Abstract
Rhodospirillum centenum is a purple photosynthetic bacterium that forms resting cyst cells when starved for nutrients. In this study, we demonstrate that chalcone synthase gene (chsA) expression is developmentally regulated, with expression of chsA increasing up to 86-fold upon induction of the cyst developmental cycle. Screening for mini-Tn5-induced mutants that exhibit elevated chsA::lacZ expression has led to the isolation of a set of R. centenum mutants that display increased chsA gene expression concomitant with constitutive induction of the cyst developmental cycle. These “hypercyst” mutants have lost the ability to regulate cyst cell formation in response to nutrient availability. Sequence analysis indicates that the mini-Tn5-disrupted genes code for a variety of factors, including metabolic enzymes and a large set of potential regulatory factors, including four gene products with homology to histidine sensor kinases and three with homology to response regulators. Several of the disrupted genes also have sequence similarity to che-like signal transduction components.
Rhodospirillum centenum is one of several bacterial species capable of cyst cell formation (1, 5). Cysts are bacterial resting cells that are extremely resistant to desiccation and mildly resistant to other environmental stresses, such as heat and UV light. Cyst formation is a complex process induced by a downshift in nutrient availability, which results in cells becoming spherical in shape, losing motility, and synthesizing a complex outer coat. The thick outer coat consists of two layers, an intine layer that consists of lipids and carbohydrates and an exine layer that consists of lipopolysaccharides and lipoproteins, which function in protecting the cell from environmental stresses (21). Cyst-forming cells also induce high levels of polyhydroxybutyrate (PHB) synthesis, resulting in the accumulation of large intracellular PHB storage granules (1, 23, 27). PHB accumulation presumably functions to provide an energy source for later stages of cyst formation and subsequent germination. In R. centenum, cysts are most commonly observed in clusters of four or more cells surrounded by a common outer coat (1, 27).
The physiology of cyst formation has been characterized in several bacterial species, including R. centenum (1, 27), Azotobacter vinelandii (23), and Azospirillum brasilense (22). However, in comparison to the detailed understanding of endospore induction in Bacillus subtilis and myxospore induction in Myxococcus xanthus, very little is known about the genetic requirements of cyst cell induction in any bacterial species. In A. vinelandii, the polymer alginate is a major component in the intine and exine layers of cysts, and mutants defective in alginate production are unable to form mature cysts (3). Several regulatory factors for alginate biosynthesis have been reported for A. vinelandii. The sensor kinase GacS (4), the response regulator AlgR (19), and the alternative sigma factor σE (18) are all required for alginate production. Strains with mutations in these alginate regulators, as well as mutations in algD, which codes for a critical enzyme in alginate biosynthesis, GDP-mannose dehydrogenase, are defective in formation of mature cysts (3). Mutations in these genes result in cells that are spherical but lack a competent outer coat. Since alginate is a major component of the A. vinelandii cyst outer coat, it is not surprising that the control of alginate biosynthesis is required for mature cyst formation. However, it is not known if these alg regulators have a more global role in the control of cyst cell formation.
If cyst formation is at all similar to endospore or myxospore formation at the molecular level, then there is likely a large hierarchy of signal transduction components that regulate encystment. In both endospore (2) and myxospore (25) formation, the formation of a resting cell requires the integration of multiple input signals through a complex signaling mechanism. We anticipate that the same may prove true in the formation of resting cyst cells. In this study, we describe the development of a genetic screen useful for the isolation of R. centenum mutants that are defective in regulating cyst cell formation. This screen has resulted in the identification of several novel regulatory elements that control cyst development in this species.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
R. centenum strain ZJC229 was the parental strain used in this study. This strain is derived from the wild-type strain (ATCC 51521), with the exception that a chsA::lacZ transcriptional fusion is inserted in an intergenic region of the chromosome. To construct this strain, the chsA promoter was PCR amplified using primers with engineered SmaI and XbaI restriction sites CHSYNSMAI, 5′-CGCCCGGGTTCACGGCCGAATAGACGCC-3′, and CHSYNXBAI, 5′-GCTCTAGAGTGTCACCGGGACAACCCG-3′. The PCR product was cloned into pZJD35, a pSP73 derivative containing a partial lacZ gene. The fusion was recombined into the che1 region of the chromosome oriented in the direction opposite to that of the che1 promoter (9) by using the gentamicin-resistant sucrose selection vector pZJD29a (28). All R. centenum strains were cultured aerobically either in liquid CENS medium (27) at 37°C or on agar-solidified CENS medium at 42°C. Escherichia coli strain S17-1 (λ pir)/pZJD17 was used for conjugation and transposition of the mini-Tn5 interposon (11). pZJD17 is derived from the previously described plasmid pUTmini-Tn5-Sm/Sp (15) with the 5′- and 3′-terminator sites deleted to decrease polarity effects caused by premature termination of transcription of downstream genes (10). Plasmid pSK(+) (Bluescript; Stratagene) and E. coli strain DH5α were used for cloning and maintaining R. centenum chromosomal DNA fragments. E. coli strains were cultured in Luria-Bertani medium at 37°C with antibiotics used when appropriate. For R. centenum, antibiotics were used at concentrations of 10 μg of spectinomycin/ml and 40 μg of kanamycin/ml; for E. coli, ampicillin and spectinomycin were used at 150 and 50 μg/ml, respectively.
