Abstract
The Escherichia coli argU10(Ts) mutation in the argU gene, encoding the minor tRNAArg species for the rare codons AGA and AGG, causes pleiotropic defects, including growth inhibition at high temperatures, as well as the Pin phenotype at 30°C. In the present study, we first showed that the codon selectivity and the arginine-accepting activity of the argU tRNA are both essential for complementing the temperature-sensitive growth, indicating that this defect is caused at the level of translation. An in vitro analysis of the effects of the argU10(Ts) mutation on tRNA functions revealed that the affinity with elongation factor Tu-GTP of the argU10(Ts) mutant tRNA is impaired at 30 and 43°C, and this defect is more serious at the higher temperature. The arginine acceptance is also impaired significantly but to similar extents at the two temperatures. An in vivo analysis of aminoacylation levels showed that 30% of the argU10(Ts) tRNA molecules in the mutant cells are actually deacylated at 30°C, while most of the argU tRNA molecules in the wild-type cells are aminoacylated. Furthermore, the cellular level of this mutant tRNA is one-tenth that of the wild-type argU tRNA. At 43°C, the cellular level of the argU10(Ts) tRNA is further reduced to a trace amount, while neither the cellular abundance nor the aminoacylation level of the wild-type argU tRNA changes. We concluded that the phenotypic properties of the argU10(Ts) mutant result from these reduced intracellular levels of the tRNA, which are probably caused by the defective interactions with elongation factor Tu and arginyl-tRNA synthetase.
In the genetic code, the 20 amino acids are each encoded by one to six synonymous codons. In unicellular organisms, some of the synonymous codons are preferentially used in the highly expressed genes, while some others are rarely used in such genes (28). This differential use of synonymous codons roughly reflects the abundance of the tRNA species recognizing each codon (15, 16). For example, the arginine codons CGU and CGC, which are used frequently in the Escherichia coli genome, are recognized by the major tRNAArg species. On the other hand, a minor tRNAArg species (tRNA4Arg), encoded by the argU gene (8), recognizes the rare arginine codons AGA and AGG (34, 44). The rare codon AGG is also decoded by another minor tRNAArg with a CCU anticodon (tRNA5Arg). The use of rare codons is not avoided in genes expressed at low or moderate levels, and the rare arginine codons are actually contained in regulatory genes and genes involved in DNA replication (28, 40).
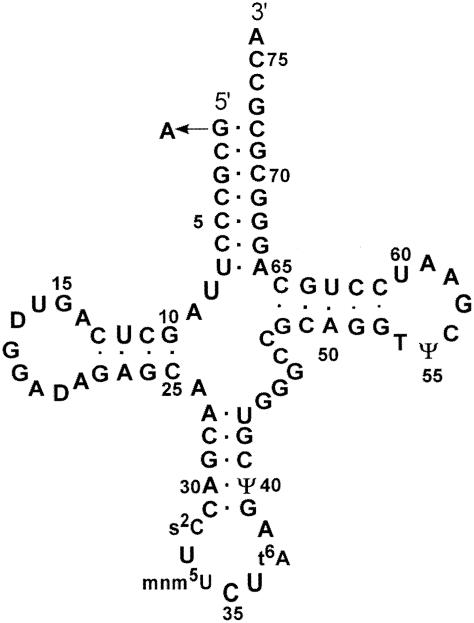
The E. coli argU10(Ts) mutation in the argU gene inhibits DNA replication, which stops cell growth at a high temperature (12), and causes the Pin phenotype at a permissive temperature (2). The argU10(Ts) defects were suggested to be due to impaired translation of the AGA and AGG codons in the mutant cells, for two reasons. First, the argU10(Ts) mutation is a G-to-A transition at the 5′ end of the argU tRNA, which generates a mismatched base pair at the terminus of the acceptor stem (2) (Fig. 1); this structural alteration may impair the tRNA function. Second, some replication genes contain a few AGA or AGG codons, while the old gene from bacteriophage P2, involved in the Pin phenotype, contains eight AGA or AGG codons (10). A reporter gene containing AGA and AGG codons can hardly be expressed in the argU10(Ts) mutant cells at a high temperature, and moderate inhibition of AGA and AGG translation was observed at the permissive temperature (2). These different levels of AGA and AGG translation inhibition explain the various phenotypes caused by the argU10(Ts) mutation. On the other hand, the molecular mechanism by which the argU10(Ts) mutation impairs AGA and AGG translation has remained obscure.
FIG. 1.
Secondary structure of the argU tRNA. The argU10(Ts) mutation, a G-to-A transition at position 1, is indicated. The modified nucleotides found in this tRNA are dihydrouridine (D), 2-thiocytidine (s2C), 5-methylaminomethyluridine (mnn5U), N6-threonine carbamoyl adenosine (t6A), pseudouridine (ψ), and 5-methyluridine (T) (17, 33).
In the present study, we first showed that the arginine-accepting activity, as well as the codon selectivity, of the argU tRNA is essential for complementation of the argU10(Ts) mutation. Then, to examine whether this mutation impairs the translational functions of the argU10 tRNA, we carried out in vitro assays and analyses of the aminoacylation levels in vivo for the argU10(Ts) mutant tRNA. We found that this mutation not only impairs the arginylation and the binding to elongation factor Tu (EF-Tu)-GTP of the argU tRNA in vitro but also seriously reduces the amount of this tRNA in the mutant cells. These findings reveal the molecular mechanism by which the argU10(Ts) mutation causes the impaired AGA and AGG translation in the mutant cells.
MATERIALS AND METHODS
Bacterial strains and gene transfer.
