Abstract
The cold shock proteins are small peptides that share a conserved domain, called the cold shock domain (CSD), that is important for nucleic acid binding. The Caulobacter crescentus genome has four csp genes that encode proteins containing CSDs. Three of these (cspA, cspB, and cspC) encode peptides of about 7 kDa and are very similar to the cold shock proteins of other bacteria. Analysis by reverse transcription-PCR of the fourth gene (cspD), which was previously annotated as encoding a 7-kDa protein, revealed that the mRNA is larger and probably encodes a putative 21-kDa protein, containing two CSDs. A search in protein sequences databases revealed that this new domain arrangement has thus far only been found among deduced peptides of α-proteobacteria. Expression of each Caulobacter csp gene was studied both in response to cold shock and to growth phase, and we have found that only cspA and cspB are induced by cold shock, whereas cspC and cspD are induced at stationary phase, with different induction rates. The transcription start sites were determined for each gene, and a deletion mapping of the cspD promoter region defined a sequence required for maximal levels of expression, indicating that regulation of this gene occurs at the transcriptional level. Deletion of cspA, but not cspD, caused a reduction in viability when cells were incubated at 10°C for prolonged times, suggesting that cspA is important for adaptation to a low temperature.
Bacterial cells face many challenges in the outward environment, being exposed to chemical and physical factors that may considerably affect their growth. Temperature is one of the most critical parameters for bacterial growth, and cells must adapt themselves fairly quickly to sudden temperature changes. Whereas a high temperature causes severe damage to the cells mainly because of protein denaturation, a low temperature may render cells nonviable because of alterations in nucleic acids and membrane lipids.
The response to a low temperature involves a change in the cell membrane lipid composition, with an increase in the proportion of unsaturated fatty acids, to keep the fluidity of the membrane at a low temperature (40). The ribosomes also adapt themselves to translate cold-specific mRNAs by incorporation of ribosomal factors that change their functional properties (24). The structure and topology of the chromosomal DNA is also affected, and its adaptation to the cold involves the induction of proteins that are nucleoid associated, such as H-NS (7).
One of the major difficulties bacteria face during a temperature downshift is the stabilization of secondary structures of nucleic acids, particularly mRNA, which prevents them from being efficiently translated. Upon cold shock, bacteria express a well-defined set of proteins to adapt the cell to the new temperature condition. The first protein described as a major protein induced upon decrease in temperature was CspA from Escherichia coli (17), an RNA chaperone which helps to destabilize the secondary structures of the RNA (23). The proposed role for CspA was also to increase mRNA translation and to render mRNA more susceptible to RNase degradation (6, 23). Eight other proteins homologous to CspA were identified in E. coli (CspB to CspI), and it was shown that CspA, CspE, and CspC, but not CspB, also act as transcription antiterminators in vitro (3), and CspD has been shown to inhibit DNA replication (56).
Homologues of cold shock proteins have not been found in archaeal genomes, and among the eubacteria they are present in most proteobacteria but not in spirochetes or cyanobacteria, the latter presenting a family of small cold-inducible proteins with RNA-binding domains similar to those found in eukaryotic proteins (36). These small proteins possess a conserved domain called the cold shock domain (CSD) that is composed of two nucleic acid-binding motifs, RNP1 and RNP2, that are crucial for the binding to single-stranded DNA and RNA (38). The CSD is also found in eukaryotic proteins, where it mediates RNA binding and interacts with other RNA-binding domains (20). The role of some of these eukaryotic proteins has been determined, showing that they are involved in coupling transcription of specific mRNAs with their translation and, in some cases, acting as transcription factors (5, 49).
Bacterial genomes contain usually many copies of csp genes, but there is a large variation in their patterns of expression. In E. coli, only four of the nine cold shock proteins are induced upon cold shock (CspA, CspB, CspG, and CspI) (47, 53), and one (CspD) is induced during stationary phase and upon nutritional starvation (51). Bacillus subtilis has three CspA-like proteins, which are all induced at a low temperature (18), and two of them (CspB and CspC) are also induced at stationary phase (21, 25). Lactobacillus plantarum has three csp genes, but only one (cspL) is highly induced at temperature downshift and stationary phase, whereas cspP and cspC are constitutively expressed (8).
The caulobacters are ubiquitous bacteria found in humid soils and in practically every aquatic environment (34). The distinct cell cycle of this bacterium, presenting a sessile phase and an obligatory motile phase, confers a good mode of dispersion through water in search for better nutritional conditions, and may also be important for adaptation to challenging situations, such as extreme environments. These bacteria were reported in frozen soil and bodies of water (1, 10), suggesting that it must be well adapted to live in low temperature. Analysis of the genome content of Caulobacter showed that it possesses four genes encoding putative small cold shock proteins (32), but their patterns of expression have not yet been determined. We have investigated here the regulation of the csp genes in response to cold shock and growth phase and determined a regulatory sequence important for expression of cspD that encodes a peptide presenting a novel CSD arrangement in bacteria. Two strains, one carrying a deletion of the cold-induced cspA gene and the other of the stationary-phase-induced cspD gene, were generated, and analyses of the phenotype indicate that cspA, but not cspD, is involved in adaptation to low temperature.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and genetic procedures.
Caulobacter crescentus was grown at 30°C in peptone-yeast extract medium or minimal M2-glucose medium (9) supplemented with kanamycin (5 μg/ml), tetracycline (1 μg/ml), or nalidixic acid (25 μg/ml) as necessary. Escherichia coli strain DH5α (Invitrogen) was used in the cloning procedures. E. coli was grown at 37°C in Luria-Bertani medium supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), or tetracycline (12.5 μg/ml) as necessary. Plasmid pRKlacZ290 (14) was introduced into Caulobacter sp. by conjugation with E. coli strain S17-1 (39).
Sequence analyses.
Protein sequence analysis was performed with the Protean program, which is included in the Lasergene DNA analysis package DNASTAR (DNASTAR, Inc., Madison, Wis.). The multiple alignments were performed with CLUSTALX (44), and searches in the sequence databases were performed with the BLAST algorithm (2).
Transcript analysis by RT-PCR.
