Abstract
Bacillus subtilis mutants classified within the ɛ (ruvA, ΔruvB, ΔrecU, and recD) and η (ΔrecG) epistatic groups, in an otherwise rec+ background, render cells impaired in chromosomal segregation. A less-pronounced segregation defect in ΔrecA and Δsms (ΔradA) cells was observed. The repair deficiency of addAB, ΔrecO, ΔrecR, recH, ΔrecS, and ΔsubA cells did not correlate with a chromosomal segregation defect. The sensitivity of ɛ epistatic group mutants to DNA-damaging agents correlates with ongoing DNA replication at the time of exposure to the agents. The Δsms (ΔradA) and ΔsubA mutations partially suppress the DNA repair defect in ruvA and recD cells and the segregation defect in ruvA and ΔrecG cells. The Δsms (ΔradA) and ΔsubA mutations partially suppress the DNA repair defect of ΔrecU cells but do not suppress the segregation defect in these cells. The ΔrecA mutation suppresses the segregation defect but does not suppress the DNA repair defect in ΔrecU cells. These results result suggest that (i) the RuvAB and RecG branch migrating DNA helicases, the RecU Holliday junction (HJ) resolvase, and RecD bias HJ resolution towards noncrossovers and that (ii) Sms (RadA) and SubA proteins might play a role in the stabilization and or processing of HJ intermediates.
Cells have evolved several mechanisms to maintain the structural and informational fidelity of their DNA and to participate in sister chromatid segregation. UV and certain chemical compounds (e.g., 4-nitroquinoline-1-oxide [4NQO] and methyl methanesulfonate [MMS]), generate deleterious obstacles to DNA replication. Stalling of the replication fork due to such obstacles or the collapse of the replication machinery with resulting unrepaired single-strand nicks or double-strand breaks (DSBs) blocks replication fork progression in all organisms (13, 21, 54). The block must be repaired or removed, and replication must be restarted. Current models for DSB repair involve the formation of Holliday junctions (HJs) that need to be resolved to allow the repaired chromosomes to separate. The Escherichia coli RuvAB (RuvABEco) helicase, together with the RuvCEco HJ-specific endonuclease, target the HJ at the stalled fork and cleave on opposite strands. If the symmetric HJs are resolved at random, crossovers and noncrossover products are generated. In circular chromosomes, the outcome will be a dimeric chromosome or two monomeric chromosomes, respectively. Dimeric chromosomes are lethal and need to be resolved before cell division. This is accomplished by bacterial Xer-like site-specific recombination systems that catalyze the resolution of the dimers (55). It has been shown in vitro that the orientation of the RuvABCEco complex determines the direction of cleavage (60), and it is proposed that the repair of broken replication forks is biased to the generation of noncrossover products (14, 41). However, in E. coli, chromosome dimers are formed by homologous recombination (HR) between sister chromosomes in about 14% of cells growing under standard laboratory conditions (46, 58).
In Bacillus subtilis, the recombination genes other than recA have been classified into six different epistatic groups (α, β, ɛ, γ, ζ, and η). Mutations in genes classified within the α (recF, recL, recO, and recR [known collectively as recFLOR] and recN), ɛ (recU, recD, and ruvA [formerly termed recB] and ruvB), and η (recG) epistatic groups markedly affect the viability of cells exposed to DNA-damaging agents, whereas mutations in genes classified within the β (addA and addB [collectively known as addAB]), γ (recH and recP), and ζ (recS) epistatic groups slightly reduce the viability of cells exposed to DNA-damaging agents (reference 16 and this study). The recA, recF, recO, recR, recN, ruvA, ruvB, and recG genes have their counterparts in E. coli in genes with identical names, whereas the addAB, recU, and sms genes have their counterpart in the recBCDEco, ruvCEco, and radAEco genes, respectively (3, 16). The B. subtilis recL, recD, recH, recP, recS, and subA genes have no obvious counterpart in genes in E. coli. The products classified within the α, β, ɛ, and η groups have their functional counterparts in the RecN-FOREco, RecBCDEco, RuvABCEco, and RecGEco products, respectively (3, 8, 10, 16, 25). The role of the functions classified within the γ and ζ epistatic groups in DNA repair and HR remains unknown (16). Unless otherwise stated, the indicated genes and products are of B. subtilis origin.
In E. coli cells, 18 to 50% of cells require replication fork reloading during a single round of chromosomal replication in the absence of any exogenous DNA-damaging agent (13, 34). Using an indirect measurement (measurement of repair centers as a measurement of blocked replication forks), we assumed that replication fork reloading might occur with a similar frequency in B. subtilis cells (25). The rate of formation of RecN-RecOF repair centers in the absence of any exogenous DNA-damaging agent was found to be about 35 and 5% in exponentially growing ΔrecA and ΔrecU cells, respectively (25).
A defect in the HJ resolvase RecU (3) (also termed penicillin-binding protein [PBP]-related factor A [designated PrfA]) or in the DNA organizer SMC complex (formed by the Smc, ScpA, and ScpB proteins) in an otherwise wild-type (wt) background, leads to the accumulation of anucleate cells (∼3 and 10%, respectively) (7, 20, 35, 42, 45, 56). The ΔrecU Δsmc double mutant does not seem to be viable. Genetic analysis of a synthetic conditional recU mutant combined with the Δsmc mutant at a permissive temperature indicated the accumulation of ∼24% anucleate cells (45). These data suggest a role for the SMC complex and RecU in chromosomal segregation. Finally, it has been shown that the recU segregation phenotype is greatly exacerbated by the additional loss of PBP1 but not by the loss of other PBPs (e.g., PBP2c or PBP4), suggesting a possible role for recU in septum formation or as a chaperone in DNA-cell wall interaction (24, 45). Furthermore, genetic evidence suggests that the Δsms (also termed ΔradA) and ΔsubA mutations partially suppress the DNA repair and recombination defect of ɛ epistatic group mutants (8).
In this paper, we analyze the effect on segregation of the different repair-deficient B. subtilis epistatic groups, as well as the putative suppression of the segregation phenotypes by the Δsms (ΔradA) and ΔsubA mutations. Our results indicate that the functions of genes classified within the ɛ and η epistatic groups, which are involved in the processing of an HJ, are required for proper chromosomal segregation in wt cells under normal growth conditions. It is likely that the replication and subsequent segregation of chromosomes bearing unrepaired DNA lesions can seriously compromise genome stability. This is consistent with the hypothesis that B. subtilis RuvAB-RecU-RecD and RecG proteins in an otherwise wt background under normal growth conditions (this work) and E. coli RuvABC proteins in UV-irradiated, rep or recBC sbcBC backgrounds (22, 36, 41) prevent dimer formation in vivo. Finally, the suppression of the segregation defect of HJ processing functions by Δsms (ΔradA) and Δsub mutations point to the role for both proteins in the stabilization or processing of branched DNA molecules.