The argU10(Ts) strain GM10 (12) and strains YT319 [ilv dnaA17(Am) rnhA-199(Am) thyA trpE9828(Am) tyrA(Am) thr], YT341 [dnaA17(Am) supF6(Ts)] (26), KN250 [supD126(Ts)] (13), KN1044 (KN250 rnhA-1::Tn3), and KN1453 [dnaA5(Ts) rnhA-1::Tn3] (27) have been described previously. Gene transfer was carried out by transduction by using phage P1vir (14).
Recombinant DNA technology, DNA sequencing, and PCR.
Standard techniques were used for isolation of plasmid or λ phage DNA, restriction endonuclease digestion, ligation, and gel electrophoresis (35). E. coli cells were transformed by using a Gene Pulser electroporation apparatus (Bio-Rad Laboratories). PCR was performed by using AmpliTaq (Takara Shuzo, Kyoto, Japan) and a DNA PJ480 thermal cycler (Perkin-Elmer Cetus). Nucleotide sequencing was performed by the dideoxy chain termination method (36) by using an AmpliTaq sequencing kit (Takara Shuzo).
Plasmids carrying the argU alleles and the argS gene.
Plasmid pDM1, carrying the wild-type argU gene, has been described previously (25), while the 4.5-kb argU10(Ts) HindIII chromosomal fragment (2) was cloned into the vector pBR322 to generate pKC1. Plasmid pAp102 was created by ligating the gene encoding β-lactamase to the 2-kb EcoRI-XbaI fragment containing the sequence essential for autonomous replication of ColIb-P9 (11). The NaeI-VspI fragment of pUC119, which includes the gene encoding the α-fragment of lacZ together with the multiple cloning site in it, was cloned into the EcoRI site of pAp102 to generate pCL1. Plasmid pCL2 consists of the 0.6-kb argU+ ClaI-SphI fragment of pDM1 cloned into the AccI-SphI sites of pCL1, and similarly, pCL3 consists of the 0.6-kbp argU10(Ts) fragment of pKC1 cloned into pCL1.
An AvaI-HindIII fragment of λ clone 12C7 from the genomic library (20), which carries the argS gene for arginyl-tRNA synthetase, was cloned into the SalI-HindIII sites of pUC119 to generate pUA1. The argS gene on the BamHI-HindIII fragment of pUA1 was ligated to the corresponding sites of the vector pACYC184 to generate pYA1.
Site-directed mutagenesis of the argU gene.
The synthetic oligomers RV (5′-CACACAGGAAACAGCTATGAC-3′) and M3 (5′-CGACGTTGTAAAACGACGGCCAG-3′) were designed as PCR primers for amplification of a fragment cloned in the multiple cloning site of pUC119 or pCL1. Site-directed mutagenesis of the argU gene was done by performing the following two successive PCR amplifications. The first amplification was carried out with the oligomer RV and a mutagenic oligomer as PCR primers and plasmid pCL2 as the template. The 0.3-kb fragment obtained was purified by agarose gel electrophoresis, and the second amplification was carried out with this fragment and the oligomer M3 as the primers and pCL2 as the template. Finally, the 0.6-kb fragment obtained was digested with EcoRI and HindIII and ligated into the corresponding sites of pCL1 or pBR322. Base substitutions were confirmed by DNA sequencing.
Preparation of the argU tRNA, EF-Tu, and arginyl-tRNA synthetase.
The wild-type and mutant argU tRNA species were overproduced in W3110 harboring pDM1 at 37°C and in GM10 harboring pKC1 at 30°C, respectively. The tRNAs were extracted from the late-log-phase cells and were fractionated by high-performance liquid chromatography on a hydroxyapatite column (HA-1000; Tosoh) (34). Finally, the argU tRNAs were purified from the column fractions by polyacrylamide gel electrophoresis (PAGE) on a denaturing 20% polyacrylamide gel. EF-Tu was prepared from a W3110 cell extract by chromatography on a DEAE-Sephadex A-50 (pH 7.5) column (1). Arginyl-tRNA synthetase was purified from W3110 harboring pUA1 by successive chromatography steps, as described previously (30).
Arginylation assay.
Arginylation of the wild-type or mutant argU tRNA (1 μM) by arginyl-tRNA synthetase (4 nM) was performed at 37°C in 18 μl of mixture A (100 mM Tris-HCl [pH 7.5], 15 mM MgCl2, 2 mM ATP, 60 μM [14C]arginine [332.1 pCi/pmol; Dupont/NEN Research Products]). Aliquots (4 μl) were withdrawn from the reaction mixture after 20 to 90 s of incubation and were immediately added to 10 μl of ice-chilled 5% trichloroacetic acid to stop the reaction. These samples were spotted onto Whatman 3MM filter disks, which were washed three times with ice-chilled 5% trichloroacetic acid and then dried to obtain radioactivity measurements with a liquid scintillation system (LSC-700; Aloka, Tokyo, Japan).
Assay of formation of a complex between arginyl-tRNA and EF-Tu-GTP.
