Abstract
The ribosomal protein L11 in bacteria is posttranslationally trimethylated at multiple amino acid positions by the L11 methyltransferase PrmA, the product of the prmA gene. The role of L11 methylation in ribosome function or assembly has yet to be determined, although the deletion of Escherichia coli prmA has no apparent phenotype. We have constructed a mutant of the extreme thermophile Thermus thermophilus in which the prmA gene has been disrupted with the htk gene encoding a heat-stable kanamycin adenyltransferase. This mutant shows no growth defects, indicating that T. thermophilus PrmA, like its E. coli homolog, is dispensable. Ribosomes prepared from this mutant contain unmethylated L11, as determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), and are effective substrates for in vitro methylation by cloned and purified T. thermophilus PrmA. MALDI-TOF MS also revealed that T. thermophilus L11 contains a total of 12 methyl groups, in contrast to the 9 methyl groups found in E. coli L11. Finally, we found that, as with the E. coli methyltransferase, the ribosomal protein L11 dissociated from ribosomes is a more efficient substrate for in vitro methylation by PrmA than intact 70S ribosomes, suggesting that methylation in vivo occurs on free L11 prior to its incorporation into ribosomes.
Extensive modification is a feature of many components of the translational apparatus, with the most common being methylation. In addition to methylated ribosomal RNAs, several ribosomal proteins are also methylated or acetylated. The most highly methylated protein component of the bacterial translational apparatus is the 50S ribosomal subunit protein L11 (2), which comprises a major part of the factor-binding region of the 50S subunit. Cryo-electron microscopic reconstructions of ribosome-EF-Tu (22, 25) and ribosome-EF-G (1) complexes demonstrated a dynamic conformation of L11 and a direct contact of the L11 region of the ribosome with both elongation factors. How, or even whether, methylation of L11 influences its function in multiple aspects of protein synthesis is not yet understood.
Escherichia coli L11 is trimethylated at three amino acid positions, specifically Ala1 (the N-terminal methionine is removed posttranslationally), Lys3, and Lys39, acquiring a total of nine methyl groups (11, 17). These methyl groups are added by a single enzyme, the L11 methyltransferase PrmA, encoded by the prmA gene (7, 27). This protein is conserved among bacteria but is absent from eukarya and archaea (4). Nevertheless, a prmA null mutant of E. coli has no detectable phenotype (27), leaving open the question of the function of this modification.
The substrate recognition mechanism of this enzyme is of particular interest because it methylates multiple positions on the same protein. Presumably, the sites of methylation exist in structurally similar contexts, although the N terminus of L11 in the X-ray crystal structure (29) is disordered, preventing a definitive answer to this question. It is also not known if PrmA produces all modifications during one binding event or if it has multiple, distinct binding states, e.g., one for each trimethylation. It is also possible that the flexible N terminus of L11 adopts several distinct conformations within the PrmA active site, facilitating the multiple modifications. The detection of mono- and dimethylated lysine residues in cloned and overproduced protein (24) leads to the speculation that the methyl groups are added sequentially in the reaction. An important step toward answering these questions would be the development of a system for the biochemical characterization of PrmA activity and a system for high-resolution structural analysis of the PrmA-L11 complex.
Here we describe the construction of a prmA null mutant of Thermus thermophilus HB8 as a source of unmethylated ribosomes to act as substrates for in vitro methylation assays using cloned and purified T. thermophilus PrmA methyltransferase. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) of ribosomal proteins from the prmA null mutant revealed that, in contrast to the 9 methyl groups on E. coli L11, T. thermophilus L11 is modified with a total of 12 methyl groups. We also describe a biochemical characterization of T. thermophilus L11 methylation in vitro. Our choice of T. thermophilus was based on the ease of genetic manipulation of this organism as well as the structural information available regarding Thermus ribosomes. Furthermore, the PrmA protein from T. thermophilus HB8 is currently being examined by X-ray crystallography, and crystals diffracting to 1.9 Å have been reported (14). This raises the possibility that a structural basis for an understanding of the recognition mechanism of this enzyme may be elucidated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
T. thermophilus strain HB8 (ATCC 27624) (20) was employed as the wild-type parental strain for construction of the prmA null mutant. T. thermophilus strains were grown in ATCC medium 1598 (Thermus enhanced medium, or TEM), either in liquid form or solidified with 2.8% (wt/vol) Difco agar. Growth rate measurements were performed in Thermus minimal medium, which is equivalent to TEM without tryptone or yeast extract and with 0.4% sucrose as a carbon source, 10 mM NH4Cl, 20 μg of lysine/ml, and 40 ng each of paraminobenzoic acid, nicotinic acid, calcium pantothenate, thiamine, pyridoxine, biotin, and cyanocobalamine/liter. Growth rate measurements were performed by use of a Klett-Summerson photometer. A pUC18 clone of the heat-stable kanamycin-adenyltransferase gene, htk, was a kind gift from H. Kagamiyama (Osaka Medical College). A T. thermophilus HB8 streptomycin-dependent mutant was a kind gift of A. Carter and V. Ramakrishnan (MRC, Cambridge, United Kingdom). The transformation of T. thermophilus was performed according to a standard method (16).
Oligonucleotides.
