Abstract
Background
High-throughput protein structure analysis of individual protein domains requires analysis of large numbers of expression clones to identify suitable constructs for structure determination. For this purpose, methods need to be implemented for fast and reliable screening of the expressed proteins as early as possible in the overall process from cloning to structure determination.
Results
88 different E. coli expression constructs for 17 human protein domains were analysed using high-throughput cloning, purification and folding analysis to obtain candidates suitable for structural analysis. After 96 deep-well microplate expression and automated protein purification, protein domains were directly analysed using 1D 1H-NMR spectroscopy. In addition, analytical hydrophobic interaction chromatography (HIC) was used to detect natively folded protein. With these two analytical methods, six constructs (representing two domains) were quickly identified as being well folded and suitable for structural analysis.
Conclusion
The described approach facilitates high-throughput structural analysis. Clones expressing natively folded proteins suitable for NMR structure determination were quickly identified upon small scale expression screening using 1D 1H-NMR and/or analytical HIC. This procedure is especially effective as a fast and inexpensive screen for the 'low hanging fruits' in structural genomics.
Keywords: structural genomics, hydrophobic interaction chromatography, homonuclear NMR, protein domains, high-throughput expression
Background
Structural genomics has gained increasing interest in recent years. The elucidation of protein structures is important to understand protein functions and thereby facilitate pharmaceutical drug development [1,2]. A prerequisite for structural analysis is the preparation of pure, natively folded protein. In high-throughput structural analysis, screening methods need to be implemented for rapid detection of suitable protein expression constructs.
NMR has been proven to be a suitable method for evaluating the structural integrity of a protein either by hetero- or homonuclear techniques [3-5]. Especially in the context of structural genomics projects acquisition of 2D 15N-HSQC spectra is widely used despite the disadvantage of having to employ 15N-minimal medium, which slows down the production process significantly. Alternatively, labelled commercial rich media can be used for protein production. However, these media are expensive.
We were attracted by the fact that a 1D 1H-NMR spectrum already contains enough information to evaluate whether a protein is folded. This enabled use of high-throughput protein expression and purification procedures based on unlabeled rich media.
As a cost-effective alternative to 1D 1H-NMR, we tested analytical HIC. In a previous work we showed that analytical HIC can be successfully applied to detect small amounts of natively folded protein upon folding screening [6]. Therefore, we asked whether this technique is useful for identifying domain constructs suitable for structural analysis. The results from 1D 1H-NMR and analytical HIC were compared.
In this work we analysed 17 human protein domains using high-throughput cloning, expression in E. coli, purification and folding analysis to obtain suitable candidates for structural analysis. Domain boundaries were predicted according to SMART http://smart.embl-heidelberg.de/[7,8] and PCR-amplified from a human cDNA library [9,10]. To express an isolated domain successfully, the expression construct needs to represent a compact folding entity. It should not contain overhanging unstructured amino acid stretches that may hamper correct folding, lead to aggregation or that may complicate NMR analysis. The domain boundary prediction is putative for domains of unknown structure. We assumed that a domain prediction by SMART might in some cases not span the complete folding entity but might miss weakly conserved residues at the ends. This could result in truncated domain constructs lacking stabilising residues at the termini. Therefore, different constructs were generated for each domain with small variations at the predicted domain boundaries. All constructs were cloned into an E. coli expression system that allows for expression of His-tag fusion proteins.
Results and Discussion
Domain selection and cloning
A human cDNA expression library from fetal brain (hEx1) [9] was screened for clones that express their inserts as soluble proteins (manuscript in preparation). Such expression clones were identified for 1,200 human proteins. These protein sequences were analysed using the SMART database of Hidden Markov Models [7,8]. The outcome was a total of 26 different protein domains (contained in 30 different proteins and represented by 41 clones) for which no three-dimensional structure was solved at that time.
For each analysed domain, we attempted to generate four constructs: one corresponding to the SMART-prediction of the respective domain and three further constructs with either five amino acid extensions at the N- or C-termini or at both. This is reflected in Table 1 by the columns 'start residue' and 'end residue'. However, some constructs could not be generated due to failure during PCR-amplification or transformation (e. g. two out of four L27 domain constructs, Table 1).
Table 1.