To prepare the wild-type and mutant argU tRNAs in the arginylated state, each tRNA species (6 pmol) was arginylated at 37°C for 10 min in mixture A containing 1 μM arginyl-tRNA synthetase. After phenol-chloroform extraction, the arginylated tRNA was precipitated with ethanol and dissolved in 5 μl of 2 mM sodium acetate buffer (pH 4.5). A 60-μl prereaction mixture containing 50 mM Tris-HCl (pH 7.5), 10 mM magnesium acetate, 150 mM NH4Cl, 50 mM β-mercaptoethanol, 60 μM GTP, 0.8 mM phosphoenolpyruvate (Sigma), 2 U of pyruvate kinase (Sigma), and EF-Tu at the concentrations indicated below was incubated on ice for 25 min in order to replace the GDP molecule bound to EF-Tu by GTP. The arginylated argU tRNA (5 μl) was then added to this mixture, and preincubation was continued for another 5 min. After the preincubation, the reaction mixture was subjected to incubation at 30 or 43°C. The final concentration of the arginyl argU tRNAs was 0.092 μM, while the final concentration of EF-Tu was 0 or 0.062 μM at 30°C and 0 or 0.15 μM at 43°C. Aliquots (11.5 μl) of the reaction mixture were withdrawn at different times and then immediately spotted onto Whatman 3MM filter disks soaked with 5% trichloroacetic acid. The disks were washed with ice-chilled 5% trichloroacetic acid. The radioactivities of the disks were measured as described above for the arginylation assay. The rate constant for deacylation (k) was calculated by using the equation k = ln2 · t1/2−1, where t1/2 is the incubation time required for 50% of the arginyl tRNA molecules to be deacylated.
In vivo analysis of the tRNA aminoacylation levels.
Strains YT319 and SF151 were each grown in Luria-Bertani medium (35) containing 0.1% glucose and 50 μg of thymine per ml at 30°C. At an optical density at 600 nm of 0.2, a portion of each culture was shifted to 43°C. Extraction of tRNA from the cell samples was carried out under acidic conditions, as described previously (46). An aliquot of each extract was used to measure the absorbance at 260 nm, and another aliquot was analyzed by PAGE in order to confirm that the ratio between the A260 and the amount of tRNA was constant for the tRNA preparations. This ratio would change if each tRNA preparation contained a different level of the contamination due to other nucleic acids. The remaining sample was subjected to acid-urea PAGE (46) to separate the aminoacylated and uncharged tRNAs. Transfer of tRNA from the gel to a Hybond-N nylon membrane (Amersham) was carried out with an electroblot apparatus (NA-1512; Nihon Eido, Tokyo, Japan), and the argU tRNA was detected by hybridization to a 32P-labeled oligodeoxyribonucleotide probe (5′-CGAACCTGCGGCCCACGAC-3′) complementary to residues 39 to 57 of the tRNA. The intensities of the bands on the autoradiogram were measured with a BAS2000 bioimaging analyzer (Fuji Photo Film, Tokyo, Japan).
An aliquot of each tRNA extract was examined for its tyrosine acceptance as an internal control for the intracellular level of the argU tRNA. The 40 μM tRNA extract was aminoacylated at 37°C for 30 min with 20 μM l-[14C]tyrosine (16.9 GBq/mmol; New England Nuclear) in a 30-μl mixture containing 100 mM Tris-Cl (pH 7.6), 15 mM MgCl2, 40 mM KCl, 1 mM dithiothreitol, 4 mM ATP, and 50 nM E. coli tyrosyl-tRNA synthetase. The tyrosine concentration used was greater than the concentration of tyrosine tRNA species in the tRNA extract. Preparation of the tyrosyl-tRNA synthetase has been described previously (18). The radioactivity of the accepted tyrosine was measured as described above for the arginylation assay.
RESULTS
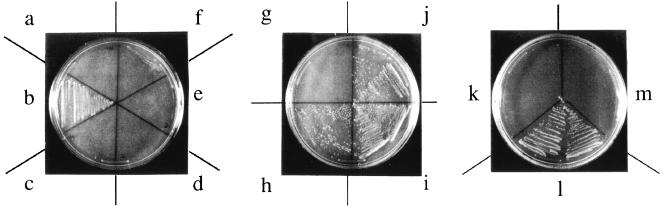
The temperature sensitivity of the argU10(Ts) mutant occurs at the level of translation. The argU10(Ts) mutant strain GM10 exhibits a leaky phenotype at high temperatures (12). To obtain mutants with a strict phenotype, we transferred the argU10(Ts) mutation into rnhA-defective backgrounds (Table 1), which evoke a nonstandard mode of DNA replication initiation (19), while the argU10(Ts) mutation inhibits DNA replication at the stage of polymerization (12). Strain SF151, which was obtained in this way, was used in the present study. The temperature-sensitive growth of SF151 can be complemented by the wild-type argU gene on a low-copy vector derived from the ColIb-P9 plasmid, whose copy number is 1.7 copies per cell (3), but it cannot be complemented by the argU10(Ts) gene on the same vector (Fig. 2a to c).
TABLE 1.
Growth of the argU10(Ts) mutants and the parental strains at 42°C
| Strain | Relevant genotype | Growth at 42°Ca |
|---|---|---|
| KN250 | supD126(Ts) | Growth |
| KN1044 | KN250 rnhA-1::Tn3 | Growth |
| KN1453 | dnaA5(Ts) rnhA-1::Tn3 | Growth |
| YT341 | dnaA17(Am) supF6(Ts) | Leaky growth |
| YT319 | dnaA17(Am) rnhA-199(Am) | Growth |
| SF1 | KN250 purE79::Tn10 argU10(Ts) | Leaky growth |
| SF5 | KN1044 purE79::Tn10 argU10(Ts) | No growth |
| SF7 | KN1453 purE79::Tn10 argU10(Ts) | No growth |
| SF9 | YT341 purE79::Tn10 argU10(Ts) | Leaky growth |
| SF131 | YT319 purE79::Tn10 argU10(Ts) | No growth |
| SF151 | YT319 argU10(Ts) | No growth |
Colonies grown at 30°C were streaked on Luria-Bertani agar, and growth was examined after incubation for 24 h at 42°C.
FIG. 2.
