Abstract
The Bacillus subtilis lmrAB operon is involved in multidrug resistance. LmrA is a repressor of its own operon, while LmrB acts as a multidrug efflux transporter. LmrA was produced in Escherichia coli cells and was shown to bind to the lmr promoter region, in which an LmrA-binding site was identified. Genome-wide screening involving DNA microarray analysis allowed us to conclude that LmrA also repressed yxaGH, which was not likely to contribute to the multidrug resistance. LmrA bound to a putative yxaGH promoter region, in which two tandem LmrA-binding sites were identified. The LmrA regulon was thus determined to comprise lmrAB and yxaGH. All three LmrA-binding sites contained an 18-bp consensus sequence, TAGACCRKTCWMTATAWT, which could play an important role in LmrA binding.
Multidrug resistance of bacteria is one of the most severe problems in the clinical treatment of infectious diseases. To date, it has been shown that many of the mechanisms for bacterial multidrug resistance require the expression of genes for drug efflux transporters (1, 12). In Bacillus subtilis, at least three genes encoding multidrug resistance efflux transporters have been characterized, namely, blt, bmr, and bmr3 (2, 14, 16). It was shown previously that bmr and blt were induced in the presence of their substrates under control of the transcriptional regulators BmrR and BltR, respectively (21, 28). Both these regulators belong to the MerR family of activators, and at least DNA binding of BmrR required interaction with a Bmr substrate, such as rhodamine 6G or tetraphenylphosphonium, as a coactivator (21, 27, 28). Recently, B. subtilis lmrB was found to encode a fourth multidrug efflux transporter belonging to the major facilitator superfamily (13). The lmrB gene is the second gene of the lmrAB operon (Fig. 1A) (10), and the first gene, lmrA, was thought to encode a transcriptional repressor of the TetR family (3).
FIG. 1.
Organization of the lmrAB operon and its promoter region. (A) Organization of the lmrAB operon. The regions cloned into plasmid pLMRA for LmrA production in E. coli and used as probes for the Northern analysis are indicated. (B) lmr promoter region. The sequences of the DNA strands corresponding to the lmr promoter region and the N-terminal part of the lmrA coding region are shown (the amino acid sequence of the coding region is indicated beneath the nucleotide sequence). The positions and orientations of PCR primers used to prepare gel retardation probes are indicated by thin horizontal arrows. The dotted lines in the arrows for the del1 and del2 primers indicate the internally deleted 36-bp stretch. The −10 and −35 regions, the transcription start site (position 1) of the promoter, and the Shine-Dalgarno sequence (SD) are enclosed in boxes. The pair of discontinuous thick horizontal arrows facing each other indicates an incomplete palindrome sequence. The vertical arrows indicate the mutation points found in strains PLR1 and 1A221.
A mutation in the lmr promoter region (lin-2) that confers lincomycin resistance in B. subtilis 1A221 through enhanced expression of lmrAB has been reported (7). Additionally, 19 spontaneous lincomycin-resistant mutants have been isolated and have also been shown to elevate the expression of lmrAB (11). Eighteen of these 19 mutants had mutations in the promoter region with no alternation in the lmrA coding region, while the remaining mutant possessed no mutation in the promoter region but had a substitution in the lmrA termination codon, which extended the C terminus for 9 amino acid residues [lmrA(stop to S)] (11, 13). In a subsequent study, two mutants selected in the presence of high concentrations of lincomycin and puromycin, PLR1 and PLR2, were isolated and shown to exhibit multidrug resistance (13). Both PLR1 and PLR2 had increased expression of lmrAB, and inactivation of lmrB in each of them abolished the multidrug resistance. PLR1 had a nucleotide substitution in the lmr promoter region, while PLR2 possessed no mutation in the promoter region but had two mutations in the lmrA coding region, lmrA(stop to S) and lmrA(Q52P) within the putative helix-turn-helix motif of the TetR family found between the 10th and 57th amino acid residues (BSORF website [http://bacillus.genome.ad.jp/]). These findings implied that LmrA might be a repressor that interacts with the promoter region of its own operon.
In this study we found that LmrA is a repressor of the lmrAB operon, and we identified its binding site in the lmr promoter region. During systematic genome-wide screening, an additional LmrA target, yxaGH, was identified, and in its putative promoter region two tandem LmrA-binding sites were found.
MATERIALS AND METHODS
Bacterial strains and plasmids.
