Abstract
Fructose-1,6-bisphosphatase (FBPase) is one of the key enzymes in gluconeogenesis. Although FBPase activity has been detected in several hyperthermophiles, no orthologs corresponding to the classical FBPases from bacteria and eukaryotes have been identified in their genomes. An inositol monophosphatase (IMPase) from Methanococcus jannaschii which displayed both FBPase and IMPase activities and a structurally novel FBPase (FbpTk) from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 have been proposed as the “missing” FBPase. For this study, using T. kodakaraensis, we took a genetic approach to elucidate which candidate is the major gluconeogenic enzyme in vivo. The IMPase/FBPase ortholog in T. kodakaraensis, ImpTk, was confirmed to possess high FBPase activity along with IMPase activity, as in the case of other orthologs. We therefore constructed Δfbp and Δimp strains by applying a gene disruption system recently developed for T. kodakaraensis and investigated their phenotypes. The Δfbp strain could not grow under gluconeogenic conditions while glycolytic growth was unimpaired, and the disruption resulted in the complete abolishment of intracellular FBPase activity. Evidently, fbpTk is an indispensable gene for gluconeogenesis and is responsible for almost all intracellular FBPase activity. In contrast, the endogenous impTk gene could not complement the defect of the fbp deletion, and its disruption did not lead to any detectable phenotypic changes under the conditions examined. These facts indicated that impTk is irrelevant to gluconeogenesis, despite the high FBPase activity of its protein product, probably due to insufficient transcription. Our results provide strong evidence that the true FBPase for gluconeogenesis in T. kodakaraensis is the FbpTk ortholog, not the IMPase/FBPase ortholog.
Fructose-1,6-bisphosphatase (FBPase; EC 3.1.3.11), which catalyzes the hydrolysis of d-fructose-1,6-bisphosphate (FBP) to d-fructose-6-phosphate (F6P) and inorganic phosphate (Pi), is a well-known key enzyme of gluconeogenesis. Along with phosphofructokinase (EC 2.7.1.11), which catalyzes the reverse reaction, the phosphorylation of F6P during glycolysis, the unidirectional FBPase regulates the flux of sugar metabolism. Therefore, FBPases have been identified and characterized from a wide variety of bacteria and eukaryotes. In most cases, the activity of FBPase is tightly controlled through various mechanisms in order to alleviate a futile cycle, which occurs by the simultaneous functioning of both FBPase and phosphofructokinase. For the yeast Saccharomyces cerevisiae, regulation at the transcriptional level (catabolite repression) (13, 39), reversible, short-term inactivation by protein modification (21), and proteolytic degradation (catabolite degradation) (13, 30, 37) have been reported. In addition, the majority of FBPases are allosterically regulated by AMP (9, 12, 18, 31, 44) and inhibited by fructose-2,6-bisphosphate (14, 18, 28).
Bacterial FBPases have been classified into three classes based on their primary structures, and eucaryal FBPases are homologous to bacterial class I enzymes. In Escherichia coli, the class I FBPase encoded by fbp has been demonstrated to fulfill the major role in gluconeogenesis (6, 7, 38), while GlpX, a class II FBPase, is not essential for gluconeogenic growth (5). In contrast, a class II FBPase in Corynebacterium glutamicum which is the sole FBPase in this bacterium has been confirmed to function in gluconeogenesis (31). Bacillus subtilis possesses a highly distinct FBPase belonging to class III which is an essential enzyme for growth on gluconeogenic carbon sources when a bypass of the FBPase reaction (10) is blocked by a mutation in the bfd gene (11). Regulation at the transcriptional level which is dependent on carbon sources has been observed for FBPase I of E. coli (27), whereas FBPase II of C. glutamicum (31) and FBPase III of B. subtilis (11) appear to be constitutive enzymes.
Recent complete genome analyses and subsequent comparative genomics have enabled us to discuss the distribution and diversity of particular genes in a variety of organisms. With respect to archaeal and bacterial (hyper)thermophiles, one intriguing finding is the absence of obvious orthologs for known FBPases in these genomes, despite the presence of the other genes involved in gluconeogenesis. Nevertheless, various lineages of (hyper)thermophiles can grow on gluconeogenic substrates, and FBPase activities have actually been detected in several strains (8, 34). These facts suggested the presence of an unknown class of FBPases in (hyper)thermophiles. This had seemed to be resolved by an unexpected finding based on protein structure. Stec et al. found that the three-dimensional architecture of inositol monophosphatase (IMPase; EC 3.1.3.25) from a hyperthermophilic archaeon, Methanococcus jannaschii (the MJ0109 product), was very similar to that of FBPase from higher eukaryotes, and indeed the MJ0109 product exhibited the dual activities of an IMPase and an FBPase (41). Moreover, the orthologs of MJ0109 from other hyperthermophiles, specifically Archaeoglobus fulgidus, Thermotoga maritima (41), and Pyrococcus furiosus (42), have also been demonstrated to possess both activities. From these catalytic properties, the MJ0109 orthologs have been considered to act as the gluconeogenic FBPase in (hyper)thermophiles and have been classified as class IV FBPases (42).
On the other hand, by purification of a protein responsible for the intracellular FBPase activity, members of our laboratory recently identified a novel candidate for the true FBPase in the sulfur-reducing hyperthermophilic euryarchaeon “Thermococcus kodakaraensis” KOD1 (1, 26, 29). The gene of the identified FBPase was designated fbpTk, and the recombinant protein actually displayed FBPase activity with a strict substrate specificity for FBP, unlike IMPase/FBPase IV orthologs. Furthermore, transcription of the gene in T. kodakaraensis cells was strongly repressed under glycolytic growth conditions with starch. These catalytic and transcriptional properties agreed well with a gluconeogenic function, although AMP did not show inhibitory effects on the activity. The primary structure of FbpTk is quite different from those of previously reported FBPases, including IMPase/FBPase IV, but shares significant homologies with hypothetical proteins that are highly conserved in (hyper)thermophiles. These facts have raised the possibility that FbpTk orthologs may be the bona fide FBPases in organisms grown under high-temperature conditions. However, to date, it still remains to be clarified which candidate, IMPase/FBPase IV, the FbpTk ortholog, or both, plays the major gluconeogenic role in (hyper)thermophiles.
For this study, we aimed to solve this question by using T. kodakaraensis, as the recently determined whole-genome sequence of strain KOD1 contains no ortholog for the classical FBPases but harbors a gene for IMPase/FBPase IV (designated impTk) along with fbpTk, as is the case for many other (hyper)thermophile genomes. There is a potent advantage in the use of T. kodakaraensis for analyses of gene function in vivo, as we have recently constructed a targeted gene disruption system for this organism (33) which is the first to be described for hyperthermophiles. In this study, we applied the gene disruption system to clarify the respective participation of impTk and fbpTk in gluconeogenesis in T. kodakaraensis. The results obtained here provide direct evidence that enables us to conclude that FbpTk, not ImpTk, is the true missing FBPase in the hyperthermophilic archaeon.