Analysis of the cspD mRNA was carried out by nonquantitative reverse transcription-PCR (RT-PCR) with total RNA from either mid-log- or stationary-phase cells treated with DNase I (amplification grade; Invitrogen) to eliminate any trace of DNA. The primers used were RT-2A and RT-2B (Table 1). Reactions were performed by using the SuperScript One-Step RT-PCR kit (Invitrogen) as recommended by the supplier. The RT-PCR conditions were as follows: 30 min at 55°C and 2 min at 94°C, followed by 35 cycles of 60 s at 94°C, 60 s at 48°C, and 60 s at 72°C, with a final cycle of 7 min at 72°C. Control reactions with only the Taq DNA polymerase were carried out to assure that no amplification was due to the presence of DNA in the samples.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| CSPA-A | CGAAACGGCTCGAGCGATG |
| CSPA-B | ATAGGATCCGTCGTTCTCAGAACCATC |
| CSPA-C | AAACTGCAGCTTCACGGTTCCAGTCGC |
| CSPA-D | TATCTGCAGCAAGCGGCCTAAAGCCCG |
| CSPA-E | AAAGGGCCCTCGCTGGTGTAGGCCTCGG |
| CSPA-PE | ATGTTCCTTGTAGGCATAGGTG |
| CSPB-A | ATAGGATCCGATCGCTCACCGCGAGAC |
| CSPB-B | GAAAAGCTTCAAGGCCTCACGCTGCGC |
| CSPB-PE | GAACCACTTTACGGTGCCG |
| CSPC-F | CGGGGTTTTTTGCGGTGCAATATCGC |
| CSPC-G | TTAGGATCCTGGCCGCCGTCCTCGGG |
| CSPC-PE | CCGTTCGCCATATTATCCTCA |
| CSPD-A | ATTGGATCCATATAACGGCTATGTTCC |
| CSPD-B | ATAGAATTCTGGTGACGATCTCGACC |
| CSPD-C | ATGGATCCACTGCCATCTTCGGC |
| CSPD-D | CCAGGATCCTGGCTTGCCCAATCAGCCC |
| CSPD-E | AAACTGCAGATTCGTCCCGAGATGATTCC |
| CSPD-F | AAAGGATCCTGGTGCGGTTCGCACGCGG |
| CSPD-G | AAAGAATTCGCTGCTCTACAGGGGTTCG |
| CSPD-PE | AAAATCGTAACCAGACATCCC |
| RT-2A | TGGTTACGATTTTGAGGACG |
| RT-2B | GTTGAACCATTTCACCTTGG |
Underlined nucleotides indicate restriction sites incorporated into oligonucleotides.
Primer extension analysis.
Oligonucleotides CSPA-PE, CSPB-PE, CSPC-PE, and CSPD-PE (Table 1), which hybridize to the beginning of the coding region of each gene were labeled with [γ-32P]ATP and used for primer extension experiments. The primers were hybridized to 50 μg of total RNA isolated from log-phase cells grown at either 30 or 10°C for 1 h and 2 h or from cells at stationary phase and then extended with the SuperScript II reverse transcriptase (Invitrogen) as recommended by the supplier. The DNA sequencing ladder was obtained by cycle sequencing with the same primer and, as the template, a plasmid containing the cloned region of each gene by using the Thermosequenase cycle sequencing kit (USB).
Cloning of the promoter regions and gene expression analysis.
The regions containing the csp genes were amplified from the C. crescentus chromosome by PCR with the various primers (see Table 1) as follows: cspB, CSPB-A and CSPB-B; cspD, CSPD-A and CSPD-B; cspA, CSPA-A and CSPA-B; and cspC, CSPC-F and CSPC-G.
PCRs were carried out with 1 μg of C. crescentus NA1000 chromosomal DNA, 50 pmol of each set of oligonucleotides (described above), 0.2 mM concentrations of each deoxynucleoside triphosphate, 1.5 mM MgCl2, 2.5 U of Taq DNA polymerase (Invitrogen), and 1× PCR buffer (supplied with the enzyme). The PCR conditions were 5 min 94°C, followed by 40 cycles of 90 s at 94°C, 1 min at 50°C, and 1 min at 72°C, with a final cycle of 7 min at 72°C. The amplified fragments were cloned into the TOPO vector from TOPO TA cloning kit for sequencing (Invitrogen) and then confirmed by DNA sequencing.
DNA fragments of the cspD regulatory region were either obtained by restriction digestion (as shown in Fig. 6) or by PCR as described above with the primer pair CSPD-A-CSPD-B, CSPD-C-CSPD-B, or CSPD-D-CSPD-B, and the sequence was confirmed by DNA sequencing. DNA fragments of the promoter regions of the cspA, cspB, and cspC genes were obtained by restriction digestion (cspA, BamHI/NcoI; cspB, EcoRI/SacI; cspC, BamHI/EcoRI). All of the fragments obtained were cloned into pRKlacZ290 (14) and introduced into C. crescentus NA1000 by conjugation. Promoter activities during cold shock and stationary phase were determined by measuring the β-galactosidase activity by the method of Miller (29).
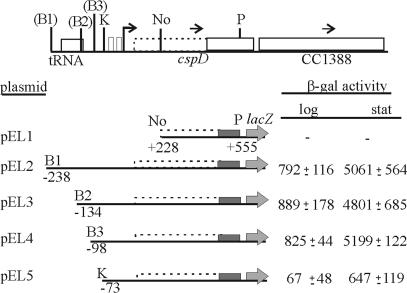
FIG. 6.
Deletion mapping of cspD promoter region. The scheme of the cspD locus is shown above, indicating the previously annotated coding region (solid line), and the proposed extended coding region (dashed line). The bent arrow indicates the transcription start sites determined by primer extension. Restriction sites are indicated as follows: B, BamHI; K, KpnI; No, NotI; P, PstI. The sites in parentheses were introduced by PCR and are not in the original sequence. The plasmids carrying the constructs were introduced into C. crescentus NA1000, and promoter activity was measured by β-galactosidase assays both in exponential-phase (log) and stationary-phase cells (stat). The results are in Miller units (29) and are the average of at least three independent assays, with the respective standard deviation.