Expression strength and solubility of the domain constructs.
| Domain | Protein ID | start residue | end residue | total cellular protein | Soluble purified protein |
| CNH | AAB48435.1 | 506/511 | 825/830 | ++ | - |
| RWD | CAB52345.1 | 8/13 | 134/140 | + | - |
| SEP-UBX | AAH02801.1 | 171/176 | 368 | ++ | ++ |
| SEP | AAH02801.1 | 171/176 | 270/275 | ++ | ++ |
| L27 | AAG34117.1 | 13 | 68/73 | ++ | ++ |
| Zpr1 | AAC33514.1 | 252/257 | 416/421 | ++ | ++ |
| Zpr1 | AAC33514.1 | 44/49 | 207/212 | ++ | ++ |
| ZnF_C3H1 | AAF28981.1 | 126/131 | 157/162 | ++ | ++ |
| G_patch | CAD38742.1 | 406/411 | 459/464 | + | + |
| G_patch | AAH07871.1 | 228/233 | 279/284 | +/- | ++ |
| G_patch | CAA68976.1 | 92/97 | 142/147 | + | ++ |
| G_patch | AAA99715.1 | 736/741 | 787/792 | +/- | + |
| OSTEO | AAA59974.1 | 14/19 | 314 | - | - |
| ZnF_RBZ | CAC28312.1 | 185 | 214 | + | ++ |
| PINT | AAB58732.1 | 316/321 | 404/409 | + | + |
| JAB_MPN | AAD03468.1 | 54 | 191 | + | - |
| JAB_MPN | AAD03465.1 | 33/38 | 172/177 | ++ | - |
| DCX | AAC31696.1 | 85/90 | 181/186 | ++ | ++ |
| DCX | AAC31696.1 | 85/90 | 305/310 | ++ | ++ |
| DCX | AAC31696.1 | 212/217 | 305/310 | ++ | ++ |
| MA3 | AAC02903.2 | 1215/1220 | 1332/1327 | ++ | - |
| eIF5C | AAC02903.2 | 1487/1492 | 1579/1583 | ++ | - |
| PRY | AAA36564.1 | 310/315 | 367/372 | ++ | +/- |
| SPRY | AAA36564.1 | 363/368 | 493/498 | ++ | - |
| TLDc | BAB13435.1 | 250/255 | 424/429 | ++ | - |
Relative amounts of purified protein were judged from SDS-PAGE. The numbers in the columns 'start residue' and 'end residue' identify the combinations of constructs tested. Bold numbers indicate that these constructs yielded more soluble expression for the respective domain than the other constructs (see Results). '-' = no expression, '+/-' = low expression, '+' = intermediate expression, '++' = strong expression. Clones are available from the RZPD, German Resource Center for Genome Research GmbH http://www.rzpd.de, refer to supplementary table Additional file: 1.
G_patch domains of four different source proteins (CAD38742.1, AAH07871.1, CAA68976.1, AAA99715.1) and JAB_MPN of two source proteins (AAD03468.1, AAD03465.1) were cloned, whereas all other domains originate from one representative protein (Table 1). Doublecortin (AAC31696.1) contains two adjacent DCX-domains (from residue 48–139 and 175–263). Constructs of the single DCX-domains as well as others covering both domains were generated. The analysed SEP domain from protein p47 (AAH02801.1) was analysed individually (residues 176–270) as well as together with its neighbouring UBX domain (residues 289–368).
Solubility of different domain constructs
Expression of the cloned constructs was tested at 37°C and 28°C. Domains fused to N-terminal His-tags were purified with metal chelate chromatography using an automated purification [11] and resulting samples were analysed with SDS-PAGE. One domain could not be expressed (OSTEO), seven could only be expressed insolubly (CNH, RWD, JAB_MPN, MA3, eIF5C, SPRY and TLDc) and one domain only yielded a small fraction of solubly expressed protein (PRY). These constructs were therefore not considered for further detection of natively folded protein (see below). The Zpr1 domain could only be expressed solubly at 28°C.
Domains predicted by SMART that were expressed insolubly might not represent complete folding entities, but might miss weakly conserved residues at the C- or N-terminus. For more reliable estimation of domain boundaries it may be helpful to consider the prediction of secondary structure in the boundary region, i.e. to avoid disruption of secondary structure elements. Such an additional bioinformatic analysis was not performed in this work but may increase the yield of domains that are expressed in a soluble, natively folded form.