MATERIALS AND METHODS
Bacterial strains.
All B. subtilis strains used in this study are listed in Table 1 and are isogenic to strain YB886 (rec+ control). A 2-kb six-cat-six cassette containing two directly repeated copies of the β site-specific recombinase target site (six) surrounding the chloramphenicol acetyltransferase gene (cat) was introduced within the coding sequences of recG and ruvAB. The disruptions were then transferred into the chromosomes of wt cells to generate ΔrecG and ΔruvAB strains. Their isogenic rec-deficient derivatives, as well as the ΔrecA ΔrecU and ΔrecU ΔrecO double mutants, were generated by a double-crossover event as previously described (1). Expression of the β gene mediated deletion of the cat gene. The attSKIN and attPBSX (62) regions were moved into wt and ΔrecU backgrounds by chromosomal transformation as previously described (2).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotypeb | Reference or source |
|---|---|---|
| YB886 | trpC2 metB5 amyE sigB37 xin-1 attSPβ | 63 |
| YB1005 | + uvrA42 | 19 |
| YB1290 | + ruvA2 (formerly recB2) | 19 |
| BG119 | + recH342 | 2 |
| BG121 | + recD41 | 2 |
| BG123 | + recU40 | 2 |
| BG127 | + recR13 | 2 |
| BG189 | + addA5 addB72 | 1 |
| BG190 | + ΔrecA | 9 |
| BG427 | + ΔrecU | 18 |
| BG651 | + ΔrecU ΔrecA | This work |
| BG425 | + ΔrecS | 18 |
| BG439 | + ΔrecO | 17 |
| BG501 | + ΔrecU Δsms (ΔradA) | 8 |
| BG545 | + ΔrecU ΔsubA | 8 |
| BG621 | + ΔrecU ΔrecO | This work |
| BG503 | + ruvA2 Δsms (ΔradA) | 8 |
| BG543 | + ruvA2 ΔsubA | 8 |
| BG547 | + recD41 Δsms (ΔradA) | 8 |
| BG547 | + recD41 ΔsubA | 8 |
| BG699 | + ΔruvAB | This work |
| BG691 | + ΔrecG | This work |
| BG745 | + ΔrecG Δsms (ΔradA) | This work |
| BG747 | + ΔrecG ΔsubA | This work |
| BG500a | + attSKIN attPBSX | This work |
| BG575a | + attSKIN attPBSX ΔrecU | This work |
The original background has a mutation in the PBSX-encoded xin gene (xin-1), whereas in the BG500 and BG575 strains the entire PBSX prophage was deleted.
+, relevant genotype is that of strain YB886 plus the indicated gene(s).
Survival studies.
Cells were grown overnight at 37°C to obtain stationary-phase cultures as previously described (2). Exponentially growing cells were obtained by inoculating overnight cultures in fresh Luria-Bertani (LB) medium and growing them to an optical density at 560 nm of 0.4 at 37°C. When indicated, chloramphenicol (CM) (20 μg/ml) was added to the exponentially growing cells and further incubated for 2 h to stop protein synthesis (11), thereby preventing new rounds of DNA replication. 4NQO was from Sigma, and MMS was from Merck. The chemical treatment (100 μM 4NQO or 10 mM MMS) of exponential- and stationary-phase recU, ruvA, ruvB, recD, recR, and uvrA mutant and wt cells was performed as previously described (11), except that LB medium was used for growing cells, and plating was done on LB agar.
Fluorescence and electron microscopy of B. subtilis cells.
Exponentially growing cells were obtained by inoculation of overnight cultures in fresh LB medium and growing them to an optical density at 560 nm of 0.4 at 37°C. The mid-log-phase cells were then fixed with 2% formaldehyde, 4′,6′-diamino-2-phenylindole (DAPI) (1 μg/ml) was added for nucleoid visualization, and cells were analyzed by fluorescence microscopy as previously described (7). For electron microscopy sectioning, cells were fixed with glutaraldehyde, treated with osmium tetroxide, and embedded in Spurr's low-viscosity medium (57).
RESULTS
Nucleoid phenotype of B. subtilis recombination mutants during exponential-phase growth.
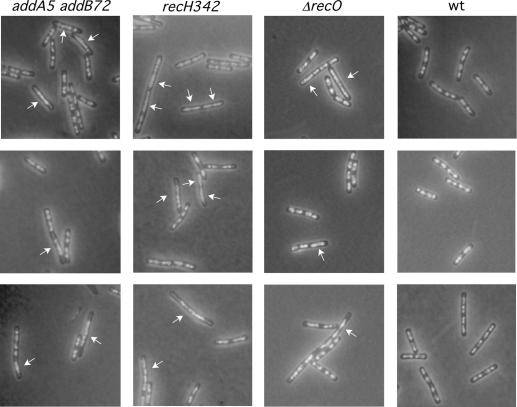
To assess the effect on nucleoid morphologies of any recombination-deficient strain in an otherwise wt background, mutant strains representative of each of the epistatic groups (α [ΔrecO and ΔrecR], β [addA5 and addB72], γ [recH342], ɛ [ΔrecU, ΔruvAB, and recD41], ζ [ΔrecS], and η [ΔrecG]) as well as the ΔrecA strain (Table 2) were collected during exponential phase, and as a measure of a segregation defect, the frequency of anucleate cells was quantified after the cells were stained with DAPI. Anucleate cells in the addA5 addB72, recH342, and ΔrecS strains were rare (Table 2). However, diffuse and “linked” nucleoids that occupied almost the whole cell were visible in 6% of addA5 addB72 cells, 10% of ΔrecO cells, and 26% of recH342 cells (Fig. 1). Very little is known about the biochemical role of RecH on DNA repair and recombination.
TABLE 2.
Anucleate cell production of recombination mutants
| Genotype of mutant strain(epistatic group) | % of anucleate cells (no. of cells counted)a |
|---|---|
| rec+ | <0.1 (1,400) |
| ΔrecA | 1 (2,240) |
| ΔrecO (α) | <0.2 (652) |
| ΔrecR (α) | <0.1 (328) |
| addA5 addB72 (β) | <0.1 (3,079) |
| recH342 (γ) | <0.1 (1,614) |
| ΔrecS (ζ) | <0.3 (338) |
| ΔrecG (η) | 7.4 (1,630) |
| ΔrecU (ɛ) | 4.4 (510) |
| recD41 (ɛ) | 4.3 (387) |
| ruvA2 (ɛ) | 4.9 (485) |
| ΔruvAB (ɛ) | 5.2 (218) |
| ΔrecU ΔrecA | <0.2 (511) |
| ΔrecU ΔrecO | 4.1 (200) |
| rec+attSKIN attPBSX | <0.1 (400) |
| ΔrecU attSKIN attPBSX | 4.5 (410) |
Cells were grown and the percentage of anucleate cells were determined as described in Materials and Methods.