Conjugation and transposition of the mini-Tn5 interposon.
The modified mini-Tn5 spectinomycin-resistant (Spr) interposon was delivered to R. centenum strain ZJC229 via conjugation with E. coli S17-1 (λ pir)/pZJD17. Plasmid pZJD17 is a suicide vector that contains an incomplete replicon from plasmid R6K, with replication dependent on the presence of the π protein encoded by the pir gene that is provided in trans on the chromosome of E. coli S17-1 (λ pir) (26). To prevent secondary transposition events, the transposase gene is located in cis outside of the transposable element. The interposon was introduced into R. centenum through a filter mating procedure (11). Briefly, R. centenum and E. coli cells were washed three times to remove antibiotics and then applied to a 0.45-μm-pore-size filter (filter no. 245-0045; Nalgene) in a 5:1 ratio of R. centenum cells to E. coli cells. Filters were placed onto a CENS plate with no antibiotics and incubated at 37°C for 4 h to allow conjugation and phenotypic segregation. The cells were then resuspended in 5 ml of CENS medium with 200-μl aliquots of the resuspension spread onto CENS plates containing 10 μg of spectinomycin/ml to select for the transposition event and 40 μg of kanamycin/ml to counterselect against the E. coli donor (R. centenum is naturally resistant to kanamycin [11]). The plates were incubated at 42°C for 72 h to allow growth of Spr transconjugants. Strains showing a “hypercyst” phenotype were identified by a distinctive altered colony morphology (dry, rippled colonies) coupled with elevated chsA::lacZ expression that was observed after addition of a 50% (vol/vol) dimethyl sulfoxide solution overlay containing 2% (wt/vol) 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Transconjugants were monitored visually and classified by the time they required to display a blue color after addition of the 2% X-Gal solution.
Recombinant DNA techniques.
Restriction and other DNA modification enzymes were purchased from New England Biolabs and used according to the manufacturer's instructions. Chromosomal DNA was isolated as described previously (24). R. centenum genomic DNA was digested with either EcoRV, EcoRI, or PstI as a restriction enzyme and ligated into the corresponding restriction enzyme sites in plasmid pSK(+). Plasmids containing a chromosomal DNA fragment that carried the mini-Tn5 Spr interposon were selected after electroporation into E. coli DH5α by growth on Luria-Bertani plates containing 150 μg of ampicillin/ml and 50 μg of spectinomycin/ml. Plasmid DNA from transformants was isolated using the QIAprep Spin Miniprep kit purchased from QIAGEN Inc., with initial sequencing of the DNA regions flanking the mini-Tn5 interposon performed using the primers 5′-CTGTTCTTCTACGGCAAG-3′ and 5′-CACAGCCAAACTATCAGG-3′, which are specific to the 5′ and 3′ ends of the interposon, respectively. Successive sequencing reactions were performed by primer walking using primers designed every ∼300 bp as needed to continue sequencing upstream and downstream of the interposon disruption. Sequencing reactions were performed using an ABI Prism sequencing kit and an ABI 3700 capillary sequencer (Applied Biosystems, Inc.) according to the manufacturer's protocol.
β-Galactosidase assays.
R. centenum strains were grown at 37°C overnight in liquid CENS medium with appropriate antibiotics. Cells were washed three times in phosphate buffer, and the final cell density was adjusted to approximately 109 cells/ml. Five microliters of cells was pipetted as colonies onto either CENS or CENBA plates. Colonies were harvested after the time indicated and resuspended in 1.5 ml of phosphate buffer. Resuspensions were sonicated to lyse cells (three times for 15 s each at 70% output power using a Microson ultrasonic cell disrupter). The total protein in the cell lysates was determined using the Advanced Protein Assay reagent (Cytoskeleton, Inc.), a colorimetric protein assay reagent, according to the manufacturer's instructions. β-Galactosidase assays were performed using 4% o-nitrophenyl-β-d-galactopyranoside (ONPG) as described previously (29). β-Galactosidase units represent micromoles of ONPG hydrolyzed per minute per milligram of protein. The assays were performed in triplicate on three separate cultures for each strain.
Desiccation resistance assays.
R. centenum colonies were harvested after 3 days of growth on CENS and resuspended in 1 ml of phosphate buffer. Resuspended cells were sonicated for 5 s at low power to disperse cellular aggregates. Total viable cell counts (vegetative cells plus cyst cells) were determined by pipetting serial dilutions on to CENS plates and incubating at 42°C for 2 to 3 days. To determine the number of desiccation-resistant cyst cells, serial dilutions were also pipetted onto 0.45-μm-pore-size filters, dried for 20 min at 22°C, and then desiccated at 42°C for 3 days. The desiccated filters were then placed onto CENS plates at 42°C to allow outgrowth of surviving cells. The total numbers of CFU before and after desiccation were determined with an analysis repeated in triplicate for each strain.