In some cases only particular constructs lead to soluble expression of a domain. For instance, the N-terminal zinc finger (Zpr1) domain of protein AAC33514.1 could only be expressed solubly when five amino acid residues were added on both N- and C-terminus (PTEIE and PHAPQ, respectively) to the in silico predicted Zpr1 sequence (Table 1). These extensions are non-conserved regions of the Zpr1 domain but are necessary for soluble expression. In contrast, the second Zpr1 domain of protein AAC33514.1 was only found in the soluble fraction when it was expressed as the shorter construct lacking the C-terminal extension (VYAPE). Three of the four analysed G_patch domains behaved in a similar manner, i. e. that particular constructs of a domain yielded more soluble protein than others (see Table 1).
Detection of natively folded protein by 1D 1H-NMR and analytical HIC
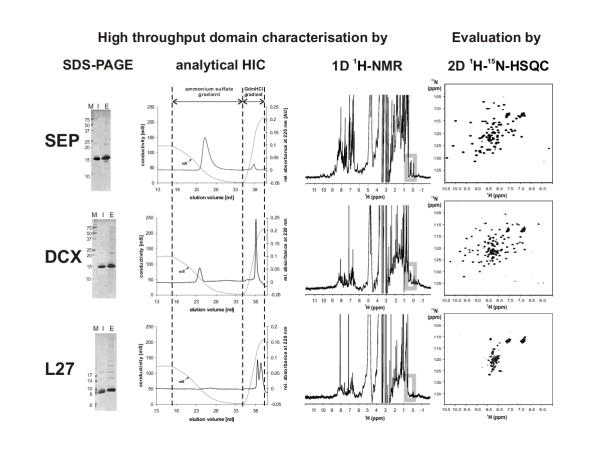
We tested, whether the amount of purified protein resulting from 1 ml cultures is sufficient to be reliably analysed for natively folded protein using 1D 1H-NMR. 1D 1H-NMR is an established and sensitive method to check for folded protein [3]. In our approach, two 70 μl EDTA eluates resulting from an automated protein purification of two 1 ml cultures of the same construct were pooled (see Materials and Methods). From these samples, 1D 1H-NMR spectra were acquired. In Figure 1, three selected examples from Table 2 are shown: (i) the SEP-domain (residues 171 – 270 from protein AAH02801.1), (ii) the N-terminal DCX-domain from protein AAC31696.1 (residues 90 – 186) and (iii) the L27-domain from protein AAG34117.1 (residues 13 – 68). The chemical shift dispersion of the proton signals is indicative for the prediction of protein folding. Large chemical shift dispersions, especially in the methyl group region between 1.0 ppm and -1.0 ppm and the amide region downfield of 8.5 ppm can be used as an indicator for protein folding. However, the signals of the amide protons occurring in this region can be broadened due to chemical exchange especially at non acidic pH. This can be a drawback when amide signals at protein concentrations close to the detection limit are to be observed. Isolated methyl group signals upfield of 0.5 ppm are further sensitive and reliable probes for folded protein. They are not affected by chemical exchange broadening. In addition their signal intensities are higher because three equivalent protons contribute rather than one proton of the amide group. The SEP- and DCX-domains show a number of signals upfield of 0.5 ppm (highlighted in the grey box in Figure 1) whereas L27 has no signals upfield of 0.5 ppm except of two signals that resulted from the buffer (spectrum not shown). A second criteria for folded protein is the amide region downfield of 8.5 ppm. In the case of SEP a number of amides are observable downfield of 8.5 ppm indicating folded protein, whereas for DCX and L27 no signals occur. In order to confirm these results SEP, DCX, and L27 were expressed with 15N labels in 400 ml scale. The domains were purified and the tags were cleaved off. Subsequently, 1D 1H-NMR spectra and 2D 1H-15N-HSQC spectra were acquired. In the methyl-group region, 1D 1H-NMR spectra from the small scale approach exhibited the same characteristics as those obtained from the 15N-labelled samples (data not shown). 2D 1H-15N-HSQC spectra (Figure 1, right side) clearly supports the findings from the 1D spectra recorded with unlabelled tagged protein domains. The peaks of SEP and DCX show a large dispersion in both dimensions indicating a folded protein. On the contrary, the signals of L27 cluster around 8.3 ppm suggesting an unfolded or only to a low extend folded protein. The absence of amide signals downfield of 8.5 ppm for the DCX domain might be due to the fact that the intensity of single amide signals fell below the detection limit under the applied conditions. An acquired 1D 1H-NMR spectrum from the more concentrated 15N-sample showed indeed amide resonances downfield of 8.5 ppm (data not shown).
Figure 1.