FIG. 1.
Nucleoid morphologies of addA5 addB72, recH342, and ΔrecO cells. Exponentially growing cells were fixed, stained with DAPI, and analyzed by fluorescence microscopy to visualize the nucleoids. White arrows point to diffuse and linked nucleoids.
The activity of ΔruvA, ΔrecU, and ΔrecG cells and the uncharacterized activity of recD41 cells, all impaired in the processing of HJs, showed a clear defect in chromosomal segregation (Table 2). The most severe segregation defect was found in ΔrecG cells: >10% of ΔrecG cells had abnormally condensed nucleoids, and ∼7% of the cells were anucleated (see below) under normal growth conditions.
In all experiments, the lysogenic prophage SKIN encoding a RusA-like HJ resolvase protein (52) was present in the genetic background used. To learn whether the RusA-like protein could play any role in chromosomal segregation, SKIN-free wt and ΔrecU strains were constructed. Similar segregation patterns were observed with SKIN-free and SKIN-containing cells (Table 2). The percentage of anucleate cells in the ΔrecU mutant that lacks bacterially encoded HJ resolvase was unaffected by the absence of the SKIN prophage (Table 2). Therefore, it is likely that the SKIN-encoded RusA-like protein either is not expressed or has no effect on chromosomal segregation under normal growth conditions.
Unlike a recAEco mutant that shows ∼10% anucleate cells (64), a ΔrecA mutant shows a moderate segregation defect (∼1% of cells) (Table 2) (28). The presence of the ΔrecA null allele in the ΔrecU background suppressed the segregation phenotype (Table 2). This is consistent with the observation that in both E. coli and B. subtilis cells, chromosome dimer formation is not observed and the Xer-like site specific recombinase is not needed in the absence of the RecA protein (6, 27, 28).
The RecOEco, RecO, and RecU proteins can catalyze D-loop formation (3, 32). A ΔrecO ΔrecU double mutant strain was constructed to assess whether the absence of DNA strand invasion could suppress the chromosomal segregation phenotype. The ΔrecO ΔrecU double mutant strain showed a segregation defect similar to that of the ΔrecU single mutant (∼4% of anucleate cells) (Table 2). These results suggest a strand-invading accessory role for both RecO and RecU proteins and confirm that RecA is primarily responsible for the formation of HJ in vivo.
The DNA damage sensitivity of recU, ruvAB, and recD cells correlates with DNA replication.
Previously, it has been shown that UV-generated DNA damages are removed by the nucleotide excision repair (NER) machinery in E. coli wt cells (12). The NER proteins are involved in the repair of UV-generated DNA damage independently of the replication state of the cell. UV-irradiated cells resume DNA synthesis after a transient inhibition by a process called replication restart that has been shown to involve recFEco, recOEco, and recREco gene products (11). These results suggest a close interplay between recombination repair and DNA replication and suggest that the failure of recFOREco and perhaps recFLOR cells arises from a defect in rescuing a stalled replication fork (12, 13, 26). To learn whether the high sensitivity of recU, ruvA, and recD cells to DNA-damaging agents also correlates with ongoing DNA replication, different assays were undertaken. First, wt, recR13, uvrA42 (uvrA42 is the counterpart to uvrBEco mutants), and recU40 cells were grown in LB medium until mid-exponential or stationary phase and exposed to 100 μM 4NQO for various times, and then the numbers of viable cells were measured. Independently of the growth phase, wt cells were resistant to the killing action of 100 μM 4NQO, whereas uvrA42 cells, deficient in NER, were sensitive (1). As previously reported, exponentially growing recU40 and recR13 cells were sensitive to DNA-damaging agents (1), but stationary-phase recU40 and recR13 cells were ∼100-fold more resistant to 4NQO (Fig. 2) than were the exponentially growing cells (1, 18). Stationary-phase ruvA2 and recD41 cells were also ∼100-fold more resistant to 100 μM 4NQO than were exponentially growing cells (8) (data not shown). Furthermore, stationary-phase recU40, ruvA2, recD41, and recR13 cells were also 80- to 100-fold more resistant to other DNA-damaging agents, such as 10 mM MMS, than were exponentially growing cells (data not shown).
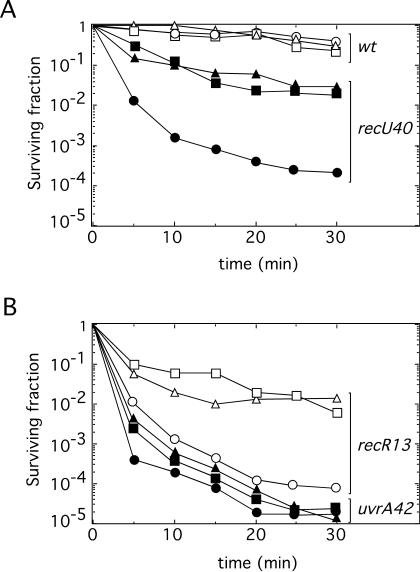
FIG. 2.
DNA damage sensitivity of recU replicating cells. The survival of wt cells (open symbols) and recU40 cells (filled symbols) (A), and of recR13 cells (open symbols) and uvrA42 control cells (filled symbols) (B) after exposure to the killing action of 100 μM 4NQO under different growth conditions is shown. Triangles, stationary-phase cells; circles, exponential-phase growing cells; squares, cells pretreated with CM (20 μg/ml) 120 min before 4NQO treatment. Survival curves represent the averages of results of three or more independent experiments.
To further confirm that the defect observed with the recU40, ruvA2, and recD41 mutants was due to a defect in replication restart recovery, wt, recU40, recR13, and uvrA42 cells were grown in LB medium until mid-exponential phase. DNA replication was reversibly halted by blocking protein synthesis with CM (20 μg/ml), the cells were then exposed to the killing action of 100 μM 4NQO for various times, and the numbers of viable cells were measured. The recU40 and recR13 cells, pretreated with CM for 120 min before exposure to 4NQO, were ∼100-fold more resistant to the DNA-damaging agent than were cells untreated with CM (Fig. 2). Similar results were observed when the ruvA2, ΔruvAB, recD41, or ΔrecO cells were pretreated with CM for 120 min before exposure to 4NQO (data not shown). Pretreated uvrA42 cells were as sensitive as untreated cells (Fig. 2B), whereas wt cells were resistant to 100 μM 4NQO exposure. Therefore, it is likely that (i) the failure of recU40, ruvA2, and recD41 cells arises from a defect in rescuing a stalled replication fork until the lesion can be removed by NER, (ii) the defect of both recREco (11) and recR13 cells correlates with ongoing DNA replication at the time of exposure to the agent, and (iii) DNA-damaged uvrA42 cells do not recover normally independently of the growth phase.