Phase-contrast microscopy.
Wild-type R. centenum and hypercyst mutants were grown as described in the present paper, and wet mounts were prepared at the stated time intervals. Individual cells were viewed with a Nikon E800 light microscope equipped with a 100× Plan Apo oil objective. Image capture was carried out with a Princeton Instruments cooled charge-coupled device camera and Metamorph imaging software, version 4.5.
Nucleotide sequence accession numbers.
The sequence information obtained in this study has been deposited in GenBank under accession numbers AY260902 to AY260905 and AY506540 to AY506543.
RESULTS
The chsA gene is a developmental reporter in R. centenum.
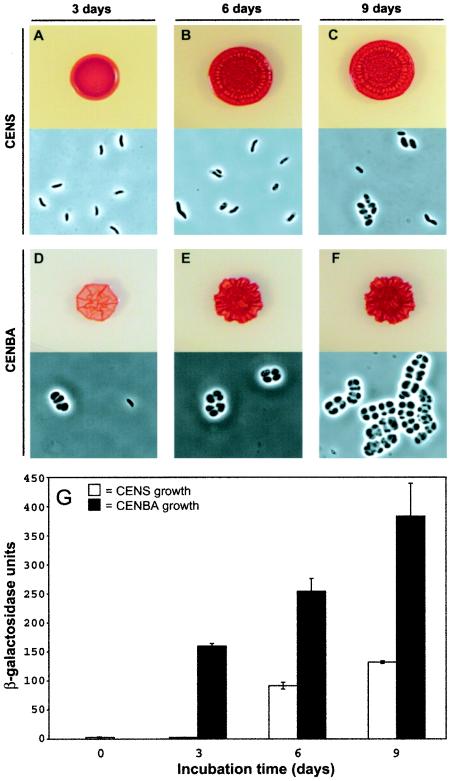
R. centenum cells are known to undergo formation of heat- and desiccation-resistant cyst cells as a consequence of nutrient deprivation or growth on energy-poor carbon sources such as butyrate (1, 27). Visual tracking of cyst development can be accomplished by observing macroscopic changes in colony morphology and/or microscopic changes in cell shape (Fig. 1A to F). As shown in Fig. 1A to C, R. centenum vegetative cells slowly enter cyst development when grown for more than 3 days on nutrient-rich CENS plates. After 3 days of growth, colonies consist of >99% vibrio-shaped vegetative cells. As the colony matures, it changes from a shiny, convex colony to one that has distinct striated and ridged features. Microscopic analysis indicates that changes in colony morphology observed at day 6 are concurrent with the onset of cyst formation, as evidenced by a change in the cell shape from vibrio-like to oblong (Fig. 1B). At day 9, the cells take on the typical features of a mature cyst cell, becoming spherical with a highly refractile outer coat surrounding clusters of cells. When grown on minimal medium with butyrate as the sole carbon source (CENBA), the cells rapidly induce cyst development with colony morphology and microscopic features typical of mature cyst cells observed in as little as 3 days (Fig. 1D to F). Longer incubations on CENBA result in larger cyst cell clusters and a higher overall percentage of cysts in the population.
FIG. 1.
The chsA promoter as a reporter for cyst formation in R. centenum. (A to F) Development of colony and cellular morphology in strain ZJC229 after 3, 6, and 9 days of growth on nutrient-rich CENS agar (A to C) and on cyst-inducing CENBA agar (D to F). (G) Corresponding β-galactosidase assays on strain ZJC229 after 0, 3, 6, and 9 days of growth on CENS (open bars) and CENBA (filled bars) media. Error bars represent standard deviations derived from assays performed in triplicate with three cultures for each time point.
To facilitate screening for cyst cell developmental mutants, we utilized the fortuitous observation that expression of the chalcone synthase gene (chsA) is developmentally regulated. The chsA gene product is homologous to proteins of the type III polyketide synthase superfamily of plant and bacterial biosynthetic enzymes. Type III polyketide synthases are necessary for the production of a wide variety of secondary metabolites (17), including flavonoid biosynthesis in higher plants (16), with flavonoids performing a number of cellular functions including UV light protection (7). The flavonoid biosynthetic pathway in plants is tightly regulated in response to a variety of developmental and environmental signals (13). The role of chalcone synthase-like polyketide synthases in bacteria is unclear; however, we have observed that chsA gene expression substantially increases as R. centenum cells enter the cyst cell developmental pathway. As indicated by Fig. 1G, β-galactosidase activity from the chsA::lacZ reporter increases over 9 days of growth on CENS medium, with the greatest increase in activity occurring between days 3 and 6, which corresponds to the onset of cyst formation as observed microscopically. When cells are grown on cyst-inducing CENBA medium, chsA activity is rapidly induced to levels 58-fold higher than that observed on CENS medium after 3 days of incubation (Fig. 1G). Again, the increase in chsA gene expression is correlated with the induction of cysts. The results of this assay indicate that chsA is a developmentally regulated gene, with increased expression from the chsA promoter occurring at an early stage in cyst formation.
Isolation of mutants with elevated chsA activity.
