Abstract
The intracellular bacterium Francisella tularensis is the causative agent of tularemia and poses a serious threat as an agent of bioterrorism. We have developed a highly effective molecular subtyping system from 25 variable-number tandem repeat (VNTR) loci. In our study, multiple-locus VNTR analysis (MLVA) was used to analyze genetic relationships and potential population structure within a global collection of 192 F. tularensis isolates, including representatives from each of the four subspecies. The VNTR loci displayed between 2 and 31 alleles with Nei's diversity values between 0.05 and 0.95. Neighbor-joining cluster analysis of VNTR data revealed 120 genotypes among the 192 F. tularensis isolates, including accurate subspecies identification. F. tularensis subsp. tularensis (type A) isolates showed great diversity at VNTR loci, while F. tularensis subsp. holarctica (type B) isolates showed much lower levels despite a much broader geographical prevalence. The resolution of two distinct clades within F. tularensis subsp. tularensis (designated A.I and A.II) revealed a previously unrecognized genetic division within this highly virulent subspecies. F. tularensis subsp. holarctica appears to have recently spread globally across continents from a single origin, while F. tularensis subsp. tularensis has a long and complex evolutionary history almost exclusively in North America. The sole non-North American type A isolates (Slovakian) were closely related to the SCHU S4 strain. Significant linkage disequilibrium was detected among VNTR loci of F. tularensis consistent with a clonal population structure. Overall, this work greatly augments the study of tularemia ecology and epidemiology, while providing a framework for future forensic analysis of F. tularensis isolates.
Francisella tularensis, the causative agent of tularemia, is one of the most highly infectious bacteria known. F. tularensis is a facultative intracellular pathogen affecting more animal species than any other known zoonotic pathogen (18). Tularemia often occurs as geographically confined outbreaks in humans and animals. Transmission to humans frequently occurs through the bite of blood-feeding arthropods, such as ticks, biting flies, or mosquitoes (6). Inhalation and the ingestion of F. tularensis can also cause disease. Historically, F. tularensis attracted attention as a biological weapon and was a subject of military research in the United States, the former Soviet Union, and Japan (8). In the post-Cold War era, however, F. tularensis is included among the top six agents showing potential for great adverse public health impact if used as a bioterrorism agent (29).
The geographical distribution of F. tularensis spans the entire Northern Hemisphere and was only recently isolated in Australia (35, 42). At present there are four recognized subspecies of F. tularensis: tularensis, holarctica, mediasiatica, and novicida (35). Although all four share most biochemical characteristics, each subspecies is predominantly associated with a specific geographical distribution. F. tularensis subsp. tularensis is predominately found in North America, yet was recently recovered in central Europe (14). F. tularensis subsp. holarctica is found over much of the Northern Hemisphere, while F. tularensis subsp. mediasiatica has only been isolated in Central Asian republics (28). F. tularensis subsp. novicida is rarely isolated and prior to 2003 appeared restricted to North America, yet was recently isolated in Australia (5, 17, 42).
Although the four subspecies show close genetic relationship, each exhibits marked variations in its virulence in mammals (38). With regard to mortality and virulence in humans, two types of tularemia remain clinically dominant and are readily distinguished: F. tularensis subsp. tularensis causes type A tularemia, and F. tularensis subsp. holarctica causes type B tularemia (6). Type A isolates cause a life-threatening disease in humans, while the less-virulent type B isolates produce milder disease.
F. tularensis exhibits highly conserved genomic sequence among strains of diverse origin. The substantial genetic similarity among Francisella subspecies makes individual strain typing difficult. All four F. tularensis subspecies are antigenically similar (35), with 99.8% identity among 16S rRNA genes (38). Although several available DNA-based typing methods can readily identify each F. tularensis subspecies, their utility for strain discrimination is limited. Repetitive element PCR (Rep-PCR) (22), arbitrary primed PCR (22), pulsed-field gel electrophoresis (12), amplified fragment length polymorphism analysis (12), insertion sequence-element probed restriction fragment length polymorphism analysis (37), and whole-genome microarray analysis (3) indicate extensive genetic homogeneity within individual F. tularensis subspecies. Importantly, all methods show concordant results consistent with current subspecies classification and suggest strains from Japan are distinct from other F. tularensis subspecies (3, 22, 37).
Although subspecies identification is clinically important, the substantial risk of laboratory-acquired infection validates that biochemical typing of this pathogen is often avoided. Biosafety level 3 facilities are required for work involving live cultures (39). Indeed, only 10 to 25 F. tularensis cells are required to infect humans by the dermal or respiratory route, and airborne laboratory-acquired infections have been frequently recorded (4, 32, 33). This has hampered the development of methods for elucidating the ecological and epidemiological prerequisites for tularemia infection. DNA-based typing systems, however, allow safe strain characterization from simple killed bacterial preparations. Such typing methods may be of significant value in identifying the natural reservoir of F. tularensis, including the elucidation of transmission routes between water, blood-feeding arthropods, and animals. Given the potential for the illegitimate use of F. tularensis, the rapid characterization of individual isolates would also prove valuable.
Variable-number tandem repeats (VNTRs) are considered high-speed molecular clocks (40) and for this reason have been used for individual strain discrimination within several bacterial species with little genomic variation, e.g., Mycobacterium tuberculosis, Bacillus anthracis, and Yersinia pestis (23, 24, 26). VNTR markers previously demonstrated individual strain discrimination among smaller collections of North American and Eurasian F. tularensis isolates (9, 21). The primary aim of this study was to develop a high-resolution typing system for F. tularensis isolates based on polymorphisms at genomic VNTR loci. Subsequently, we evaluated deeper genetic relationships among F. tularensis isolates by utilizing VNTR markers with a range of individual diversity levels. Application of this typing system on a set of 192 globally diverse F. tularensis isolates showed great discriminatory power yet retained the capacity for isolate resolution at the taxonomic level.
MATERIALS AND METHODS
Genomic analysis.
Genomic sequence data were analyzed for the presence of tandem repeats using the software programs Tandem Repeats Finder (2), REPuter (25), Gene Quest of the DNAstar package (Lasergene, Inc., Madison, Wis.), and an in-house modification of Sputnik (C. Abajian; http://abajian.net/sputnik/). The genomic sequence of F. tularensis subsp. tularensis strain SCHU S4 (type A) was provided by the European Sequencing Consortium (Swedish Defence Research Agency and Umeå University, Umea, Sweden; Walter Reed Army Institute of Research, Washington, D.C.; Uppsala University, Uppsala, Sweden; Defense Science and Technology Laboratories, Porton Down, United Kingdom). The preliminary genomic sequence of F. tularensis subsp. holarctica live vaccine strain (type B) was provided by Lawrence Livermore National Laboratory, Livermore, Calif. More recently, the SCHU S4 and the live vaccine strain genomic sequences have been completed and made available as assembled contiguous DNA sequences at the websites http://artedi.ebc.uu.se/Projects/Francisella/ and http://bbrp.llnl.gov/bbrp/html/microbe.html. The positions of VNTR loci relative to protein-coding regions in the SCHU S4 genome sequence were annotated using the software Artemis release 5 (cutoff set to 100 codons) (30).
F. tularensis isolate DNA.
This study included 192 geographically diverse F. tularensis isolates from throughout North America (n = 91) and the following nine Eurasian countries: Sweden (n = 52), Finland (n = 10), Russia (n = 9), Ukraine (n = 3), Czech Republic (n = 8), France (n = 2), Spain (n = 1), Norway (n = 2), Japan (n = 7), Slovakia (n = 3), and Central Asian Republics (n = 4) (see Fig. 4). All isolates, with the exception of 36 phenotypically untyped isolates from California, were subjected to biochemical characterization by institutions performing the primary isolation. A genomic locus specific for the identification of F. tularensis subsp. holarctica (22) was amplified across all isolates (see Ft-M19 in Tables 1 and 3, below).
