Abstract
Cymbomonas tetramitiformis is a peculiar green alga that unites in one cell the abilities of photosynthesis and phagocytosis, which makes it a very useful model for the study of the evolution of plastid endosymbiosis. We have pondered over this issue and propose an evolutionary scenario of trophic strategies in eukaryotes, including primary and secondary plastid endosymbioses. C. tetramitiformis is a prototroph, just like the common ancestor of Archaeplastida was, and can synthesize most small organic molecules contrary to other eukaryotic phagotrophs, e.g. some metazoans, amoebozoans, and ciliates, which have not evolved tight endosymbiotic relationships. In order to establish a permanent photosynthetic endosymbiont they do not have to become prototrophs, but have to acquire the genes necessary for plastid retention via horizontal (including endosymbiotic) gene transfer. Such processes occurred successfully in the ancestors of eukaryotes with permanent secondary plastids and thus led to their great diversification. The preservation of phagocytosis in Cymbomonas (and some other prasinophytes as well) seems to result from nutrient deficiency in their oligotrophic habitats. This forces them to supplement their diet with phagocytized prey, in contrasts to the thecate amoeba Paulinella chromatophora, which also successfully transformed cyanobacteria into permanent organelles. Although Paulinella endosymbionts were acquired very recently in comparison to primary plastids, Paulinella has lost the ability to phagocytose, most probably due to the fact that it inhabits nutrient-rich environments, which renders the phagotrophy nonessential.
Keywords: Cymbomonas, Endosymbiosis, Green algae, Mixotrophy, Paulinella, Phagocytosis
Introduction
About 1.5 billion years ago, a phagotrophic eukaryote acquired a cyanobacterium that was subsequently transformed into the two-membrane-bounded photosynthetic organelle called a primary plastid (Parfrey et al. 2011; McFadden 2014; Zimorski et al. 2014). Before the transformation, however, the cyanobacteria were hunted and phagocytized by their prospective eukaryotic host. This initially hostile interaction triggered the evolution of Archaeplastida (Plantae), a supergroup of eukaryotes that comprises three photosynthetic lineages: glaucophytes (Glaucophyta), red algae (Rhodophyta), and green algae & higher plants (Viridiplantae) (De Clerck et al. 2012; Löffelhardt 2014; Mackiewicz and Gagat 2014).
Recently, a draft genome sequence of Cymbomonas tetramitiformis, a marine green alga with a peculiar trophic strategy, has been published (Burns et al. 2015). This early diverging prasinophyte not only performs photosynthesis, as the other archaeplastidians do, but also feeds by phagocytosis like a typical heterotroph and the presumed Archaeplastida progenitor (Moestrup et al. 2003; Maruyama and Kim 2013). Interestingly, bacterivorous prasinophytes have also been found in the genera of Pterosperma, Halosphaera, Pyramimonas, and Micromonas (Inouye et al. 1990; O’Kelly 1992; Bell and Laybourn-Parry 2003; McKie-Krisberg and Sanders 2014). Due to their mixotrophy, these green algae may represent an ancestral state of Archaeplastida evolution and thus help to understand the process of endosymbiosis and plastid evolution in general.
The ancestral nature of C. tetramitiformis is not only supported by its phylogenetic position among prasinophytes and by its feeding strategy, but also by the fact that its genome carries genes involved in lipid-A and peptidoglycan metabolism, two components of the bacterial cell wall derived from the cyanobacterial endosymbiont (Moestrup et al. 2003; Leliaert et al. 2012; Maruyama and Kim 2013; Burns et al. 2015). These genes, although uncommon, are also present in some other representatives of green plants (e.g. the green alga Micromonas pusilla, the moss Physcomitrella patens), and their phylogenies indicate that they are evolutionarily conserved among Viridiplantae species and have originated from a common ancestor (Burns et al. 2015). Genes involved in peptidoglycan (but not lipid-A synthesis) were also found in Glaucophyta, however, their plastids have additionally retained a clear peptidoglycan layer between the outer and inner plastid membranes (Pfanzagl et al. 1996; Jackson et al. 2015). Interestingly, Hirano et al. (2016) have recently visualized a peripherally distributed peptidoglycan layer in P. patens plastids using confocal microscopy, and Sato and colleagues (University of Tokyo, personal communication) have pinpointed its location to the intermembrane space applying a new method involving electron microscopy and density analysis of digital images at the resolution of 1 pixel. Their results indicate that the peptidoglycan layer might still be present in other Viridiplantae species, including C. tetramitiformis, and that it was lost independently at least three times in the Archaeplastida evolution: once in red algae, and twice in the green lineage, i.e. in chlorophytes and land plants (Takano and Takechi 2010).