Mutants that alter the ability of R. centenum cells to undergo cyst development can be isolated by screening for mutants that fail to undergo a change in colony morphology. However, screening for a lack of colony morphology changes over a 2- to 3-week period can be rather time-consuming. Furthermore, screening for mutants that exhibit an inability to form cysts can identify a very wide range of mutants, such as regulatory mutants that are unable to induce cyst development and strains with mutations in the structural components of the cyst cell outer coat. In order to more readily obtain regulatory mutants, we instead screened for mutants that prematurely form cysts under rich growth conditions (termed hypercyst mutants).
To isolate mutants defective in regulating cyst formation, we mutagenized R. centenum strain ZJC229, which contains a chromosomally encoded chsA::lacZ fusion, with a modified mini-Tn5 transposon in which the kanamycin resistance (Kmr) gene was replaced by a Spr gene that has no Ω terminator sites (10). To screen for cyst developmental mutants, transposon-mutagenized cultures of R. centenum were plated onto CENS with appropriate antibiotics and incubated at 42°C for 3 days. To monitor β-galactosidase activity, plates were then overlaid with a 2% X-Gal solution. Transconjugants were qualitatively classified for the time required to produce a blue color. With this assay, wild-type control colonies turn dark blue within 20 min of the addition of the overlay, while mutant strains with increased β-galactosidase activity were readily observable as colonies that turned dark blue after 5, 10, or 15 min of incubation.
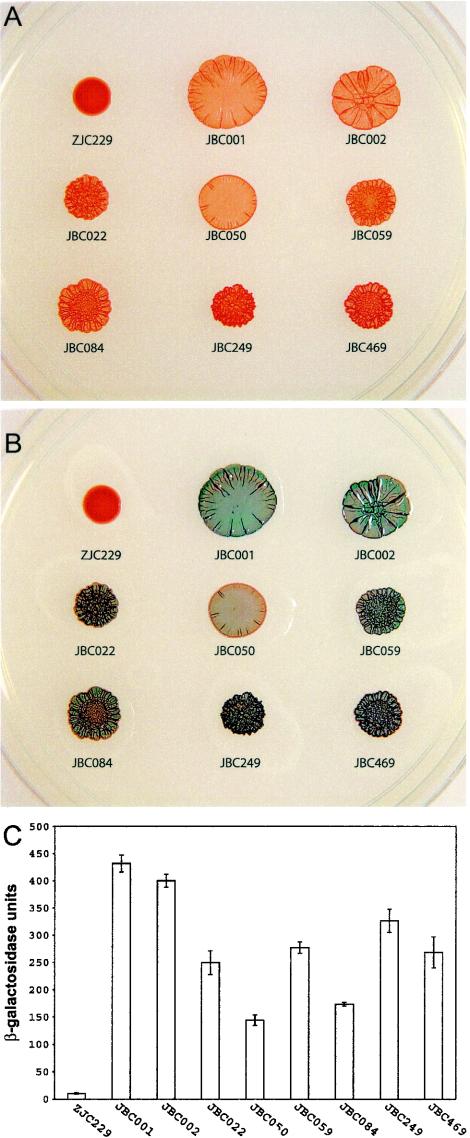
By this method, 150 mutants that exhibited elevated chsA::lacZ expression were independently isolated from a pool of over 10,000 transposition events. Surprisingly, all but three of these mutants also exhibited a ridged colony phenotype indicative of cyst cell production during, or soon after, the formation of visible colonies. As shown in Fig. 2A, hypercyst mutant strains exhibit a dramatic ridged colony morphology after incubation at 42°C for 72 h, which is contrasted by the smooth colony phenotype exhibited by the parent (ZJC229). After being overlaid with 2% X-Gal for 10 min, the hypercyst colonies rapidly develop a deep blue color indicative of the increased β-galactosidase activity of the chsA::lacZ reporter relative to the parent strain (Fig. 2B). To quantitate the level of chsA::lacZ expression in hypercyst cells, we performed β-galactosidase activity assays on cells that were harvested after 72 h of growth on CENS medium. As shown in Fig. 2C, the β-galactosidase activity of the hypercyst strains ranges from 14- to 42-fold higher than that of the parent cells, which confirms the results seen using X-Gal overlays on plates.
FIG. 2.
Colony morphology and csh::lacZ expression patterns of hypercyst mutants. Strains of R. centenum were grown aerobically in liquid CENS medium, harvested, washed, and pipetted onto agar-solidified CENS medium in 5-μl aliquots. (A) Distinct colony morphologies observed after 3 days of incubation at 42°C. (B) The same colonies 10 min after being overlaid with 2% X-Gal. The increased β-galactosidase activities are easily observed by the rapid production of a blue color in hypercyst colonies. (C) β-Galactosidase activities of the parent strain ZJC229 and hypercyst mutants after 3 days of growth on CENS medium. Error bars represent standard deviations derived from assays performed in triplicate with three cultures for each strain.
Analysis of hypercyst mutants.
