Abstract
FtsZ is the major cytoskeletal component of the bacterial cell division machinery. It forms a ring-shaped structure (the Z ring) that constricts as the bacterium divides. Previous in vivo experiments with green fluorescent protein-labeled FtsZ and fluorescence recovery after photobleaching have shown that the Escherichia coli Z ring is extremely dynamic, continually remodeling itself with a half time of 30 s, similar to microtubules in the mitotic spindle. In the present work, under different experimental conditions, we have found that the half time for fluorescence recovery of E. coli Z rings is even shorter (∼9 s). As before, the turnover appears to be coupled to GTP hydrolysis, since the mutant FtsZ84 protein, with reduced GTPase in vitro, showed an ∼3-fold longer half time. We have also extended the studies to Bacillus subtilis and found that this species exhibits equally rapid dynamics of the Z ring (half time, ∼8 s). Interestingly, null mutations of the FtsZ-regulating proteins ZapA, EzrA, and MinCD had only modest effects on the assembly dynamics. This suggests that these proteins do not directly regulate FtsZ subunit exchange in and out of polymers. In B. subtilis, only 30 to 35% of the FtsZ protein was in the Z ring, from which we conclude that a Z ring only 2 or 3 protofilaments thick can function for cell division.
FtsZ is a structural homolog of tubulin found in virtually all prokaryotes, where it is the major cytoskeletal protein in cell division. It forms a ring structure (the Z ring) at the midpoint of the cell and, for both Escherichia coli and Bacillus subtilis, is required for the recruitment of all of the subsequent proteins involved in the synthesis of the new septum (7). FtsZ has many features in common with eukaryotic tubulin, including a very similar tertiary structure, GTPase activity, and the ability to assemble into protofilaments in vitro (6, 14, 19, 20, 30). It is generally assumed that the Z ring in vivo consists of some arrangement of polymerized FtsZ protofilaments. Proper localization of the Z ring to mid-cell is accomplished by at least two mechanisms, (i) nucleoid occlusion and (ii) inhibition of polar FtsZ assembly by the Min system (MinCD and DivIVA in B. subtilis and MinCDE in E. coli) (7, 11, 18). In addition to the Min proteins, other potential regulators of FtsZ polymerization have been identified. In B. subtilis, lack of the negative regulator EzrA lowers the FtsZ concentration required for Z-ring formation in vivo and gives rise to extraneous, mislocalized FtsZ structures (13). In addition, a positive regulator, ZapA, was found to promote FtsZ polymerization and bundling in vitro, and deletion of its gene led to death of cells with abnormally low FtsZ levels (10).
Recent results have shown that the Z ring is a surprisingly dynamic structure (28), with subunits turning over at a rate similar to that of microtubules in mitotic spindles of eukaryotic cells (2, 25). However, it was unclear whether this dynamic behavior is unique to E. coli or occurs in other species. Another question is whether the rapid dynamics are intrinsic to FtsZ assembly or are regulated by other proteins, in particular, the proteins described above. We have therefore extended our fluorescence recovery after photobleaching (FRAP) studies to examine Z-ring dynamics in another species, B. subtilis, and have investigated the effect of known FtsZ-regulating proteins.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli and B. subtilis strains used in this study are listed in Table 1. The B. subtilis strains were all derivatives of the wild-type PY79 strain and contained an integrated chromosomal copy of ftsZ-gfp (in addition to the genomic ftsZ gene) under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter. Mutants were constructed by transforming Pspac-ftsZ-gfp strain JDB401 with genomic DNA from strains PL990 (ΔminCD::spc) (12a), PL867 (ΔezrA::spc) (13), FG345 (ΔzapA-yshB::tet) (10), and FG196 (ΔminD::kan). The E. coli strains contained either plasmid pJSB150 (ftsZ-gfp) or pJSB151 (ftsZ84-gfp). Plasmids pJSB150 and pJSB151 are derivatives of pJSB2, an expression plasmid based on pBAD18 but containing an altered multicloning site and a less efficient Shine-Dalgarno sequence and with the ampicillin resistance replaced by chloramphenicol resistance (27).