The presence of lipid-A and peptidoglycan metabolism among members of green plants, and the mixed mechanism of feeding found in bacteriovorous prasinophytes (a sine qua non for plastid acquisition) suggest that the Viridiplantae clade might be the earliest branch of the Archaeplastida as some phylogenies indicate (e.g. the trees based on rRNA operon genes), although this contrasts with the prevailing belief that the root of the supergroup lies within Glaucophyta (see Mackiewicz and Gagat 2014 and references therein, and Jackson et al. 2015). The latter hypothesis is based on trees of nuclear genes and is supported by the presence of many ancestral traits typical of cyanobacteria preserved in glaucophyte plastids, including the peptidoglycan layer and unstacked thylakoid membranes with phycobilisomes (Mackiewicz and Gagat 2014; Jackson et al. 2015). Since these and other ancient characteristics represent preserved rather than derived traits, they do not lend decisive support for the Viridiplantae-first or Glaucophyta-first evolutionary scenarios. This issue could be resolved by phylogenetic analyses but their results are still controversial (see Mackiewicz and Gagat 2014 and references therein). More sequence data is required from glaucophytes and other poorly studied eukaryotic lineages, as well as new computational methods to eliminate stochastic and systematic errors in phylogenies (Mackiewicz and Gagat 2014). Nevertheless, the Cymbomonas case can provide interesting insight into the acquisition of permanent plastids and endosymbiosis in general.
Gene transfer and plastid establishment
It is assumed that the ancestor of Archaeplastida was a unicellular eukaryote with phagotrophic and prototrophic abilities, i.e. it could phagocytize and synthesize small organic molecules (e.g. amino acids, coenzymes, and vitamins) required for its growth, respectively (Fig. 1; Martin and Muller 1998; Martin 2005; Burns et al. 2015). These features helped it to acquire and retain endosymbionts, and in the long term to become fully independent of heterotrophy, i.e. evolve into a strict phototroph (photoautotroph). C. tetramitiformis is well-suited to this concept as a transitional form resorting to phagotrophy under phosphate-limited conditions (Paasch et al. 2016). Analyses of its genome have revealed that it has retained genes related to both phagocytic feeding and synthesis of small molecules (Burns et al. 2015). In contrast, heterotrophic phagotrophs lost the latter genes and became auxotrophic, i.e. they cannot synthesize some organic compounds needed for their growth and must obtain them through their diet. According to Burns et al. (2015), these losses prevented, for example, some metazoans, amoebozoans, and ciliates from establishing permanent relationships with their transient photosynthetic endosymbionts (Fig. 1). However, this “transiency” does not have to mean an evolutionary dead end at all. Eukaryotes that have lost genes involved in the synthesis of some small compounds can obtain them by horizontal (and also endosymbiotic) gene transfer (Timmis et al. 2004) according to the “you are what you eat” hypothesis (Doolittle 1998), the “shopping bag” model (Larkum et al. 2007), and the “minor mistargeting” mechanism (Martin 2010).
Fig. 1.
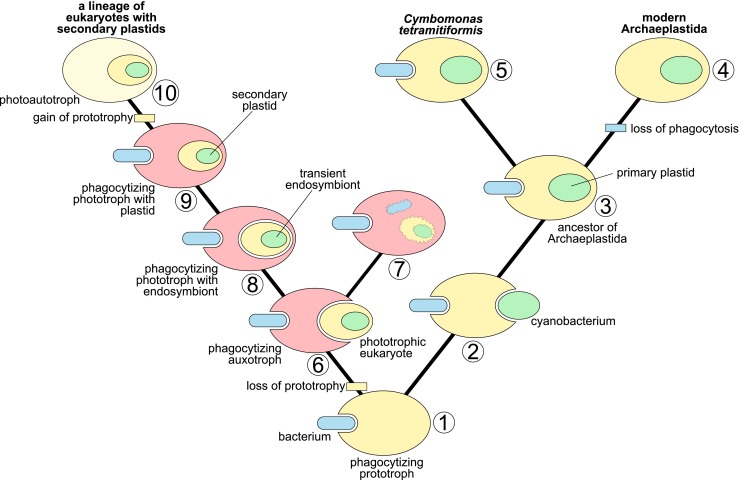
Evolution of trophic strategies in eukaryotes in the context of plastid endosymbiosis and phylogenetic position of Cymbomonas tetramitiformis. It is assumed that the early eukaryotes were prototrophs feeding via phagocytosis (1). This trophic strategy played an important role in the acquisition of a cyanobacterium (2), which was transformed into a primary plastid in the ancestor of Archaeplastida (3). Modern members of this supergroup lost the ability to phagocytose (4) with the exception of some marine prasinophytes including C. tetramitiformis (5). On the other hand, heterotrophic eukaryotes lost genes involved in the synthesis of small molecules and became auxotrophic phagotrophs (6). Some of their descendants are still auxotrophic phagotrophs (7), however, others have learned to profit from the prolonged upkeep of their photosynthetic prey (transient endosymbionts) and have become phototrophs (8). These phagocytizing phototrophs could, in the long term, acquire necessary genes via horizontal (including endosymbiotic) gene transfer to tighten the relationships with their endosymbionts following in footsteps of many secondary plastid-containing algae (9). Finally, they could reacquire the lost genes for small compounds, abandon phagotrophy, and become strict phototrophs (i.e. photoautotrophs) (10). The horizontal (including endosymbiotic) gene transfer plays an important role also at all presented stages of the model