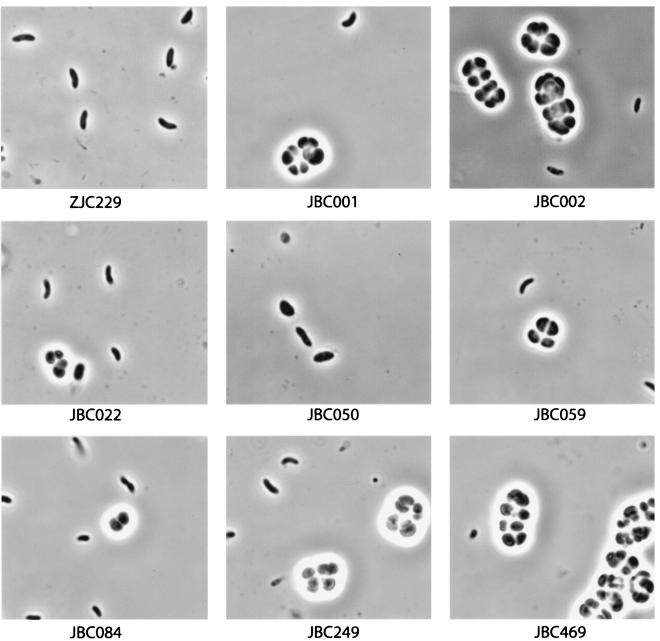
Hypercyst mutant colonies were examined microscopically by preparing wet mounts after 3 days of growth and checking for the presence of cysts with phase-contrast microscopy (Fig. 3). All of the hypercyst strains exhibited cells at various stages of cyst formation, which is contrasted by the parent cell line that shows no microscopically observable cysts. Strains JBC001, JBC002, JBC059, JBC249, and JBC469 exhibited large quantities of multicelled cyst clusters indicative of mature cysts. These strains also have the highest level of chsA::lacZ activity. Strains JBC022, JBC050, and JBC084 have only a moderate hypercyst phenotype, with the majority of the cells retaining motile vegetative cell characteristics and only a small percentage of cells being in the early stages of cyst formation (i.e., nonmotile oblong-shaped cells containing intracellular PHB granules). Not surprisingly, these cell lines also have lower chsA::lacZ expression than that observed with the other hypercyst mutants. Upon further incubation, strains JBC022, JBC050, and JBC084 also develop mature cyst cells much more quickly than the wild type. Thus, these mutants are not locked in one particular stage of cyst formation; rather, these mutants are defective in the timing of cyst cell differentiation.
FIG. 3.
Phase-contrast microscopy of wet mounts of ZJC229 and hypercyst strains after 3 days of growth on nutrient-rich, agar-solidified CENS medium. The mutant strains display various stages of cyst cell development, whereas the wild-type (ZJC229) cells remain vegetative.
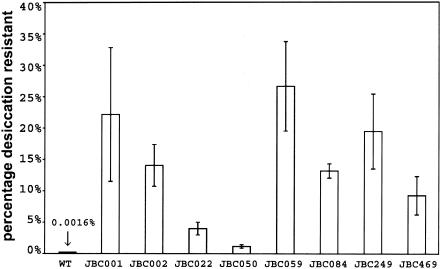
Microscopic quantitation of the percentage of cyst cells in a colony is problematic due to the presence of large multicelled clusters that tend to aggregate. Consequently, we used an assay that measures the number of cyst cells by resistance to desiccation (1) to quantitate the levels of cyst formation in these strains (Fig. 4). Under nutrient-rich growth conditions, only a fraction of the cells from the parental strain (0.0016% ± 0.0002%) survive 3 days of desiccation. Survivability is greatly increased in the hypercyst mutant strains, ranging from 1.2% of the total viable cell count in JBC050 to 26.6% in JBC059. This observation indicates a 750- to 16,000-fold increase in cyst formation in these strains. Indeed, the upper range of cyst formation in these mutants is comparable to the level of cyst formation observed with wild-type cells that have been induced to form cysts by growth on butyrate (21.2% ± 3.8%). The level of cyst formation in the hypercyst mutants also correlates well with the level of chsA activity; specifically, strains with higher levels of chs activity exhibit greater derepression of cyst formation. Additionally, this assay reveals two important traits of hypercyst mutant strains: (i) that the cyst cells observed microscopically are in fact mature, indicating that the process of cyst formation in the mutants is similar to that in the wild type, and (ii) that these strains are capable of germination and vegetative outgrowth after 3 days of desiccation, indicating that the cyst cells formed by these strains are not trapped in a cyst cell state. Although outgrowth is markedly slower than in the wild type, these strains can form isolated colonies which attain the same ratio of cyst/vegetative cells observed prior to desiccation (J. E. Berleman and C. E. Bauer, unpublished data).
FIG. 4.
Desiccation resistance of wild-type R. centenum (ZJC229) and hypercyst mutants after 3 days of growth on nutrient rich, agar-solidified CENS medium. Error bars represent standard deviations derived from assays performed with three cultures for each strain.
Identification of transposon-disrupted genes.
