Abstract
Mycoplasma hyopneumoniae is the most significant bacterial pathogen of the respiratory tract of swine. p65 is an immunodominant surface lipoprotein of M. hyopneumoniae that is specifically recognized during disease. Analysis of the translated amino acid sequence of the gene encoding p65 revealed similarity to the GDSL family of lipolytic enzymes. To examine the lipolytic activity of p65, the gene was cloned and expressed in Escherichia coli after truncation of the prokaryotic lipoprotein signal sequence and mutagenesis of the mycoplasma TGA tryptophan codons. After treatment with thrombin, the recombinant glutathione S-transferase (GST)-p65 protein yielded a 66-kDa fusion protein cleavage product corresponding in size to the mature p65 protein. The esterase activity of recombinant GST-p65 was indicated by the formation of a cleared zone on tributyrin agar plates and the hydrolysis of p-nitrophenyl esters of caproate (pNPC) and p-nitrophenyl esters of palmitate (pNPP). Lipase activity was indicated by the hydrolysis of the artificial triglyceride 1,2-O-dilauryl-rac-glycero-3-glutaric acid resorufin ester. Using pNPC and pNPP as substrates, recombinant GST-p65 had optimal activity between pHs 9.2 and 10.2 and at a temperature higher than 39°C. Calcium ions did not increase the activity of recombinant GST-p65. Rabbit anti-p65 antibodies inhibited the activity of recombinant GST-p65 and also inhibited the growth of M. hyopneumoniae in vitro. Examination of the kinetic parameters of recombinant GST-p65 for the hydrolysis of pNPC and pNPP indicated a preference for the shorter fatty acid chain of pNPC. The physiological and/or pathogenic role of mycoplasma lipolytic enzymes has not been determined, but they are likely to play an important role in mycoplasmas' nutritional requirements for long-chain fatty acids and may reduce the function of lung surfactants in mycoplasma-induced respiratory diseases. This is the first report of the lipolytic activity of a lipid-modified surface immunogen of a mycoplasma.
Mycoplasma hyopneumoniae is the causative agent of porcine enzootic pneumonia (25). The high prevalence of this disease incurs substantial economic losses in the pig industry throughout the world. In the absence of a cell wall, proteins at the cell surface are essential for the survival and colonization of mycoplasmas within the host. A comparatively large proportion of the mycoplasma genome encodes proteins that are putatively modified by the addition of a lipid moiety, to permit attachment to the cell membrane and exposure to the extracellular milieu (5, 11, 17, 40). These surface-exposed lipoproteins are among the primary targets of the host humoral immune response. Despite the pathogenic significance of M. hyopneumoniae, there is limited understanding of the major immunogens or their functions. Further understanding of the functional role of these proteins is likely to be essential to improving the control of mycoplasmoses.
Previous work by Wise and Kim (47) identified three lipoproteins in M. hyopneumoniae strain J. These lipoproteins had relative molecular masses of 44, 50, and 65 kDa, equivalent in size to the major surface antigens identified in earlier studies (49). p65 is used in the serological diagnosis of M. hyopneumoniae infections and is a potential candidate for subunit vaccination (24, 47). The complete nucleotide sequences of the p65 gene from M. hyopneumoniae strains J and 232A (GenBank accession numbers AAB67173 and AAB70214, respectively) code for a unique conserved, amino-terminal Gly-Asp-Ser-Leu (GDSL) motif associated with members of a novel family of lipolytic enzymes (42). The conserved serine residue comprises part of the active site catalytic triad formed by the amino acids Ser, His, and Asp (2, 31). In the true family of lipolytic enzymes, the active-site consensus motif is defined by the pentapeptide Gly-X-Ser-X-Gly (GXSXG) (7). Prokaryotic and eukaryotic members of the GDSL family are classified by the identification of five conserved blocks with a high degree of amino acid homology and similar relative location (42). The term lipolytic enzymes collectively describes lipases (EC 3.1.1.3), esterases (EC 3.1.1.1), and also phospholipases (EC 3.1.4.3); however, lipases and esterases are differentiated by their preference for long-chain acylglycerols (at least 10 carbon atoms) and specificity for short-chain acylglycerols (at most 10 carbon atoms), respectively (21).
Although the mycoplasma membrane is rich in essential enzymes, lipase activity has been detected only in the soluble fraction of disrupted cells from Mycoplasma gallisepticum, Acholeplasma laidlawii (37), and mycoplasmas isolated from human saliva (6). Esterase activity has also been detected by the histochemical staining of crude lysates of 22 Mycoplasma and Acholeplasma species (30). Lipolytic activity in M. hyopneumoniae has not previously been detected. Mycoplasmas are fatty acid auxotrophs, and lipolytic enzymes are thus likely to play an essential role in their nutritional requirement for long-chain fatty acids (33). In several pathogenic bacterial species, lipases have also been suspected to be potential virulence factors (21). The indirect role of various metabolic processes in the pathogenesis of mycoplasmas is a recurring theme in studies of host pathogen interactions; however, the role of lipases in the physiology and pathogenicity of mycoplasmas has not been investigated.
In order to determine whether p65 is a lipolytic enzyme, the corresponding gene was cloned and expressed in Escherichia coli, and the lipolytic activity and other biochemical properties of the purified recombinant glutathione S-transferase (GST)-p65 protein were examined.
MATERIALS AND METHODS
Bacterial strains and media.
M. hyopneumoniae strain LKR (26) was grown at 37°C in either Friis broth or Friis agar (containing 1% bacteriological agar) (12). Cells were harvested at the late logarithmic phase of growth. Plasmid-transformed E. coli JM109 was grown at 37°C in Luria-Bertani (LB) broth or on agar containing 50 μg of ampicillin/ml (39).
DNA cloning, sequencing, and expression of the p65 gene.