MATERIALS AND METHODS
Strains and growth conditions.
The strains and plasmids used for this study are listed in Table 1. T. kodakaraensis KOD1 and its derivatives were cultivated under strictly anaerobic conditions at 85°C in a rich growth medium (ASW-YT) or a synthetic medium (ASW-AA) (33). ASW-YT medium (pH 6.6) was composed of 0.8× artificial seawater (ASW), 5.0 g of yeast extract/liter, 5.0 g of tryptone/liter, and 2.0 g of elemental sulfur/liter. ASW-AA medium consisted of 0.8× ASW, 20 amino acids (total, 2,125 mg/liter) as carbon sources, modified Wolfe's trace minerals, a vitamin mixture (total, 0.44 mg/liter), and 2.0 g of elemental sulfur/liter (the pH was adjusted to 6.9 with NaOH). The growth properties of Δfbp and Δimp strains of T. kodakaraensis were investigated in ASW-AA medium containing 5.0 g of soluble starch (Nacalai Tesque, Kyoto, Japan)/liter or 5.0 g of sodium pyruvate (Nacalai Tesque)/liter after preculture in ASW-YT medium. The preparation of plate medium and cultivation of the cells on it were performed as described previously (33).
TABLE 1.
Strains and plasmids used for this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Stratagene (La Jolla, Calif.) |
| BL21-CodonPlus(DE3)-RIL | E. coli B F−ompT hsdS(rB− mB−) dcm+ Tetrgal λ(DE3) endA Hte (argU ileY leuW Camr) | Stratagene |
| T. kodakaraensis | ||
| KOD1 | Wild type | 1, 26 |
| KW128 | ΔpyrF ΔtrpE::pyrF | Unpublished data |
| Δfbp-8J | KW128 Δfbp::trpE | This study |
| Δimp-2A | KW128 Δimp::trpE | This study |
| Plasmids | ||
| pUC118 | Ampr general cloning vector | Takara (Kyoto, Japan) |
| pET-21a(+) | Ampr general expression vector | Novagen (Madison, Wis.) |
| pET-fbp | pET-21a(+) derivative; fbp | 29 |
| pET-imp | pET-21a(+) derivative; imp | This study |
| pUD | pUC118 derivative; pyrF marker cassette (PpyrF::pyrF) | 33 |
| pUMT2 | pUC118 derivative; trpE marker cassette (PpyrF::trpE) | This study |
| pUDFbp | pUC118 derivative; Δfbp::trpE | This study |
| pUDImp | pUC118 derivative; Δimp::trpE | This study |
E. coli strains DH5α and BL21-CodonPlus(DE3)-RIL, used for general DNA manipulation and heterologous gene expression, respectively, were routinely cultivated at 37°C in Luria-Bertani (LB) medium (32).
Overexpression of impTk and fbpTk genes in E. coli and purification of recombinant enzymes.
The expression vector for impTk was constructed as follows. A DNA fragment containing the imp-coding region (771 bp) was amplified from the genomic DNA of T. kodakaraensis KOD1 by a PCR using primers EIMP-R and EIMP-F (5′-GGGGTGATCATATGGAGTTTAACTGGAGTGAG-3′ and 5′-GCTCAGGGAATTCCCGCCGCTCAAAACTG-3′ [the underlined sequences indicate NdeI and EcoRI sites, respectively]). The amplified DNA fragment (885 bp) was inserted into pUC118 at the HincII site. After confirmation of the absence of unintended mutations in the sequence, the NdeI-EcoRI restriction fragment was inserted into the pET-21a(+) expression vector (Novagen) at the corresponding sites. E. coli strain BL21-CodonPlus(DE3)-RIL was transformed with the resulting plasmid, pET-imp (6,262 bp). The transformant was cultivated at 37°C in LB medium containing 100 μg of ampicillin/ml until the cell density (optical density at 660 nm [OD660]) reached about 0.5. The culture was then supplemented with 0.1 mM (final concentration) isopropyl-1-thio-β-d-galactopyranoside to induce overexpression and was incubated for a further 14 h at 17°C.
The cells were harvested by centrifugation, resuspended in 100 mM Tris-HCl buffer (pH 8.0), and then sonicated. After centrifugation (8,000 × g, 30 min), the soluble cell extract was heat treated for 30 min at 80°C to remove thermolabile proteins derived from the host, followed by centrifugation (8,000 × g, 30 min). The supernatant was applied to a Resource Q anion exchange column (6 ml) (Amersham Biosciences, Buckinghamshire, United Kingdom), and the recombinant protein was eluted with a linear gradient of NaCl (0 to 1.0 M) in 50 mM Tris-HCl (pH 8.0) with a flow rate of 2.0 ml/min. The resulting fractions were combined and concentrated by use of Centricon YM-30 columns (Millipore, Bedford, Mass.) and then were further purified through a Superdex 200 HR 10/30 gel filtration column (Amersham Biosciences) with a mobile phase of 50 mM Tris-HCl (pH 8.0) containing 0.15 M NaCl at a flow rate of 0.6 ml/min. The molecular mass of native ImpTk was determined from a calibration curve constructed with the standard proteins ferritin (440 kDa), catalase (232 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and RNase A (13.7 kDa). The protein concentration was determined with the Bio-Rad (Hercules, Calif.) protein assay system, with bovine serum albumin as a standard.
Overexpression of fbpTk in E. coli and purification of the recombinant protein were performed as described previously (29).
IMPase and FBPase assay.