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype | Reference | Source |
|---|---|---|---|
| B. subtilis | |||
| JDB401 | amyE::Pspac-ftsZ-gfp cat | 1 | J. Dworkin |
| FG487 | ΔzapA-yshB::tet amyE::Pspac-ftsZ-gfp cat | This study | F. Gueiros-Filho |
| FG489 | ΔezrA::spc amyE::Pspac-ftsZ-gfp cat | This study | F. Gueiros-Filho |
| FG522 | ΔminD::kan amyE::Pspac-ftsZ-gfp cat | This study | F. Gueiros-Filho |
| FG524 | ΔminCD::spc amyE::Pspac-ftsZ-gfp cat | This study | F. Gueiros-Filho |
| E. coli | |||
| BW27783 | lacIqrrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 Δ(araFGH) Φ(ΔaraEp PCP8-araE) | 12 | J. Keasling |
| JFL101 | F−ilv deo ara(Am) lac-125(Am) galU42(Am) tyrT(SupFtsA81) recA ftsZ84(Ts) | 16 | J. Lutkenhaus |
| WM1032 | MG1655 Δlac ΔminCDE::kan | 29 | W. Margolin |
Media and growth conditions.
For the B. subtilis strains, overnight cultures were diluted 100- or 200-fold in Luria-Bertani (LB) medium or LB plus 5 μg of chloramphenicol per ml. They were allowed to recover for ∼1 h at 37°C with shaking and then induced with 50 μM IPTG until mid-log phase (2 to 3 h) prior to viewing in the microscope. E. coli BW27783(pJSB150) was grown similarly, but in LB plus 34 μg of chloramphenicol per ml, and was induced with 0.00005 to 0.0002% arabinose. Strain WM1032(pJSB150) was diluted and grown in LB plus 30 μg of kanamycin per ml plus 34 μg of chloramphenicol per ml at room temperature (∼24°C) until early log phase (optical density, 0.05 to 0.1) and then induced with 0.001% arabinose for 2 to 3 h before microscopy. To avoid salt rescue of the ftsZ84 phenotype, strain JFL101(pJSB151) was grown in MOPS+ medium (28) containing 0.5% glycerol and 34 μg of chloramphenicol per ml at 30°C until early log phase (optical density, 0.05 to 0.1) and then induced with 0.001% arabinose for 2 to 3 h before microscopy. For all strains, the final 30 min or more of shaking prior to microscopy was done at room temperature. Also, during FRAP experiments requiring multiple slides, cultures were rediluted with continued shaking at room temperature in order to maintain a log-phase culture. To check that the green fluorescent protein (GFP) remained attached to the FtsZ protein, we performed Western blot assays with a polyclonal anti-GFP antibody (Clontech). The B. subtilis fusion protein showed no visible degradation, and in E. coli <5% of the GFP was cleaved off. This is an important control for our measurement of the relative amounts of FtsZ in the ring and cytoplasm.
Microscopy and FRAP.
Slides were prepared by (i) melting a solution of 1% agarose in MOPS+ medium-0.5% glycerol, (ii) pipetting 50 μl onto a siliconized glass slide with two pieces of laboratory tape as spacers, and (iii) dropping a plain glass slide on top to create a thin agarose pad. After separating the two slides, 5 μl of culture was pipetted onto the agarose pad on the plain glass slide and then covered with a number 1 coverslip that was fixed to the slide with 2 drops of melted Valap (1:1:1 petrolatum-lanolin-paraffin). Cells were imaged and FRAP experiments were performed on a Nikon TE300 inverted fluorescence microscope equipped with a laser source with Metamorph software as previously described (17, 28). Unless otherwise noted, all images and FRAP measurements were made at room temperature (24 to 26°C). Exposures were 350 to 400 ms, and laser bleach pulses were 35 to 45 ms. Time series were typically taken over 1 to 5 min with exposures every 2 to 10 s, depending on the speed of recovery.
Measurements of integrated fluorescence intensity in the bleached and other regions at each time point were made in Metamorph and exported to Excel 2000. For each measured region, background intensity was subtracted and a correction factor was applied for overall photobleaching of the sample during observation (determined from intensity over time of a control cell in the same field). Recovery half times were determined by performing a least-squares fit of the intensity of the bleached region over time to the single-exponential equation F∞ − F(t) = [F∞ − F0] e−kt, where F∞ is the fluorescence of the bleached region after maximal recovery, F(t) is the fluorescence at time t, and F0 is the initial fluorescence just after bleaching (t = 0). Fitting was performed with the Solver function of Excel and allowing F∞, F0, and the half-time, (t1/2 t1/2 = ln 2/k), to vary.