The uptake of foreign DNA is of great importance for the expansion of metabolic capabilities and as a result can prepare the host to develop a more stable relationship with its transient endosymbiont. There is no doubt that gene transfer was one of the key processes behind the primary plastid establishment (Timmis et al. 2004; Keeling and Palmer 2008; Keeling 2009). Ample evidence for such transfers is found in the highly reduced plastid genomes of Archaeplastida and their mosaic nuclear genomes (Martin et al. 2002; Reyes-Prieto et al. 2006; Deusch et al. 2008; Green 2011). The latter genomes include not only 10–20 % genes of cyanobacterial origin, but also sequences acquired from α-proteobacteria and chlamydia (Martin et al. 2002; Reyes-Prieto et al. 2006; Deusch et al. 2008). The mitochondrial ancestor is responsible for the α-proteobacterial share, however, the gene transfer from chlamydia is much less expected. Recently, it has sparked a very interesting debate and the formulation of the “ménage à trois” hypothesis (Ball et al. 2013). According to Ball et al. (2013) a chlamydia-like symbiont facilitated the acquisition of the cyanobacterial endosymbiont. The chlamydial infection was to provide the enzymes necessary for the host to be able to use the ADP-glucose produced in cyanobacterial photosynthesis, which was inaccessible for its UDP-glucose-dependent glucan synthases (Ball et al. 2013). Moreover, chlamydial effector proteins and transporters could have also helped cyanobacteria to evade the host defence mechanisms and thus proliferate in the cytoplasm (Ball et al. 2016). When all the necessary genes were transferred to the host nuclear genome, a metabolic link that tightened the primary endosymbiosis was forged making the chlamydial co-symbiont redundant (Ball et al. 2013; see also Deschamps 2014 for discussion).
Horizontal (including endosymbiotic) gene transfer from various sources has also facilitated the acquisition of secondary and tertiary plastids (Fig. 1, Larkum et al. 2007), i.e. green and red alga-derived organelles of heterokonts (Bowler et al. 2008; Deschamps and Moreira 2012), chlorarachniophytes (Curtis et al. 2012; Yang et al. 2014), haptophytes (Miller and Delwiche 2015), cryptophytes (Curtis et al. 2012), and dinoflagellates (Patron et al. 2006; Wisecaver et al. 2013; Burki et al. 2014). Interestingly, many members of these lineages still retain the phagotrophic ability like C. tetramitiformis does, despite being obligatory phototrophs (Urban 1998; Stoecker et al. 2006; Burkholder et al. 2008; Jeong et al. 2010; Hansen 2011; Unrein et al. 2014; Mitra et al. 2016). The fact that phagotrophic protists often carry many foreign genes suggests that the feeding strategy plays an important role in gene acquisition (Doolittle 1998; Yue et al. 2013; Grant and Katz 2014).
One can argue that a single gene, e.g. involved in the synthesis of small compounds, may not constitute a unit of selection but only the entire set of genes engaged in a metabolic pathway. However, reacquisition of a lost pathway does not have to mean building it from scratch because its ‘remnants’ or homologous genes may function in the host metabolic network. This reasoning is well supported by the phylogenetic mosaicism of metabolic pathways, which, however, can also be explained by gene replacement events, and the general flexibility and robustness of the metabolic network (Curtis et al. 2012; Deschamps and Moreira 2012; Reyes-Prieto and Moustafa 2012; Qiu et al. 2013; Burki et al. 2014; Yang et al. 2014).