Growth of SF151 transformed with the following plasmids: vector pCL1 (a); the argU+ plasmid pCL2 (b); the argU(Ts) plasmid pCL3 (c); the argU alleles with U20, G20, and C20 (d, e, and f, respectively), each cloned in pCL1; vector pCL1 together with the ArgRS-overproducing plasmid pYA1 (g); the argU alleles with U20, G20, and C20 (h, i, and j, respectively), each together with pYA1; vector pBR322 (k); the argU+ plasmid pDM1 (l); and pBR322 carrying the argU gene with a CCT anticodon (m). Colonies transformed at 30°C were streaked on Luria-Bertani agar containing ampicillin (20 μg/ml) and thymine (50 μg/ml) (a to f), chloramphenicol (25 μg/ml) in addition to ampicillin (20 μg/ml) and thymine (50 μg/ml) (g to j), and ampicillin (50 μg/ml) and thymine (50 μg/ml) (k to m) and were then incubated for 24 h at 42°C.
We examined whether the translational properties of the argU tRNA is necessary for complementation of the argU10(Ts) mutant. The argU tRNA recognizes the AGA and AGG codons (34, 44). The TCT anticodon sequence in the argU gene was replaced by CCT, so that it could recognize only the AGG codon. It was previously reported that this argU tRNA (CCU) does not complement the temperature-sensitive growth of strain GM10 (44); a similar result was obtained with the SF151 strain (Fig. 2k to m). This observation showed that the ability of the argU tRNA to recognize the AGA codon is necessary for the complementation.
Then we carried out complementation experiments using the argU tRNA variants with low arginine-accepting activities. Since A20 in the D loop of E. coli tRNAArg is required specifically for arginylation (23, 24), the base substitutions in this position only impair the arginine-accepting activity. None of the argU gene variants with C, G, and T at position 20 complemented the temperature sensitivity of SF151 (Fig. 2d to f). The complementing activities of these variants were recovered when E. coli arginyl-tRNA synthetase (ArgRS) was overproduced in SF151, while overproduction of ArgRS alone did not complement SF151 (Fig. 2g to j). This observation indicates that the arginylation of the argU tRNA is a prerequisite for the argU+ complementing activity.
These results unambiguously show that the argU10(Ts) phenotype is caused at the level of translation. In order to examine the possibility that any deleterious property of the mutant tRNA causes the argU10(Ts) phenotype, we analyzed the cell growth of the W3110 strains harboring pDM1 and pKC1, which overproduced the wild-type and mutant argU tRNA species, respectively. These overproducing strains exhibited similar doubling times (1.3 h), as determined on the basis of the optimal density at 600 nm of the culture, at 43°C. Since the overproduction of the mutant tRNA hardly affected the cell growth of the wild-type strain, it is unlikely that the temperature sensitivity of the argU10(Ts) mutant results from any possible deleterious property of the tRNA.
Effects of the argU10(Ts) mutation on the translational function of the argU tRNA.
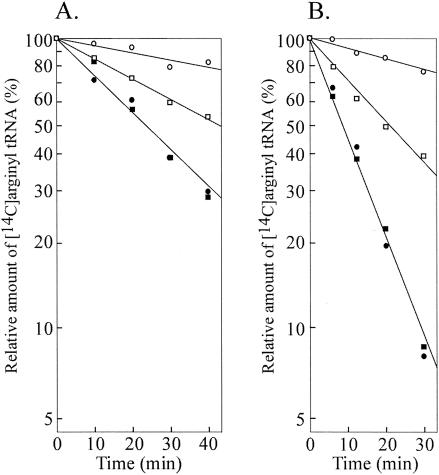
We prepared argU10(Ts) tRNA from the mutant cells that overproduced this mutant tRNA from a multicopy plasmid. Similarly, the wild-type argU tRNA was prepared from wild-type cells overproducing it. The affinity of the arginylated tRNA for EF-Tu-GTP was analyzed on the basis of protection from deacylation by EF-Tu-GTP (22, 32, 47). In the absence of EF-Tu-GTP, the arginylated molecules of the argU10(Ts) tRNA were deacylated at a rate similar to that of the arginylated molecules of the wild-type argU tRNA at both 30 and 43°C (Fig. 3). The rate constants for these deacylations were 2.9 × 10−2 and 8.7 × 10−2 min−1 at 30 and 43°C, respectively.
FIG. 3.
Deacylation of the arginylated molecules of the wild-type argU tRNA (○ and •) and the argU10(Ts) tRNA (□ and ▪) in the presence (○ and □) or absence (• and ▪) of EF-Tu at 30°C (A) and 43°C (B). Samples were withdrawn after incubation for 0, 10, 20, 30, and 40 min at 30°C and after incubation for 0, 6, 12, 20, and 30 min at 43°C. The relative amounts of arginyl-tRNA are plotted on a log scale against the incubation time.
The addition of EF-Tu-GTP to the reaction mixture at 30°C significantly decreased the deacylation rate for the wild-type argU tRNA, and the rate constant was 7.7 × 10−3 min−1 (Fig. 3A). The deacylation was also suppressed strongly at 43°C, and the rate constant was 8.7 × 10−3 min−1 (Fig. 3B). For the argU10(Ts) mutant tRNA, the protection from deacylation by EF-Tu-GTP was significantly weaker even at 30°C (rate constant, 1.7 × 10−2 min−1) and was further impaired at 43°C (rate constant, 3.3 × 10−2 min−1). Thus, the deacylation rate was increased by 13 and 94% for the argU and argU10(Ts) tRNAs, respectively. These findings indicate that the interaction between the argU10(Ts) tRNA and EF-Tu-GTP are impaired at both 30 and 43°C, and the defective interaction was more serious at the higher temperature.
