Abstract
Rickettsia typhi, the causative agent of murine typhus, is an obligate intracellular bacterium with a life cycle involving both vertebrate and invertebrate hosts. Here we present the complete genome sequence of R. typhi (1,111,496 bp) and compare it to the two published rickettsial genome sequences: R. prowazekii and R. conorii. We identified 877 genes in R. typhi encoding 3 rRNAs, 33 tRNAs, 3 noncoding RNAs, and 838 proteins, 3 of which are frameshifts. In addition, we discovered more than 40 pseudogenes, including the entire cytochrome c oxidase system. The three rickettsial genomes share 775 genes: 23 are found only in R. prowazekii and R. typhi, 15 are found only in R. conorii and R. typhi, and 24 are unique to R. typhi. Although most of the genes are colinear, there is a 35-kb inversion in gene order, which is close to the replication terminus, in R. typhi, compared to R. prowazekii and R. conorii. In addition, we found a 124-kb R. typhi-specific inversion, starting 19 kb from the origin of replication, compared to R. prowazekii and R. conorii. Inversions in this region are also seen in the unpublished genome sequences of R. sibirica and R. rickettsii, indicating that this region is a hot spot for rearrangements. Genome comparisons also revealed a 12-kb insertion in the R. prowazekii genome, relative to R. typhi and R. conorii, which appears to have occurred after the typhus (R. prowazekii and R. typhi) and spotted fever (R. conorii) groups diverged. The three-way comparison allowed further in silico analysis of the SpoT split genes, leading us to propose that the stringent response system is still functional in these rickettsiae.
The rickettsiae are small (ca. 0.4 by 0.9 μm), gram-negative, aerobic, coccobacillary, α-proteobacteria. They are obligate intracellular parasites with a life cycle that involves both vertebrate and invertebrate hosts. Rickettsiae depend on hematophagous arthropods as vectors and primary reservoir, although small mammals, such as rats and opossums, also serve as amplifying hosts (8, 10). Rickettsia are classified into two groups; the spotted fever group (SFG), which includes R. conorii, R. sibirica, and R. rickettsii, and the typhus group (TG), which includes R. prowazekii and R. typhi. Two rickettsiae (R. prowazekii and R. rickettsii) are on the list of agents of the Antiterrorism and Effective Death Penalty Act of 1996 because they are stable, highly infectious agents that cause severe disease (7). Both Japan, during World War II, and the Soviet Union, during the Cold War, investigated the use of rickettsiae as biological weapons (9, 35, 65).
Rickettsia typhi is the causative agent of murine typhus (endemic typhus). Infection with R. typhi causes fever, headache, and myalgia and leads to disseminated, multisystem disease, including infection of the brain, lung, liver, kidney, and heart endothelia, lymphohistiocytic vasculitis of the central nervous system, diffuse alveolar damage and hemorrhage, interstitial pneumonia, pulmonary edema, interstitial myocarditis and nephritis, portal triaditis, and cutaneous, mucosal, and serosal hemorrhages (63, 64). The nonspecificity and nonuniformity of symptoms and the lack of specific diagnostic tests that are effective during the acute stage of the illness often lead to misdiagnosis, delaying appropriate treatment. Although the mortality rate is low (1% of reported cases), in severe cases R. typhi can cause meningoencephalitis, interstitial pneumonia, and disseminated vascular lesions (12). Without specific treatment, 99% of those infected will clear the disease within weeks, making proper accounting of R. typhi infections difficult (28).
Although R. typhi can be transmitted to the mammalian host by the bite of an infected flea or louse (the rat flea Xenopsylla cheopis, the cat flea Ctenocephalides felis, or the rat louse Polyplax spinulos), the more important mechanism of inoculation is through the feces of the vector. R. typhi multiplies in the epithelium of the flea midgut, is shed in the feces, and is then deposited during feeding. Organisms in the feces enter the host through irritated, abraded skin. The bacterium is then hematogenously spread and ultimately invades endothelial cells (12). Transmission can also occur via inhalation of aerosolized fecal particles. To enter the host cell, R. typhi induces phagocytosis by an unknown mechanism. Once within the cell, the organisms rapidly escape the phagosome, multiply within the cytoplasm, and then exit the host cell by burst lysis, allowing subsequent spread to other endothelial cells (65).
R. typhi is one of the leading causes of rickettsioses in the world. Although distributed worldwide, it is most common in warm coastal areas with large rat populations (12). Prior to World War II, 2,000 to 5,000 cases were reported annually in the United States. Due to intensive efforts to control rat fleas, as well as rat populations, the number of incidences has dropped to 9 to 72 cases per year from 1980 to 1998 (12, 28). Outbreaks continue to occur both around the world and in the United States. Murine typhus is considered endemic to the Hawaiian Islands with five to six cases per year reported. However, 47 cases were reported in 2002 leading the Hawaii Department of Health to institute active surveillance measures and mandatory reporting of R. typhi test requests. Due to the nonspecific clinical signs associated with murine typhus, it is expected that the actual number of cases was greater than those reported (40).
Rickettsial organisms have comparatively small genomes (1.1 to 1.3 Mb) that have arisen through reductive evolution as they developed dependence on the host cell for necessary functions (6). As a result, their genomes are littered with pseudogenes. The rickettsiae have a close evolutionary relationship with the progenitor of the mitochondria (5). The genomes of R. prowazekii (1.11 Mb) Madrid E, an attenuated strain of the causative agent of epidemic typhus, and R. conorii (1.27 Mb), the causative agent of boutonneuse fever and a member of the SFG of rickettsiae, have been sequenced previously (5, 52). A draft genome of R. sibirica, the causative agent of North Asian tick typhus and a member of the SFG, has recently been deposited in GenBank (accession no. NZ_AABW01000001) as has the unpublished complete sequence of R. rickettsii (accession number AADJ01000001).
We present here the complete genome of R. typhi and compare it to previously completed rickettsial genomes. We anticipate that the complete genome sequence of R. typhi will enhance the opportunities for investigation of virulence factors, pathogenesis, attenuation, and novel targets for antimicrobial therapy or blocking of pathogenic pathways. Of the five rickettsial genomes now available, this is only the second in the TG, the other three being SFG organisms. This second TG sequence will allow insight into phenotypic differences between R. prowazekii and R. typhi by genome comparison. This is likely to yield important leads regarding the study of pathogenicity since the case-fatality rate of louse-borne typhus fever is an order of magnitude greater than that of murine typhus.
MATERIALS AND METHODS
Bacterial growth and DNA isolation.
R. typhi strain Wilmington (ATCC VR-144) was cultivated in the yolk sacs of chicken embryos. The rickettsial inoculum was adjusted so that most embryos died ca. 4 to 5 days after inoculation. Infected eggs were incubated at 37°C until the embryos died. The yolk sacs were then harvested and frozen at −80°C for later isolation of bacteria. The yolk sacs were homogenized and diluted in 10 volumes of sucrose phosphate glutamate (SPG) buffer (0.22 M sucrose, 0.01 M potassium phosphate [pH 7.0], 0.005 M potassium glutamate) (16). The suspension was centrifuged at 200 × g for 10 min to remove debris, and then the supernatant was centrifuged at 10,000 × g for 20 min to pellet the rickettsial cells. The aqueous phase and fat layer were discarded, and the pellet was resuspended in SPG buffer. The suspension was sonicated three times at 40 W for 10 s on ice. The bacteria were further purified by two rounds of discontinuous renografin gradient centrifugation by using 32, 36, and 42% steps (66) at 87,275 × g for 60 min. Light bands containing intact organisms were collected and mixed with 2 volumes of SPG. The suspension was centrifuged at 11,400 × g for 20 min, and the pellet containing the purified rickettsiae was suspended in SPG buffer for storage. The rickettsial DNA was extracted from the purified bacteria by using an IsoQuick nucleic acid extraction kit (ORCA Research, Inc., Bothell, Wash.) according to the manufacturer's instructions.
Library construction and sequencing.
Genomic DNA was sheared to a size of ∼2 kb by nebulization (CIS-US, Inc., Bedford, Mass.) and cloned into a derivative of pUC18 by the double adapter method (2). These clones were used for whole genome shotgun DNA sequencing by using dye terminator chemistry, and data were collected on ABI 3700 sequencers (Applied Biosystems, Foster City, Calif.).
Sequence assembly.
The quality for each base sequenced was determined by PHRED (26, 27). Initial assembly of 21,831 whole-genome shotgun reads by using PHRAP (P. Green, unpublished data) gave 78 contigs. Linkages among contigs were built based on the read-pair information, and gaps between contigs were filled by template walking with the existing clone templates. Template walking reduced the assembly to a total of 17 contigs. Further ordering of the contigs was accomplished by comparison of the contigs to the finished genomes of R. prowazekii and R. conorii. Gaps between contigs were then closed by sequencing products from PCRs across these regions. Low-quality regions on the contigs were sequenced by template walking or by sequencing of PCR products. In the final assembly, the quality of each base was greater than or equal to a PHRAP value of 30. The average PHRAP value for the entire genome is 87.6.
Identical repeats in the R. typhi genome were detected by REPFIND (14), and BLASTN (1) was used for the identification of nonidentical repeats. A 124-kb inversion of the R. typhi sequence was observed compared to the finished genomes of R. prowazekii and R. conorii. The inversion was examined by PCR with two pairs of primers across each junction region and confirmed to be authentic. In addition, the frameshift within dnaX (RT0856) and a premature stop codon within the tRNA nucleotidyltransferase gene (RT0619) were verified by resequencing the regions.
Gene identification and annotation.