Deletion of cspA and cspD and viability tests.
To delete the coding region of cspA, two fragments containing the region upstream and downstream of the gene, were amplified by PCR with primers CSPA-B/CSPA-C and CSPA-D/CSPA-E, respectively (Table 1), and ligated in tandem to the suicide vector pNPTS138. This 1.0-kb ApaI/BamHI fragment contains the flanking regions of the gene without the deleted region. The same was done for the cspD coding region, with the primer pairs CSPD-D-CSPD-E and CSPD-F-CSPD-G, generating a 1.6-kb PstI/EcoRI fragment. The pNTPS138 vectors were then introduced into C. crescentus NA1000 by conjugation with E. coli S17-1, and the genes were deleted by allelic exchange after double recombination. The deletions were confirmed by PCR amplification with primers flanking each gene and by Southern blots.
Determination of survival at low temperature was performed as follows. Cells were grown at 30°C up to mid-log phase and then transferred to 10°C, with agitation. Samples of each culture were taken at different time points, and viability tests were carried out by determination of the number of CFU. The relative survival was calculated as the number of CFU of the mutant strains at each time point divided by the number of CFU of strain NA1000 at the same points, considering that the absorbance at 600 nm for all cultures were identical.
RESULTS
Sequence analysis of open reading frames (ORFs) containing the CSD.
There are four genes encoding small cold shock proteins similar to E. coli CspA in the genome of Caulobacter (32). The genes were arbitrarily named as follows: cspA (CC2903), cspB (CC0665), cspC (CC2623), and cspD (CC1387). Three of the peptides (CspA, CspB, and CspC) showed a higher degree of similarity to each other and to E. coli CspA and possess one CSD, which harbors the nucleic acid-binding motifs RNP1 and RNP2 (Fig. 1A). These proteins have similar predicted molecular mass of 7 kDa but have different pIs, being CspB and CspC acidic (pI 5.74 and 4.82, respectively) and CspA neutral (pI 7.16).
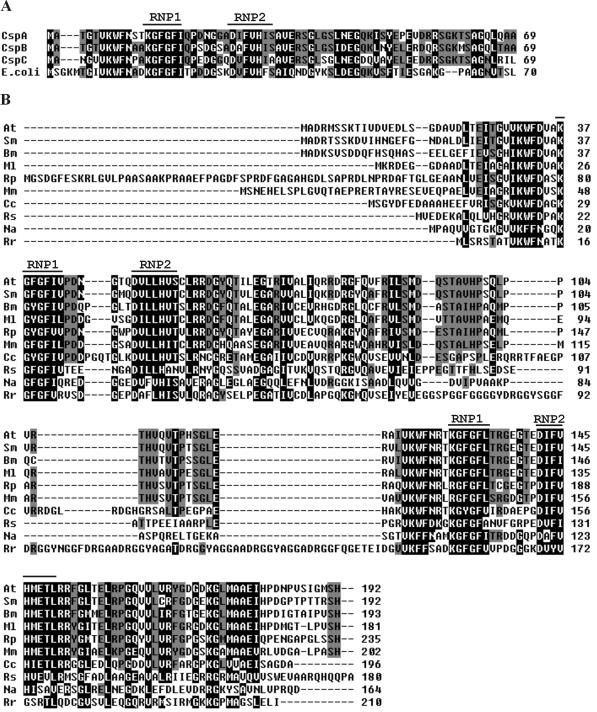
FIG. 1.
(A) Amino acid sequence comparison of the three C. crescentus genes containing one CSD with E. coli cspA (GenBank no. AAB18533). (B) Amino acid sequence comparison of the proposed new annotation of cspD from C. crescentus (Cc) with other predicted α-Proteobacteria peptides showing two CSDs. Above the sequences are indicated the two RNA-binding motifs (RNP1 and RNP2). Residues shaded in black indicate conserved residues present in at least 90% of the proteins; those shaded in dark gray are present in at least 60% of the proteins. The GenBank no. and abbreviations are as follows: Agrobacterium tumefaciens (At; AAK87573), Sinorhizobium meliloti (Sm; CAC46297), Brucella melitensis (Bm; AAL51912), Mesorhizobium loti (Ml; BAB47810), Rhodopseudomonas palustris (Rp; ZP_00012063), Magnetospirillum magnetotacticum (Mm; ZP_00049605), Rhodobacter sphaeroides (Rs; ZP_00006193), Novosphingobium aromaticivorans (Na; ZP_00093526), and Rhodospirillum rubrum (Rr; ZP_00013462).
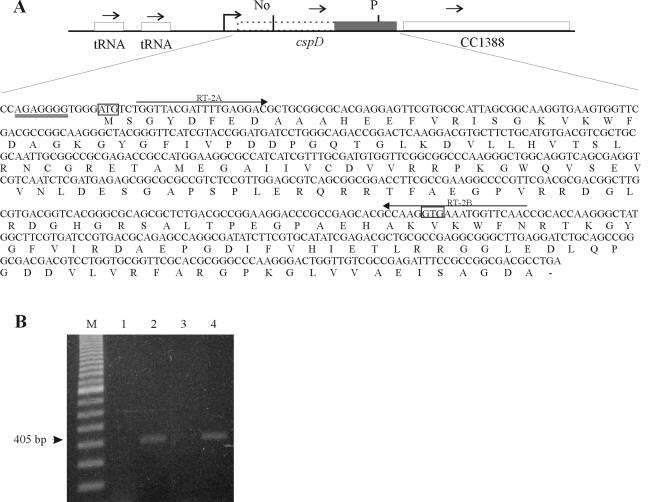
The original genome annotation of the fourth gene (cspD) identified a coding region corresponding to 192 bp, but a more detailed sequence analysis showed that the coding region is probably 588 bp long, coding for a putative protein of 21.5 kDa and a predicted pI of 5.94 (Fig. 2A). In order to investigate whether the cspD coding region is larger than what was previously determined, we performed an RT-PCR experiment, with primers that hybridize with the beginning of the proposed longer ORF and the beginning of the annotated cspD, respectively (Fig. 2A). A specific product was obtained, indicating that there is an mRNA encompassing the whole predicted ORF and suggesting that cspD could encode a 21.5-kDa protein (Fig. 2B). This putative protein contains two CSDs of 70 residues separated by a nonconserved region of 52 residues. A search in protein databases revealed that other proteins with the same domain structure are found in α-Proteobacteria but not in other eubacteria (Fig. 1B). Interestingly, we could not find any similar peptide in the two Rickettsia species that have their complete genome sequence determined.