RESULTS
Assembly dynamics of B. subtilis and E. coli Z rings are equally rapid.
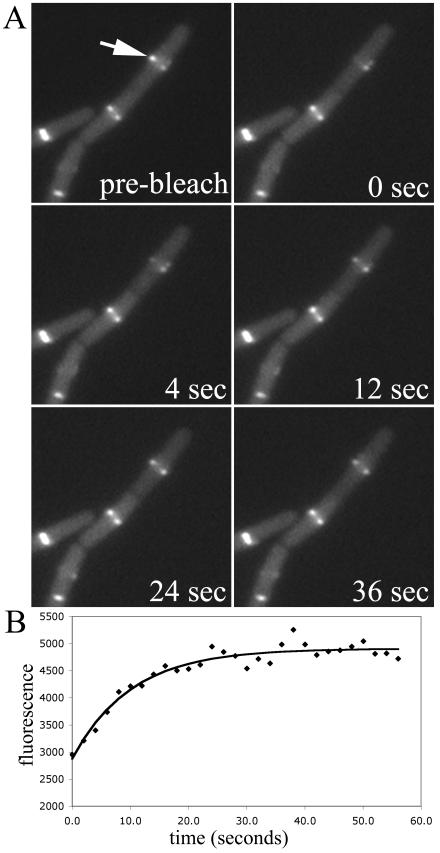
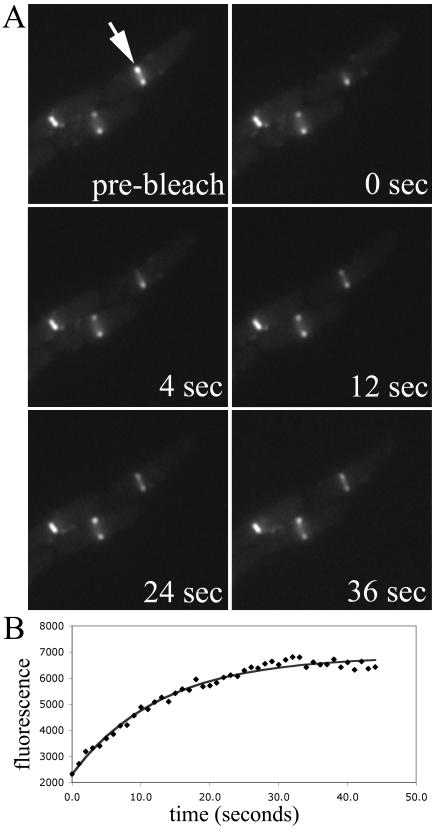
In order to test whether the rapid subunit exchange observed in E. coli Z rings also occurs in other bacterial species, we performed FRAP experiments with B. subtilis strain JDB401, which expresses an FtsZ-GFP fusion at relatively low levels, along with the normal level of endogenous FtsZ (13). Figure 1A shows selected time points of a typical FRAP image series in which we bleached one-half of the Z ring, and Fig. 1B shows the corresponding time course of fluorescence recovery in the bleached region. Fitting the corrected fluorescence intensity to the equation given in Materials and Methods gave a half time of recovery of 7.0 s for the series shown. This shows that Z rings of other bacterial species do in fact exhibit extremely rapid dynamics. The average half time of recovery for JDB401 cells was 8 ± 3 s (n = 19). Because the 8-s half time observed for B. subtilis was much shorter than the 30-s half time previously reported for E. coli, we also performed FRAP with E. coli BW27783(pJSB150) cells under the same conditions (agarose pad immobilization, LB medium, ∼25°C). E. coli BW27783 cells were chosen because of their more uniform level of expression from pBAD within a population of E. coli cells (12). A typical FRAP time series for these cells is shown in Fig. 2. Surprisingly, the E. coli Z rings exhibited the same rapid turnover as those of B. subtilis, with an average half time of 9 ± 3 s (n = 10). This exchange is about threefold faster than that observed previously for E. coli (28). The different exchange rates in the two studies could not be wholly attributed to any one factor, but the method of immobilization (agarose pad in the present study, polylysine-coated coverslips in the previous one) had the greatest effect (data not shown). The exact method of data analysis may also be a factor.
FIG. 1.
