Abstract
The OmpA (or heat-modifiable) protein is a major structural component of the outer membranes of gram-negative bacteria. The protein contains eight membrane-traversing β-strands and four surface-exposed loops. The genetic diversity and molecular evolution of OmpA were investigated in 31 Mannheimia (Pasteurella) haemolytica, 6 Mannheimia glucosida, and 4 Pasteurella trehalosi strains by comparative nucleotide sequence analysis. The OmpA proteins of M. haemolytica and M. glucosida contain four hypervariable domains located at the distal ends of the surface-exposed loops. The hypervariable domains of OmpA proteins from bovine and ovine M. haemolytica isolates are very different but are highly conserved among strains from each of these two host species. Fourteen different alleles representing four distinct phylogenetic classes, classes I to IV, were identified in M. haemolytica and M. glucosida. Class I, II, and IV alleles were associated with bovine M. haemolytica, ovine M. haemolytica, and M. glucosida strains, respectively, whereas class III alleles were present in certain M. haemolytica and M. glucosida isolates. Class I and II alleles were associated with divergent lineages of bovine and ovine M. haemolytica strains, respectively, indicating a history of horizontal DNA transfer and assortative (entire gene) recombination. Class III alleles have mosaic structures and were derived by horizontal DNA transfer and intragenic recombination. Our findings suggest that OmpA is under strong selective pressure from the host species and that it plays an important role in host adaptation. It is proposed that the OmpA protein of M. haemolytica acts as a ligand and is involved in binding to specific host cell receptor molecules in cattle and sheep. P. trehalosi expresses two OmpA homologs that are encoded by different tandemly arranged ompA genes. The P. trehalosi ompA genes are highly diverged from those of M. haemolytica and M. glucosida, and evidence is presented to suggest that at least one of these genes was acquired by horizontal DNA transfer.
Mannheimia (Pasteurella) haemolytica is a commensal of cattle, sheep, and other ruminants, but it also causes bovine and ovine pneumonic pasteurellosis (infecting the respiratory tract), which is responsible for considerable economic losses to the cattle and sheep industries (6, 20, 21). The organism consists of genetically distinct subpopulations that are differentially adapted to and elicit disease in either cattle or sheep (11, 14). M. haemolytica possesses various putative virulence determinants (23), including a transferrin receptor (38, 59) and a leukotoxin (26, 49) which are specific for ruminant transferrin (38, 59) and lymphoid cells (4, 7, 27, 49), respectively, and are thought to contribute to the organism's host specificity. However, the molecular basis of host adaptation in the bovine and ovine lineages of M. haemolytica remains largely unclear. Mannheimia glucosida was previously recognized as the A11 serotype of M. haemolytica and represents a heterogeneous group of organisms that are mainly opportunistic sheep pathogens with low virulence (1, 11). Pasteurella trehalosi was previously recognized as the T biotype of M. haemolytica (51) and is associated exclusively with sheep, in which it causes a systemic disease that is pathologically distinct from pneumonic pasteurellosis (21).
The OmpA protein is an integral component of the outer membranes of gram-negative bacteria and is highly conserved (3). The protein has characteristic heat-modifiable properties (3), is present at a high copy number (>105/cell) (22), and is immunogenic (31, 45, 53, 60). Functions that have been attributed to OmpA include the maintenance of outer membrane integrity and cell shape (52), the action of a bacteriophage receptor (10, 34, 35), a role in conjugation (48), and resistance to the bactericidal effect of serum (58). However, OmpA is also involved in adherence to host tissues in Chlamydia spp. (36), Escherichia coli (44, 55), Haemophilus influenzae (24), and Pasteurella multocida (9). For E. coli and H. influenzae, the host cell receptor molecules have been identified (24, 42, 43). The 35-kDa OmpA protein of E. coli consists of an N-terminal transmembrane domain (19 kDa) and a C-terminal globular periplasmic domain (16 kDa) (2). The three-dimensional structure of the transmembrane domain has been determined by X-ray crystallography and nuclear magnetic resonance spectroscopy, and it consists of eight membrane-traversing antiparallel β-strands and four relatively long, mobile, hydrophilic surface-exposed loops (2, 39, 40). The periplasmic domain interacts with the underlying peptidoglycan and confers upon OmpA its role in maintaining the structural integrity of the outer membrane (18). The ompA gene of M. haemolytica has been cloned and sequenced and the immunological properties of OmpA have been investigated (31, 60). However, very little is known about the role of OmpA in the pathogenesis of bovine and ovine pneumonic pasteurellosis.
The OmpA proteins of bovine and ovine M. haemolytica isolates have previously been shown to exhibit interstrain molecular mass heterogeneity that correlates with the host of origin (14). The inference is that OmpA is somehow involved in the adaptation of bovine and ovine M. haemolytica strains to cattle and sheep, respectively (i.e., in host specificity). Horizontal DNA transfer and intragenic recombination have played important roles in the evolution of the leukotoxin structural gene (lktA) (17) and associated activation (lktC) and transport (lktB and lktD) genes (13) of M. haemolytica. In particular, certain M. haemolytica strains contain large segments of leukotoxin DNA that have been derived from M. glucosida and P. trehalosi (17). Similar evolutionary processes may have been involved in the diversification of OmpA, and the objective of the present study was to investigate nucleotide sequence variations of the ompA genes of bovine and ovine strains of M. haemolytica and of M. glucosida and P. trehalosi. In particular, we wanted to determine how these variations relate to the observed molecular mass heterogeneity and host association of OmpA in M. haemolytica (14) and to ascertain the evolutionary influence, if any, of M. glucosida and P. trehalosi. The nucleotide sequences were used to infer the evolutionary history of the ompA gene in these three species, to detect past recombination events involving the ompA gene, and to assess the action of natural selection on amino acid diversity in the different OmpA domains. To accomplish these goals, we sequenced the ompA genes of representative strains of M. haemolytica, M. glucosida, and P. trehalosi and used statistical tools to analyze these data from an evolutionary perspective.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The ompA genes from 31 M. haemolytica, 6 M. glucosida, and 4 P. trehalosi isolates were sequenced. The 41 strains were well characterized in previous studies (11-17) and were selected to represent specific evolutionary lineages, capsular serotypes, and hosts of origin. Some properties of these isolates are presented in Table 1. The strains were stored at −85°C in 50% (vol/vol) glycerol in brain heart infusion broth and were grown on blood agar (brain heart infusion agar containing 5% [vol/vol] sheep's blood) by overnight incubation at 37°C. Liquid cultures were prepared by inoculating a few colonies into 15 ml of brain heart infusion broth and incubating them overnight at 37°C with shaking at 120 rpm.
TABLE 1.