Even assuming that all the genes involved in a metabolic pathway are required for plastid establishment, their gain may be as simple as the acquisition of a single gene if the transfer sources are bacteria or unicellular eukaryotes with a simple organization of their genomes. Genes associated with metabolic pathways in prokaryotes are often organised into operons, which highly increases the probability of their simultaneous transfer to a potential host. Such a transfer was described in the case of the parasitic nematode Heterodera glycines, which acquired the genes for the synthesis of vitamin B6 from bacteria (Craig et al. 2008). This particular example shows that an entire metabolic pathway can be acquired, even in multicellular organisms.
Although horizontally transferred genes involved in the synthesis of small compounds can transform an auxotroph into a prototroph, it is important to emphasize that prototrophy is not required to maintain a permanent endosymbiont. This trophic ability is only necessary to become a true photoautotroph (strict phototroph). Accordingly, many phototrophic protists with permanent plastids simultaneously feed on eukaryotic microorganisms or bacteria, gain nutrients by other endocytic processes, and perform osmotrophy (Urban 1998; Stoecker et al. 2006; Burkholder et al. 2008; Jeong et al. 2010; Hansen 2011; Unrein et al. 2014; Mitra et al. 2016).
Acquired phototrophy in the light of the “luggage” hypothesis
The lack of permanent endosymbionts can also be explained by the “luggage” hypothesis proposed by Wouters et al. (2009) on the pages of “Symbiosis” nearly seven years ago. It states that as long as symbionts or plastid donors are present and abundant in the host environment, the most favoured host strategy will be preying on them. Because the prey can be easily acquired from the environment, there is no selective pressure to evolve energy-expensive mechanisms for their permanent upkeep. Therefore some metazoans, amoebozoans, and ciliates may host only transient endosymbionts (Burns et al. 2015). In the case of ciliates, nearly one fourth is estimated to apply this cost-effective strategy. Their kleptoplastids or endosymbionts provide photosynthetic metabolites and oxygen, enabling them to grow faster and find refuge in hypoxic or anoxic waters (Stoecker et al. 2009). On the other hand, the fewer symbionts or plastid donors in the environment, the more likely the host is to hold them as permanent luggage. The hosts that are able to maintain endosymbionts or kleptoplastids for a longer time are favoured, for example, in dynamic environments or when the host finds itself in a new environment (see next section for an example; Wouters et al. 2009).
The only obstacle to establish a permanent endosymbiont or plastid in a unicellular eukaryote is, perhaps, exemplified by foraminiferans. They transmit their endosymbionts vertically during asexual reproduction but the large size of most endosymbionts constrains them from entering gametes. Therefore, the endosymbionts have to be reacquired after gametogamy, resulting in a cyclic nature of the endosymbiotic relationship (Nowack and Melkonian 2010). This kind of interaction is indeed an evolutionary dead end from the endosymbiotic point of view, contrary to auxotrophy suggested by Burns et al. (2015). Given evolutionary time and possibilities, i.e. gene donors, the evolution of endosymbiotic relationships is possible if there is a selective pressure favouring them. The mechanisms driving transient endosymbionts into permanent plastids are described in the “luggage” hypothesis. Although endosymbiosis is sometimes treated as an example of non-Darwinian evolution via the inheritance of acquired characteristics (e.g., the acquisition of new genes), it still comes under the law of natural selection, resulting in the survival (and reproduction) of those best adapted to the environment.
The case of Paulinella chromatophora
Besides C. tetramitiformis, another interesting model of endosymbiosis is the thecate amoeba Paulinella chromatophora, which harbours two photosynthetic cyanobacteria-derived bodies (called chromatophores), acquired independently of archaeplastidian primary plastids (Bodył et al. 2012; Nowack 2014). The chromatophores are deeply integrated with the host and a substantial number of their genes has been transferred to the host nuclear genome (Nowack et al. 2011). Consequently, a transport system to import these host-encoded proteins has evolved in chromatophores’ membranes (Bodył et al. 2010; Mackiewicz et al. 2012a, b; Nowack and Grossman 2012; Gagat and Mackiewicz 2014). Therefore, the chromatophores should be considered true cell organelles, though they represent an early step of organelle transformation. The case of Paulinella fits well into the “luggage” hypothesis as there are marine heterotrophic Paulinella species feeding on bacteria (including cyanobacteria) and two photosynthetic P. chromatophora strains, FK01 and CCAC 0185, found in brackish and freshwater environments worldwide. That suggests the change of the habitat and consequently the change of the prey supply to be the cause behind the chromatophores’ establishment. Interestingly, another photosynthetic Paulinella (P. longichromatophora) has recently been discovered in marine sand flats on the western coast of Korea (Kim and Park 2016). This species groups significantly with the two other photosynthetic Paulinella on both nuclear and chromatophore rDNA trees indicating a single acquisition of a cyanobacterium by their ancestor. However, the two freshwater Paulinella are not monophyletic because Paulinella FK01 is closer related to the marine P. longichromatophora than to the CCAC 0185 strain. It seems that P. longichromatophora returned secondarily to the sea if we assume that the ancestor of the three amoebas lived in freshwater (Kim and Park 2016).