B. subtilis strain 168 (trpC2) was used as the wild-type strain. B. subtilis strain 1A221 (lin-2) (7), a lincomycin-resistant mutant, was obtained from the Bacillus Genetic Stock Center (Columbus, Ohio). B. subtilis strains PLR1 [trpC2 Plmr(A-1T) lmrA(stop to S)], PLR2 [trpC2 lmrA(Q52P stop to S)], and PLR3 (= PLR2/lmrB::cat) [trpC2 lmrA(Q52P stop to S) lmrB::cat] were produced in a previous study (13). B. subtilis strains YXAGd (trpC2 yxaG::pMUTIN2) and YXAHd (trpC2 yxaH::pMUTIN2) are two of the pMUTIN2 integrants (20) of strain 168 constructed in the course of a functional analysis in the B. subtilis genome project (24) (BSORF website). Strains PLR2 and PLR3 were transformed with DNA from YXAGd, and this was followed by selection for erythromycin resistance, which yielded strains PLR4 [trpC2 lmrA(Q52P stop to S) yxaG::pMUTIN2] and PLR5 [trpC2 lmrA(Q52P stop to S) lmrB::cat yxaG::pMUTIN2], respectively. Strains PLR6 [trpC2 lmrA(Q52P stop to S) yxaH::pMUTIN2] and PLR7 [trpC2 lmrA(Q52P stop to S) lmrB::cat yxaH::pMUTIN2] were constructed as described above by using DNA of YXAHd. Escherichia coli strain JM109 and plasmid pUC18 (22) were used to clone and express lmrA as described below. B. subtilis cells were grown on tryptose blood agar base (Difco) plates supplemented with 0.18% glucose at 30°C and in Luria-Bertani (LB) liquid medium (18) at 37°C with shaking. E. coli cells were grown on LB medium plates at 37°C and in TGA liquid medium (9) at 37°C with shaking. Ampicillin (50 μg/ml), erythromycin (0.3 μg/ml), and lincomycin (100 μg/ml) were added to culture media for selection and growth of mutants and transformants as required.
Production of LmrA in E. coli cells.
For production of LmrA in E. coli cells, plasmid pLMRA was constructed as follows. A 0.6-kb PCR fragment, which contained the entire reading frame of lmrA with its corresponding ribosome binding site (Fig. 1A), was amplified from B. subtilis strain 168 genomic DNA by using a pair of primers, primers lmrAE and lmrAB (Table 1), which were designed to generate EcoRI and BamHI sites at the head and tail of the fragment, respectively. This fragment was trimmed with EcoRI and BamHI and then ligated with the arm of plasmid pUC18 that had been cleaved with the same enzymes. E. coli JM109 was transformed with the ligated DNA described above to obtain ampicillin-resistant colonies on LB plates. Plasmid DNA was extracted from one of the transformants, and its correct construction was confirmed by nucleotide sequencing in order to produce plasmid pLMRA. JM109 cells carrying pLMRA were grown in TGA medium containing 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce lmrA under control of the pUC18-borne lac promoter.
TABLE 1.
PCR primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| 1mrAE | CCGGAATTCGATTGCAATTCAGTAAGGTGG (EcoRI site is underlined) |
| 1mrAB | CGCGGATCCATGCTTTAGCTGTTGTTTCC (BamHI site is underlined) |
| MK1 | CAGTCAAAAGATCAGTCAGC |
| MK2 | TGCCGTAATACCCTTGCAGC |
| d1 | AAAAAGCCGGGTAGCTGCC |
| d2 | GATTTTCTCACGGGAATCTC |
| d3 | AACTCATAACATTCCCACCTTA |
| d4 | GCAATCAAAAATATGTGACTGG |
| d5 | CTTGATTCAATCAATCATCAATTGTCAAG |
| d3R | TAAGGTGGGAATGTTATGAGTT |
| d4R | CCAGTCACATATTTTTGATTGC |
| d5R | CTTGACAATTGATGATTGAATCAAG |
| de11 | TTCTTGACAATTGATGATTGGCAATTCAGTAAGGTGGGAA |
| de12 | TTCCCACCTTACTGAATTGCCAATCATCAATTGTCAAGAA |
| PyxaG1 | GGTGAGGAAAAAAGGGTAGC |
| PyxaG2 | CTCCGGAGCAAATAAGGCAT |
| NlmrA1 | ACGGGGCTGAACCAGATTAT |
| NlmrA2 | CAGGAATGCAGCTGGAGATA |
| NlmrB1 | GTATTCGGATTCAGCAACGC |
| NlmrB2 | ATCATCAAGCAGGTGTGCAG |
| NyxaG1 | TAGATGAAGCAAAGCCTGCA |
| NyxaG2 | AAAGCGCAAAGCCTGAGGCT |
| NyxaH1 | TGCGGCAGCAAAATCACGGT |
| NyxaH2 | CAATAGGCTGCCACGACAGA |
Gel retardation and DNase I footprinting experiments.
Gel retardation experiments were performed as described previously (23). A protein extract was prepared from strain JM109 cells carrying either plasmid pLMRA or plasmid pUC18 grown in the presence of 1 mM IPTG. For analysis of the lmr promoter region, DNA probes (Fig. 2A) were amplified and labeled by PCR in the presence of [α-32P]dCTP (ICN Biomedicals) by using B. subtilis strain 168 genomic DNA as a template and specific primer pairs (Fig. 1B and Table 1). A probe carrying an internal deletion of a 36-bp region carrying the incomplete palindrome (Fig. 1B) was prepared as follows. A DNA fragment with the deletion was amplified by recombinant PCR (8) from DNA of strain 168 by using a flanking primer pair (primers MK1 and MK2) and an internal overlapping primer pair (primers del1 and del2) (Fig. 1B and Table 1). The correct deletion was confirmed by DNA sequencing. The fragment was used as a template for subsequent PCR in the presence of [α-32P]dCTP by employing the pair of flanking primers described above to obtain a labeled probe with the internal deletion. To confirm the putative LmrA-binding sites predicted as described below, labeled probes designed to carry each of the putative sites were prepared. The PyxaG probe (see Fig. 5A), an example of such a probe, was a PCR fragment derived from strain 168 DNA that was amplified and labeled by using the specific primer pair PyxaG1 and PyxaG2 (Table 1). Each of the labeled probes (0.02 pmol) was combined with various amounts of E. coli protein extract in a reaction mixture (25 μl) (10 mM Tris-Cl [pH 7.6], 1 mM Na-EDTA, 0.1 mM dithiothreitol, 2 μg of bovine serum albumin per μl) in the presence of 3.3 μg of fragmented salmon sperm DNA as described previously (23), and then the mixture was subjected to 5% polyacrylamide gel electrophoresis. For the competition assay, specific competitors were synthesized in the form of PCR fragments amplified from DNA of strains 168, 1A221, and PLR1 with the MK1-MK2 primer pair. Various concentrations of these competitors were added to the assay mixture to compete for LmrA binding with a fixed amount of the labeled probe prepared from strain 168 DNA by using the same primer pair.