Properties of 31 M. haemolytica, 6 M. glucosida, and 4 P. trehalosi isolates
| Isolate | ETa | Capsular serotype | Host species | ompA allele | No. of nucleotides | No. of amino acids | Molecular mass of protein (Da) | GenBank accession no. |
|---|---|---|---|---|---|---|---|---|
| M. haemolytica | ||||||||
| PH2 | 1 | A1 | Bovine | ompA1.1 | 1,137 | 379 | 40,460 | AY244653 |
| PH30 | 1 | A1 | Bovine | ompA1.1 | 1,137 | 379 | 40,460 | |
| PH376 | 1 | A6 | Bovine | ompA1.1 | 1,137 | 379 | 40,460 | |
| PH346 | 1 | A12 | Ovine | ompA2.1 | 1,119 | 373 | 39,778 | AY244658 |
| PH540 | 2 | A1 | Bovine | ompA1.2 | 1,137 | 379 | 40,472 | AY244654 |
| PH338 | 3 | A9 | Ovine | ompA2.1 | 1,119 | 373 | 39,778 | |
| PH388 | 4 | A7 | Ovine | ompA2.1 | 1,119 | 373 | 39,778 | |
| PH50 | 5 | A5 | Ovine | ompA2.1 | 1,119 | 373 | 39,778 | |
| PH56 | 5 | A8 | Ovine | ompA2.2 | 1,119 | 373 | 39,794 | AY244659 |
| PH238 | 5 | A9 | Ovine | ompA2.1 | 1,119 | 373 | 39,778 | |
| PH8 | 6 | A1 | Ovine | ompA2.1 | 1,119 | 373 | 39,778 | |
| PH398 | 7 | A1 | Ovine | ompA2.1 | 1,119 | 373 | 39,778 | |
| PH284 | 8 | A6 | Ovine | ompA2.1 | 1,119 | 373 | 39,778 | |
| PH232 | 9 | A6 | Ovine | ompA2.1 | 1,119 | 373 | 39,778 | |
| PH66 | 10 | A14 | Ovine | ompA2.1 | 1,119 | 373 | 32,919 | |
| PH706 | 11 | A16 | Ovine | ompA2.1 | 1,119 | 373 | 32,919 | |
| PH296 | 12 | A7 | Ovine | ompA4.1 | 1,104 | 368 | 39,151 | AY244662 |
| PH396 | 13 | A7 | Ovine | ompA4.1 | 1,104 | 368 | 39,151 | |
| PH484 | 14 | A7 | Ovine | ompA4.1 | 1,104 | 368 | 39,151 | |
| PH588 | 15 | A13 | Ovine | ompA4.2 | 1,104 | 368 | 39,179 | AY244663 |
| PH494 | 16 | A2 | Bovine-like | ompA1.4 | 1,137 | 379 | 40,528 | AY244656 |
| PH550 | 17 | A2 | Bovine | ompA1.5 | 1,125 | 375 | 40,062 | AY244657 |
| PH196 | 18 | A2 | Bovine | ompA3.1 | 1,122 | 374 | 39,911 | AY244661 |
| PH526 | 19 | A2 | Ovine | ompA2.3 | 1,119 | 373 | 39,798 | |
| PH598 | 20 | A2 | Ovine | ompA2.3 | 1,119 | 373 | 39,798 | |
| PH202 | 21 | A2 | Bovine | ompA1.3 | 1,137 | 379 | 40,460 | AY244655 |
| PH470 | 21 | A2 | Bovine | ompA1.3 | 1,137 | 379 | 40,460 | |
| PH278 | 21 | A2 | Ovine | ompA2.3 | 1,119 | 373 | 39,798 | AY244660 |
| PH372 | 21 | A2 | Ovine | ompA2.3 | 1,119 | 373 | 39,798 | |
| PH292 | 22 | A2 | Ovine | ompA2.3 | 1,119 | 373 | 39,798 | |
| PH392 | 22 | A2 | Ovine | ompA2.3 | 1,119 | 373 | 39,798 | |
| M. glucosida | ||||||||
| PH344 | 1 | A11 | Ovine | ompA5.1 | 1,104 | 368 | 39,135 | AY244664 |
| PH498 | 3 | A11 | Ovine | ompA5.1 | 1,104 | 368 | 39,135 | |
| PH240 | 5 | A11 | Ovine | ompA7.1 | 1,104 | 368 | 39,233 | AY244666 |
| PH496 | 7 | UG3 | Ovine | ompA6.1 | 1,104 | 368 | 39,233 | AY244665 |
| PH574 | 10 | UG3 | Ovine | ompA7.1 | 1,104 | 368 | 39,233 | |
| PH290 | 16 | UG3 | Ovine | ompA6.1 | 1,104 | 368 | 39,233 | |
| P. trehalosi | ||||||||
| PH68 (NCTC 11550) | 19 | T3 | Ovine | ompA8.1 | 1,095 | 365 | 38,633 | AY582755 |
| ompA10.1 | 1,083 | 361 | 37,978 | |||||
| PH246 (NCTC 10626) | 2 | T4 | Ovine | ompA8.2 | 1,095 | 365 | 38,633 | AY582756 |
| ompA10.2 | 1,083 | 361 | 37,978 | |||||
| PH252 (NCTC 10641) | 4 | T10 | Ovine | ompA8.3 | 1,095 | 365 | 38,662 | AY582757 |
| ompA10.3 | 1,083 | 361 | 37,978 | |||||
| PH254 (NCTC 10370T) | 15 | T15 | Ovine | ompA9.1 | 1,095 | 365 | 38,844 | AY582758 |
| ompA10.4 | 1,083 | 361 | 37,978 |
SDS-PAGE analysis of P. trehalosi OmpA proteins.
The preparation of P. trehalosi outer membrane proteins (OMPs) and their analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) have been described previously (16). The OmpA proteins of P. trehalosi were identified by heating the OMP samples at 80, 90, and 100°C for 5 min prior to SDS-PAGE. The molecular masses of individual proteins were calculated with Labworks image acquisition and analysis computer software.
Preparation of chromosomal DNAs.
Cells from 1.0-ml overnight liquid cultures were harvested by centrifugation for 1 min at 13,000 × g and were washed once in sterile distilled H2O. DNAs were prepared by use of an InstaGene Matrix kit (Bio-Rad) according to the manufacturer's instructions and were stored at −20°C.
PCR amplification and DNA sequence analysis.
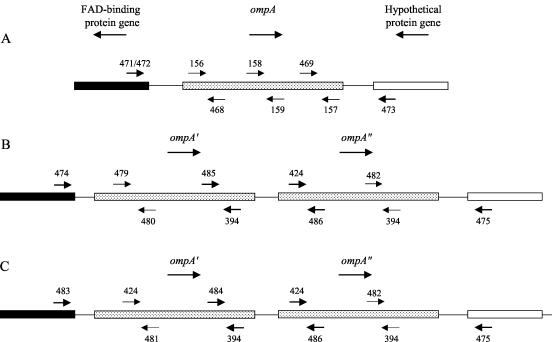
The ompA gene was found to be located at contigs 87 and 125 of the M. haemolytica genome sequence (http://www.hgsc.bcm.tmc.edu/microbial/Mhaemolytica/) by a BLAST analysis with the GenBank M. haemolytica ompA sequence (accession no. AF133259). The flanking open reading frames were identified by use of the Lasergene Editseq (DNAstar, Inc.) software application, and the identities of these genes were determined by a BLAST analysis against the GenBank database. The homologous genes were identified in Actinobacillus actinomycetemcomitans, H. influenzae, and P. multocida, and the four sequences were aligned with the Lasergene Megalign (DNAstar, Inc.) software application. Two forward and two reverse universal primers were designed within conserved regions of the four aligned sequences corresponding to each of the two flanking genes for preliminary PCR testing with representative strains of M. haemolytica, M. glucosida, and P. trehalosi. Successfully amplified bands were partially sequenced by use of the same primers, and a second set of internal primers specific for each individual strain was designed. In this way, the primer pairs 471-473 and 472-473 were designed for the amplification of ompA from M. haemolytica and M. glucosida strains, respectively (Fig. 1A). During the course of these preliminary experiments, difficulties were encountered in sequencing the ompA gene of P. trehalosi, despite the fact that definite PCR products were obtained. The results led us to suspect that two tandem ompA genes were present. Consequently, it became necessary to adopt a more complex sequencing strategy in which the ompA genes were amplified and sequenced as three separate overlapping fragments (Fig. 1B and C). The primer pairs 474-394, 485-486, and 424-475 were used for P. trehalosi strains PH68, PH246, and PH252 (Fig. 1B), and the primer pairs 483-394, 484-486, and 424-475 were used for strain PH254 (Fig. 1C). Both strands of ompA were sequenced, and internal sequencing primers were designed as sequence data became available (Fig. 1). The primers were designed with the computer program Primer Designer (version 2.0) and were synthesized by Sigma-GenoSys (Cambridge, United Kingdom). Full details of the PCR and sequencing primers are provided in Table 2.
FIG. 1.
Locations and numerical designations of PCR amplification and DNA sequencing primers for M. haemolytica and M. glucosida strains (A), P. trehalosi strains PH68, PH246, and PH252 (B), and P. trehalosi strain PH254 (C). The relative positions of the primers are represented by arrows; the primers used for PCR amplification are indicated by bold arrows. Arrows beneath gene names indicate the direction of transcription.
TABLE 2.
