier (5 h after treatment) than the one developed after pathogen inoculation (72 h after infection). However, systemic expression of these genes was only observed in C. lagenarium infected plants.
Small GTP-binding proteins form a large family of nucleotide triphosphatases whose activity is related to the binding, hydrolysis and release of guanosine triphosphate [34]. MAP kinase cascade represents an important downstream effector pathway for RAS in most cells [35] and recent studies have shown that MAP kinases are activated in plants in response to pathogen attack and wounding [36,37]. Small GTP-binding proteins have been shown to be involved in important cell mechanisms like cell division, transduction of external signals across the plasma membrane, endocytosis and exocytosis, cell death in plants and establishment of plant defense reaction [34,38-42]. The β-1,4-N-Acetylglucosaminyltranferase III catalyses the addition of N-acetylglucosamine to the β-mannoside of the tri-mannose core in N-glycans, resulting in the suppression of further processing and elongation. In mammals, N-acetylglucosaminyltransferase III expression is associated with differentiation, cell adhesion and tumor progression [43,44]. It was shown recently to interfere with epidermal growth factor signaling and H2O2-induced activation of protein kinase C [45,46]. N-linked glycans were shown to be widely distributed in plants and highly expressed at the cell surface, which might suggest a putative function in cell/cell communication [47]. To our knowledge β-1,4-N-Acetylglucosaminyltranferase has never been described in the context of a plant-pathogen interaction. Several cDNAs or gene encoding enzymes involved in the biosynthesis of complex N-linked glycans have been cloned from plants. They include N-acetylglucosaminyltranferase I cDNAs from A. thaliana, potato and tobacco [48], N-acetylglucosaminyltransferase II cDNA from A. thaliana [49] and defense-related glucosyl transferase gene from tomato [50]. GTP-binding proteins and N-acetylglucosaminyltranferases are involved in important mechanisms, notably signaling pathways and therefore seem of high interest. The putative PR protein, which has been first identified in barley after fungal infection [32] has no known function.
Only PR-8 and CPR genes were induced by wounding but not systemically. PR-8 and CPR genes were not induced by P. putida BTP1 treatment, which triggers induced systemic resistance (ISR) in cucumber. ISR is phenotypically similar to SAR but the mechanisms of this resistance were shown to be different: although this was not tested with P. putida BTP1, in other experimental systems ISR was shown to be dependent on NPR/NIM1 function, but does not involve salicylic acid, nor PR protein accumulation [51]. Our results correlate with those of Pieterse et al. who showed that induced systemic resistance in A. thaliana was independent of PR genes expression [51]. In the same way, ISR also seems to be independent on CGT and CRG expression, as these genes were not induced by P. putida BTP1.
Conclusions
Among different SAR inducers tested on cucumber, BTH proved to be very efficient. Applied by spraying, BTH induced PR-8 mRNA accumulation and allowed effective local and systemic protection against C. lagenarium. Using RT-PCR differential display, we identified four BTH-inducible genes, including a putative ras-related GTP binding protein (CRG), a translation releasing factor 2 (CTR), a putative β-1,4-N-acetylglucosaminyltranferase III (CGT) and a putative pathogenesis related protein (CPR). Genes expression studies confirmed the local induction of CGT and CPR after BTH treatment or C. lagenarium infection and their systemic induction in response to C. lagenarium infection. Moreover CPR was locally induced by wounding. The significant level of expression of CGT and CPR genes during defense responses suggests a potential role for the gene products in the SAR pathway or in the state of resistance in cucumber.
Methods
Organisms and growth conditions
Cucumber plants (Cucumis sativus cv. Marketer) were grown in compost at 22°C with a 16 h light photoperiod.
Cultures of Colletotrichum lagenarium were maintained on malt agar at 25 °C. Spores suspensions in water supplemented with 0.01 % tween 80 were prepared from four- to six-week-old cultures grown in petri plates. Suspensions were filtered through cheesecloth and the spore concentration determined with a hemacytometer.
Chemical treatments
All chemicals were obtained from Sigma except for BTH (provided by Novartis). Chemicals were dissolved in water and the pH adjusted to 6.6 with NaOH except for thiamine, BaCl2 and BTH. Infiltration experiments: cotyledons of two-week-old plants were infiltrated through a vein until the whole leaf was impregnated with the solution. Spraying experiments: the two first true leaves of four-week-old cucumber plants were sprayed with +/- 3 ml solution/plant. The upper leaves were protected with a plastic sheet when systemic resistance was to be tested.