Genomic DNA was purified from M. hyopneumoniae by a method described previously (28). The DNA sequence of the p65 gene from M. hyopneumoniae strains J (GenBank accession number AAB67175) and 232A (GenBank accession number AAB70214) were used to design oligonucleotide primers for the PCR amplification of the p65 gene from strain LKR. The p65 gene was amplified downstream of the predicted lipoprotein acylation signal sequence, and the mycoplasma TGA tryptophan codons were mutagenized to TGG by overlap extension PCR to enable the full-length expression of the p65 gene in E. coli. Briefly, separate PCRs were performed with 1 μl of genomic DNA template for each oligonucleotide primer pair (BAMH1F-1WR, 1WF-2WR, 2WF-3WR, and 3WF-SAL1R) (Table 1) in a reaction volume of 50 μl containing 2 mM MgSO4, a 100 μM concentration of each deoxynucleoside triphosphate, a 0.4 μM concentration of each primer, and 1.5 U of Platinum Taq thermopolymerase (Life Technologies, Inc.). Touchdown PCRs were performed with a thermocycler (Hybaid) under the following conditions. For primer pair BAMH1F-1WR, 95°C for 5 min; then 18 cycles of 95°C for 1 min, 65°C lowered to 55°C (approximately 1.25°C every two cycles) for 1 min, and 68°C for 2 min; followed by 25 cycles of 95°C for 1 min, 55°C for 1 min, and 68°C for 2 min; with a final extension at 68°C for 10 min. For primer pairs 1WF-2WR, 2WF-3WR, and 3WF-SAL1R, 95°C for 5 min; then 18 cycles of 95°C for 1 min, 57.5°C lowered to 47.5°C (1.25°C every two cycles) for 1 min, and 68°C for 2 min; followed by 25 cycles of 95°C for 1 min, 47.5°C for 1 min, and 68°C for 2 min; with a final extension at 68°C for 10 min. The PCR products were purified with the QIAquick gel extraction kit (QIAGEN), and approximately equimolar amounts of all four PCR products were used as templates in an overlap extension touchdown PCR with the oligonucleotide primer pair BAMH1F-SAL1R (Table 1) under the following conditions: 95°C for 5 min; then 18 cycles of 95°C for 1 min, 57.5°C lowered to 47.5°C (1.25°C every two cycles) for 1 min, and 68°C for 2 min; followed by 25 cycles of 95°C for 1 min, 55°C for 1 min, and 68°C for 2 min; with a final extension at 68°C for 10 min. The final PCR product was purified with the QIAquick PCR purification kit (QIAGEN), digested with BamHI and SalI, and ligated into the expression vector pGEX-4T-1 (Amersham Pharmacia Biotech) according to the manufacturer's instructions. E. coli JM109 cells were transformed with the ligation mixture, and clones containing the mutagenized p65 gene were selected. The DNA sequence of the mutagenized recombinant p65 gene construct in E. coli was determined with the BigDye terminator cycle sequencing reaction kit (Applied Biosystems) and the oligonucleotides 1WR, 2WF, 2WR, 3WF, 3WR, and SAL1R. The expression of the recombinant GST-p65 fusion protein in E. coli was induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 2 mM. The recombinant GST-p65 protein was purified by affinity chromatography with a glutathione-Sepharose column (Amersham Pharmacia Biotech) and dialyzed against phosphate-buffered saline overnight at 4°C. The GST-fusion protein was removed by enzymatic cleavage with thrombin (Amersham Pharmacia Biotech) according to the manufacturer's instructions. The thrombin cleavage products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western immunoblotting as described previously (8).
TABLE 1.
Oligonucleotides used in this study
| Oligo-nucleotide | Sequencea (5′-3′) |
|---|---|
| BAMH1F | CTTCggatccCTTATTTCAGCTGGTTGgTTGC |
| 1WR | TTAAGAAGGTAAAGcCAGTTTTCTGTTGTTGTCC |
| 1WF | GACAACAACAGAAAACTGgCTTTACCTTCTTAA |
| 2WR | TATTATACTTGCcCATTTTTTAAATTCATTAAAAATTGC |
| 2WF | TAATGAATTTAAAAAATGgGCAAGTATAATAAAACC |
| 3WR | ATCAGAATCATTcCAAATACTTTTATCATAGAC |
| 3WF | CTATGATAAAAGTATTTGgAATGATTCTG |
| SAL1R | TAGgtcgacTATAATTATAAGTTATTAAAAAATTAGTTTG |
Lowercase letters indicate differences in nucleotide sequences from published sequences. These changes were introduced to create a restriction endonuclease cleavage site or to mutate a TGA tryptophan codon to TGG or to mutate the TGT cysteine codon to TGG.
Preparation of immune sera.
Purified, thrombin-cleaved recombinant p65 protein was used to immunize New Zealand White rabbits as described previously (14). Briefly, thrombin-cleaved recombinant p65 was separated by SDS-PAGE and lightly stained with Coomassie brilliant blue. The stained thrombin-cleaved recombinant p65 protein band was excised, and the polyacrylamide gel slice was emulsified in an equal volume of Freund's complete adjuvant. Three further booster immunizations were given at 1-month intervals with the thrombin-cleaved recombinant p65 polyacrylamide gel slice emulsified in Freund's incomplete adjuvant. Serum was collected 10 days after each booster, and the titer was assessed by Western immunoblotting. Antibodies were purified by affinity chromatography with a protein A-Sepharose column (Amersham Pharmacia Biotech) as described previously (39) and sterilized by passage through a 0.22-μm-pore-size filter. Immune serum was heat inactivated by incubation at 57°C for 20 min.
Assays for enzymatic activity.
The esterase and lipase activities of recombinant GST-p65 were examined on LB agar plates prepared by the emulsion of tributyrin (Sigma) or olive oil (Sigma) with molten LB broth agar medium to a final concentration of 1% (vol/vol). Activity was indicated by the formation of a clear zone around filter disks containing 200 μg of recombinant GST-p65 after incubation at 37°C for 2 days. For the specific detection of lipase activity, 2 μM recombinant GST-p65 was incubated at room temperature with 100 μg of 1,2-O-dilauryl-rac-glycero-3-glutaric acid resorufin ester (Sigma) (20) in 1 ml of TT buffer (100 mM Tris-HCl [pH 7.8] and 0.2% Triton X-100). The release of resorufin and the subsequent change in optical density at 572 nm (ΔOD572/min) was detected with an Ultrospec 4050 spectrophotometer (LKB). The hydrolase activity of recombinant GST-p65 was further examined on LB agar plates containing skim milk (1% [wt/vol]), egg yolk (1 egg yolk per 800 ml), or sheep blood erythrocytes (7% [vol/vol]) incubated at 37°C with filter disks containing 200 μg of recombinant GST-p65. Proteinase K, phospholipase A2 from porcine pancreas (Sigma), phospholipase B from Vibrio species (Sigma), phospholipase C from Clostridium perfringens (Sigma), and phospholipase D from Streptomyces chromofuscus type VI (Sigma) and Pseudomonas aeruginosa were used as positive controls.