Details of oligonucleotide primers used for PCR and sequencing
| Target species or primer no.a | Direction | Sequence (5′-3′) | Corresponding nucleotide positionsb |
|---|---|---|---|
| M. haemolytica and M. glucosida | |||
| 471 | Forward | CCAGTTGCGGTACTTCAG | |
| 472 | Forward | CCAGTTGAGGTACTTCAG | |
| 156 | Forward | CTCAAGCAGCTCCACAAG | 50-67 |
| 158 | Forward | TAGGTGCTGGTCTTGAGT | 515-532 |
| 469 | Forward | TTAGATGCAGCACACGCT | 820-837 |
| 473 | Reverse | GCTGGTTAAGGCTCTCTA | |
| 157 | Reverse | GTTTGCTTCACCGTAACC | 1020-1003 |
| 159 | Reverse | GAGCAGCTAATTCAGGAG | 559-542 |
| 468 | Reverse | GTAACCACCGAATACACC | 225-208 |
| P. trehalosi | |||
| 474 | Forward | GTATCAGTGGCAAGCGAA | |
| 479 | Forward | GTGATGGTCCAACTGCTT | 494-511 |
| 485 | Forward | TCGACTTTGGTAAAGCA | 1061-1077 |
| 424 | Forward | GGTGCTAAAGCTGGTTGG | 1710-1727 |
| 482 | Forward | CCAGTTGCTGAGCCAGA | 2286-2302 |
| 475 | Reverse | TATGCAAGCTGGCTAAGG | |
| 394 | Reverse | AGCGTGTGCTGCATCTAA | 2405-2388 |
| 486 | Reverse | CCGTATTTACCACCGTT | 1783-1767 |
| 394 | Reverse | AGCGTGTGCTGCATCTAA | 1122-1105 |
| 480 | Reverse | TGCCACGAACACGACCAA | 619-602 |
| 483 | Forward | CTCGGCATAACTATCAGC | |
| 424 | Forward | GGTGCTAAAGCTGGTTGG | 415-432 |
| 484 | Forward | TCCAGTAGCAGCTCCTGA | 1002-1019 |
| 481 | Reverse | GAACGCGACCGAAGTAGT | 613-596 |
The positions of the primers are shown in Fig. 1.
PCR fragments containing the complete or partial ompA gene were amplified from chromosomal DNAs by use of a Taq DNA polymerase kit (Boehringer Mannheim) according to the manufacturer's instructions. PCRs were carried out in a Perkin-Elmer 480 DNA thermal cycler using the following amplification parameters: denaturation at 94°C for 45 s, annealing at 56°C for 45 s, and extension at 72°C for 2 min. Thirty cycles were performed, and a final extension step of 72°C for 10 min was used. The production of PCR amplicons of the expected sizes was confirmed by agarose gel electrophoresis, and the DNAs were purified with a Qiaquick PCR purification kit (Qiagen, Chatsworth, Calif.). DNAs were finally eluted in 30 μl of sterile distilled H2O and stored at −20°C. Sequence reactions were performed with an ABI Prism Big Dye Terminator cycle sequencing kit (Applied Biosystems) in a GeneAmp PCR System 9700 (Applied Biosystems) thermal cycler. Sequence analysis was performed with an Applied Biosystems 377 DNA sequencer (University of Glasgow Sequencing Service).
Analysis of nucleotide and protein sequence data.
Nucleotide sequence data were analyzed and edited with Seqed (Applied Biosystems) and Lasergene (DNAstar, Inc.) sequence analysis software. Phylogenetic and molecular evolutionary analyses were conducted with Mega, version 2.1 (30), in conjunction with alignment programs written by T. S. Whittam (Michigan State University). Statistical analyses for clustering of polymorphic sites were performed by the maximum chi-square method (50) with the computer program Maxchi (32, 47). Secondary structure predictions were performed with the Psipred secondary structure prediction method (25; http://bioinf.cs.ucl.ac.uk/psipred/) and the SAM-T99 sequence alignment and modeling system (28; http://www.cse.ucsc.edu/research/compbio/HMM-apps). The M. haemolytica and M. glucosida OmpA sequences were also aligned and compared with E. coli OmpA three-dimensional structural models (MMDB 16249 and PDB IG90 [2]; MMDB 9208 and PDB 1BXW [40]) by use of the computer program Cn3D (http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the ompA sequences obtained in this study are provided in Table 1.
RESULTS
Nucleotide and amino acid variation.
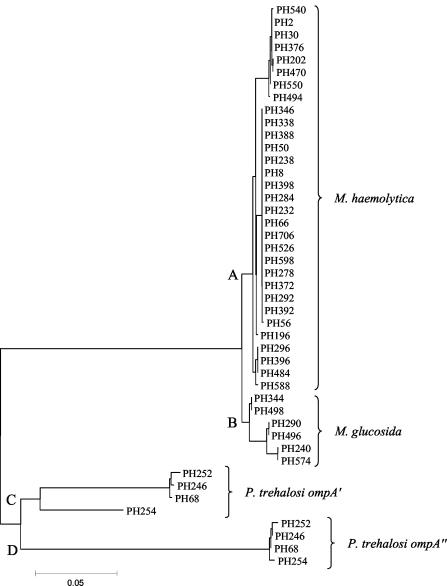
The ompA genes from 31 M. haemolytica isolates representing 12 capsular serotypes and 22 electrophoretic types (ETs) previously defined by multilocus enzyme electrophoresis (11) were sequenced. The ompA genes from six isolates representing different ETs of M. glucosida (11) and from four isolates representing each of the capsular serotypes T3, T4, T10, and T15 of P. trehalosi (12) were also sequenced. PCR errors were shown to be insignificant by duplicate amplification and sequencing of ompA for strains PH2, PH376, and PH344. M. haemolytica and M. glucosida isolates contain a single ompA gene, but the P. trehalosi strains were shown to possess two tandemly arranged ompA genes, ompA′ and ompA" (Fig. 1). The phylogenetic relationships of the ompA sequences are shown in Fig. 2. The ompA genes of the M. haemolytica and M. glucosida strains represent two distinct but closely related lineages, A and B. However, the ompA′ and ompA" genes of P. trehalosi represent two lineages, C and D, that are as divergent from each other as they are from the ompA genes of M. haemolytica and M. glucosida (Fig. 2). In addition, the ompA′ gene of strain PH254 shows considerable divergence from that of strains PH68, PH246, and PH252.
FIG. 2.
Neighbor-joining tree representing the phylogenetic relationships of the ompA genes of 31 M. haemolytica, 6 M. glucosida, and 4 P. trehalosi strains constructed with the Jukes-Cantor correction for synonymous changes.
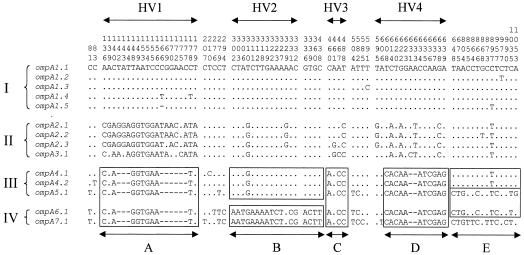
The ompA genes of the M. haemolytica and M. glucosida isolates varied from 1,104 to 1,137 nucleotides in length, and the predicted proteins varied from 368 to 379 amino acids in length and from 39,135 to 40,528 Da in molecular mass (Table 1). Since these proteins contain a putative signal sequence of 19 amino acids (1,855 Da) (60), the predicted molecular masses of the putative mature proteins varied from 37,280 to 38,673 Da. Fourteen unique ompA sequences, each representing a distinct allele, were identified among the M. haemolytica and M. glucosida isolates (Fig. 3). The alleles were assigned to seven subclasses, ompA1 to ompA7, based on their overall sequence similarities, and individual alleles within each subclass were designated ompA1.1, ompA1.2, etc. The ompA1- to ompA4-type alleles were associated exclusively with M. haemolytica, whereas the ompA5- to ompA7-type alleles were associated only with M. glucosida. The subclasses were grouped into four major classes, I to IV, which represent distinct phylogenetic lineages (discussed below). Class I consists of ompA1-type alleles, class II consists of ompA2- and ompA3-type alleles, class III consists of ompA4- and ompA5-type alleles, and class IV consists of ompA6- and ompA7-type alleles (Fig. 3). There were 82 (7.2%) polymorphic nucleotide sites and 33 (8.7%) variable inferred amino acid positions among the 14 alleles. Pairwise differences in nucleotide and inferred amino acid sequences between representative pairs of alleles ranged from 1 to 60 nucleotide sites and 1 to 23 amino acid positions.