The IMPase activities of the recombinant proteins were determined by measuring the release of free Pi from d-myo-inositol-1-monophosphate (IMP) (Sigma, St. Louis, Mo.) by the Malachite Green procedure (17). The reaction was performed in a 100-μl mixture containing 50 mM Tris-HCl (pH 8.0), 20 mM MgCl2, 2 mM IMP, and protein solution. The assay mixture without the substrate was preincubated at 85°C for 3 min, and the reaction was started by the addition of IMP. After 1 min at the same temperature, the reaction was stopped by rapid cooling on ice for 5 min. Next, 20 μl of 0.2% Tween 20 and 500 μl of Malachite Green-ammonium molybdate solution (5 ml of 35% HCl, 562 mg of (NH4)6Mo7 · 4H2O [Wako Pure Chemicals, Osaka, Japan], and 75 mg of Malachite Green [Sigma] per 50 ml) were added to a portion of the reaction mixture (95 μl). The increase in liberated Pi was determined by measuring the increase in absorbance at 620 nm derived from the formation of a complex between molybdophosphoric acid and Malachite Green. The spontaneous increase in released Pi due to the thermal decomposition of IMP was subtracted from the datum of each experiment. To determine the activities in cell extracts from T. kodakaraensis strains, we cultivated cells in ASW-YT medium supplemented with 5.0 g of soluble starch/liter or 5.0 g of sodium pyruvate/liter at 85°C for 15 h and prepared cell extracts from cells in early stationary phase as described previously (33). IMPase activity in the extracts was measured by the Malachite Green procedure as described above, with modifications in the concentration of IMP (4 mM) and the reaction time (15 min). Alternatively, NAD-dependent inositol dehydrogenase was applied as a specific coupling enzyme (19). In this discontinuous assay, the first reaction with the extracts was performed at 85°C as described above. The second reaction, containing the coupling enzyme, was carried out in the presence of 200 mM NaCl, 0.2 U of inositol dehydrogenase (Sigma), and 4 mM NAD+ in the reaction mixture. After 5 min at 25°C, the amount of NADH was determined by measuring the increase in the absorbance at 340 nm.
FBPase activities of the recombinant proteins and those of cell extracts were measured by a spectrophotometric assay coupled with glucose-6-phosphate isomerase and NADP-dependent glucose-6-phosphate dehydrogenase (29). NADPH was quantified by measuring the increase in the absorbance at 340 nm. An experiment without FBP was used as a blank value, and the thermal decomposition of FBP was also considered in each measurement. One unit of activity was defined as 1 μmol of product produced per min for both assays.
Construction of disruption vectors pUDImp and pUDFbp.
Two disruption vectors, pUDImp and pUDFbp, were constructed for the targeted disruption of imp and fbp genes, respectively, in T. kodakaraensis by homologous double-crossover recombination. First, we prepared a trpE marker cassette in which trpE is oriented downstream of a putative promoter region for pyrF. The trpE coding region was amplified from the genomic DNA of T. kodakaraensis KOD1 by the use of primers MTRP-R and MTRP-F (5′-GGGCATATGCCTCTCAAAAAGCTGAAGCCCGTTGAC-3′ and 5′-GGGGGATCCTCATTCCCTCACCCCCAGCGCCTTCAGA-3′ [the underlined sequences indicate NdeI and BamHI sites, respectively]). A fragment digested with NdeI and BamHI was inserted into pUD harboring the pyrF marker cassette (33) at the corresponding sites to replace the coding region of pyrF with trpE. The pyrF promoter-trpE fusion was again amplified from the resulting plasmid to introduce PvuII sites at both ends with primers PJPRO-R2 and PJTRP-F2 (5′-CAGCTGCCGCAACGCGCATTTTGCTC-3′ and 5′-CAGCTGTCATTCCCTCACCCCCAGCGCCTTCAGA-3′ [the underlined sequences indicate PvuII sites]), followed by insertion into pUC118 at the HincII site. The resulting plasmid harboring the trpE marker cassette was designated pUMT2 (4,591 bp).
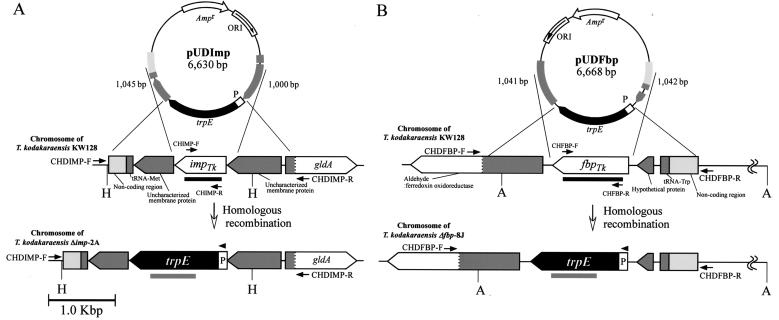
For construction of the two disruption vectors, pUDImp and pUDFbp, two DNA fragments of imp and fbp along with their flanking regions (about 1,000 bp) for homologous recombination were amplified from the genomic DNA of T. kodakaraensis KOD1 by the use of primers PIMP-R and PIMP-F for pUDImp (5′-TAACGAGTGCCTTTCCAGTAAG-3′ and 5′-AGTGCTCCCTTTCTTTTGACTTTC-3′) and PFBP-R and PFBP-F for pUDFbp (5′-CTCTTGATAGACGGGCAGAGAAGTGTGG-3′ and 5′-GCATCTGCCAGTTGAGAATCGGGACGAAGTCGCCC-3′). Each fragment was subcloned into pUC118 at the HincII site. DNA fragments including the homologous regions and the entire pUC118 plasmid, but not the target gene coding regions, were amplified with primers PDDIMP-R and PDDIMP-F for pUDImp (5′-CTGGAGGCGATGAAGGATGAGCC-3′ and 5′-AATATCACCCCAGCAAGGCTAT-3′) and PDDFBP-R and PDDFBP-F for pUDFbp (5′-TTTCCAGCCCTTTTCTGTTCATTTTACCC-3′ and 5′-GGCAACCACCGGTATTTTTAACCTCT-3′). The trpE marker cassette, excised from pUMT2 by PvuII digestion (1,423 bp), was ligated with each resulting fragment to give pUDImp (6,630 bp) and pUDFbp (6,668 bp) (Fig. 1). In these plasmids, the trpE marker cassette replaced the entire coding region of fbp (1,128 bp) and most of imp (a 27-bp sequence at the 3′ terminus of imp was retained in order to not disturb a downstream open reading frame and its putative ribosome-binding sequence overlapping with imp), with the same orientation as the original genes.
FIG. 1.
Schematic drawing of pUDImp (A) and pUDFbp (B) for disruption of imp and fbp in T. kodakaraensis KW128. The homologous regions between the circular DNAs and the chromosome of T. kodakaraensis are shaded. Restriction site abbreviations: A, ApaI; H, HindIII. The bold gray bar below the trpE gene indicates the region spanned by the trpE probe for Southern blot analyses. The bold black bars below the imp and fbp genes indicate the regions spanned by the imp and fbp probes used for Northern blot analyses.
Transformation of T. kodakaraensis.