FIG. 4.
Isolate identification within individual subgroups of the NJ dendrogram. The origin (including state, province, and country) and year are specified to the right of each branch or clade. Reference strains from the American Type Culture Collection (ATCC) and Gamaleya Institute of Epidemiology and Microbiology (GIEM) are identified in boxes. Branch lengths among clades are not to scale, due to space constraints. Major clades containing type A and type B F. tularensis isolates were arbitrarily designated A.I and A.II and B.I to B.V, respectively. Filled circles indicate isolates from geographically confined tularemia outbreaks.
TABLE 1.
VNTR loci genomic locations and primers
| Marker | Genomic locationa | Forward primer sequence | Reverse primer sequence |
|---|---|---|---|
| Ft-M1 | 277650-959 | GAGCTGGTCAAGTTTATTTAAGTA | CAGGATTGCTTGAACATGATA |
| Ft-M2 | 1886647-7109 | TTTATGATAAGGATGATTTAAAACAAAATA | GCTTAAATCTCGCAATACCATGTAAT |
| Ft-M3 | 308585-9016 | GTTTTCACGCTTGTCTCCTATCA | CAAAAGCAACAGCAAAATTCACAAA |
| Ft-M4 | 316972-7405 | AAAAGGGCGGGTTACTGAGG | GTATCAAATAGCGCAAAAATAACTGC |
| Ft-M5 | 1649600-807 | TAGGCATGACAAACCCTGCTAT | CAGCTCGAACTCCGTCATAC |
| Ft-M6 | 1442674-984 | TTTTGGGTTTTCTCTAAACATTTCTA | CAATTCAGCGAAACCCTATCTTA |
| Ft-M7 | 1868734-940 | ATTGGGTGATTTGGATGGTTG | CAGCTCGAACTCCGTCATACb |
| Ft-M8 | 8155-440 | AGTAATCTAGCCAAGGTAATA | CAGCTCGAACTCCGTCATACb |
| Ft-M9 | 3930-4177 | AAGGACCTATTTTTACATCAGT | CAGCTCGAACTCCGTCATACb |
| Ft-M10 | 1283441-4086 | GTTGGCGAACCTAAAATAATAGC | CAGCTCGAACTCCGTCATACb |
| Ft-M11 | 1628132-523 | AAACCTACAATCAACATCTGACAAT | TTGTTATATTAACCTCATCAGTTCAATTTA |
| Ft-M12 | 801242-586 | CGCTAGATGGTGCTGATACTATCTT | CCTGCTAGAAAACCCATATTTACAT |
| Ft-M13 | 1169264-588 | TTTGCAACTACTAGGTGTGGAGAT | TTGATATTCCAAATGATCAAGTTTT |
| Ft-M14 | 1390169-610, 1783513-954 | ACCGCCATCTTTTCTATCATAAT | AACCTTAAGTGATAAATATAACCCAAAA |
| Ft-M15 | 1473082-303 | GCATGGACATGAGTGTCTATGGCGTAGATC | GATAAAGGAATGTTTTAAATAATGTGATGTTTTGCATC |
| Ft-M16 | 176810-7030 | AGGAAAGCATACCCAACATTATT | CCAAAGATCGCCGTGATT |
| Ft-M17 | 1010288-638 | GCTATAGCAGTAAATGTAGGCTCAA | ACATATCGGTGGATCACTATCAA |
| Ft-M18 | 1483015-386 | AACAGCCTTCAAACCACCTT | CATAAAATACAGCTTCAATAAACAATCTT |
| Ft-M19 | 1524117-432 | TCCGGTTGGATAGGTGTTGGATT | AGGCGGAGATCTAGGAACCTTT |
| Ft-M20 | 736003-257 | GCATAACTTTTGAGACAATTGGTGCAGATGATC | GACCGCCAGTATATGCTTGACCTTGACTCC |
| Ft-M21 | 1572002-404 | CCACAGCTAGCCAGACCAAAT | AGTTTGGCGCGAGCTAAT |
| Ft-M22 | 603279-519 | GTCAAAATCTCAAGATGAGCAAATATTTGAATGGT | GGAGTTTTTTCTCGTCCGCTGTTAGTGATTT |
| Ft-M23 | 620493-927 | TGAGATGTGGAACTTTATAGGTTCAA | TGTAAACTAAAAGATAACTAATGGCAATTT |
| Ft-M24 | 685645-6103 | ATACGGTCCTAATAATATTCCTGTCA | ATTCATTTATAGATGCCTTTGTTACC |
| Ft-M25 | 864491-641 | GTGGTCTTTTAAGCGTCTTAGCAAGCTCGAC | GGGTACCCATCCCATATGTAAGTACAAATGTAGC |
Location of the DNA amplified by PCR in the chromosome of F. tularensis strain SCHU S4.
Identical reverse primer sequence used to target VNTR locus within multicopy F. tularensis insertion sequence element, ISFtu1. Forward primers at these loci targeted alternate genomic locations to include the 16-bp VNTR locus.
TABLE 3.
VNTR allelic distribution among F. tularensis subspecies
| VNTR locus | VNTR copy no. in 192 isolates of the following genetic groupsa:
|
|||||
|---|---|---|---|---|---|---|
| F. tularensis subsp. tularensis A.I (n = 31) | F. tularensis subsp. tularensis A.II (n = 14) | F. tularensis subsp. holarctica B.I to B.IV (n = 132) | F. tularensis subsp. holarctica from Japan B.V (n = 7) | F. tularensis subsp. mediasiatica (n = 4) | F. tularensis subsp. novicida (n = 4) | |
| Ft-M1 | 3 | 3 | 3 | 3 | 4b | 3 |
| Ft-M2 | 4-34 | 19-25 | 2 | 6-26 | 2 | 2 |
| Ft-M3 | 9-29 | 7-16 | 8-28 | 3 | 25-28 | 30-43 |
| Ft-M4 | 3 | 3 | 4-5 | 4 | 3 | 3-4 |
| Ft-M5 | 2-6 | 2 | 2 | 2 | 6-10 | 2 |
| Ft-M6 | 4-6 | 4-7 | 4-7 | 3-5 | 2-3 | 4 |
| Ft-M7 | 2-6 | 2 | 2 | 2 | 2 | 2 |
| Ft-M8 | 2-6 | 1-2 | 2 | 1-4 | 1 | —c |
| Ft-M9 | 2-15 | 2 | 2 | 2 | 3 | 2-7 |
| Ft-M10 | 2-18 | 1-2 | 2 | 4-8 | 2 | 2 |
| Ft-M11 | 5 | 5 | 5 | 5 | 3 | 4-5 |
| Ft-M12 | 2 | 1-2 | 2 | 2 | 1-2 | 2 |
| Ft-M13 | 2 | 1 | 1-2 | 1 | 1 | 2-2.5 |
| Ft-M14 | 3 | 3 | 3 | 3 | 2 | 3 |
| Ft-M15 | 2 | 3 | 3 | 3 | 3 | 3 |
| Ft-M16 | 1-2 | 1-2 | 1-2 | 1 | 1 | 1 |
| Ft-M17 | 3 | 2 | 2 | 2 | 2 | 2 |
| Ft-M18 | 4 | 2 | 2-4 | 2 | 2 | 2c |
| Ft-M19 | 2 | 2 | 1 | 1 | 2 | 2 |
| Ft-M20 | 3 | 22-25d | 3-4 | 11-18 | 6-7 | 31-39 |
| Ft-M21 | 3-7 | 3-4 | 3-5 | 3-5 | 3 | 3 |
| Ft-M22 | 2 | 3 | 3-4 | 4 | 8-14 | 5 |
| Ft-M23 | 1-2 | 1 | 1 | 1 | 1 | 1-1.5 |
| Ft-M24 | 1 | 1 | 1-2 | 1 | 1 | 1 |
| Ft-M25 | 5 | 5 | 4 | 5 | 5 | 5 |
Genetic group corresponds to the results of the NJ analysis displayed in Fig. 3. Array size is the repeat copy number calculated from PCR amplicon size scores representing each strain and VNTR locus. Numerical values represent the smallest and largest number of repeats found in a given locus.