FIG. 2.
Determination of the existence of a longer cspD transcript. (A) The scheme indicates the region of the cspD gene, showing the new proposed coding region (dotted lines), and the originally annotated coding region (dark box). Below is shown the sequence of the proposed cspD coding region, indicating the position of the two primers used in the RT-PCR (arrows). The new (ATG) and annotated (GTG) start sites are boxed. Ribosomal binding site is double underlined. (B) RNA was isolated from mid-log phase cells (lanes 1 and 2) and from cells at 24 h after entry into stationary phase (lanes 3 and 4) and treated with DNase I previous to the experiment. RT-PCR was performed with a pair of oligonucleotides—one that hybridizes close to the ATG of the proposed longer cspD ORF and one at the beginning of the annotated cspD coding region. Control reactions, carried out with Taq DNA polymerase but without reverse transcriptase, yielded no amplified bands (lanes 1 and 3), confirming that there is no contamination of DNA in the samples. The expected 405-nt fragment obtained for both samples is indicated by an arrow.
These proteins share extensive similarity in their CSDs, but the amino terminus and the region between the two CSDs show high divergence, with the Caulobacter, Rhodopseudomonas, and Rhodospirillum proteins having insertions in these regions. An analysis of the CspD sequence with respect to backbone chain flexibility indicated that the region between residues 87 and 128 is highly flexible. A longer insertion at this same relative position was also seen in a Rhodospirillum homolog (Fig. 1B), suggesting that there may be less selective pressure on this interdomain region than on the CSDs.
The next ORF, CC1388, encodes a conserved 184-amino-acid protein, which possesses a domain of unknown function (DUF192) when analyzed by the PFAM program (4). The same genetic organization of cspD and CC1388 found in C. crescentus was observed in other α-Proteobacteria, except for M. magnetotacticum, R. rubrum, and N. aromaticivorans, in which the CC1388 homolog is found elsewhere in the genome.
Expression of csp genes in response to cold shock.
It was observed for several bacteria that the expression of some homologues of CspA increases with cold shock, whereas other homologues are not induced under this condition. To determine whether the C. crescentus csp genes are induced by cold shock, the promoter region of each gene was cloned upstream of a lacZ gene in a reporter plasmid, and expression was analyzed by β-galactosidase activity (Fig. 3). It should be noted, however, that with reporter genes the results are only indicative of the time and extent of the induction, and some variation may occur, as reported by Goldenberg et al. for the cspA promoter (16). Figure 3B shows that expression of fusions carrying cspA or cspB promoters was increased by cold shock. On the other hand, expression of fusions carrying cspC or cspD promoters did not show any increase under the same conditions, indicating that these genes are not cold induced, similarly to E. coli cspC, cspD, or cspE genes (51, 55).
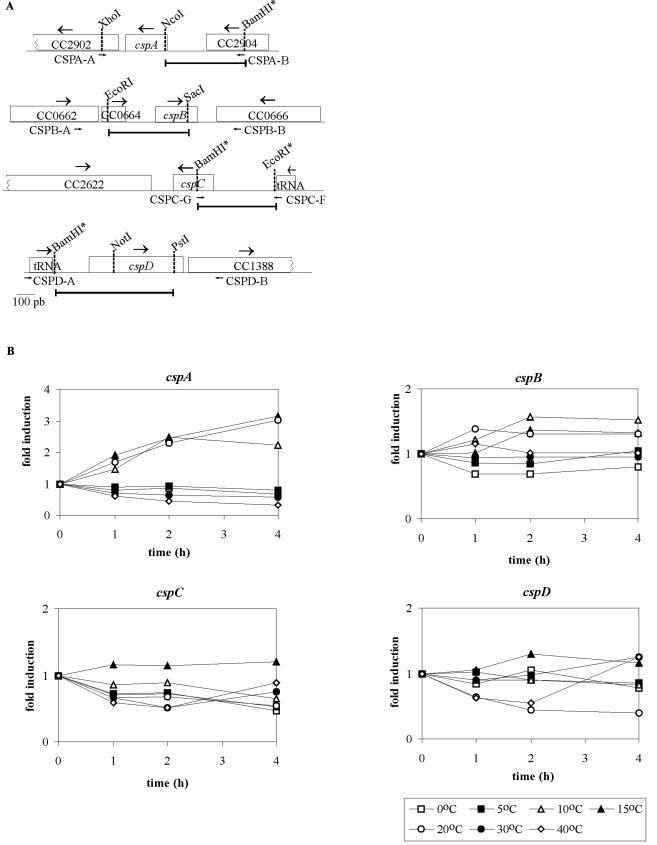
FIG.3.
(A) Schematic representation of the csp genes. The DNA fragments cloned to the pRKlacZ290 vector in the transcriptional fusions are indicated by bars. The small arrows indicate the primers used to amplify each region from the genome. Some restriction sites are indicated, and the sites inserted by PCR are labeled with an asterisk. (B) Analysis of the cold induction of the csp genes. Cells harboring the transcription fusions of each gene were grown at 30°C up to mid-log phase and were then transferred to different temperatures. Expression of each construct was measured by β-galactosidase activity assays (29) at sequential time points, and the results are shown as relative measurements of induction.
In order to evaluate the temperatures for which the cspA and cspB gene expression is maximized, we tested the levels of transcription at several temperatures ranging from 0 to 40°C (Fig. 3B). We observed that there is no induction at temperatures lower than 10°C (0 and 5°C) or higher than 20°C (30 and 40°C) and that both genes are induced at 10, 15, and 20°C. The cspA gene showed higher levels of induction, and expression was still going up after 4 h at 15 and 20°C, whereas the peak of expression of cspB was at 2 h and remained stable after that.