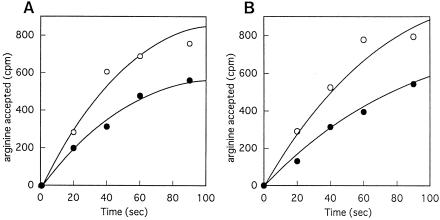
On the other hand, an arginylation assay was performed with a tRNA concentration of 1 μM, because the Km for the arginylation of E. coli tRNAArg is 0.5 to 2.5 μM (21, 37, 45). Furthermore, in vitro arginylation assays showed that the arginylation rate of the argU10(Ts) tRNA was reduced to some extent at both 30 and 43°C compared with that of the wild-type argU tRNA (Fig. 4). The levels of this reduction were similar at these temperatures.
FIG. 4.
Arginylation of the wild-type (○) and mutant (•) argU tRNA at 30°C (A) and 43°C (B). Samples were withdrawn after 20, 40, 60, and 90 s of incubation.
The argU10(Ts) mutation impaired both arginylation and complex formation with EF-Tu-GTP at 30 and 43°C, and only the latter defect exhibited temperature sensitivity.
argU10(Ts) mutation significantly reduces the arginylation level and abundance of the argU10(Ts) tRNA in vivo.
We then investigated the arginylation levels of the argU10(Ts) tRNA in the mutant cells. The wild-type and argU10(Ts) mutant strains (YT319 and SF151, respectively) were grown at 30°C, and a portion of each culture was shifted to 43°C. The tRNA fractions were extracted from these strains before and 60 min after this temperature shift. Strain YT319 continued to grow exponentially, while SF151 stopped growing 60 to 90 min after the shift.
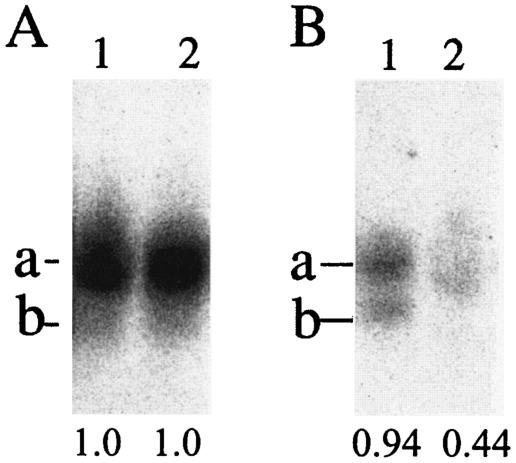
Figure 5 shows the arginylation levels of the argU tRNAs in these strains before and after the temperature shift. The amount of tRNA extract analyzed in each lane was twofold larger for the analysis of the argU10(Ts) tRNA (Fig. 5B) than for the analysis of the argU tRNA (Fig. 5A), because the intracellular level of this mutant tRNA was much lower than that of the wild-type tRNA. The level of tyrosine acceptance, reflecting the intracellular level of tRNATyr, was determined for the same amount of each tRNA extract as an internal control for the cellular level of the argU tRNAs. It was found that most of the argU tRNA molecules were charged with arginine, and only 5% of them were deacylated in the YT319 cells at 30 and 43°C (Fig. 5A). The total amount of this wild-type tRNA was not affected by the temperature increase, and the level of tyrosine acceptance was constant as well.
FIG. 5.
argU tRNA arginylation levels in the wild-type cells (A) and mutant cells (B) before and 60 min after the temperature upshift (lanes 1 and 2, respectively). The tRNA extracts analyzed in each lane by acid-urea gel electrophoresis were 2.5- and 5-μg extracts in panels A and B, respectively. The positions of the arginylated and uncharged tRNAs on the gel are indicated by a and b, respectively. The argU tRNA was detected by hybridization to a 32P-labeled specific probe after transfer of tRNAs from the gel onto a nylon membrane. The values below the lanes are the relative levels of tyrosine acceptance for the tRNA extracts at the same concentration.
On the other hand, 30% of the argU10(Ts) tRNA molecules were uncharged in the SF151 cells at 30°C, and, in addition, the total amount of this mutant tRNA in the charged and uncharged states was only one-tenth the amount of the wild-type argU tRNA in YT319. By shifting the temperature to 43°C, the amount of the tRNA was much more drastically reduced to a trace level (Fig. 5B). The levels of tyrosine acceptance were similar for YT319 and SF151 at 30°C, whereas the temperature shift reduced the level for SF151 alone by 53%. This finding suggests that there was overall degradation of tRNAs within 60 min after the shift, although the SF151 cells stopped growing 60 to 90 min after the shift. Since the cellular level of the argU10(Ts) tRNA was reduced even at 30°C and the reduction at 43°C was more drastic than the observed decrease in the level of tyrosine acceptance, the reduced levels for the mutant tRNA did not seem to result simply from the overall tRNA degradation, and another explanation is required.
DISCUSSION
The argU10(Ts) mutation was isolated as a mutation that inhibits DNA synthesis at a high temperature and was therefore originally designated dnaY10(Ts) (12). This mutation occurs in the gene encoding the minor tRNAArg for the AGA and AGG codons, the argU tRNA, and confers pleiotropic effects at the permissive and nonpermissive temperatures, including inhibition of translation of AGA and AGG codons (2). Genetic complementation experiments have shown that the ability of the argU tRNA to recognize AGA codons is required to complement the argU10(Ts) temperature sensitivity, suggesting that the argU10 (Ts) phenotype is caused at the level of translation (2, 44). The present study provided further evidence for this claim by showing that the arginine-accepting ability of the argU tRNA, another translational activity, is also crucial for the complementation.