Glimmer (23) and GeneMark (48) were used independently to predict open reading frames (ORFs). For GeneMark, we used the complete genomes of R. conorii and R. prowazekii as training models. We also used BLASTX to search for proteins that the hidden Markov model programs missed. This proved advantageous since it allowed us to discover pseudogenes not predicted by the other programs. The annotation process was performed in four stages. The first was an automated step, in which we compared the entire sequence to many different databases including GenBank nr (67), SwissProt (15), COGs (58), EXProt (61), and Pfam (11). These results were then loaded into a database for use by annotators. The next stage was a double-blind annotation in which regions of the genome were assigned to different annotators for analysis. After each ORF had been annotated by two different persons, we reconciled the two annotations. This was followed by a final review and cleanup of the annotation. Visualization of gene predictions and sequence markers was performed by using the Genboree system (www.genboree.org).
We created a set of rules to keep the annotation as consistent as possible since 10 individuals were involved. First, each identified region was assigned a class: rRNA, tRNA, ncRNA, protein, frameshift, pseudogene, other, sequencing error, or error. An ORF was considered a frameshift if a single base change would restore the frame to encode a complete a protein that matched a known protein. Pseudogenes were defined as regions with one or more stops, typically in-frame, that interrupted the reading frame. These were typically detected by BLASTX, and the ends were identified by alignment of the DNA with the analogous regions in R. conorii and R. prowazekii. The “other” class was used as a flag for the annotator to reinspect the ORF and seek assistance in determining gene organization or function. The sequencing error class was used when the sequence quality was low, leading to ambiguity, or when additional sequencing of a region was required. The class “error” (not used in this study) was to be used where our data conflicted with known biological data. For proteins with an identified biochemical function and an Enzyme Commission (EC) number, we used the official EC name as provided by ExPasy (http://us.expasy.org/). An example were the tRNA amino acid ligases, which are commonly known as amino acyl-tRNA synthetases. We also included known alternative gene names to allow for text searching on previously used names. Additional descriptive information was added when useful (e.g., a subunit designation). We used the term subunit instead of chain in all cases of multiprotein functional units. For proteins where there was not enough evidence to be certain of the designation, we chose to use the terms possible and probable (as opposed to putative, predicted, etc.) to designate those that we believed were likely to be correct (probable) and those for which we were less confident (possible). Gene names followed the Demerec standard (24), except for duplicated proteins where the gene name was followed by a number to indicate its order in the genome (e.g., iscA1 and iscA2) or in cases where meaning would be lost by removing the number that had been added by previous groups (e.g., virBN and scaN). Genes whose closest nonrickettsial matches were eukaryotic genes were designated “XYZ-like.”
Predicted proteins with unknown functions were placed in one of three categories. A “hypothetical protein” is an ORF that has no database matches in any other organism. A “conserved hypothetical protein” is defined as an ORF that has matches to other proteins of unknown function in organisms outside of the rickettsiae. Proteins that have significant matches only to other rickettsial sequences are labeled “rickettsial conserved hypothetical proteins.”
Database submission.
The annotated R. typhi genome sequence has been deposited in GenBank and assigned accession number AE017197. The annotated genome and supplemental data are also available at http://www.hgsc.bcm.tmc.edu/microbial/Rtyphi.
RESULTS
Genome anatomy.
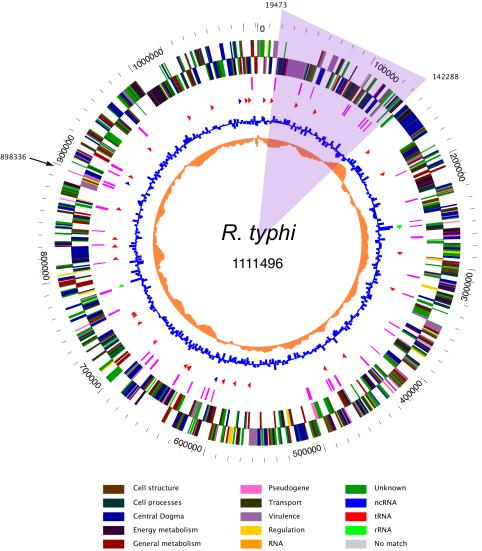
The genome of R. typhi is a single circular chromosome consisting of 1,111,496 bp with an average G+C content of 28.9% (Table 1). The origin of replication (near base pair number 1) was predicted based on this region being one of the switching points of G+C skew (Fig. 1) and by searching for potential DnaA-binding sequences. In addition, this region contains sequences found at replication origins of other bacteria (see below). A 124-kb inversion with respect to the R. prowazekii and R. conorii genomes (see below), shaded in Fig. 1, disrupts the continuity of the appearance of the G+C skew, making this criterion less clear than in other bacterial genomes. As in the other sequenced rickettsiae, the putative origin of replication in R. typhi is not located at the dnaA region (coordinate 762,664) but lies ca. 350 kb from dnaA between uroporphyrinogen decarboxylase (hemE) and a conserved hypothetical protein gene.
TABLE 1.
Comparison of genome statistics between R. typhi, R. prowazekii, R. conorii, R. sibirica, and R. rickettsii
| Organism | Genome size (bp) | No. of genes | Coding region (% of genome)a | % G+Cb for the:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Genome | Coding regions | Noncoding regions | First codon position | Second codon position | Third codon position | ||||
| R. typhi | 1,111,496 | 877 | 76.27 | 28.92 | 30.56 | 23.55 | 41.11 | 31.69 | 18.39 |
| R. prowazekii | 1,111,523 | 872 | 76.24 | 29.00 | 30.59 | 23.61 | 41.07 | 31.75 | 18.44 |
| R. conorii | 1,268,755 | 1,412 | 81.45 | 32.44 | 32.98 | 30.05 | 42.53 | 32.42 | 23.58 |
| R. sibiricac | 1,250,021 | 1,234 | 77.76 | 32.47 | 32.90 | 30.94 | 42.85 | 32.50 | 23.35 |
| R. rickettsiid | 1,257,710 | NA | NA | 32.47 | NA | NA | NA | NA | NA |
The coding regions include ORFs for proteins, rRNAs, tRNAs and ncRNAs except for R. sibirica.
The G+C content for the first, second, and third codon positions was calculated from protein-encoding ORFs.
The numbers calculated for R. sibirica are based on available protein-encoding ORFs (accession no. NZ_AABW01000001) only. They do not include ORFs for rRNA, tRNA, or snRNA, which are not available.
The annotations for R. rickettsii (accession no. AADJ01000001) are not available (NA) in GenBank.
FIG. 1.
Circular genome map of R. typhi. The outer circle shows the predicted coding regions on the plus strand, color-coded by functional category. The second circle shows the predicted coding regions on the minus strand. The third circle shows the location of pseudogenes. The fourth circle shows tRNAs (red), rRNAs (blue), and ncRNAs (green). The fifth circle shows the %G+C content. The innermost circle shows the G+C skew. The shaded region illustrates the inversion with respect to R. prowazekii, while the arrow denotes the 12-kb insertion in R. prowazekii. Base pair 1 was set at the first base pair of the first ORF after the origin of replication.
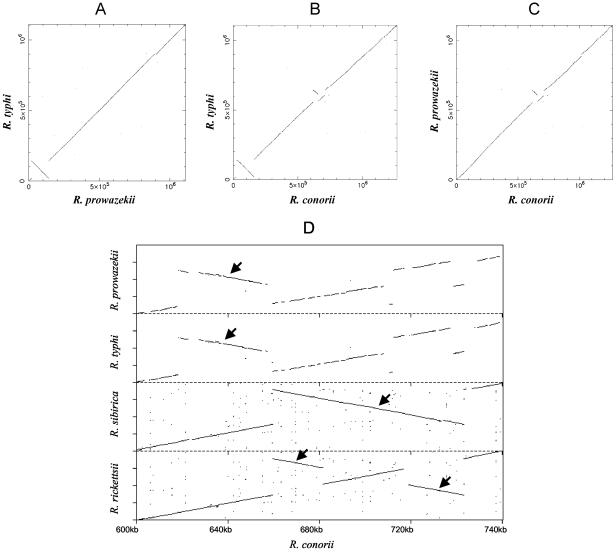
The region from 19 to 142 kb in the R. typhi genome is inverted with respect to R. prowazekii and R. conorii (Fig. 2) (5, 52) and to R. sibirica and R. rickettsii (results not shown). The latter three strains are all in the SFG, whereas R. prowazekii, like R. typhi, is in the TG. Thus, it is likely this is a recent inversion, occurring after the divergence of R. typhi and R. prowazekii. No repetitive sequences were found near the junctions of the inversion, and no gene was deleted in the inverted region or in the flanking junction regions.
FIG. 2.
Nucleotide alignment among R. typhi, R. prowazekii, R. conorii, R. sibirica, and R. rickettsii. Each line represents an alignment obtained by BLASTN (E ≤ 1e−10). (A) Nucleotide alignment between R. typhi and R. prowazekii genomes; (B) nucleotide alignment between R. typhi and R. conorii genomes; (C) nucleotide alignment between R. prowazekii and R. conorii genomes; (D) nucleotide alignment of the region from 600 to 740 kb of R. conorii with the corresponding regions of R. prowazekii, R. typhi, R. sibirica, and R. rickettsii. Inversions are indicated by arrows.
A second region of multiple rearrangements was located from 612 to 644 kb (Fig. 2). This region is similar in the two TG bacteria but differs from the SFG bacteria. Thus, the inversion likely occurred after the divergence of TG and SFG rickettsiae but before the divergence of R. typhi and R. prowazekii. Analysis of the inversion junction regions in the TG bacteria showed one short repeat in R. typhi (33 bp [three copies]) and two short repeats in R. prowazekii (46 and 28 bp [two and three copies, respectively]). Although the TG had a similar structure in this region, the SFG bacteria were all different, indicating continued rearrangements occurring after their divergence (Fig. 2). The R. rickettsii region could have been derived by an additional inversion from the R. sibirica region, suggesting that it was the most recent derivative. Comparison of R. sibirica and R. conorii shows an 81-kb inversion in the 660- to 743-kb region of R. conorii which contains 47 ORFs. This region is next to where the TG bacteria have inversions (corresponding to 618 to 657 kb on R. conorii), indicating that this region is a hot spot for rearrangements. Examination of the inversion junction regions in R. conorii revealed three repeats (114, 73, and 30 bp [two, three, and two copies, respectively]) in the junction region at around 618 kb and seven repeats (114, 96, 92, 74, 54, 42, and 33 bp [55, 62, 44, 22, 3, 11, and 4 copies, respectively]) in the junction region at around 657 kb. The sequences of these repeats do not show sequence similarity to any known transposon or IS sequence. It is possible that repeats at the inversion junction regions contributed to the inversions that occurred in R. typhi, R. prowazekii, and R. sibirica.