ISR induction
Prior to planting, disinfected cucumber seeds were soaked for 10 min in a Pseudomonas putida BTP1 suspension (4.108 CFU/ml in 0.85% NaCl). Control seeds were soaked in 0.85% NaCl. Cucumber seeds were sown in 10 cm-pots containing sterilized potting soil previously mixed with a P. putida BTP1 inoculum to a final concentration of 3.107 CFU/g or with an equal volume of sterile water for control plants. Cucumber were germinated at 25°C with a 16 h light photoperiod. Six and 12 days after sowing, 20 ml of a bacterial suspension at 108 CFU/ml were added as a drench to the roots. Control plants were watered with 20 ml of 0.85% NaCl.
C. lagenarium infection
Plants were placed in a humidity chamber 48 hours before infection experiments. The first true leaf was inoculated with 10 drops (10 μl) of a conidial suspension of C. lagenarium (106 spores/ml). In challenge experiments, inoculation was performed 5 (or 7) days after chemical treatment on the second or third leaf.
Plant RNA extraction and analysis
The extraction of total RNA from cucumber leaves (5 plants per treatment) was performed using a phenol/SDS method [52]. Northern hybridization of RNA fractionated by agarose-formaldehyde gel electrophoresis [52] was performed with a α-32P PR-8, CPR, CGT or CRG cDNA probe (Random Primed DNA labeling kit, Roche).
mRNA Differential Display
The mRNA differential display was performed using the differential display kit from Eurogentec (Belgium) and following the instructions of the supplier. PCR products were labeled with α-35S dATP, separated by electrophoresis on denaturing 6% polyacrylamide gels and visualized by autoradiography. PCR reactions showing differentially displayed bands were repeated twice on two different RNA samples in order to reduce the number of false-positives.
Cloning and sequencing
Differentially displayed bands were excised and eluted during 30 min in 100 μl of H2O followed by boiling during 10 min, then precipitated and suspended in 10 μl of H2O. A second PCR amplification was performed on 4 μl of DNA following the same protocol than for the differential display itself. Products were cloned in pGEM-T vector (Promega) and manually sequenced by the dideoxy sequencing method. Available public databases (i.e. EMBL) were searched for homology with our sequences using the GCG software package (Blastall, Fasta).
Slot-Blot hybridisation
PCR amplified DNA from four to ten clones of each differentially displayed band was blotted on a positively charged nylon membrane using a filtration manifold and following the instructions of the supplier (Hoefer). Blots were hybridized with the 32P labeled RT-PCR corresponding products from either BTH or H2O treated plants.
cDNA library construction and screening
mRNA from cucumber plants sprayed with 10 μM BTH (leaves were sampled 24 h after treatment) were purified with the PolyATractmRNA Isolation System III from Promega. The cDNA library was performed with the SMART cDNA Library Construction Kit from Clontech following the instructions of the supplier for cDNA synthesis by primer extension. The library was screened with 32P labeled cDNA probes. cDNA sequencing was performed at Genome Express, France. Available public databases (i.e. EMBL) were searched for homology with our sequences using the GCG software package (Blastall, Fasta).
Authors' contributions
CB carried out the plants treatments, the molecular biology studies and the database research and drafted the manuscript. MO supplied the pathogen and participated in the challenge and ISR induction experiments. JD and PT participated in the design and coordination of the study. All authors read and approved the manuscript.
Supplementary Material
PR-8 mRNA accumulation in cucumber leaves in response to different concentrations of BTH. Total RNA was extracted from cucumber leaves 24 h and 48 h after spraying with water, 10 μM BTH, 30 μM BTH, 100 μM BTH or from untreated leaves (UT). RNA was fractionated by electrophoresis on agarose gel. Northern blot was probed with α-32P labeled PR-8 cDNA. Loading of equal amounts of RNA was confirmed by ethidium bromide (Et Br) staining.
Acknowledgments
Acknowledgements
This work was supported by a grant from the National Funds for Scientific Research (FNRS, Belgium) (Program FRFC n° 2.4.570.00).
Contributor Information
Catherine Bovie, Email: Catherine.Bovie@ulg.ac.be.
Marc Ongena, Email: ongena.m@fsagx.ac.be.