FRAP of an FtsZ ring in wild-type B. subtilis cells. (A) Time-lapse series of fluorescence images showing the time course of recovery. The arrow shows the half-ring to be bleached. (B) Intensity of the photobleached region over time. The data points have been corrected for background and photobleaching during the observation period. The points represent the fluorescence data, and the solid line is the predicted recovery curve given the first-order rate constant, k, determined by least-squares fitting of the data to the equation given in Materials and Methods. The recovery half time for this series was 7.0 s.
FIG. 2.
FRAP of an FtsZ ring in wild-type E. coli BW27783(pJSB150) cells. (A) Time-lapse series of fluorescence images. (B) Intensity of the photobleached region over time. The half time of recovery for the series shown was 8.9 s.
A contrast-enhanced movie showing disassembly and reassembly of Z rings in dividing E. coli BW27783(pJSB150) cells at 37°C can be accessed at http://www.cellbio.duke.edu/Faculty/Erickson/. The overall time scale is 10 min, with frames taken at 10-s intervals. An interesting feature of this movie is that individual FtsZ-GFP foci can be seen translocating across the cells, into and out of the constricting and reforming Z rings.
We also found, on the basis of integrated fluorescence intensities in unbleached cells, that the proportion of FtsZ protein in the Z ring is the same, approximately 30 to 35%, in both E. coli and B. subtilis cells, and we did not observe any obvious correlation between the degree of constriction of rings and their rate of recovery (data not shown). Because of the size of the focused laser beam and the small volume of bacterial cells, we unavoidably bleached a significant portion of the FtsZ-GFP pool with the laser photobleaching pulse. Thus, on the basis of this study, we are not able to draw any conclusions about percent recovery and mobile or immobile fractions. The graphs of fluorescence recovery over time in Fig. 1 to 4 are not normalized.
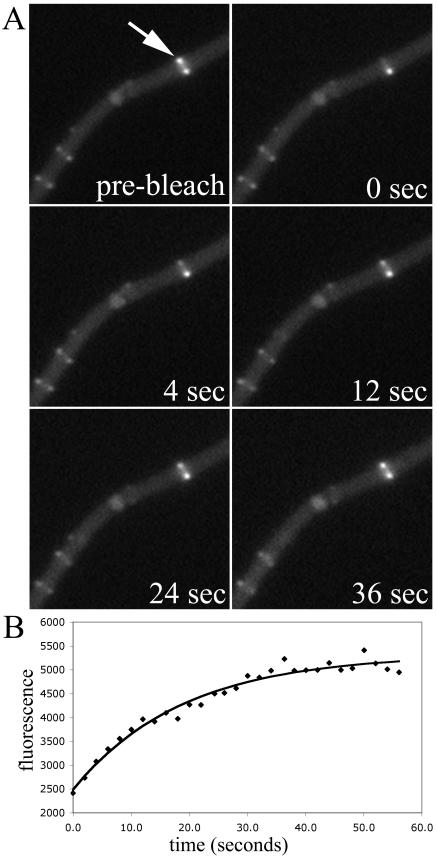
FIG. 4.
FRAP of FtsZ rings in B. subtilis cells lacking minCD. (A) Time-lapse series of fluorescence images. (B) Intensity of the photobleached region over time. Recovery half time, 10.7 s.
Effect of FtsZ-regulating proteins on Z-ring turnover.
To determine whether the rapid dynamics observed are due to the action of known FtsZ-interacting and -regulating proteins, we performed FRAP with strains containing null mutations of several genes. We will subsequently refer to E. coli BW27783 and B. subtilis JDB401 as the wild-type strains since they served as our control strains for comparison with the mutants.