It is noteworthy to mention that contrary to its heterotrophic relatives and also to mixotrophic prasinophytes, Paulinella has lost the ability to phagocytose (see Nowack et al. (2011 and references therein). However, taking into account the time that has passed since the primary endosymbiosis (~1500 million years) and Paulinella endosymbiosis (~60 – 140 million years), the opposite should be expected (Nowack et al. 2008; Parfrey et al. 2011; Delaye et al. 2016). It is interesting to ponder which selective pressure could be responsible for the maintenance of phagocytosis among prasinophytes. Experimental data indicates that the opportunity to receive additional carbon compounds is not the answer, but combined with the acquisition of other nutrients, such as for example: nitrogen, phosphorus, and iron, it may constitute the selective advantage, especially in oligotrophic (low-nutrient) environments that offer little to sustain life (Flynn et al. 2013; Mitra et al. 2016; Paasch et al. 2016). Such conditions, in general, support mixotrophy as a dominant way of feeding. In accordance with this, another marine phagotrophic prasinophyte Micromonas CCMP2099 increases its grazing on bacteria in low light and poor nutrient conditions (McKie-Krisberg and Sanders 2014). In contrast, P. chromatophora thrives in small eutrophic (nutrient-rich) ponds. In such environments, phototrophs can easily acquire simple compounds by alternative to phagocytosis endocytic processes and osmotrophy, and therefore do not need to resort to phagotrophy. Nutrient-rich conditions have been shown to drive protist diversification into photoautotrophs and heterotrophs with specialized trophic strategies (Troost et al. 2005).
Acknowledgments
We are grateful to dr Andrzej Bodył for his valuable comments. This work was supported by KNOW funding and Foundation for Polish Science to PG (no. START 20.2015).
Authors’ contributions
Authors contributed equally to this publication.
Compliance with ethical standards
Competing interests
Authors have no competing interests.
References
- Ball SG, et al. Metabolic effectors secreted by bacterial pathogens: essential facilitators of plastid endosymbiosis? Plant Cell. 2013;25:7–21. doi: 10.1105/tpc.112.101329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball SG, Bhattacharya D, Weber AP. Pathogen to powerhouse. Science. 2016;351:659–660. doi: 10.1126/science.aad8864. [DOI] [PubMed] [Google Scholar]
- Bell EM, Laybourn-Parry J. Mixotrophy in the antarctic phytoflagellate, Pyramimonas gelidicola (Chlorophyta; Prasinophyceae) J Phycol. 2003;39:644–649. doi: 10.1046/j.1529-8817.2003.02152.x. [DOI] [Google Scholar]
- Bodył A, Mackiewicz P, Stiller JW. Comparative genomic studies suggest that the cyanobacterial endosymbionts of the amoeba Paulinella chromatophora possess an import apparatus for nuclear-encoded proteins. Plant Biol (Stuttg) 2010;12:639–649. doi: 10.1111/j.1438-8677.2009.00264.x. [DOI] [PubMed] [Google Scholar]
- Bodył A, Mackiewicz P, Gagat P. Organelle evolution: paulinella breaks a paradigm. Curr Biol. 2012;22:R304–R306. doi: 10.1016/j.cub.2012.03.020. [DOI] [PubMed] [Google Scholar]
- Bowler C, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Burkholder JM, Glibert PM, Skelton HM. Mixotrophy, a major mode of nutrition for harmful algal species in eutrophic waters. Harmful Algae. 2008;8:77–93. doi: 10.1016/j.hal.2008.08.010. [DOI] [Google Scholar]
- Burki F, Imanian B, Hehenberger E, Hirakawa Y, Maruyama S, Keeling PJ. Endosymbiotic gene transfer in tertiary plastid-containing dinoflagellates Eukaryot. Cell. 2014;13:246–255. doi: 10.1128/EC.00299-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JA, Paasch A, Narechania A, Kim E. Comparative genomics of a bacterivorous green alga reveals evolutionary causalities and consequences of phago-mixotrophic mode of nutrition. Genome Biol Evol. 2015;7:3047–3061. doi: 10.1093/gbe/evv144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig JP, Bekal S, Hudson M, Domier L, Niblack T, Lambert KN. Analysis of a horizontally transferred pathway involved in vitamin B6 biosynthesis from the soybean cyst nematode Heterodera glycines. Mol Biol Evol. 2008;25:2085–2098. doi: 10.1093/molbev/msn141. [DOI] [PubMed] [Google Scholar]