Size scores unique to an F. tularensis subspecies are marked in boldface. Notably, Japanese isolates showed scores distinct from those of Eurasian and North American F. tularensis subsp. holarctica isolates.
DNA did not amplify from all strains.
PCR fragment sizes were in some strains due to an additional VNTR sequence present adjacent to the 12-bp VNTR.
Bacterial thermolysates were obtained from the Swedish Defence Research Agency (n = 116), the California Department of Health (n = 43), Centers for Disease Control and Prevention, Fort Collins, Colo. (n = 24), the Oklahoma Department of Health (n = 3), and the Arizona Department of Health (n = 6). For the analysis of epidemiological concordance of our VNTR typing system, isolates representing distinct tularemia epidemics within restricted geographical areas and specific time periods were included. Selection criteria for epidemic isolates included a maximum geographical spread of 50 km from the suspected location of infection and occurrence within a 2-month window. Five distinct outbreaks were included: Maricopa County, year 2000 (n = 6); Ljusdal area, Sweden, 1995 (n = 7) and 1998 (n = 6); Västerdal river area, Sweden, 1995 (n = 6); and Oulu area, Finland, 1997 (n = 6). In addition, we included isolates representing outbreaks over a greater geographical area and time: Sweden, 2000 (n = 7); Czech Republic, 1995 (n = 8), as well as older outbreak isolates for which the presumed location for infection was uncertain; and Sweden, 1981 (n = 9).
PCR amplification of VNTR markers.
Flanking molecular primers were developed around 52 potential VNTR loci using the Consed (13) and Primer Select of the DNAstar package (Lasergene, Inc.) computer programs. Molecular primers were designed with annealing temperatures from 56 to 61°C. Individual annealing temperatures for flanking primers were within 2°C of each other. A set of diverse F. tularensis strains representing each of the four subspecies was used to screen for VNTR polymorphisms. Twenty-five primer sets amplified polymorphic VNTR loci (Table 1). The physical distribution of the 25 VNTR markers throughout the genome of strain SCHU S4 is illustrated in Fig. 1.
FIG. 1.
VNTR marker location within the physical map of F. tularensis subsp. tularensis SCHU S4 genome. Positions are given with reference to the predicted origin of replication set at position 0.
PCR amplification of the 25 variable loci (including Ft-M19, a F. tularensis subsp. holarctica-specific direct repeat locus) was carried out as follows: 2 mM MgCl2, 1× PCR buffer, 0.1 mM deoxynucleoside triphosphates, 1 μM R110, R6G, or carboxytetramethyl rhodamine phosphoramide fluorescent-labeled dUTPs (Perkin-Elmer Biosystems), 0.5 U of Taq polymerase (Life Technologies, Inc., Rockville, Md.), 1.0 μl of template DNA, 0.5 μM forward primer, 0.5 μM reverse primer, and filtered sterile water to a volume of 12.5 μl. The reaction mixtures were incubated at 94°C for 5 min and then cycled at 94°C for 30 s, 56 or 61°C for 30 s, and 72°C for 30 s for 35 cycles, with a final incubation of 72°C for 5 min. Reagents used in the PCRs were obtained from Life Technologies, Inc. An annealing temperature of 64°C was used for the Ft-M19 primer set.
Automated genotyping.
Fluorescently labeled amplicons were sized by denaturing polyacrylamide gel electrophoresis in an ABI 377 DNA sequencer. PCR product was diluted threefold and mixed 1:1 with equal parts of a 5:1 formamide-dextran blue dye and size standard prior to electrophoresis under ABI filter set A. Amplicon size scores (in base pairs) were determined using Bioventures Custom ROX 1000 size standards and GeneScan analysis software (24). Controls with SCHU S4 and live vaccine strain DNAs were included with each GeneScan analysis to ensure that fragment sizes correlated to sequence data. In this study, we only occasionally experienced a stutter phenomenon. This is a phenomenon believed to result from in vitro slippage of the polymerase during PCR and was only observed for Ft-M3 and Ft-M2. However, correct sizing was routine due to the controls and distinct band intensities.
Sequence verification of VNTR loci.
To verify repeat copy number variation, PCR products representing at least two alternate allelic states were sequenced for each VNTR marker. PCR products were purified by using MicroSpin columns (Amersham Biosciences, Uppsala, Sweden) and sequenced on the ABI 377 platform using forward and reverse amplification primers and the Big Dye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, Calif.). Amplicon size scores (±2 bp) were translated into multiples of VNTR length and used to predict repeat copy number for each locus and isolate (see Table 3, below).
Phylogenetic and statistical analyses.
At each VNTR locus, amplicon size scores obtained from all isolates were coded as discrete characters (i.e., a, b, c, etc.). PAUP4a software was used to generate a pair-wise distance matrix and apply the neighbor-joining (NJ) algorithm (D. Swofford, Sinauer Associates, Inc., Sunderland, Mass.). Nei's diversity index (D) was calculated as follows: [1 − Σ(allele frequencies)2] for each marker across the total number of individual genotypes (n = 120) (41). Diversity differences between clades were tested by comparing the genetic distance means with significance based upon 10,000 random permutations of the data.
VNTR marker linkage disequilibrium.
Nonrandom association (linkage disequilibrium) among the 25 VNTR markers was evaluated by calculating the index of association (IA) (36). Linkage analysis was performed using the MultiLocus 1.3.b software and the LIAN 3.1 software (1,000 iterations). The standardized index of association (sIA) (15, 16) and a modification of IA (r̄d) (1) were calculated for individual isolates and individual genotypes. Separate linkage analyses were performed on two data sets representing the two major subspecies (F. tularensis subsp. tularensis and F. tularensis subsp. holarctica) and using isolates from limited geographical areas. Comparative groups included F. tularensis subsp. tularensis isolates (n = 13) from California (1982 to 1994) collected within a range of 660 km and F. tularensis subsp. holarctica isolates (n = 35) recovered from central Sweden (1985 to 2000) within a range of 440 km.
Nucleotide sequence accession number.
The VNTR regions from the genome of F. tularensis subsp. tularensis strain SCHU S4 were assigned GenBank accession numbers AY522354, AY522355 (Ft-M1 and Ft-M2), AY522356 to AY522361 (Ft-M4 through Ft-M9), AY522362 to AY522369 (Ft-M11 through FT-M18), and AY522370 to AY522375 (Ft-M20 through Ft-M25). Sequences from strain SCHU S4 at loci Ft-M3, Ft-M10, and Ft-M19 were previously assigned the accession numbers AF356777, AF357005, and AF524865, respectively.
RESULTS
VNTR marker attributes.