Further comparison of the TG bacteria shows R. typhi is missing a 12-kb segment of DNA (at 898 kb) compared to R. prowazekii (Fig. 2). As a result, nine R. prowazekii ORFs (RP709 to RP717) are not present in the R. typhi chromosome. This region is also absent from R. conorii (Fig. 2), as well as R. sibirica and R. rickettsii (not shown). This unique R. prowazekii DNA may have been acquired by horizontal transfer, since three of the R. prowazekii pseudogenes within this region (RP710, RP711, and RP717) share similarity with transposon proteins.
Like R. prowazekii, the R. typhi genome has very few repetitive sequences (5). Five major classes of repeats were found in the R. typhi genome. The repeated sequences in R. typhi and R. prowazekii have similarities in sequence and locations within their genomes, although they are not similar to other known repeat sequences.
Proteome comparisons.
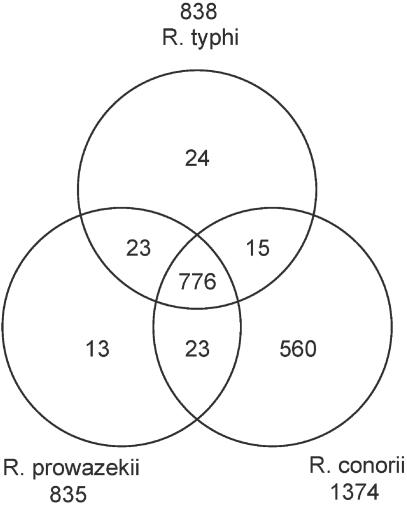
Most predicted proteins in the R. typhi genome have matches to ORFs in R. prowazekii and R. conorii (Fig. 3), so the exceptions (Table 2) deserve attention in determining the distinguishing characteristics of these three organisms. Most of the unique ORFs encode proteins of unknown function. A total of 24 R. typhi ORFs do not match any ORFs in either R. conorii or R. prowazekii. All but two of these are hypothetical proteins with no database matches. One is a rickettsial conserved hypothetical protein (RT0301) that had been previously annotated in R. typhi (accession number CAC33749) and matches hypothetical proteins in R. rickettsii and R. montanensis. The other is a conserved hypothetical protein found within the 23S rRNA gene that matches only a cell-wall-associated hydrolase of Vibrio vulnificus.
FIG. 3.
Shared proteins between R. typhi, R. prowazekii and R. conorii. A total of 776 proteins can be found in all three organisms; 24 are unique to R. typhi, 23 are shared only between R. typhi and R. prowazekii, and 15 are found only in R. typhi and R. conorii.
TABLE 2.
Genes with restricted distribution in rickettsial genomes
| Category and RT no. | Function | Position
|
Gene | Description | RP no. | RC no. | |
|---|---|---|---|---|---|---|---|
| Start | Stop | ||||||
| R. typhi ORFs not found in R. conorii or R. prowazekii | |||||||
| RT0148 | Unknown | 189321 | 189461 | None | Hypothetical protein | None | None |
| RT0154 | Unknown | 195647 | 195811 | None | Hypothetical protein | None | None |
| RT0155 | Unknown | 196020 | 196154 | None | Hypothetical protein | None | None |
| RT0184 | Unknown | 233126 | 233230 | None | Hypothetical protein | None | None |
| RT0199 | Unknown | 255711 | 255848 | None | Hypothetical protein | None | None |
| RT0201 | Unknown | 258437 | 258853 | None | Conserved hypothetical protein | None | None |
| RT0301 | Unknown | 382095 | 383057 | None | Rickettsial conserved hypothetical protein | None | None |
| RT0364 | Unknown | 461966 | 462193 | None | Hypothetical protein | None | None |
| RT0418 | Unknown | 529419 | 529619 | None | Hypothetical protein | None | None |
| RT0480 | Unknown | 614027 | 614167 | None | Hypothetical protein | None | None |
| RT0499 | Unknown | 640260 | 640412 | None | Hypothetical protein | None | None |
| RT0505 | Unknown | 645384 | 645518 | None | Hypothetical protein | None | None |
| RT0525 | Unknown | 668721 | 668858 | None | Hypothetical protein | None | None |
| RT0538 | Unknown | 686867 | 687124 | None | Hypothetical protein | None | None |
| RT0542 | Unknown | 694039 | 694113 | None | Hypothetical protein | None | None |
| RT0601 | Unknown | 778825 | 778974 | None | Hypothetical protein | None | None |
| RT0607 | Unknown | 790366 | 790560 | None | Hypothetical protein | None | None |
| RT0664 | Unknown | 841537 | 841782 | None | Hypothetical protein | None | None |
| RT0685 | Unknown | 868875 | 868988 | None | Hypothetical protein | None | None |
| RT0708 | Unknown | 906635 | 906880 | None | Hypothetical protein | None | None |
| RT0751 | Unknown | 955938 | 956084 | None | Hypothetical protein | None | None |
| RT0772 | Unknown | 984575 | 984706 | None | Hypothetical protein | None | None |
| RT0796 | Unknown | 1018722 | 1018862 | None | Hypothetical protein | None | None |
| RT0835 | Unknown | 1066262 | 1066381 | None | Hypothetical protein | None | None |
| R. typhi ORFs found in R. conorii but not in R. prowazekii | |||||||
| RT0034 | Virulence | 47136 | 47423 | virB3 | Probable type IV secretory system protein VirB3 | None | RC0140 |
| RT0172 | General metabolism | 217326 | 218498 | None | Carboxypeptidase | None | RC0228 |
| RT0213 | Unknown | 275036 | 275173 | None | Rickettsial conserved hypothetical protein | None | RC0298 |
| RT0270 | Unknown | 341040 | 341273 | None | Rickettsial conserved hypothetical protein | None | RC1041 |
| RT0314 | Unknown | 397194 | 397355 | None | Rickettsial conserved hypothetical protein | None | RC0444 |
| RT0373 | Unknown | 474114 | 474401 | None | Rickettsial conserved hypothetical protein | None | RC0530 |
| RT0520 | Unknown | 660968 | 661153 | None | Rickettsial conserved hypothetical protein | None | RC0767 |
| RT0563 | Unknown | 728779 | 728994 | None | Rickettsial conserved hypothetical protein | None | RC0876 |
| RT0610 | Unknown | 794494 | 794634 | None | Rickettsial conserved hypothetical protein | None | RC0961 |
| RT0623 | Unknown | 813926 | 814159 | None | Rickettsial conserved hypothetical protein | None | RC0453 |
| RT0665 | Unknown | 842115 | 842354 | None | Rickettsial conserved hypothetical protein | None | RC1041 |
| RT0670 | Unknown | 848273 | 848619 | None | Rickettsial conserved hypothetical protein (frameshift 848487-848490) | None | RC1027 |
| RT0672 | Unknown | 850822 | 851025 | None | Rickettsial conserved hypothetical protein | None | RC1029 |
| RT0742 | Unknown | 941930 | 942133 | None | Rickettsial conserved hypothetical protein | None | RC1172 |
| RT0872 | Unknown | 1104639 | 1104791 | None | Rickettsial conserved hypothetical protein | None | RC1362 |
| R. typhi ORFs found in R. prowazekii but not in R. conorii | |||||||
| RT0026 | Energy metabolism | 27080 | 28237 | ackA | Acetate kinase | RP110 | None |
| RT0033 | Virulence | 44546 | 46963 | virB4a | VirB4 protein precursor | RP103 | None |
| RT0035 | Unknown | 48098 | 48742 | engB | Probable GTP-binding protein EngB | RP102 | None |
| RT0156 | Unknown | 196185 | 196379 | None | Rickettsial conserved hypothetical protein | RP164 | None |
| RT0160 | Unknown | 199110 | 199382 | None | Rickettsial conserved hypothetical protein | RP169 | None |
| RT0268 | Unknown | 337676 | 338362 | None | Rickettsial conserved hypothetical protein | RP277 | None |
| RT0269 | Unknown | 338385 | 339527 | None | Rickettsial conserved hypothetical protein | RP278 | None |
| RT0325 | Unknown | 412632 | 413753 | None | Rickettsial conserved hypothetical protein | RP335 | None |
| RT0326 | General metabolism | 413782 | 415005 | None | Probable glycosyltransferase | RP337 | None |
| RT0327 | Unknown | 415002 | 416213 | None | Possible glycosyltransferase | RP337 | None |
| RT0328 | Unknown | 416200 | 417237 | None | Conserved hypothetical protein | RP338 | None |
| RT0330 | Virulence | 418166 | 419182 | None | WaaG-like sugar transferase | RP340 | None |
| RT0341 | Unknown | 434960 | 435208 | None | Conserved hypothetical protein | RP351a | None |
| RT0362 | Virulence | 458514 | 459878 | None | Sec7-domain containing protein transport protein | RP374 | None |
| RT0426 | Unknown | 535490 | 536665 | None | Rickettsial conserved hypothetical protein | RP439 | None |
| RT0463 | Virulence | 593138 | 594691 | None | Possible lipopolysaccharide 1,2-glucosyltransferase WaaJ | RP476 | None |
| RT0549 | Transport | 710632 | 710895 | None | Possible Na+/H+ antiporter | RP561a | None |
| RT0615 | Regulation | 804351 | 805103 | spoTb | Guanosine-3,5-bis(diphosphate) 3-pyrophosphohydrolase SpoTb | PG624 | None |
| RT0616 | Regulation | 805109 | 805450 | spoTc | Guanosine-3,5-bis(diphosphate) 3-pyrophosphohydrolase SpoTc | PG625 | None |
| RT0710 | Unknown | 907724 | 908002 | None | Rickettsial conserved hypothetical protein | RP723 | None |
| RT0764 | Central dogma | 968047 | 969195 | metK | Methionine adenosyltransferase | RP777 | None |
| RT0793 | Unknown | 1012772 | 1013206 | None | Conserved hypothetical protein | RP806 | None |
| RT0808 | General metabolism | 1027687 | 1029441 | phbC | Poly-β-hydroxybutyrate polymerase | RP820 | None |
Fifteen ORFs common to R. typhi and R. conorii are absent in R. prowazekii. Thirteen are conserved hypothetical proteins, one is a thermostable carboxypeptidase, and one is the type IV secretion system outer membrane component VirB3.