To identify the mini-Tn5-disrupted loci in hypercyst mutants, we cloned Tn5-disrupted genes and then sequenced out from the transposon by primer walking. Several of the sequenced mutants had disruptions in metabolism-related genes (Table 1). In this case, the hypercyst phenotype is likely due to an inability to consume available nutrients. Four independent insertions (in strains JBC050, JBC082, JBC119, and JBC265) mapped to the same pyruvate phosphate dikinase gene, ppdK. Pyruvate phosphate dikinase catalyzes the conversion of pyruvate to phosphoenolpyruvate, hydrolyzing an ATP in the process. Pyruvate is a major carbon source in CENS medium, so it is likely that these disruptions cause a defect in pyruvate metabolism, which leads to carbon starvation and subsequent induction of cyst formation. Indeed, growth of these strains on carbon sources other than pyruvate does not result in a hypercyst phenotype. Strain JBC457 carries a transposon disruption of the gene encoding dihydroorotate dehydrogenase, an enzyme involved in pyrimidine biosynthesis. Thus, the status of the nucleotide pool may be another signal leading up to encystment. Strains JBC493 and JBC503 contain disruptions of the gene encoding apolipoprotein n-acyltransferase, which is an enzyme involved in modifying outer membrane proteins. In M. xanthus, environmental factors that disrupt the outer membrane, such as glycerol, can stimulate myxospore formation (20); thus, it may be that integrity of the outer membrane is also required to repress cyst formation in R. centenum. Strain JBC290 has a disruption of a gene encoding an ABC transporter, and strains JBC249, JBC278, and JBC452 have disruptions in genes with no known homologs. These results indicate that there are potentially several metabolic pathways which are essential for maintaining the vegetative cell state and that, when disrupted, result in constitutive cyst cell formation.
TABLE 1.
Genes disrupted by mini-Tn5 mutagenesis of R. centenum that give rise to a hypercyst phenotype
| Strain | Gene name | Description or gene product |
|---|---|---|
| Regulatory class | ||
| JBC001 | cstS1 | Sensor kinase-response regulator hybrid |
| JBC188 | cstS1 | Sensor kinase-response regulator hybrid |
| JBC084 | cstS2 | Sensor kinase |
| JBC002 | cstS3 | Response regulator-sensor kinase hybrid |
| JBC478 | cheA3 | CheA-like sensor kinase |
| JBC174 | cheY2 | Response regulator |
| JBC067 | cheB2 | CheB-like methylesterase 1 |
| JBC469 | cheB2 | CheB-like methylesterase 1 |
| JBC059 | cheB3 | CheB-like methylesterase 2 |
| JBC022 | mcp3 | Chemoreceptor |
| Metabolic class | ||
| JBC050 | ppdK | Pyruvate phosphate dikinase |
| JBC082 | ppdK | Pyruvate phosphate dikinase |
| JBC119 | ppdK | Pyruvate phosphate dikinase |
| JBC265 | ppdK | Pyruvate phosphate dikinase |
| JBC290 | abcA | ABC transporter |
| JBC457 | pyrD | Dihydroorotate dehydrogenase |
| JBC493 | cutE | Apolipoprotein n-acyltransferase |
| JBC503 | cutE | Apolipoprotein n-acyltransferase |
| Novel class | ||
| JBC249 | No homolog | |
| JBC278 | No homolog | |
| JBC452 | No homolog |
We also identified 10 independent insertions in eight genes with products that contain homology to regulatory or signal transduction elements. Four genes that code for proteins with homology to histidine kinases have been identified (in strains JBC001, JBC188, JBC084, JBC002, and JBC478). Interestingly, all of these kinases are predicted to be cytosolic proteins based on a Kyte and Doolittle (14) hydrophobicity profile, suggesting that intracellular signals may be sensed by these components (Berleman and Bauer, unpublished). Strains JBC001 and JBC188 have disruptions in a gene predicted to encode a sensor kinase-response regulator hybrid with conserved GAF, PAS, and PAC (6) domains at the N-terminal sensing domain. JBC084 contains a disruption in a gene coding for sensor kinase that also has a PAS domain. JBC002 disrupts a gene predicted to encode an unusual hybrid protein with an N-terminal response regulator domain and a C-terminal histidine kinase domain. Sequencing upstream and downstream of these disrupted sensor kinase genes has not revealed the presence of genetically linked response regulators that contain identifiable DNA-binding domains. Additionally, the mini-Tn5 disruptions in strains JBC001 (cstS1), JBC188 (cstS1), JBC084 (cstS2), and JBC002 (cstS3) as well as the four independent insertions in ppdK are not likely to have a polar effect on the transcription of downstream genes. In each of these cases, the gene immediately downstream is located on the cDNA strand and thus should be transcribed unobtrusively despite the presence of a transposon in a neighboring gene.
Interestingly, strain JBC478 contains a disruption of a gene which codes for a CheA-like histidine kinase, and strain JBC174 has a disruption of a gene that codes for a CheY-like receiver domain-only protein. We have also identified two CheB-like response regulators that have an N-terminal receiver domain and a C-terminal methylesterase domain (in strains JBC067, JBC469, and JBC059). Additionally, strain JBC022 has a disruption of a gene with homology to methyl-accepting chemotaxis proteins. Thus, these hypercyst strains have disruptions of components necessary to form a nearly complete che-like signal transduction cascade. Since che-like genes are frequently organized as operons, it is possible that some of these disruptions are polar. Interestingly, none of these disruptions map to the che locus that was previously shown to control chemotaxis and phototaxis in this organism (10).