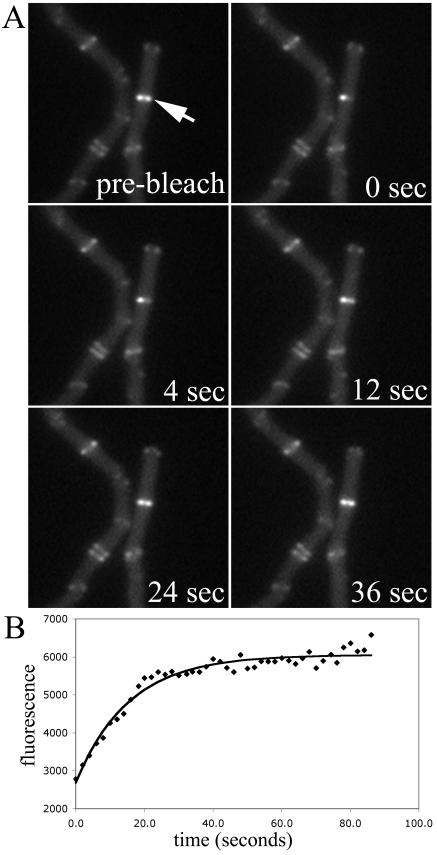
The first mutant that we tested was a B. subtilis strain, FG487, that lacks the protein ZapA, a positive FtsZ assembly factor (10). As previously reported, the FtsZ structures in this mutant appeared normal. We had expected that the absence of the stabilizing factor might increase assembly dynamics, but the average recovery half time was similar to that of the wild type, at 10 ± 4 s (n = 12). The next strain we examined was FG489, shown in Fig. 3, which lacks the negative regulator of ring assembly, EzrA (13). As previously reported, cells often showed extraneous, mislocalized rings and also residual Z-ring material at previous division sites. In this case, the absence of the destabilizing factor did give a longer recovery half time of 14 ± 5 s (n = 13), which was statistically significant (Table 2). However, the absolute difference is not very large, less than a factor of 2. The final two B. subtilis mutant strains examined were FG522, which lacks MinD, and FG524, which lacks both MinC and MinD. As expected, these cells showed extra, mislocalized Z rings and some double rings in FG524 (Fig. 4). Similar to the results obtained with the ΔzapA and ΔezrA mutant strains, neither FG522 nor FG524 showed a large difference in Z-ring stability, with average recovery half times of 12 ± 6 (n = 8) and 10 ± 2 (n = 5) s. In general for all of these strains, we chose single, medial rings for the FRAP measurements. In the case of the ΔezrA and ΔminCD strains, we also examined several mislocalized rings, and these showed dynamic behavior similar to that of the medial rings (data not shown).
FIG. 3.
FRAP of FtsZ rings in B. subtilis cells lacking ezrA. (A) Time-lapse series of fluorescence images. (B) Intensity of the photobleached region over time. Recovery half time, 13.0 s.
TABLE 2.
Summary of B. subtilis and E. coli FRAP measurements
| Strain | Recovery half time ± SD (s) |
|---|---|
| B. subtilis | |
| JDB401 | 8 ± 3 |
| FG487 (ΔzapA) | 10 ± 4 |
| FG489 (ΔezrA) | 14 ± 5a |
| FG522 (ΔminD) | 12 ± 6 |
| FG524 (ΔminCD) | 10 ± 2b |
| E. coli | |
| BW27783 | 9 ± 3 |
| JFL101 ftsZ84 | 30 ± 10c |
| WM1032 (ΔminCDE) | 19 ± 3d |
Significantly different from wild type (P < 0.001).
Significantly different from wild type (P < 0.05).
Significantly different from wild type (P < 0.0001).
Significantly different from wild type (P < 0.0005).
We also investigated Z-ring stability in two mutant E. coli strains. The first, JFL101(pJSB151), carries the ftsZ84 mutation (G105S), which was shown previously to reduce the turnover of the Z ring by a factor of about 10, consistent with its reduced GTPase activity in vitro (28). Under our present experimental conditions this strain gave an approximately three- to fourfold decrease in the turnover of Z rings (t1/2 = 30 ± 10 s; n = 9). The second E. coli strain, WM1032(pJSB150), lacks the MinC, MinD, and MinE proteins. Similar to B. subtilis, most of these cells had extraneous, polar Z-ring structures. In this case, however, the absence of the MinCDE system had a more pronounced effect on Z-ring turnover, with a recovery half time of 19 ± 3 s (n = 3), approximately twice that of the wild type. Table 2 summarizes the average recovery times for all of the strains and their statistical significances.
Finally, we attempted to measure FRAP of the cytoplasmic pool of FtsZ to gain information about its polymerization state. The recovery was too fast for accurate measurement. When a spot was bleached in the cytoplasm on one side of the bacterium, that side showed a uniform decrease in fluorescence in ∼0.5 s, and the fluorescence was equilibrated on the two sides in ∼1 s (data not shown).
DISCUSSION
The rapid assembly dynamics of the Z ring were virtually identical for E. coli and B. subtilis, suggesting that rapid exchange of subunits is a conserved property of Z rings in most or all bacterial species. This continuous turnover of subunits may therefore play a role in the mechanism of Z-ring function. It is, however, important to remember that the mutant FtsZ84 protein appears to divide normally at 30°C, despite a threefold reduction in dynamics. This suggests that the dynamics of FtsZ have evolved to exceed what may be essential for division and that some other factor is rate limiting for division and cell cycle timing.