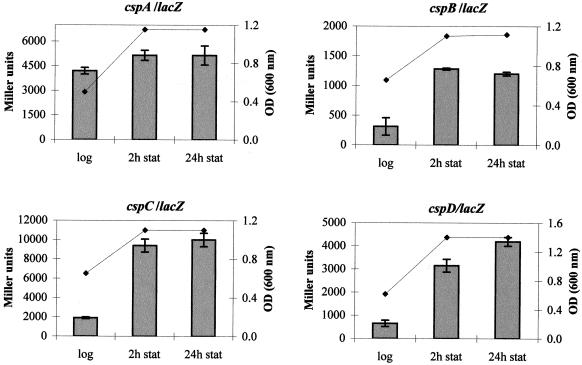
These results prompted us to determine whether the genes are induced at stationary phase, as with the cspD gene in E. coli (51). There is a great increase in enzyme activity at the onset of stationary phase when cells carry promoter fusions of cspB (3.8-fold), cspC (5.3-fold), and cspD (6.5-fold) (Fig. 4) and a very small increase (1.2-fold) with the promoter fusion of cspA. Once at stationary phase, the expression of these genes did not significantly change even 24 h after the onset of this phase, with the exception of the cspD/lacZ fusion, which showed a small increase after this time.
FIG. 4.
Growth-phase-dependent expression of the csp genes. Expression of the csp genes was determined from cells harboring the respective promoter fusions at mid-log phase and at 2 and 24 h after entry into stationary phase. The β-galactosidase activity is expressed in Miller units (29), and growth was monitored by measuring the optical density (OD) at 600 nm.
Analysis of transcription start sites.
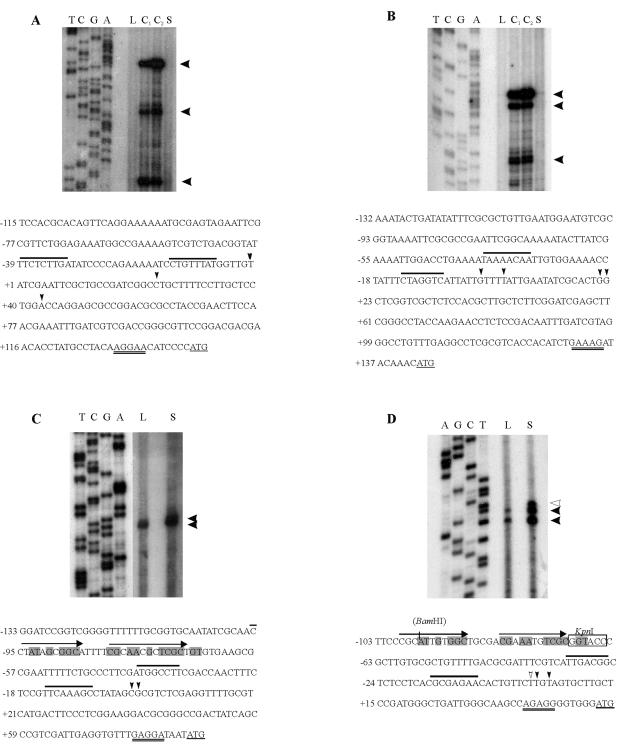
The transcription start sites of the csp genes were determined by primer extension analyses (Fig. 5). All genes showed multiple start sites, and the cspA and cspB transcripts were greatly induced after 2 h at 10°C (Fig. 5A and B), whereas cspC and cspD were induced at stationary phase (Fig. 5C and D). Despite the fact that the cspB/lacZ fusion showed an increased β-galactosidase activity at stationary phase (Fig. 4), we could not detect an increase in the cspB transcript, suggesting that the results may be due to some interference of the fusion to lacZ. Transcripts from cspA and cspB were detected from RNA of cells growing at 30°C at longer exposure times (not shown). If we consider the major transcripts of each gene, the results showed that the 5′-untranslated regions of the cold-induced cspA and cspB genes are longer (127 and 143 nucleotides [nt], respectively) than those of the genes that are not cold induced (cspC [64 nt] and cspD [45 nt]). Long 5′-untranslated regions are found in several cold-induced genes and have a role in regulation of gene expression (11, 12, 16, 47, 48). The −35 and −10 sequences of the major transcripts share low similarity among the promoters, but there are AT-rich regions upstream of the cspB −35 region, similar to what was reported for E. coli cspA (16, 30).
FIG. 5.
Determination of the transcription start sites of csp genes. Primer extension analysis was carried out with total RNA from exponential-phase cells (L) or stationary-phase cells (S) at 30°C or incubated at 10°C for 1 h (C1) or 2 h (C2). The primers were end labeled with 32P and extended with reverse transcriptase to determine the transcription start sites and were also used in DNA sequencing reactions (shown on the left). Below each panel is shown the respective regulatory regions: cspA (A), cspB (B), cspC (C), and cspD (D). Black arrowheads indicate the transcription start sites, and a white arrowhead indicates the stationary-phase start site of cspD. The most upstream start sites were arbitrarily chosen as position +1, and the −35/−10 sequences are overlined (for clarity, only the most upstream promoters are indicated). The start codons are underlined, and the ribosome-binding sites are double underlined. In panel D, the KpnI restriction site used for the transcription fusion pEL5 (Fig. 6) is boxed, and the position of the BamHI restriction site introduced by PCR in construct pEL4 is shown in parentheses. In panels C and D, the arrows indicate two imperfect direct repeats, and the shaded nucleotides indicate the regulatory sequence of cspD that is also found in cspC.
For the cspD gene, the −35 (TTGACGG) and −10 (GCGAGAAC) regions follow the consensus proposed for promoter regions of Caulobacter housekeeping genes (27). Two start sites were observed when RNA isolated from exponential-phase cells was used, with the downstream signal being the more intense (Fig. 5D), and three signals were observed when stationary-phase RNA was used—two corresponding to the log-phase RNA and a new one located upstream. All three bands were more intense in stationary phase than in the log phase, indicating that the induction observed at this growth phase is due to an increase in mRNA level whose transcription initiates from the same promoter.