DISCUSSION
This study demonstrates that mutants can readily be obtained that derepress induction of cyst formation in R. centenum. Several of the isolated mutants form cysts on rich growth medium at a remarkable 26.6% of the total viable cell count. Microscopic analysis and desiccation resistance assays determined that these strains are forming mature cysts, indicating that once initiated, the process of cyst formation occurs normally in these strains. Since cysts are resting cells, it may appear paradoxical that these hypercyst mutants are capable of growth. However, it has previously been noted that a single R. centenum vegetative cell is capable of forming a cyst cluster containing >4 cells per cluster (1), indicating that division is possible even after the commitment to cyst cell differentiation has been made. Furthermore, cyst cells isolated from hypercyst strains are capable of rapid germination on nutrient-rich medium. Consequently, even though these hypercyst mutants rapidly form cysts under vegetative conditions, the cysts that are formed are not locked in a resting cell state that would render them inviable. Instead, these cells germinate and quickly reenter the cyst developmental pathway.
The number of genes that can be identified by using this screen is likely small (<25) based on the frequency of isolating hypercyst mutants and on the identification of multiple independent insertions in 5 of the 15 genes that were isolated in this study. Interestingly, eight of these genes appear to disrupt potential regulatory factors based on homology to sensor kinases, response regulators, and che signal transduction components. All of the identified sensor kinases are predicted to be cytosolic proteins, based on the absence of identifiable membrane spanning domains. This possibility indicates that repression of cyst formation may respond to intracellular levels of metabolic intermediates rather than to extracellular signals from the environment. All of the sensor kinase mutants are also capable of effectively growing on minimal growth medium (Berleman and Bauer, unpublished), so it seems unlikely that the identified sensor kinases are simply controlling induction or repression of metabolic pathways that secondarily results in the induction of cyst formation. Rather, we suspect that formation of cysts involves multiple input signals not unlike that of sporulation in B. subtilis, which utilizes five known sensor kinases to control induction of spore formation (8).
Another intriguing aspect to the genes identified is the number of che-like homologs that were disrupted in this study. Specifically, hypercyst mutants were obtained that contained disruptions in genes which code for two different CheB homologs, a CheY homolog, a CheA homolog, and a methyl-accepting chemoreceptor. Interestingly, these Che-like proteins, which are involved in cyst formation, are not the same as those encoded by the previously identified che operon that controls chemotaxis and phototaxis in this species (10). Further analysis of hypercyst-related che loci is ongoing as part of a more detailed study of the roles of distinct che loci in R. centenum. Thus far we have obtained no evidence that the hypercyst che loci identified by this study are involved in chemotaxis or phototaxis (Berleman and Bauer, unpublished). We therefore suspect that cyst formation in R. centenum may involve a che-like signal transduction cascade similar to that reported to control myxospore formation in M. xanthus (12). In M. xanthus, it has been shown that disruption of various che-like loci also results in overproduction of myxospores under conditions where myxospore formation would normally be repressed. It has also been demonstrated that a CheA-like protein interacts with an NtrC-like DNA-binding protein in M. xanthus, suggesting that there may be direct control of gene expression by the Che-like components. Clearly, additional studies need to be undertaken with the R. centenum Che-like hypercyst components to determine if these gene products are functioning in a similar manner. If so, then there would be an intriguing similarity between the mechanisms of the induction of cyst formation in R. centenum and the induction of myxospore formation in M. xanthus.
Finally, the screen described in this study can easily be modified to isolate mutants that have decreased instead of increased chsA expression. Indeed, such a screen has been used to successfully isolate numerous mutants that have delayed or inhibited ability to form cysts (Berleman and Bauer, unpublished). The mutants identified by such a screen appear to represent a much larger set of genes that includes those that are involved in various stages of mature cyst formation. Thus, the use of chsA gene expression as a reporter for the formation of cysts appears to function well for the isolation of mutants involved in the regulation of cyst induction as well as mutants that are defective in synthesizing structural components of the developing cyst cell. The challenge in the future will be to characterize both classes of mutants as well as to obtain an understanding of their specific roles in cyst development and survivability with regard to environmental stresses.
Acknowledgments
J.E.B. was supported by National Institutes of Health training grant GM007757.