Previously, a direct correlation between increased damage sensitivity of recFOREco cells and DNA replication has been established (11). Therefore, it is likely that the rescue of arrested replication forks in exponentially growing cells occurs via HR in both E. coli cells (11, 12) and B. subtilis cells (Fig. 2).
Nucleoid and cell morphology phenotypes of recU, recD, and ruvA cells during exponential-phase growth.
To investigate whether the chromosome segregation defect may be due to a defect in replication fork progression, the wt strain and its isogenic derivatives (ΔrecU, recU40, ruvA2, and recD41 cells) were grown to mid-exponential phase in rich medium and either stained with DAPI, fixed, and visualized by fluorescence microscopy or fixed, processed, and visualized by electron microscopy.
Previously, it was shown that 3 to 5% of ΔrecU cells have a chromosomal segregation phenotype (45). A similar chromosomal segregation defect was observed with the ruvA, ΔruvAB, and recD mutants (Fig. 3; Table 2). This observation is consistent with the classification of ruvA, ruvB, and recD in the same epistatic group as recU (3). An absence of DAPI-stained material was observed for 3 to 5% of the ruvA2 and recD41 cells, whereas <0.05% of wt cells were anucleate under identical growth conditions (Fig. 3A). In addition to a higher abundance of cells showing no nucleoids, a high proportion of ΔrecU, recU40, ruvA2, and recD41 cells had defects in nucleoid structure. The one or two normally compact, condensed, and regular nucleoid bodies seen in fixed wt cells often appeared as highly condensed nucleoids asymmetrically located in recU40, ruvA2, and recD41 cells (Fig. 3A). ΔrecU, ruvA2, and recD41 cells had many more nucleoids of much higher DNA content and with large cytoplasmic spaces free of nucleoid bodies than did wt cells. Similar results were obtained when the ΔruvAB strain was analyzed.
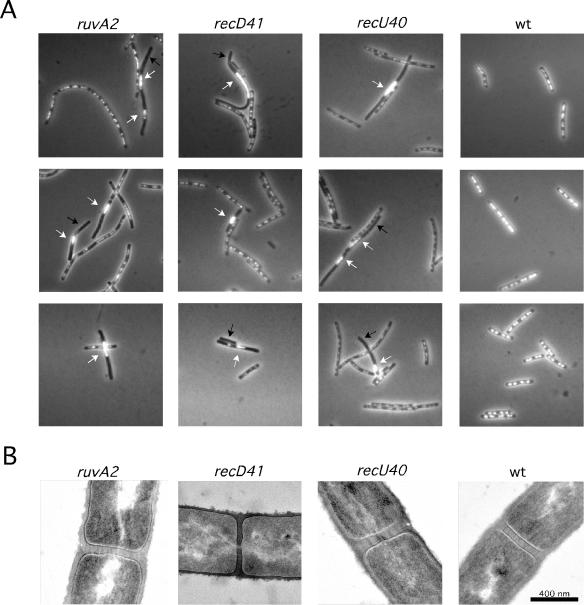
FIG. 3.
recU, recD, and ruvA mutations produce anucleate cells and aberrant nucleoids. (A) Exponentially growing cells were fixed, stained with DAPI, and analyzed by fluorescence microscopy to visualize the nucleoids. Black arrows indicate anucleate cells, whereas white arrows show aberrant and misplaced nucleoids. (B) Electron micrographs of cross-sectioned processed cells. The nucleoids appear as light material in the cytoplasm.
The chromosomal segregation defect of recU40, recD41, and ruvA2 cells was more apparent when cells were visualized by electron microscopy, and nucleoids that appear bisected by the septum were observed (Fig. 3B).
The recU gene maps upstream and forms an operon with ponA, which encodes PBP1. As shown in Fig. 3, the absence of the genetically unlinked recU, ruvAB, and recD genes has the same profound effect on both chromosomal structure and segregation. It is likely, therefore, that the segregation defects observed with recU, recD, and ruvAB cells are unlinked to PBP1 and therefore not due to the PBP1 defect in septation and its localization at sites of cell division (45).
The ruvA2 and recD41 segregation defect is partially suppressed in Δsms (ΔradA) cells.
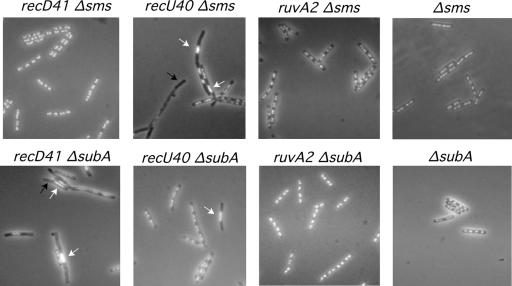
Previously it was shown that Sms (RadA), the counterpart of RadAEco, and SubA proteins play an active role in recombinational repair, most likely through the stabilization and/or processing of branched DNA molecules or blocked replication forks (5, 8). Mutations in both proteins partially suppress the recombination defect of mutations in proteins expressed by genes of the ɛ epistatic group (8). To learn whether the chromosomal segregation defect of ΔrecU, recD41, and ruvA2 cells may be also suppressed by mutations in the sms (radA) and subA genes, we constructed double mutant strains and investigated their segregation phenotypes. The Δsms (ΔradA), ΔsubA, ΔrecU Δsms (ΔradA), recD41 Δsms (ΔradA), ruvA2 Δsms (ΔradA), ΔrecU ΔsubA, recD41 ΔsubA, and ruvA2 ΔsubA cells were grown to mid-exponential phase in rich medium and stained with DAPI and either fixed and visualized by fluorescence microscopy (Fig. 4) or fixed, processed, and visualized by electron microscopy (data not shown). The Δsms (ΔradA) strain contains a low number of anucleate cells (∼0.5% of total cells). The chromosomal segregation defect observed with ruvA2 and recD41 cells was partially suppressed if the Δsms (ΔradA) mutation was present in the background (Fig. 4). In contrast, the ΔrecU segregation defect was not suppressed by the presence of the Δsms (ΔradA) mutation.