T. kodakaraensis strain KW128 (ΔpyrF ΔtrpE::pyrF), in which tryptophan auxotrophy can be complemented by the selectable marker trpE (unpublished results), was used for the targeted disruption of fbp or imp. The transformation of KW128 was performed as reported previously (33), with slight modifications. Approximately 4 × 108 cells at the late exponential phase were harvested, resuspended in 200 μl of 0.8× ASW, and kept on ice for 30 min. After treatment with DNA and a successive heat shock, the cells were incubated in 1.5 ml of ASW-YT medium at 85°C for 2 h. The cells that were harvested and resuspended in 200 μl of 0.8× ASW were then directly spread on a selective synthetic plate medium that lacked tryptophan (ASW-AAW−) (33), with a supplementation of 5.0 g of soluble starch/liter. After cultivation for 5 to 8 days at 85°C, the transformants grown on the plate medium were analyzed by colony PCR or PCR with the genomic DNA as a template and with primers CHDIMP-R and CHDIMP-F for Δimp candidates (5′-TCTCTACCAGCTATTTCCTTCGTTTTTGGG-3′ and 5′-AACGTCGCGCAGGAAACTTTTGGAAAAAGC-3′) and CHDFBP-R and CHDFBP-F for Δfbp candidates (5′-TTGAATGTCTTCTTGATGTTGGCCTGATGCGG-3′ and 5′-TCTTGATCCTCTCTTCTTTCGGGATGTAGG-3′) as primer sets that anneal outside of the homologous regions (Fig. 1A and B, respectively). Control experiments without any exogenous DNA gave no tryptophan prototrophs. The gene disruptant candidates were purified by repeated selection on ASW-AAW− plate medium with starch and then were further analyzed. In order to confirm the complete deletion of the respective target regions, primer sets that annealed within the target genes were applied (CHIMP-R and CHIMP-F for imp [5′-GGACTAACGTGAGCGGAGACGTAACAAAGT-3′ and 5′-ACTCCCTTCCCCTTTCGTCCGTTACTATTC-3′] and CHFBP-R and CHFBP-F for fbp [5′-AGGATGTTCTTTCAAAAGCAGTCGAAGATG-3′ and 5′-CGCATGTATTCGGTTATCTCAAGGGCCTTCTGGCGGG-3′]) (Fig. 1).
Hybridization analyses.
Southern blot analysis was performed with 5.0 μg of genomic DNA digested with HindIII for Δimp-2A and ApaI for Δfbp-8J. Total RNAs were isolated from cells of T. kodakaraensis at the early exponential phase by use of an RNeasy Midi kit (Qiagen, Hilden, Germany). For Northern blot analysis, 30 μg of RNA was applied. DNAs or RNAs were separated by agarose gel electrophoresis according to standard procedures (32) and were transferred onto positively charged nylon membranes (Roche Diagnostics, Mannheim, Germany) by vacuum blotting. The preparation of specific probes, hybridization, and signal detection were performed with a DIG-DNA labeling and detection kit (Roche Diagnostics) according to the instructions from the manufacturer. The primer sets used for the preparation of trpE, imp, and fbp probes (Fig. 1) were PROTRP-R plus PROTRP-F (5′-TCCATCATCGGGGGGAAGATCGAAGAGC-3′ and 5′-CGAACGCGTTTTTCCCCTCATCGAGTT-3′), CHIMP-R plus CHIMP-F, and CHFBP-R plus CHFBP-F, respectively. The Northern hybridization experiments were performed with a common RNA-blotted membrane to which the fbp probe was applied after stripping of the initial imp probe.
RESULTS
FBPase and IMPase activities of recombinant Fbp and Imp from T. kodakaraensis.
Along with fbpTk, previously identified as a gene for a novel candidate for the true FBPase in hyperthermophiles (29), T. kodakaraensis possesses a gene (impTk) corresponding to the IMPase/FBPase IV genes PF2014 from P. furiosus (53.8% identity at the protein level) (42), MJ0109 from M. jannaschii (48.8% identity), AF2372 from A. fulgidus (36.4% identity), and TM1415 from Thermotoga maritima (31.9% identity) (41). To compare the respective activities of FbpTk and ImpTk, we overexpressed each gene in E. coli and individually purified the recombinant proteins to apparent homogeneity, as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). The native molecular mass of ImpTk was determined to be 54 kDa by gel filtration column chromatography. According to the subunit molecular mass determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (33 kDa) and that deduced from the primary structure (28.0 kDa), a homodimeric subunit assembly was indicated for ImpTk, as in the cases of PF2014, MJ0109, and AF2372. FbpTk has been determined to be a homooctamer (29).
We then measured the phosphatase activities of the recombinant enzymes toward FBP and IMP at 85°C (Table 2). As reported for orthologs from other hyperthermophiles, ImpTk was a bifunctional enzyme exhibiting a high FBPase activity as well as IMPase activity. In fact, the FBPase activity (52.6 U/mg) was much higher than the IMPase activity (9.5 U/mg), as was observed for the ortholog from the closely related archaeon P. furiosus (42). In contrast, FbpTk has been reported to display strict substrate specificity for FBP (29), and it actually exhibited only negligible activity on IMP. Using an enzyme-coupled assay, we confirmed that both enzymes released the 1-phosphate group of FBP regioselectively to generate F6P. The FBPase activity of ImpTk followed Michaelis-Menten kinetics at 85°C, without homotropic allosteric properties or substrate inhibition. The specific activity of ImpTk towards FBP was 2.5-fold higher than that of FbpTk (18.9 U/mg). A kinetic analysis of the FBPase reaction of ImpTk indicated that the enzyme exhibited a higher affinity for the substrate and a larger turnover number than FbpTk previously examined at 95°C (29) (Table 2). Even at the lower assay temperature, ImpTk exhibited a higher catalytic efficiency in the FBPase reaction, with a kcat/Km value of 1,170 s−1 mM−1, which was >6.5-fold higher than that of FbpTk.
TABLE 2.
IMPase and FBPase activities and kinetic parameters of recombinant ImpTk and FbpTkd
| Protein | IMPase activityac (U/mg) | FBPase activitybc (U/mg) | Km for FBP (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|---|
| ImpTk | 9.5 ± 0.3 | 52.6 ± 0.3 | 0.02 ± 0.00 | 23.4 ± 0.4 | 1,170 |
| FbpTk | 0.05 ± 0.01 | 18.9 ± 0.7 | 0.1d | 17d | 170d |
IMPase activity was determined by measuring released Pi by the Malachite Green method with 2 mM IMP at 85°C.
FBPase activity was determined by a coupling assay with 2 mM FBP at 85°C.
Means and standard deviations were obtained from three independent experiments.
Previously determined at 95°C (29).
Construction of Δimp and Δfbp strains of T. kodakaraensis.