The regulatory region of cspD was further analyzed by cloning several promoter fragments containing progressive deletions each in front of a lacZ reporter gene (Fig. 6). Deletion analysis showed that there is no promoter activity downstream of the NotI site (pEL1), which is 225 bp upstream of the annotated start codon. Fragments comprising the region from the upstream tRNA gene to the PstI site drive the maximal values of β-galactosidase activity in the log phase and also a great induction (∼5.5-fold) in the stationary phase. These levels of expression are observed for all constructs that contain the region downstream of position −98 (pEL2, pEL3, and pEL4). The activity of pEL5 is much lower than that of pEL4, although the promoter region is present in this construction, which suggests that the region between positions −98 and −73 is necessary for maximal cspD expression in both log and stationary phases. The sequence found in this region comprises two imperfect direct repeats (Fig. 5D), and a similar sequence was also found upstream of the cspC gene (Fig. 5C), whose promoter fusion showed similar levels of β-galactosidase activity during stationary phase. These results indicate that this region may be a regulatory site involved in the maximal levels of expression of cspD, but it is not involved in the growth-phase-specific induction.
Analysis of cspA and cspD mutants.
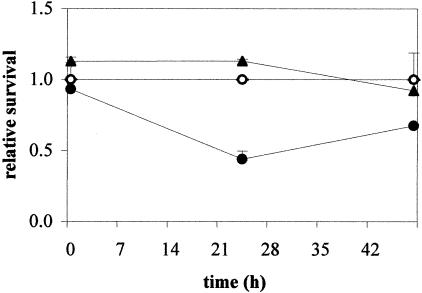
cspA is the major cold-induced csp gene, and it probably has a role in adaptation of Caulobacter to low temperature, whereas the cspD gene could be more important in adaptation to stationary phase. In order to verify this hypothesis, two mutant strains were generated in which the cspA gene and cspD genes were deleted (NA1000 [ΔcspA] and NA1000 [ΔcspD], respectively). The strains were tested for survival at low temperature (10°C) and at stationary phase. Neither strain showed any defect in survival at stationary phase (results not shown), and when cells were incubated for prolonged times at 10°C, the parental NA1000 strain and the cspD mutant did not show any decrease in survival (Fig. 7). On the other hand, the cspA mutant showed a decrease in survival after 24 h at 10°C and kept that reduced viability after 48 h. These results suggest that cspA is important for Caulobacter to withstand low temperature, whereas the role of cspD is probably not directly related to cold survival.
FIG. 7.
Viability of the mutant strains at 10°C. Cells of the parental strain NA1000 (○), NA1000 (ΔcspA) (•), and NA1000 (ΔcspD) (▴) were grown at 30°C up to early log phase and then transferred to 10°C. Aliquots were taken before (0 h) and 24 or 48 h after incubation at 10°C, and serial dilutions were plated to determine the number of CFU. Survival rates were determined relative to NA1000 at each time point.
DISCUSSION
The cold shock response in bacteria involves the activation of several genes important for adjusting the essential cellular processes to the new temperature. The best-studied cold shock genes are those encoding small proteins (7 kDa), which are very conserved among bacteria (33), playing a major role during cold shock adaptation but also important under normal growth conditions. The prototype of this family, the CspA protein from E. coli, is composed of five antiparallel β-strands and presents a very efficient folding (31, 35, 37). The finding of eukaryotic proteins sharing sequence similarity with the bacterial cold shock proteins showed that the small cold shock proteins are constituted of a single domain, the CSD, which is conserved from bacteria to humans (20). CSDs have also been determined to be integral components of larger proteins in eukaryotes (22).
We show here that C. crescentus has four predicted peptides that present the CSD; three of them belong to the E. coli CspA family of 7-kDa proteins (CspA, CspB, and CspC), and one of them belongs to a novel class of bacterial proteins that possesses two CSDs (CspD). Despite the modular nature of the CSD, which has been clear for some time (20), proteins with this two-domain structure have not yet been described. One possible reason for this could be that we have identified this particular arrangement in predicted proteins only from proteobacteria of the α subdivision (Fig. 1B), whose genome sequences only recently became available.
The role of Caulobacter CspD and these novel α-Proteobacteria proteins possessing a double CSD structure in the cell is still unknown. A protein with five CSDs in humans, the UNR protein, has been described (22) that was found to bind single-stranded DNA and RNA with high affinity and double-stranded DNA with lower affinity (13); only three of the domains are sufficient to confer the same affinity for RNA as does the full-length protein (45). The interaction of Unr with a second protein, the gene regulator ALL-1, requires two CSDs, suggesting that this double-domain arrangement could be necessary for protein-protein interaction (26). Another interesting observation is that some bacterial cold shock proteins are able to form dimers in vitro (28, 56), although the physiological relevance of this is still unclear. It is tempting to speculate that in some cold shock proteins from α-Proteobacteria, dimerization of the CSD was ensured by encoding the two domains within the same polypeptide.
We showed that the fusions containing the promoter regions of cspA or cspB are highly induced by a temperature downshift from 10 to 20°C, whereas those containing the cspC and cspD promoters are not. It has been demonstrated that the expression of E. coli CspA is regulated at the transcriptional level during cold shock and that the expression of β-galactosidase under control of its promoter was increased three- to fivefold upon a decrease in temperature (16, 43, 46). The cold inducibility of E. coli CspA, as well as B. subtilis CspB, is also the result of increased mRNA stability at a low temperature (6, 12, 15, 25). The presence of a sequence called upstream box in the long 5′-untranslated region and a sequence downstream of the initiation codon (called the downstream box) seem to increase translation efficiency in E. coli (11, 30, 41, 54). Although C. crescentus cspA and cspB genes have long 5′-untranslated regions, no sequences similar to the consensus for E. coli boxes could be found. The use of transcriptional fusions prevents the regulation at the level of translation, since the reporter gene has its own translation signals; therefore, the increase in expression observed for Caulobacter cspA and cspB promoter fusions is a result of transcription and/or mRNA stability.