Philippe Thonart, Email: P.Thonart@ulg.ac.be.
Jacques Dommes, Email: J.Dommes@ulg.ac.be.
References
- Heath MC. Hypersensitive response related death. Plant Mol Biol. 2000;44:321–334. doi: 10.1023/A:1026592509060. [DOI] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Wlilliams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Métraux J-P, Ryals JA. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klessig DF, Malamy J. The salicylic acid signal in plants. Plant Mol Biol. 1994;26:1439–1458. doi: 10.1007/BF00016484. [DOI] [PubMed] [Google Scholar]
- Métraux J-P, Signer H, Ryals J, Ward E, Wyss-Benz M, Gaudin J, Rascdorf K, Schmid E, Blum W, Inverardi B. Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science. 1990;250:1004–1006. doi: 10.1126/science.250.4983.1004. [DOI] [PubMed] [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I. Salicylic acid : A likely endogenous signal in the resistance response of tobacco to viral infection. Science. 1990;250:1002–1004. doi: 10.1126/science.250.4983.1002. [DOI] [PubMed] [Google Scholar]
- Uknes S, Winter AM, Delaney T, Vernooij B, Morse A, Friedrich L, Nye G, Potter S, Ward E, Ryals J. Biological induction of systemic acquired resistance in Arabidopsis. Mol Plant-Microbe Interact. 1993;6:692–698. [Google Scholar]
- Delaney TP, Uknes S, Vernoij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gur-Rella M, Kessmann H, Ward E, Ryals J. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science. 1993;261:754–756. doi: 10.1126/science.261.5122.754. [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon S, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 1994;6:1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney TP, Friedrich L, Ryals JA. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc Natl Acad Sci U S A. 1995;92:6602–6606. doi: 10.1073/pnas.92.14.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C, DeLong C, Glaze S, Liu E, Fobert PR. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell. 2000;12:279–290. doi: 10.1105/tpc.12.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J-M, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol Plant-Microbe Interact. 2000;13:191–202. doi: 10.1094/MPMI.2000.13.2.191. [DOI] [PubMed] [Google Scholar]
- Fan W, Dong X. In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell. 2002;14:1377–1389. doi: 10.1105/tpc.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam R, Desveaux D, Spickler C, Michnick SW, Brisson N. Direct visualization of protein interactions in plant cells. Nat Biotechnol. 2001;19:769–772. doi: 10.1038/90831. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Koorneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux J-P, Brown R, Kazan K, Van Loon LC, Dong X, Pieterse CMJ. NPR1 modulates cross-talk between salicylate and jasmonate-dependent defense pathways through a novel fonction in the cytosol. Plant Cell. 2003;15:760–770. doi: 10.1105/tpc.009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. Genes controlling expression of defense responses in Arabidopsis-2001 status. Curr Opin Plant Biol. 2001;4:301–308. doi: 10.1016/S1369-5266(00)00177-1. [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Chen W, Estes B, Chang H-S, Métraux J-P, Zhu T, Katagiri F. Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 2003;34:217–228. doi: 10.1046/j.1365-313x.2003.01717.x. [DOI] [PubMed] [Google Scholar]
- Kessmann H, Staub T, Hofmann C, Maetzke T, Herzog J, Ward E, Uknes S, Ryals J. Induction of systemic acquired resistance in plants by chemicals. Annu Rev Phytopathol. 1994;32:439–459. doi: 10.1146/annurev.py.32.090194.002255. [DOI] [PubMed] [Google Scholar]
- Malamy J, Sanchez-Casas P, Henning J, Guo A, Klessig DF. Dissection of the salicylic acid signaling pathway in tobacco. Mol Plant-Microbe Interact. 1996;9:474–482. [Google Scholar]
- Friedrich L, Lawton K, Ruess W, Masner P, Specker N, Gut Rella M, Meier B, Dincher S, Staub T, Uknes S, Métraux J-P, Kessman H, Ryals J. A benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 1996;10:61–70. doi: 10.1046/j.1365-313X.1996.10010061.x. [DOI] [Google Scholar]
- Görlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel K-H, Oostendorp M, Staub T, Ward E, Kessmann H, Ryals J. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell. 1996;8:629–643. doi: 10.1105/tpc.8.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 1996;10:71–82. doi: 10.1046/j.1365-313X.1996.10010071.x. [DOI] [PubMed] [Google Scholar]
- Benhamou N, Bélanger RR. Induction of systemic resistance to Pythium damping-off in cucumber plants by benzothiadiazole: ultrastructure and cytochemistry of the host response. Plant J. 1998;14:13–21. doi: 10.1046/j.1365-313X.1998.00088.x. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van Loon LC. Salicylic acid-independent plant defence pathways. Trends Plant Sci. 1999;4:52–58. doi: 10.1016/S1360-1385(98)01364-8. [DOI] [PubMed] [Google Scholar]
- Kuc J. Induced immunity to plant disease. Bioscience. 1982;32:854–860. [Google Scholar]
- Lawton KA, Beck J, Potter S, Ward E, Ryals J. Regulation of cucumber class III chitinase gene expression. Mol Plant-Microbe Interact. 1994;7:48–57. doi: 10.1094/mpmi-7-0048. [DOI] [PubMed] [Google Scholar]
- Dalisay RF, Kuc JA. Persistence of reduced penetration by Colletotrichum lagenarium into cucumber leaves with induced systemic resistance and its relation to enhanced peroxidase and chitinase activities. Physiol Mol Plant Pathol. 1995;47:329–338. doi: 10.1006/pmpp.1995.1062. [DOI] [Google Scholar]
- Narusaka Y, Narusaka M, Horio T, Ishii H. Comparison of local and systemic induction of acquired disease resistance in cucumber plants treated with benzothiadiazoles or salicylic acid. Plant Cell Physiol. 1999;40:388–395. doi: 10.1093/oxfordjournals.pcp.a029554. [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Métraux J-P, Burkhart W, Moyer M, Dincher S, Middlesteadt W, Williams S, Payne G, Carnes M, Ryals J. Isolation of a complementary DNA encoding a chitinase with structural homology to a bifunctional lysosyme / chitinase. Proc Natl Acad Sci U S A. 1989;86:896–900. doi: 10.1073/pnas.86.3.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen JB, Hammerschmidt R, Zook MN. Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv syringae. Plant Physiol. 1991;97:1342–1347. doi: 10.1104/pp.97.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutidamrongphan W, Mackinnon G, Manners JM, Scott KJ. Sequence of a near-full length cDNA clone for a mRNA of barley induced by fungal infection. Nucleic Acids Res. 1989;17:9478. doi: 10.1093/nar/17.22.9478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena M, Daayf F, Jacques P, Thonart P, Benhamou N, Paulitz TC, Cornélis P, Koedam N, Bélanger RR. Protection of cucumber against Pythium root rot by fluorescent pseudomonads: predominant role of induced resistance over siderophores and antibiosis. Plant Pathol. 1999;48:66–76. doi: 10.1046/j.1365-3059.1999.00315.x. [DOI] [Google Scholar]
- Irani K, Goldschimidt-Clermont PJ. Ras, superoxide and signal transduction. Biochem Pharmacol. 1998;55:1339–1346. doi: 10.1016/S0006-2952(97)00616-3. [DOI] [PubMed] [Google Scholar]
- Ridley AJ. The GTP-binding protein Rho. Int J Biochem Cell Biol. 1997;29:1225–1229. doi: 10.1016/S1357-2725(97)00052-6. [DOI] [PubMed] [Google Scholar]
- Zhang S, Klessig DF. The tobacco wounding-activated mitogen-activated protein kinase is encoded by SIPK. Proc Natl Acad Sci. 1998;95:7225–7230. doi: 10.1073/pnas.95.12.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang S, Klessig DF. Molecular cloning and characterization of a tobacco MAP kinase kinase that interacts with SIPK. Mol Plant-Microbe Interact. 2000;13:118–124. doi: 10.1094/MPMI.2000.13.1.118. [DOI] [PubMed] [Google Scholar]
- Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM. Modulation of cell proliferation by heterodimeric G protein in Arabidopsis. Science. 2001;292:2066–2069. doi: 10.1126/science.1059040. [DOI] [PubMed] [Google Scholar]