FIG. 2.
Gel retardation analysis of LmrA binding to the lmr promoter region. (A) Probe settings. The positions and orientations of the PCR primers used for probe preparation are indicated schematically. The pair of horizontal arrows facing each other indicates the incomplete palindrome sequence. The probe designations are indicated on the left, and the thick horizontal lines and dotted lines indicate the stretches present and deleted in each of the probes, respectively. On the right, the results of gel retardation assay are summarized (Yes and No indicate LmrA binding and no LmrA binding, respectively). (B) Interaction between LmrA and the MK1-MK2 probe. The MK1-MK2 probe (0.02 pmol) was mixed with a protein extract of JM109 cells carrying plasmid pLMRA to obtain a reaction mixture (25 μl) (lane 2, 7.5 μg; lane 3, 3.8 μg; lane 4, 1.9 μg; lane 5, 0.9 μg) or pUC18 (lane 6, 7.5 μg) grown in the presence of 1 mM IPTG, and without the extract (lane 1). The positions of LmrA-probe complexes (bound) and free probe (free) are indicated on the right. (C) Deletion analysis. Gel retardation experiments were performed like the experiments described above. Each of the probes indicated was mixed with an extract of JM109 cells carrying pLMRA (lanes 2, 7.5 μg; lanes 3, 3.8 μg) or pUC18 (lanes 4, 7.5 μg) or with no extract (lanes 1). The position of LmrA-probe complexes is indicated by arrows.
FIG. 5.
Organization of the yxaGH operon and gel retardation analysis of LmrA binding to the putative promoter region. (A) Organization of the yxaGH operon. The regions corresponding to the PyxaG probe for gel retardation (see panel B) and the yxaG and yxaH probes for Northern analyses are indicated. (B) Gel retardation analysis of LmrA binding to the putative yxaGH promoter region. The conditions for the experiments and the lane assignments are the same as those described in the legend to Fig. 2B, except that the PyxaG probe was used.
DNase I footprinting experiments were performed as described previously (23). The same protein extracts that were used in the gel retardation experiments described above were employed. The probes for DNase I footprinting experiments were prepared by PCR amplification of fragments of strain 168 DNA by using the specific primer pair MK1-MK2 (for the lmr promoter region) and PyxaG1-PyxaG2 (for the yxaGH promoter region). Prior to PCR amplification, the 5′ termini of the primers were labeled by using a Megalabel kit (Takara Shuzo) with [γ-32P]ATP (Amersham) so that either the coding strand or the noncoding strand was labeled.
DNA microarray analysis and GRASP-DNA search for putative LmrA binding sites.
To screen for additional LmrA targets, we employed a strategy analogous to that used in a previous study involving combined DNA microarray and gel retardation analyses (26). DNA microarray analysis was performed as described previously (26). B. subtilis strains PLR2 and 168 were grown in LB liquid medium, and then the cells were harvested in the middle of the logarithmic phase at an optical density at 600 nm of 0.5 and disrupted to extract total RNA by vigorous shaking with glass beads in the presence of sodium dodecyl sulfate and phenol (25). The two RNA samples were converted to cDNAs, and this was followed by differential labeling with a fluorescent dye, Cy3 or Cy5 (15). The differentially labeled cDNAs were mixed and hybridized to a glass slide microarray on which probe DNAs of 4,005 B. subtilis genes and control DNAs had been spotted (25). After washing, the microarray was scanned with a GMS 418 array scanner (Affymetrix/Genetic MicroSystems) to generate two images of dye-specific fluorescence. The signals in these images were quantified by using ImaGene software (version 4.1; Biodiscovery). Subsequently, the signal data set was processed as described previously (26) to identify genes exhibiting altered expression profiles, and then putative transcription units containing such genes were deduced to be candidates for additional LmrA targets.
For prediction of putative LmrA-binding sites, a web-based application, GRASP-DNA, was used (19; http://www2.genomatica.com/grasp-dna/). The 36-bp sequence carrying the incomplete palindrome of the lmr promoter region (Fig. 1B) was used as a query sequence. GRASP-DNA automatically built a weight matrix based on the query, found sequences matching the matrix within the B. subtilis genome, and reported such sequences together with their adjacent genes. Among the reported sequences, those associated with at least one gene within the deduced LmrA target candidates were selected as putative LmrA-binding sites.
Northern hybridization.