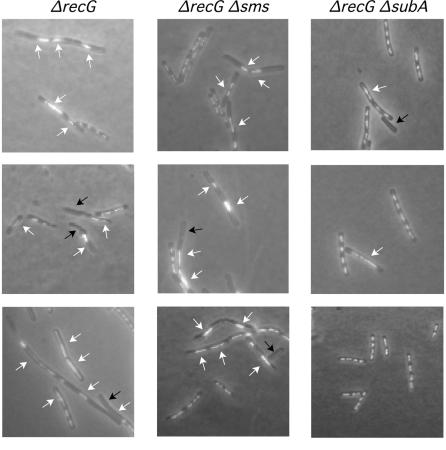
FIG. 4.
Effect of the Δsms (ΔradA) and ΔsubA suppressors in the segregation defect of the ɛ epistatic group mutants. Exponentially growing cells were fixed, stained with DAPI, and analyzed by fluorescence microscopy to visualize the nucleoids. Black arrows indicate anucleate cells, whereas white arrows show aberrant and misplaced nucleoids.
Anucleate cells in the ΔsubA strain were rare. The presence of the ΔsubA mutation suppressed the segregation phenotype of ΔruvA cells, but ΔsubA did not suppress the segregation defect of ΔrecU and recD cells (Fig. 4). This finding is consistent with the observation that ΔsubA partially overcomes the repair defect of ruvA2 cells but fails to suppress the recombinational defect of recD41 cells (8).
The ΔsubA mutation partially suppressed DNA repair and segregation phenotypes of ΔrecG cells.
The RuvABEco and RecGEco helicases, in concert with the HJ endonuclease RuvCEco, are involved in the formation and processing of branched recombination intermediate structures (38, 40). Above, we showed that ruvA, ΔruvAB, ΔrecU, and recD cells have a segregation defect that can be, in some cases, partially suppressed either in Δsms (ΔradA), ΔsubA, or both genetic backgrounds. To determine whether ΔrecG cells show any segregation and DNA repair phenotype and if the Δsms (ΔradA) or the ΔsubA null mutation has any influence in the segregation pattern of ΔrecG cells, ΔrecG single and double mutant strains (ΔrecG Δsms [ΔradA] and ΔrecG ΔsubA mutants) were constructed and analyzed.
The ΔrecG strain failed to form colonies in the presence of 20 μg of MMS/ml. ΔsubA and Δsms (ΔradA) strains formed colonies in the presence of 250 μg of MMS/ml, and the wt strain formed colonies in the presence of 300 μg of MMS/ml (8). The ΔrecG Δsms (ΔradA) strains failed to form colonies in the presence of 20 μg of MMS/ml, whereas the ΔrecG ΔsubA double mutant strain was able to form colonies in the presence of 250 μg of MMS/ml. Therefore, it is likely that the ΔsubA mutation partially suppresses the recombinational defect of ΔrecG cells.
The absence of DAPI-stained material was observed for ∼7% of exponentially growing ΔrecG cells, and >30% of these cells had abnormally condensed nucleoids (Fig. 5). Similar results were observed with ΔrecG Δsms (ΔradA) cells (Fig. 5). The chromosomal segregation defect observed with ΔrecG cells was partially suppressed if the ΔsubA mutation was present in the background. The presence of the ΔsubA null allele in the ΔrecG background reduced the number of anucleated cells to ∼1% (Fig. 5). This finding is consistent with the observed partial ΔsubA suppression of the DNA repair defect of ΔrecG cells.
FIG. 5.
The recG segregation defect is suppressed by the absence of the SubA product. Exponentially growing cells were fixed, stained with DAPI, and analyzed by fluorescence microscopy to visualize the nucleoids. Black arrows indicate anucleate cells, whereas white arrows show aberrant and misplaced nucleoids.
DISCUSSION
Chromosomal segregation in presynaptic stage mutants is not affected.
Several models to overcome the block of the replication fork progression have been proposed, depending on the nature of the lesion that encounters the replication fork (13, 14, 21, 26, 41, 54). RecBCDEco (counterpart of AddAB) processes DSBs to produce the single-stranded DNA required for homologous pairing by the RecAEco protein, and the RecFOREco proteins (counterparts of RecFLOR) load RecAEco on single-stranded gaps and accelerate DNA strand exchange (13, 26, 40). Both processes lead to the formation of an HJ that will be resolved with a specific polarity (60). Some authors have proposed that RecBCD-dependent DSB repair leads to crossing over and subsequent dimerization and that RecFOREco-dependent gap repair will not lead to crossover (14, 15). Based on the viability of E. coli rep mutants in the absence of XerC or dif, other authors have proposed that recombination events at arrested forks generally do not lead to the formation of dimers (41). Furthermore, it has been described that about half of the dimers appear to arise through RecBCD-dependent events in E. coli cells, while the other half arise from RecFOR-dependent recombination events (58). In order to clarify which of the recombination proteins could be involved in the formation of crossover or noncrossover events in B. subtilis, representatives of each of the described epistatic groups were examined by fluorescence microscopy after DAPI staining. The recO and recR (representatives of the α epistatic group), addAB (β), recH (γ), and recS (ζ) mutant cells did not show any chromosomal segregation phenotypes. We favor the hypothesis that recombination events catalyzed by RecFLOR, AddAB, RecH-RecP, and RecS might occur predominantly in the absence of crossing over. This hypothesis is consistent with the observation that RecFOREco-dependent recombination events occur in the absence of crossing over (14) and with the viability of E. coli rep mutants in the absence of XerC (41). Alternatively, all presynaptic proteins can be considered RecA accessory proteins, and mutations in only one of the genes will never lead to a strong segregation phenotype.
Genes classified within the ɛ epistatic group are required for replication fork repair and chromosomal segregation.
Genetic and biochemical evidence suggests that the genes classified within the ɛ epistatic group (ruvAB, recU, and recD) are involved in DNA repair and HR (3, 8, 18). We show here that the recU, ruvA, ruvB, and recD gene products are involved in recombinational repair of replicating cells and in proper chromosomal segregation. Furthermore, the results presented suggest a postsynaptic role for the unknown activity associated with the recD41 mutation.