Twenty-three ORFs common to R. typhi and R. prowazekii are absent in R. conorii. Seven are rickettsial conserved hypothetical proteins, and three are conserved hypothetical proteins. The others include an acetate kinase, a GTP-binding protein EngB and two proteins likely involved in type IV secretion. Only one type IV secretion substrate-binding protein (VirB4) is found in R. conorii, but two were annotated in R. typhi, suggesting that R. typhi and R. prowazekii may transport a substrate that R. conorii does not. The Sec7 domain-containing protein, a possible substrate of the type IV secretion system, also has no R. conorii homolog. A group of three sugar transferases found within 5.4 kb of each other and possibly functioning in lipopolysaccharide synthesis were among the proteins with no R. conorii homolog. In addition, the SpoTb and SpoTc proteins, which were annotated as pseudogenes in R. prowazekii (see below), are absent in R. conorii. Finally, R. typhi and R. prowazekii share an Na+/H+ antiporter, a methionine adenosyltransferase, and a poly-β-hydroxybutyrate polymerase that are absent in R. conorii.
Seventeen hypothetical proteins that were present in the R. prowazekii genome but missing in R. typhi were identified. One contained an ankyrin domain (41), suggesting that it could provide a unique adhesive or structural feature to R. prowazekii. Sixteen additional R. prowazekii genes are present in R. typhi as pseudogenes. Eight of these are pseudogenes of hypothetical, conserved hypothetical, or rickettsial conserved hypothetical proteins. Six are pseudogenes of cytochrome c oxidase proteins (discussed below). The R. prowazekii Surf1 and Sco2 proteins were also found as pseudogenes in R. typhi.
The recently released whole-genome shotgun sequence of R. sibirica (accession number NZ_AABW01000001) was also compared to R. typhi, R. prowazekii, and R. conorii. R. sibirica was most similar to R. conorii in genome size, ORF sequences, codon G+C composition (Table 1), and ORF order. All predicted R. sibirica ORFs had hits to R. conorii ORFs. However, 131 R. conorii ORFs did not have a match in R. sibirica.
Noncoding RNA genes.
A total of 39 noncoding RNA genes have been identified, and a single set of rRNA genes was present. As in the other rickettsiae, the 16S rRNA gene was separated from the 23S and 5S rRNA genes. The coordinates were initially identified by locating highly conserved sequence and secondary structural features that are proximal to the terminus of each rRNA. Final extension to predicted start and stop sites relied on distances from these conserved features seen in Escherichia coli, where the ends have been experimentally verified. Although an exhaustive search was not undertaken, three additional noncoding RNA genes were identified. These were genes for tmRNA, which is associated with trans-translation, the RNA component of RNase P, and the 4.5S RNA, which is associated with the signal recognition particle. All of these genes are present in the other rickettsial genomes, but not all were annotated. Thirty-three tRNA genes were initially identified with tRNAscan-SE (47) and subsequently verified for compliance with known tRNA structural features. The R. typhi tRNA genes have annotated equivalents in the other rickettsiae, although the assigned amino acid acceptor is different in two cases (RT510 [Val-GAC versus RP523, Phe-GAA] and RT0757 [Met-CAT versus RP770, Ile-CAT]). All amino acids are represented, but no tRNA genes were found that would be expected to read the proline codons CCC and CCU. These codons are present in R. typhi, so it is possible that the rickettsiae have a proline tRNA (gene) of unusual structure. The three arginine tRNA genes have the anticodons ACG, CCG, and UCU. It is possible the A of the ACG anticodon may be converted to inosine to allow wobble reading of three codons (CGU, CGC, and CGA). Finally, there are three methionine tRNA genes. One of these was identified as the initiator based on the presence of the characteristic mispair in the CCA stem. Since no tRNA was found that could obviously decode AUA, it is possible that one of the methionine tRNAs may be an isoleucine tRNA.
Transcriptional control.
Like R. prowazekii and R. conorii, R. typhi encodes four subunits of RNA polymerase core enzyme (α, β, β′, and ω) and two predicted sigma factors, the major housekeeping sigma factor, σ70, and a heat shock sigma factor, σ32, encoded by rpoH. Madan Babu (49) suggested that loss of expressed sigma factor genes can initiate the accumulation of pseudogenes by eliminating entire sets of coregulated genes. No additional sigma factor-like sequences were detected in R. typhi, suggesting that most expressed genes are coregulated at this level.
No repressor-like proteins were found to be encoded by the R. typhi genome and a limited repertoire of other transcription factors that can respond to changes in the environment were present. The genome potentially contains four two-component signal transduction systems. An EnvZ-OmpR system (RT0412, RT0413), like those that function in osmoregulation in other bacteria, was found. An NtrY-NtrX-like two-component system (RT0603 and RT0550) involved in nitrogen metabolism in some organisms was also observed. The envZ-ompR genes are adjacent, as is common for such sensor kinase and response regulator systems, but the ntrY and ntrX genes are separated by more than 90 kb. Two additional, nonadjacent, response regulator genes and two sensor histidine kinase genes were also annotated in the R. typhi genome. One of the response regulators, CzcR (RT0061), is similar to the Caulobacter crescentus CtrA protein, which binds the Caulobacter origin of replication at TTAAN7TTAA sequences and represses DNA replication in swarmer cells (53). CtrA binds to similar sites in the R. prowazekii origin of replication and CzcR can complement CtrA function in C. crescentus (17). Presumably, the R. typhi CzcR protein has a similar DNA-binding activity since at least four TTAAN7TTAA-like sequences map within the putative origin of replication region. The histidine kinase partner for CzcR is uncharacterized but may be encoded by one of the two “unpaired” sensor kinases annotated in the R. typhi genome (RT0221 and RT0452). The last response regulator (RT0229) is similar to putative response regulators in the α-proteobacteria; its histidine kinase partner is also unknown. Beyond transcription initiation, R. typhi encodes proteins involved in transcript elongation (NusA, NusB, NusG, and GreA) and termination (Rho), as do R. prowazekii and R. conorii.
Pathogenesis.
Little is known about the genetic determinants required for the rickettsiae to invade a host and cause disease. Analysis of the finished genome has suggested 42 genes that may be involved in virulence (Table 3). These include hemolysins, enzymes that allow intracellular survival, and phospholipases that permit escape of the organism from the phagolysosome. Hemolysis is thought to play a role in the pathogenesis (53, 68) of TG but not SFG of rickettsiae. R. prowazekii has two potential hemolysin genes, tlyA and tlyC (5), and the cloned tlyC from R. typhi can complement hemolytic activity in a nonhemolytic Proteus mirabilis (54). Both tlyA (RT0543) and tlyC (RT0725) were found in R. typhi. Another protein implicated in the virulence of R. prowazekii is the dinucleoside polyphosphate hydrodolase, InvA. This protein is thought to function by hydrolyzing toxic dinucleoside oligophosphates within the host cell to produce ATP, thus providing an environment more conducive to growth (30, 31). ORF RT0228 is 98% identical to the R. prowazekii InvA. Oxidative stress due to reactive oxygen species has been implicated in both host cell injury caused by Rickettsia and in host defense to infection (65). ORF RT0523, sodB, is an ortholog of an iron-associated superoxide dismutase that is predicted to neutralize reactive oxygen species, allowing survival in the host cell. It has been suggested that the rickettsiae use phospholipase A2 to exit the phagosome and the cell (69). A putative phospholipase-like protein (RT0590) with significant similarity to the patatin family of proteins was found in the genome. Patatin is the main storage protein found in potatoes but has also been found to have phospholipase A activity (39, 43). It is possible that this protein acts as a phospholipase in R. typhi. In R. conorii, the pldA gene has been shown to encode a protein with phospholipase D activity (55). An ortholog of pldA is present in R. typhi (RT0807). Outer membrane proteins, which are in contact with the host milieu and could function in virulence, have also been identified. These include omp1 (RT0150), a predicted antigenic lipoprotein precursor nlpD (RT0291), and an ortholog of a Salmonella enterica serovar Typhimurium virulence factor mviN (RT0579) (20). Further, 12 lipopolysaccharide biosynthesis genes have been identified. Most are genes predicted to be involved in lipid A and core biosynthesis. Finally, we have identified 16 genes associated with a type IV secretion system and 5 genes of an autotransporter family that also may be involved in the pathogenicity of R. typhi. These latter groups of genes will be discussed in more detail below.
TABLE 3.