All oligonucleotides were obtained from Qiagen. The oligonucleotides used for cloning prmA and for constructing the prmA null mutant (TLK90) were as follows: Tth prmA-1, 5′-GATATACATATGTGGGTTTACCGGCTTAAGGGC-3′; Tth prmA-2, 5′-GCTCGAATTCAGTGGTGGTGGTGGTGGTGCCTCCCGTAGGCGAGGAGG-3′); Tth prmA-3, 5′-CCAGGTACCGTGTGGGTTTACCGGCTTAAGGGC-3′; Tth prmA-4, 5′-GGTCTGCAGGCCGGGCTCAATGACCAAGGGG-3′; Tth prmA-5, 5′-CCACTGCAGCCCCCAGGCGGAGGCGAACGCCAAGCGG-3′; Tth prmA-6, 5′-GGTAAGCTTCTACCTCCCGTAGGCGAGGAGG-3′; Tth htk-1, 5′-CCACTGCAGGGTACCCGTTGACGGCGGATATGG-3′; and Tth htk-2, 5′-GGTCTGCAGCGTAACCAACATGATTAAC-3′. Oligonucleotides for cloning prmA were based on the published T. thermophilus HB8 sequence (GenBank accession number AB103400). Oligonucleotides used for diagnostic PCRs were as follows: primer A, 5′-GTGTGGGTTTACCGGCTTAAGGGC-3′; primer B, 5′-GCGGGGGCGCACCCCGTTCCG-3′; primer C, 5′-GGCCCTCGCCCGCCACCTCCGCCCCGGGG-3′; primer D, same as Tth prmA-6; primer E, 5′-CCCCGGGGCGGAGGTGGCGGGCGAGGGCC-3′; primer F, same as Tth htk-1; and primer G, same as Tth htk-2.
Purification of ribosomes.
Ribosomes from wild-type HB8 and strain TLK90 were purified essentially as previously described (5). Ribosomes were stored at −80°C in a buffer containing 10 mM HEPES-KOH (pH 7.6), 10 mM MgCl2, 50 mM NH4Cl, 5 mM β-mercaptoethanol, and 10% (vol/vol) glycerol.
MALDI analysis.
All MALDI-MS experiments were done on a Reflex IV reflectron MALDI-TOF mass spectrometer (Bruker Daltonics, Billerica, Mass.) equipped with a nitrogen laser, as previously described (23). For all protein analyses, saturated sinapinic acid (SA; Fluka, Milwaukee, Wis.) in 33% aqueous acetonitrile plus 0.1% trifluoroacetic acid was used as the matrix. Samples were prepared by mixing 1 μl of acidified sample solution (approximately 1.8 pmol of ribosome) with 9 μl of matrix. Protein mass spectra were obtained in the positive ion mode at an acceleration voltage of 20 kV by accumulating 300 laser shots. All samples were analyzed under identical instrumental parameters. Acids and organic solvents were high-performance liquid chromatography grade or higher.
Overexpression and purification of T. thermophilus HB8 PrmA.
C-terminally His6-tagged PrmA was purified from E. coli BL21(DE3) containing the plasmid clone pET30bprmA (see Results for a description of the construction of this plasmid). Cultures were grown to mid-logarithmic phase, and expression of the enzyme was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM final concentration). Growth was continued for 2 h, after which the cells were harvested, washed, and lysed by passage through a French press (20,000 lb/in2) in a buffer containing 50 mM Tris-HCl (pH 7.6), 500 mM NaCl, 5 mM imidazole, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, and 10% (vol/vol) glycerol. After a DNase I treatment, the lysates were cleared by centrifugation at 20,000 × g for 20 min. The supernatants were heated to 65°C for 30 min to denature E. coli proteins, cleared once again by centrifugation, and then gently mixed with a Ni-nitrilotriacetic acid (Ni-NTA) slurry (Qiagen) for 2 h at 4°C. The Ni-NTA resin bound with PrmA was then collected by centrifugation, washed twice with a buffer containing 50 mM Tris-HCl (pH 7.6), 500 mM NaCl, 20 mM imidazole, and 1 mM DTT, and then mixed gently for 1 h at 4°C to elute the protein in a buffer containing 50 mM Tris-HCl (pH 7.6), 500 mM NaCl, 500 mM imidazole, and 1 mM DTT. The eluate was concentrated and diluted multiple times by the use of Centricon spin columns into a storage buffer containing 10 mM HEPES-KOH (pH 7.6), 10 mM MgCl2, 50 mM NH4Cl, 5 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride. The protein concentration was determined by measuring the absorbance at 280 nm (1 mg of His6-PrmA/ml gives an absorbance at 280 nm of 2.025).
In vitro methylation assays.
Methylation assays were carried out at 50°C in a final volume of 70 μl and contained 50 nM ribosomes (intact or RNase A treated), 100 nM PrmA, and 100 nM S-adenosyl-l-[methyl-3H]methionine (3H-SAM; specific activity, 4.37 × 1011 Bq/mmol) in a buffer containing 10 mM HEPES-KOH (pH 7.6), 10 mM MgCl2, 50 mM NH4Cl, and 5 mM β-mercaptoethanol or in buffer A (50 mM Tris-HCl [pH 7.6], 600 mM KCl, 10 mM EDTA, 10 mM β-mercaptoethanol). Reactions were started by the addition of 3H-SAM, and 20-μl samples were removed at each time point and placed into 1 ml of 10% trichloroacetic acid. The samples were heated to 100°C for 10 min and then passed through Whatman GF/A glass microfiber filters. The filters were washed three times with 3 ml of 5% trichloroacetic acid each time and then were dried, and radioactivity was estimated by liquid scintillation spectrometry.