We created and used SF151, an argU10(Ts) mutant with strict temperature sensitivity, in order to judge the complementation by the argU tRNA variants unambiguously. Since SF151 has the dnaA rnh double mutation, it is defective in the initiation of DNA replication in the normal mode and probably circumvents this defect by starting the replication from unusual sites (19). Since the argU10(Ts) mutation inhibits DNA replication at a stage of polymerization, this defect in the normal mechanism for the replication initiation appears to enhance the temperature sensitivity of this mutant.
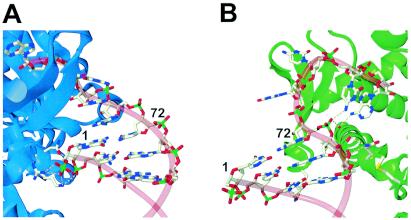
By using in vitro assays, the argU10(Ts) mutation was found to reduce both the arginylation efficiency and the affinity to EF-Tu-GTP to some extent. This mutation generates a mismatched base pair, A1-C72, at the terminus of the acceptor stem (2). Base pairing at this acceptor stem terminus is important in the binding of an aminoacyl-tRNA to EF-Tu-GTP (7, 38, 39). In the crystal structure of the aminoacyl-tRNA-EF-Tu-GTP ternary complex (29), EF-Tu binds to the sugar-phosphate backbone of residue 1 of the tRNA but not to the side of residue 72 (Fig. 6A). Base pairing between these residues places the aminoacylated 3′ end of the tRNA in the correct position to facilitate its accommodation by EF-Tu-GTP. On the other hand, ArgRS interacts with the side of residue 72 but not with the side of residue 1 in the crystal structure of the yeast ArgRS-tRNAArg complex (4) (Fig. 6B). The mismatched base pair at these positions probably destabilizes the helical structure around residue 72 that facilitates the interaction with ArgRS. This probably also occurs with a prokaryotic ArgRS because of its structural similarity to yeast ArgRS (41). Because of these contrasting binding manners of EF-Tu-GTP and ArgRS, the stability of the acceptor stem at its terminus seems to be more crucial for the binding of EF-Tu-GTP, and this may explain our observation that the interaction between the argU10(Ts) tRNA and this factor alone exhibits temperature sensitivity.
FIG. 6.
Interactions of the G1-C72 base pair at the acceptor stem terminus with EF-Tu-GTP (28) (A) and yeast ArgRS (4) (B). EF-Tu and ArgRS are represented by blue and green ribbons, respectively. Nucleotide residues are represented by sticks, and the phosphate-sugar backbone is outlined by pink tubes. The amino acid residues of EF-Tu (A) and ArgRS (B) that interact with residues 1 to 2 and 71 to 72, respectively, are also represented by balls and sticks.
These defects in arginylation and binding to EF-Tu-GTP can explain the uncharged fraction of the argU10(Ts) tRNA in the mutant cells at 30°C and probably underlie the reduced intracellular levels of this tRNA at 30 and 43°C. In the translation process, tRNA molecules successively interact with several cellular components, such as aminocyl-tRNA synthetase, EF-Tu-GTP, and ribosomes, and are thus protected from degradation by RNases. The argU10(Ts) tRNA is therefore more susceptible to RNases than its wild-type counterpart because of its defective arginine acceptance and binding to EF-Tu-GTP. Furthermore, this mutant tRNA is exposed to the nucleases to a larger extent at 43°C than at 30°C because of the further reduction in its affinity for EF-Tu-GTP at the higher temperature.
Single base substitutions confer temperature sensitivity on the amber suppressor tRNATyr (42, 43) and tRNATrp (6) from E. coli. These base substitutions, which occur in the central part of the tRNA, destabilize the tRNA structure (5, 42) and have thus been thought to make the mutant tRNAs more susceptible to RNases. The efficiencies of tryptophanylation are similar for the mutant tRNATrp and its wild-type counterpart (5). On the other hand, the argU10(Ts) tRNA has the mutation at an extremity, and the exposure of this tRNA to RNases is probably due to the defects in the arginylation and the binding to EF-Tu-GTP rather than to destabilization of the structure.
The different in vivo levels of the argU10(Ts) tRNA at 30 and 43°C explain the different extents to which the translation of AGA and AGG codons in the mutant cells is inhibited at these temperatures. The argU tRNA has a UCU anticodon, where the uridine in the first position is modified to 5-methylaminomethyluridine (34), and thus it recognizes both the AGA and AGG codons (44). The AGG codon is also translated by another minor tRNAArg species with a CCU anticodon, while this argU tRNA is the only tRNAArg species that decodes AGA. Therefore, the inhibited translation of the genes with AGA and AGG codons in the arg10(Ts) mutant is probably mainly due to inhibition of AGA translation, although it is not clear yet if the reduced argU tRNA levels affect the efficiency of AGG codon translation.
The reduced argU10(Ts) tRNA level at 30°C explains the Pin phenotype displayed by the argU10(Ts) mutant at this temperature. This phenotype is exhibited by E. coli when expression of the P2 old gene, with eight AGA or AGG codons (10), is inhibited (9). Interestingly, although the in vivo amount of the argU10(Ts) tRNA in the arginylated form is only 5% of the amount of the argU tRNA in wild-type cells, the expression of the E. coli genes essential for cell growth hardly seems to be impaired at this temperature.