The effects of various physical and chemical conditions on the activity of recombinant GST-p65 were determined by monitoring the release of p-nitrophenyl (pNP) from p-nitrophenyl caproate (pNPC; C6) and p-nitrophenyl palmitate (pNPP; C16) (Sigma). Unless otherwise stated, approximately 0.185 μM recombinant GST-p65 was incubated at room temperature with 15 nmol of pNPC or pNPP in a microtiter plate containing 200 μl of TT buffer. To examine the effect of pH, TT buffer was adjusted to pH 5.2, 6.2, 7.2, 8.2, 9.2, and 10.2. To examine the effect of calcium ions, calcium chloride was added to a final concentration of 1 or 5 mM, or EDTA was added to a final concentration of 5 mM. To examine the effect of temperature, reaction mixtures were incubated at 33, 37, and 39°C. The effect of anti-p65 antibodies was examined at dilutions of 10−1, 10−2, and 10−3. Each reaction was repeated in triplicate and the OD414 was measured with a Labsystems Multiskan MS spectrophotometer. The OD414 was measured every minute to examine the effect of pH, calcium ions, and anti-p65 antibodies and every 5 min to examine the effect of temperature. The molar absorption coefficient of pNP at 414 nm was 68 × 105 M−1. One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmol of pNP per min. Specific activity was defined as the enzymatic activity per mg of recombinant GST-p65 and was adjusted by a factor of 0.72 to account for the contribution to mass of the 26-kDa GST fusion protein. The hydrolysis of pNPC and pNPP by recombinant GST-p65 was examined in triplicate at substrate concentrations of 9, 13, 17, 28, and 58 μM for 10 min at room temperature in 200 μl of TT buffer. The kinetic parameters, Km and Vmax, were determined by linear regression analysis of the Lineweaver-Burk double-reciprocal plots. Each recombinant GST-p65 molecule was assumed to contain only one binding site. The type VII lipase of Candida rugosa (Sigma) was used as a positive control in all assays. GST was expressed and affinity purified from E. coli transformed with pGEX-4T-1 in a manner identical to that described for the recombinant GST-p65 protein and was used as a negative control in all assays.
Growth inhibition.
The growth inhibitory effect of anti-p65 antibodies was examined by monitoring the growth of M. hyopneumoniae in FF2 broth cultures containing a 1/50 or 1/100 dilution of the purified, filtered p65 antibodies. A sample of each culture was removed after 0, 18, 42, 50, 77, 101, and 122 h of incubation, and a count of viable mycoplasmas was determined by limiting dilution titration as the number of color-changing units per milliliter by the method of Meynell and Meynell (29). Purified antibodies from nonimmunized rabbits were used as negative controls.
Sequence analysis.
The ClustalX program was used to align the translated p65 sequence from M. hyopneumoniae strain LKR with that of strains J (GenBank accession no. AAB67173) and 232A (GenBank accession no. AAB70214). Protein sequence similarity searches were performed with the BLAST program (1). The GDSL-like lipase family signature sequences were identified by using the PROSITE database (9).
RESULTS
Cloning and expression of the p65 gene.
All three tryptophan codons identified by sequence analysis of the p65 gene from M. hyopneumoniae strains J and 232A used the trinucleotide TGA and were mutagenized to TGG by overlap extension PCR to allow the full-length expression of p65. The portion of the gene encoding the mature form of p65 was cloned into expression vector pGEX4T-1 with the amino-terminal cysteine replaced by a tryptophan residue. SDS-PAGE analysis of the purified recombinant GST-p65 fusion protein demonstrated the expected band of approximately 92 kDa (Fig. 1A, lane 1). Thrombin cleavage of recombinant GST-p65 produced two bands of approximately 26 and 66 kDa (Fig. 1A, lane 2), corresponding in size to the GST fusion partner and the full-length mature p65 gene product, respectively. Rabbit anti-GST sera detected only bands corresponding in size to the recombinant GST-p65 (92 kDa) and thrombin-cleaved GST (26 kDa) in Western blots (Fig. 1B, lanes 1 and 2). Rabbit anti-p65 sera bound only to bands corresponding in size to recombinant GST-p65 (92 kDa) and the p65 gene product (66 kDa) after thrombin cleavage (Fig. 1B, lanes 3 and 4). Western blot analysis of whole-cell proteins from M. hyopneumoniae with rabbit anti-p65 sera detected a protein equivalent in size to the full-length mature p65 gene product (66 kDa) (Fig. 1B, lane 5). Furthermore, the M. hyopneumoniae protein detected by rabbit anti-p65 sera partitioned exclusively into the hydrophobic phase following Triton X-114 fractionation and was shown to be susceptible to proteolytic cleavage after the treatment of intact whole M. hyopneumoniae cells with an increasing concentration of trypsin (results not shown). Collectively, these observations confirm that p65 is a surface-exposed membrane protein and confirms the results obtained by Wise and Kim (47) with mouse monoclonal antibodies to p65.
FIG. 1.
Expression, purification, and cleavage of recombinant GST-p65 and immunostaining with rabbit antisera. Recombinant GST-p65 was expressed in E. coli and purified by affinity chromatography. (A) Coomassie brilliant blue-stained gel of purified recombinant GST-p65 (lane 1) and thrombin-cleaved recombinant p65 fusion products (lane 2) separated by SDS-10% PAGE together with molecular mass markers (Novex). (B) Rabbit anti-GST and anti-p65 sera were used to immunostain the purified recombinant GST-p65 protein (lanes 1 and 3) and thrombin-cleaved recombinant p65 fusion products (lanes 2 and 4). M. hyopneumoniae strain LKR whole cells were immunostained with rabbit anti-p65 sera (lane 5). All protein samples were separated by SDS-10% PAGE together with prestained molecular mass markers (New England BioLabs) and transferred for Western blotting.
Sequence analysis.
The cloned p65 gene fragment from M. hyopneumoniae was 1.791 kbp in length and coded for a mature peptide of 597 amino acids with a predicted molecular mass of 67.6 kDa. The calculated molecular weight was consistent with that determined by SDS-PAGE (Fig. 1A, lane 2). The nucleotide and translated amino acid sequences of the p65 gene of M. hyopneumoniae strain LKR were nearly identical to those of the equivalent gene from strain 232A, with the exception of the absence of the amino acids threonine and leucine at positions 342 and 343, respectively, in the p65 prolipoprotein from strain 232A. Differences between the amino acid sequence of p65 from M. hyopneumoniae type strains J and LKR were concentrated at the carboxyl terminus, and most substitutions involved similar amino acid residues. The amino acid substitutions identified in the p65 prolipoprotein of strain LKR compared to those on strain J were isoleucine replaced by valine at position 250; serine replaced by threonine at position 262; serine replaced by glycine at positions 564, 579, and 596; aspartate replaced by asparagine at position 615; glutamate replaced by aspartate at position 623; and proline replaced by serine at position 624. One dissimilar substitution, a change from lysine to methionine, was identified at position 581. These differences in the amino acid sequences of p65 were not at residues predicted to be functionally significant.