List of possible virulence factors
| RT no. | Gene | Function |
|---|---|---|
| RT0003 | wzt | O-antigen export system ATP-binding protein |
| RT0004 | wzm | O-antigen export system permease protein |
| RT0006 | lpxA | Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase |
| RT0008 | lpxD | UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase |
| RT0015 | sca1 | Cell surface antigen |
| RT0028 | VirB6-like protein of the type IV secretion system | |
| RT0029 | VirB6-like protein of the type IV secretion system | |
| RT0030 | VirB6-like protein of the type IV secretion system | |
| RT0031 | VirB6-like protein of the type IV secretion system | |
| RT0032 | VirB6-like protein of the type IV secretion system | |
| RT0033 | virB4a | VirB4 protein precursor |
| RT0034 | virB3 | Probable type IV secretory system protein VirB3 |
| RT0048 | waaA | 3-Deoxy-d-manno-octulosonic-acid transferase |
| RT0052 | sca2 | Cell surface antigen |
| RT0150 | omp1 | Outer membrane protein |
| RT0228 | invA | Dinucleoside polyphosphate hydrolase |
| RT0246 | lpxC | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase |
| RT0277 | trbG | Probable VirB9-like conjugal transfer protein precursor |
| RT0278 | VirB8-like protein of type IV secretion system | |
| RT0280 | VirB8-like protein of the Type IV secretion system | |
| RT0281 | virB9 | VirB9 protein precursor of the type IV secretion system |
| RT0282 | virB10 | VirB10 protein of the type IV secretion system |
| RT0283 | virB11 | VirB11 protein of the type IV secretion system |
| RT0284 | virD4 | VirD4 protein of the type IV secretion system |
| RT0291 | nlpD | Outer membrane antigenic lipoprotein B precursor |
| RT0311 | lpxB | Lipid-A-disaccharide synthase |
| RT0330 | WaaG-like sugar transferase | |
| RT0334 | WaaG-like glycosyltransferase | |
| RT0362 | Sec7-domain containing protein transport protein | |
| RT0438 | sca3 | Cell surface antigen |
| RT0463 | Possible lipopolysaccharide 1,2-glucosyltransferase WaaJ | |
| RT0485 | sca4 | Cell surface antigen |
| RT0523 | sodB | Superoxide dismutase |
| RT0543 | tlyA | Hemolysin A |
| RT0579 | MviN-like protein | |
| RT0590 | Patatin-like protein | |
| RT0699 | ompB | Rickettsial outer membrane protein B |
| RT0704 | lpxL | Lipid A biosynthesis lauroyl acyltransferase |
| RT0705 | lpxK | Tetraacyldisaccharide 4′-kinase |
| RT0725 | tlyC | Hemolysin C |
| RT0771 | virB4b | VirB4 protein precursor |
| RT0807 | pldA | Phospholipase D |
| RT0821 | Rickettsial 17-kDa surface antigen precursor |
Actin-based motility.
Previous studies (36) have shown that R. typhi undergoes actin-based intracellular motility like the SFG rickettsiae. However, R. typhi seems to be deficient (or inefficient) in its directional control of motility since its movement occurs in circular paths with many changes in speed and direction, unlike the SFG rickettsiae, which tend to move in straight paths with constant speeds (37). It has been shown that the actin tail formed by R. typhi is shorter and more hook-shaped than those formed by the SFG rickettsiae (62). Ogata et al. originally identified a protein in R. conorii (RC0909) that has a domain organization similar to the actin organizing protein, ActA, of Listeria monocytogenes, and they hypothesized that this protein may play a role in rickettsial actin tail formation (52, 69). Further work showed that this protein, now called RickA, can activate Arp2/3 and induce actin polymerization (33). We were unable to identify a homolog of RickA in the R. typhi genome, and the absence of this protein may explain the actin tail defects and inefficient motility seen in R. typhi.
Antibiotic resistance.
Doxycycline is the antibiotic of choice for treating R. typhi, since the organism is resistant to many other antibiotics, such as beta-lactams, gentamicin, and trimethoprim-sulfamethoxazole (56). Analysis of the genome has provided some potential mechanisms for resistance to these antibiotics, both specific and nonspecific.
One metallo-β-lactamase ortholog was found in R. typhi (RT0428) and in the other two finished rickettsial genome sequences. Rickettsiae produce a peptidoglycan cell wall containing penicillin-binding proteins (R. typhi has six penicillin-binding proteins) and show relative resistance to penicillin and other beta-lactam antimicrobial agents (50).
Several orthologs of multidrug transporters were found. An ortholog (RT0146) of the emrB multidrug transporter (46) was found in the R. typhi genome. In E. coli, EmrB confers resistance to nalidixic acid. It also shares sequence similarity with QacA, a multidrug-resistant pump of Staphylococcus aureus that belongs to a family that includes tetracycline resistance pumps of gram-positive bacteria. An ortholog of the EmrB regulator, EmrA, was also found in R. typhi, R. prowazekii, and R. conorii. It is unlikely that EmrB functions as a tetracycline pump since the rickettsiae are sensitive to tetracycline. Nor is it likely that it acts as a quinolone efflux pump since this class of antibiotics are moderately active against most rickettsiae (50). In addition, two copies of bcr gene orthologs (RT0591 and RT0693) were found each in R. typhi, R. prowazekii, and R. conorii. In E. coli, Bcr confers resistance to diketopiperazine antibiotics (13), usually used in therapy of Vibrio and Mycobacterium infections. Like EmrB, Bcr also resembles a multidrug transporter. Besides the potential EmrB and Bcr transporters, 10 additional potential ABC transporters were annotated in the R. typhi genome. Resistance to trimethoprim-sulfamethoxazole is likely due to the absence of enzymes of the folic acid pathways, since synthesis of folic and folinic acids are the target of these antibiotics. Gentamicin resistance is likely due to the fact that it remains extracellular, whereas the rickettsiae are intracellular pathogens.
Two other possible resistance genes, the acriflavin resistance gene (acrD, RT0161) and a tellurite resistance gene (terC, RT0774) were found in the R. typhi genome. These two genes are also present in R. prowazekii and R. conorii. Although the clinical relevance of these resistances is unknown, the possible presence of resistance to acriflavin (a mutagen) and tellurite (a reducing agent) in all three rickettsial genomes may suggest that they provide a selective advantage to the organism.
Autotransporters.
Autotransporters are proteins that are responsible for their own secretion across the outer membrane of gram-negative bacteria. The carboxy-terminal β-domain is thought to form a β-barrel outer membrane porin that allows the amino-terminal passenger domain to pass through it in an unfolded state. The secreted passenger domain is then released from the transporter as a functional mature protein (38). Analysis of the R. typhi genome revealed the presence of five previously described autotransporter proteins: Sca1 (RT0015), Sca2 (RT0052), Sca3 (RT0438), Sca4 (RT0485), and OmpB (RT0699) (5, 52). Work in the SFG rickettsiae showed that both OmpB and a related protein, OmpA (not found in the TG), play a role in adhesion to host cells (45, 60). Although no function of the Sca proteins was demonstrated, it was suggested that some autotransporters have adhesion or protease activities and could play a role in rickettsial virulence (38, 69). A characteristic of bacterial adhesins that bind to eukaryotic extracellular matrix proteins is the presence of amino acid repeats (57). A 50-residue repeat occurs six times within the carboxy-terminal portion of the Sca2 passenger domain, suggesting that this protein may be involved in binding of R. typhi to host cells.
In R. typhi, each of the four Sca proteins is found within a single predicted ORF, while at least one Sca protein has degraded into multiple ORFs in both R. conorii and R. prowazekii. Sca1, Sca2, and Sca4 are intact in R. conorii, but portions of the Sca3 sequence are found in two different ORFs (RC0630 and RC0631). The R. typhi Sca1 sequence corresponds to three different ORFs in R. prowazekii (RP016, RP017, and RP018), whereas the Sca2-like sequence is found in four different ORFs (RP081, RP082, RP083, and RP084). Sca1 and Sca2 contain only the β-domain and may function to transport other proteins across the outer membrane (69). Sca4 is found in two large ORFs in R. prowazekii (RP498 and RP499).
Type IV secretion components.
Several components of a type IV secretion system (19) found in R. conorii (52), R. prowazekii (5), and other α-proteobacteria, were present in R. typhi. Type IV secretion systems use a complex of transmembrane proteins and a pilus to export protein or DNA substrates across bacterial and eukaryotic cell membranes and into target cells (18, 19).
Fifteen ORFs related to the Agrobacterium tumefaciens VirB type IV secretion system were found in R. typhi. These include orthologs to the inner membrane components VirB4 (RT0033 and RT0771) and VirB11 (RT0283). In A. tumefaciens, VirB4 binds to the DNA substrate since it is translocated across the membrane (19); the presence of two VirB4 proteins in R. typhi may indicate versatility in substrate selection. The substrate-binding protein VirD4 (19) was also present (RT0283). The periplasmic components VirB6, VirB8, and VirB10 (RT0282) were identified. These included two ORFs resembling VirB8 (RT0278 and RT0280), in addition to five different ORFs with similarity to VirB6 (RT0028, RT0029, RT0030, RT0031, and RT0032). VirB6 functions in the stabilization of VirB5 and VirB7 (36), neither of which was found in R. typhi. VirB6 also stabilizes VirB3 (RT0034) (36), which was found in R. typhi. Multiple VirB6-like ORFs may be the result of inactivation of one or more genes or the development of new functions not yet described. Outer membrane components VirB3 and VirB9 (RT0277 and RT0281) were also found, but an ortholog of VirB7, which bridges the pilus to the transmembrane complex, was not (36). Neither VirB2 nor VirB5, the major and minor pilus components, were identified (19). Because R. typhi live within the host cell cytoplasm, there may be no need to secrete substrates across a eukaryotic host cell membrane, hence the lack of pilus components.
A potential protein substrate for a type IV secretion system was also found. R. typhi has a Sec7 domain-containing protein (RT0362) that is similar to RalF in Legionella pneumophila. RalF is secreted by the L. pneumophila dot/icm type IV secretion system (51). Along with a R. prowazekii ortholog, these are the only three known examples of Sec7 family proteins in bacteria (51). Sec7 proteins, named for Saccharomyces cerevisiae Sec7p, are guanine-nucleotide exchange factors and cofactors for ADP-ribosylation factors (ARFs), which function in the regulation of membrane traffic and organelle structure in eukaryotic cells (25, 42). The similarity to the L. pneumophila RalF protein suggests that RT0362 may be secreted by the R. typhi type IV system. In Legionella, the function of RalF is related to recruiting host ARFs to bacterium-containing phagosomes (51) but, since rickettsiae do not live in phagosomes, the role of the RalF-like protein in R. typhi is unknown.