Twenty-five informative VNTR markers revealed genetic relationships among 192 globally diverse F. tularensis isolates (Table 2). The 25 variable repeat motifs displayed repeat sizes from 2 bp (Ft-M25) to 23 bp (Ft-M23) in length (Table 2). A 16-bp repeat structure was previously identified within an insertion sequence element designated ISFtu1 SSTR16 (21). Within the SCHU S4 sequence, a total of 50 copies of ISFtul containing this repeat motif was observed. In this study, markers Ft-M5 and Ft-M7-10 represented five variable copies of this 16-bp motif, each located within a distinct ISFtu1. Allele states at these loci were most variable among the F. tularensis subsp. tularensis isolates (Table 3). Previous studies indicated Ft-M19 contains a 30-bp sequence present in isolates of F. tularensis subsp. tularensis, mediasiatica, and novicida but absent in isolates of F. tularensis subsp. holarctica (22). In this study, amplicon sizing confirmed previous findings. All investigated F. tularensis subsp. holarctica isolates (n = 139) had a 30-bp deletion at this locus (Table 3). DNA sequencing of this locus in multiple geographically diverse F. tularensis subsp. holarctica isolates consistently showed deletion of an internal 17-bp region and one out of two flanking 13-bp direct repeats (22). The location of each VNTR locus was mapped in the SCHU S4 genomic sequence. The VNTR loci analyzed here appeared to be widely distributed across the SCHU S4 genome (Fig. 1), consistent with their assumed status as independent variable markers. Of the 25 VNTR markers analyzed, 12 were located within predicted open reading frames (Table 2).
TABLE 2.
VNTR attributes
| VNTR marker | Repeat motif | Repeat size (nt)a | Genomic locationb | Repeat copy no., strain SCHU S4 | Allele no. | Diversity (D)c |
|---|---|---|---|---|---|---|
| Ft-M1 | AAT | 3 | I (−76) | 3 | 2 | 0.06 |
| Ft-M2 | TAAATA | 6 | G (+12) | 4 | 20 | 0.58 |
| Ft-M3d | AATAAGGAT | 9 | G (+1401) | 25 | 31 | 0.95 |
| Ft-M4 | TTGTT | 5 | G (+55) | 3 | 5 | 0.65 |
| Ft-M5e | TTTCTACAAATATCTT | 16 | I (−21) | 3 | 6 | 0.28 |
| Ft-M6d | TTGGTGAACTTTCTTGCTCTT | 21 | G (+1160) | 4 | 6 | 0.60 |
| Ft-M7e | TTTCTACAAATATCTT | 16 | I (−21) | 4 | 6 | 0.24 |
| Ft-M8e | TTTCTACAAATATCTT | 16 | I (−21) | 4 | 6 | 0.42 |
| Ft-M9e | TTTCTACAAATATCTT | 16 | I (−21) | 4 | 7 | 0.35 |
| Ft-M10de | TTTCTACAAATATCTT | 16 | I (−21) | 18 | 15 | 0.57 |
| Ft-M11 | AATTATAAAT | 10 | I (−113) | 5 | 3 | 0.10 |
| Ft-M12 | TAGCTTTTTT | 10 | I (−113) | 2 | 2 | 0.05 |
| Ft-M13 | CTCCAGGACCAA | 12 | G (+1174) | 2 | 3 | 0.40 |
| Ft-M14 | TCATTA | 6 | G (+67) | 3 | 2 | 0.06 |
| Ft-M15 | ATACTT | 6 | G (+32) | 2 | 2 | 0.33 |
| Ft-M16 | TAAAAGTAAG | 10 | I (+551) | 2 | 2 | 0.34 |
| Ft-M17 | TATTTA | 6 | G (+484) | 3 | 2 | 0.33 |
| Ft-M18d | CATTAA | 6 | I (−52) | 4 | 3 | 0.41 |
| Ft-M19 | TAAATTTCTCATA | 13 | I (−20) | 2 | 2 | 0.47 |
| Ft-M20 | ATTATTTTGATC | 12 | G (+1964) | 3 | 15 | 0.47 |
| Ft-M21 | TCAATTA | 7 | G (+586) | 3 | 6 | 0.41 |
| Ft-M22 | AAAAAT | 6 | G (+2254) | 2 | 5 | 0.66 |
| Ft-M23 | AAGTAGCATTGTCACGACCTCCT | 23 | I (+1864) | 2 | 3 | 0.22 |
| Ft-M24 | ATAAATTATTTATTTTGATTA | 21 | I (−93) | 1 | 3 | 0.51 |
| Ft-M25d | GT | 2 | G (+525) | 5 | 2 | 0.49 |
| Average | 11.0 | 4.5 | 6.4 | 0.4 |
Indicates repeat size in nucleotides.
G or I indicates repeat motif is located within an open reading frame (genic) or between two open reading frames (intergenic), respectively. Distance to the presumed translation start is indicated in parentheses (+ indicates downstream and − indicates upstream location).
Individual marker diversity (D) was calculated as 1 − Σ(allele frequency)2 and based upon allele frequencies of 120 distinct genotypes among 192 F. tularensis isolates.
Specifies previously identified VNTR loci Ft-M3 (previous designations SSTR9 and Ft-V4), Ft-M10 (SSTR16 and Ft-V2), Ft-M18 (Ft-V3), Ft-M6 (Ft- V1), Ft-M25 (Ft-V5) (9, 21).
Indicates 16-bp VNTR sequence located inside an F. tularensis multicopy insertion element, ISFtul.
To verify that PCR fragment size variation resulted from differences in repeat copy number, at least two different alleles were sequenced for each marker (data not shown). In one case, variation in amplicon size for a VNTR marker (Ft-M20) was explained by two additional VNTR sequences in some isolates, i.e., some isolates contained an additional distinct tandem repeat sequence at the locus Ft-M20 (Table 3). For many markers (Ft-M1, Ft-M3, Ft-M11, Ft-M14, Ft-M15, Ft-M17, Ft-M19, Ft-M20, Ft-M22, and Ft-M25), one or more subspecies-specific PCR fragment sizes were detected. Although it is possible that more alleles would be detected for some VNTR loci if more isolates were analyzed, the data suggest VNTR typing of F. tularensis is feasible for rapid taxonomic classification at the subspecies level (Table 3).
The calculated diversity value (D) reflects the discriminatory capacity of a VNTR marker and is a function of individual allele frequency and the number of alleles a marker detects. In this context, each sequence at a given locus with a different number of tandem repeats is designated as a distinct allele. The number of alleles ranged from 2 for Ft-M1 (among others) to 31 for Ft-M3 (Fig. 2; Table 2). D values for the VNTR markers in this study ranged from 0.05 for Ft-M12 to 0.95 for Ft-M3, with an overall average diversity index of 0.40 (Table 2; Fig. 2). The hypervariable marker Ft-M3 (D = 0.95) previously demonstrated strong discriminatory power among North American (Ft-V4 [9]) and Scandinavian F. tularensis isolates (SSTR9 [21]).
FIG. 2.
Number of alleles and allelic diversity detected at 25 VNTR markers. Bars indicate allele number, and filled diamonds indicate Nei's diversity index (D), calculated as 1 − Σ(allele frequency)2 analyzed across 120 F. tularensis genotypes and 192 isolates.
Genetic relationships.
Phylogenetic analysis using the NJ method revealed 120 individual genotypes among the 192 F. tularensis isolates analyzed. The VNTR data provided a high level of strain discrimination as well as accurate subspecies classification (Fig. 3). Analysis of 16S ribosomal gene sequences (10, 38) and a whole-genome comparison (3) indicated F. tularensis subsp. novicida shows the greatest amount of genetic divergence compared to other F. tularensis subspecies. For this reason, the F. tularensis subsp. novicida isolates were used to root the dendrogram in Fig. 3. The subspecies specificity achieved with VNTR analysis corroborated the clustering of F. tularensis subspecies observed by IS element-probed restriction fragment length polymorphism analysis (37), whole-genome microarray analysis (3), and Rep-PCR (22).
FIG. 3.