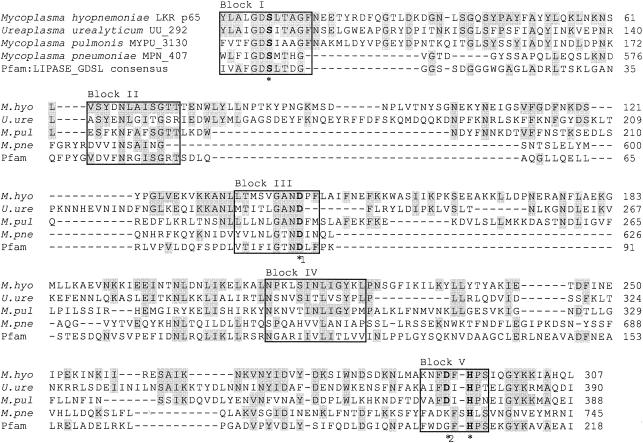
The search for protein family signature sequences and conserved domains in the PROSITE database identified five conserved sequence blocks of high amino acid similarity and similar relative position to the novel family of GDSL lipase/acylhydrolase proteins (Fig. 2) (42). In comparison to the true lipases (2), the consensus motif of the GDSL family of lipolytic enzymes is located much closer to the amino terminus within the first conserved sequence block. In M. hyopneumoniae, the active-site catalytic triad proposed by Brumlik and Buckley (3) is formed by the amino acids serine, aspartate, and histidine located at positions 18, 143, and 294 in blocks I, III, and V, respectively (Fig. 2).
FIG. 2.
Multiple-sequence alignment of M. hyopneumoniae strain LKR p65 with putative mycoplasma GDSL-like lipase/acylhydrolase proteins. The conserved sequence blocks are in boxes and identical amino acids are shaded in grey. Amino acid residues forming the putative catalytic triad (*) are printed in boldface. The active-site aspartate residue proposed by Brumlik and Buckley (3) for the A. hydrophila lipase/acylhydrolase is indicated (*1) together with the possible alternate active site aspartate residue (*2) proposed by Arpigny and Jaeger (2). The GDSL consensus motif is located in block I. Numbers on the right indicate amino acid positions.
BLAST searches of GenBank with the p65 sequence revealed closest identity to conserved hypothetical proteins from Mycoplasma pulmonis (MYPU_3130) (23%) and Ureaplasma urealyticum (UU_292) (27%) (Fig. 2). The GDSL-like motif and conserved sequence blocks in Mycoplasma pneumoniae MPN_407 have also recently been annotated (Fig. 2). The carboxyl-terminal location of the GDSL motif of M. pneumoniae MPN_407 is a novel characteristic. It is notable that the sequence similarity between the mycoplasma GDSL-like lipase/acylhydrolase proteins is restricted to the conserved domains and signature sequences. M. pulmonis MYPU_3130, U. urealyticum UU_292, and M. pneumoniae MPN_407 vary considerably in size (323, 208, and 101 kDa, respectively) and do not encode a prokaryotic lipoprotein acylation signal sequence. GDSL-like lipase/acylhydrolase proteins have not been annotated in the genomic sequences of Mycoplasma genitalium, Mycoplasma penetrans, or M. gallisepticum and could not be detected by BLAST search with the Pfam GDSL lipase/acylhydrolase consensus sequence. Interestingly, the acyl carrier protein (AcpP) and the acyl carrier protein phosphodiesterase (AcpD) are located immediately upstream of M. pneumoniae MPN_407 and M. pulmonis MYPU_3130, respectively. The first 88 amino acids encoded by the predicted open reading frame (ORF) immediately downstream of M. hyopneumoniae p65 from strain J (GenBank accession no. AF013714) showed 77% identity and 90% similarity with a 115-kDa ABC transporter ATP-binding protein homolog of Mycoplasma hyorhinis.
Enzymatic activity of p65.
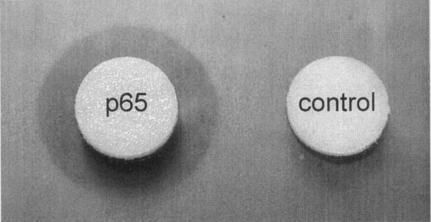
Filter disks impregnated with recombinant GST-p65 produced a zone of clearance when incubated on esterase indicator plates containing the short-chain acylglycerol tributyrin (Fig. 3). A zone of clearance was also observed when filter disks impregnated with thrombin-cleaved recombinant p65 were used. No zone of clearance was produced by GST (Fig. 3). The cleared zone produced by recombinant GST-p65 was significantly small and appeared only after 48 h of incubation, whereas the cleared zone produced by the type VII lipase of C. rugosa was significantly larger and appeared after 12 h of incubation. Although lipases will hydrolyze ester substrates of short-chain acylglycerols (at most 10 carbon atoms), they are specifically defined by their ability to hydrolyze long-chain acylglycerols (at least 10 carbon atoms) (20). Specific and sensitive detection of lipolytic activity can be achieved by the spectrophotometric detection of resorufin released from the artificial triglyceride 1,2-O-dilauryl-rac-glycero-3-glutaric acid resorufin ester (20, 22). The release of resorufin was detected following incubation with recombinant GST-p65, but the rate of release was significantly lower than with the type VII lipase of C. rugosa (results not shown). No resorufin was released following incubation with purified GST.
FIG. 3.
Halo formation around a filter disk containing recombinant GST-p65 on esterase indicator plates after 48 h of incubation at 37°C. The negative control contained only purified GST.
The enzymatic activity of recombinant GST-p65 was further examined on agar plates containing skim milk, egg yolk, or sheep blood erythrocytes. No zones of clearance were observed following incubation for more than 120 h (results not shown), indicating that recombinant GST-p65 does not have proteolytic, hemolytic, or phospholipase activity. The phospholipase activity of recombinant GST-p65 was also examined by thin-layer chromatographic analysis of the end products of phosphatidylcholine and lysophosphatidylcholine hydrolysis. No free fatty acids could be detected following incubation with recombinant GST-p65 for more than 48 h (results not shown).
Biochemical characterization of p65 lipase.