The promoter fusions of three of the genes (cspB, cspD, and cspC) showed similar degrees of induction of β-galactosidase when cells entered stationary phase, but the promoter fusions of cspC and cspD genes did not show any increase in expression at a low temperature. Other CspA homologues in E. coli were described that are not induced by cold shock (51, 55) and, among them, cspD is induced during stationary phase. The Caulobacter cspD gene is regulated at the transcriptional level, since the presence of an upstream regulatory region is essential for maximal levels of expression. A sequence similar to this activator sequence is also present in the promoter region of the cspC gene, but it is not found in the regulatory region of another stationary-phase-induced gene, katG (42). Since this element is not responsible for the growth phase regulation, the stationary-phase induction observed could be a result of both transcriptional regulation and increased mRNA stability. In B. subtilis, two of three small cold shock-induced proteins, CspB and CspC, are also induced in the stationary phase and were shown to be essential for adaptation to this phase (20, 25). Since the Caulobacter cspC and cspD genes are induced in the stationary phase, their role is probably related more specifically to adapting the cell to survive long periods of growth arrest. The environmental signals that trigger the expression of these two genes are still not determined, but they might respond to the nutritional status of the cell, as described for CspA and CspD from E. coli (52).
Gene knockout of Caulobacter cspA and cspD showed that these genes are not essential at 30°C, but the cspA strain shows a lower survival rate during prolonged growth at 10°C. The phenotype observed is consistent but not severe, indicating that the lack of a single csp gene is not very deleterious to the cell. Cells carrying deletions of individual E. coli csp genes or even a triple deletion (ΔcspA ΔcspB ΔcspG) were also shown to be viable, but a combination of four deletions (ΔcspA ΔcspB ΔcspG ΔcspE) presented a cell division defect at a low temperature (50). It was shown that when E. coli cells carry a double or triple csp deletion, there is a compensatory induction of the remaining csp homologues (50). In B. subtilis, double cspB/cspC or cspB/cspD deletions show a reduction in growth rate at both 15 and 37°C and lower viability at stationary phase (19). Although the Caulobacter cspD gene is induced at stationary phase, it is not essential for viability at this phase. The knockout of the other two genes, as well as obtaining double mutations, will enable us to determine the respective role of each gene in response to cold shock and stationary-phase survival.
Acknowledgments
We thank Luis C. Ferreira and Beny Spira for critical reading of the manuscript, and M. R. K. Alley for pNPTS138.
This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP grant 2002/05762-1). During the course of this work, E.A.S.L. was supported by fellowship from FAPESP. M.V.M. is partly supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
REFERENCES
- 1.Abyzov, S. S., I. N. Mitskevich, M. N. Poglazova, N. I. Barkov, V. Y. Lipenkov, N. E. Bobin, B. B. Koudryashov, V. M. Pashkevich, and M. V. Ivanov. 2001. Microflora in the basal strata at Antarctic ice core above the Vostok lake. Adv. Space Res. 28:701-706. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 27:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bae, W., B. Xia, M. Inouye, and K. Severinov. 2000. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. USA 97:7784-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The PFAM protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouvet, P., and A. P. Wolffe. 1994. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell 77:931-941. [DOI] [PubMed] [Google Scholar]
- 6.Brandi, A., P. Pietroni, C. O. Gualerzi, and C. L. Pon. 1996. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol. Microbiol. 9:231-240. [DOI] [PubMed] [Google Scholar]
- 7.Dersch, P., S. Kneip, and E. Bremer. 1994. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K-12 to a cold environment. Mol. Gen. Genet. 245:255-259. [DOI] [PubMed] [Google Scholar]
- 8.Derzelle, S., B. Hallet, K. P. Francis, T. Ferain, J. Delcour, and P. Hols. 2000. Changes in cspL, cspP, and cspC mRNA abundance as a function of cold shock and growth phase in Lactobacillus plantarum. J. Bacteriol. 182:5105-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ely, B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372-384. [DOI] [PubMed] [Google Scholar]
- 10.Eriksson, M., G. Dalhammar, and W. W. Mohn. 2002. Bacterial growth and biofilm production on pyrene. FEMS Microbiol. Ecol. 40:21-27. [DOI] [PubMed] [Google Scholar]
- 11.Etchegaray, J. P., and M. Inouye. 1999. A sequence downstream of the initiation codon is essential for cold shock induction of cspB of Escherichia coli. J. Bacteriol. 181:5852-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, L., W. Jiang, W. Bae, and M. Inouye. 1997. Promoter-independent cold-shock induction of cspA and its derepression at 37°C by mRNA stabilization. Mol. Microbiol. 23:355-364. [DOI] [PubMed] [Google Scholar]
- 13.Ferrer, N., A. Garcia-Espana, M. Jeffers, and A. Pellicer. 1999. The unr gene: evolutionary considerations and nucleic acid-binding properties of its long isoform product. DNA Cell Biol. 18:209-218. [DOI] [PubMed] [Google Scholar]
- 14.Gober, J. W., and L. Shapiro. 1992. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol. Biol. Cell 3:913-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldenberg, D., I. Azar, and A. B. Oppenheim. 1996. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol. Microbiol. 19:241-248. [DOI] [PubMed] [Google Scholar]
- 16.Goldenberg, D., L. Azar, A. R. Oppenheim, A. Brandi, C. L. Pon, and C. O. Gualerzi. 1997. Role of Escherichia coli cspA promoter sequences and adaptation of translational apparatus in the cold shock response. Mol. Gen. Genet. 256:282-290. [DOI] [PubMed] [Google Scholar]
- 17.Goldstein, J., N. S. Pollitt, and M. Inouye. 1990. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:283-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graumann P., K. Schroder, R. Schmid, and M. A. Marahiel. 1996. Cold shock stress-induced proteins in Bacillus subtilis. J. Bacteriol. 178:4611-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graumann P., T. M. Wendrich, M. H. Weber, K. Schroder, and M. A. Marahiel. 1997. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol. Microbiol. 25:741-756. [DOI] [PubMed] [Google Scholar]
- 20.Graumann, P. L., and M. A. Marahiel. 1998. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 23:286-290. [DOI] [PubMed] [Google Scholar]