FIG. 3.
Distribution of polymorphic nucleotide sites among the 14 ompA alleles of M. haemolytica and M. glucosida. Allele designations are shown to the left of each sequence. Roman numerals I to IV represent the major allele classes. The numbers above the sequences (read vertically) represent the positions of polymorphic nucleotide sites. The dots represent sites where the nucleotides match those of the first (topmost) sequence. Gaps are indicated by dashes. Boxes highlight identical, or nearly identical, segments of DNA (A to E) in class III and IV alleles. HV1 to HV4 represent the hypervariable domains.
The ompA′ and ompA" genes of the P. trehalosi isolates each consisted of four unique sequences that represented distinct alleles (Fig. 2 and Table 1). The ompA′ alleles were assigned to two subclasses, ompA8 (alleles ompA8.1 to ompA8.3) and ompA9 (allele ompA9.1), whereas the ompA" alleles consisted of a single subclass, ompA10 (alleles ompA10.1 to ompA10.4). The ompA′ alleles were 1,095 nucleotides long and the predicted proteins were 365 amino acids long, with molecular masses of 38,633 to 38,844 Da (Table 1). Since the putative signal sequence is 20 amino acids (1,918 Da), the molecular masses of the predicted mature proteins varied from 36,714 to 36,926 Da. The ompA" alleles were 1,083 nucleotides long and the predicted proteins were 361 amino acids long, with a molecular mass of 37,978 Da; the mature proteins were predicted to have a molecular mass of 36,060 Da. There were 10 (0.9%) polymorphic nucleotide sites and a single variable amino acid position among the three ompA8-type alleles and 9 (0.8%) polymorphic nucleotide sites (no amino acid changes) among the four ompA10-type alleles. The ompA9.1 allele differed from the ompA8.1 to ompA8.3 alleles at 129 to 136 nucleotide sites and 39 to 40 amino acid positions. Pairwise differences between the P. trehalosi ompA′ and ompA" alleles and those of M. haemolytica and M. glucosida ranged from 216 to 265 (ompA′) and 295 to 307 (ompA") nucleotides and from 84 to 102 (ompA′) and 118 to 121 (ompA") amino acids, respectively. Pairwise differences between ompA′ and ompA" alleles ranged from 213 to 231 nucleotides and 83 to 84 amino acids.
The majority of polymorphic nucleotide and inferred amino acid sites in the ompA genes of M. haemolytica and M. glucosida occur in four hypervariable domains located within the surface-exposed loops.
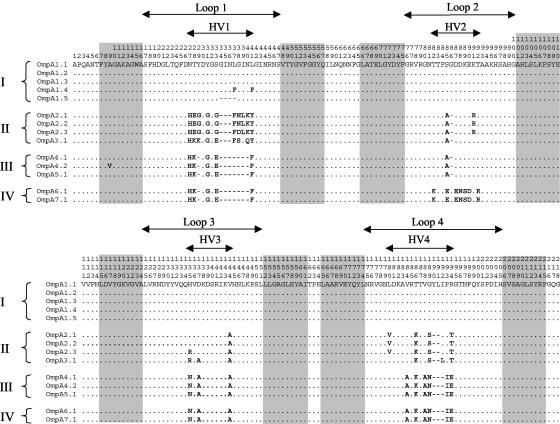
The locations of the polymorphic nucleotide sites and variable inferred amino acid positions within the ompA genes of M. haemolytica and M. glucosida were strikingly nonrandom in their distribution (Fig. 3 and 4). The majority of polymorphic sites occurred within four hypervariable regions, HV1 to HV4, located in the transmembrane domain. The hypervariable domains together consisted of 53 of 153 (35%) polymorphic nucleotide sites and 29 of 51 (57%) variable amino acid positions. In contrast, the remainder of the ompA gene was highly conserved and contained only 29 of 985 (3%) polymorphic nucleotide sites and 4 of 328 (1%) variable inferred amino acid positions. Domains HV1, HV2, and HV4 were characterized by amino acid deletions and/or insertions that accounted for the molecular mass variation of OmpA described above (Fig. 4).
FIG. 4.
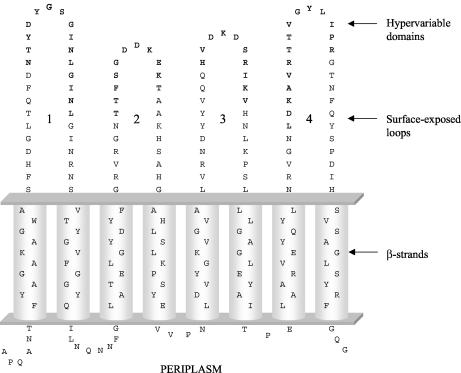
Distribution of variable inferred amino acid sites in the N-terminal transmembrane domains of the 14 OmpA proteins of M. haemolytica and M. glucosida. Protein designations are shown to the left of each sequence. Roman numerals I to IV represent the major allele classes. The numbers above the sequences (read vertically) represent amino acid positions. The dots represent sites where the amino acids match those of the first (topmost) sequence. Gaps are indicated by dashes. HV1 to HV4 represent the hypervariable domains within surface-exposed loops 1 to 4. Shaded regions represent predicted membrane-spanning β-strand structures.
The locations of the four hypervariable regions within the M. haemolytica and M. glucosida OmpA proteins, in relation to the β-strands and surface-exposed loops of the transmembrane domain, were identified by secondary structure prediction (25, 28), alignment with the three-dimensional structural models of the E. coli OmpA protein (2, 40), and comparison with the proposed secondary structure model of the P5 (OmpA) protein of H. influenzae (57; also results not shown). In this way, the four hypervariable domains of the M. haemolytica and M. glucosida OmpA proteins were shown to be located at the distal ends of the surface-exposed loop regions of the eight-stranded β-barrel structure (Fig. 4 and 5).
FIG. 5.
Proposed secondary structure of N-terminal transmembrane domain of the OmpA proteins of M. haemolytica and M. glucosida. The sequence is based on OmpA1.1 of strain PH2 (see Fig. 4). The hypervariable domains are shown in bold.
Evidence for assortative (entire gene) recombination of ompA among divergent lineages of M. haemolytica and M. glucosida.
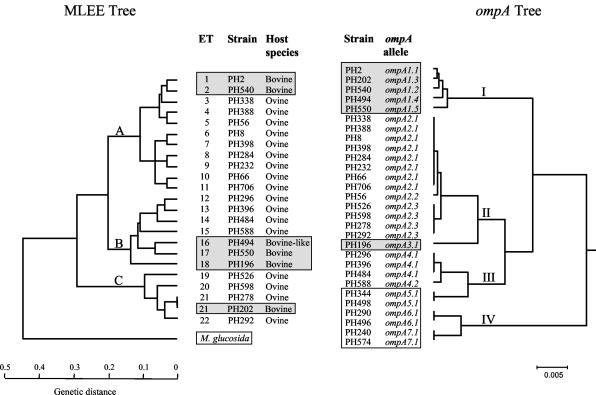
Assortative (entire gene) recombination leads to the presence of identical, or almost identical, alleles in strains that are genetically divergent. The evolutionary relationships of the M. haemolytica and M. glucosida ompA alleles with respect to the genetic relationships of the strains of origin based on multilocus enzyme electrophoresis data (11) are shown in Fig. 6. The ompA alleles were represented by four distinct lineages, I to IV, which correspond to the four classes described above. The association of identical, or almost identical, alleles with divergent lineages of M. haemolytica provides strong evidence that these alleles have undergone horizontal transfer and assortative (entire gene) recombination.
FIG. 6.
(Left) Phylogenetic relationships of 178 M. haemolytica and 16 M. glucosida strains based on electrophoretically demonstrable variation of 18 housekeeping enzymes (11). (Right) Phylogenetic relationships of ompA genes from 31 M. haemolytica and 6 M. glucosida strains. Both trees were constructed by the unweighted pair group method using arithmetic averages. Shaded boxes represent the bovine M. haemolytica isolates and ompA alleles. White boxes represent the M. glucosida isolates and ompA alleles.