An important observation is that the fraction of FtsZ incorporated into the ring, ∼30%, is the same for both species. On the basis of an estimation of 15,000 FtsZ molecules per E. coli cell (15), if one-third of that protein is in the ring, the E. coli Z ring would be an average of 6 protofilaments wide (28). In a cell ready for division (about 1.4 times as long as an average cell) it could be just over 8 protofilaments thick. However, in B. subtilis, it has been estimated that there are only 5,000 FtsZ molecules per cell (8). Thus, in B. subtilis we estimate the Z ring to be only 2 protofilaments wide on average, or about 3 protofilaments at the time of constriction. E. coli strain MC1061 was likewise reported to have only 5,000 molecules of FtsZ when growing in minimal medium (24). Assuming that it also has only ∼30% of its FtsZ in the Z ring, this E. coli B/r K strain can achieve cell division with a ring only 2 to 3 protofilaments thick.
It has been hypothesized that Z-ring dynamics are the result of a balance of stabilizing and destabilizing factors (10, 22), and in this study we tested several potential modulators of this behavior. It is not very surprising that assembly dynamics are not altered by a lack of ZapA, given that the null strain has almost no phenotype in terms of septation. It also has only a small effect on the distribution of FtsZ between the ring and the cytoplasm (wild type, 30% ± 4%; ΔzapA mutant, 25% ± 4%; F. Gueiros-Filho, unpublished observations). However, we have not tested the effect of zapA deletion in the absence of EzrA or with lowered levels of FtsZ. EzrA, which has a more noticeable phenotype when deleted, did affect the dynamics more significantly and in the direction expected. However, recovery half times from this strain were also more variable and the small absolute difference from the wild type is of questionable significance. Removing the Min proteins from B. subtilis also produced only a small increase in the turnover half time, suggesting that direct inhibition of FtsZ assembly in the Z ring is not a primary function of B. subtilis MinC. In E. coli, however, Z rings of the ΔminCDE strain were stabilized approximately twofold, almost as much as those formed of FtsZ84 with its decreased GTPase activity. This suggests that in E. coli, MinC(DE) might directly interact with already formed Z rings to inhibit or actively disassemble FtsZ polymers.
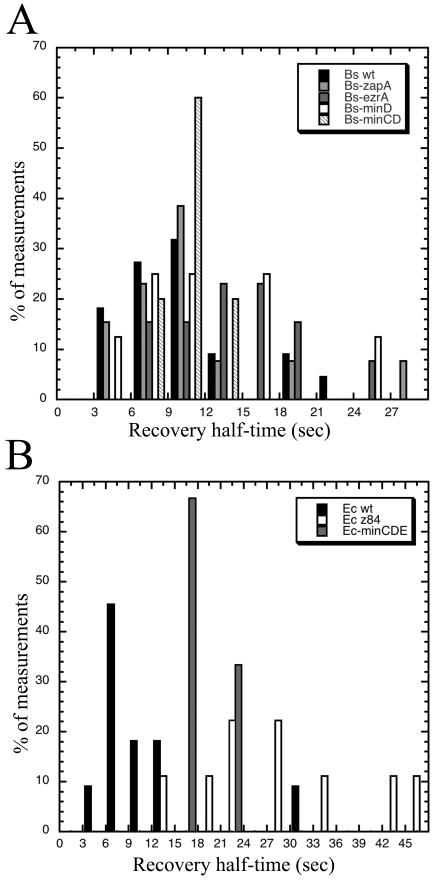
The recovery times showed a large variation for each strain examined (Fig. 5), with half times ranging from 4 s to more than 20 s even in the wild-type strains. The few cells with long recovery times (>20 s) were considered possibly unhealthy and were excluded from the wild-type average recovery times. Given this large variability in the populations of wild-type cells, we suggest that the slightly altered dynamics in the mutant B. subtilis strains may not represent an important functional difference. The simplest interpretation is that the dynamic behavior of the Z ring is an intrinsic property of FtsZ polymers and is only slightly altered by known regulatory proteins. It is notable, however, that E. coli strains with min deleted showed a more pronounced increase in recovery half times. This may represent a direct effect on Z-ring stability, but the stabilization was still only a factor of 2.