High-throughput domain characterisation and evaluation by 2D 1H-15N-HSQC spectroscopy. Analytical results for the SEP-domain (residues 171 – 270 from protein AAH02801.1), the N-terminal DCX domain from protein AAC31696.1 (residues 90 – 186) and the L27-domain from protein AAG34117.1 (residues 13 – 68) are shown. Samples for SDS-PAGE, analytical HIC and 1D 1H-NMR spectra were obtained from two separate automated small-scale purifications of 1 ml cultures. 2D 1H-15N-HSQC spectra resulted from the purification of a 400 ml culture. SDS-PAGE lanes: M: marker; I: NiNTA-bound protein eluted with imidazole for analytical HIC; E: NiNTA-bound protein eluted with EDTA for 1D 1H-NMR analysis.
Table 2.
Analysis of soluble constructs with 1D 1H-NMR and analytical HIC.
| Domain | Protein ID | start residue | end residue | 1H 1D-NMR | analytical HIC |
| SEP | AAH02801.1 | 171 | 270 | + | + |
| SEP | AAH02801.1 | 176 | 270 | + | + |
| DCX | AAC31696.1 | 90 | 181 | + | +/- |
| DCX | AAC31696.1 | 90 | 186 | + | + |
| DCX | AAC31696.1 | 85 | 181 | + | + |
| DCX | AAC31696.1 | 85 | 186 | + | + |
| DCX | AAC31696.1 | 212 | 310 | - | - |
| DCX | AAC31696.1 | 217 | 310 | n. d. | - |
| DCX | AAC31696.1 | 217 | 305 | n. d. | - |
| DCX | AAC31696.1 | 85 | 305 | +/- | - |
| DCX | AAC31696.1 | 90 | 305 | +/- | - |
| DCX | AAC31696.1 | 90 | 310 | - | - |
| DCX | AAC31696.1 | 85 | 310 | - | - |
| SEP-UBX | AAH02801.1 | 171 | 368 | + | + |
| SEP-UBX | AAH02801.1 | 176 | 368 | - | +/- |
| PINT | AAB58732.1 | 316 | 409 | n. d. | - |
| PINT | AAB58732.1 | 321 | 404 | n. d. | - |
| PINT | AAB58732.1 | 316 | 404 | n. d. | - |
| PINT | AAB58732.1 | 321 | 409 | n. d. | - |
| G_patch | CAD38742.1 | 406 | 464 | n. d. | - |
| G_patch | AAH07871.1 | 228 | 284 | n. d. | - |
| G_patch | AAH07871.1 | 233 | 284 | n. d. | +/- |
| G_patch | CAA68976.1 | 91 | 141 | n. d. | - |
| G_patch | CAA68976.1 | 91 | 146 | - | - |
| G_patch | CAA68976.1 | 96 | 141 | - | - |
| G_patch | CAA68976.1 | 96 | 146 | - | +/- |
| G_patch | AAA99715.1 | 736 | 787 | n. d. | - |
| Zpr1 | AAC33514.1 | 252 | 416 | n. d. | - |
| Zpr1 | AAC33514.1 | 257 | 416 | n. d. | - |
| Zpr1 | AAC33514.1 | 44 | 212 | n. d. | - |
| ZnF_C3H1 | AAF28981.1 | 131 | 157 | n. d. | - |
| ZnF_C3H1 | AAF28981.1 | 126 | 162 | n. d. | - |
| ZnF_C3H1 | AAF28981.1 | 131 | 162 | - | - |
| ZnF_RBZ | CAC28312.1 | 185 | 214 | n. d. | - |
| L27 | AAG34117.1 | 13 | 68 | - | - |
| L27 | AAG34117.1 | 13 | 73 | - | - |
For column '1D 1H-NMR': '-' = unfolded, '+/-' = presumably folded, '+' = folded, 'n. d' = not detectable (protein amount insufficient for detection). For column 'analytical HIC': '-' = no peak upon ammoniumsulfate gradient, but protein elutes only during GdmHCl gradient, '+/-' = very small or broad peak observed upon ammoniumsulfate gradient, '+' = distinct peak observed upon ammoniumsulfate gradient.
In summary, NMR-signals in the methyl-group region seem to be a reliable and sensitive indicator for folded protein. In addition, the amide proton signals can provide useful information but are not as strong as proton signals from methyl-groups. Because most protein samples analysed in this work were rather dilute, resonance signals from amid protons might in most cases not be detectable. The occurrence of proton signals in one of the two chemical shift regions is sufficient to detect folded protein.