Northern blot analysis was performed as described previously (24). RNA samples were prepared in the same way that samples were prepared for the DNA microarray analysis. The probes were PCR products, corresponding to parts of lmrA, lmrB, yxaG, and yxaH (see Fig. 1A and 5A), that were amplified from B. subtilis strain 168 genomic DNA by using specific primer pairs and were labeled by using a BcaBest labeling kit (Takara Shuzo) with [α-32P]dCTP. The primer pairs employed were NlmrA1-NlmrA2 (for lmrA), NlmrB1-NlmrB2 (for lmrB), NyxaG1-NyxaG2 (for yxaG), and NyxaH1-NyxaH2 (for yxaH) (Table 1).
Assay for sensitivity to drugs and heavy metal ions.
The sensitivity of B. subtilis strains to drugs and heavy metal ions was assayed as described previously (13). Cellular growth was monitored by measuring the optical density at 540 nm of cells grown in LB medium containing various concentrations of a drug or heavy metal ion. The concentration of the drug or ion that resulted in 50% growth inhibition was determined.
RESULTS
LmrA produced in E. coli binds specifically to the lmr promoter region.
A DNA fragment containing the coding region of B. subtilis lmrA with its corresponding ribosome-binding site (Fig. 1A) was cloned into the multiple-cloning site of plasmid pUC18 to produce plasmid pLMRA, which placed lmrA under control of the lac promoter (22). When E. coli strain JM109 cells carrying pLMRA were grown in the presence of IPTG, we observed an extra protein band for the crude extract of the cells exhibiting the size (about 20 kDa) expected for LmrA (data not shown). The crude extract containing LmrA was used in gel retardation assays to examine the DNA binding of LmrA. As shown in Fig. 2B, the MK1-MK2 probe (Fig. 2A) carrying the entire lmr promoter region gave a distinct retarded band with the extract containing LmrA (Fig. 2B). The amount of this retarded band increased as the concentration of LmrA increased, while no such retarded band was observed with the most concentrated extract of JM109 cells carrying pUC18 that did not contain LmrA. Therefore, the retarded band most likely represented a specific LmrA-probe complex.
Identification of the LmrA-binding site.
To identify the LmrA-binding site within the MK1-MK2 probe, a series of deleted probes (Fig. 2A) were prepared to examine the LmrA binding by the gel retardation assay. As shown in Fig. 2C, four probes (MK1-d1, MK1-d2, MK1-d3, and d5R-MK2) gave distinct retarded bands for LmrA-DNA complexes, while the other four probes (MK1-d4, MK1-d5, d3R-MK2, and d4R-MK2) did not. However, the MK1-d4 probe exhibited specific tailing in the presence of LmrA, implying that there was a weak interaction with LmrA (Fig. 2C). These results suggested that the region from position −36 to position 20 (position 1 is the transcription initiation nucleotide [11]) present in both of the shortest LmrA-interacting probes, MK1-d4 and d5R-MK2, could be required for the LmrA interaction (Fig. 2A). When the sequence of this region was examined, a 36-bp sequence comprising an incomplete palindrome sequence from position −18 to position 18 was found (Fig. 1). An internal deletion that eliminated the 36-bp region (del probe in Fig. 2A) abolished the LmrA binding completely (Fig. 2C). Furthermore, DNase I footprinting of LmrA in the lmr promoter region revealed that the regions from position −13 to position 11 of the noncoding strand (Fig. 3A, left gel) and from position −12 to position 12 of the coding strand (Fig. 3A, right gel) were protected from DNase I by the LmrA binding, and the protected area was located in the middle of the region containing the palindrome sequence (Fig. 3B). Addition of an excess amount of lincomycin (10 mM) failed to abolish the LmrA binding (Fig. 3A, lane 6), suggesting that this drug does not act as an inducer that antagonizes the interaction between LmrA and DNA.
FIG. 3.
DNase I footprinting of LmrA in the lmr promoter region. (A) DNase I footprinting of the noncoding (left gel) and coding (right gel) strands of the DNA of the lmr promoter region. The 5′-labeled probe (0.04 pmol) was combined in a reaction mixture (50 μl) with protein extract (lanes 2, 4, and 6, 15.0 μg; lanes 3 and 5, 7.5 μg) prepared from JM109 cells carrying plasmid pLMRA (lanes 2, 3, and 6) or pUC18 (lanes 4 and 5) or with no extract (lane 1). The mixture loaded in lane 6 also contained 10 mM lincomycin. After DNase I digestion, samples were analyzed by sequencing 6% polyacrylamide gel electrophoresis. Areas protected by LmrA binding are indicated on the right of each gel, and the nucleotide sequences are shown. Lanes G, A, T, and C contained ladders created by dideoxy sequencing reactions with the corresponding 5′-labeled primers. (B) Summary of the DNase I footprint assay. The nucleotide sequences of the noncoding and coding strands of the lmr promoter region are shown. The −10 and −35 regions are underlined, and the transcription start site (position 1) and the Shine-Dalgarno sequence (SD) are enclosed in boxes. The facing horizontal arrows indicate the incomplete palindrome sequence. The protected areas found in the noncoding and coding strands are indicated by open boxes above and below the sequences, respectively.
Mutations in the LmrA-binding site lowered LmrA affinity.