- Curtis BA, et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 2012;492:59–65. doi: 10.1038/nature11681. [DOI] [PubMed] [Google Scholar]
- De Clerck O, Bogaert KA, Leliaert F (2012) Chapter two - diversity and evolution of algae: primary endosymbiosis. In: Gwenaël P (ed) Advances in botanical research, vol 64. Academic Press. doi:10.1016/B978-0-12-391499-6.00002-5
- Delaye L, Valadez-Cano C, Pérez-Zamorano B. How really ancient is Paulinella chromatophora? PLoS Currents Tree of Life. 2016 doi: 10.1371/currents.tol.e68a099364bb1a1e129a17b4e06b0c6b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps P. Primary endosymbiosis: have cyanobacteria and Chlamydiae ever been roommates? Acta Soc Bot Pol. 2014;83:291–320. doi: 10.5586/asbp.2014.048. [DOI] [Google Scholar]
- Deschamps P, Moreira D. Reevaluating the green contribution to diatom genomes. Genome Biol Evol. 2012;4:683–688. doi: 10.1093/gbe/evs053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deusch O, et al. Genes of cyanobacterial origin in plant nuclear genomes point to a heterocyst-forming plastid ancestor. Mol Biol Evol. 2008;25:748–761. doi: 10.1093/molbev/msn022. [DOI] [PubMed] [Google Scholar]
- Doolittle FW. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998;14:307–311. doi: 10.1016/S0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- Flynn KJ, et al. Misuse of the phytoplankton–zooplankton dichotomy: the need to assign organisms as mixotrophs within plankton functional types. J Plankton Res. 2013;35:3–11. doi: 10.1093/plankt/fbs062. [DOI] [Google Scholar]
- Gagat P, Mackiewicz P. Protein translocons in photosynthetic organelles of Paulinella chromatophora. Acta Soc Bot Pol. 2014;83:399–407. doi: 10.5586/asbp.2014.053. [DOI] [Google Scholar]
- Grant JR, Katz LA. Phylogenomic study indicates widespread lateral gene transfer in Entamoeba and suggests a past intimate relationship with parabasalids. Genome Biol Evol. 2014;6:2350–2360. doi: 10.1093/gbe/evu179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BR. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66:34–44. doi: 10.1111/j.1365-313X.2011.04541.x. [DOI] [PubMed] [Google Scholar]
- Hansen PJ. The role of photosynthesis and food uptake for the growth of marine mixotrophic dinoflagellates. J Eukaryot Microbiol. 2011;58:203–214. doi: 10.1111/j.1550-7408.2011.00537.x. [DOI] [PubMed] [Google Scholar]
- Hirano T, et al. Moss chloroplasts are surrounded by a peptidoglycan wall containing D-amino acids. Plant Cell. 2016;28:1521–1532. doi: 10.1105/tpc.16.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye I, Hori T, Chihara M. Absolute configuration analysis of the flagellar apparatus of Pterosperma cristatum (prasinophyceae) and consideration of its phylogenetic position. J Phycol. 1990;26:329–344. doi: 10.1111/j.0022-3646.1990.00329.x. [DOI] [Google Scholar]
- Jackson C, Clayden S, Reyes-Prieto A (2015) The Glaucophyta: the blue-green plants in a nutshell. Acta Soc Bot Pol 84. doi:10.5586/asbp.2015.020
- Jeong HJ, Yoo YD, Kim JS, Seong KA, Kang NS, Kim TH. Growth, feeding and ecological roles of the mixotrophic and heterotrophic dinoflagellates in marine planktonic food webs. Ocean Sci J. 2010;45:65–91. doi: 10.1007/s12601-010-0007-2. [DOI] [Google Scholar]
- Keeling PJ (2009) Role of horizontal gene transfer in the evolution of photosynthetic eukaryotes and their plastids. In: Gogarten MB, Gogarten JP, Olendzenski LC (eds) Horizontal gene transfer: genomes in flux. Humana Press. doi:10.1007/978-1-60327-853-9_29 [DOI] [PubMed]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- Kim S, Park MG. Paulinella longichromatophora sp. nov., a new marine photosynthetic testate amoeba containing a chromatophore. Protist. 2016;167:1–12. doi: 10.1016/j.protis.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Larkum AW, Lockhart PJ, Howe CJ. Shopping for plastids. Trends Plant Sci. 2007;12:189–195. doi: 10.1016/j.tplants.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O. Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci. 2012;31:1–46. doi: 10.1080/07352689.2011.615705. [DOI] [Google Scholar]