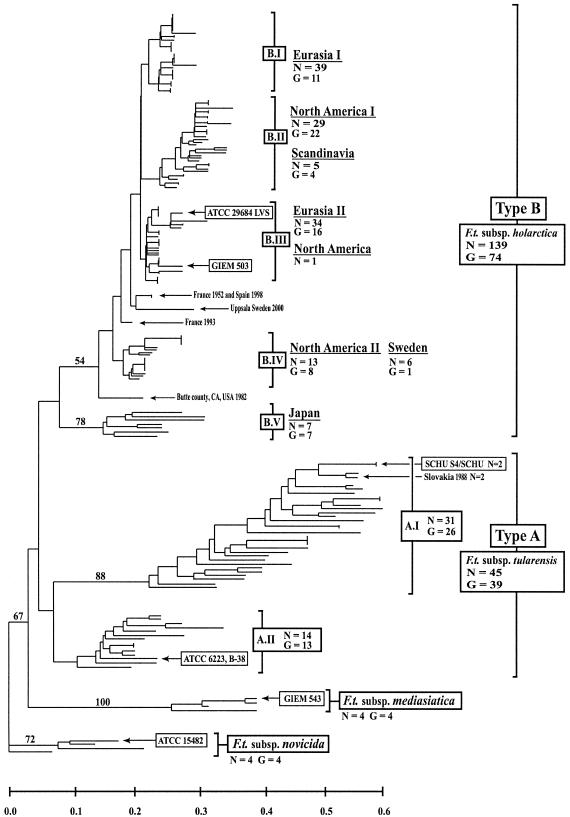
Genetic relationships among global F. tularensis isolates based on allelic differences at 25 VNTR markers. The NJ dendrogram was rooted with F. tularensis subsp. novicida isolates. Geographical origin, number of isolates (n), number of genotypes (G), and F. tularensis subspecies are indicated. Reference strains from American Type Culture Collection (ATCC) and Gamaleya Institute of Epidemiology and Microbiology (GIEM) are specified in boxes. Major clades containing type A and type B F. tularensis isolates were arbitrarily designated A.I and A.II and B.I to B.V, respectively. Bootstrap values greater than 50% are shown and were calculated using 1,000 iterations. Scale bar represents genetic distance.
The 45 geographically diverse F. tularensis subsp. tularensis isolates exhibited 39 unique genotypes and grouped into two distinct clusters, designated A.I (n = 31) and A.II (n = 14) (Fig. 3). Compared to other dendrogram subdivisions, F. tularensis subsp. tularensis genotypes showed great genetic diversity, with long branch lengths and topologically distant genotypic clades (Fig. 3). VNTR analysis revealed a significant difference in genetic diversity between F. tularensis subsp. tularensis and F. tularensis subsp. holarctica genotypes. The average pair-wise genetic distance (x) was 0.40 for F. tularensis subsp. tularensis, which was significantly higher than the average for F. tularensis subsp. holarctica (x = 0.16; P < 0.0001). Clade A.I includes 29 North American isolates and two Slovakian isolates. This clade contains the highly virulent F. tularensis subsp. tularensis laboratory strain SCHU S4, a SCHU strain phenotypic variant, and a Canadian isolate. Both SCHU lab strains showed 100% marker identity and clustered immediately distant to the Slovakian isolates (Fig. 3 and 4). Isolates from Kansas (n = 2), Massachusetts (n = 3), and South Dakota (n = 2) showed 100% marker identity within group A.I (Fig. 4). Four phenotypically untyped California isolates represented the most distinct genotypes present in this group (Fig. 4). Longer branch lengths illustrated the great genetic diversity within this group (Fig. 3). Group A.II contains the F. tularensis species type strain, ATCC 6223, a single Canadian isolate, and 12 North American F. tularensis subsp. tularensis isolates (Fig. 3 and 4). Other North American genotypes within this group included isolates from Nevada (n = 2), nine phenotypically untyped isolates from California, and a single isolate from Wyoming (Fig. 4). Clade A.II was significantly (P < 0.0001) less diverse than clade A.I, with an average pair-wise genetic distances of 0.20 and 0.32, respectively.
Overall, F. tularensis subsp. holarctica genotypes (G = 74) grouped into five distinct clades, arbitrarily designated B.I to B.V (Fig. 3). F. tularensis subsp. holarctica isolates were included from multiple geographically distinct tularemia outbreaks in North America and Eurasia. Among nonoutbreak isolates, F. tularensis subsp. holarctica isolates (n = 139) exhibited fewer overall genotypes (G = 74), in contrast to the diverse F. tularensis subsp. tularensis genotypes (n = 45; G = 39) (Fig. 4). North American genotypes within the B.II and B.IV clades appeared more diverse with greater genetic distance among genotypes compared to Eurasian genotypes within clades B.I, B.II, and B.III (Fig. 3). Among 43 isolates from California, only 7 were previously characterized by biochemical methods and found to be type B isolates. Accordingly, all seven of these isolates were typed here as F. tularensis subsp. holarctica (Fig. 4, B.II and B.IV).
VNTR analysis revealed identical or few genotypes among outbreak isolates (Fig. 4). Specifically, identical allele states for all 25 loci were observed within North American F. tularensis subsp. tularensis and F. tularensis subsp. holarctica tularemia epidemics from Arizona (n = 6; group B.IV), South Dakota (n = 2, A.I), and Kansas (n = 2, A.I), and within Scandinavian epidemics from Sweden (Ljusdal, 1995, n = 7, and 1998, n = 4, B.I; Västerdal river, 1995, n = 3, B.III) and Finland (Oulu, 1997, n = 3, B.III, and n = 2, B.III) (Fig. 4). Distinct genotypes within an outbreak were identified from Ljusdal 1998, Västerdal river 1995, and Oulu 1997, suggesting distinct populations of F. tularensis may circulate during a tularemia outbreak. The low number of outbreak isolates and the low variability among F. tularensis subsp. holarctica in general did, however, prevent a thorough analysis of isolate populations within an outbreak. Because a single locus (Ft-M3) accounted for all individual genotypes observed within an outbreak, conclusions remain weakly supported. However, if a stepwise repeat contraction or extension at the locus Ft-M3 were assumed, two distinct F. tularensis populations were present during the 1995 outbreak in the Västerdal river area: repeat copy numbers were 12 (two isolates) or 22 to 23 (four isolates). In the Ljusdal 1998 and Oulu 1997 outbreaks, repeat copy numbers were similar within an outbreak: 10 to 11 and 21 to 23, respectively. The single genotype of a former outbreak in Ljusdal in 1995 displayed an Ft-M3 copy number of 10. Three isolates obtained from a fatal human case of tularemia were typed (34). The isolates, recovered from upper and lower lung tissue and from blood, were identical at all 25 VNTR loci (Massachusetts 2000, n = 3, A.I) (Fig. 4). In addition, the 25 VNTR loci showed no variation among pairs of laboratory-related isolates, including representatives of the SCHU strain (n = 2, group A.I) and isolates from laboratory-acquired tularemia from Nevada (n = 2, group A.II) (Fig. 4). To further examine the stability of the markers, we included isolates from an animal passage experiment. A Swedish F. tularensis subsp. holarctica isolate typed identical to the isolate recovered after serial passage in eight laboratory animals (mice, guinea pig, and hare) (Sweden, n = 2, group B.IV) (Fig. 4).