Nonlabeled DNA fragments corresponding to the MK1-MK2 probe were amplified by PCR from DNA of strains 168, PLR1, and 1A221. The PLR1 and 1A221 fragments possessed mutations at the −1 position (A to T) and at both position −1 (A to C) and position 15 (G to T), respectively (Fig. 1). Specific competitors in the form of various amounts of each of the three fragments were added to gel retardation assay mixtures containing fixed amounts of the protein extract and the labeled MK1-MK2 probe. As shown in Fig. 4, the PCR fragment amplified from strain 168 DNA was able to compete with the labeled probe to decrease LmrA-probe complex formation as the amount was increased. Neither the fragment derived from PLR1 DNA nor the fragment derived from 1A221 DNA was found to be as competitive as the fragment derived from 168 DNA. The results suggested that the mutations of PLR1 and 1A221 could lower the LmrA-binding affinity.
FIG. 4.
Competition assay of LmrA binding between the wild-type DNA and mutated DNA of the lmr promoter region. The MK1-MK2 probe (0.02 pmol) was combined in the gel retardation assay reaction mixture (25 μl) with an extract (3.8 μg) of JM109 cells carrying plasmid pLMRA (lanes 2 to 14) or pUC18 (lane 15) or with no extract (lane 1). The specific competitors comprised nonlabeled PCR fragments corresponding to the MK1-MK2 probe amplified from DNA of strains 168 (WT) (lanes 3 to 6), PLR1 (lanes 7 to 10), and 1A221 (lanes 11 to 14), and the following amounts were added to the reaction mixture: 0.02 pmol (1×) (lanes 3, 7, and 11), 0.04 pmol (2×) (lanes 4, 8, and 12), 0.08 pmol (4×) (lanes 5, 9, and 13), and 0.16 pmol (8×) (lanes 6, 10, and 14). The positions of the LmrA-probe complex (bound) and free probe (free) are indicated by arrows.
yxaGH operon is in the LmrA regulon.
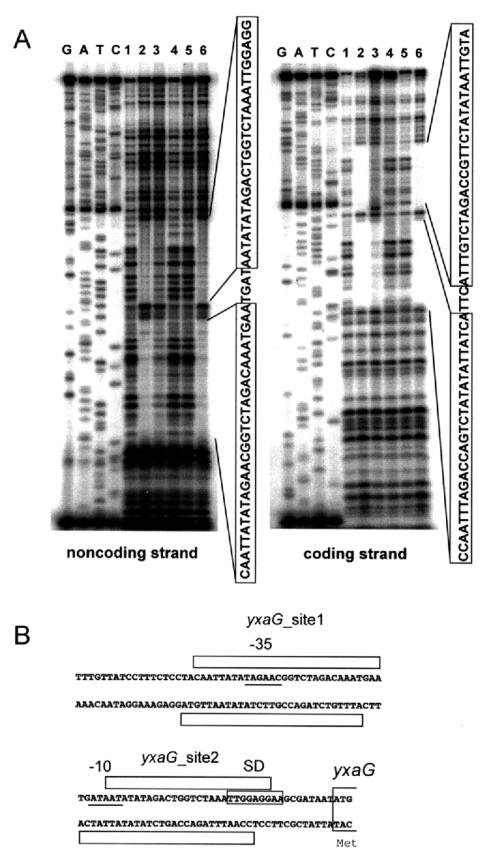
The mutated LmrA of strain PLR2 had completely lost repressor activity (13). In order to screen for additional LmrA candidate targets, DNA microarray analysis was carried out to compare the transcriptomes in strain PLR2 and 168 cells (data available at http://www.genome.ad.jp/kegg/expression/). On the other hand, a GRASP-DNA search (19) by using the 36-bp sequence carrying the incomplete palindrome sequence found in the lmr promoter region as a query was performed. This combined search revealed some additional candidates associated with a putative LmrA-binding site, such as yxaG. Gel retardation analyses to examine the interaction between LmrA and each of the putative binding sites revealed that LmrA strongly bound to a putative promoter region of the yxaGH operon (Fig. 5) but not to the other regions under the conditions which we used (data not shown). Northern analysis was performed to confirm that the levels of both the yxaGH and lmrAB transcripts were increased in PLR2; the yxaG and yxaH genes were cotranscribed as a 2.5-kb transcript (data not shown). These results indicated that yxaGH was an additional LmrA target operon. DNase I footprint analysis of LmrA in the yxaGH promoter region revealed two tandem LmrA-binding sites, yxaG sites 1 and 2 (Fig. 6). yxaG site 2 corresponded to the putative site predicted in the GRASP-DNA search, while the search failed to predict site 1 because of its lower level of similarity to the query sequence (Fig. 7). Lincomycin did not interfere with the LmrA binding to either of these sites (Fig. 6A, lane 6). Putative −35 (TAGAAC) and −10 (GATAAT) sequences separated by a 17-bp spacer were found within the region covered by LmrA (Fig. 6B). The two newly identified LmrA-binding sites of the yxaGH promoter region did not possess the palindrome sequence found in the binding site of the lmr promoter region. Instead, all three binding sites were found to share a conserved 18-bp consensus sequence, TAGACCRKTCWMTATAWT (Fig. 7). This 18-bp sequence is located within and close to the middle of the 36-bp incomplete palindrome sequence of the lmr promoter region. In all three cases, the 18-bp sequence is located within the area protected from DNase I by LmrA binding. Conservation of this 18-bp sequence was not seen for the other putative binding sites predicted by the GRASP-DNA search, and LmrA did not interact with any of the probes carrying the other putative sites (data not shown). Moreover, another GRASP-DNA search involving all three conserved 18-bp sequences as queries failed to predict any more additional putative binding sites (data not shown). These results suggested that the conserved 18-bp region might play an important role in LmrA binding and that LmrA could regulate lmrAB and yxaGH exclusively within the genome.