In lineage I, alleles ompA1.1 to ompA1.5, which differ from each other at only one to three nucleotide sites (Fig. 3), were associated exclusively with bovine isolates (serotypes A1, A2, and A6) of ETs 1 and 2 (lineage A), 16 and 17 (lineage B), and 21 (lineage C) (Fig. 6 and Table 1). In lineage II, alleles ompA2.1 to ompA2.3, which also differ from each other at only one to three nucleotide sites (Fig. 3), were associated exclusively with ovine isolates (serotypes A1, A2, A5 to A8, A9, A12, A14, and A16) of ETs 3 to 11 (lineage A) and 19 to 22 (lineage C) (Fig. 6 and Table 1). Interestingly, the majority of ovine isolates representing lineage A (ETs 3 to 11) possessed ompA2.1, whereas all of the isolates representing lineage C (ETs 19 to 22) possessed ompA2.3; these two alleles differ at two nucleotide sites (Fig. 3). In lineage III, alleles ompA4.1 and ompA4.2 differ from each other at only two nucleotide sites. However, ompA4.1 alleles were associated exclusively with ovine serotype A7 isolates of ETs 12 to 14 (lineage B), whereas ompA4.2 was associated with an ovine serotype A13 isolate of ET 15 (lineage B).
There was also some evidence of assortative recombination in M. glucosida since the ompA5.1 alleles of lineage III were associated with isolates of ETs 1 and 3, and the ompA6.1 and ompA7.1 alleles of lineage IV were present in strains of ETs 7 and 16 and ETs 5 and 10, respectively (Fig. 6 and Table 1).
Evidence for intragenic recombination among ompA alleles of M. haemolytica and M. glucosida.
Intragenic recombination leads to the formation of linked runs of nucleotides within a sequence whose ancestry is different from other nucleotides in the same sequence (50), i.e., the sequence has a mosaic structure. A visual inspection of the distribution of polymorphic nucleotide sites among the class I and II alleles revealed no evidence of intragenic recombination involving these two groups (Fig. 3). In contrast, there was clear visible evidence of mosaicism within the class III and IV alleles because runs of nucleotides representing recombinant segments were present. For example, segments A, C, and D were identical in all five class III and IV alleles, but the sequence of segment B was very different in ompA4.1, ompA4.2, and ompA5.1 compared to that in ompA6.1 and ompA7.1 (Fig. 3). In addition, the sequences of segment E were very different in ompA4.1 and ompA4.2, in ompA5.1 and ompA6.1, and in ompA7.1.
A pairwise comparison of the ompA sequences by the maximum chi-square method identified the locations of significant breakpoints (kmax) that represent the end points of recombinant segments in class II, III, and IV alleles. A comparison of alleles ompA5.1 and ompA6.1 identified a region from nucleotides 276 to 438 that differs at 11% of the nucleotide sites and is embedded in a region that is almost identical in the two alleles (Fig. 3). A comparison of alleles ompA2.1 and ompA4.1 identified a region from nucleotides 640 to 1119 that is identical in both alleles, whereas the region from nucleotides 1 to 639 differs at 3% of the nucleotide sites.
Synonymous and nonsynonymous substitution rates among ompA alleles of M. haemolytica and M. glucosida.
To determine how the level of selective constraint varies along the ompA genes of M. haemolytica and M. glucosida, we estimated the numbers of synonymous substitutions per synonymous site (dS) and nonsynonymous substitutions per nonsynonymous site (dN) (37) and calculated the dS/dN ratios for each of the hypervariable domains and for the combined conserved regions (Table 3). A high dS/dN ratio indicates that natural selection at the molecular level is purifying (conservative), acting against mutations resulting in amino acid replacement. Conversely, a dS/dN ratio of <1 indicates that selection is diversifying and favors amino acid replacement. The dS values were four to seven times higher for the hypervariable domains HV1, HV2, and HV4 than for the conserved regions, whereas the dN values were 2 to 3 orders of magnitude higher for the same domains than for the conserved regions (Table 3). The dS/dN ratio for the conserved regions of ompA was relatively high, at 35.17, whereas the corresponding values for the hypervariable domains HV1, HV2, and HV4 ranged from 0.52 to 0.77. These data provide strong evidence of a selective constraint against amino acid replacement in the conserved parts of the gene and of diversifying selection in the hypervariable regions.
TABLE 3.
Sequence diversity and substitution rates for hypervariable domains HV1 to HV4 and for conserved regions of the ompA genes of 31 M. haemolytica, 6 M. glucosida, and 4 P. trehalosi isolates
| Domain | Sequence diversity (%)
|
dSa | dNa | dS/dN | |
|---|---|---|---|---|---|
| Nucleotide | Amino acid | ||||
| HV1 | 44 | 67 | 0.1396 ± 0.1015 | 0.4064 ± 0.1433 | 0.71 |
| HV2 | 42 | 72 | 0.0831 ± 0.0601 | 0.1586 ± 0.0605 | 0.52 |
| HV3 | 13 | 30 | 0.0000 ± 0.0000 | 0.0691 ± 0.0404 | |
| HV4 | 33 | 53 | 0.1518 ± 0.1244 | 0.1954 ± 0.0814 | 0.77 |
| Conserved regions | 3 | 1 | 0.0211 ± 0.0046 | 0.0006 ± 0.0004 | 35.17 |
dS is the number of synonymous substitutions per 100 synonymous sites; dN is the number of nonsynonymous substitutions per 100 nonsynonymous sites. Values are means ± standard deviations.
Evidence for independent transcription and expression of two OmpA proteins in P. trehalosi.
The upstream intragenic regions of ompA′ and ompA" were 213 and 200 nucleotides long, respectively, in contrast to the downstream intragenic region of ompA", which was only 56 nucleotides long. Putative ribosome-binding sequences and promoter sites were identified upstream of the start codons of ompA′ and ompA", and potential inverted repeat terminator sequences were present downstream of the stop codons of ompA′ and ompA" (results not shown). The existence of putative ribosome-binding, promoter, and terminator sequences associated with ompA′ and ompA" suggests that both genes are independently transcribed.
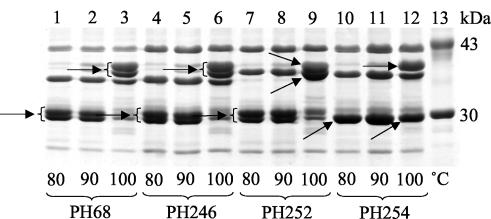
The expression of two OmpA proteins, OmpA′ and OmpA", by P. trehalosi was confirmed by SDS-PAGE analysis of OMPs after heating the samples at 80, 90, and 100°C (Fig. 7). The OmpA protein undergoes a characteristic shift from low- to high-molecular-mass forms after heating at 100°C. Two low-molecular-mass proteins (29 to 30 kDa) were clearly visible for strains PH68, PH246, and PH252 at 80 and 90°C, whereas these were transformed to two high-molecular-mass proteins (37 to 39 kDa) at 100°C (Fig. 7, lanes 1 to 9, arrows). However, only one low-molecular-mass band was present in strain PH254 at 80 and 90°C (Fig. 7, lanes 10 and 11, arrow), although two bands, of 29 and 39 kDa, were present at 100°C (Fig. 7, lane 12, arrows). The most probable explanation for this difference is that the single 29-kDa band present at 80 and 90°C consists of both proteins (OmpA′ and OmpA"), which in this strain differ in their heat modification properties. Thus, at 100°C the 29-kDa band presumably corresponds to the OmpA′ protein OmpA9.1 (since this band and protein were not present in any of the other isolates), whereas the 39-kDa band represents the OmpA" protein OmpA10.4.
FIG. 7.