A G1-to-A1 transition occurs in the tRNATyr and tRNAGly mutants of E. coli and Salmonella enterica serovar Typhimurium, respectively. The mutant tRNATyr can be charged with glutamine (43), because a weak base pair, including a mismatched base pair, and U35, which is also present in tRNATyr, are recognized by glutaminyl-tRNA synthetase (33). The mutant tRNAGly promotes frameshifting, and the reduced affinities for EF-Tu-GTP and/or certain ribosomal components may underlie this phenomenon (31). Although the possibility of either mischarging or frameshifting was not investigated for the argU10(Ts) tRNA, it is unlikely that a deleterious property conferred by the argU10(Ts) mutation causes the temperature sensitivity, because the overproduction of the argU10(Ts) tRNA hardly affects cell growth.
In conclusion, the functional defects of the argU10(Ts) tRNA reduce the cellular abundance of this tRNA to different extents at various temperatures, and this is the molecular basis for the pleiotropic effects of the argU10(Ts) mutation.
Acknowledgments
We thank Kiyoshi Mizobuchi for the gift of plasmid pAp102 and the E. coli genomic library and Yota Murakami for providing strains YT319 and YT341.
This work was supported by the RIKEN Structural Genomics/Proteomics Initiative, National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Arai, K., M. Kawakami, and Y. Kaziro. 1972. Studies on polypeptide elongation factors from Escherichia coli. J. Biol. Chem. 247:7029-7037. [PubMed] [Google Scholar]
- 2.Chen, K.-S., T. C. Peters, and J. R. Walker. 1990. A minor arginine tRNA mutant limits translation preferentially of a protein dependent on the cognate codon. J. Bacteriol. 172:2504-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clewell, D. B., and D. R. Helinski. 1970. Existence of the colicinogenic factor-sex factor ColIb-P9 as a supercoiled circular DNA-protein relaxation complex. Biochem. Biophys. Res. Commun. 41:150-156. [DOI] [PubMed] [Google Scholar]
- 4.Delagoutte, B., D. Moras, and J. Cavarelli. 2000. tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrate binding. EMBO J. 19:5599-5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenberg, S. P., and M. Yarus. 1980. The structure and aminoacylation of a temperature-sensitive tRNATrp (Escherichia coli). J. Biol. Chem. 255:1128-1137. [PubMed] [Google Scholar]
- 6.Eisenberg, S. P., L. Soll, and M. Yarus. 1979. The purification and sequence of a temperature-sensitive tryptophan tRNA. J. Biol. Chem. 254:5562-5566. [PubMed] [Google Scholar]
- 7.Fischer, W., T. Doi, M. Ikehara, E. Ohtsuka, and M. Sprinzl. 1985. Interaction of methionine-specific tRNAs from Escherichia coli with immobilized elongation factor Tu. FEBS Lett. 192:151-154. [DOI] [PubMed] [Google Scholar]
- 8.Garcia, G. M., P. K. Mar, D. A. Mullin, J. R. Walker, and N. E. Prather. 1986. The E. coli dnaY gene encodes an arginine transfer RNA. Cell 45:453-459. [DOI] [PubMed] [Google Scholar]
- 9.Ghisotti, D., S. Zangrossi, and G. Sironi. 1983. An Escherichia coli gene required for bacteriophage P2-λ interference. J. Virol. 48:616-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haggård-Ljungquist, E., V. Barreiro, R. Calendar, D. M. Kurnit, and H. Cheng. 1989. The P2 old gene: sequence, transcription and translational control. Gene 85:25-33. [DOI] [PubMed] [Google Scholar]
- 11.Hama, C., T. Takizawa, H. Moriwaki, Y. Urasaki, and K. Mizobuchi. 1990. Organization of the replication control region of plasmid ColIb-P9. J. Bacteriol. 172:1983-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henson, J. M., H. Chu, C. A. Irwin, and J. R. Walker. 1979. Isolation and characterization of dnaX and dnaY temperature-sensitive mutants of Escherichia coli. Genetics 92:1041-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horiuchi, T., and T. Nagata. 1973. Mutations affecting growth of the Escherichia coli cell under a condition of DNA polymerase I-deficiency. Mol. Gen. Genet. 123:89-110. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda, H., and J. Tomizawa. 1965. Transducing fragments in generalized transduction by phage P1. I. Molecular origin of the fragments. J. Mol. Biol. 14:85-109. [DOI] [PubMed] [Google Scholar]
- 15.Ikemura, T. 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 146:1-21. [DOI] [PubMed] [Google Scholar]
- 16.Kanaya, S., Y. Yamada, Y., Kudo, and T. Ikemura. 1999. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238:143-155. [DOI] [PubMed] [Google Scholar]
- 17.Kiesewetter, S., W. Fischer, and M. Sprinzl. 1987. Sequences of three minor tRNAArg from E. coli. Nucleic Acids Res. 15:3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiga, D., K. Sakamoto, K. Kodama, T. Kigawa, T. Matsuda, T. Yabuki, M. Shirouzu, Y. Harada, H. Nakayama, K. Takio, Y. Hasegawa, Y. Endo, I. Hirao, and S. Yokoyama. 2002. An engineered Escherichia coli tyrosyl-tRNA synthetase for site-specific incorporation of an unnatural amino acid into proteins in eukaryotic translation and its application in a wheat germ cell-free system. Proc. Natl. Acad. Sci. USA 99:9715-9723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kogoma, T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61:212-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohara, Y., K. Akiyama, and K. Isono. 1987. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell 50:495-508. [DOI] [PubMed] [Google Scholar]