FIG. 6.
DNase I footprinting of LmrA in the putative yxaGH promoter region. (A) DNase I footprinting in the yxaGH promoter region. The conditions for the experiments and the lane assignments are the same as those described in the legend to Fig. 3A, except for the probes (see text).Protected areas are enclosed in boxes in the nucleotide sequence on the right of each gel. (B) Summary of the DNase I footprint assaying. The nucleotide sequences of both strands of the yxaGH promoter region are shown. The putative −10 and −35 regions are underlined, and the putative Shine-Dalgarno sequence (SD) is enclosed in a box. The protected areas of yxaG site 1 and yxaG site 2 are indicated by open boxes.
FIG. 7.
Alignment of the nucleotide sequences of the three LmrA-binding sites. The 36-bp nucleotide sequences of both strands of the three LmrA-binding sites (lmrA site, yxaG site 1, and yxaG site 2) are aligned. To optimize the alignment, the sequences of the lmrA site are oriented so that transcription occurs from left to right, while those of yxaG site 1 and yxaG site 2 are in the opposite orientation. The nucleotide positions conserved in two of the three sites are indicated by asterisks on the lines labeled lmrA vs site2 (between the lmrA site and yxaG site 2), lmrA vs site1 (between the lmrA site and yxaG site 1), and site2 vs site1 (between yxaG site 2 and yxaG site 1). Beneath the alignment, the 18-bp consensus sequence for the three sites is indicated by asterisks that indicate the positions conserved in the three sites. The protected areas found in the DNase I footprinting experiments are enclosed in boxes. The pair of horizontal arrows facing each other above the sequence of the lmrA site indicates the incomplete palindrome sequence. The dotted line beneath the upper sequence of yxaG site 2 indicates that this sequence was predicted to be a putative LmrA-binding site during the GRASP-DNA search. The vertical arrows indicate the mutation points found in strains PLR1 and 1A221.
Functional analysis of lmrB and yxaGH.
Since LmrA is a repressor of lmrAB for multidrug resistance, we investigated the possibility that the second LmrA target, yxaGH, also contributed to drug resistance. Functional analysis of lmrB, yxaG, and yxaH was carried out in order to determine the extents of involvement of these genes in resistance to drugs and heavy metal ions. The growth rates of B. subtilis strains 168, PLR2, PLR3, PLR4, PLR5, PLR6, and PLR7 (Table 2) were compared in the presence of various chemicals (chloramphenicol, ampicillin, carbenicillin, erythromycin, lincomycin, puromycin, novobiocin, kanamycin, neomycin, spectinomycin, streptomycin, ofloxacin, norfloxacin, levofloxacin, tosufloxacin, mitomycin C, daunorubicin, doxorubicin, rifampin, verapamil, bicyclomycin, acriflavine, acridine orange, crystal violet, ethidium bromide, rhodamine 6G, proflavine, pyronin Y, Cd2+, Hg2+, Zn2+, and Co2+). Mutant PLR2 (which expresses both lmrB and yxaGH) was found to exhibit multidrug resistance, as reported previously (13) (data for lincomycin and puromycin are shown in Table 2), while PLR3 (which possesses the lmrB inactivation locus [lmrB::cat] but expresses yxaGH) was found to have almost completely lost the resistance phenotype. These results suggest that yxaGH might not contribute to multidrug resistance. In addition, the pMUTIN2 integration disrupting either yxaG or yxaH (PLR4 and PLR6 in a PLR2 background and PLR5 and PLR7 in a PLR3 background) did not cause a significant change in the sensitivity to puromycin (Table 2) or the other chemicals tested (data not shown) except for erythromycin, lincomycin, and Hg2+. The pMUTIN2-borne erm gene (20) conferred resistance to macrolides such as erythromycin and lincomycin; thus, the 50% inhibitory concentrations of lincomycin for the pMUTIN2 integrants could not be determined (Table 2) (at least 70% of the cells were viable at the highest concentration tested, 2 mg/ml). Unexpectedly, the Hg2+ sensitivity of the cells was elevated after disruption of either yxaG or yxaH only in a PLR2 background (PLR4 and PLR6), in which the lmrA mutation allowed lmrB expression (Table 2).
TABLE 2.
Growth inhibition of B. subtilis strains in the presence of lincomycin, puromycin, and Hg2+
| Strain | Relevant genotype | IC50a
|
||
|---|---|---|---|---|
| Lincomycin (μg/ml) | Puromycin (μg/ml) | Hg2+ (μM) | ||
| 168 | Wild type | 8.5 ± 2.9 | 14.4 ± 5.1 | 7.2 ± 1.4 |
| PLR2 | 1mrA(Q52P stop to S) | 106.6 ± 25.3 | 474.6 ± 57.7 | 6.6 ± 1.3 |
| PLR3 | 1mrA(Q52P stop to S) 1mrB::cat | 3.8 ± 0.5 | 7.3 ± 1.4 | 8.3 ± 2.1 |
| PLR4 | 1mrA(Q52P stop to S) yxaG::pMUTIN2 | NDb | 464.6 ± 23.8 | 3.4 ± 1.1 |
| PLR5 | 1mrA(Q52P stop to S) 1mrB::cat yxaG::pMUTIN2 | ND | 9.2 ± 0.8 | 9.5 ± 0.7 |
| PLR6 | 1mrA(Q52P stop to S) yxaH::pMUTIN2 | ND | 468.4 ± 18.4 | 3.4 ± 1.1 |
| PLR7 | 1mrA(Q52P stop to S) 1mrB::cat yxaH::pMUTIN2 | ND | 9.5 ± 0.9 | 10.2 ± 0.3 |
IC50, concentration of the drug or ion that gave 50% growth inhibition. The values are mean ± standard deviations for at least four independent measurements.