- Schiene K, Puhler A, Niehaus K. Transgenic tobacco plants that express an antisense construct derived from a Medicago sativa cDNA encoding a Rac-related small GTP-binding protein fail to develop necrotic lesions upon elicitor infiltration. Mol Gen Genet. 2000;263:761–770. doi: 10.1007/s004380000248. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K. The small GTP-binding protein Rac is a regulator of cell death in plants. Proc Natl Acad Sci U S A. 1999;96:10922–10926. doi: 10.1073/pnas.96.19.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Ohashi Y. Involvement of small GTP-binding proteins in defense signal-transduction pathways of higher plants. Proc Natl Acad Sci U S A. 1995;92:4138–4144. doi: 10.1073/pnas.92.10.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Huckelhoven R. A small GTP-binding host protein is required for entry of powdery mildew fungus into epidermal cells of barley. Plant Physiol. 2002;128:1447–1454. doi: 10.1104/pp.010805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara Y, Yoshimura M, Miyoshi E, Nishikawa A, Sultan AS, Toyosawa S, Ohnishi A, Suzuki M, Yamamura K-I, Ijuhin N, Taniguchi N. Ectopic expression of N-acetylglucosaminyltransferase III in transgenic hepatocytes disrupts apolipoprotein B secretion and induces aberrant cellular morphology with lipid storage. Proc Natl Acad Sci U S A. 1998;95:2526–2530. doi: 10.1073/pnas.95.5.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P. Biological consequences of overexpressing or eliminating N-acetylglucosaminyltransferase-TIII in the mouse. Biochim Biophys Acta. 2002;1573:363–368. doi: 10.1016/S0304-4165(02)00404-X. [DOI] [PubMed] [Google Scholar]
- Sato Y, Takahashi M, Shibukawa Y, Jain SK, Hamaoaka R, Miyagawa J-I, Yaginuma Y, Honke K, Ishikawa M, Tanigushi N. Overexpression of N-acetylglucosaminyltransferase III enhances the epidermal growth factor-induced phosphorylation of ERK in HeLaS3 cells by up-regulation of the internalization rate of the receptors. J Biol Chem. 2001;276:11956–11962. doi: 10.1074/jbc.M008551200. [DOI] [PubMed] [Google Scholar]
- Shibukawa Y, Takahashi M, Laffont I, Honke K, Taniguchi N. Down-regulation of hydrogen peroxide-induced PKCδ activation in N-acetylglucosaminyltransferase III-transfected HeLaS3 cells. J Biol Chem. 2003;278:3197–3203. doi: 10.1074/jbc.M207870200. [DOI] [PubMed] [Google Scholar]
- Fitchette-Lainé AC, Gomord V, Cabanes M, Michalski J-C, Saint Macary M, Foucher B, Cavelier B, Hawes C, Lerouge P, Faye L. N-glycans harboring the Lewis a epitope are expressed at the surface of plant cells. Plant J. 1997;12:1411–1417. doi: 10.1046/j.1365-313x.1997.12061411.x. [DOI] [PubMed] [Google Scholar]
- Wenderoth I, von Schaewen A. Isolation and characterization of plant N-acetylglucosaminyltransferase I (GntI) cDNA sequences. Functional analyses in the Arabidopsis cgl mutant and in antisense plants. Plant Physiol. 2000;123:1097–1108. doi: 10.1104/pp.123.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Steinkellner H, Boren M, Altmann F, Mach L, Glossl J, Mucha J. Molecular cloning of cDNA encoding N-acetylglucosaminyltransferase II from Arabidopsis thaliana. Glycoconj J. 1999;16:787–791. doi: 10.1023/A:1007127815012. [DOI] [PubMed] [Google Scholar]
- O'Donnell PJ, Truesdale MR, Calvert CM, Dorans A, Roberts MR, Bowles DJ. A novel tomato gene that rapidly responds to wound- and pathogen-related signals. Plant J. 1998;14:137–142. doi: 10.1046/j.1365-313X.1998.00110.x. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, van Wees SCM, Hoffland E, van Pelt JA, van Loon LC. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell. 1996;8:1225–1237. doi: 10.1105/tpc.8.8.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in molecular biology. USA: Wiley Press; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PR-8 mRNA accumulation in cucumber leaves in response to different concentrations of BTH. Total RNA was extracted from cucumber leaves 24 h and 48 h after spraying with water, 10 μM BTH, 30 μM BTH, 100 μM BTH or from untreated leaves (UT). RNA was fractionated by electrophoresis on agarose gel. Northern blot was probed with α-32P labeled PR-8 cDNA. Loading of equal amounts of RNA was confirmed by ethidium bromide (Et Br) staining.