Coomassie blue-stained SDS-PAGE OMP profiles of P. trehalosi strains PH68 (lanes 1 to 3), PH246 (lanes 4 to 6), PH252 (lanes 7 to 9), and PH254 (lanes 10 to 12) after heating at 80°C (lanes 1, 4, 7, and 10), 90°C (lanes 2, 5, 8, and 11), and 100°C (lanes 3, 6, 9, and 12). Molecular mass standards (ovalbumin, 43 kDa; carbonic anhydrase, 30 kDa) are shown in lane 13. Only the relevant part of the gel is shown. Arrows indicate the low- and high-molecular-mass forms of the two OmpA proteins in each strain. The transition is clearly seen between 90 and 100°C. Only one band is present in strain PH254 at 80 and 90°C, whereas two bands occur at 100°C (see the text for further explanation).
DISCUSSION
OmpA structure and function in M. haemolytica.
Three-dimensional structural analyses of the E. coli OmpA protein have revealed that the transmembrane domain consists of eight highly conserved membrane-spanning regions and four relatively long, mobile, hydrophilic surface-exposed loops (2, 39, 40). The OmpA proteins of M. haemolytica and M. glucosida contain four discrete hypervariable domains that are located at the distal ends of the surface-exposed loops (Fig. 5). Similar hypervariable regions correspond to the surface-exposed loops of the OmpA (P5) protein of H. influenzae (19, 57). Different selection pressures and evolutionary constraints operate on different parts of the molecule, since the patterns of synonymous and nonsynonymous nucleotide substitution rates vary throughout the ompA gene. Amino acid replacement is highly constrained within the conserved regions of OmpA (dS/dN = 35.17) because these parts of the molecule correspond to the membrane-spanning and periplasmic domains and cannot tolerate excessive amino acid change. In contrast, diversifying selection operates on the hypervariable domains of the surface-exposed loops because the number of nonsynonymous nucleotide substitutions greatly exceeds the number of synonymous substitutions (dS/dN = 0.52 to 0.77 for domains HV1, HV2, and HV4). Similar high rates of nonsynonymous nucleotide substitutions also occur in the four surface-exposed loops of the OmpA (P5) protein of H. influenzae (19). Amino acid diversity within the surface-exposed loops is clearly advantageous to M. haemolytica and suggests that these parts of the molecule play an important role in some aspect of this pathogen's biology.
An increasing body of evidence from other pathogens indicates that OmpA functions as a ligand, is involved in binding to specific host cell receptor molecules, and plays a role in adherence and colonization (9, 24, 33, 42, 44, 46, 55). The exclusive association of the OmpA1- and OmpA2-type proteins with bovine and ovine strains of M. haemolytica, respectively, together with evidence that the ompA1- and ompA2-type genes have undergone horizontal transfer between strains of divergent phylogenetic lineages, indicates that OmpA is under strong selective pressure from the host and plays an important role in host-pathogen relationships. The evidence from other pathogens suggests that the OmpA protein of M. haemolytica acts as a ligand and participates in binding to specific host cell receptor molecules in the upper respiratory tracts of cattle and sheep. We propose that the OmpA1 and OmpA2 proteins are involved in binding to bovine and ovine receptors, respectively, and that they play important roles in host specificity. In a similar way, receptor binding of the variable loop regions of the related Opa protein determines tissue tropism in Neisseria (5, 41, 56). Although there is no direct evidence to support this hypothesis, a host-specific ligand-like function would account for the variation in the surface-exposed loop regions between the bovine class I and ovine class II proteins and also for the amino acid conservation within each class. Clearly, the surface-exposed loops of OmpA need to be different in bovine and ovine strains if cattle and sheep have different receptor molecules, but they would also need to be conserved within each class if they are involved in binding to specific bovine (class I) or ovine (class II) receptor molecules. It is thought that the long, mobile, surface-exposed loops of OmpA are required for interactions with other structures, such as cell receptors and bacteriophages, and that otherwise there is no evolutionary advantage to maintaining them (39, 40). The locations of the four hypervariable domains, HV1 to HV4, at the distal ends of the corresponding loops (Fig. 5) provide further evidence to support the hypothesis that these regions are involved in receptor recognition and binding. Confirmation of this hypothesis will require the production of genetically modified strains and the development of appropriate in vitro adherence assays.
Evolution of OmpA in M. haemolytica and M. glucosida.
The class I, II, and IV ompA alleles are associated exclusively with bovine M. haemolytica, ovine M. haemolytica, and M. glucosida strains, respectively. These three groups of alleles have very different nucleotide sequences (Fig. 3) and have evolved independently, by point mutations and the accumulation of insertions and/or deletions, since their divergence from a common ancestor. There is no evidence that intragenic recombination has occurred among these three groups of alleles. In contrast, the class III alleles are associated with strains of both M. haemolytica and M. glucosida and have mosaic structures that have been derived, by horizontal DNA transfer and intragenic recombination, from class II and IV alleles. A comparison of the M. glucosida ompA5.1 allele with the M. glucosida ompA6.1 and ompA7.1 alleles suggested that ompA5.1 was derived from an ompA6.1-like allele by the acquisition of segment B from an M. haemolytica class II allele (Fig. 3). The similarity of the M. haemolytica ompA4.1 and ompA4.2 alleles to the M. glucosida ompA5.1 allele (Fig. 3) also suggests that ompA4.1 and ompA4.2 were acquired by horizontal transfer from M. glucosida. The occurrence of ompA4.1 and ompA4.2 within phylogenetically related serotype A7 and A13 strains of ETs 12 to 14 and 15 (Fig. 6), respectively, indicates that they have a common evolutionary origin. Serotype A7 and A13 M. haemolytica strains appear to represent a distinct clonal complex that has undergone frequent recombination with M. glucosida because serotype A7 and A13 M. haemolytica strains share features of their OMP and lipopolysaccharide profiles with M. glucosida isolates (14), and the lktA alleles of serotype A7 and A13 M. haemolytica strains contain recombinant segments that have been derived from M. glucosida isolates (17).
The association of identical, or nearly identical, class I and II ompA alleles with divergent phylogenetic lineages of M. haemolytica (Fig. 6) suggests that horizontal transfer and assortative (entire gene) recombination have been important factors in the evolution of ompA. The data also provide further evidence to support the view that host switching of strains from cattle to sheep and vice versa has contributed to these evolutionary events and to the emergence of new strains (13, 17). The horizontal transfer of class I ompA1-type alleles has occurred independently on different occasions within lineages A, B, and C. Bovine A1 and A6 strains of ETs 1 and 2 may have evolved from ovine A1 and A6 strains after transmission of the latter from sheep to cattle and the subsequent acquisition of ompA1-type alleles from bovine A2 isolates of ET 16, 17, or 21. Bovine A1 and A6 strains are more closely related to ovine isolates of the same serotypes than they are to bovine A2 strains (11), and they also share very similar lktA alleles (17). The horizontal transfer of class II ompA2-type alleles has occurred on numerous occasions within lineages A and C. The presence of identical ompA2.1 alleles in ovine A1, A5 to A7, A9, A12, A14, and A16 strains of ETs 3 to 11 (lineage A) suggests that ompA2.1 has undergone multiple and recent horizontal gene transfer and recombination events between divergent strains (11). The horizontal transfer of ompA2.1 has taken place so recently that there has been insufficient time for point mutations to accumulate. In contrast, the capsular polysaccharide antigens within these lineages are extremely diverse (there are at least eight capsular serotypes) and are clearly subject to strong diversifying selection, presumably due to the host immune response. Therefore, the capsular polysaccharide antigens and OmpA proteins of these strains are subject to very different selection pressures that are presumably related to their different functions.
The presence of divergent ompA alleles in closely related bovine (ompA1.3) and ovine (ompA2.3) serotype A2 isolates representing ET 21 provides clues about the evolutionary histories of these strains. One possibility is that the ovine strains evolved from ancestral bovine isolates after transmission of the latter from cattle to sheep. Subsequently, these bovine-derived strains acquired, by horizontal DNA transfer, ompA2-type alleles from ovine isolates, and after other evolutionary changes, evolved into the present-day ovine-adapted strains represented in lineage ET 21. Additional evidence to support this hypothesis comes from a comparative sequence analysis of the lktA gene (17). Thus, evidence for both the lktA and ompA genes suggests that strains of a common evolutionary origin have diverged and become adapted to different host species. The trigger for this divergence appears to have been the transmission of isolates from cattle to sheep and vice versa, which is probably linked to the domestication of these species (17).