The recombinational repair of stalled or collapsed replication forks leads to the production and resolution of an HJ. In both E. coli and B. subtilis, the HJ resolvases RuvC and RecU, respectively, bind and resolve the HJ (3, 61). Depending on the particular binding orientation, RuvCEco or RecU can resolve the symmetric HJs to crossover or noncrossover status. The defect of ΔrecU, ΔruvAB, and recD41 mutations in chromosomal segregation might be a consequence of their inability to bias HJ resolution toward noncrossovers. In that case, the crossover product will produce a dimeric chromosome. Alternatively, the dimer is formed because the HJ remains unresolved in both ruvABCEco cells (41) and ruvAB recU recD cells (this work). In both cases, dimers need to be resolved before cell division can occur. In E. coli and B. subtilis cells, specific site-specific recombinase systems, the XerCD/FtsK and CodVRipX/SpoIIIE complexes, respectively, act at dif to ensure the resolution of dimeric chromosomes (6, 27, 48, 49). This is consistent with the observations that for both E. coli and B. subtilis, the segregation defect of xerCEco Δdif and ΔripX mutants is suppressed by inactivation of the RecA protein (6, 27, 28) and that the absence of the RecA protein also suppresses the segregation defect of ΔrecU cells. We propose that the HJs made in the absence of the RecA protein are resolved to noncrossovers. This proposal is consistent with the observation that chromosome dimer formation (crossovers) is prevented in ΔrecA ΔrecU mutants, in repEco ruvABCEco ΔdifEco recAEco or priAEco recAEco mutants, or in UV-irradiated ruvCEco recAEco cells (22, 36, 41).
Previously, it has been shown that the Δsms (ΔradA) and ΔsubA mutations partially suppress the DNA repair defect of genes classified within the ɛ epistatic group (8). As shown in Fig. 4, the Δsms (ΔradA) mutation suppressed the segregation phenotype of ruvA2 and recD41 cells but failed to suppress the segregation defect of ΔrecU cells. We propose that, in the absence of the Sms (RadA) and RuvAB or RecD proteins, the branch migration RecG protein bound to an HJ intermediate will dictate the RecU resolution of the HJs in a way that should allow replication restart and noncrossover formation. This proposal is consistent with the observation that in E. coli, the sms (radA) and ruv mutations are synergistic with the recG mutation (5, 30). Alternatively, as previously proposed by McGlynn and Lloyd (38) for E. coli cells, the RecG protein in the sms (radA) ruvAB background would reestablish the fork ready for PriA-dependent reloading of the replisome. The Sms (RadA) protein shares a significant degree of identity with the RecA protein at its central region and with the Lon protease at its C-terminal region and plays a role in recombinational repair (5, 8). At present the biochemical activity(ies) associated with the Sms (RadA) protein remains to be elucidated.
The recG gene product is required for chromosomal segregation.
The RecGEco protein plays an essential role in the processing of recombination intermediates in E. coli cells (38, 40). Unlike the recGEco mutation that confers moderate sensitivity to DNA-damaging agents (31), the ΔrecG mutation markedly affects the viability of cells exposed to 20 μg of MMS/ml (M. C. Cozar and H. Sanchez, personal communication). Furthermore, ΔrecG cells show a chromosomal segregation phenotype (Fig. 4), suggesting that the recG mutant failed to repair stalled or collapsed replication forks. Furthermore, as observed with E. coli cells, if positive supercoiling is allowed to accumulate ahead of the replication fork, the forks may be converted to HJs, which have to be converted back to forks if replication is to be completed (43, 47). Hence, in both ruvAB recU (recD) and recG cells, replication should be stalled and anucleate cells should accumulate.
It has been suggested that the Sms (RadA) and SubA proteins are involved in the formation, stabilization, or processing of branched DNA molecules or blocked replication forks (5, 8). Here, we show that the ΔsubA mutation also partially suppresses the DNA repair and segregation phenotypes of ΔrecG cells, but the Δsms (ΔradA) mutation suppresses neither the DNA repair nor the segregation defect of ΔrecG cells. Interestingly, the ΔsubA mutation suppresses the DNA repair and segregation phenotypes of both previously described branch-migrating DNA helicases (RuvAB and RecG).
What is the role of the SubA protein? The subA and mfd genes form an operon (4, 8), and a subA counterpart in E coli is apparently absent. SubA shares a low degree of identity with the UvrA protein, and Mfd shares a significant degree of identity with the RecG and PriA proteins (4, 8, 33). Both Mfd and MfdEco proteins recognize a stalled RNA polymerase (RNAP) at UV-induced lesions in the template DNA, dissociate RNAP from the DNA, and recruit UvrA to the site of damage, thereby facilitating excision repair of the transcribed strand (4, 44, 51). RNAP molecules stalled at lesions in the DNA are major obstacles to replication fork progression, and RuvABCEco is required to promote the rescue of the stalled replication forks (39, 50, 59). With E. coli, it has been shown that elevation of ppGpp levels or certain RNAP mutations improves the survival of UV-irradiated RuvABC mutants, probably by minimizing stalling of RNAP at lesions (39).
PriAEco loads the replisome at recombination intermediates to rescue arrested forks (29, 37). Although a mutation in the helicase motif of PriAEco reduces the ability of ruv mutants to survive DNA damage, it suppresses the DNA repair defect in recG cells (23). Since (i) the ΔsubA mutation suppresses the phenotype of mutations (ruvA2, ΔruvAB, and ΔrecG) in genes encoding the major branch migrating helicases and (ii) ruvAB and recG suppressors in E. coli are helicase-defective proteins, we hypothesize that Mfd alone or in concerted action with another factor(s) may recognize branched structures and translocate in such structures in the presence of SubA. This hypothesis is consistent with our previous failure to detect Mfd-specific binding to HJs and promotion of branch migration (4) and with the fact that the DNA translocation motifs of RecGEco and MfdEco are conserved (33). However, E. coli mfd recG and mfd ruvAB cells were two- to threefold-more UV sensitive than the recG or ruvAB cells (53). Furthermore, we predict that the low degree of identity of SubA with UvrA might correspond to the domain of interaction with Mfd. At present, the MfdEco-interacting domain in UvrAEco remains unknown.
A direct effect due to the absence of Mfd in the ΔsubA strain can be ruled out because (i) the downstream mfd gene is under the control of an inducible promoter in the ΔsubA cells (8) and (ii) the Δmfd mutation increased the sensitivity to DNA-damaging agents of ΔrecU cells (4), whereas the ΔsubA mutation partially suppressed its defect (8). Alternatively, the subA gene might code for an Mfd repressor. However, a suppression of the ΔrecU segregation defect was observed with ΔsubA cells, even in the absence of induction, which will render low levels of Mfd. At present, the biochemical activity(ies) associated with the SubA protein remains to be elucidated.
Acknowledgments
This research was partially supported by grants BMC2003-00150 and BCM2003-01969 from MCT-DGI to J.C.A. and S.A. B.C. was the recipient of a fellowship from MCT-DGI (BMC2000-0548), and S.A. is supported by the Ramón y Cajal program.
We thank H. Sanchez for communicating unpublished results.