The results of all screened protein domains are summarised in Table 2. Out of 36 tested proteins samples, 19 contained sufficient protein to observe 1H protein signals within 1 hour and 23 min. For nine samples, folded proteins were detected. We assigned a '+' for folded protein whenever we could detect NMR signals either in the amide and/or methyl-group region within the chemical shift ranges described above. Two domain constructs were annotated with '+/-' because 1H signals occurred in the respective regions but with very low intensities. For 10 samples protein 1H signals are present but neither amide signals downfield of 8.3 ppm nor methyl-group resonances upfield of 0.5 ppm could be observed (classified with '-'). We found that the minimal protein concentration to get an evaluation by NMR in 1 h and 23 min on a 600 MHz spectrometer equipped with a 5 mm triple resonance probe head is about 3 μM (15 μg in 500 μl in case of a 10 kDa protein). This value corresponds to an expression level of about 15 mg recombinant protein per 1 l of culture. This level was not achieved for the entries 'n.d.' in Table 2, although it is an average yield for recombinant protein expression in E. coli. The sensitivity can further be improved or the measurement time can be reduced by using CryoProbe technology. Alternatively, expression can be performed in 5 ml scale in 24 deep-well plates.
A similar approach to check the folding of proteins was used before [4]. In this work, a 15N labelled 50 ml set up was used, followed by purification, tag removal and 2D 1H-15N-HSQC evaluation on a 60 μM sample. In contrast, we used an unlabelled expression for up to 46 clones in parallel in a 1 ml scale, a single purification step and a 1D 1H-NMR on 3 μM samples. Our method is less expensive and very fast, but only useful for proteins with small tags like His- or Strep-tag. In cases where larger affinity-tags for purification are used (e.g. GST-tag), an on-column fusion cleavage as described by Woestenenk et al. [4] should be performed. The outcome is a classification into folded or unfolded proteins leading to a decision which construct is most promising for structure determination.
As a cost-effective alternative to 1D 1H-NMR, we tested analytical HIC, which we previously used to detect small amounts of natively folded protein upon folding screening [6]. Ammonium sulfate was added to all samples, protein domains were bound to a HIC-resin and eluted in a decreasing ammonium sulfate gradient (see Figure 1). Misfolded proteins can be distinguished from native protein because they (i) precipitate upon the addition of ammonium sulfate at lower concentrations, (ii) elute erratically from the column and not in a distinct peak due to conformational heterogeneity, or (iii) bind very strongly to the column material. In the latter case, tightly bound proteins might only be eluted upon addition of strong denaturants like GdmHCl. Such a behaviour can be observed with the L27-domain that was shown to be unfolded using NMR (see Fig. 1). Compact, natively folded proteins are assumed to elute in a distinct peak upon a decreasing ammonium sulfate gradient. Some constructs of the SEP- and the DCX-domain behaved in this manner. These were the constructs that yielded natively folded protein shown by NMR analysis (see above and Fig. 1). In the case of the DCX-domain, an additional fraction of the protein is eluted during the GdmHCl-gradient. This could be due to two species present upon expression: a compactly folded one that is eluted in the decreasing ammonium sulfate gradient; and a misfolded one that binds tightly to the HIC resin and is only eluted with high concentrations of GdmHCl. However, an evaluation of the signals obtained from 2D 1H-15N-HSQC did not support this assumption. No additional signals to the expected number of signals calculated from amino acid sequence were observed. Another explanation would be that a fraction of the DCX-domain is unfolded upon addition of ammonium sulfate or upon binding to the HIC resin. The detection limit for analytical HIC is 3 – 5 μg protein, corresponding to 0.3 – 0.5 nmol for a 10 kDa protein.
Analytical HIC has a minor drawback: the optimal ammonium sulfate concentration, i. e. high enough to bind the analysed protein to the HIC resin but not too high to precipitate the protein, is not known or may not exist within a batch of different proteins. In our case 1 M ammonium sulfate seemed to be adequate for all constructs. Adjusting the concentration to 2 M ammonium sulfate was accompanied with precipitation in almost all samples. In a general screening procedure it might be necessary to test two concentrations (e. g. 1 M and 2 M).