Other transporters.
In addition to the autotransporters, drug efflux pumps, and VirB secretion system, R. typhi has proteins that function in transporting substances across the cell membrane. There were five copies of an ADP/ATP translocase, a transporter that allows R. typhi to acquire host cell ATP (5). As in other rickettsiae, there are multiple copies of the osmosensory proline-betaine transporter, ProP. We found 7 copies of ProP in R. typhi compared to 11 copies in R. conorii and 7 copies in R. prowazekii (52). Transport proteins for cell membrane and cell wall components were found, including three copies of the murein peptide permease AmpG, two heme exporters CcmAB (RT0781 and RT0259), and two O-antigen exporter proteins (RT0002 and RT0003). Also found was a homolog of the SAM transporter of R. prowazekii (RT0056) that has recently been shown to transport S-adenosylmethionine into the bacterial cell (59). A variety of other transport proteins, including seven possible ABC transport systems, were found. These transporters include a potential zinc/manganese transporter (RT0013 and RT0612), a magnesium transporter (RT0572), potassium transporter (RT0798), a glutamine transporter (RT0118, RT0139, and RT0859), a putrescine/ornithine transporter (RT0470), a ribonucleotide transporter (RT0040), and a sodium/proton symporter (RT0549). Four members of the Tol family—TolC (RT0216), TolB (RT0293), TolQ (RT0299), and TolR (RT0300)—were identified. Transporters with less clear possible functions were given generic descriptions such as metal transporter and cation transporter. Still others with no known or postulated substrate were also found. In addition, a PSORT-B (29) analysis of coding regions identified 39 hypothetical, conserved hypothetical, or rickettsial conserved hypothetical proteins that could be membrane localized. Some of these may also function in transport.
Stringent response.
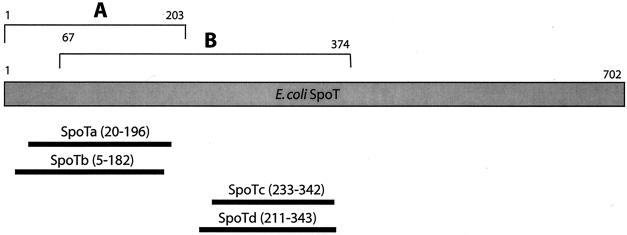
In response to starvation and other stress signals, bacteria increase their intracellular concentration of GDP 3′-diphosphate and GTP 3′-diphosphate, or (p)ppGpp, in a process called the stringent response (21, 22). This leads to a downregulation of transcription and translation. In E. coli, the RelA and SpoT proteins modulate (p)ppGpp levels in the cell. No intact R. typhi ORF with significant similarity to either RelA or SpoT was found, although four small ORFs with significant similarity to SpoT were found. These ORFs align to two different regions of SpoT (Fig. 4) and have been previously described and classified as pseudogenes or “split genes”, named spoTa, spoTb, spoTc, and spoTd (4, 5, 52). Andersson and Andersson suggested that spoT underwent a gene duplication event and then each copy degraded into two pseudogenes (4). They postulated that spoTa and spoTd arose from one copy and that spoTb and spoTc arose from the other. In agreement with this, amino acid sequences of the R. typhi SpoTa and SpoTd roughly correspond to the same region of SpoT from E. coli, whereas SpoTb and SpoTc both correspond to a different region of SpoT. The authors of that study compared the lengths and arrangements of these four ORFs in R. prowazekii, R. typhi, R. rickettsii, and R. montanesis and concluded that they were not functional, although the study also indicated that SpoT proteins corresponding to the amino-terminal catalytic domain could be functional (4).
FIG. 4.
Relation between R. typhi SpoT fragments and the catalytic domains of SpoT. Each R. typhi SpoT fragment is aligned with a full-length SpoT polypeptide from E. coli. All numbers refer to the E. coli amino acids. A, (p)ppGpp degradation domain; B, (p)ppGpp synthetase domain.
We suggest that the SpoT proteins in R. typhi may be functional and have chosen to adopt the term “split gene” as proposed by Ogata et al. (52), thus classifying them as coding ORFs instead of pseudogenes. The E. coli SpoT protein has a two-domain structure, with an amino-terminal (p)ppGpp degradation domain and a separate, but overlapping, (p)ppGpp synthetase domain (32). The degradation domain consists of no more than the first 203 amino acids, whereas the synthetase domain lies between residues 67 to 374 (32). The R. typhi SpoTa and SpoTb proteins align within the first 200 amino acids of E. coli SpoT (SpoTa, amino acids 20 to 196; SpoTb, amino acids 5 to 182) and thus, if translated, could code for functional (p)ppGpp hydrolases. The R. typhi SpoTc and SpoTd proteins correspond to the (p)ppGpp synthetase domain (SpoTc, amino acids 233 to 342; SpotD, amino acids 211 to 343), so these proteins could synthesize (p)ppGpp. These proteins may not function singly but may instead form complexes, e.g., SpoTa/SpoTd and SpoTb/SpoTd, to become active.
Further support of SpoT functionality is the high level of conservation of the genes among the Rickettsia species. In agreement with previous results, we found the most conserved ORF to be spoTa and the least conserved ORF to be spoTd (4). The DNA sequences of spoTa are 95% identical between R. typhi and R. prowazekii and 91% identical between R. typhi and R. conorii, whereas the spoTd genes are 86% identical between R. typhi and R. prowazekii and 90% identical between R. typhi and R. conorii. The spoTd genes vary in length, with an R. typhi homolog of 573 bp, an R. conorii gene 607 bp, and an R. prowazekii homolog of only 264 bp. The sequences for spoTb and spoTc are 98 and 100% identical between R. typhi and R. prowazekii, respectively, although homologs were not found in R. conorii. With the exception of spoTd, these levels of identity rank with those of highly conserved genes such as the 54 shared ribosomal proteins (94.8% identity between R. typhi and R. prowazekii and 91% identity between R. typhi and R. conorii), while the overall conservation rate between the genomes is 94.3% identity between R. typhi and R. prowazekii and 88.8% identity between R. typhi and R. conorii. Thus, there appears to be selective pressure on the spoT gene sequences.
Several additional lines of evidence suggest that the SpoTa to -d ORFs may be functional in R. typhi. First, two other components of the stringent response pathway, Ndk (nucleoside diphosphokinase) and GppA (pppGpp-5′-phosphohydrolase), are found in the genome. Ndk is involved in the addition of phosphates to nucleoside diphosphates and should be necessary even in the absence of the stringent response, whereas GppA is active only on GTP derivatives. Thus, without a ppGpp synthetase there would be no reason to maintain the gppA function. Second, Ogata et al. reported that in R. conorii some split genes may be transcribed and retain functionality (52). Lastly, there is evidence that the rickettsiae do possess a functional stringent response since bacteria in blood-starved ticks downregulate protein synthesis (68, 69).
Pseudogenes.
The rickettsiae are well known for containing DNA that is no longer capable of producing a functional protein (pseudogenes) (3). We discovered 41 sequences that appear to be pseudogenes (Table 4). Although several pseudogenes matched proteins in other bacteria, most appear only in other rickettsiae (rickettsial conserved hypothetical proteins). For example, R. typhi is missing the entire cytochrome c oxidase pathway. Both R. prowazekii and R. conorii contain intact electron transport systems, including the genes necessary for NADH dehydrogenase (complex I), succinate dehydrogenase (complex II), cytochrome reductase (complex III), and cytochrome oxidase (complex IV) (5, 52). Many bacteria contain more than one terminal oxidase for aerobic respiration under differing oxygen concentrations (44). Both R. prowazekii and R. conorii contain the genes coxABC, which encode a putative cytochrome c oxidase, and cydAB, which encode a putative cytochrome d ubiquinol oxidase. This situation is similar to Mycobacterium smegmatis in which an aa3-type cytochrome c oxidase is expressed during normal aerobic growth and a cydAB-encoded bd-type oxidase is expressed during low oxygen conditions (44). In R. typhi, all three genes encoding cytochrome c oxidase (coxCAB; RTPG10, RTPG22, and RTPG23) have decayed and are likely nonfunctional. In addition, the genes encoding the three cytochrome c oxidase assembly proteins, CoxW, CtaG, and CtaB (RTPG14, RTPG15, and RTPG18), have also decayed. As a result, it appears that R. typhi retains only one terminal oxidase for aerobic respiration. The remaining oxidase, CydAB (RT0207 and RT0208), is predicted to be of the d-type that is commonly expressed during low-oxygen conditions. Such cytochrome d oxidases generally have a higher affinity for oxygen than do cytochrome c oxidases and thus mediate the transfer of electrons to water at a slower rate (34). This may lead to physiological effects that play a role in the attenuated disease manifestations of R. typhi compared to R. prowazekii.
TABLE 4.