A direct method for examining the in vivo function of a particular gene is to analyze the phenotypic changes displayed in corresponding knockout strains. We have previously reported targeted gene disruption in T. kodakaraensis, which is expected to be a powerful tool for research on hyperthermophilic archaea. The gene targeting system was applied here to disrupt imp or fbp in order to clarify which gene is mainly responsible for gluconeogenesis in T. kodakaraensis. For this purpose, we adopted T. kodakaraensis strain KW128 (ΔpyrF ΔtrpE::pyrF) and trpE as a host strain and a selectable marker, respectively. The host strain, KW128, shows tryptophan auxotrophy due to the replacement of trpE with pyrF on the chromosome, and the exogenous trpE gene can be used as a marker that can complement the auxotrophy (unpublished results). Two disruption plasmids, pUDImp and pUDFbp, were constructed for the targeted disruption of impTk and fbpTk, respectively (Fig. 1). These vectors harbored the trpE marker cassette between the upstream and downstream flanking regions (about 1,000 bp) of the respective gene of interest. The host strain was individually transformed with each plasmid as described in Materials and Methods. The resulting transformants were isolated on a selective plate medium without tryptophan and with supplementation of starch. The transformation efficiencies were determined to be 120 and 213 transformants/μg of DNA/4 × 108 cells for the disruption of impTk and fbpTk, respectively. After a second round of selection, we isolated a candidate strain for each disruption and designated them the Δimp-2A and Δfbp-8J strains, respectively.
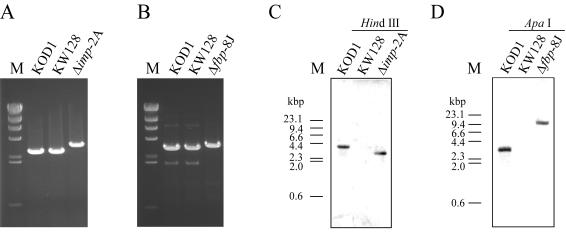
The genotypes of Δimp-2A and Δfbp-8J were confirmed by PCR, sequencing, and Southern blot analyses. PCR analyses of the Δimp-2A and Δfbp-8J strains with primer sets that annealed outside of the homologous regions (Fig. 1) resulted in the amplification of fragments corresponding to the loci of Δimp::trpE (3,499 bp) and Δfbp::trpE (3,575 bp), respectively, which were formed by double-crossover recombination (Fig. 2A and B). In both cases, no amplification was observed by PCRs using primer sets that annealed within the respective target genes (data not shown), indicating a complete deletion of the target genes and the absence of contaminant strains harboring the target genes. The replacement of imp and fbp with the trpE marker in the respective transformants was also confirmed by sequencing analysis of the targeted regions. Furthermore, we performed a Southern blot analysis with a trpE probe (Fig. 1). As shown in Fig. 2C and D, the wild-type KOD1 strain showed single signals deriving from the endogenous trpE gene in the trp operon, while no positive signal could be detected in the host strain KW128 with a ΔtrpE::pyrF genotype. For the Δimp-2A and Δfbp-8J strains, the signals could be detected with expected mobilities corresponding to a trpE insertion within the targeted regions (2.8 kbp for Δimp-2A [Fig. 2C] and 11.4 kbp for Δfbp-8J [Fig. 2D]). The absence of other signals confirmed the unique occurrence of the desired gene replacement without unintended nonhomologous recombination in both disruptants.
FIG. 2.
(A) Amplification of imp locus from T. kodakaraensis KOD1, KW128, and Δimp-2A, with CHDIMP-R and CHDIMP-F as primers. (B) Amplification of fbp locus from T. kodakaraensis KOD1, KW128, and Δfbp-8J, with CHDFBP-R and CHDFBP-F as primers. (C) Southern blot analysis using the trpE probe with genomic DNAs of KOD1, KW128, and Δimp-2A digested with HindIII. (D) Southern blot analysis using the trpE probe with genomic DNAs of KOD1, KW128, and Δfbp-8J digested with ApaI. The region corresponding to the trpE probe is indicated in Fig. 1. M, DNA size marker (HindIII-digested λ DNA).
Growth properties of the disruptants.
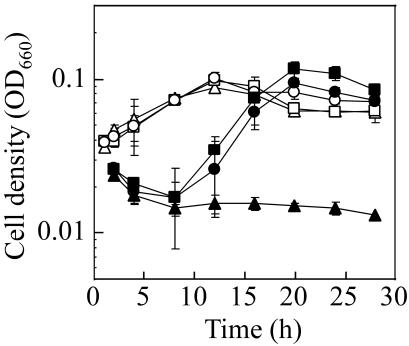
The host strain and the constructed disruptants were preincubated in an ASW-YT medium followed by cultivation in synthetic ASW-AA medium containing 20 amino acids supplemented with soluble starch (glycolytic conditions) or pyruvate (gluconeogenic conditions) to investigate the contribution of the impTk and/or fbpTk gene to gluconeogenesis. As shown in Fig. 3, all of the strains showed comparable growth on starch as a glycolytic substrate. On the other hand, under gluconeogenic conditions with pyruvate and 20 amino acids, the Δfbp-8J strain was not able to grow, in contrast to the unimpaired growth of KW128 and Δimp-2A. The same results were also obtained for cultivation in ASW-AA medium without the addition of pyruvate, in which the amino acids can be utilized as gluconeogenic substrates. PCR analyses confirmed that the gluconeogenic growth of Δimp-2A was not due to contamination by any imp+ strain, such as the host strain (data not shown). These results clearly demonstrate that fbp is an essential gene for the growth of T. kodakaraensis under gluconeogenic conditions, whereas imp is not only irrelevant to gluconeogenesis, but is also incapable of complementing the defect of fbp.
FIG. 3.
Growth properties of T. kodakaraensis KW128, Δimp-2A, and Δfbp-8J under glycolytic (open symbols) or gluconeogenic (closed symbols) conditions. The cells were cultured in ASW-AA medium supplemented with soluble starch or pyruvate at 85°C. Symbols: open circles, KW128 with starch; open squares, Δimp-2A with starch; open triangles, Δfbp-8J with starch; closed circles, KW128 with pyruvate; closed squares, Δimp-2A with pyruvate; closed triangles, Δfbp-8J with pyruvate. Error bars represent standard deviations for repeated independent experiments.
Enzyme assay in cell extracts from disruptants.