REFERENCES
- 1.Alonso, J. C., A. C. Stiege, and G. Luder. 1993. Genetic recombination in Bacillus subtilis 168: effect of recN, recF, recH, and addAB mutations on DNA repair and recombination. Mol. Gen. Genet. 239:129-136. [DOI] [PubMed] [Google Scholar]
- 2.Alonso, J. C., R. H. Tailor, and G. Lüder. 1988. Characterization of recombination-deficient mutants of Bacillus subtilis. J. Bacteriol. 170:3001-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayora, S., B. Carrasco, E. Doncel, R. Lurz, and J. C. Alonso. 2004. Bacillus subtilis RecU protein cleaves Holliday junctions and anneals single-stranded DNA. Proc. Natl. Acad. Sci. USA 101:452-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayora, S., F. Rojo, N. Ogasawara, S. Nakai, and J. C. Alonso. 1996. The Mfd protein of Bacillus subtilis 168 is involved in both transcription-coupled DNA repair and DNA recombination. J. Mol. Biol. 256:301-318. [DOI] [PubMed] [Google Scholar]
- 5.Beam, C. E., C. J. Saveson, and S. T. Lovett. 2002. Role for radA/sms in recombination intermediate processing in Escherichia coli. J. Bacteriol. 184:6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blakely, G., S. Colloms, G. May, M. Burke, and D. Sherratt. 1991. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 3:789-798. [PubMed] [Google Scholar]
- 7.Britton, R. A., D. C. Lin, and A. D. Grossman. 1998. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 12:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrasco, B., S. Fernández, K. Asai, N. Ogasawara, and J. C. Alonso. 2002. Effect of the recU suppressors sms and subA on DNA repair and homologous recombination in Bacillus subtilis. Mol. Genet. Genom. 266:899-906. [DOI] [PubMed] [Google Scholar]
- 9.Ceglowski, P., G. Lüder, and J. C. Alonso. 1990. Genetic analysis of recE activities in Bacillus subtilis. Mol. Gen. Genet. 222:441-445. [DOI] [PubMed] [Google Scholar]
- 10.Chedin, F., and S. C. Kowalczykowski. 2002. A novel family of regulated helicases/nucleases from Gram-positive bacteria: insights into the initiation of DNA recombination. Mol. Microbiol. 43:823-834. [DOI] [PubMed] [Google Scholar]
- 11.Courcelle, J., C. Carswell-Crumpton, and P. C. Hanawalt. 1997. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl. Acad. Sci. USA 94:3714-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courcelle, J., A. K. Ganesan, and P. C. Hanawalt. 2001. Therefore, what are recombination proteins there for? Bioessays 23:463-470. [DOI] [PubMed] [Google Scholar]
- 13.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 14.Cromie, G. A., and D. R. Leach. 2000. Control of crossing over. Mol. Cell 6:815-826. [DOI] [PubMed] [Google Scholar]
- 15.Cromie, G. A., C. B. Millar, K. H. Schmidt, and D. R. Leach. 2000. Palindromes as substrates for multiple pathways of recombination in Escherichia coli. Genetics 154:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández, S., S. Ayora, and J. C. Alonso. 2000. Bacillus subtilis homologous recombination: genes and products. Res. Microbiol. 151:481-486. [DOI] [PubMed] [Google Scholar]
- 17.Fernández, S., Y. Kobayashi, N. Ogasawara, and J. C. Alonso. 1999. Analysis of the Bacillus subtilis recO gene: RecO forms part of the RecFLOR function. Mol. Gen. Genet. 261:567-573. [DOI] [PubMed] [Google Scholar]
- 18.Fernández, S., A. Sorokin, and J. C. Alonso. 1998. Genetic recombination in Bacillus subtilis 168: effects of recU and recS mutations on DNA repair and homologous recombination. J. Bacteriol. 180:3405-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedman, B. M., and R. E. Yasbin. 1983. The genetics and specificity of the constitutive excision repair system of Bacillus subtilis. Mol. Gen. Genet. 190:481-486. [DOI] [PubMed] [Google Scholar]
- 20.Graumann, P. L., R. Losick, and A. V. Strunnikov. 1998. Subcellular localization of Bacillus subtilis SMC, a protein involved in chromosome condensation and segregation. J. Bacteriol. 180:5749-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haber, J. E. 1999. DNA recombination: the replication connection. Trends Biochem. Sci. 24:271-275. [DOI] [PubMed] [Google Scholar]
- 22.Ishioka, K., A. Fukuoh, H. Iwasaki, A. Nakata, and H. Shinagawa. 1998. Abortive recombination in Escherichia coli ruv mutants blocks chromosome partitioning. Genes Cells 3:209-220. [DOI] [PubMed] [Google Scholar]
- 23.Jaktaji, R. P., and R. G. Lloyd. 2003. PriA supports two distinct pathways for replication restart in UV-irradiated Escherichia coli cells. Mol. Microbiol. 47:1091-1100. [DOI] [PubMed] [Google Scholar]
- 24.Jedrzejas, M. J., and W. J. Huang. 2003. Bacillus species proteins involved in spore formation and degradation: from identification in the genome, to sequence analysis, and determination of function and structure. Crit. Rev. Biochem. Mol. Biol. 38:173-198. [DOI] [PubMed] [Google Scholar]
- 25.Kidane D., H. Sanchez, J. C. Alonso, and P. L. Graumann. 2004. Visualization of DNA double-strand break repair in live bacteria reveals dynamic recruitment of Bacillus subtilis RecF, RecO and RecN proteins to distinct sites on the nucleoids. Mol. Microbiol. 52:1627-1639. [DOI] [PubMed] [Google Scholar]
- 26.Kowalczykowski, S. C. 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25:156-165. [DOI] [PubMed] [Google Scholar]
- 27.Kuempel, P. L., J. M. Henson, L. Dircks, M. Tecklenburg, and D. F. Lim. 1991. dif, a recA-independent recombination site in the terminus region of the chromosome of Escherichia coli. New Biol. 3:799-811. [PubMed] [Google Scholar]