- 21.Graumann, P. L., and M. A. Marahiel. 1999. Cold shock proteins CspB and CspC are major stationary-phase-induced proteins in Bacillus subtilis. Arch. Microbiol. 171:135-138. [DOI] [PubMed] [Google Scholar]
- 22.Jacquemin-Sablon, H., G. Triqueneaux, S. Deschamps, M. le Maire, J. Doniger, and F. Dautry. 1994. Nucleic acid binding and intracellular localization of unr, a protein with five cold shock domains. Nucleic Acids Res. 22:2643-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, W., Y. Hou, and M. Inouye. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196-202. [DOI] [PubMed] [Google Scholar]
- 24.Jones, P. G., and M. Inouye. 1996. RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol. Microbiol. 21:1207-1218. [DOI] [PubMed] [Google Scholar]
- 25.Kaan, T., B. Jurgen, and T. Schweder. 1999. Regulation of the expression of the cold shock proteins CspB and CspC in Bacillus subtilis. Mol. Gen. Genet. 262:351-354. [DOI] [PubMed] [Google Scholar]
- 26.Leshkowitz, D., O. Rozenblatt, T. Nakamura, T. Yano, F. Dautry, C. M. Croce, and E. Canaani. 1996. ALL-1 interacts with unr, a protein containing multiple cold shock domains. Oncogene 13:2027-2031. [PubMed] [Google Scholar]
- 27.Malakooti, J., and B. Ely. 1995. Principal sigma subunit of the Caulobacter crescentus RNA polymerase. J. Bacteriol. 177:6854-6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayr, B., T. Kaplan, S. Lechner, and S. Scherer. 1996. Identification and purification of a family of dimeric major cold shock protein homologs from the psychrotrophic Bacillus cereus WSBC 10201. J. Bacteriol. 178:2916-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 30.Mitta, M., L. Fang, and M. Inouye. 1997. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription, and the downstream box in the coding region for its cold-shock induction. Mol. Microbiol. 26:321-335. [DOI] [PubMed] [Google Scholar]
- 31.Newkirk, K., W. Feng, W. Jiang, R. Tejero, S. D. Emerson, M. Inouye, and G. T. Montelione. 1994. Solution NMR structure of the major cold shock protein (CspA) from Escherichia coli: identification of a binding epitope for DNA. Proc. Natl. Acad. Sci. USA 91:5114-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, C. M. Fraser, and J. Eisen. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phadtare, S., A. Janivette, and M. Inouye. 1999. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 2:175-180. [DOI] [PubMed] [Google Scholar]
- 34.Poindexter, J. S. 1981. The caulobacters: ubiquitous unusual bacteria. Microbiol. Rev. 45:123-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reid, K. L., H. M. Rodriguez, B. J. Hillier, and L. M. Gregoret. 1998. Stability and folding properties of a model beta-sheet protein, Escherichia coli CspA. Protein Sci. 7:470-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato, N. 1995. A family of cold-regulated RNA-binding protein genes in the cyanobacterium Anabaena variabilis M3. Nucleic Acids Res. 23:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schindelin, H., W. Jiang, M. Inouye, and U. Heinemann. 1994. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 91:5119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroder, K., P. Graumann, A. Schnuchel, T. A. Holak, and M. A. Marahiel. 1995. Mutational analysis of the putative nucleic acid-binding surface of the cold-shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single-stranded DNA containing the Y-box motif. Mol. Microbiol. 16:699-708. [DOI] [PubMed] [Google Scholar]
- 39.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 40.Sinensky, M. 1974. Homeoviscous adaptation: a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci. USA 71:522-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sprengart, M. L., E. Fuchs, and A. G. Porter. 1996. The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J. 15:665-674. [PMC free article] [PubMed] [Google Scholar]
- 42.Steinman, H. M., F. Fareed, and L. Weinstein. 1997. Catalase-peroxidase of Caulobacter crescentus: function and role in stationary-phase survival. J. Bacteriol. 179:6831-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanabe, H., J. Goldstein, M. Yang, and M. Inouye. 1992. Identification of the promoter region of the Escherichia coli major cold shock gene, cspA. J. Bacteriol. 174:3867-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Triqueneaux, G., M. Velten, P. Franzon, F. Dautry, and H. Jacquemin-Sablon. 1999. RNA binding specificity of Unr, a protein with five cold shock domains. Nucleic Acids Res. 27:1926-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vasina, J. A., and E. Baneyx. 1996. Recombinant protein expression at low temperatures under the transcriptional control of the major Escherichia coli cold shock promoter cspA. Appl. Environ. Microbiol. 62:1444-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, N., K. Yamanaka, and M. Inouye. 1999. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J. Bacteriol. 181:1603-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willimsky, G.,H. Bang, G. Fischer, and M. A. Marahiel. 1992. Characterization of cspB, a Bacillus subtilis inducible cold shock gene affecting cell viability at low temperatures. J. Bacteriol. 174:6326-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolffe, A. P. 1994. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays 1 6:245-251. [DOI] [PubMed] [Google Scholar]
- 50.Xia, B., H. Ke, and M. Inouye. 2001. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 40:179-188. [DOI] [PubMed] [Google Scholar]
- 51.Yamanaka, K., and M. Inouye. 1997. Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J. Bacteriol. 179:5126-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamanaka, K., and M. Inouye. 2001. Induction of CspA, an Escherichia coli major cold-shock protein, upon nutritional upshift at 37°C. Genes Cells 6:279-290. [DOI] [PubMed] [Google Scholar]
- 53.Yamanaka, K., L. Fang, and M. Inouye. 1998. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol. Microbiol. 27:247-255. [DOI] [PubMed] [Google Scholar]
- 54.Yamanaka, K., M. Mitta, and M. Inouye. 1999. Mutation analysis of the 5′ untranslated region of the cold shock cspA mRNA of Escherichia coli. J. Bacteriol. 181:6284-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamanaka, K., T. Mitani, T. Ogura, H. Niki, and S. Hiraga. 1994. Cloning, sequencing, and characterization of multicopy suppressors of a mukB mutation in Escherichia coli. Mol. Microbiol. 13:301-312. [DOI] [PubMed] [Google Scholar]
- 56.Yamanaka, K., W. Zheng, E. Crooke, Y. H. Wang, and M. Inouye. 2001. CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherichia coli. Mol. Microbiol. 39:1572-1584. [DOI] [PubMed] [Google Scholar]