Cell extracts of the host strain and the two disruptants were prepared from cells grown in a nutrient-rich ASW-YT medium with starch or pyruvate. FBPase activity in the extracts was determined by a coupled assay. The host strain KW128 showed an FBPase activity of 0.37 ± 0.02 U/mg under gluconeogenic conditions, while the activity could not be observed under glycolytic conditions (<0.01 U/mg), probably due to the strict glucose repression of the gene, as reported previously (29). Likewise, there was no detectable FBPase activity in the two disruptants in the presence of starch. With respect to the cells grown on pyruvate, Δimp-2A cells exhibited FBPase activity (0.34 ± 0.01 U/mg) comparable to that in the host strain, whereas activity could not be detected in Δfbp-8J cells under the same conditions. These results indicate that most of the FBPase activity within the cells derives from the fbpTk product and also imply that no other candidate for FBPase is present in this organism. Furthermore, the results were also in agreement with the growth properties of each strain described above. Note that the Δfbp-8J strain, which did not show gluconeogenic growth in the synthetic medium, could grow in the rich ASW-YT medium without supplementation of starch. This is presumed to be due to the presence of some glycolytic substrates in yeast extract, a component of the medium, to an extent that they sustain the growth of strains that are deficient in gluconeogenesis. We further examined IMPase activity in the cell extracts by measuring the release of Pi from IMP by the Malachite Green method. We found that the levels of IMPase activity were below the detection limit (<0.01 U/mg) in all extracts, even those from strains KW128 and Δfbp-8J, which harbored the intact imp gene. With cell extracts, this method was hampered to some extent by the high levels of IMP-independent Pi that were present and produced during incubation at a high temperature. We therefore examined an alternative assay using NAD-dependent inositol dehydrogenase as a specific coupling enzyme. Even with this second method, the same results were obtained. The low levels of IMPase activity indicate that the contribution of ImpTk to the total amount of intracellular FBPase activity is negligible, or at most very limited, regardless of the high catalytic efficiency for the FBPase reaction observed for recombinant ImpTk.
Transcriptional analysis of impTk and fbpTk.
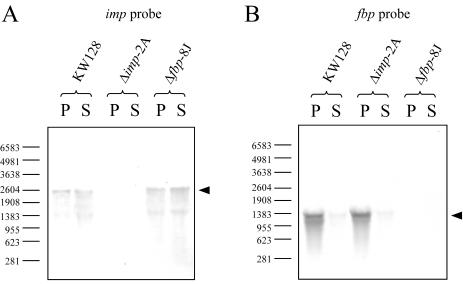
The transcriptional profiles of impTk and fbpTk were investigated by Northern blot analysis. The regions spanned by specific probes for impTk and fbpTk are displayed in Fig. 1. Total RNAs were isolated from cells grown in rich ASW-YT medium with starch or pyruvate. A positive signal was not detected for the imp probe with RNA from Δimp-2A (Fig. 4A) or for the fbp probe with Δfbp-8J RNA (Fig. 4B), consistent with the complete deletion of the respective target genes by homologous recombination.
FIG. 4.
Northern blot analysis with imp probe (A) and fbp probe (B). Total RNAs were isolated from cells of strains KW128, Δimp-2A, and Δfbp-8J grown in ASW-YT medium supplemented with pyruvate (P) or starch (S). The regions corresponding to the respective probes are indicated in Fig. 1. Each lane contained 30 μg of total RNA. The signal intensities between the panels cannot be directly compared due to the prolonged chromogenic reaction time (>10 times longer) for panel A compared to that for panel B. Numbers on the left are lengths of RNA size markers (in bases).
Compared with the clear signals for fbp with KW128 and Δimp-2A cells grown under gluconeogenic conditions, the signals with cells grown on starch were highly reduced, as shown in Fig. 4B. This observation confirmed that the previously described transcriptional repression of fbp found in the wild-type cells (29) also occurred in these strains and coincided with the results of the enzyme assay described above. Although IMPase activity could hardly be detected in the cell extracts, the use of the imp probe enabled us to identify transcripts of the gene in KW128 and Δfbp-8J cells (Fig. 4A). However, the signals were extremely weak and were detectable only after a prolonged chromogenic reaction (>10 times longer than that applied for the fbp probe), indicating much lower levels of transcription of impTk than of fbpTk. This weak transcription was estimated to be a primary reason for the low intracellular IMPase activity and the inability of the gene to complement the defect of fbp. Unlike fbpTk, the transcription of impTk was constitutive, without regulation dependent on the carbon source.
The signal length for fbpTk (1.3 kb) corresponded to a monocistronic transcript from the 1,128 bp fbpTk gene (Fig. 4B), while the 2.6-kb signal length for impTk suggested a tricistronic transcription of impTk (771 bp) together with an upstream (810 bp) and a downstream (621 bp) gene encoding uncharacterized membrane proteins, both of which are probably unrelated to sugar metabolism (deduced from T. kodakaraensis genome analysis). The monocistronic transcription of fbpTk implies that the phenotype of the Δfbp-8J strain can be attributed to the disruption of the fbpTk gene per se, most likely ruling out polar effects accompanied by the gene disruption. We also observed that in the Δimp-2A and Δfbp-8J strains, no remarkable enhancement of transcription occurred for one gene in the absence of the other. This fact and the different transcriptional profiles of these genes demonstrate that the transcriptional regulation of fbpTk and impTk are independent from one another.
DISCUSSION
The results obtained in this study indicate the following points. (i) fbpTk is no doubt an indispensable gene for gluconeogenesis, and almost all FBPase activity within the cells derives from fbpTk. (ii) impTk was unable to complement the defect of fbpTk in Δfbp-8J cells, demonstrating that the gene does not participate in gluconeogenesis, in spite of the fact that the recombinant protein of impTk exhibited a higher kcat/Km value for FBP than that of fbpTk. These results clearly provide evidence that the true FBPase for gluconeogenesis in the hyperthermophilic archaeon T. kodakaraensis is the structurally divergent FBPase encoded by fbpTk, not IMPase/FBPase IV. The transcriptional profile of fbpTk was also in good agreement with the physiological function elucidated here. Although the protein product is a nonallosteric enzyme (29), the observed transcriptional regulation of the gene allows fbpTk to play an important role in controlling the flux of gluconeogenesis. As described previously, the divergent FBPase is not unique to T. kodakaraensis; its orthologs (COG1980) are highly conserved in hyperthermophiles (29). In contrast, the IMPase/FBPase IV orthologs (COG0483) are widely distributed in organisms that are unrelated in terms of domain classification and growth temperature. As mentioned above, none of the 17 (hyper)thermophiles whose genomes have been sequenced (including T. kodakaraensis) harbor a classical FBPase. Among them, the FbpTk ortholog is present in 16 strains, including Aquifex aeolicus and Thermoanaerobacter tengcongensis, belonging to the domain Bacteria, and 13 of the 16 strains also possess IMPase/FBPase IV. The results obtained in this study imply that the FbpTk orthologs in these (hyper)thermophiles most likely fulfill the gluconeogenic role in vivo. Therefore, we propose that orthologs of FbpTk should be classified as class V FBPases, representing the true gluconeogenic FBPases of (hyper)thermophiles. At present, the bacterium Thermotoga maritima is the only exception that exhibits hyperthermophily without an obvious FBPase V ortholog. Since an IMPase/FBPase IV ortholog is present in its genome, that protein may function as the gluconeogenic FBPase in Thermotoga maritima, or alternatively, a further divergent class of FBPase may exist in the organism. The FBPase V ortholog is also absent from the three mesophilic archaea Halobacterium sp. strain NRC-1, Methanosarcina acetivorans, and Methanosarcina mazei. However, these archaea possess orthologs for classical FBPases (FBPase I in Halobacterium sp. and FBPase II in Methanosarcina species) that can fulfill this step in gluconeogenesis. These facts imply that FBPase V is a (hyper)thermophile-specific enzyme rather than an archaeal enzyme. It has been reported that COG1980 of FBPase V is one of the most striking COGs (clusters of orthologous groups of proteins) whose presence is biased toward hyperthermophiles, after reverse gyrase (22). It can be speculated that FBPase V has structural features that limit efficient functioning of the protein to high temperatures, and thereby the enzyme is replaced by the structurally distinct FBPases in mesophiles, or vice versa.