- 28.Lemon, K. P., I. Kurtser, and A. D. Grossman. 2001. Effects of replication termination mutants on chromosome partitioning in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 98:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, J., L. Xu, S. J. Sandler, and K. J. Marians. 1999. Replication fork assembly at recombination intermediates is required for bacterial growth. Proc. Natl. Acad. Sci. USA 96:3552-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloyd, R. G. 1991. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J. Bacteriol. 173:5414-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lloyd, R. G., and C. Buckman. 1991. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J. Bacteriol. 173:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luisi-DeLuca, C. 1995. Homologous pairing of single-stranded DNA and superhelical double-stranded DNA catalyzed by RecO protein from Escherichia coli. J. Bacteriol. 177:566-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahdi, A. A., G. S. Briggs, G. J. Sharples, Q. Wen, and R. G. Lloyd. 2003. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 22:724-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maisnier-Patin, S., K. Nordstrom, and S. Dasgupta. 2001. RecA-mediated rescue of Escherichia coli strains with replication forks arrested at the terminus. J. Bacteriol. 183:6065-6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mascarenhas, J., J. Soppa, A. V. Strunnikov, and P. L. Graumann. 2002. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 21:3108-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCool, J. D., and S. J. Sandler. 2001. Effects of mutations involving cell division, recombination, and chromosome dimer resolution on a priA2::kan mutant. Proc. Natl. Acad. Sci. USA 98:8203-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGlynn, P., A. A. Al-Deib, J. Liu, K. J. Marians, and R. G. Lloyd. 1997. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J. Mol. Biol. 270:212-221. [DOI] [PubMed] [Google Scholar]
- 38.McGlynn, P., and R. G. Lloyd. 2002. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 18:413-419. [DOI] [PubMed] [Google Scholar]
- 39.McGlynn, P., and R. G. Lloyd. 2000. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101:35-45. [DOI] [PubMed] [Google Scholar]
- 40.McGlynn, P., and R. G. Lloyd. 2002. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 3:859-870. [DOI] [PubMed] [Google Scholar]
- 41.Michel, B., G. D. Recchia, M. Penel-Colin, S. D. Ehrlich, and D. J. Sherratt. 2000. Resolution of Holliday junctions by RuvABC prevents dimer formation in rep mutants and UV-irradiated cells. Mol. Microbiol. 37:180-191. [DOI] [PubMed] [Google Scholar]
- 42.Moriya, S., E. Tsujikawa, A. K. Hassan, K. Asai, T. Kodama, and N. Ogasawara. 1998. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol. 29:179-187. [DOI] [PubMed] [Google Scholar]
- 43.Olavarrieta, L., M. L. Martinez-Robles, J. M. Sogo, A. Stasiak, P. Hernandez, D. B. Krimer, and J. B. Schvartzman. 2002. Supercoiling, knotting and replication fork reversal in partially replicated plasmids. Nucleic Acids Res. 30:656-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park, J. S., M. T. Marr, and J. W. Roberts. 2002. E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109:757-767. [DOI] [PubMed] [Google Scholar]
- 45.Pedersen, L. B., and P. Setlow. 2000. Penicillin-binding protein-related factor A is required for proper chromosome segregation in Bacillus subtilis. J. Bacteriol. 182:1650-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perals, K., H. Capiaux, J.-B. Vincourt, J. M. Louarn, D. J. Sherratt, and F. Cornet. 2001. Interplay between recombination, cell division and chromosome structure during dimer resolution in Escherichia coli. Mol. Microbiol. 39:904-913. [DOI] [PubMed] [Google Scholar]
- 47.Postow, L., C. Ullsperger, R. W. Keller, C. Bustamante, A. V. Vologodskii, and N. R. Cozzarelli. 2001. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 276:2790-2796. [DOI] [PubMed] [Google Scholar]
- 48.Sciochetti, S. A., P. J. Piggot, and G. W. Blakely. 2001. Identification and characterization of the dif site from Bacillus subtilis. J. Bacteriol. 183:1058-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sciochetti, S. A., P. J. Piggot, D. J. Sherratt, and G. Blakely. 1999. The ripX locus of Bacillus subtilis encodes a site-specific recombinase involved in proper chromosome partitioning. J. Bacteriol. 181:6053-6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selby, C. P., and A. Sancar. 1994. Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol. Rev. 58:317-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selby, C. P., and A. Sancar. 1993. Molecular mechanism of transcription-repair coupling. Science 260:53-58. [DOI] [PubMed] [Google Scholar]
- 52.Sharples, G. J. 2001. The X philes: structure-specific endonucleases that resolve Holliday junctions. Mol. Microbiol. 39:823-834. [DOI] [PubMed] [Google Scholar]
- 53.Sharples, G. J., and L. M. Corbett. 1997. Recombination is unaffected by mutation of E. coli mfd. Microbiology 143:690-691. [DOI] [PubMed] [Google Scholar]
- 54.Sherratt, D. J. 2003. Bacterial chromosome dynamics. Science 301:780-785. [DOI] [PubMed] [Google Scholar]
- 55.Sherratt, D. J., I. F. Lau, and F. X. Barre. 2001. Chromosome segregation. Curr. Opin. Microbiol. 4:653-659. [DOI] [PubMed] [Google Scholar]
- 56.Soppa, J., K. Kobayashi, M. F. Noirot-Gros, D. Oesterhelt, S. D. Ehrlich, E. Dervyn, N. Ogasawara, and S. Moriya. 2002. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 45:59-71. [DOI] [PubMed] [Google Scholar]
- 57.Spurr, A. R. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26:31-43. [DOI] [PubMed] [Google Scholar]
- 58.Steiner, W. W., and P. L. Kuempel. 1998. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J. Bacteriol. 180:6269-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trautinger, B. W., and R. G. Lloyd. 2002. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J. 21:6944-6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Gool, A. J., N. M. Hajibagheri, A. Stasiak, and S. C. West. 1999. Assembly of the Escherichia coli RuvABC resolvasome directs the orientation of Holliday junction resolution. Genes Dev. 13:1861-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.West, S. C. 1996. The RuvABC proteins and Holliday junction processing in Escherichia coli. J. Bacteriol. 178:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Westers, H., R. Dorenbos, J. M. Van Dijl, J. Kabel, T. Flanagan, K. M. Devine, F. Jude, S. J. Seror, A. C. Beekman, E. Darmon, C. Eschevins, A. De Jong, S. Bron, O. P. Kuipers, A. M. Albertini, H. Antelmann, M. Hecker, N. Zamboni, U. Sauer, C. Bruand, D. S. Ehrlich, J. C. Alonso, M. Salas, and W. J. Quax. 2003. Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol. Biol. Evol. 20:2076-2090. [DOI] [PubMed] [Google Scholar]
- 63.Yasbin, R. E., P. I. Fields, and B. J. Andersen. 1980. Properties of Bacillus subtilis 168 derivatives freed of their natural prophages. Gene 12:155-159. [DOI] [PubMed] [Google Scholar]
- 64.Zyskind, J. W., A. L. Svitil, W. B. Stine, M. C. Biery, and D. W. Smith. 1992. RecA protein of Escherichia coli and chromosome partitioning. Mol. Microbiol. 6:2525-2537. [DOI] [PubMed] [Google Scholar]