Application of the 25 VNTR markers in this study provided an opportunity to test for linkage disequilibrium within the F. tularensis population under study. In essence, we evaluated whether two isolates identical at one locus would be more likely to show such identity at another. Detection of significant linkage disequilibrium is an indication of a clonal population structure. Two subsets of isolates were used for calculating the IA such as to minimize effects of geographical and niche separation. Thirteen Californian F. tularensis subsp. tularensis isolates and genotypes showed significant linkage disequilibrium (sIA = 0.186, r̄d = 0.379; P < 0.001). Thirty-five Swedish F. tularensis subsp. holarctica isolates showed significant disequilibrium (sIA = 0.032; r̄d = 0.167; P < 0.001), while linkage statistics for the corresponding 16 Swedish genotypes were skewed away from randomness (sIA = 0.011; rbarD = 0.056; P = 0.120). Calculations based on the full data set of 120 worldwide F. tularensis genotypes showed significant disequilibrium (sIA = 0.361; r̄d = 0.242; P < 0.001).
DISCUSSION
In this work we have employed whole genome sequences to identify rapidly evolving loci and to use their highly diverse nature to describe the genetic structure of F. tularensis. The multiple-locus approach allowed us to, first, uniquely identify 120 genotypes among 192 isolates spanning all the recognized subspecies and, then, to identify subspecies and clonal populations within them. We noted a major subdivision within the highly virulent F. tularensis subsp. tularensis that may indicate a unique ecological niche or in the least a significant evolutionary separation in spite of geographical overlap. This particular subspecies was highly diverse in our analysis, suggesting a rich and relatively long evolutionary history. In contrast, the less-virulent F. tularensis subsp. holarctica has lower diversity, more consistent with a recent derivation from a common ancestor. Likely, F. tularensis subsp. holarctica has recently expanded across the globe, while F. tularensis subsp. tularensis has a longer evolutionary history in North America. A highly virulent pathogen may not be as generally fit for geographical dissemination as a more moderate one. Whether the spread of F. tularensis subsp. holarctica was from the New World to the Old, or vice versa, is problematic. Isolates from North America can generally be distinguished from those in Europe and Asia, and North American isolates are slightly more diverse. While New and Old World F. tularensis subsp. holarctica isolates are highly similar, their distinct separation argues against a continual exchange of this pathogen across continents. The unique F. tularensis subsp. holarctica isolates from Japan are somewhat intermediate to F. tularensis subsp. tularensis and the other F. tularensis subsp. holarctica isolates. This suggests that there is still a great deal of unsampled F. tularensis diversity to be discovered. The finding of great diversity within the small set of F. tularensis subsp. mediasiatica isolates is in line with such reasoning.
Finally, we note that two isolates of F. tularensis subsp. tularensis were recovered from mites within a field surveillance program in Slovakia in Central Europe (14). This finding is remarkable, and any interpretation will remain speculative. The isolates might be remnants of an ancient Eurasian population of F. tularensis subsp. tularensis, or the highly virulent subspecies has relatively recently become mobile and is now invading a geographic region that was previously devoid of it. An ancient population separation seems unlikely, because of its close genetic affinity to North American isolates. Indeed, the high-resolution nature of the F. tularensis MLVA identified a common laboratory strain (SCHU S4) as a close relative to the Slovakian type A isolates (Fig. 3 and 4). One explanation would be that these isolates are a consequence of past human activities that led to the establishment of this highly virulent subspecies in nature.
High-resolution subtyping system.
Biochemical and virulence-based characterization of F. tularensis is difficult and tedious, while current DNA-based typing methods lack individual strain discrimination. The present VNTR typing system is diagnostic for F. tularensis subspecies identification and provides much greater discriminatory power among individual F. tularensis isolates compared to other DNA-based typing systems (3, 7, 12, 22, 37). Conveniently, the method requires only very small amounts of simple DNA preparations.
The present typing system targets VNTR loci over a wide range of mutation rates. The accurate classification of F. tularensis isolates at the subspecies level is achieved by use of several slowly evolving loci. In the NJ analysis (Fig. 3), major clades were separated by fixed allelic differences at such loci (Table 3). The observed fixed allelic differences may provide a novel and rapid means for unambiguous characterization of isolates at the subspecies level. Unfortunately, the limited number of available isolates from Japan and isolates representing F. tularensis subsp. mediasiatica and F. tularensis subsp. novicida prevented a more exhaustive analysis of fixed allelic differences. Possibly, some observed fixed differences among clades may have resulted from the small sample number of isolates from Japan, F. tularensis subsp. mediasiatica and F. tularensis subsp. novicida. Other VNTR loci, especially Ft-M3, are more rapidly evolving and were used here to identify individual isolates within major clades. Importantly, the variations at these loci still showed concordance to classical epidemiological data. This study did not address the potential functionality of VNTR sequences in F. tularensis genomes. Although functional constraints on a highly variable locus such as Ft-M3 appear relaxed, we have not proved this to be the case. Nor has it been proved that less variable loci undergo stronger functional restrictions, limiting the number of observed alleles. It will, however, be an interesting subject for further studies to examine whether some of the observed fixed allelic differences among subspecies might have an explanation related to different ecological or virulence constraints acting on different subspecies. Taken together, the current VNTR typing system allows for standardized detection of a broad range of genetic diversity within F. tularensis. Such capacity provides a foundation for future development of customizable molecular genetic typing systems.
Phylogenetic structure.
While the use of VNTR variation in a phylogenetic analysis may not be ideal, it represents a viable alternative when the taxonomic units are young and there is a lack of variability in other types of sequences (3, 22, 31, 37). The concordance of genetic relationships of isolates by VNTR analysis with traditional phenotyping strongly argues that there is a phylogenetic signal in F. tularensis VNTR loci. Still, local inconsistencies in the genetic relationships of F. tularensis isolates may occur from homoplasy and the use of rapidly evolving sequences. VNTR data within taxonomic subdivisions revealed a great range of VNTR-based genetic diversity (Fig. 3; Table 3). In particular, the identification of two genetically distinct groups among type A isolates represents a previously uncharacterized pattern of genetic diversity within this subspecies. This finding should stimulate further investigation of A.I and A.II group isolates with regard to virulence, geographic origin, and ecological niches. F. tularensis subsp. tularensis isolates within the A.I group exhibit the greatest level of genetic diversity when compared with other genotype groups. The A.I group genotypes included 31 isolates from 11 U.S. states, Canada, and Central Europe (Fig. 3 and 4). The less-diverse A.II group contained 14 isolates from Canada (n = 1) and four U.S. states: California, Wyoming, Utah, and Nevada (Fig. 4). In our sample of type A isolates, the source of an isolate (i.e., geographical, species, or environmental origin) was not clearly related to the A.I. or A.II type. However, we noted that a high proportion of isolates originating in western U.S. states were typed as A.II. Hence, with regard to geographical prevalence and genotype diversity, the A.II group appears distinct from A.I isolates. We also note that the A.II isolates showed less diversity at VNTR loci than A.I isolates. Within type A isolates, the A.I and A.II subdivision may reflect two unique populations which differ in virulence or unknown biogeographical characteristics. Alternatively, the less-diverse A.II group may have been clonally derived from an A.I group ancestor, and their differentiation is purely neutral. Future comparative sequence analyses and biological studies will likely determine the validity and evolutionary background of this genetic division as well as the deeper evolutionary relationships among various F. tularensis subspecies.
Interestingly, the avirulent F. tularensis type strain ATCC 6223 clustered within the A.II group (Fig. 3). A recent microarray analysis (3) indicated strain ATCC 6223 has lost parts of its genome in relation to other F. tularensis type A isolates (which included some A.II group isolates). Genetic data indicate strain ATCC 6223 appears more unique than previously thought and, as a type strain, may not adequately represent the F. tularensis species. Such results emphasize the benefit of using multiple approaches when characterizing bacterial isolates. Interestingly, the clustering of ATCC 6223 among fully virulent type A strains by VNTR analysis conforms with the clinical history of tularemia. Edward Francis isolated the strain in 1920 from the inguinal node of a 7-year-old girl in Millard County, Utah (11). This isolate was repeatedly propagated on artificial media at the Francis laboratory and subsequently lost its virulence (20). Because it was safe, it has since been used for safe antigen production for tularemia diagnostics.