ND, not determined under the assay conditions employed, due to the presence of the pMUTIN2-borne erm gene (20), which is known to confer resistance to lincomycin, as well as erythromycin, on the cells.
DISCUSSION
In this study the LmrA-binding site for repression of the lmr promoter was identified by gel retardation and DNase I footprint analyses (Fig. 2 and 3). DNA microarray and gel retardation analyses revealed that yxaGH acted as an additional LmrA target (Fig. 5). Two tandem LmrA-binding sites were identified within the putative yxaGH promoter region in the DNase I footprinting experiments (Fig. 6). All three LmrA-binding sites were found to possess an 18-bp consensus sequence, TAGACCRKTCWMTATAWT (Fig. 7). However, the results of gel retardation competition assays (Fig. 4) suggested that strain 1A221 DNA carrying the double mutations at positions −1 (A to C) and 15 (G to T) could have slightly lower affinity for LmrA than the PLR1 DNA with the single mutation at position −1 (A to T) has (Fig. 4), and thus the mutation at position 15 outside the 18-bp sequence (Fig. 7) might also be involved in LmrA binding. In addition, the MK1-d4 probe, including the 18-bp sequence, failed to yield a distinct LmrA-probe complex, as judged by the gel retardation experiment (Fig. 2). Nevertheless, the MK1-d4 probe band exhibited tailing in the presence of LmrA (Fig. 2), implying that an incomplete interaction may have occurred. Therefore, it is likely that tight binding of LmrA to the lmr promoter region requires not only the 18-bp sequence but also some extended region. A further point for consideration is that the tandem LmrA-binding sites in the yxaGH promoter region were found to be located close to each other (Fig. 6). It is possible that they might enhance the affinity of LmrA binding in a cooperative fashion. More precise studies are needed to clarify these findings.
None of the broad range of chemicals which we tested (including lincomycin) interfered with the interaction between LmrA and the three binding sites identified above (Fig. 3 and 6) (data not shown). Therefore, at present, no factors other than mutations have been found to induce the LmrA-repressed genes. However, it is very possible that an unidentified inducer could be responsible for the inactivation of the repressor function of LmrA, allowing expression of LmrA targets. Furthermore, it is possible that LmrA-repressed genes might be induced only by mutations in the binding sites and/or the lmrA coding region, as found for spontaneous drug-resistant mutants PLR1, PLR2, 1A221, etc. (11, 13). A similar situation has been reported for the Streptomyces coelicolor A3(2) pqrAB operon regarding paraquat resistance (6). PqrA is a TetR family repressor of its own operon, and PqrB is a putative efflux transporter of paraquat. Mutations in pqrA fully induced pqrAB transcription, while paraquat could only slightly induce transcription, but the precise mechanisms underlying the regulation were not clarified.
Our results provided no evidence for involvement of yxaGH in multidrug resistance. However, the Hg2+ sensitivity of the cells was elevated after disruption of either yxaG or yxaH only in a PLR2 background (PLR4 and PLR6), in which the lmrA mutation allowed lmrB expression (Table 2). At present, we are unable to properly explain why yxaGH is involved in Hg2+ resistance only with lmrB expression. Very recently, we noticed that YxaG is an iron-containing quercetin 2,3-dioxygenese, which converts the flavonol quercetin into 2-protocatechuoylphloroglucinol carboxylic acid and carbon monoxide (4, 5). Quercetin is one of the most abundant natural flavonoids inhibiting bacterial DNA gyrase that induces DNA cleavage (17), and thus YxaG might function in the detoxification of this compound. Our results clearly indicated that yxaGH forms an operon, and YxaH, a putative membrane protein with nine transmembrane segments (BSORF website), might act in a cooperative manner with YxaG as a drug exporter. In future investigations we will focus on these possibilities.
Acknowledgments
We thank M. Kitagawa, H. Shimizu, and M. Yukawa for technical assistance. We are also grateful to A. Wipat, University of Newcastle-upon-Tyne, for critical reading of the manuscript.
This work was supported by a grant-in-aid for scientific research on priority areas from the Ministry of Education, Science and Sports and Culture of Japan.