The described high-throughput analysis procedure (1D 1H-NMR and/or analytical HIC) allowed us to generate a priority list for constructs for structural analysis (Table 2). The SEP domain and the N-terminal DCX domain of doublecortin were selected for structural analysis. However, the NMR-structure of the N-terminal DCX-domain was solved recently by Kim et al. [12]. In this study the authors noted that the C-terminal DCX-domain of doublecortin expressed in E. coli is thermodynamically unstable with a pronounced surface hydrophobicity as assayed by ANS (8-anilino-1-naphtalene-sulphonic acid) binding. This is also reflected in our observations with analytical HIC as this domain could only be eluted with high amounts of GdmHCl (data not shown). Correspondingly, 1D 1H-NMR suggested that the C-terminal DCX domain of doublecortin is not folded (Table 2). Kim et al. recorded a 1H-15N-HSQC and observed significant line-broadening which presumably is due to conformational disorder on the millisecond timescale and hampered structure solution [12].
The SEP-UBX construct covering amino acids 171 – 368 was found to be folded according to both analytical methods, 1D 1H-NMR and analytical HIC. Accordingly, large scale preparation of this construct and detailed biophysical analysis indicated that this protein is compact and folded: (i) the hydrodynamic diameter determined with dynamic light scattering was 4 nm, (ii) thermal unfolding measured by differential scanning calorimetry revealed a transition temperature of 54°C and (iii) a Fourier-transform infrared spectrum indicated a mostly β-sheet fold (Frank Niesen, unpublished data).
In most cases, the data from 1D 1H-NMR and analytical HIC have good correlation (Table 2). Only minor deviations could be observed. For example, the G_Patch-domains that were presumably folded according to analytical HIC. A 2D 1H-15N-HSQC spectrum showed, however, that the chemical shift dispersion of the peaks is small suggesting an unfolded species present. This shows that analytical HIC can lead to 'false positive' signals.
Altogether, only two domains out of 17 were found suitable for NMR-structure analysis. This low yield reflects the typical behaviour of eukaryotic proteins/domains expressed in E. coli. Therefore, it is important to generate a large set of constructs for a particular domain with different boundaries as well as from different source proteins. Applying high-throughput protein production coupled to a biophysical method (1D 1H-NMR) and/or to a widely used chromatographic method (HIC), we were able to distinguish suitable from non-suitable proteins for detailed structural investigations.
Conclusions
We demonstrate here, how a large number of E. coli expression constructs for human domains can be quickly analysed to detect natively folded protein suitable for structural analysis. Upon expression in 96-well format (1 ml culture volume), constructs were purified with metal chelate chromatography using an automated purification procedure. Samples that yielded enough purified protein were successfully analysed for natively folded protein using 1D 1H-NMR (detection limit about 15 μg for 10 kDa domain) and analytical HIC (detection limit about 4 μg). Hence, identification of suitable constructs for structural analysis is facilitated without time consuming and expensive large-scale preparations. In most cases, the data obtained from both analyses approaches, 1D 1H-NMR and analytical HIC provided complementary results. HIC is the more cost-effective and more sensitive approach, but may be more error-prone.
Methods
Cloning
PCR reactions have been performed using the Expand High Fidelity system (Roche Diagnostics) according to manufacturers' instructions. PCR products were restriction digested using restriction sites introduced into the primers, ligated into the vector pQTEV (GenBank Accession Number AY243506) and transformed into E. coli SCS-1 cells carrying the pRosetta (Novagen) helper plasmid using a chemotransformation method [13]. All cloning steps were carried out in a parallel 96-well format where appropriate.
Protein expression in 96 deep-well microplates
A 96 deep-well microplate with 2 ml cavities was filled with 0.1 ml 2xYT-Medium supplemented with 2% glucose, 100 μg/ml ampicillin and 15 μg/ml kanamycin. Pre-cultures were inoculated with 96 expression clones using a custom made steel replicator carrying 96 × 6 cm long pins. The plate was sealed tightly and bacteria were grown overnight at 37°C. 0.9 ml pre-warmed SB-medium was added. The plate was covered with a lid allowing air exchange and a 3 h incubation at 37°C was performed. Protein expression was induced for 3 h at 37°C, or alternatively at 28°C, by adding IPTG to 1 mM final concentration. Cells were harvested by centrifugation at 4°C at 2,000 g for 10 min and frozen at -80°C.