Global representatives of F. tularensis subsp. holarctica exhibit little genetic diversity, although they are the most geographically diverse F. tularensis subspecies. Comparatively, F. tularensis subsp. tularensis and F. tularensis subsp. holarctica differ greatly in host specificity and ecology (19), and VNTR data demonstrate the two subspecies show marked differences in genetic diversity. Representative genotypes among F. tularensis subsp. tularensis isolates appear much more diverse compared to those of F. tularensis subsp. holarctica (Fig. 2). This is represented first by the two distinct groups (A.I and A.II) within F. tularensis subsp. tularensis and, secondly, by the relatively great diversity within each subtype.
Although a few instances of similarity occur among North American and Scandinavian F. tularensis subsp. holarctica genotypes (clades B.I, B.III, and B.IV), our data, overall, indicate North American and Eurasian F. tularensis subsp. holarctica isolates are genetically distinct. Interestingly, North American type B genotypes are slightly more diverse compared to Eurasian type B genotypes. A model assuming that increased genetic diversity reflects evolutionary time would suggest an ancestral F. tularensis subsp. holarctica clone evolved within North American reservoirs and subsequently spread transglobally. A genetic bottleneck associated with the spread to Eurasia would result in a severe reduction in the original F. tularensis subsp. holarctica genotype population, thereby limiting the observed genetic diversity. If so, this represents a rare occurrence of a human and animal pathogen spreading from the New World to the Old.
Although representatives of Japanese F. tularensis subsp. holarctica show the F. tularensis subsp. holarctica phenotype, much genetic evidence suggests they represent a fifth F. tularensis subspecies. Previous studies involving IS element analysis (37), microarray analysis (3), 16S rRNA probe analysis (31), repetitive element PCR analysis (22), and biochemical analysis (28) indicate Japanese F. tularensis subsp. holarctica variants are distinct from other F. tularensis subsp. holarctica representatives. Accordingly, VNTR data show a clear genetic division between Japanese F. tularensis subsp. holarctica variants and all other F. tularensis subsp. holarctica isolates. Further studies on a greater number of Japanese isolates will validate their distinct taxonomic status (27), such as F. tularensis subsp. japonica, as opposed to their current grouping with other members of F. tularensis subsp. holarctica. The present analysis does, however, support their loose genetic affinity with other F. tularensis subsp. holarctica isolates (Fig. 3).
Several DNA-based methods indicated that F. tularensis subsp. mediasiatica shares great genetic affinity with F. tularensis subsp. tularensis (3, 22). However, these conclusions were based upon only a minimal number of F. tularensis subsp. mediasiatica isolates that were available for genetic analysis. Though VNTR data do not support this genetic association, it is not inconsistent, either (Fig. 3). This subspecies is thought to extend throughout most of Central Asia, and it is hopeful that future studies on a larger sample of isolates may better describe this subspecies' geographical prevalence and unique genetic characteristics.
F. tularensis was traditionally considered endemic only within the Northern Hemisphere, with F. tularensis subsp. tularensis restricted to North America. The present report of human tularemia in Australia (F. tularensis subsp. novicida) and the recent isolation of F. tularensis subsp. tularensis in Central Europe indicate that the geographical distribution of F. tularensis is more pervasive than originally thought or that the pathogen is spreading (14, 42). Natural foci of F. tularensis may be underrepresented both in Eurasia and the Southern Hemisphere, and it is hopeful that future analyses may reveal a more comprehensive description of this species' true geographical distribution. The availability of a high-resolution subtyping method will allow the evaluation of future isolates for affinity across great geographic distances.
Population structure.
Our VNTR typing system was used to estimate the population structure of F. tularensis. Long branch lengths are indicative of clonal expansions that can also be evaluated by calculating linkage disequilibrium across loci. This analysis indicated a nonrandom association of the VNTR loci when isolates from two restricted geographical areas were investigated. This finding suggests the two major subspecies of F. tularensis, F. tularensis subsp. holarctica and F. tularensis subsp. tularensis, display predominantly clonal population structures and little evidence of horizontal DNA transfer at these loci. As should be expected, significant linkage disequilibrium was detected in the analysis encompassing all isolates and subspecies. There is reason to be cautious in the interpretation of linkage data when ecological niche and/or geographical separation of lineages is not excluded (36). In fact, the intracellular lifestyle of F. tularensis, as well as geographical separation, could be the reason for the absence of horizontal DNA transfer. Regardless, our data support that F. tularensis is a predominantly clonal pathogen. Previous studies identified insertion sequences present in multiple copies across the four subspecies (21, 37). Such findings suggest there might be opportunities for recombination events involving IS elements in an individual subspecies as well as across subspecies. These types of events must be viewed with caution, as they are very susceptible to convergent evolution and may not be homologous. Somewhat surprisingly, previous analysis of a limited number of F. tularensis isolates disclosed a remarkable stability of the genomic positions of individual IS elements in all isolates of a subspecies (37). This finding suggested that these elements may be inactive with regard to recombination. Our present results targeting VNTR loci give further evidence for a remarkable stability of the F. tularensis genome, including little horizontal gene transfer among various F. tularensis lineages.
Forensic analysis.
When used as a biological threat agent, F. tularensis poses a serious public health risk (8). Its effective development and use by State-sponsored bioweapons programs (8) may be a harbinger of future terrorist activities, much as was recently illustrated with the anthrax letter attacks. Detailed information about natural populations will greatly assist in distinguishing what is natural from events that are not. Rapid high-resolution subtyping provides a crucial tool in understanding natural population structure, while filling a forensic role in the event of a biological attack. This is illustrated by the characterization of F. tularensis subsp. tularensis recently isolated in Slovakia. In this highly diverse subspecies, the great similarity between a laboratory strain (SCHU S4) and a Slovakian isolate was unexpected and merits further investigation. The ongoing genome sequencing of one of the Slovakian type A isolates will likely further explain their origin.
Acknowledgments
This work was supported by funding from the Department of Energy's CBNP program, the National Institutes of Health's program on pathogen evolution, the Cowden Endowment in Microbiology, the Swedish Medical Research Council, the Medical Faculty, Umeå University, and the Swedish Defence Research Agency.
We thank numerous scientific colleagues for kindly providing strains to the Francisella Strain Collection in Umeå, Sweden. We thank Jane Wong (California Department of Health), Michael Lytle and Kristy Bradley (Oklahoma Department of Health), and Powell Gammill (Arizona Department of Health) for providing isolate DNAs. In addition, this work would have been impossible without timely access to the preliminary LVS and SCHU S4 genomic sequences. For the LVS sequence, we thank Emilo Garcia and Patrick Chain at Lawrence Livermore National Laboratory. We thank the F. tularensis strain SCHU S4 genome sequencing consortium. Finally, we thank David Wagner for assistance on the statistical analysis of data.
REFERENCES
- 1.Agapow, P. M., and A. Burt. 2001. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes. 1:101-102. [Google Scholar]
- 2.Benson, G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broekhuijsen, M., P. Larsson, A. Johansson, M. Byström, U. Eriksson, E. Larsson, R. G. Prior, A. Sjöstedt, R. W. Titball, and M. Forsman. 2003. Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis. J. Clin. Microbiol. 41:2924-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke, D. S. 1977. Immunization against tularemia: analysis of the effectiveness of live Francisella tularensis vaccine in prevention of laboratory-acquired tularemia. J. Infect. Dis. 135:55-60. [DOI] [PubMed] [Google Scholar]
- 5.Clarridge, J. E., III, T. J. Raich, A. Sjöstedt, G. Sandström, R. O. Darouiche, R. M. Shawar, P. R. Georghiou, C. Osting, and L. Vo. 1996. Characterization of two unusual clinically significant Francisella strains. J. Clin. Microbiol. 34:1995-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross, J. T., and R. L. Penn. 2000. Francisella tularensis (tularemia), p. 2393-2402. In G. L. Mandell, J. E. Bennet, and R. Dolin (ed.), Mandell, Douglas and Bennet's principles and practice of infectious diseases, 5th ed., vol. 2. Churchill Livingstone, Philadelphia, Pa.