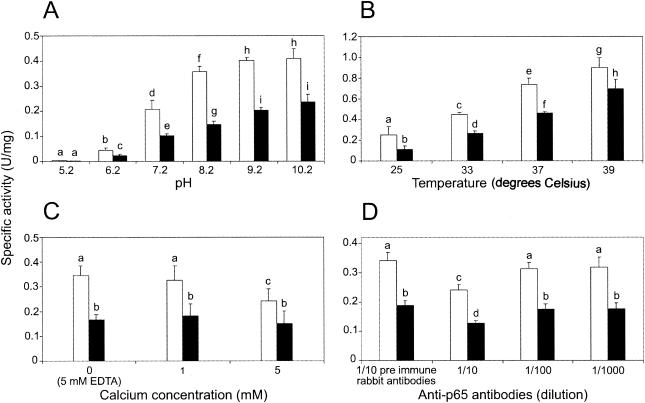
The effect of pH, temperature, calcium, and rabbit anti-p65 antibodies on the specific activity of recombinant GST-p65 was determined by microtiter plate assay with pNPC and pNPP. In all parallel experiments, the specific activity of recombinant GST-p65 was significantly greater with pNPC as a substrate (Fig. 4). An increase in pH and temperature produced a concomitant increase in the mean specific activity of recombinant GST-p65 with either pNPC or pNPP as a substrate (Fig. 4A and B). At pH 10.2, the mean specific activity of recombinant GST-p65 was estimated to be 0.41 or 0.22 U/mg with pNPC or pNPP as a substrate, respectively. An increase in pH from 9.2 to 10.2 did not produce a significant change in the mean specific activity of recombinant GST-p65 when either substrate was used, and thus the optimum pH was estimated to be between pH 9.2 and 10.2. The mean specific activity of recombinant GST-p65 was greatest at 39°C and was estimated to be 0.90 or 0.69 U/mg with pNPC or pNPP as a substrate, respectively. The optimum temperature for the activity of recombinant GST-p65 using either substrate was higher than 39°C. As noted previously (23), the stability of both pNP substrates decreased substantially above pH 8.0 and above 30°C. The rate of pNP release from pNPC and pNPP in the absence of recombinant GST-p65 was subtracted from all parallel experiments in the presence of recombinant GST-p65 to compensate for differences in the stabilty of either substrate under each condition.
FIG. 4.
Effects of pH (A), temperature (B), calcium ion concentration (C), and anti-p65 antibodies (D) on the specific activity of recombinant GST-p65 with pNPP (black columns) or pNPC (white columns) as a substrate. The means and standard errors of results of triplicate assays are shown. Columns labeled with different letters indicate significant differences (Student's t test; P < 0.05).
At a calcium concentration of 1 mM, the mean specific activity of recombinant GST-p65 was not significantly different from that of the control reaction conducted in the presence of 5 mM EDTA and was estimated to be 0.32 or 0.18 U/mg with pNPC or pNPP as a substrate, respectively (Fig. 4C). At a calcium concentration of 5 mM, the mean specific activity of recombinant GST-p65 was estimated to be 0.24 or 0.15 U/mg with pNPC or pNPP as a substrate, respectively. Compared to the control reaction, the mean specific activity of recombinant GST-p65 in the presence of 5 mM calcium was found to be significantly lower with pNPC as the substrate but was not significantly different with pNPP as the substrate. No significant increase in the specific activity of recombinant GST-p65 was detected in the presence of zinc at a concentration of 1 or 5 mM or in the presence of zinc and calcium, each at a concentration of 1 or 5 mM (results not shown). The mean specific activity of recombinant GST-p65 was found to be significantly lower in the presence of purified anti-p65 antibodies at a dilution of 1/10 and was estimated to be 0.18 or 0.12 U/mg with pNPC or pNPP as a substrate, respectively (Fig. 4D). No significant difference was detected in the mean specific activity of recombinant GST-p65 in reactions with either substrate conducted in the presence of purified antibody from nonimmunized rabbits at a dilution of 1/10 or purified anti-p65 antibodies at a dilution of 1/100 or 1/1,000 (Fig. 4D).
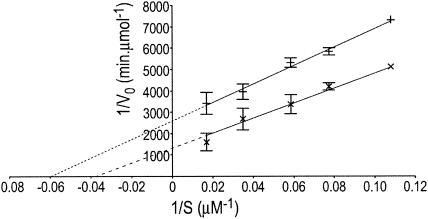
The hydrolysis of pNPC and pNPP by recombinant GST-p65 followed Michaelis-Menten kinetics. The Lineweaver-Burk double-reciprocal plots illustrate the relationship between V0 and S given by the equation 1/V0 = (Km/Vmax)S + 1/Vmax, where V is the velocity of the enzyme activity and S is the concentration of the substrate (Fig. 5). The relationships between V0 and S with pNPC or pNPP as a substrate were derived by linear regression and are given by the equations 1/V0 = 34984.3/S + 1315.8 and 1/V0 = 43056.5/S + 2593.8, respectively. The equations were solved to determine the kinetic parameters Km and kcat (Table 2). The Michaelis-Menten constant (Km) was greater for the hydrolysis of pNPC (26.6 μM) than for the hydrolysis of pNPP (16.6 μM), indicating that recombinant GST-p65 binds more strongly to the pNP ester of the short-chain fatty acid. The catalytic activity (kcat) of recombinant GST-p65 was also greater for pNPC (20.85 min−1) than for pNPP (10.42 min−1). Furthermore, pNPC was the preferred substrate for recombinant GST-p65, as indicated by the higher kcat/Km value for pNPC (0.78 min−1 μM−1) than for pNPP (0.62 min−1 μM−1).
FIG. 5.
Lineweaver-Burk double-reciprocal plot of the lipolytic activity of recombinant GST-p65 with pNPC (×) or pNPP (+) as a substrate. The means and standard errors of results of triplicate assays are shown. The relationships between V0 and S were derived by linear regression and are given by the equations 1/V0 = 34984.3/S + 1315.8 and 1/V0 = 43056.5/S + 2593.8 with pNPC or pNPP as a substrate, respectively. Note that at high-substrate concentrations, the errors were very small.
TABLE 2.
Kinetic parameters of M. hyopneumoniae recombinant GST-p65 for the hydrolysis of p-nitrophenyl esters of fatty acids
| p-Nitrophenyl estera | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) |
|---|---|---|---|
| Caproate (C6) | 26.6 | 20.85 | 0.78 |
| Palmitate (C16) | 16.6 | 10.42 | 0.62 |
Acyl chain length is indicated in parentheses.
Growth inhibition.
The growth of M. hyopneumoniae was significantly inhibited by purified anti-p65 antibodies. The concentrations of M. hyopneumoniae after 77 h of incubation in anti-p65 antibodies at dilutions of 1/50 and 1/100 were 10- and 5-fold lower, respectively, than in cultures incubated in the presence of antibodies from nonimmunized rabbits. This difference was consistent throughout the exponential phase of growth. Anti-p65 antibodies at a dilution of 1/50 also extended the lag phase from 18 to 50 h. Similar results were obtained when the experiment was repeated.