RESULTS
Construction of T. thermophilus prmA null mutant indicates that PrmA-directed methylation is dispensable for growth.
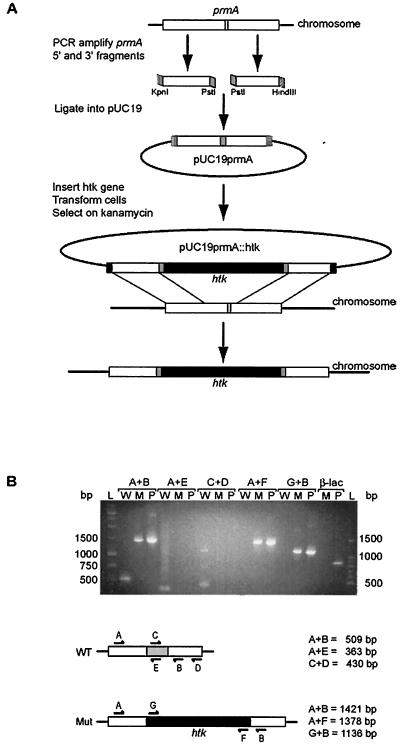
The T. thermophilus HB8 prmA gene has been identified by others based on its homology with the E. coli gene (14) (GenBank accession number AB103400). Thus, a null mutant allele of prmA was constructed by gene disruption (Fig. 1) in which wild-type T. thermophilus HB8 was transformed with a plasmid (pUC19prmA::htk) containing the T. thermophilus prmA gene interrupted by a heat-stable kanamycin resistance gene, htk. To ensure that any resulting gene product would not be functional, the region of the prmA gene that encodes the SAM binding motif was deleted. For the construction of pUC19prmA::htk, the 5′ end of the T. thermophilus prmA coding region was amplified by a PCR using a forward primer containing a KpnI restriction site (Tth prmA-3) and a reverse primer containing a PstI restriction site (Tth prmA-4), giving a product of 306 bp. The 3′ end of the T. thermophilus prmA coding region was amplified by a PCR using a forward primer containing a PstI restriction site (Tth prmA-5) and a reverse primer containing a HindIII restriction site (Tth prmA-6), giving a product of 319 bp. Both products were appropriately digested and ligated simultaneously into KpnI- and HindIII-digested pUC19 to form plasmid pUC19prmA.
FIG. 1.
Construction of T. thermophilus prmA null mutant. (A) Approximately 165 bp of the wild-type prmA gene (white) containing the SAM binding motif (light gray) was replaced by the heat-stable kanamycin resistance gene (htk; black). KpnI restriction sites are indicated by vertical lines, HindIII restriction sites are indicated by horizontal lines, and PstI restriction sites are indicated in gray. The resulting T. thermophilus strain was designated TLK90. (B) Diagnostic PCRs to confirm disruption of the prmA gene in strain TLK90. Genomic DNA from either wild-type HB8 (W), the null mutant (M), or plasmid pUC19prmA::htk (P) was used as a template for PCRs. The primer sets used for each reaction (A to G) are indicated and shown schematically along with the expected product sizes. The region of the prmA gene that was deleted from the null mutant is represented schematically in gray, and the htk gene is indicated in black. Amplification of the β-lactamase gene (β-lac) from the plasmid template but not from TLK90 genomic DNA template confirmed the absence of this plasmid-derived ampicillin resistance marker gene in the null mutant. Molecular weight DNA ladders are shown (L), and their sizes are indicated in base pairs.
A DNA fragment containing the coding region of the htk gene was amplified by PCR from a pUC18 clone (13) by the use of primers containing PstI restriction sites (Tth htk-1 and Tth htk-2), giving a product of 1,093 bp. This DNA fragment was digested with PstI and inserted into the PstI site of pUC19prmA to form pUC19prmA::htk (Fig. 1A). Plasmid constructs were verified by sequencing, and in order to avoid problems caused by transcriptional polarity, only constructs containing the htk gene inserted into the vector in the same orientation as the prmA gene were used for transformations. The final plasmid construct was introduced into T. thermophilus HB8 by standard transformation, using 5 μg of pUC19prmA::htk and plating on solid TEM containing kanamycin (100 μg/ml). Since pUC19-derived plasmids do not replicate in Thermus spp., kanamycin resistance results from integration of the plasmid into the chromosome at a site of homology. Gene replacement occurs upon resolution of the cointegrate. Transformants arose only on plates containing cells that received plasmid DNA. Genomic DNAs from several transformants were prepared, and the absence of the full-length prmA gene, the absence of the pUC19 plasmid vector, and the insertion of the htk gene were verified by diagnostic PCR and sequencing, as indicated in Fig. 1B. A single transformant, designated TLK90, was used in all subsequent experiments.
The viability of kanamycin-resistant transformants which contained no intact prmA gene indicates that this gene is dispensable. Moreover, the T. thermophilus prmA::htk null mutant exhibited no growth defect, with wild-type HB8 and TLK90 having doubling times in Thermus minimal medium of 172 ± 10 and 166 ± 11 min, respectively. This result is consistent with the phenotype of a prmA null mutant of E. coli, which showed no detectable physiological defect (27). We can also conclude, therefore, that L11 methylation by PrmA does not contribute to the thermostability of T. thermophilus ribosomes in vivo, as has been suggested for other modifications of the translational apparatus in thermophilic organisms (15).
MALDI-TOF MS of ribosomes.