- Löffelhardt W (2014) The single primary endosymbiotic event. In: Endosymbiosis. Springer
- Mackiewicz P, Gagat P. Monophyly of Archaeplastida supergroup and relationships among its lineages in the light of phylogenetic and phylogenomic studies. Are we close to a consensus? Acta Soc Bot Pol. 2014;83:263–280. doi: 10.5586/asbp.2014.044. [DOI] [Google Scholar]
- Mackiewicz P, Bodyl A, Gagat P. Possible import routes of proteins into the cyanobacterial endosymbionts/plastids of Paulinella chromatophora. Theory Biosci. 2012;131:1–18. doi: 10.1007/s12064-011-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz P, Bodyl A, Gagat P. Protein import into the photosynthetic organelles of and its implications for primary plastid endosymbiosis. Symbiosis. 2012;58:99–107. doi: 10.1007/s13199-012-0202-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W (2005) Woe is the tree of life. In: Sapp J (ed) Microbial Phylogeny and evolution: concepts and controversies. Oxford University Press
- Martin W. Evolutionary origins of metabolic compartmentalization in eukaryotes. Philos Trans R Soc Lond [Biol] 2010;365:847–855. doi: 10.1098/rstb.2009.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Muller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S, Kim E. A modern descendant of early green algal phagotrophs. Curr Biol. 2013;23:1081–1084. doi: 10.1016/j.cub.2013.04.063. [DOI] [PubMed] [Google Scholar]
- McFadden GI. Origin and evolution of plastids and photosynthesis in eukaryotes. Cold Spring Harb Perspect Biol. 2014;6:a016105. doi: 10.1101/cshperspect.a016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKie-Krisberg ZM, Sanders RW. Phagotrophy by the picoeukaryotic green alga Micromonas: implications for Arctic Oceans. ISME J. 2014;8:1953–1961. doi: 10.1038/ismej.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JJ, Delwiche CF. Phylogenomic analysis of Emiliania huxleyi provides evidence for haptophyte–stramenopile association and a chimeric haptophyte nuclear genome. Mar Genomics. 2015;21:31–42. doi: 10.1016/j.margen.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Mitra A, et al. Defining planktonic protist functional groups on mechanisms for energy and nutrient acquisition: incorporation of diverse mixotrophic strategies. Protist. 2016;167:106–120. doi: 10.1016/j.protis.2016.01.003. [DOI] [PubMed] [Google Scholar]
- Moestrup Ø, Inouye I, Hori T. Ultrastructural studies on Cymbomonas tetramitiformis (Prasinophyceae). General structure, scale microstructure, and ontogeny. Can J Bot. 2003;81:657–671. doi: 10.1139/b03-055. [DOI] [Google Scholar]
- Nowack EC. Paulinella chromatophora – rethinking the transition from endosymbiont to organelle. Acta Soc Bot Pol. 2014;83:387–397. doi: 10.5586/asbp.2014.049. [DOI] [Google Scholar]
- Nowack EC, Grossman AR. Trafficking of protein into the recently established photosynthetic organelles of Paulinella chromatophora. Proc Natl Acad Sci U S A. 2012;109:5340–5345. doi: 10.1073/pnas.1118800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack ECM, Melkonian M. Endosymbiotic associations within protists. Philos Trans R Soc Lond [Biol] 2010;365:699–712. doi: 10.1098/rstb.2009.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack EC, Melkonian M, Glockner G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr Biol. 2008;18:410–418. doi: 10.1016/j.cub.2008.02.051. [DOI] [PubMed] [Google Scholar]
- Nowack ECM, Vogel H, Groth M, Grossman AR, Melkonian M, Glöckner G. Endosymbiotic gene transfer and transcriptional regulation of transferred genes in Paulinella chromatophora. Mol Biol Evol. 2011;28:407–422. doi: 10.1093/molbev/msq209. [DOI] [PubMed] [Google Scholar]