Detection of folded protein using analytical HIC
For analysis with HIC, protein domains were purified as described [11]. 15 μl of the 80 μl eluates (containing 500 mM imidazole) were analysed with Coomassie-stained SDS-PAGE, using a modified protocol to separate small proteins [15]. The ammonium sulfate concentration of the remaining eluate was adjusted to 1 M using a 2 M stock solution under pipett mixing in 96-well format. Samples were filtrated through a 96 well 0.22 μm PVDF membrane (Millipore) and applied to a POROS HP2 column (0.83 ml bed volume, Applied Biosystems), equilibrated in 20 mM Tris-HCl, pH 7.7, 1 M ammonium sulfate. Chromatography was performed at 8°C (Vision workstation, Applied Biosystems). The column was washed with equilibration buffer and a gradient to 0 M ammonium sulfate was performed over 10 column volumes. Finally, a gradient from 0 M to 5 M GdmHCl over 5 column volumes was performed to elute tightly bound, presumably misfolded, protein. The flow rate was 3.5 ml/min; each run took approximately 15 min. Absorbance at 280 nm and 220 nm was monitored in parallel to detect eluted proteins. All shown chromatograms were baseline corrected.
Detection of folded protein using analytical 1D 1H-NMR and 2D 1H-15N-HSQC spectroscopy
Due to the perturbing effect of Tris-HCl and imidazole resonance signals during analysis with 1D 1H-NMR, the automated purification protocol [11] was slightly modified. Before elution with 80 μl 100 mM EDTA in 20 mM NaHPO4, pH 7.0, 50 mM NaCl, NiNTA-beads were washed using the same buffer without EDTA to remove Tris-HCl. Eluates were analysed with SDS-PAGE (10 μl) and 1D 1H-NMR (70 μl). Concentrations of purified protein were determined according to Bradford [14]. All constructs were run in duplicate. Duplicates were pooled for NMR analysis if SDS-PAGE indicated the same product. As a negative control (buffer spectrum), two 1 ml cultures from SCS1 cells containing no expression vector were subjected to the same purification protocol. The resulting 140 μl were made up to 500 μl with NMR-buffer stock solution to a final concentration of 20 mM K2HPO4/KH2PO4, pH 7.0, 50 mM NaCl and 10% D2O. NMR measurements were carried out at 15°C with a Bruker DRX600 spectrometer in standard configuration equipped with triple resonance probe head and a Bruker Sample changer. NMR-data were processed using the XWINNMR software (Bruker Biospin GmbH).
For the 1D NMR experiments 3072 scans were accumulated with a relaxation delay of 1.2 s. 8 K complex data points were acquired with a sweep of 10000 Hz. Water suppression was achieved by WATERGATE [16].
For expression of 15N labeled proteins, E. coli was grown on M9 minimal medium with 15NH4Cl (0.5 g/l) in 400 ml scale. Protein expression was induced for 4 h at 30°C by adding 1 mM IPTG. Cells were collected and desintegrated using a FRENCH® Press (SLM Aminco). Purification of the soluble His-tagged domains was achieved via a 1.7 ml MC-POROS column (Applied Biosystems) that had been previously loaded with Ni2+, TEV cleavage in the presence of 1 mM β-mercaptoethanol (β-mercaptoethanol in this concentration range does not interfere with MC-POROS resin) and a second metal chelating chromatography. The protein in the flow-through was concentrated to 0.5 ml using Vivaspin 15, 5,000 MWCO (Vivascience) and washed with 20 mM phosphate buffer, pH 7.0 and 50 mM NaCl. D2O was added (10% final concentration) before 2D 1H-15N-HSQC [17] spectra were measured at 15°C, recording 256 × 1024 complex points in the indirect (15N) and acquisition (1H) dimensions. Four scans were acquired per increment with a relaxation delay of 1.2 s.
Abbreviations
EDTA, ethylenediaminetetraacetic acid; GdmHCl, guanidine hydrochloride; HIC, hydrophobic interaction chromatography; HSQC, heteronuclear single quantum coherence; IPTG, isopropyl β-D-1-thiogalactopyranoside, NMR, nuclear magnetic resonance; TEV, tobacco etch virus
Authors' contributions
CS: protein expression, protein purification, SDS-PAGE and analysis with HIC, data evaluation; DL: evaluation of NMR spectra; VS: production of the expression clones, sequence analysis; ML: large scale expression for 2D 1H-15N-HSQC; BSch: recording/processing NMR-spectra; BS: identification of domains/preparation of the target list; IL: identification of domains/preparation of the target list; KB: project initiation, identification of domains/preparation of the target list, data processing, AD: project initiation, NMR-analysis
Supplementary Material
List of RZPD clone IDs to order the used expression clones at the German Resource Center for Genome Research GmbH http://http//:www.rzpd.de
Acknowledgments
Acknowledgments
We are thankful to Frank Götz, Thrung Pham and Thomas Grund for large-scale expression and purification of the analysed SEP-UBX construct and Frank Niesen for biophysical analysis of this construct. Furthermore, we thank Joshi Mangesh for recording 1D 1H-NMR spectra and Grant Langdon for critical revision of the manuscript. This work was funded by the German Federal Ministry of Education and Research (BMBF) through the "Leitprojektverbund Proteinstrukturfabrik".