Pseudogenes found in R. typhi and their functional counterparts in R. prowazekii and R. conorii
| RT no. | Position
|
Description of functional counterpart | RP no. | RC no. | |
|---|---|---|---|---|---|
| Start | Stop | ||||
| RTPG01 | 19015 | 19631 | tRNA/rRNA methyltransferase | None | RC0160 |
| RTPG02 | 47435 | 48207 | Acetylglutamate kinase | None | RC0139 |
| RTPG03 | 61590 | 61776 | RP088 | RP088 | None |
| RTPG04 | 103201 | 103580 | RP052 | RP052 | None |
| RTPG05 | 105249 | 105804 | RP050/RC0076 | RP050 | RC0076 |
| RTPG06 | 126004 | 126468 | RC0049 | None | RC0049 |
| RTPG07 | 129317 | 129872 | Sco2 protein precursor | RP031 | RC0042 |
| RTPG08 | 134057 | 134355 | Rickettsial conserved protein with coenzyme PQQ synthesis protein C domain | None | RC0033 |
| RTPG09 | 198021 | 198258 | Conserved hypothetical protein | RP167 | RC0209 |
| RTPG10 | 229960 | 230742 | Cytochrome c oxidase subunit III | RP191 | RC0240 |
| RTPG11 | 253368 | 253540 | Rickettsial conserved hypothetical protein | None | RC0276 |
| RTPG12 | 258881 | 259212 | Agrobacterium protein | None | None |
| RTPG13 | 271206 | 272364 | Rickettsial conserved hypothetical protein | None | RC0295 |
| RTPG14 | 314921 | 315914 | Cytochrome c oxidase assembly protein CoxW | RP257 | RC0343 |
| RTPG15 | 373523 | 374000 | Cytochrome c oxidase assembly protein CtaG | RP304 | RC0408 |
| RTPG16 | 383166 | 383538 | Rickettsial conserved hypothetical protein | None | RC0425 |
| RTPG17 | 408613 | 408790 | Rickettsial conserved hypothetical protein | RP331 | None |
| RTPG18 | 425149 | 425995 | Protoheme IX farnesyltransferase (cytochrome c oxidase assembly protein CtaB) | RP346 | RC0469 |
| RTPG19 | 462673 | 463315 | Rickettsial conserved hypothetical protein | None | RC0519 |
| RTPG20 | 470138 | 471491 | Rickettsial conserved hypothetical protein | RP382 | RC0526 |
| RTPG21 | 473514 | 474113 | Cell filamentation protein | None | RC0529 |
| RTPG22 | 496379 | 497698 | Cytochrome c oxidase subunit I | RP405 | RC0553 |
| RTPG23 | 497853 | 498935 | Cytochrome c oxidase subunit II | RP406 | RC0555 |
| RTPG24 | 605974 | 606376 | Rickettsial conserved hypothetical protein | RP488 | RC0732 |
| RTPG25 | 617447 | 617602 | Hypothetical protein RC0669 | None | RC0669 |
| RTPG26 | 650083 | 650663 | Hypothetical protein RC0754 | None | RC0754 |
| RTPG27 | 680777 | 680989 | Hypothetical protein RP543 | RP543 | None |
| RTPG28 | 687125 | 688408 | Hypothetical protein RC0815 | None | RC0815 |
| RTPG29 | 693501 | 693801 | Hypothetical protein RC0819 | None | RC0819 |
| RTPG30 | 753627 | 754173 | 3-Oxoacid coenzyme A-transferase subunit B | None | RC0906 |
| RTPG31 | 777824 | 778728 | Mg/Co transport protein RC0943 | None | RC0943 |
| RTPG32 | 831141 | 831549 | Hypothetical protein RC1011 | None | RC1011 |
| RTPG33 | 846656 | 846872 | Hypothetical protein RC1025 | None | RC1025 |
| RTPG34 | 872509 | 872815 | Hypothetical protein RC1065 | None | RC1065 |
| RTPG35 | 897902 | 898363 | Bacterioferritin comigratory protein | None | RC1090 |
| RTPG36 | 900174 | 900814 | Rickettsial conserved hypothetical protein | None | RC1093 |
| RTPG37 | 900807 | 901141 | Rickettsial conserved hypothetical protein | None | RC1094 |
| RTPG38 | 913123 | 913788 | Surf1-like protein | RP733 | RC1113 |
| RTPG39 | 920761 | 921226 | Rickettsial conserved hypothetical protein | None | RC1129 |
| RTPG40 | 932587 | 932807 | Hypothetical protein RC1157 | None | RC1157 |
| RTPG41 | 1105415 | 1105657 | Hypothetical protein RC1365 | None | RC1365 |
DISCUSSION
The genome of R. typhi is nearly identical to its close relative R. prowazekii and highly similar to R. conorii and other SFG bacteria. The few differences between the two TG rickettsiae include a 12-kb insertion in the genome of R. prowazekii, a large inversion close to the origin of replication with no loss of genes in the region, and the fact that R. typhi has lost the complete cytochrome c oxidase system. In addition, R. typhi has several pseudogenes for which functional homologs are found in R. prowazekii. The similarity of the genome sequence of R. typhi to the TG rickettsiae further illustrates the delineation between the TG and SFG rickettsiae. Based on comparisons of these three genomes, it appears that rickettsiae may have a functional stringent response system that utilizes split genes of the SpoT protein as previously described. Further comparisons that include R. sibirica and R. rickettsii, as briefly mentioned here, should provide additional insight into the evolutionary relationship between these important pathogens.
The rickettsiae appear to maintain nonfunctional DNA for a much longer period than do other bacteria. It has been proposed that slow-growth times can account for this. However, there are many slow-growing bacteria that do not accumulate pseudogenes. It is possible that the rickettsiae retain this DNA for a reason or that their pathway for removing nonfunctional DNA is different. By comparing this genome to others, which lose nonfunctional genes faster, we may discover the mechanisms by which organisms lose DNA. It is interesting that most of the pseudogenes in R. typhi were found in regions with deletions in R. prowazekii and R. typhi with respect to R. conorii, and in fact many appear to be remnants of the deletion event. The pseudogenes may be functional in other rickettsiae or indicative of regions that are not functional in any of the rickettsiae.
The high degree of similarity between R. typhi and R. prowazekii illustrates the small differences that can affect virulence in different hosts. Thus, despite their close genetic relatedness, R. prowazekii is highly pathogenic in humans, whereas R. typhi is a milder pathogen in humans. Future studies should identify which of the differences in these genomes accounts for these phenotypes.
Acknowledgments
This research was supported by a grant from the National Institute of Allergy and Infectious Disease (AI49040). Michael P. McLeod was supported in part by a training grant from the Keck Center for Computational Biology (NLM grant LM07093). George E. Fox was supported in part by a Ruth L. Kirschstein National Research Service Award (F33-HGO2551).
We are grateful to Virginia Vera, Baize Montgomery, and Regina Wleczyk for technical assistance. We thank Manuel Gonzalez-Garay, Andrei Volkov, Andrew R. Jackson, and Aleksandar Milosavljevic for hosting annotation on the Genboree server.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, B., M. A. Wentland, J. Y. Ricafrente, W. Liu, and R. A. Gibbs. 1996. A “double adaptor” method for improved shotgun library construction. Anal. Biochem. 236:107-113. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, J. O., and S. G. Andersson. 1999. Insights into the evolutionary process of genome degradation. Curr. Opin. Genet. Dev. 9:664-671. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, J. O., and S. G. Andersson. 2001. Pseudogenes, junk DNA, and the dynamics of Rickettsia genomes. Mol. Biol. Evol. 18:829-839. [DOI] [PubMed] [Google Scholar]
- 5.Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. S. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. [DOI] [PubMed] [Google Scholar]
- 6.Andersson, S. G. E., and C. G. Kurland. 1998. Reductive evolution of resident genomes. Trends Microbiol. 6:263-268. [DOI] [PubMed] [Google Scholar]
- 7.Atlas, R. 1998. Biological weapons pose challenge for microbiology community. ASM News 64:383-389. [Google Scholar]
- 8.Azad, A. F., and C. B. Beard. 1998. Rickettsial pathogens and their arthropod vectors. Emerg. Infect. Dis. 4:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azad, A. F., and S. Radulovic. 2003. Pathogenic rickettsiae as bioterrorism agents. Ann. N. Y. Acad. Sci. 990:734-738. [DOI] [PubMed] [Google Scholar]
- 10.Azad, A. F., S. Radulovic, J. A. Higgins, B. H. Noden, and J. M. Troyer. 1997. Flea-borne rickettsioses: ecologic considerations. Emerg. Infect. Dis. 3:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baxter, J. D. 1996. The typhus group. Clin. Dermatol. 14:271-278. [DOI] [PubMed] [Google Scholar]
- 13.Bentley, J., L. S. Hyatt, K. Ainley, J. H. Parish, R. B. Herbert, and G. R. White. 1993. Cloning and sequence analysis of an Escherichia coli gene conferring bicyclomycin resistance. Gene 127:117-120. [DOI] [PubMed] [Google Scholar]
- 14.Betley, J. N., M. C. Frith, J. H. Graber, S. Choo, and J. O. Deshler. 2002. A ubiquitous and conserved signal for RNA localization in chordates. Curr. Biol. 12:1756-1761. [DOI] [PubMed] [Google Scholar]
- 15.Boeckmann, B., A. Bairoch, R. Apweiler, M. C. Blatter, A. Estreicher, E. Gasteiger, M. J. Martin, K. Michoud, C. O'Donovan, I. Phan, S. Pilbout, and M. Schneider. 2003. The SWISS-PROT protein knowledge base and its supplement TrEMBL in 2003. Nucleic Acids Res. 31:365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bovarnick, M. R., J. C. Miller, and J. C. Snyder. 1950. The influence of certain salts, amino acids, sugars and proteins on the stability of rickettsiae. J. Bacteriol. 59:509-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brassinga, A. K., R. Siam, W. McSween, H. Winkler, D. Wood, and G. T. Marczynski. 2002. Conserved response regulator CtrA and IHF binding sites in the alpha-proteobacteria Caulobacter crescentus and Rickettsia prowazekii chromosomal replication origins. J. Bacteriol. 184:5789-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burns, D. L. 1999. Biochemistry of type IV secretion. Curr. Opin. Microbiol. 2:25-29. [DOI] [PubMed] [Google Scholar]
- 19.Burns, D. L. 2003. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6:29-34. [DOI] [PubMed] [Google Scholar]
- 20.Carsiotis, M., B. Stocker, D. Weinstein, and A. O'Brien. 1989. A Salmonella typhimurium virulence gene linked to flg. Infect. Immun. 57:3276-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cashel, M., and K. E. Rudd. 1987. The stringent response, p. 1410-1438. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed.