- O’Kelly CJ. Flagellar apparatus architecture and the phylogeny of “green” algae. In: Menzel D, editor. The cytoskeleton of the algae. Boca Raton: CRC Press; 1992. [Google Scholar]
- Paasch AE, Burns JA, Kim E. Retention of bacterivory in the dominantly photoautotrophic green alga Cymbomonas tetramitiformis is influenced by phosphate limitation. Protistology. 2016;10:55–56. [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci U S A. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron NJ, Waller RF, Keeling PJ. A tertiary plastid uses genes from two endosymbionts. J Mol Biol. 2006;357:1373–1382. doi: 10.1016/j.jmb.2006.01.084. [DOI] [PubMed] [Google Scholar]
- Pfanzagl B, et al. Primary structure of cyanelle peptidoglycan of Cyanophora paradoxa: a prokaryotic cell wall as part of an organelle envelope. J Bacteriol. 1996;178:332–339. doi: 10.1128/jb.178.2.332-339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Price DC, Weber APM, Facchinelli F, Yoon HS, Bhattacharya D. Assessing the bacterial contribution to the plastid proteome. Trends Plant Sci. 2013;18:680–687. doi: 10.1016/j.tplants.2013.09.007. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Moustafa A. Plastid-localized amino acid biosynthetic pathways of Plantae are predominantly composed of non-cyanobacterial enzymes. Sci Report. 2012;2:955. doi: 10.1038/srep00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Prieto A, Hackett JD, Soares MB, Bonaldo MF, Bhattacharya D. Cyanobacterial contribution to algal nuclear genomes is primarily limited to plastid functions. Curr Biol. 2006;16:2320–2325. doi: 10.1016/j.cub.2006.09.063. [DOI] [PubMed] [Google Scholar]
- Stoecker DK, Tillmann U, Granéli E (2006) Phagotrophy in harmful algae. In: Granéli E, Turner JT (eds) Ecology of harmful algae. Springer
- Stoecker DK, Johnson MD, de Vargas C, Not F. Acquired phototrophy in aquatic protists. Aquat Microb Ecol. 2009;57:279–310. doi: 10.3354/ame01340. [DOI] [Google Scholar]
- Takano H, Takechi K. Plastid peptidoglycan. Biochim Biophys Acta. 2010;1800:144–151. doi: 10.1016/j.bbagen.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Troost T, Kooi B, Kooijman S. Ecological specialization of mixotrophic plankton in a mixed water column. Am Nat. 2005;166:E45–E61. doi: 10.1086/432038. [DOI] [PubMed] [Google Scholar]
- Unrein F, Gasol JM, Not F, Forn I, Massana R. Mixotrophic haptophytes are key bacterial grazers in oligotrophic coastal waters. ISME J. 2014;8:164–176. doi: 10.1038/ismej.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban T. Phagotrophy by a plastidic haptophyte, Prymnesium patelliferum. Aquat Microb Ecol. 1998;14:155–160. doi: 10.3354/ame014155. [DOI] [Google Scholar]
- Wisecaver JH, Brosnahan ML, Hackett JD. Horizontal gene transfer is a significant driver of gene innovation in dinoflagellates. Genome Biol Evol. 2013;5:2368–2381. doi: 10.1093/gbe/evt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters J, Raven JA, Minnhagen S, Janson S. The luggage hypothesis: comparisons of two phototrophic hosts with nitrogen-fixing cyanobacteria and implications for analogous life strategies for kleptoplastids/secondary symbiosis in dinoflagellates. Symbiosis. 2009;49:61–70. doi: 10.1007/s13199-009-0020-3. [DOI] [Google Scholar]
- Yang Y, Matsuzaki M, Takahashi F, Qu L, Nozaki H. Phylogenomic analysis of “red” genes from two divergent species of the “green” secondary phototrophs, the Chlorarachniophytes, suggests multiple horizontal gene transfers from the red lineage before the divergence of extant Chlorarachniophytes. PLoS One. 2014;9:e101158. doi: 10.1371/journal.pone.0101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Sun G, Hu X, Huang J. The scale and evolutionary significance of horizontal gene transfer in the choanoflagellate Monosiga brevicollis. BMC Genomics. 2013;14:1–10. doi: 10.1186/1471-2164-14-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimorski V, Ku C, Martin WF, Gould SB. Endosymbiotic theory for organelle origins. Curr Opin Microbiol. 2014;22:38–48. doi: 10.1016/j.mib.2014.09.008. [DOI] [PubMed] [Google Scholar]