FIG. 5.
Histograms of Z-ring fluorescence recovery times for wild-type (wt) versus mutant cells of B. subtilis (Bs) (A) and E. coli (Ec) (B).
We tried to address the question of whether cytoplasmic FtsZ is monomeric or assembled into protofilaments. In vitro studies with E. coli FtsZ indicate a critical concentration of ∼1 μM under physiological solution conditions (9), so we expect most of the cytoplasmic FtsZ (∼8 μM for E. coli, assuming that 70% of the total is in the cytoplasm) to be assembled into protofilaments. When we did FRAP with cytoplasmic FtsZ in strain BW27783, we found that it was completely re-equilibrated across the bleached half of the cell in ∼0.5 s and across both sides of the cell in ∼1 s. This seems somewhat slower than the ∼0.5-s time for equilibration of monomeric GFP (4), but our instrument did not have a time resolution that could accurately measure a diffusion coefficient. If the equilibration time is slower than for GFP it would suggest that cytoplasmic FtsZ is assembled into protofilaments, but with the present data we cannot make a clear distinction between monomers and short polymers of FtsZ. We attempted to use fluorescence correlation spectroscopy to determine a diffusion coefficient for the cytoplasmic form(s) of FtsZ, but we found that the cellular FtsZ-GFP pool was photobleached too rapidly to obtain useful measurements.
The 8- to 9-s half time we observed for turnover of the FtsZ ring in B. subtilis and E. coli is similar to the fastest turnover in the eukaryotic cytoskeleton, specifically, the mitotic apparatus of cultured mammalian cells (26). Turnover of tubulin is generated by dynamic instability, which involves extended cycles of assembly and disassembly at the microtubule ends (3, 5). The assembly dynamics of FtsZ are much less well understood. Recent work in our laboratory suggests that FtsZ protofilaments assemble cooperatively by addition of subunits to the ends, with an assembly time similar to the in vivo turnover rate (Y. Chen et al., submitted for publication). Novel experiments are needed to confirm this and establish a detailed assembly mechanism.
Finally, the rapid dynamics has implications for the structure of the Z ring. We have estimated that the Z ring is approximately 6 to 8 or 2 to 3 protofilaments wide for E. coli and B. subtilis, but we have not resolved whether there are a few very long protofilaments or many short ones. We will postulate that assembly occurs by addition of subunits to the ends at a rate of 5 μM−1 s−1. This is the diffusion-limited rate for protein-protein association (21) and is in agreement with our recent in vitro kinetic studies (Chen et al., submitted). If the concentration of free subunits is 1 μM, which is the critical concentration determined in vitro (9), then protofilaments would grow at a rate of five subunits per s. This would mean that protofilaments could grow only 45 subunits long during the 9-s turnover half time, or 90 subunits long during the entire recovery time. A protofilament 45 subunits long would be about 200 nm long, much shorter than the 3,000-nm circumference of the bacterium. This estimate of protofilament length is similar to the average protofilament length of 23 subunits for E. coli FtsZ measured in vitro (23). Thus, both in vitro studies and the rapid assembly dynamics observed in vivo suggest that the Z ring is an assembly of many short protofilaments.
Acknowledgments
This work was supported by NIH grants GM 066014 to H. P. Erickson and GM 18568 to R. Losick. F. J. Gueiros-Filho was a Helen Hay Whitney Foundation postdoctoral fellow during part of this work.
We especially thank the laboratory of E. D. Salmon, University of North Carolina, for the use of the FRAP microscope and R. Losick, Harvard University, for support of F.J.G.-F. and comments on the manuscript. We also thank J. Lutkenhaus, J. D. Keasling, P. A. Levin, and W. Margolin for providing bacterial strains and J. Stricker for construction of pJSB150 and pJSB151.