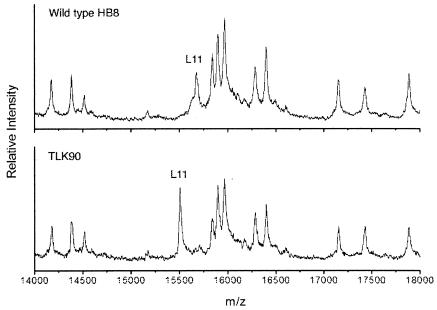
MALDI-TOF MS was performed in order to ascertain the methylation state of the ribosomal protein L11 in ribosomes from wild-type T. thermophilus HB8 and the prmA::htk null mutant (TLK90; Fig. 2). The molecular mass values obtained for unmethylated and methylated L11 were 15,506.2 and 15,675.1 Da, respectively. The mass value for unmodified L11 was precisely the same as that predicted from the gene sequence (GenBank accession number X81375). The mass difference of 169 Da is equivalent to a total of 12 methyl groups, which is 3 more than the 9 methyl groups introduced by the E. coli enzyme.
FIG. 2.
MALDI-TOF MS of 70S ribosomes from T. thermophilus strain HB8 (top) and prmA::htk null mutant TLK90 (bottom). The portions of the spectra containing proteins in the mass range of 14,000 to 18,000 Da are shown. The peaks corresponding to L11 show masses of 15,675.1 and 15,506.2 Da for the wild type and the mutant, respectively. The mass difference of 169 Da is equivalent to the mass of 12 methyl groups.
Disruption of the prmA gene does not suppress streptomycin dependence caused by a mutation in ribosomal protein S12.
The involvement of the ribosomal protein L11 in the interaction of the ribosome with EF-Tu (22, 25), plus the observation that mutations in L11 and in the L11-binding site of 23S rRNA suppress nonsense mutations (18, 26), suggested the possibility that the absence of L11 methylation by PrmA might affect translational accuracy. Such effects can in some instances be manifested as a suppression of streptomycin dependence due to mutant alleles of rpsL, encoding the ribosomal protein S12 (3). Indeed, an early study reported that ribosomes from a strain that had spontaneously reverted from streptomycin dependence contained L11 with an altered electrophoretic mobility (10). This is perhaps one of the few aspects of the E. coli prmA null mutant that has not been previously examined. We therefore introduced the prmA::htk null allele into a mutant of strain T. thermophilus HB8 carrying a P90L streptomycin dependence mutation in ribosomal protein S12 (5a) by transformation with chromosomal DNA from strain TLK90. Kanamycin-resistant transformants were sequenced to confirm both the presence of mutated rpsL and the disruption of prmA and were screened for the ability to grow in the absence of streptomycin. All transformants tested retained the streptomycin dependence phenotype, suggesting that a strong error-prone phenotype is not produced by the absence of L11 methylation.
Cloning of T. thermophilus HB8 prmA gene and purification of PrmA enzyme.
The coding region of the prmA gene was amplified by PCR from a genomic DNA template by use of a forward primer, Tth prmA-1, containing an NdeI restriction site, and a reverse primer, Tth prmA-2, containing an EcoRI restriction site and the complement of a stop codon followed by a (GTG)6 repeat encoding a His6 tag. The resulting DNA fragment was inserted into the NdeI and EcoRI sites of plasmid pET30b to form plasmid pET30bprmA. Plasmid constructions were verified by sequencing.
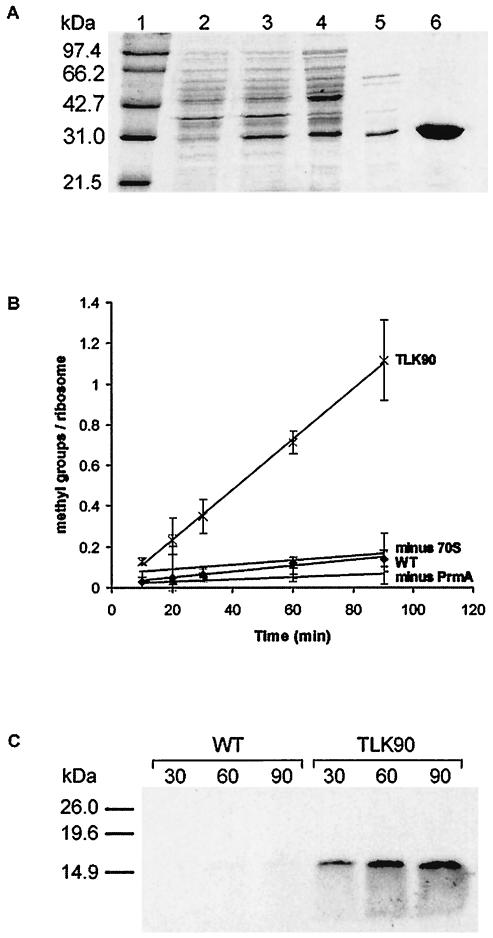
PrmA expression was induced with 1 mM IPTG. Heat treatment of the crude extract resulted in the denaturation and precipitation of the E. coli proteins without affecting the integrity of T. thermophilus PrmA, which is heat stable. Further purification was achieved by Ni-NTA chromatography. The fractionation of the PrmA extract is demonstrated in Fig. 3A. From this gel, we estimate that purification was achieved to >95%. The apparent molecular mass of the cloned protein was 29 kDa, which is in good agreement with the 27.6 kDa deduced from its gene sequence (14) plus approximately 1 kDa for the His6 tag.
FIG. 3.