Despite the high catalytic efficiency of the ImpTk protein for the FBPase reaction in vitro, the enzyme cannot fulfill a gluconeogenic function in vivo. Northern blot analysis revealed that impTk was transcribed in T. kodakaraensis, as in the case of IMPase/FBPase IV in P. furiosus (42). However, the constitutive transcription of impTk was estimated to be very weak. IMPase activity was also trivial in all strains examined. At least under the conditions examined in this study, the short supply of ImpTk protein is the main reason why the protein is unable to function as a gluconeogenic enzyme in vivo. However, we cannot exclude the possibility that the FBPase activity of the weakly expressed protein is specifically suppressed by an unknown mechanism in the cell.
IMPase in mammalian cells supplies myo-inositol from IMP to synthesize phosphatidylinositols together with CDP-diacylglycerol. In E. coli, the SuhB protein, which is orthologous to eukaryotic IMPase, actually exhibits IMPase activity (25), but the major role of this protein in vivo is suggested to be the posttranscriptional control of gene expression (15, 16, 40, 43). On the other hand, IMPase in some hyperthermophiles has been thought to be related to the biosynthesis of di-myo-inositol-1,1′-phosphate (DIP), which is quite different from its role in mammalian counterparts. DIP has been found in various kinds of hyperthermophiles, e.g., Pyrococcus (23, 35), Thermococcus (20), Methanococcus (4), and Thermotoga (24). This unique compatible solute was presumed to serve as an osmolyte against extracellular stresses such as high salinity or a high growth temperature. Two DIP biosynthesis pathways have been proposed, both of which share a common first step in IMP formation from glucose-6-phosphate by IMP synthase (EC 5.5.1.4). DIP was generated from myo-inositol and CDP-inositol in Methanococcus igneus (3), while the coupling of two IMP molecules in an NTP-dependent manner generated DIP and Pi in Pyrococcus woesei (36). IMPase activity is required for the production of myo-inositol from IMP in the former pathway. Although such DIP accumulation has not yet been examined in T. kodakaraensis, there is a possibility that imp may function in DIP biosynthesis. The independent transcriptional regulation of impTk from that of fbpTk does not contradict this supposition. If this were the case, the transcription of imp in T. kodakaraensis might be up-regulated in response to extracellular stresses. Alternatively, ImpTk, with its broad substrate specificity, might dephosphorylate other compounds in different pathways, or as in the case of E. coli SuhB (2), it may display a distinct function that is unrelated to its apparent phosphatase activity. Further detailed analyses of the Δimp-2A strain will help to clarify the function of the archaeal IMPase.
Acknowledgments
This study was supported by a grant-in-aid for scientific research to T. I. (no. 14103011) and was partly supported by a grant-in-aid for JSPS fellows to T.S. (no. 15005649) from the Ministry of Education, Science, Sports, Culture, and Technology.
REFERENCES
- 1.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea, in press. [DOI] [PMC free article] [PubMed]
- 2.Chen, L., and M. F. Roberts. 2000. Overexpression, purification, and analysis of complementation behavior of E. coli SuhB protein: comparison with bacterial and archaeal inositol monophosphatases. Biochemistry 39:4145-4153. [DOI] [PubMed] [Google Scholar]
- 3.Chen, L., E. T. Spiliotis, and M. F. Roberts. 1998. Biosynthesis of di-myo-inositol-1,1′-phosphate, a novel osmolyte in hyperthermophilic archaea. J. Bacteriol. 180:3785-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciulla, R. A., S. Burggraf, K. O. Stetter, and M. F. Roberts. 1994. Occurrence and role of di-myo-inositol-1,1′-phosphate in Methanococcus igneus. Appl. Environ. Microbiol. 60:3660-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donahue, J. L., J. L. Bownas, W. G. Niehaus, and T. J. Larson. 2000. Purification and characterization of glpX-encoded fructose 1,6-bisphosphatase, a new enzyme of the glycerol 3-phosphate regulon of Escherichia coli. J. Bacteriol. 182:5624-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraenkel, D. G. 1967. Genetic mapping of mutations affecting phosphoglucose isomerase and fructose diphosphatase in Escherichia coli. J. Bacteriol. 93:1582-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraenkel, D. G., and B. L. Horecker. 1965. Fructose-1,6-diphosphatase and acid hexose phosphatase of Escherichia coli. J. Bacteriol. 90:837-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs, G., H. Winter, I. Steiner, and E. Stupperich. 1983. Enzymes of gluconeogenesis in the autotroph Methanobacterium thermoautotrophicum. Arch. Microbiol. 136:160-162. [Google Scholar]
- 9.Fujita, Y., and E. Freese. 1979. Purification and properties of fructose-1,6-bisphosphatase of Bacillus subtilis. J. Biol. Chem. 254:5340-5349. [PubMed] [Google Scholar]
- 10.Fujita, Y., and E. Freese. 1981. Isolation and properties of a Bacillus subtilis mutant unable to produce fructose-bisphosphatase. J. Bacteriol. 145:760-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita, Y., K. Yoshida, Y. Miwa, N. Yanai, E. Nagakawa, and Y. Kasahara. 1998. Identification and expression of the Bacillus subtilis fructose-1,6-bisphosphatase gene (fbp). J. Bacteriol. 180:4309-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gancedo, C., M. L. Salas, A. Giner, and A. Sols. 1965. Reciprocal effects of carbon sources on the levels of an AMP-sensitive fructose-1,6-diphosphatase and phosphofructokinase in yeast. Biochem. Biophys. Res. Commun. 20:15-20. [DOI] [PubMed] [Google Scholar]
- 13.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gancedo, J. M., M. J. Mazón, and C. Gancedo. 1982. Kinetic differences between two interconvertible forms of fructose-1,6-bisphosphatase from Saccharomyces cerevisiae. Arch. Biochem. Biophys. 218:478-482. [DOI] [PubMed] [Google Scholar]