P. trehalosi produces two OmpA homologs.
The discovery that P. trehalosi produces two OmpA homologs encoded by different tandemly arranged ompA genes was not entirely unexpected because similar findings have been described for Aeromonas salmonicida and Haemophilus ducreyi (8, 29). The presence of putative ribosome-binding, promoter, and termination sequences associated with each of the ompA genes also provides indirect evidence that they are independently transcribed (8, 54). A retrospective examination of the OMP profiles of a wide range of P. trehalosi isolates (16) indicated that the expression of two OmpA homologs is common in this species. The identification of tandem ompA genes in another bacterial species suggests that this phenomenon is more widespread in gram-negative bacteria than was previously thought. It also seems likely that a common underlying mechanism is responsible for generating tandem ompA genes in certain bacterial species and that the expression of two OmpA homologs provides a selective advantage to these organisms.
It has been suggested that the two ompA genes of A. salmonicida and H. ducreyi have arisen by gene duplication (8, 29). Based on the relatively low level of homology between the two OmpA proteins of A. salmonicida, Costello et al. (8) concluded that such a gene duplication event occurred in the distant evolutionary past. However, our data suggest an alternative possibility. The presence of very different ompA′-type alleles in strains PH68, PH246, and PH252 (ompA8.1 to -8.3) compared to that in strain PH254 (ompA9.1) and the occurrence of very similar ompA"-type alleles in the same strains (ompA10.1 to -10.4) indicate that one or the other (or both) of the ompA8- or ompA9-type alleles has been acquired by horizontal transfer. The low frequency of occurrence of the ompA9.1 allele suggests that this allele has replaced an ompA8-type allele, not vice versa, by horizontal gene transfer. In addition, the low level of similarity between the ompA′ and ompA" alleles, in contrast to the high degree of similarity among the ompA8- and ompA10-type alleles, is consistent with acquisition by horizontal transfer rather than by gene duplication. If one of the ompA genes had arisen by duplication in the distant past, we would also expect to see more divergence among alleles representing each of the ompA types, but this is not the case. Therefore, horizontal DNA transfer, rather than gene duplication, might account for the second ompA gene in this and other bacterial species.
Finally, it has previously been shown that large segments of DNA from the lktA genes of M. glucosida and P. trehalosi have become incorporated by intragenic recombination into the lktA gene of certain M. haemolytica serotypes (17). In the case of ompA, there was evidence for recombinational exchange involving M. glucosida and M. haemolytica but not for P. trehalosi and M. haemolytica. Therefore, we conclude that M. glucosida has been involved in frequent recombinational exchanges with certain M. haemolytica strains but that recombination between P. trehalosi and M. haemolytica is much less common.
Acknowledgments
This study was supported by a Wellcome Trust University Award to R. L. Davies (053669/Z/98/Z). I. Lee was supported by an Overseas Research Students' award.
REFERENCES
- 1.Angen, O., R. Mutters, D. A. Caugant, J. E. Olsen, and M. Bisgaard. 1999. Taxonomic relationships of the [Pasteurella] haemolytica complex as evaluated by DNA-DNA hybridizations and 16S rRNA sequencing with proposal of Mannheimia haemolytica gen. nov., comb. nov., Mannheimia granulomatis comb. nov., Mannheimia glucosida sp. nov., Mannheimia ruminalis sp. nov. and Mannheimia varigena sp. nov. Int. J. Syst. Bacteriol. 49:67-86. [DOI] [PubMed] [Google Scholar]
- 2.Arora, A., F. Abildgaard, J. H. Bushweller, and L. K. Tamm. 2001. Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat. Struct. Biol. 8:334-338. [DOI] [PubMed] [Google Scholar]
- 3.Beher, M. G., C. A. Schnaitman, and A. P. Pugsley. 1980. Major heat-modifiable outer membrane protein in gram-negative bacteria: comparison with the OmpA protein of Escherichia coli. J. Bacteriol. 143:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berggren, C. A., C. S. Baluyut, R. R. Simonson, W. J. Bemrick, and S. K. Maheswaran. 1981. Cytotoxic effects of Pasteurella haemolytica on bovine neutrophils. Am. J. Vet. Res. 42:1383-1388. [PubMed] [Google Scholar]
- 5.Bos, M. P., D. Hogan, and R. J. Belland. 1999. Homologue scanning mutagenesis reveals CD66 receptor residues required for neisserial Opa protein binding. J. Exp. Med. 190:331-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowland, S. L., and P. E. Shewen. 2000. Bovine respiratory disease: commercial vaccines currently available in Canada. Can. Vet. J. 41:33-48. [PMC free article] [PubMed] [Google Scholar]
- 7.Clinkenbeard, K. D., D. A. Mosier, A. L. Timko, and A. W. Confer. 1989. Effects of Pasteurella haemolytica leukotoxin on cultured bovine lymphoma cells. Am. J. Vet. Res. 50:271-275. [PubMed] [Google Scholar]
- 8.Costello, G. M., R. Vipond, and S. MacIntyre. 1996. Aeromonas salmonicida possesses two genes encoding homologs of the major outer membrane protein, OmpA. J. Bacteriol. 178:1623-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dabo, S. M., A. W. Confer, and R. A. Quijano-Blas. 2003. Molecular and immunological characterization of Pasteurella multocida serotype A:3 OmpA: evidence of its role in P. multocida interaction with extracellular matrix molecules. Microb. Pathog. 35:147-157. [DOI] [PubMed] [Google Scholar]
- 10.Datta, D. B., B. Arden, and U. Henning. 1977. Major proteins of the Escherichia coli outer cell envelope membrane as bacteriophage receptors. J. Bacteriol. 131:821-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, R. L., S. Arkinsaw, and R. K. Selander. 1997. Evolutionary genetics of Pasteurella haemolytica isolates recovered from cattle and sheep. Infect. Immun. 65:3585-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies, R. L., S. Arkinsaw, and R. K. Selander. 1997. Genetic relationships among Pasteurella trehalosi isolates based on multilocus enzyme electrophoresis. Microbiology 143:2841-2849. [DOI] [PubMed] [Google Scholar]
- 13.Davies, R. L., S. Campbell, and T. S. Whittam. 2002. Mosaic structure and molecular evolution of the leukotoxin operon (lktCABD) in Mannheimia (Pasteurella) haemolytica, Mannheimia glucosida, and Pasteurella trehalosi. J. Bacteriol. 184:266-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, R. L., and W. Donachie. 1996. Intraspecific diversity and host specificity within Pasteurella haemolytica based on variation of capsular polysaccharide, lipopolysaccharide and outer-membrane proteins. Microbiology 142:1895-1907. [DOI] [PubMed] [Google Scholar]
- 15.Davies, R. L., B. J. Paster, and F. E. Dewhirst. 1996. Phylogenetic relationships and diversity within the Pasteurella haemolytica complex based on 16S rRNA sequence comparison and outer membrane protein and lipopolysaccharide analysis. Int. J. Syst. Bacteriol. 46:736-744. [DOI] [PubMed] [Google Scholar]
- 16.Davies, R. L., and M. Quirie. 1996. Intra-specific diversity within Pasteurella trehalosi based on variation of capsular polysaccharide, lipopolysaccharide and outer-membrane proteins. Microbiology 142:551-560. [DOI] [PubMed] [Google Scholar]
- 17.Davies, R. L., T. S. Whittam, and R. K. Selander. 2001. Sequence diversity and molecular evolution of the leukotoxin (lktA) gene in bovine and ovine strains of Mannheimia (Pasteurella) haemolytica. J. Bacteriol. 183:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Mot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these proteins with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 19.Duim, B., L. D. Bowler, P. P. Eijk, H. M. Jansen, J. Dankert, and L. Van Alphen. 1997. Molecular variation in the major outer membrane protein P5 gene of nonencapsulated Haemophilus influenzae during chronic infections. Infect. Immun. 65:1351-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank, G. H. 1989. Pasteurellosis of cattle, p. 197-222. In C. F. Adlam and J. M. Rutter (ed.), Pasteurella and pasteurellosis. Academic Press, London, United Kingdom.