(A) Purification of cloned His6-tagged T. thermophilus L11 methyltransferase, PrmA, from E. coli BL21(DE3) carrying plasmid pET30bprmA. Lane 1, molecular size markers (sizes are indicated); lane 2, whole-cell lysate prior to induction; lane 3, whole-cell lysate after 2-h induction with IPTG; lane 4, cleared lysate after DNase treatment; lane 5, supernatant after 30-min incubation of lysate at 65°C; lane 6, purified PrmA after Ni-NTA purification, desalting, and concentration. (B). In vitro methylation of ribosomes from wild-type HB8 and the prmA null mutant, TLK90. Methylation assays were performed over a 90-min time course. ♦, ribosomes from wild-type HB8 (WT); ×, ribosomes from TLK90; +, reaction without ribosomes (minus 70S); −, ribosomes from TLK90 without the enzyme (minus PrmA). The results shown were obtained from at least three independent experiments. (C) 3H-labeled products after in vitro methylation of ribosomes from wild-type HB8 (WT) and the prmA null mutant (TLK90). Methylation assays were performed, with samples being removed after 30, 60, and 90 min and electrophoresed through a sodium dodecyl sulfate-15% polyacrylamide gel (1 pmol equivalent of ribosomes per lane). After fixing in a 10% methanol-acetic acid solution, the gels were incubated for 20 min in Enlightning rapid autoradiography enhancer (Perkin-Elmer), dried, and exposed to film for 72 h. The positions of molecular mass markers on the gel are indicated.
Methylation of ribosomes from prmA::htk null mutant by cloned and purified PrmA.
Ribosomes from the prmA::htk null mutant were examined for the ability to act as a substrate for cloned PrmA in vitro. As shown in Fig. 3B, ribosomes from wild-type T. thermophilus HB8 did not act as efficient substrates because they contain methylated L11. In contrast, ribosomes from the prmA::htk null mutant TLK90 incorporated 3H-radiolabeled methyl groups in a PrmA-dependent manner. Under the conditions used, the reaction proceeded at an initial rate of 2.07 × 10−4 methyl groups incorporated/ribosome/s for approximately 90 min, after which the amount of 3H detected actually began to decrease, probably due to the aggregation and/or precipitation of reaction components after such a long incubation at 50°C. Since T. thermophilus L11 is methylated in vivo with 12 methyl groups and since a maximum of only 1.1 methyl groups was incorporated per ribosome, only very partial methylation of fully assembled L11 was observed. The slow time course of methyl group incorporation into wild-type ribosomes (2.3 × 10−5/ribosome/s) was not significantly different from that observed in the absence of ribosomes and may have been due to the detection of 3H-SAM bound to the PrmA enzyme. Alternatively, this low-level incorporation of methyl groups into wild-type ribosomes may have been due to the presence of under-methylated ribosomes in the reaction. This 3H incorporation, however, was not due to an exchange of methyl groups on L11 by PrmA, as the addition of a vast excess of unlabeled SAM to the reaction did not lead to a decrease in the amount of 3H incorporated (data not shown). These results indicate that ribosomes from the null mutant are substantially less methylated than ribosomes from the wild-type parental strain, presumably within L11.
To confirm that L11 is the sole ribosomal component that is methylated by PrmA, we subjected ribosomes to electrophoresis through a sodium dodecyl sulfate-15% polyacrylamide gel after in vitro methylation with PrmA (Fig. 3C). Ribosomes from TLK90 contained a single 3H-methylated protein of a size corresponding to that of L11 (∼15.5 kDa). Furthermore, the extent of methylation increased with the reaction time, consistent with the results shown in Fig. 3B. In contrast, ribosomes from wild-type HB8 did not contain significant amounts of 3H-methylated protein, even after 90 min. Importantly, no other mass differences between ribosomal proteins from wild-type HB8 and TLK90 were detected by MALDI-TOF MS (data not shown), indicating that L11 is the only ribosomal protein target for methylation by PrmA.
Dissociated L11 is methylated more efficiently than ribosome-assembled L11.