- 15.Inada, T., and Y. Nakamura. 1995. Lethal double-stranded RNA processing activity of ribonuclease III in the absence of SuhB protein of Escherichia coli. Biochimie 77:294-302. [DOI] [PubMed] [Google Scholar]
- 16.Inada, T., and Y. Nakamura. 1996. Autogenous control of the suhB gene expression of Escherichia coli. Biochimie 78:209-212. [DOI] [PubMed] [Google Scholar]
- 17.Itaya, K., and M. Ui. 1966. A new micromethod for the colorimetric determination of inorganic phosphate. Clin. Chim. Acta 14:361-366. [DOI] [PubMed] [Google Scholar]
- 18.Kelley-Loughnane, N., S. A. Biolsi, K. M. Gibson, G. Lu, M. J. Hehir, P. Phelan, and E. R. Kantrowitz. 2002. Purification, kinetic studies, and homology model of Escherichia coli fructose-1,6-bisphosphatase. Biochim. Biophys. Acta 1594:6-16. [DOI] [PubMed] [Google Scholar]
- 19.Kwok, F., and S. C. L. Lo. 1994. Development of a continuous coupled enzymatic assay for myo-inositol monophosphatase. J. Biochem. Biophys. Methods 29:173-178. [DOI] [PubMed] [Google Scholar]
- 20.Lamosa, P., L. O. Martins, M. S. Da Costa, and H. Santos. 1998. Effects of temperature, salinity, and medium composition on compatible solute accumulation by Thermococcus spp. Appl. Environ. Microbiol. 64:3591-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenz, A. G., and H. Holzer. 1980. Rapid reversible inactivation of fructose-1,6-bisphosphatase in Saccharomyces cerevisiae by glucose. FEBS Lett. 109:271-274. [DOI] [PubMed] [Google Scholar]
- 22.Makarova, K. S., Y. I. Wolf, and E. V. Koonin. 2003. Potential genomic determinants of hyperthermophily. Trends Genet. 19:172-176. [DOI] [PubMed] [Google Scholar]
- 23.Martins, L. O., and H. Santos. 1995. Accumulation of mannosylglycerate and di-myo-inositol-phosphate by Pyrococcus furiosus in response to salinity and temperature. Appl. Environ. Microbiol. 61:3299-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martins, L. O., L. S. Carreto, M. S. Da Costa, and H. Santos. 1996. New compatible solutes related to di-myo-inositol-phosphate in members of the order Thermotogales. J. Bacteriol. 178:5644-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuhisa, A., N. Suzuki, T. Noda, and K. Shiba. 1995. Inositol monophosphatase activity from the Escherichia coli suhB gene product. J. Bacteriol. 177:200-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morikawa, M., Y. Izawa, N. Rashid, T. Hoaki, and T. Imanaka. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh, M. K., L. Rohlin, K. C. Kao, and J. C. Liao. 2002. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277:13175-13183. [DOI] [PubMed] [Google Scholar]
- 28.Pilkis, S. J., and T. H. Claus. 1991. Hepatic gluconeogenesis/glycolysis: regulation and structure/function relationships of substrate cycle enzymes. Annu. Rev. Nutr. 11:465-515. [DOI] [PubMed] [Google Scholar]
- 29.Rashid, N., H. Imanaka, T. Kanai, T. Fukui, H. Atomi, and T. Imanaka. 2002. A novel candidate for the true fructose-1,6-bisphosphatase in archaea. J. Biol. Chem. 277:30649-30655. [DOI] [PubMed] [Google Scholar]
- 30.Regelmann, J., T. Schüle, F. S. Josupeit, J. Horak, M. Rose, K. D. Entian, M. Thumm, and D. H. Wolf. 2003. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol. Biol. Cell 14:1652-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rittmann, D., S. Schaffer, V. F. Wendisch, and H. Sahm. 2003. Fructose-1,6-bisphosphatase from Corynebacterium glutamicum: expression and deletion of the fbp gene and biochemical characterization of the enzyme. Arch. Microbiol. 180:285-292. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schäfer, T., and P. Schönheit. 1993. Gluconeogenesis from pyruvate in the hyperthermophilic archaeon Pyrococcus furiosus: involvement of reactions of the Embden-Meyerhof pathway. Arch. Microbiol. 159:354-363. [Google Scholar]
- 35.Scholz, S., J. Sonnenbichler, W. Schäfer, and R. Hensel. 1992. Di-myo-inositol-1,1′-phosphate: a new inositol phosphate isolated from Pyrococcus woesei. FEBS Lett. 306:239-242. [DOI] [PubMed] [Google Scholar]
- 36.Scholz, S., S. Wolff, and R. Hensel. 1998. The biosynthesis pathway of di-myo-inositol-1,1′-phosphate in Pyrococcus woesei. FEMS Microbiol. Lett. 168:37-42. [Google Scholar]
- 37.Schüle, T., M. Rose, K. D. Entian, M. Thumm, and D. H. Wolf. 2000. Ubc8p functions in catabolite degradation of fructose-1,6-bisphosphatase in yeast. EMBO J. 19:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sedivy, J. M., F. Daldal, and D. G. Fraenkel. 1984. Fructose bisphosphatase of Escherichia coli: cloning of the structural gene (fbp) and preparation of a chromosomal deletion. J. Bacteriol. 158:1048-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sedivy, J. M., and D. G. Fraenkel. 1985. Fructose bisphosphatase of Saccharomyces cerevisiae: cloning, disruption and regulation of the FBP1 structural gene. J. Mol. Biol. 186:307-319. [DOI] [PubMed] [Google Scholar]
- 40.Shiba, K., K. Ito, and T. Yura. 1984. Mutation that suppresses the protein export defect of the secY mutation and causes cold-sensitive growth of Escherichia coli. J. Bacteriol. 160:696-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stec, B., H. Yang, K. A. Johnson, L. Chen, and M. F. Roberts. 2000. MJ0109 is an enzyme that is both an inositol monophosphatase and the ‘missing’ archaeal fructose-1,6-bisphosphatase. Nat. Struct. Biol. 7:1046-1050. [DOI] [PubMed] [Google Scholar]
- 42.Verhees, C. H., J. Akerboom, E. Schiltz, W. M. de Vos, and J. van der Oost. 2002. Molecular and biochemical characterization of a distinct type of fructose-1,6-bisphosphatase from Pyrococcus furiosus. J. Bacteriol. 184:3401-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yano, R., H. Nagai, K. Shiba, and T. Yura. 1990. A mutation that enhances synthesis of σ32 and suppresses temperature-sensitive growth of the rpoH15 mutant of Escherichia coli. J. Bacteriol. 172:2124-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y., J. Y. Liang, S. Huang, and W. N. Lipscomb. 1994. Toward a mechanism for the allosteric transition of pig kidney fructose-1,6-bisphosphatase. J. Mol. Biol. 244:609-624. [DOI] [PubMed] [Google Scholar]