REFERENCES
- 1.Ahmed, M., C. M. Borsch, S. S. Taylor, N. Vazquez-Laslop, and A. A. Neyfakh. 1994. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506-28513. [PubMed] [Google Scholar]
- 2.Ahmed, M., L. Lyass, P. N. Markham, S. S. Taylor, N. Vazquez-Laslop, and A. A. Neyfakh. 1995. Two highly similar multidrug transporters of Bacillus subtilis whose expression is differentially regulated. J. Bacteriol. 177:3904-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aramaki, H., N. Yagi, and M. Suzuki. 1995. Residues important for the function of a multihelical DNA binding domain in the new transcription factor family of Cam and Tet repressors. Protein Eng. 8:1259-1266. [DOI] [PubMed] [Google Scholar]
- 4.Barney, B. M., M. R. Schaab, R. LoBrutto, and W. A. Francisco. 2004. Evidence for a new metal in a known active site: purification and characterization of an iron-containing quercetin 2,3-dioxygenase from Bacillus subtilis. Protein Expr. Purif. 35:131-141. [DOI] [PubMed] [Google Scholar]
- 5.Bowater, L., S. A. Fairhurst, V. J. Just, and S. Bornemann. 2004. Bacillus subtilis YxaG is a novel Fe-containing quercetin 2,3-dioxygenase. FEBS Lett. 557:45-48. [DOI] [PubMed] [Google Scholar]
- 6.Cho, Y. H., E. J. Kim, H. J. Chung, J. H. Choi, K. F. Chater, B. E. Ahn, J. H. Shin, and J. H. Roe. 2003. The pqrAB operon is responsible for paraquat resistance in Streptomyces coelicolor. J. Bacteriol. 185:6756-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldthwaite, C., D. Dubnau, and I. Smith. 1970. Genetic mapping of antibiotic resistance markers in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 65:96-103. (Erratum, 65:771.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.
- 9.Kaempfer, R. O., and B. Magasanik. 1967. Effect of infection with T-even phage on the inducible synthesis of beta-glactosidase in Escherichia coli. J. Mol. Biol. 27:453-468. [DOI] [PubMed] [Google Scholar]
- 10.Kumano, M., A. Tamakoshi, and K. Yamane. 1997. A 32 kb nucleotide sequence from the region of the lincomycin-resistance gene (22 degrees-25 degrees) of the Bacillus subtilis chromosome and identification of the site of the lin-2 mutation. Microbiology 143:2775-2782. [DOI] [PubMed] [Google Scholar]
- 11.Kumano, M., M. Fujita, K. Nakamura, M. Murata, R. Ohki, and K. Yamane. 2003. Lincomycin resistance mutations in two regions immediately downstream of the −10 region of lmr promoter cause overexpression of a putative multidrug efflux pump in Bacillus subtilis mutants. Antimicrob. Agents Chemother. 47:432-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomovskaya. O., K. Lewis, and A. Matin. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murata, M., S. Ohno, M. Kumano, K. Yamane, and R. Ohki. 2003. Multidrug resistant phenotype of Bacillus subtilis spontaneous mutants isolated in the presence of puromycin and lincomycin. Can. J. Microbiol. 49:71-77. [DOI] [PubMed] [Google Scholar]
- 14.Neyfakh, A. A., V. E. Bidnenko, and L. B. Chen. 1991. Efflux-mediated multidrug resistance in Bacillus subtilis: similarities and dissimilarities with the mammalian system. Proc. Natl. Acad. Sci. USA 88:4781-4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogura, M., H. Yamaguchi, K. Yoshida, Y. Fujita, and T. Tanaka. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29:3804-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohki, R., and M. Murata. 1997. bmr3, a third multidrug transporter gene of Bacillus subtilis. J. Bacteriol. 179:1423-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plaper, A., M. Golob, I. Hafner, M. Oblak, T. Solmajer, and R. Jerala. 2003. Characterization of quercetin binding site on DNA gyrase. Biochem. Biophys. Res. Commun. 306:530-536. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Schilling, C. H., L. Held, M. Torre, and M. H. Saier, Jr. 2000. GRASP-DNA: a web application to screen prokaryotic genomes for specific DNA-binding sites and repeat motifs. J. Mol. Microbiol. Biotechnol. 2:495-500. [PubMed] [Google Scholar]
- 20.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 21.Vazquez-Laslop, N., P. N. Markham, and A. A. Neyfakh. 1999. Mechanism of ligand recognition by BmrR, the multidrug-responding transcriptional regulator: mutational analysis of the ligand-binding site. Biochemistry 38:16925-16931. [DOI] [PubMed] [Google Scholar]
- 22.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strain: nucleotide sequences of M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida, K., T. Shibayama, D. Aoyama, and Y. Fujita. 1999. Interaction of a repressor and its binding sites for regulation of the Bacillus subtilis iol divergon. J. Mol. Biol. 285:917-929. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida, K., I. Ishio, E. Nagakawa, Y. Yamamoto, M. Yamamoto, and Y. Fujita. 2000. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology 146:573-579. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida, K., K. Kobayashi, Y. Miwa, C. M. Kang, M. Matsunaga, H. Yamaguchi, S. Tojo, M. Yamamoto, R. Nishi, N. Ogasawara, T. Nakayama, and Y. Fujita. 2001. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 29:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida, K., H. Yamaguchi, M. Kinehara, Y. Ohki, Y. Nakaura, and Y. Fujita. 2003. Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA-box. Mol. Microbiol. 49:157-165. [DOI] [PubMed] [Google Scholar]
- 27.Zheleznova, E. E., P. N. Markham, A. A. Neyfakh, and R. G. Brennan. 1997. Preliminary structural studies on the multi-ligand-binding domain of the transcription activator, BmrR, from Bacillus subtilis. Protein Sci. 6:2465-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheleznova, E. E., P. N. Markham, A. A. Neyfakh, and R. G. Brennan. 1999. Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell 96:353-362. [DOI] [PubMed] [Google Scholar]