- 21.Lin, S. X., J. P. Shi, X. D. Cheng, and Y. L. Wang. 1988. Arginyl-tRNA synthetase from Escherichia coli, purification by affinity chromatography, properties, and steady-state kinetics. Biochemistry 27:6343-6348. [DOI] [PubMed] [Google Scholar]
- 22.Louie, A., S. Ribeiro, B. R. Reid, and F. Jurnak. 1984. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J. Biol. Chem. 259:5010-5016. [PubMed] [Google Scholar]
- 23.McClain, W. H., and K. Foss. 1988. Changing the acceptor identity of a transfer RNA by altering nucleotides in a “variable pocket.” Science 241:1804-1807. [DOI] [PubMed] [Google Scholar]
- 24.McClain, W., H., K. Foss, R. A. Jenkins, and J. Schneider. 1990. Nucleotides that determine Escherichia coli tRNAArg and tRNALys acceptor identities revealed by analyses of mutant opal and amber suppresser tRNAs. Proc. Natl. Acad. Sci. USA 87:9260-9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullin, D. A., G. M. Garcia, and J. R. Walker. 1984. An E. coli DNA fragment 118 base pairs in length provides dnaY+ complementing activity. Cell 37:669-674. [DOI] [PubMed] [Google Scholar]
- 26.Murakami, Y., H. Ohmori, T. Yura, and T. Nagata. 1987. Requirement of the Escherichia coli dnaA gene function for ori-2-dependent mini-F plasmid replication. J. Bacteriol. 169:1724-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagata, T., Y. Murakami, and M. Imai. 1988. Requirement of the Escherichia coli dnaA gene function for integrative suppression of dnaA mutations by plasmid R100-1. Mol. Gen. Genet. 213:163-165. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nissen, P., M. Kjeldgaard, S. Thirup, G. Polekhnica, L. Reshetnikova, B. F. C. Clark, and J. Nyborg. 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270:1464-1472. [DOI] [PubMed] [Google Scholar]
- 30.Nureki, O., T. Kohno, K. Sakamoto, T. Miyazawa, and S. Yokoyama. 1993. Chemical modification and mutagenesis studies on zinc binding of aminoacyl-tRNA synthetase. J. Biol. Chem. 268:15368-15373. [PubMed] [Google Scholar]
- 31.O'Mahoney, D. J., B. H. Mims, S. Thompson, E. J. Murgola, and J. F. Atkins. 1989. Glycine tRNA mutants with normal anticodon loop size cause −1 frameshifting. Proc. Natl. Acad. Sci. USA 86:7979-7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pingoud, A., C. Urbanke, G. Krauss, F. Peters, and G. Maas. 1977. Ternary complex formation between elongation factor Tu, GTP, and aminoacyl-tRNA: an equilibrium study. Eur. J. Biochem. 78:403-409. [DOI] [PubMed] [Google Scholar]
- 33.Rould, M. A., J. J. Perona, D. Söll, and T. A. Steitz. 1989. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 Å resolution. Science 246:1135-1142. [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto, K., G. Kawai, T. Niimi, T. Satoh, M. Sekine, Z. Yamaizumi, S. Nishimura, T. Miyazawa, and S. Yokoyama. 1993. A modified uridine in the first position of the anticodon of a minor species of arginine tRNA, the argU gene product, from Escherichia coli. Eur. J. Biochem. 216:369-375. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulman, L. H., and H. Pelka. 1988. The anticodon contains a major element of the identity of arginine transfer RNA. Science 246:1595-1597. [DOI] [PubMed] [Google Scholar]
- 38.Schulman, L. H., H. Pelka, and R. M. Sundari. 1974. Structural requirements for recognition of Escherichia coli initiator and non-initiator transfer ribonucleic acids by bacterial T factor. J. Biol. Chem. 249:7102-7110. [PubMed] [Google Scholar]
- 39.Seong, B., and U. L. RajBhandary. 1987. Mutants of Escherichia coli formylmethionine tRNA: a single base change enables initiator tRNA to act as an elongator in vitro. Proc. Natl. Acad. Sci. USA 84:8859-8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharp, P., and W.-H. Li. 1986. Codon usage in regulatory genes in Escherichia coli does not reflect selection for ‘rare’ codons. Nucleic Acids Res. 14:7737-7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimada, A., O. Nureki, M. Goto, S. Takahashi, and S. Yokoyama. 2001. Structural and mutational studies of the recognition of the arginine tRNA-specific major identity element, A20, by arginyl-tRNA synthetase. Proc. Natl. Acad. Sci. USA 98:13537-13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimura, Y., and H. Ozeki. 1973. Genetic study on transfer RNA. Adv. Biophys. 4:191-226. [PubMed] [Google Scholar]
- 43.Smith, J. D., and J. E. Celis. 1973. Mutant tyrosine transfer RNA that can be charged with glutamine. Nat. New Biol. 243:66-71. [PubMed] [Google Scholar]
- 44.Spanjaard, R. A., K. Chen, J. R. Walker, and J. van Duin. 1990. Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: assigning a codon to argU tRNA and T4 tRNAArg. Nucleic Acids Res. 18:5031-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura, K., H. Himeno, H. Asahara, T. Hasegawa, and M. Shimizu. 1992. In vitro study of E. coli tRNAArg and tRNALys identity elements. Nucleic Acids Res. 20:2335-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varshney, U., C.-P. Lee, and U. L. RajBhandary. 1991. Direct analysis of aminoacylation levels of tRNAs in vivo. J. Biol. Chem. 266:24712-24718. [PubMed] [Google Scholar]
- 47.Wagner, T., and M. Sprinzl. 1980. The complex formation between Escherichia coli aminoacyl-tRNA, elongation factor Tu and GTP. Eur. J. Biochem. 108:213-221. [DOI] [PubMed] [Google Scholar]