Contributor Information
Christoph Scheich, Email: scheich@molgen.mpg.de.
Dietmar Leitner, Email: leitner@fmp-berlin.de.
Volker Sievert, Email: sievert@molgen.mpg.de.
Martina Leidert, Email: leidert@fmp-berlin.de.
Brigitte Schlegel, Email: schlegel@fmp-berlin.de.
Bernd Simon, Email: simon@embl-heidelberg.de.
Ivica Letunic, Email: letunic@embl-heidelberg.de.
Konrad Büssow, Email: buessow@molgen.mpg.de.
Anne Diehl, Email: diehl@fmp-berlin.de.
References
- Buchanan SG, Sauder JM, Harris T. The promise of structural genomics in the discovery of new antimicrobial agents. Current Pharmaceutical Design. 2002;8:1173–1188. doi: 10.2174/1381612023394809. [DOI] [PubMed] [Google Scholar]
- Zhang C, Kim SH. Overview of structural genomics: from structure to function. Current Opinion in Chemical Biology. 2003;7:28–32. doi: 10.1016/S1367-5931(02)00015-7. [DOI] [PubMed] [Google Scholar]
- Rehm T, Huber R, Holak TA. Application of NMR in structural proteomics: screening for proteins amenable to structural analysis. Structure. 2002;10:1613–8. doi: 10.1016/S0969-2126(02)00894-8. [DOI] [PubMed] [Google Scholar]
- Woestenenk EA, Hammarström M, Härd T, Berglund H. Screening methods to determine biophysical properties of proteins in structural genomics. Anal Biochem. 2003;318:71–79. doi: 10.1016/S0003-2697(03)00162-3. [DOI] [PubMed] [Google Scholar]
- Gronenborn AM, Clore GM. Rapid screening for structural integrity of expressed proteins by heteronuclear NMR spectroscopy. Protein Science. 1996;5:174–177. doi: 10.1002/pro.5560050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheich C, Niesen F, Seckler R, Büssow K. An automated in vitro folding screen applied to a human dynactin subunit. Protein Science. 2004;13:370–380. doi: 10.1110/ps.03304604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Research. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Research. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büssow K, Nordhoff E, Lubbert C, Lehrach H, Walter G. A human cDNA library for high-throughput protein expression screening. Genomics. 2000;65:1–8. doi: 10.1006/geno.2000.6141. [DOI] [PubMed] [Google Scholar]
- Büssow K, Cahill D, Nietfeld W, Bancroft D, Scherzinger E, Lehrach H, Walter G. A method for global protein expression and antibody screening on high-density filters of an arrayed cDNA library. Nucleic Acids Research. 1998;26:5007–5008. doi: 10.1093/nar/26.21.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheich C, Sievert V, Büssow K. Comparison of His-tag and GST-tag affinity chromatography by automated protein purification. BMC Biotechnol. 2003;3:12. doi: 10.1186/1472-6750-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Cierpicki T, Derewenda U, Krowarsch D, Feng YY, Devedjiev Y, Dauter Z, Walsh CA, Otlewski J, Bushweller JH, Derewenda ZS. The DCX-domain tandems of doublecortin and doublecortin-like kinase. Nature Structural Biology. 2003;10:324–333. doi: 10.1038/nsb918. [DOI] [PubMed] [Google Scholar]
- Inoue H, Nojima H, Okayama H. Abstract High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–8. doi: 10.1016/0378-1119(90)90336-P. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Okajima T, Tanabe T, Yasuda T. Nonurea sodium dodecyl sulfate-polyacrylamide gel electrophoresis with high-molarity buffers for the separation of proteins and peptides. Anal Biochem. 1993;211:293–300. doi: 10.1006/abio.1993.1272. [DOI] [PubMed] [Google Scholar]
- Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–5. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Kay LE, Keifer P, Saarinen P. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–5. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of RZPD clone IDs to order the used expression clones at the German Resource Center for Genome Research GmbH http://http//:www.rzpd.de