REFERENCES
- 1.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. [DOI] [PubMed] [Google Scholar]
- 2.Cassimeris, L. U., R. A. Walker, N. K. Pryer, and E. D. Salmon. 1987. Dynamic instability of microtubules. Bioessays 7:149-154. [DOI] [PubMed] [Google Scholar]
- 3.Desai, A., and T. J. Mitchison. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83-117. [DOI] [PubMed] [Google Scholar]
- 4.Elowitz, M. B., M. G. Surette, P. E. Wolf, J. B. Stock, and S. Leibler. 1999. Protein mobility in the cytoplasm of Escherichia coli. J. Bacteriol. 181:197-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erickson, H. P., and E. T. O'Brien. 1992. Microtubule dynamic instability and GTP hydrolysis. Annu. Rev. Biophys. Struct. Biol. 21:145-166. [DOI] [PubMed] [Google Scholar]
- 6.Erickson, H. P., D. W. Taylor, K. A. Taylor, and D. Bramhill. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. USA 93:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feucht, A., I. Lucet, M. D. Yudkin, and J. Errington. 2001. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40:115-125. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez, J. M., M. Jimenez, M. Velez, J. Mingorance, J. M. Andreu, M. Vicente, and G. Rivas. 2003. Essential cell division protein FtsZ assembles into one monomer-thick ribbons under conditions resembling the crowded intracellular environment. J. Biol. Chem. 278:37664-37671. [DOI] [PubMed] [Google Scholar]
- 10.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harry, E. J. 2001. Bacterial cell division: regulating Z-ring formation. Mol. Microbiol. 40:795-803. [DOI] [PubMed] [Google Scholar]
- 12.Khlebnikov, A., K. A. Datsenko, T. Skaug, B. L. Wanner, and J. D. Keasling. 2001. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241-3247. [DOI] [PubMed] [Google Scholar]
- 12a.Levin, P. A., J. J. Shim, and A. D. Grossman. 1998. Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis J. Bacteriol. 180:6048-6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levin, P. A., I. G. Kurtser, and A. D. Grossman. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 96:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203-206. [DOI] [PubMed] [Google Scholar]
- 15.Lu, C., J. Stricker, and H. P. Erickson. 1998. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima—quantitation, GTP hydrolysis, and assembly. Cell Motil. Cytoskelet. 40:71-86. [DOI] [PubMed] [Google Scholar]
- 16.Lutkenhaus, J. F., H. Wolf-Watz, and W. D. Donachie. 1980. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J. Bacteriol. 142:615-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maddox, P. S., K. S. Bloom, and E. D. Salmon. 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margolin, W. 2001. Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 4:647-652. [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee, A., K. Dai, and J. Lutkenhaus. 1993. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc. Natl. Acad. Sci. USA 90:1053-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nogales, E., S. G. Wolf, and K. H. Downing. 1998. Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391:199-203. [DOI] [PubMed] [Google Scholar]
- 21.Northrup, S. H., and H. P. Erickson. 1992. Kinetics of protein-protein association explained by Brownian dynamics computer simulation. Proc. Natl. Acad. Sci. USA 89:3338-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romberg, L., and P. A. Levin. 2003. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu. Rev. Microbiol. 57:125-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romberg, L., M. Simon, and H. P. Erickson. 2001. Polymerization of Ftsz, a bacterial homolog of tubulin. is assembly cooperative? J. Biol. Chem. 276:11743-11753. [DOI] [PubMed] [Google Scholar]
- 24.Rueda, S., M. Vicente, and J. Mingorance. 2003. Concentration and assembly of the division ring proteins FtsZ, FtsA, and ZipA during the Escherichia coli cell cycle. J. Bacteriol. 185:3344-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salmon, E. D., and P. Wadsworth. 1986. Fluorescence studies of tubulin and microtubule dynamics in living cells, p. 377-403. In D. L. Taylor, A. S. Waggoner, R. F. Murphy, F. Lanni, and R. R. Birge (ed.), Applications of fluorescence in the biomedical sciences. Alan R. Liss, Inc., New York.
- 26.Saxton, W. M., D. L. Stemple, R. J. Leslie, E. D. Salmon, M. Zavortink, and J. R. McIntosh. 1984. Tubulin dynamics in cultured mammalian cells. J. Cell Biol. 99:2175-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stricker, J., and H. P. Erickson. 2003. In vivo characterization of Escherichia coli ftsZ mutants: effects on Z-ring structure and function. J. Bacteriol. 185:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stricker, J., P. Maddox, E. D. Salmon, and H. P. Erickson. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. USA 99:3171-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun, Q., and W. Margolin. 2001. Influence of the nucleoid on placement of FtsZ and MinE rings in Escherichia coli. J. Bacteriol. 183:1413-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, X., and J. Lutkenhaus. 1993. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol. Microbiol. 9:435-442. [DOI] [PubMed] [Google Scholar]