DISCUSSION
Putative lipase genes have been identified in the complete genomic sequences of all mycoplasma species studied so far (5, 11, 13, 17, 32, 40). Although most lipase genes code for the conserved pentapeptide GXSXG motif associated with the family of true lipolytic enzymes (2), the amino acid sequence of p65 of M. hyopneumoniae had closest identity with a novel family of lipolytic enzymes in which the active-site serine residue is located within a conserved, amino-terminal GDSL motif (42). This classification was further defined by the identification of five conserved sequence blocks that shared high amino acid identity and similar relative positions with other members of the GDSL family. Homologous structures were also identified by multiple-sequence alignment of p65 with GDSL-like proteins from the complete genomic sequences of U. urealyticum, M. pulmonis, and M. pneumoniae. Although members of the GDSL family in a number of prokaryotes and eukaryotes have been putatively annotated, homologous proteins in mycoplasmas have not been reported. Furthermore, the enzymatic function of most GDSL family members has yet to be determined.
Although the amino acid residues belonging to the catalytic triad in M. hyopneumoniae could be identified, Brumlik and Buckley (3) could not definitively assign the putative aspartate residue in Aeromonas hydrophila lipase/acyltransferase to the active site; the reported loss in lipase activity associated with the mutagenesis of aspartate to asparagine correlated with the impaired secretion of the lipase and may thus be due to incorrect folding of the protein and its subsequent degradation by proteases (2). An alternative aspartate residue located three positions upstream from the active-site histidine has been identified by analysis of the three-dimensional structure of an esterase from bovine brain (18). In p65 from M. hyopneumoniae, an aspartate residue located two positions upstream from the active-site histidine may alternatively form part of the catalytic triad (Fig. 2). An aspartate residue located at a similar relative position could also be identified in the GDSL-like lipase/acylhydrolase sequences of U. urealyticum and M. pulmonis (Fig. 2). Interestingly, this third acidic residue is replaced by tryptophan in the catalytic site of an esterase from Streptomyces scabiei (44). Further work is still needed to establish the common structure of the catalytic site of the GDSL family of lipolytic enzymes.
The initial identification of the esterase activity of recombinant GST-p65 on tributyrin agar plates was supported by the demonstration that rabbit anti-p65 antibodies specifically inhibited the esterase activity of recombinant GST-p65 against pNP esters of fatty acids. Definitive evidence of the lipase and esterase activity of recombinant GST-p65 was subsequently demonstrated by the release of resorufin from the artificial triglyceride 1,2-O-dilauryl-rac-glycero-3-glutaric acid resorufin ester. While a number of other enzymatic functions have also been associated with members of the GDSL family, recombinant GST-p65 had no detectable protease, hemolysin, or phospholipase activity. Phospholipase activity in association with membrane-bound proteins of several mycoplasma species has been detected (38, 43) and has been demonstrated for a recent member of the GDSL family of lipolytic enzymes (10). However, phospholipids are generally poor substrates for lipolytic enzymes (41). Although a lipase gene has also been cloned and characterized from Mycoplasma mycoides subsp. mycoides LC (35), our study is the first report of lipolytic activity associated with an exposed lipid-modified mycoplasma antigen. Despite the relative abundance and diversity of lipoprotein genes in the sequenced mycoplasma genomes, the only other mycoplasma lipoprotein identified that is predicted to show enzymatic activity is subunit b of the F0F1-type ATPase of M. pneumoniae (34). However, the definitive function of this lipoprotein has not been determined.
Lipolytic enzymes are typically characterized by their ability to catalyze the hydrolysis of a wide range of fatty acid esters. In particular, aliphatic chain length specificity is an important means of differentiating between lipase and esterase activity. The comparatively higher levels of esterase activity of recombinant GST-p65 were initially indicated by its relatively higher specific activity when pNPC was used as a substrate under all the reaction conditions tested. Kinetic studies subsequently indicated that recombinant GST-p65 has a greater specificity (kcat/Km) for the relatively short aliphatic chain of pNPC. The kcat/Km values for the hydrolysis of pNPC and pNPP were 0.78 and 0.62 min−1 μM−1, respectively. In comparison, the kcat/Km values for the hydrolysis of pNP esters of fatty acids from oleate (C18) to acetate (C2) by a novel recombinant E. coli esterase ranged from 4.7 to <0.078 min−1 μM−1 (23). Many lipases are further defined by their relative increase in activity in the presence of emulsified substrates, due to a conformational change that exposes the active-site residues at the lipid-water interface (21). Although interfacial activation in all bacterial lipases has not been observed, true esterases are not activated by the interfacial area presented to the enzyme when the substrate forms an emulsion and will degrade only monomeric substrates (21). All enzyme assays with recombinant GST-p65 were conducted with substrates emulsified in Triton X-100, which thus indirectly suggests that recombinant GST-p65 is not a true esterase. While it is possible that the increase in activity of recombinant GST-p65 at high pH and temperature is associated with the concomitant increase in emulsification efficiency, empirical results indicate that recombinant GST-p65 is able to degrade monomeric substrates. Thus, the activity of recombinant GST-p65 is not predicted to be dependent on the emulsification of the substrate.
The biochemical properties of a mycoplasma lipolytic enzyme have only previously been characterized for a partially purified lipase from M. gallisepticum (37). Although the biochemical properties of many bacterial lipolytic enzymes have been examined previously, the diversity of materials and methods used often prevents the direct comparison of results. With pNP esters of fatty acids, both the lipase and esterase activities of recombinant GST-p65 were estimated to be optimal at >39°C and pH 9.2. In comparison, the activity of the partially purified M. gallisepticum lipase on triglycerides was shown to be optimal at 37°C and a pH between 7.5 and 8.0, depending on the nature of the substrate. Although the pH optima may vary substantially, most serine hydrolases show little or no activity below pH 5, and the specific activity of recombinant GST-p65 decreased substantially below pH 7.2. Calcium ions have previously been shown to increase the lipolytic activity of A. laidlawii (37) and are known to function in the structural stabilization and activation of many lipolytic enzymes. However, recombinant GST-p65 did not require calcium, zinc, or other divalent cations for activity. Similarly, the presence or absence of calcium, magnesium, or manganese ions did not affect the activity of the partially purified M. gallisepticum lipase. Although high concentrations of calcium ions appeared to inhibit the enzymatic activity of recombinant GST-p65, this is likely to be an indirect affect and has not previously been reported for other bacterial lipases. Anti-p65 antibodies significantly reduced the specific activity of recombinant GST-p65, and thus the complement-independent growth inhibition of M. hyopneumoniae by anti-p65 antibodies may have been caused by a reduction in the lipolytic activity of the p65 enzyme. It is possible that anti-p65 antibodies mediated changes in the expression of the p65 gene that resulted in growth inhibition through the loss of phenotype. The expression of vlhA by M. gallisepticum has been shown to be influenced by the presence of anti-pMGA monoclonal antibodies or polyclonal serum (27).