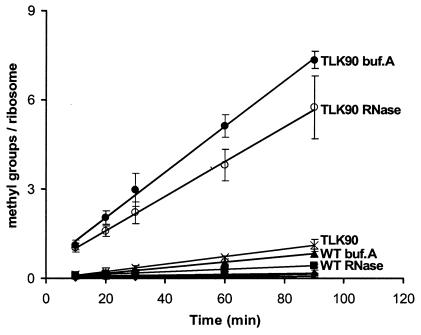
It has been shown that high salt and low magnesium concentrations stimulate the methylation of E. coli L11 in crude extracts, perhaps suggesting that free L11 is methylated more efficiently than ribosome-assembled L11 (8). Consistent with this, an early study found that free L11 obtained by LiCl extraction of 50S subunits prepared from methionine-starved cells was methylated approximately twice as efficiently as intact 50S subunits (6). However, methionine starvation is likely to result in ribosomes that contain a heterogeneous population of L11 with various degrees of methylation, and since the methylation state of L11 could potentially affect its interaction with the ribosome, it is possible that under-methylated L11 was preferentially extracted from the subunits, thereby enriching the pool of methyl group acceptors. More recently, it was proposed that the N-terminal domain of PrmA, which contains the moderately conserved PrmA motif, may be structurally similar to RNA-binding proteins exhibiting the ferredoxin-like fold (4). Based on this, it was suggested that PrmA may interact with a larger ribonucleoprotein complex, presumably an assembly intermediate, rather than free L11. Thus, the issue of whether PrmA acts in vivo on fully assembled 50S ribosomal subunits or on free L11 prior to assembly remains unresolved. We therefore compared the PrmA-directed methylation of intact 70S ribosomes with methylation of either ribosomes dissociated in a buffer containing high salt but lacking magnesium (buffer A) or ribosomes treated with RNase A (Fig. 4). The methylation of TLK90 ribosomes in buffer A occurred to a substantially greater extent than that of intact ribosomes (7.3 methyl groups/L11 protein compared to 1.1 methyl groups/L11 protein for intact ribosomes). Similarly, methylation was also stimulated by RNase treatment (5.7 methyl groups/L11 protein), although to a lesser extent than ribosomes dissociated in buffer A. Since L11 protects a 61-nucleotide rRNA fragment from RNase cleavage in E. coli ribosomes (21), treatment with RNase A may produce L11-rRNA ribonucleoprotein complexes that are less efficient substrates for methylation than free L11. In both cases, the reactions proceeded for approximately 90 min before the amount of 3H detected began to decrease, as noted earlier. However, due to the accelerated reaction rates, the reactions proceeded to >60.0% completion during the 90-min time course, taking into account the 12 methyl groups present in T. thermophilus L11. It is noteworthy that the dissociation of wild-type ribosomes in buffer A or by RNase treatment also led to a modest increase in the incorporation of 3H-labeled methyl groups, further supporting the possibility that the reactions contained a population of under-methylated wild-type ribosomes. Taken together, these results suggest that PrmA preferentially recognizes free L11 prior to its incorporation into 50S subunits in vivo.
FIG. 4.
In vitro methylation of ribosomes from wild-type HB8 and the prmA null mutant (TLK90). The ribosomes were either intact, dissociated in a buffer containing a high salt concentration but lacking magnesium (buffer A), or dissociated by a treatment with RNase A. Methylation assays were performed for a 90-min time course. •, ribosomes from TLK90 in buffer A (TLK90 buf.A); ○, ribosomes from TLK90 treated with RNase A (TLK90 RNase); ×, ribosomes from TLK90; ▴, ribosomes from wild-type HB8 in buffer A (WT buf.A); ▪, ribosomes from wild-type HB8 treated with RNase A (WT RNase). Clustered near the bottom of the plot are ribosomes from wild-type HB8 (WT; filled diamonds), a reaction without ribosomes (+), and ribosomes from TLK90 without the enzyme (−). The results shown were obtained from at least three independent experiments.
DISCUSSION
In this study, we have described the construction of a T. thermophilus prmA null mutant and the in vitro methylation of under-methylated ribosomes and ribosomal protein L11 by cloned and purified T. thermophilus PrmA. As previously observed for E. coli (27), the T. thermophilus PrmA enzyme is dispensable, despite its phylogenetic conservation (4).
The purified PrmA protein is capable of methylating T. thermophilus L11 to a stoichiometry in excess of seven methyl groups per protein molecule. Direct evidence that the sites of methylation are the same as those in E. coli is lacking, but the high degree of conservation between both the L11 proteins and the methyltransferases is highly suggestive that this may be the case. Indeed, in a clone of E. coli carrying overexpressed, plasmid-borne T. thermophilus L11, the third lysine appeared to carry a mixture of mono- and dimethylations (24), indicating that E. coli PrmA was capable of recognizing T. thermophilus L11 as a substrate. The lysine at position 39, the most C-terminal of the methylated residues in E. coli, is conserved in T. thermophilus L11, with a very high level of surrounding amino acid conservation. Finally, as in E. coli L11, in which the N-terminal alanine is trimethylated, T. thermophilus L11 is also N-terminally blocked (24).
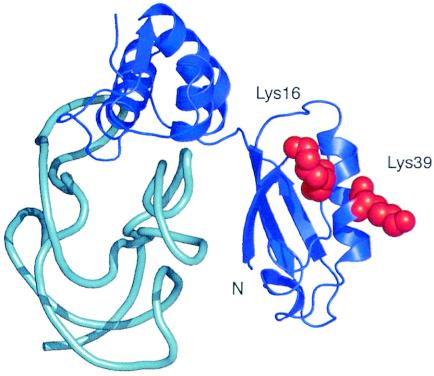
The MALDI-TOF MS data presented here indicate that T. thermophilus L11 contains three additional methyl groups, consistent with trimethylation of an additional lysine. An alignment of the E. coli and T. thermophilus L11 sequences revealed the presence of a lysine residue at position 16 of the T. thermophilus protein that is a methionine in the E. coli homolog. Lys16 and Lys39 in the Thermotoga maritima L11-rRNA cocrystal structure (29) are located on the same side of the N-terminal domain of L11, raising the possibility that Lys16 is methylated by PrmA in T. thermophilus (Fig. 5). The N-terminal seven residues of L11 are disordered in the crystal structure, precluding any conclusion about their proximity to Lys16 and Lys39. It is possible, however, that they could reach the same side of the N-terminal domain containing Lys16 and Lys39. This would, in principle, allow PrmA to methylate all four positions during one binding event. Whether the difference in the extent of methylation between the T. thermophilus and E. coli proteins is biologically relevant or simply fortuitous is unknown at this point.
FIG. 5.
Three-dimensional structure of Thermotoga maritima ribosomal protein L11 (blue) and its associated binding site on 23S rRNA (cyan) as determined by X-ray crystallography (29). Amino acid residues Lys16 and Lys39 are shown with red space-filling diagrams to indicate their mutual proximity. The N-terminal seven amino acid residues of L11, including Ala1 and Lys3, are disordered in the structure and are not shown.