REFERENCES
- 1.Berleman, J. E., and C. E. Bauer. 2004. Characterization of cyst cell formation in the purple photosynthetic bacterium Rhodospirillum centenum. Microbiology 150:383-390. [DOI] [PubMed] [Google Scholar]
- 2.Burkholder, W. F., and A. D. Grossman. 2000. Regulation of the initiation of endospore formation in Bacillus subtilis, p. 151-166. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 3.Campos, M., J. M. Martínez-Salazar, L. Lloret, S. Moreno, C. Núñez, G. Espín, and G. Soberón-Chávez. 1996. Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J. Bacteriol. 178:1793-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castañeda, M., J. Sánchez, S. Moreno, C. Núñez, and G. Espín. 2001. The global regulators GacA and σS form part of a cascade that controls alginate production in Azotobacter vinelandii. J. Bacteriol. 183:6787-6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favinger, J., R. Stadtwald, and H. Gest. 1989. Rhodospirillum centenum, sp. nov., a thermotolerant cyst-forming anoxygenic photosynthetic bacterium. Antonie Leeuwenhoek 55:291-296. [DOI] [PubMed] [Google Scholar]
- 6.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 7.Iwashina, T. 2003. Flavonoid function and activity to plants and other organisms. Biol. Sci. Space 17:24-44. [DOI] [PubMed] [Google Scholar]
- 8.Jiang, M., W. Shao, M. Perego, and J. A. Hoch. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38:535-542. [DOI] [PubMed] [Google Scholar]
- 9.Jiang, Z., and C. E. Bauer. 1997. Analysis of a chemotaxis operon from Rhodospirillum centenum. J. Bacteriol. 179:5712-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang, Z.-Y., H. Gest, and C. E. Bauer. 1997. Chemosensory and photosensory perception in purple photosynthetic bacteria utilize common signal transduction components. J. Bacteriol. 179:5720-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang, Z.-Y., B. G. Rushing, Y. Bai, H. Gest, and C. E. Bauer. 1998. Isolation of Rhodospirillum centenum mutants defective in phototactic colony motility by transposon mutagenesis. J. Bacteriol. 180:1248-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirby, J. R., and D. R. Zusman. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 100:2008-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubasek, W. L., F. M. Ausubel, and B. W. Shirley. 1998. A light-independent developmental mechanism potentiates flavonoid gene expression in Arabidopsis seedlings. Plant Mol. Biol. 37:217-223. [DOI] [PubMed] [Google Scholar]
- 14.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 15.Lorenzo, C., E. Howard, and R. Nagel. 1990. Studies on Tn10 transposition and excision in DNA-repair mutants of Salmonella typhimurium. Mutat. Res. 232:99-104. [DOI] [PubMed] [Google Scholar]
- 16.Martin, C. R. 1993. Structure, function, and regulation of the chalcone synthase. Int. Rev. Cytol. 147:233-284. [DOI] [PubMed] [Google Scholar]
- 17.Moore, B. S., C. Hertweck, J. N. Hopke, M. Izumikawa, J. A. Kalaitzis, G. Nilsen, T. O'Hare, J. Piel, P. R. Shipley, L. Xiang, M. B. Austin, and J. P. Noel. 2002. Plant-like biosynthetic pathways in bacteria: from benzoic acid to chalcone. J. Nat. Prod. 65:1956-1962. [DOI] [PubMed] [Google Scholar]
- 18.Moreno, S., R. Nájera, J. Guzmán, G. Soberón-Chávez, and G. Espín. 1998. Role of alternative σ factor AlgU in encystment of Azotobacter vinelandii. J. Bacteriol. 180:2766-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Núñez, C., S. Moreno, G. Soberón-Chávez, and G. Espín. 1999. The Azotobacter vinelandii response regulator AlgR is essential for cyst formation. J. Bacteriol. 181:141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Connor, K. A., and D. R. Zusman. 1997. Starvation-independent sporulation in Myxococcus xanthus involves the pathway for beta-lactamase induction and provides a mechanism for competitive cell survival. Mol. Microbiol. 24:839-850. [DOI] [PubMed] [Google Scholar]
- 21.Pope, L. M., and O. Wyss. 1970. Outer layers of the Azotobacter vinelandii cyst. J. Bacteriol. 102:234-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadasivan, L., and C. A. Neyra. 1987. Cyst production and brown pigment formation in aging cultures of Azospirillum brasilense ATCC 29145. J. Bacteriol. 169:1670-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadoff, H. L. 1975. Encystment and germination in Azotobacter vinelandii. Bacteriol. Rev. 39:516-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., and P. MacCallum. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Shimkets, L. J. 2000. Growth, sporulation, and other tough decisions, p. 277-284. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 26.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vitro genetic engineering: transposition mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 27.Stadtwald-Demchick, R., F. R. Turner, and H. Gest. 1990. Physiological properties of the thermotolerant photosynthetic bacterium, Rhodospirillum centenum. FEMS Microbiol. Lett. 67:139-144. [Google Scholar]
- 28.Swem, L. R., B. J. Kraft, D. L. Swem, A. T. Setterdahl, S. Masuda, D. B. Knaff, J. M. Zaleski, and C. E. Bauer. 2003. Signal transduction by the global regulator RegB is mediated by a redox-active cysteine. EMBO J. 22:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young, D.A., C. E. Bauer, J. C. Williams, and B. L. Marrs. 1989. Genetic evidence for superoperonal organization of genes for photosynthetic pigments and pigment-binding proteins in Rhodobacter capsulatus. Mol. Gen. Genet. 218:1-12. [DOI] [PubMed] [Google Scholar]