The GDSL family of lipolytic enzymes comprises a diverse group of proteins of various sizes and functions, and several reports have highlighted their potential role as bacterial virulence factors (10, 46). The primary metabolic function of the GDSL lipase homologs M. pulmonis MYPU_3130 and M. pneumoniae MPN_407 is suggested by the presence of an ORF encoding an acyl carrier protein homolog immediately upstream of these ORFs. Similarly, the identification of an ORF coding for an ABC transporter ATP-binding protein homolog downstream of the p65 gene suggests that it is part of an active transport system. Exogenous fatty acids produced by the action of mycoplasma lipases have been predicted to be an important substrate for the biosynthesis of lipoproteins, phospholipids, and glycolipids (33). The process of acquisition of complex nutrients is thought to play an indirect role in mycoplasma pathogenesis and is a common theme in studies of the intimate relationship of the mycoplasma with the host cell surface. The lesions associated with mycoplasmoses appear to be primarily the result of host immune reactions and inflammatory responses, rather than due to the direct toxic effects of mycoplasma cell components and reactive metabolic by-products (36). Free fatty acids are known to modulate a number of immune parameters (4, 15, 48), and thus the liberation of free fatty acids by lipolytic enzymes may induce a local immune response. However, it is important to note that mycoplasmas are not protected by a cell wall and are thus susceptible to lysis at high concentrations of long-chain fatty acids. The inhibition of growth of M. hyopneumoniae by anti-p65 antibodies suggests a primary physiological role for p65. It is possible that the lipolytic function of p65 may reduce the function of surfactants in pneumonic lungs (45). In P. aeruginosa, the synergistic effect of a lipase and a phospholipase can result in the complete hydrolysis of a major lung surfactant in vitro (19). Surfactant proteins help maintain normal lung function and are also involved in the induction of the mycoplasmacidal activity of alveolar macrophages (16).
Despite the apparent abundance of genes for lipid-modified proteins in the complete genomic sequences of all mycoplasma species studied so far, the functions of most of these proteins are yet to be determined. The functional classification of mycoplasma proteins is an important adjunct to the current emphasis on genomics-based research. These studies have shown that p65, a lipid-modified, major immunodominant surface antigen of M. hyopneumoniae, is a functional member of the GDSL family of lipolytic enzymes. Further work is required to investigate the physiological and pathogenic significance of p65 in vitro and in vivo.
Acknowledgments
This work was supported by funding from the Australian Research Council and Bioproperties Australia Pty. Ltd.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arpigny, J. L., and K. E. Jaeger. 1999. Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343:177-183. [PMC free article] [PubMed] [Google Scholar]
- 3.Brumlik, M. J., and J. T. Buckley. 1996. Identification of the catalytic triad of the lipase/acyltransferase from Aeromonas hydrophila. J. Bacteriol. 178:2060-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buttke, T. M., and M. A. Cuchens. 1984. Inhibition of lymphocyte proliferation by free fatty acids. II. Toxicity of stearic acid towards phytohaemagglutinin-activated T cells. Immunology 53:507-514. [PMC free article] [PubMed] [Google Scholar]
- 5.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. Rocha, and A. Blanchard. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, B. C., and P. Pease. 1967. Lipolytic activity by oral pleuropneumonia-like (Mycoplasma) organisms. J. Gen. Microbiol. 47:171-174. [DOI] [PubMed] [Google Scholar]
- 7.Derewenda, Z. S., and U. Derewenda. 1991. Relationships among serine hydrolases: evidence for a common structural motif in triacylglyceride lipases and esterases. Biochem. Cell Biol. 69:842-851. [DOI] [PubMed] [Google Scholar]
- 8.Duffy, M. F., A. H. Noormohammadi, N. Baseggio, G. F. Browning, and P. F. Markham. 1998. Immunological and biochemical characterisation of membrane proteins, p. 267-278. In R. A. J. Nicholas (ed.), Methods in molecular biology: Mycoplasma protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 9.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farn, J. L., R. A. Strugnell, P. A. Hoyne, W. P. Michalski, and J. M. Tennent. 2001. Molecular characterization of a secreted enzyme with phospholipase B activity from Moraxella bovis. J. Bacteriol. 183:6717-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, J. L. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J.-F. Tomb, B. A. Dougherty, K. F. Bott, P.-C. Hu, T. S. Lucier, S. N. Petterson, H. O. Smith, C. A. Hutchinson III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 12.Friis, N. F. 1975. Some recommendations concerning primary isolation of Mycoplasma suipneumoniae and Mycoplasma flocculare a survey. Nord. Vet. Med. 27:337-339. [PubMed] [Google Scholar]
- 13.Glass, J. I., E. J. Lefkowitz, J. S. Glass, C. R. Heiner, E. Y. Chen, and G. H. Cassell. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407:757-762. [DOI] [PubMed] [Google Scholar]
- 14.Harlow, E., and D. P. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Hawley, H. P., and G. B. Gordon. 1976. The effects of long chain free fatty acids on human neutrophil function and structure. Lab. Investig. 34:216-222. [PubMed] [Google Scholar]
- 16.Hickman-Davis, J., J. Gibbs-Erwin, J. R. Lindsey, and S. Matalon. 1999. Surfactant protein A mediates mycoplasmacidal activity of alveolar macrophages by production of peroxynitrite. Proc. Natl. Acad. Sci. USA 96:4953-4958.10220400 [Google Scholar]
- 17.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho, Y. S., L. Swenson, U. Derewenda, L. Serre, Y. Wei, Z. Dauter, M. Hattori, T. Adachi, J. Aoki, H. Arai, K. Inoue, and Z. S. Derewenda. 1997. Brain acetylhydrolase that inactivates platelet-activating factor is a G-protein-like trimer. Nature 385:89-93. [DOI] [PubMed] [Google Scholar]