- 22.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 23.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demerec, M., E. A. Adelberg, A. J. Clark, and P. E. Hartman. 1966. A proposal for a uniform nomenclature in bacterial genetics. Genetics 54:61-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donaldson, J. G., and C. L. Jackson. 2000. Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 12:475-482. [DOI] [PubMed] [Google Scholar]
- 26.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 27.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 28.Fergie, J. E., K. Purcell, and D. Wanat. 2000. Murine typhus in South Texas children. Pediatr. Infect. Dis. J. 19:535-538. [DOI] [PubMed] [Google Scholar]
- 29.Gardy, J. L., C. Spencer, K. Wang, M. Ester, G. E. Tusnady, I. Simon, S. Hua, K. deFays, C. Lambert, K. Nakai, and F. S. Brinkman. 2003. PSORT-B: improving protein subcellular localization prediction for gram-negative bacteria. Nucleic Acids Res. 31:3613-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaywee, J., S. Radulovic, J. A. Higgins, and A. F. Azad. 2002. Transcriptional analysis of Rickettsia prowazekii invasion gene homolog (invA) during host cell infection. Infect. Immun. 70:6346-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaywee, J., W. Xu, S. Radulovic, M. J. Bessman, and A. F. Azad. 2002. The Rickettsia prowazekii invasion gene homolog (invA) encodes a nudix hydrolase active on adenosine (5′)-pentaphospho-(5′)-adenosine. Mol. Cell Proteomics 1:179-183. [DOI] [PubMed] [Google Scholar]
- 32.Gentry, D. R., and M. Cashel. 1996. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol. Microbiol. 19:1373-1384. [DOI] [PubMed] [Google Scholar]
- 33.Gouin, E., C. Egile, P. Dehoux, V. Villiers, J. Adams, F. Gertler, R. Li, and P. Cossart. 2004. The RickA of Rickettsia conorii activates the Arp2/3 complex. Nature 427:457-461. [DOI] [PubMed] [Google Scholar]
- 34.Govantes, F., A. V. Orjalo, and R. P. Gunsalus. 2000. Interplay between three global regulatory proteins mediates oxygen regulation of the Escherichia coli cytochrome days oxidase (cydAB) operon. Mol. Microbiol. 38:1061-1073. [DOI] [PubMed] [Google Scholar]
- 35.Handelman, S., and K. Alibek. 2000. Biohazard. Delta, New York, N.Y.
- 36.Hapfelmeier, S., N. Domke, P. C. Zambryski, and C. Baron. 2000. VirB6 is required for stabilization of VirB5 and VirB3 and formation of VirB7 homodimers in Agrobacterium tumefaciens. J. Bacteriol. 182:4505-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinzen, R. A. 2003. Rickettsial actin-based motility: behavior and involvement of cytoskeletal regulators. Ann. N. Y. Acad. Sci. 990:535-547. [DOI] [PubMed] [Google Scholar]
- 38.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 39.Holk, A., S. Rietz, M. Zahn, H. Quader, and G. F. E. Scherer. 2002. Molecular identification of cytosolic, patatin-related phospholipases A from Arabidopsis with potential functions in plant signal transduction. Plant Physiol. 130:90-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoskinson, S., M. Lipetz, C. Sherer, S. Fraser, E. Taniguchi, A. Tice, D. Behling, B. Tanabe, D. Kwock, P. Effler, L. Pang, E. Brown, L. Granville, K. Mills, D. Sasaki, M. Ching Lee, H. Matsubayashi, W. Warashina, A. Ueno, C. Takekuma, J. Haruno, S. Oshiro, P. Kitsutani, J. McQuiston, C. Paddock, J. Sumner, and J. Krebs. 2003. Murine typhus-Hawaii, 2002. Morb. Mortal. Wkly. Rep. 52:1224-1226. [PubMed] [Google Scholar]
- 41.Hryniewicz-Jankowska, A., A. Czogalla, E. Bok, and A. F. Sikorsk. 2002. Ankyrins, multifunctional proteins involved in many cellular pathways. Folia Histochem. Cytobiol. 40:239-249. [PubMed] [Google Scholar]
- 42.Jackson, C. L., and J. E. Casanova. 2000. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 10:60-67. [DOI] [PubMed] [Google Scholar]
- 43.Jimenez-Atienzar, M., J. Cabanes, F. Gandia-Herrero, J. Escribano, F. Garcia-Carmona, and M. Perez-Gilabert. 2003. Determination of the phospholipase activity of patatin by a continuous spectrophotometric assay. Lipids 38:677-682. [DOI] [PubMed] [Google Scholar]
- 44.Kana, B. D., E. A. Weinstein, D. Avarbock, S. S. Dawes, H. Rubin, and V. Mizrahi. 2001. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J. Bacteriol. 183:7076-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, H., and D. H. Walker. 1998. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb. Pathogen. 24:289-298. [DOI] [PubMed] [Google Scholar]
- 46.Lomovskaya, O., and K. Lewis. 1992. Emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 89:8938-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madan Babu, M. 2003. Did the loss of sigma factors initiate pseudogene accumulation in Mycobacterium leprae? Trends Microbiol. 11:59-61. [DOI] [PubMed] [Google Scholar]
- 50.Miyamura, S., T. Ohta, and A. Tamura. 1989. Comparison of in vitro susceptibilities of Rickettsia prowazekii, R. rickettsii, R. sibirica, and R. tsutsugamushi to antimicrobial agents. Nippon Saikingaku Zasshi. 44:717-721. [DOI] [PubMed] [Google Scholar]
- 51.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 52.Ogata, H., S. Audic, P. Renesto-Audiffren, P. E. Fournier, V. Barbe, D. Samson, V. Roux, P. Cossart, J. Weissenbach, J. M. Claverie, and D. Raoult. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093-2098. [DOI] [PubMed] [Google Scholar]
- 53.Quon, K. C., B. Yang, I. J. Domian, L. Shapiro, and G. T. Marczynski. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. USA 95:120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Radulovic, S., J. M. Troyer, M. S. Beier, A. O. T. Lau, and A. F. Azad. 1999. Identification and molecular analysis of the gene encoding Rickettsia typhi hemolysin. Infect. Immun. 67:6104-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renesto, P., P. Dehoux, E. Gouin, L. Touqui, P. Cossart, and D. Raoult. 2003. Identification and characterization of a phospholipase D superfamily gene in rickettsiae. J. Infect. Dis. 188:1276-1283. [DOI] [PubMed] [Google Scholar]
- 56.Rolain, J. M., L. Stuhl, M. Maurin, and D. Raoult. 2002. Evaluation of antibiotic susceptibilities of three rickettsial species including Rickettsia felis by quantitative PCR DNA assay. Antimicrob. Agents Chemother. 46:2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimoji, Y., Y. Ogawa, M. Osaki, H. Kabeya, S. Maruyama, T. Mikami, and T. Sekizaki. 2003. Adhesive surface proteins of Erysipelothrix rhusiopathiae bind to polystyrene, fibronectin, and type I and IV collagens. J. Bacteriol. 185:2739-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 59.Tucker, A. M., H. H. Winkler, L. O. Driskell, and D. O. Wood. 2003. S-Adenosylmethionine transport in Rickettsia prowazekii. J. Bacteriol. 185:3031-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uchiyama, T. 1999. Role of major surface antigens of Rickettsia japonica in the attachment to host cell, p. 182-188. In J. Kazar and D. Raoult (ed.), Rickettsiae and rickettsial diseases. Publishing House of the Slovak Academy of Sciences, Bratislava, Czechoslovakia.
- 61.Ursing, B. M., F. H. van Enckevort, J. A. Leunissen, and R. J. Siezen. 2002. EXProt: a database for proteins with an experimentally verified function. Nucleic Acids Res. 30:50-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Kirk, L. S., S. F. Hayes, and R. A. Heinzen. 2000. Ultrastructure of Rickettsia rickettsii actin tails and localization of cytoskeletal proteins. Infect. Immun. 68:4706-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker, D. H., H. M. Feng, S. Ladner, A. N. Billings, S. R. Zaki, D. J. Wear, and B. Hightower. 1997. Immunohistochemical diagnosis of typhus rickettsioses using an anti-lipopolysaccharide monoclonal antibody. Mod. Pathol. 10:1038-1042. [PubMed] [Google Scholar]
- 64.Walker, D. H., F. M. Parks, T. G. Betz, J. P. Taylor, and J. W. Muehlberger. 1989. Histopathology and immunohistologic demonstration of the distribution of Rickettsia typhi in fatal murine typhus. Am. J. Clin. Pathol. 91:720-724. [DOI] [PubMed] [Google Scholar]
- 65.Walker, D. H., G. A. Valbuena, and J. P. Olano. 2003. Pathogenic mechanisms of diseases caused by Rickettsia. Ann. N. Y. Acad. Sci. 990:1-11. [DOI] [PubMed] [Google Scholar]
- 66.Weiss, E., J. C. Coolbaugh, and J. C. Williams. 1975. Separation of viable Rickettsia typhi from yolk sac and L cell host components by renografin density gradient centrifugation. Appl. Microbiol. 30:456-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wheeler, D. L., D. M. Church, A. E. Lash, D. D. Leipe, T. L. Madden, J. U. Pontius, G. D. Schuler, L. M. Schriml, T. A. Tatusova, L. Wagner, and B. A. Rapp. 2002. Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res. 30:13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wike, D. A., and W. Burgdorfer. 1972. Plaque formation in tissue cultures by Rickettsia rickettsii isolated directly from whole blood and tick hemolymph. Infect. Immun. 6:736-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu, X.-J., and D. H. Walker. 2003. The order Rickettsiales. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. (release 3.12 ed.). Springer-Verlag, New York, N.Y.