- 7.de la Puente-Redondo, V. A., N. G. del Blanco, C. B. Gutierrez-Martin, F. J. Garcia-Pena, and E. F. Rodriguez Ferri. 2000. Comparison of different PCR approaches for typing of Francisella tularensis strains. J. Clin. Microbiol. 38:1016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennis, D. T., T. V. Inglesby, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, M. Layton, S. R. Lillibridge, J. E. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, and K. Tonat. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285:2763-2773. [DOI] [PubMed] [Google Scholar]
- 9.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisella tularensis strain typing using multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsman, M., G. Sandström, and A. Sjöstedt. 1994. Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int. J. Syst. Bacteriol. 44:38-46. [DOI] [PubMed] [Google Scholar]
- 11.Francis, E. 1921. Tularemia. I. The occurrence of tularemia in nature as a disease of man. Public Health Rep. 36:1731-1753. [Google Scholar]
- 12.Garcia Del Blanco, N., M. E. Dobson, A. I. Vela, V. A. De La Puente, C. B. Gutierrez, T. L. Hadfield, P. Kuhnert, J. Frey, L. Dominguez, and E. F. Rodriguez Ferri. 2002. Genotyping of Francisella tularensis strains by pulsed-field gel electrophoresis, amplified fragment length polymorphism fingerprinting, and 16S rRNA gene sequencing. J. Clin. Microbiol. 40:2964-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 14.Gurycova, D. 1998. First isolation of Francisella tularensis subsp. tularensis in Europe. Eur. J. Epidemiol. 14:797-802. [DOI] [PubMed] [Google Scholar]
- 15.Haubold, B., and R. R. Hudson. 2000. Lian 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847-848. [DOI] [PubMed] [Google Scholar]
- 16.Haubold, B., M. Travisano, P. B. Rainey, and R. R. Hudson. 1998. Detecting linkage disequilibrium in bacterial populations. Genetics 150:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollis, D. G., R. E. Weaver, A. G. Steigerwalt, J. D. Wenger, C. W. Moss, and D. J. Brenner. 1989. Francisella philomiragia comb. nov. (formerly Yersinia philomiragia) and Francisella tularensis biogroup novicida (formerly Francisella novicida) associated with human disease. J. Clin. Microbiol. 27:1601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopla, C. E., and A. K. Hopla. 1994. Tularemia, p. 113-126. In G. W. Beran (ed.), Handbook of zoonoses, 2nd ed. CRC Press, Boca Raton, Fla.
- 19.Jellison, W. L. 1974. Tularemia in North America, 1930-1974. University of Montana Foundation, Missoula.
- 20.Jellison, W. L. 1972. Tularemia: Dr. Edward Francis and his first 23 isolates of Francisella tularensis. Bull. Hist. Med. 46:477-485. [PubMed] [Google Scholar]
- 21.Johansson, A., I. Göransson, P. Larsson, and A. Sjöstedt. 2001. Extensive allelic variation among Francisella tularensis strains in a short-sequence tandem repeat region. J. Clin. Microbiol. 39:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson, A., A. Ibrahim, I. Göransson, U. Eriksson, D. Gurycova, J. E. Clarridge III, and A. Sjöstedt. 2000. Evaluation of PCR-based methods for discrimination of Francisella species and subspecies and development of a specific PCR that distinguishes the two major subspecies of Francisella tularensis. J. Clin. Microbiol. 38:4180-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klevytska, A. M., L. B. Price, J. M. Schupp, P. L. Worsham, J. Wong, and P. Keim. 2001. Identification and characterization of variable-number tandem repeats in the Yersinia pestis genome. J. Clin. Microbiol. 39:3179-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurtz, S., and C. Schleiermacher. 1999. REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics 15:426-427. [DOI] [PubMed] [Google Scholar]
- 26.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsufjev, N. G., and I. S. Meshcheryakova. 1982. Infraspecific taxonomy of tularemia agent Francisella tularensis McCoy et Chapin. J. Hyg. Epidemiol. Microbiol. Immunol. 26:291-299. [PubMed] [Google Scholar]
- 28.Olsufjev, N. G., and I. S. Meshcheryakova. 1983. Subspecific taxonomy of Francisella tularensis. Int. J. Syst. Bacteriol. 33:872-874. [Google Scholar]
- 29.Rotz, L. D., A. S. Khan, S. R. Lillibridge, S. M. Ostroff, and J. M. Hughes. 2002. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 8:225-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 31.Sandström, G., A. Sjöstedt, M. Forsman, N. V. Pavlovich, and B. N. Mishankin. 1992. Characterization and classification of strains of Francisella tularensis isolated in the central Asian focus of the Soviet Union and in Japan. J. Clin. Microbiol. 30:172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saslaw, S., and S. Carhart. 1961. Studies with tularemia vaccines in volunteers. III. Serological aspects following intracutaneous or respiratory challenge in both vaccinated and non-vaccinated volunteers. Am. J. Med. Sci. 241:689-699. [PubMed] [Google Scholar]
- 33.Saslaw, S., and H. N. Carlisle. 1961. Studies with tularemia vaccines in volunteers. IV. Brucella agglutinins in vaccinated and non-vaccinated volunteers challenged with Pasteurella tularensis. Am. J. Med. Sci. 242:166-172. [DOI] [PubMed] [Google Scholar]
- 34.Shapiro, D. S., and D. R. Schwartz. 2002. Exposure of laboratory workers to Francisella tularensis despite a bioterrorism procedure. J. Clin. Microbiol. 40:2278-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sjöstedt, A. 2003. Family XVII, Francisellaceae. Genus I, Francisella, p. 111-113. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer-Verlag, New York, N.Y.
- 36.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas, R., A. Johansson, B. Neeson, K. Isherwood, A. Sjöstedt, J. Ellis, and R. W. Titball. 2003. Discrimination of human pathogenic subspecies of Francisella tularensis by using restriction fragment length polymorphism. J. Clin. Microbiol. 41:50-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Titball, R. W., A. Johansson, and M. Forsman. 2003. Will the enigma of Francisella tularensis virulence soon be solved? Trends Microbiol. 11:118-123. [DOI] [PubMed] [Google Scholar]
- 39.U.S. Department of Health and Human Services. 1999. Biosafety in microbiological and biomedical laboratories (BMBL), 4th ed. Revision date, 6 April 2002. U.S. Government Printing Office, Washington, D.C. [Online.] http://www.cdc.gov/od/ohs/.
- 40.van Belkum, A. 1999. The role of short sequence repeats in epidemiologic typing. Curr. Opin. Microbiol. 2:306-311. [DOI] [PubMed] [Google Scholar]
- 41.Weir, B. S. 1990. Genetic data analysis: methods for discrete population genetic data analysis. Sinauer Associates, Inc., Sunderland, Mass. [DOI] [PubMed]
- 42.Whipp, M. J., J. M. Davis, G. Lum, J. de Boer, Y. Zhou, S. W. Bearden, J. M. Petersen, M. C. Chu, and G. Hogg. 2003. Characterization of a novicida-like subspecies of Francisella tularensis isolated in Australia. J. Med. Microbiol. 52:839-842. [DOI] [PubMed] [Google Scholar]