- 19.Jaeger, K. E. 1994. Extrazellulare Enzyme von Pseudomonas aeruginosa als Virulenzfaktoren. Immun. Infekt. 22:177-180. [PubMed] [Google Scholar]
- 20.Jaeger, K. E., B. W. Dijkstra, and M. T. Reetz. 1999. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol. 53:315-351. [DOI] [PubMed] [Google Scholar]
- 21.Jaeger, K. E., S. Ransac, B. W. Dijkstra, C. Colson, M. van Heuvel, and O. Misset. 1994. Bacterial lipases. FEMS Microbiol. Rev. 15:29-63. [DOI] [PubMed] [Google Scholar]
- 22.Johnston, J. L., S. J. Billington, V. Haring, and J. I. Rood. 1995. Identification of fimbrial assembly genes from Dichelobacter nodosus: evidence that fimP encodes the type-IV prepilin peptidase. Gene 161:21-26. [DOI] [PubMed] [Google Scholar]
- 23.Kanaya, S., T. Koyanagi, and E. Kanaya. 1998. An esterase from Escherichia coli with a sequence similarity to hormone-sensitive lipase. Biochem. J. 332:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, M. F., M. B. Heidari, S. J. Stull, M. A. McIntosh, and K. S. Wise. 1990. Identification and mapping of an immunogenic region of Mycoplasma hyopneumoniae p65 surface lipoprotein expressed in Escherichia coli from a cloned genomic fragment. Infect. Immun. 58:2637-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobisch, M., and N. F. Friis. 1996. Swine mycoplasmoses. Rev. Sci. Tech. Off. Int. Epizoot. 15:1569-1605. [DOI] [PubMed] [Google Scholar]
- 26.Lloyd, L. C., and J. R. Etheridge. 1981. The pathological and serological response induced in pigs by parenteral inoculation of Mycoplasma hyopneumoniae. J. Comp. Pathol. 91:77-83. [DOI] [PubMed] [Google Scholar]
- 27.Markham, P. F., M. D. Glew, G. F. Browning, K. G. Whithear, and I. D. Walker. 1998. Expression of two members of the pMGA gene family of Mycoplasma gallisepticum oscillates and is influenced by pMGA-specific antibodies. Infect. Immun. 66:2845-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markham, P. F., M. D. Glew, K. G. Whithear, and I. D. Walker. 1993. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect. Immun. 61:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meynell, G. G., and E. Meynell. 1970. Theory and practice in experimental bacteriology, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 30.O'Brien, S. J., J. M. Simonson, M. W. Grabowski, and M. F. Barile. 1981. Analysis of multiple isoenzyme expression among twenty-two species of Mycoplasma and Acholeplasma. J. Bacteriol. 146:222-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ollis, D. L., E. Cheah, M. Cygler, B. Dijkstra, F. Frolow, S. M. Franken, M. Harel, S. J. Remington, I. Silman, and J. Schrag. 1992. The alpha/beta hydrolase fold. Protein Eng. 5:197-211. [DOI] [PubMed] [Google Scholar]
- 32.Papazisi, L., T. S. Gorton, G. Kutish, P. F. Markham, G. F. Browning, D. K. Nguyen, S. Swartzell, A. Madan, G. Mahairas, and S. J. Geary. 2003. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain R(low). Microbiology 149:2307-2316. [DOI] [PubMed] [Google Scholar]
- 33.Pollack, J. D., M. V. Williams, and R. N. McElhaney. 1997. The comparative metabolism of the mollicutes (Mycoplasmas): the utility for taxonomic classification and the relationship of putative gene annotation and phylogeny to enzymatic function in the smallest free-living cells. Crit. Rev. Microbiol. 23:269-354. [DOI] [PubMed] [Google Scholar]
- 34.Pyrowolakis, G., D. Hofmann, and R. Herrmann. 1998. The subunit b of the F0F1-type ATPase of the bacterium Mycoplasma pneumoniae is a lipoprotein. J. Biol. Chem. 273:24792-24796. [DOI] [PubMed] [Google Scholar]
- 35.Rawadi, G., J. L. Lalanne, and D. Roulland-Dussoix. 1995. Cloning and characterization of the lipase operon from Mycoplasma mycoides subspecies mycoides LC. Gene 158:107-111. [DOI] [PubMed] [Google Scholar]
- 36.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rottem, S., and S. Razin. 1964. Lipase activity of mycoplasma. J. Gen. Microbiol. 37:123-134. [DOI] [PubMed] [Google Scholar]
- 38.Salman, M., and S. Rottem. 1995. The cell membrane of Mycoplasma penetrans: lipid composition and phospholipase A1 activity. Biochim. Biophys. Acta 1235:369-377. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sasaki, Y., J. Ishikawa, A. Yamashita, K. Oshima, T. Kenri, K. Furuya, C. Yoshino, A. Horino, T. Shiba, T. Sasaki, and M. Hattori. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 30:5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simons, J. W., M. D. van Kampen, S. Riel, F. Gotz, M. R. Egmond, and H. M. Verheij. 1998. Cloning, purification and characterisation of the lipase from Staphylococcus epidermidis—comparison of the substrate selectivity with those of other microbial lipases. Eur. J. Biochem. 253:675-683. [DOI] [PubMed] [Google Scholar]
- 42.Upton, C., and J. T. Buckley. 1995. A new family of lipolytic enzymes? Trends Biochem. Sci. 20:178-179. [DOI] [PubMed] [Google Scholar]
- 43.van Golde, L. M., R. N. McElhaney, and L. L. van Deenen. 1971. A membrane-bound lysophospholipase from Mycoplasma laidlawii strain B. Biochim. Biophys. Acta 231:245-249. [DOI] [PubMed] [Google Scholar]
- 44.Wei, Y., J. L. Schottel, U. Derewenda, L. Swenson, S. Patkar, and Z. S. Derewenda. 1995. A novel variant of the catalytic triad in the Streptomyces scabies esterase. Nat. Struct. Biol. 2:218-223. [DOI] [PubMed] [Google Scholar]
- 45.Wichert, P., and A. Wilke. 1976. Alveolar stability and phospolipid content in normal pig lungs and in pig lungs with Mycoplasma pneumonia. Scand. J. Respir. Dis. 57:25-30. [PubMed] [Google Scholar]
- 46.Wilhelm, S., J. Tommassen, and K. E. Jaeger. 1999. A novel lipolytic enzyme located in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 181:6977-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wise, K. S., and M. F. Kim. 1987. Major membrane surface proteins of Mycoplasma hyopneumoniae selectively modified by covalently bound lipid. J. Bacteriol. 169:5546-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yaqoob, P. 2003. Lipids and the immune response: from molecular mechanisms to clinical applications. Curr. Opin. Clin. Nutr. Metab. Care 6:133-150. [DOI] [PubMed] [Google Scholar]
- 49.Young, T. F., and R. F. Ross. 1987. Assessment of antibody response of swine infected with Mycoplasma hyopneumoniae by immunoblotting. Am. J. Vet. Res. 48:651-656. [PubMed] [Google Scholar]