- 21.Gilmour, N. J. L., and J. S. Gilmour. 1989. Pasteurellosis of sheep, p. 223-261. In C. F. Adlam and J. M. Rutter (ed.), Pasteurella and pasteurellosis. Academic Press, London, United Kingdom.
- 22.Hancock, R. E. W. 1991. Bacterial outer membranes: evolving concepts. ASM News 57:175-182. [Google Scholar]
- 23.Highlander, S. K. 2001. Molecular genetic analysis of virulence in Mannheimia (Pasteurella) haemolytica. Front. Biosci. 6:1128-1150. [DOI] [PubMed] [Google Scholar]
- 24.Hill, D. J., M. A. Toleman, D. J. Evans, S. Villullas, L. Van Alphen, and M. Virji. 2001. The variable P5 proteins of typeable and non-typeable Haemophilus influenzae target human CEACAM1. Mol. Microbiol. 39:850-862. [DOI] [PubMed] [Google Scholar]
- 25.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 26.Kaehler, K. L., R. J. Markam, C. C. Muscoplat, and D. W. Johnson. 1980. Evidence of cytocidal effects of Pasteurella haemolytica on bovine peripheral blood mononuclear leukocytes. Am. J. Vet. Res. 41:1690-1693. [PubMed] [Google Scholar]
- 27.Kaehler, K. L., R. J. F. Markham, C. C. Muscoplat, and D. W. Johnson. 1980. Evidence of species specificity in the cytocidal effects of Pasteurella haemolytica. Infect. Immun. 30:615-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karplus, K., C. Barrett, and R. Hughey. 1998. Hidden Markov models for detecting remote protein homologies. Bioinformatics 14:846-856. [DOI] [PubMed] [Google Scholar]
- 29.Klesney-Tait, J., T. J. Hiltke, I. Maciver, S. M. Spinola, J. D. Radolf, and E. J. Hansen. 1997. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J. Bacteriol. 179:1764-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 31.Mahasreshti, P. J., G. L. Murphy, J. H. Wyckoff III, S. Farmer, R. E. Hancock, and A. W. Confer. 1997. Purification and partial characterization of the OmpA family of proteins of Pasteurella haemolytica. Infect. Immun. 65:211-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGraw, E. A., J. Li, R. K. Selander, and T. S. Whittam. 1999. Molecular evolution and mosaic structure of alpha, beta, and gamma intimins of pathogenic Escherichia coli. Mol. Biol. Evol. 16:12-22. [DOI] [PubMed] [Google Scholar]
- 33.Millman, K. L., S. Tavaré, and D. Dean. 2001. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J. Bacteriol. 183:5997-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morona, R., M. Klose, and U. Henning. 1984. Escherichia coli K12 outer membrane protein as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J. Bacteriol. 159:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morona, R., C. Kramer, and U. Henning. 1985. Bacteriophage receptor area of outer membrane protein OmpA of Escherichia coli K12. J. Bacteriol. 164:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 38.Ogunnariwo, J. A., and A. B. Schryvers. 1990. Iron acquisition in Pasteurella haemolytica: expression and identification of a bovine-specific transferrin receptor. Infect. Immun. 58:2091-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pautsch, A., and G. E. Schulz. 2000. High-resolution structure of the OmpA membrane domain. J. Mol. Biol. 298:273-282. [DOI] [PubMed] [Google Scholar]
- 40.Pautsch, A., and G. E. Schulz. 1998. Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 5:1013-1017. [DOI] [PubMed] [Google Scholar]
- 41.Popp, A., C. Dehio, F. Grunert, T. F. Meyer, and S. D. Gray-Owen. 1999. Molecular analysis of neisserial Opa protein interactions with the CEA family of receptors: identification of determinants contributing to the differential specificities of binding. Cell. Microbiol. 1:169-181. [DOI] [PubMed] [Google Scholar]
- 42.Prasadarao, N. V. 2002. Identification of Escherichia coli outer membrane protein A receptor on human brain microvascular endothelial cells. Infect. Immun. 70:4556-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prasadarao, N. V., C. A. Wass, and K. S. Kim. 1996. Endothelial cell GlcNAcβ1-4GlcNAc epitopes for outer membrane protein A enhance traversal of Escherichia coli across the blood-brain barrier. Infect. Immun. 64:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prasadarao, N. V., C. A. Wass, J. N. Weiser, M. F. Stins, S. Huang, and K. S. Kim. 1996. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64:146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puohiniemi, R., M. Karvonen, J. Vuopio-Varkila, A. Muotiala, I. M. Helander, and M. Sarvas. 1990. A strong antibody response to the periplasmic C-terminal domain of the OmpA protein of Escherichia coli is produced by immunization with purified OmpA or with whole E. coli or Salmonella enterica serovar Typhimurium bacteria. Infect. Immun. 58:1691-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy, M. S., J. M. Bernstein, T. F. Murphy, and H. S. Faden. 1996. Binding between outer membrane proteins of nontypeable Haemophilus influenzae and human nasopharyngeal mucin. Infect. Immun. 64:1477-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reid, S. D., R. K. Selander, and T. S. Whittam. 1999. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J. Bacteriol. 181:153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schweizer, M., and U. Henning. 1977. Action of a major outer cell envelope membrane protein in conjugation of Escherichia coli K-12. J. Bacteriol. 129:1651-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shewen, P. E., and B. N. Wilkie. 1982. Cytotoxin of Pasteurella haemolytica acting on bovine leukocytes. Infect. Immun. 35:91-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, J. M. 1999. The detection and measurement of recombination from sequence data. Genetics 153:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sneath, P. H. A., and M. Stevens. 1990. Actinobacillus rossii sp. nov., Actinobacillus seminis sp. nov., nom. rev., Pasteurella bettii sp. nov., Pasteurella lymphangitidis sp. nov., Pasteurella mairi sp. nov., and Pasteurella trehalosi sp. nov. Int. J. Syst. Bacteriol. 40:148-153. [DOI] [PubMed] [Google Scholar]
- 52.Sonntag, I., H. Schwartz, Y. Hirota, and U. Henning. 1978. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J. Bacteriol. 136:280-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tagawa, Y., M. Haritani, H. Ishikawa, and N. Yuasa. 1993. Characterization of a heat-modifiable outer membrane protein of Haemophilus somnus. Infect. Immun. 61:1750-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Throm, R. E., J. A. Al-Tawfiq, K. R. Fortney, B. P. Katz, A. F. Hood, C. A. Slaughter, E. J. Hansen, and S. M. Spinola. 2000. Evaluation of an isogenic major outer membrane protein-deficient mutant in the human model of Haemophilus ducreyi infection. Infect. Immun. 68:2602-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torres, A. G., and J. B. Kaper. 2003. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect. Immun. 71:4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Virji, M., D. Evans, A. Hadfield, F. Grunert, A. M. Teixeira, and S. M. Watt. 1999. Critical determinants of host receptor targeting by Neisseria meningitidis and Neisseria gonorrhoeae: identification of opa adhesiotopes on the N-domain of CD66 molecules. Mol. Microbiol. 34:538-551. [DOI] [PubMed] [Google Scholar]
- 57.Webb, D. C., and A. W. Cripps. 1998. Secondary structure and molecular analysis of interstrain variability in the P5 outer-membrane protein of non-typable Haemophilus influenzae isolated from diverse anatomical sites. J. Med. Microbiol. 47:1059-1067. [DOI] [PubMed] [Google Scholar]
- 58.Weiser, J. N., and E. C. Gotschlich. 1991. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect. Immun. 59:2252-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu, R. H., S. D. Gray-Owen, J. Ogunnariwo, and A. B. Schryvers. 1992. Interaction of ruminant transferrins with transferrin receptors in bovine isolates of Pasteurella haemolytica and Haemophilus somnus. Infect. Immun. 60:2992-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeng, H., K. Pandher, and G. L. Murphy. 1999. Molecular cloning of the Pasteurella haemolytica pomA gene and identification of bovine antibodies against PomA surface domains. Infect. Immun. 67:4968-4973. [DOI] [PMC free article] [PubMed] [Google Scholar]