To date, no convincing role for PrmA has been demonstrated. Previous studies with the E. coli null mutant exhaustively examined phenotypic effects, but none were found (27). Thus, the absence of L11 methylation does not appear to affect either growth physiology or ribosome function in any detectable way. Even L11 itself is not absolutely essential, although L11 null mutants of Bacillus megaterium (9), Bacillus subtilis (28), and several Streptomyces species (19) are severely debilitated. The facts that PrmA is ubiquitous in the bacterial clade, is highly conserved, and engages in an energetically costly process all suggest an important function, despite two independent findings of its dispensability in phylogenetically divergent genera, Escherichia (27) and Thermus (this study). Continued investigation of this protein is therefore warranted. The inference that PrmA preferentially recognizes free L11 suggests a role for this interaction during 50S subunit assembly. There are several examples for which ribosome modification is dispensable while the existence of the modifying enzyme is essential for assembly. Most notably, the 23S rRNA pseudouridine synthase RluD is indispensable, even though mutagenesis of the catalytic residue does not create a phenotype (12). Thus, the dispensability of PrmA does not exclude its participation in assembly, as it is feasible that its role is redundant and can be performed by other components of the assembly apparatus. This issue could be resolved genetically by identifying synthetically lethal mutants derived from the T. thermophilus prmA null mutant described here.
The T. thermophilus prmA null mutant will provide a source of unmethylated ribosomes for future investigations. Of particular interest are the aspects of L11-ribosome interaction that interfere with PrmA-directed methylation. It is not yet clear whether the preference of PrmA for free L11 is due to conformational differences between the free and ribosome-bound forms of L11 or if there are steric clashes between PrmA and other components of the ribosome. We have previously isolated thiostrepton-resistant L11 mutants of T. thermophilus HB8 which exhibit a more open conformation within the ribosome (5). It is possible that the L11 methylation sites of such mutants may be more accessible to PrmA. In order to address this question fully, more information regarding the interaction between PrmA and L11 is needed. Ultimately, such studies would be greatly aided by high-resolution structural information, such as crystal structures of thermophilic PrmA alone or a cocrystal structure of a PrmA-L11 complex.
ADDENDUM IN PROOF
The T. thermophilus HB8 PrmA crystal structure has now been solved (PDB entry 1UFK) and released by Kamanishi and colleagues.
Acknowledgments
This work was supported by National Institutes of Health grants GM19756 (to A.E.D.) and GM58843 (to P.A.L.).
We are grateful to members of the Dahlberg laboratory for critical reading of the manuscript, to H. Kagamiyama of Osaka Medical College for providing the htk clone, and to A. Carter and V. Ramakrishnan for providing us with the T. thermophilus HB8 rpsL P90L mutant strain prior to publication.
REFERENCES
- 1.Agrawal, R. K., J. Linde, J. Sengupta, K. H. Nierhaus, and J. Frank. 2001. Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation. J. Mol. Biol. 311:777-787. [DOI] [PubMed] [Google Scholar]
- 2.Alix, J. H. 1992. Extrinsic factors in ribosome assembly, p. 173-184. In K. H. Nierhaus, A. R. Subramanian, V. A. Erdmann, F. Franceschi, and B. Wittmann-Liebold (ed.), The translational apparatus. Plenum Publishing, New York, N.Y.
- 3.Allen, P. N., and H. F. Noller. 1991. A single base substitution in 16S ribosomal RNA suppresses streptomycin dependence and increases the frequency of translational errors. Cell 66:141-148. [DOI] [PubMed] [Google Scholar]
- 4.Bujnicki, J. M. 2000. Sequence, structural, and evolutionary analysis of prokaryotic ribosomal protein L11 methyltransferases. Acta Microbiol. Pol. 49:19-28. [PubMed] [Google Scholar]
- 5.Cameron, D. M., J. Thompson, S. T. Gregory, P. E. March, and A. E. Dahlberg. 2004. Thiostrepton-resistant mutants of Thermus thermophilus. Nucleic Acids Res. 32:3220-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Carter, A. 2002. Ph.D. thesis. Cambridge University, Cambridge, United Kingdom.
- 6.Chang, F. N., L. B. Cohen, I. J. Navickas, and C. N. Chang. 1975. Purification and properties of a ribosomal protein methylase from Escherichia coli Q13. Biochemistry 14:4994-4998. [DOI] [PubMed] [Google Scholar]
- 7.Colson, C., J. Lhoest, and C. Urlings. 1979. Genetics of ribosomal protein methylation in Escherichia coli. III. Map position of two genes, prmA and prmB, governing methylation of proteins L11 and L3. Mol. Gen. Genet. 169:245-250. [DOI] [PubMed] [Google Scholar]
- 8.Colson, C., and H. O. Smith. 1977. Genetics of ribosomal protein methylation in Escherichia coli. Mol. Gen. Genet. 154:167-173. [DOI] [PubMed] [Google Scholar]
- 9.Cundliffe, E., P. Dixon, M. Stark, G. Stoffler, R. Ehrlich, M. Stoffler-Meilicke, and M. Cannon. 1979. Ribosomes in thiostrepton-resistant mutants of Bacillus megaterium lacking a single 50S subunit protein. J. Mol. Biol. 132:235-252. [DOI] [PubMed] [Google Scholar]
- 10.Dabbs, E. R. 1978. Mutational alterations in 50 proteins of the Escherichia coli ribosome. Mol. Gen. Genet. 165:73-78. [DOI] [PubMed] [Google Scholar]
- 11.Dognin, M. J., and B. Wittmann-Liebold. 1980. Purification and primary structure determination of the N-terminal blocked protein, L11, from Escherichia coli ribosomes. Eur. J. Biochem. 112:131-151. [DOI] [PubMed] [Google Scholar]
- 12.Gutgsell, N. S., M. D. Del Campo, S. Raychaudhuri, and J. Ofengand. 2001. A second function for pseudouridine synthases: a point mutant of RluD unable to form pseudouridines 1911, 1915, and 1917 in Escherichia coli 23S ribosomal RNA restores normal growth to an RluD-minus strain. RNA 7:990-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto, Y., T. Yano, S. Kuramitsu, and H. Kagamiyama. 2001. Disruption of Thermus thermophilus genes by homologous recombination using a thermostable kanamycin-resistant marker. FEBS Lett. 506:231-234. [DOI] [PubMed] [Google Scholar]
- 14.Kaminishi, T., H. Sakai, C. Takemoto-Hori, T. Terada, N. Nakagawa, N. Maoka, S. Kuramitsu, M. Shirouzu, and S. Yokoyama. 2003. Crystallization and preliminary X-ray diffraction analysis of ribosomal protein L11 methyltransferase from Thermus thermophilus HB8. Acta Crystallogr. D 59:930-932. [DOI] [PubMed] [Google Scholar]
- 15.Kowalak, J. A., J. J. Dalluge, J. A. McCloskey, and K. O. Stetter. 1994. The role of posttranscriptional modification in stabilization of transfer RNA from hyperthermophiles. Biochemistry 33:7869-7876. [DOI] [PubMed] [Google Scholar]
- 16.Koyama, Y., T. Hoshino, N. Tomizuka, and K. Furukawa. 1986. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 166:338-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lederer, F., J. H. Alix, and D. Hayes. 1977. N-Trimethylalanine, a novel blocking group, found in E. coli ribosomal protein L11. Biochem. Biophys. Res. Commun. 77:470-480. [DOI] [PubMed] [Google Scholar]
- 18.Murgola, E. J., F. T. Pagel, K. A. Hijazi, A. L. Arkov, W. Xu, and S. W. Zhao. 1995. Variety of nonsense suppressor phenotypes associated with mutational changes at conserved sites in Escherichia coli ribosomal RNA. Biochem. Cell Biol. 73:925-931. [DOI] [PubMed] [Google Scholar]
- 19.Ochi, K. 1990. Streptomyces relC mutants with an altered ribosomal protein ST-L11 and genetic analysis of a Streptomyces griseus relC mutant. J. Bacteriol. 172:4008-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oshima, T., and K. Imahori. 1974. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int. J. Syst. Bacteriol. 24:102-112. [Google Scholar]
- 21.Schmidt, F. J., J. Thompson, K. Lee, J. Dijk, and E. Cundliffe. 1981. The binding site for ribosomal protein L11 within 23S ribosomal RNA of Escherichia coli. J. Biol. Chem. 256:12301-12305. [PubMed] [Google Scholar]
- 22.Stark, H., M. V. Rodnina, J. Rinke-Appel, R. Brimacombe, W. Wintermeyer, and M. van Heel. 1997. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389:403-406. [DOI] [PubMed] [Google Scholar]
- 23.Suh, M.-J., and P. A. Limbach. 2004. Investigation of methods suitable for the matrix-assisted laser desorption/ionization mass spectrometric analysis of proteins from ribonucleoprotein complexes. Eur. J. Mass Spectrom. 10:89-99. [DOI] [PubMed] [Google Scholar]
- 24.Triantafillidou, D., M. Simitsopoulou, F. Franceschi, and T. Choli-Papadopoulou. 1999. Structural and functional studies on the overproduced L11 protein from Thermus thermophilus. J. Protein Chem. 18:215-223. [DOI] [PubMed] [Google Scholar]
- 25.Valle, M., J. Sengupta, N. K. Swami, R. A. Grassucci, N. Burkhardt, K. H. Nierhaus, R. K. Agrawal, and J. Frank. 2002. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 21:3557-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Dyke, N., and E. J. Murgola. 2003. Site of functional interaction of release factor 1 with the ribosome. J. Mol. Biol. 330:9-13. [DOI] [PubMed] [Google Scholar]
- 27.Vanet, A., J. A. Plumbridge, M. F. Guerin, and J. H. Alix. 1994. Ribosomal protein methylation in Escherichia coli: the gene prmA, encoding the ribosomal protein L11 methyltransferase, is dispensable. Mol. Microbiol. 14:947-958. [DOI] [PubMed] [Google Scholar]
- 28.Wienen, B., R. Ehrlich, M. Stoffler-Meilicke, G. Stoffler, I. Smith, D. Weiss, R. Vince, and S. Pestka. 1979. Ribosomal protein alterations in thiostrepton- and micrococcin-resistant mutants of Bacillus subtilis. J. Biol. Chem. 254:8031-8041. [PubMed] [Google Scholar]
- 29.Wimberly, B. T., R. Guymon, J. P. McCutcheon, S. W. White, and V. Ramakrishnan. 1999. A detailed view of a ribosomal active site: the structure of the L11-RNA complex. Cell 97:491-502. [DOI] [PubMed] [Google Scholar]